Abstract
Lithium has been a drug for bipolar disorders (BD) for over 70 years; however, its usage has been limited by its narrow therapeutic window (between 0.6 and 1.2 mM). Understanding the cellular distribution of lithium ions (Li+) in patient cells will offer deep insight into this limitation, but selective imaging of Li+ in living cells under biomedically relevant concentration ranges has not been achieved. Herein, we report in vitro selection and development of a Li+-specific DNAzyme fluorescent sensor with >100-fold selectivity over other biorelevant metal ions. This sensor allows comparative Li+ visualization in HeLa cells, human neuronal progenitor cells (NPCs), and neurons derived from BD patients and healthy controls. Strikingly, we detected enhanced accumulation of Li+ in cells derived from BD patients compared with healthy controls in differentiated neurons but not NPCs. These results establish the DNAzyme-based sensor as a novel platform for biomedical research into BD and related areas using lithium drugs.
Short abstract
A highly Li+ selective DNAzyme sensor for live imaging of Li+ at a biomedical relevant concentration range reveals bipolar disorder patient-derived neurons can accumulate more Li+ than healthy neurons can.
Introduction
Bipolar disorder (BD) is one of the most prevalent mental health disorders worldwide, affecting nearly 2.8% of the adult U.S. population.1 This disease results in not only emotional hardships to individuals and their families but also considerable costs to society. The annual economic cost of BD in the U.S. alone is estimated to be $195 billion for 2018, with 25% representing direct costs related to care, frequent psychiatric interventions, and the presence of comorbid medical/psychiatric conditions being the primary drivers of cost.2 BD typically requires medication, and lithium remains the most effective mood stabilizing medicine for long-term management.3 It was fortuitously discovered by psychiatrist John Cade in 1949 and was confirmed in later clinical trials4 and is considered one of the first-line drugs for BD and mania as a mood stabilizer.5 With the low cost and high efficacy toward BD treatment, the application of lithium was expanded to other diseases in recent years. For example, lithium has also been found to display neuroprotective properties that can attenuate the effects of traumatic brain injury (TBI).6 Despite its advantages in treating BD and TBI, lithium treatment is complicated by its extremely narrow therapeutic window of 0.6–1.2 mM and bimodal response from patients.7 For example, lithium toxicity, a concentration only slightly above the therapeutic window has severe side effects including tremors, renal failure, hypothyroidism, and cognitive impairment.8 Moreover, only one-third of the BD patients are lithium responders, and the rest of patients respond to lithium therapy partially or not at all.9 To address these issues, it is important to understand the mechanism of lithium-based therapy, particularly the concentration of lithium ions (Li+) within neural cells after treatment. This is because the cellular concentration of Li+ may not directly correlate with the dosage of lithium administered to the patient and can be changed depending on the condition and unique physiology of different patients. Through comparison of Li+ distribution and enrichment differences in neurons between BD patients and healthy individuals, we may provide insights into lithium responsiveness and identify more effective and safer prescriptions for patients.
For many years, several methods have been developed to detect the spatial distributions and concentrations of Li+ in biological samples. Among them, instrumental analyses by atomic absorption spectroscopy and ion mass spectrometry are the most commonly used methods to measure the concentration of Li+ in biological samples, including revealing a heterogeneous distribution in rat/mouse brain.10,11 However, these methods require sample fixation, which makes it difficult to obtain temporal information on Li+ distribution in living cells or tissues. To overcome this limitation, 7Li NMR has been used to map Li+ in BD patients’ brains; yet, this method involves time-consuming data collection and has a low resolution,12 making it difficult to image Li+ in living cells or tissues with high spatial and temporal resolution.
Fluorescent sensors are excellent choices to image Li+ in live cells or tissues, as they provide sensitive detection with high spatiotemporal resolution. Despite demonstrated success in using fluorescent sensors to detect divalent metal ions such as Ca2+,13 Zn2+,14 and Cu2+,15,16 effective sensors for detecting monovalent metal ions, especially Li+, is quite limited.17−19 Among the fluorescent Li+ sensors, sensors based on organic crown, such as chiral diaza-9-crown-3 derivatives, [2.1.1]-cryptates, KLI-1 and KLI-2, methyl-1-aza-12-crown-4, and 1-(9-anthryl)-4-ferrocenyl-2-aza-1,3-butadiene, have received the most attention.20−25 However, they have limited selectivity over other competing monovalent ions, such as Na+ and K+, which are present in biological systems in much higher concentrations than that of Li+, and most of them have limited solubility in water and thus do not work well under physiological conditions. To our knowledge, Sabrina Heng lithium (SHL), based on merocyanine and spiropyran isomers changing upon Li+ binding, was the first and is the only intracellular Li+ optical sensor reported.26 However, its cellular detection range (137 mM Li+ without Na+) is beyond the window of physiological relevant concentrations. Therefore, despite the importance and high demand for imaging Li+ in living cells and tissues, it is still a major challenge to design fluorescent sensors with high sensitivity and selectivity for Li+ over other monovalent and multivalent metal ions that work under physiological conditions in human neural cells.
To meet this challenge, we and others have taken advantage of DNAzymes (deoxyribozymes or catalytic DNA) as a general platform for developing metal ion sensors.27 First discovered in 1994, DNAzymes are metalloenzymes that recruit metal ions as a cofactor for catalysis.28−30 Among them, RNA-cleaving DNAzymes are of particular interest for metal ion sensing due to their fast reaction rate, high metal ion cofactor selectivity, and ease of converting the cleavage into a detectable signal by the catalytic beacon strategy.31−36 Unlike screening the rationally designed small molecule or genetically encoded protein sensors, in vitro selection allows selection of DNAzymes with desired sensitivity and specificity for a metal ion of interest using a much larger library of DNA molecules containing up to 1015 different sequences. Additionally, prior knowledge of the necessary metal ion binding sites or specific metal–DNA interaction is not required, and the “winner” can be amplified using polymerase chain reaction.37−42 Moreover, through the in vitro selection and reselection process, metal ion binding affinity and selectivity can be optimized by tuning the stringency of selection pressure, such as varying the time of the reaction, manipulating the concentrations of the metal ion, and introducing counter selection to remove the DNAzymes that depend on competing metal ions.39,43 Finally, DNA synthesis is cost-effective with a variety of useful modifications, and its biocompatibility makes DNAzyme-based sensors excellent tools for live-cell imaging of metal ions. As a result, many metal-specific DNAzymes have been isolated and converted into sensors for their respective metal ion cofactors44,45 and have recently been delivered into cells for monitoring UO22+,46 Pb2+,28,47 Zn2+,48,49 Mg2+,50−52 and Na+53,54 in living cells or organisms. Despite the progress, no DNAzyme has been reported to have high selectivity toward Li+, likely because Na+ and K+ are present in biological samples and buffers in much higher concentrations.
In this report, we describe the in vitro selection and characterization of an RNA-cleaving DNAzyme with high selectivity (>100-fold) for Li+ over other competing metal ions. This Li+-specific DNAzyme was transformed into a biocompatible fluorescent sensor and achieved intracellular Li+ detection in living cells. We also established the sensing system for comparative metal ion visualization in human neuronal cells derived from iPSCs with the goal of determining whether there is differential Li+ accumulation in living neurons from BD patients and healthy individuals. As a result, we observed a higher Li+ accumulation in neurons derived from BD patient cells compared to differentiated neurons from healthy donors or from human neural progenitor cells. The work reported here opens a new avenue for the selective detection of Li+ in living BD neurons and thus provides the opportunity for further studies that uncover the physiological response to lithium therapy.
Results and Discussion
In Vitro Selection of Li+-Specific DNAzyme
To acquire highly selective Li+-specific DNAzymes, we carried out in vitro selection using methods reported previously.46,49,53 The selection used a 95-mer oligonucleotide (nt) containing a 35-nt random sequence, flanked by conserved primer-binding regions (Supporting Information, Figure S1). At the putative cleavage site, a single guanosine ribonucleotide (rG marked in red) was incorporated in the 5′-conserved region. The selection pools were incubated with selection buffer (200 mM LiCl, 20 mM MOPS, 0.5 mM EDTA, pH 7.4) for 2 h at room temperature. DNAzymes that could catalyze cleavage of the rG in the presence of Li+ were separated from the bulk pool by denaturing PAGE and regenerated by PCR amplification for the next round selection. To increase the stringency and search for more efficient DNAzymes, the incubation time was decreased in the later rounds from 2 h in Round 1 to 0.5 h in Round 10 (Supporting Information, Table S2). The selection progress and enrichment of Li+-responsive DNAzymes in the pool were monitored with gel-based activity assays (Supporting Information, Figure S2). Noticeable activity of the pool was observed by Round 6 and was further increased in Rounds 7 and 8. Therefore, the DNA molecules in Round 8 were cloned and then sequenced (Supporting Information, Figure S2). Analysis of 26 resulting sequences from 100 colonies revealed the high convergence of the Round 8 selection pool and identified 13 distinct sequences (Supporting Information, Figure S3).
Four sequences with Li+-dependent activity were identified from these 13 sequences by activity tests, and of the four, three had a consensus sequence carrying only 2–3 nucleotide variations (Supporting Information, Figure S4). A potential secondary structure of one Li+-DNAzyme was analyzed by UNAfold for further validation (Supporting Information, Figure S5). To identify the catalytic core sequence responsible for the Li+-dependent activity, the above DNAzyme sequence was subjected to truncation and conversion from a cis-cleaving form to trans-cleaving forms, and the resulting DNAzymes were tested for activity (see Figures S5 and S6). As a result, the length of the catalytic core region was shortened from 35 nt in the selection pool to 15 nt in the trans-Li+ DNAzyme.
To obtain a faster Li+ specific DNAzyme with a lower detection limit, reselection was performed with a partially randomized pool with an approximately 30% mutation rate during chemical synthesis of the DNAzyme pool in the trans-Li+ DNAzyme catalytic core and flanking nucleotides; i.e., 70% of the nucleotide is the same as the parent trans-Li+ DNAzyme catalytic core sequence at each position with 10% of each of the three remaining nucleotides being incorporated (Supporting Information, Figure S7). To increase the reaction rate of the Li+-selective DNAzyme, the reaction time was gradually decreased from 3 h to 1 min during 20 rounds of reselection with 70 mM LiCl in 20 mM MOPS pH 7.4 (Supporting Information, Table S3). Sequencing of the cultivated DNA pool after 20 rounds of reselection revealed that the most active sequence, named “cis-20-4” DNAzyme, was enriched, accounting for approximately 65% of the selection pool (Supporting Information, Figures S8 and S9).
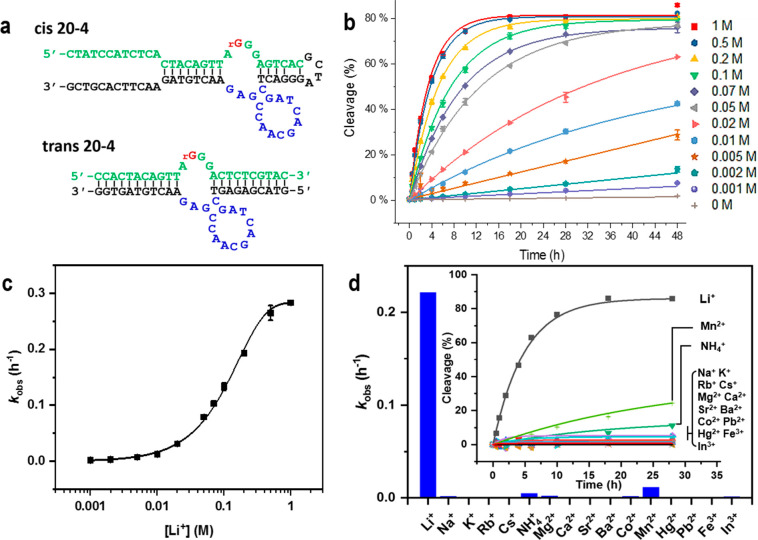
Although the cis-20-4 DNAzyme had only a two-base difference in comparison with the DNAzyme generated from the first selection (Supporting Information, Figure S10), it showed 7–10 times higher activity (Supporting Information, Figure S11). To facilitate the conversion to DNAzyme sensors, the cis-20-4 DNAzyme was truncated to become the trans-20-4 DNAzyme while maintaining the activity of the cis-20-4 DNAzyme (Figure 1a). Under 100 mM of Li+ at 20 °C, the trans-20-4 DNAzyme displayed an initial observed rate constant (kobs) of 0.14 ± 0.01 h–1 in the selection buffer (Figure 1b, 100 mM group: green color reverse triangle). Overall, to the best of our knowledge, the 20-4 DNAzyme is the first Li+ specific RNA-cleaving catalytic nucleic acids with substantial activity.
Figure 1.
Activity of the Li+ specific DNAzyme. (a) The sequence of the Li+-specific DNAzyme (named 20-4) from in vitro selection in the cis- and trans- form. The ribonucleotide cleaving site is shown in red, the substrate region is in green, the catalytic core in the enzyme region is in blue, and other binding arm and linker regions are in black. (b) The activity of the trans-20-4 DNAzyme under different Li+ concentrations from a 32P radiolabeled PAGE assay. Data are shown as the mean ± SD n = 3 for each group. (c) The dynamic response of the trans-20-4 DNAzyme under different Li+ concentrations. Data are shown as the mean ± SD n = 3 for each group. (d) The selectivity of the trans-20-4 DNAzyme over other monovalent (200 mM), divalent (4 mM), and trivalent (0.4 mM) metal ions. Inset: the trace of fraction of cleaved product in response to different competing metal ions.
Sensitivity and Selectivity of the 20-4 DNAzyme
To further evaluate the sensitivity of the trans-20-4 DNAzyme for Li+ detection under various concentrations, polyacrylamide gel electrophoresis (PAGE)-based activity assays were performed (Figure 1b–d). The trans-20-4 DNAzyme showed a significant response to Li+ concentrations as low as 1 mM compared with the background ribonucleotide cleavage with extended incubation longer than 28 h (Figure 1b). The observed rate of ribonucleotide site cleavage increased with increasing concentrations of Li+, until saturation at ∼1 M Li+, with an apparent dissociation constant (Kd) of 110 ± 1.7 mM (Figure 1c).
To determine the selectivity of the DNAzyme for Li+ over other metal ions, we monitored ribonucleotide cleaving activity to 15 different metal ions. More than 100-fold selectivity was detected from the trans-20-4 DNAzyme in response to Li+ over other metal ions (200 mM for monovalent, 4 mM for divalent, 0.4 mM for trivalent ions), except for Mn2+ and NH4+ (Figure 1d, Supporting Information, Table S4). Given that NH4+ is present in the low millimolar concentration range and Mn2+ is in the tens of micromolar range in living cells,55 this selectivity is sufficient for the 20-4 DNAzyme to work in biological systems, such as living cells and blood serum, without interference from other biorelevant metal ions.
DNAzyme-Based Fluorescent Sensor for Li+ Detection
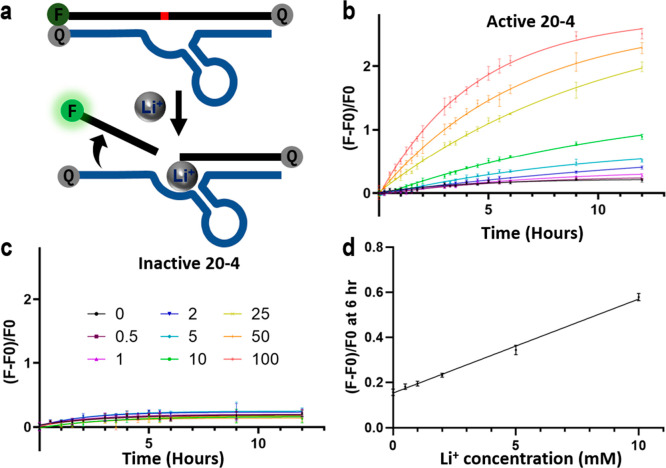
To convert the Li+-dependent catalytic activity of the 20-4 DNAzyme into a turn-on fluorescence response, we next designed a sensor based on the “catalytic beacon” approach by labeling the substrate strand of the 20-4 DNAzyme with a 6-carboxyfluorescein fluorophore (FAM) at its 5′ end and the enzyme strand of the 20-4 DNAzyme with an Iowa Black FQ quencher at its 3′ end.56,57 In addition, a second quencher was added at the 3′ end of the substrate strand to minimize background fluorescence33 (Figure 2a). Since the length of binding arms is critical for full hybridization and complete quenching before cleavage, as well as the successful dehybridization of the fluorophore labeled strand after cleavage, we did a careful design of the length and sequences of the binding arms (Figure 2a, Supporting Information, Figure S12).
Figure 2.
In vitro performance of the trans-20-4 Li+ fluorescence sensor. (a) Design of the Li+ DNAzyme catalytic beacon. (b, c) Fluorescence increase of (b) the active sensor and (c) the inactive sensor over time at different Li+ concentrations under a molecular crowding environment. Data are plotted as the mean ± SD n = 3 for each group. (d) The linear response range of normalized fluorescence intensity from the active sensor at the 6-h time point corresponding to the Li+ concentration under a molecular crowding environment. Data are shown as mean ± SD n = 3 for each group.
To evaluate the DNAzyme sensor, we tested the fluorescence intensity in the presence of Li+ at different concentrations. The sensor displayed an increasing fluorescence signal with increasing concentrations of Li+ (Figure 2b). The limit of detection was determined to be 2.2 mM or 15.7 ppm (3σ/slope) in the selection buffer (Supporting Information, Figures S13 and S14). When adding 25% PEG8000 as a molecular crowding agent to better mimic the environment in cells, the limit of detection was lowered to 1.1 mM or 7.9 ppm (3σ/slope), with a dynamic range up to 10 mM (Figure 2d), which overlaps with the biomedical Li+ concentration in the therapeutic window. To rule out artifacts in the detection due to nonspecific DNA cleavage, an inactive 20-4 DNAzyme that had the same binding arm sequence but a reversed catalytic core sequence which rendered the DNAzyme inactive in the presence of Li+ was tested as a negative control. No elevated fluorescence signals were detected with the inactive DNAzyme when the Li+ concentration was increased, indicating the specificity of this detection (Figure 2c, Supporting Information, Figure S15).
Intracellular Li+ Imaging Using the 20-4 Li+ DNAzyme
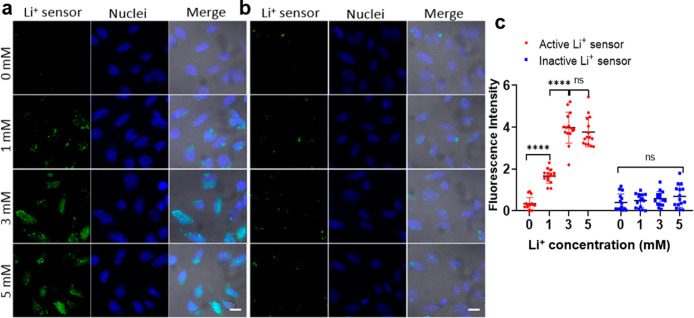
To investigate the ability of this novel 20-4 Li+ DNAzyme sensor in detecting intracellular Li+ in living cells, we chose the HeLa CCL-2 adenocarcinoma cell line, a widely used model for living cell sensor development, as a proof-of-concept cell model for Li+ sensor optimization and validation. According to a cell toxicity assay, HeLa cells could tolerate up to 5 mM Li+ treatment for 12 h (Supporting Information, Figure S16). Thus, we collected cell images under different Li+ concentrations ranging from 0 to 5 mM (Figure 3a, b). The sensor showed minimal background fluorescence after its delivery, indicating that most of the Li+ DNAzyme sensor remained intact during the delivery process. After the HeLa cells were washed to remove excess probes and transfection reagent in the culture medium, the cells were incubated in different concentrations of Li+ with media. Since Li+ is orally delivered in patient treatment and influence the cells through the elevation of blood Li+ levels, increasing the Li+ level in cell culture media is a practical way to mimic drug treatment of cells and adjust the intracellular Li+ concentration. The influx of Li+ from the extracellular medium caused a gradual increase of the fluorescence inside cells (Figure 3a). Statistical data quantifying the fluorescence intensity, which indicates sensor response (Figure 3c), showed that Li+ could be detected in cells when the environmental Li+ concentration was higher than 1 mM. The fluorescence intensity increased in the 3 mM group but did not increase further after 3 mM. This result could be caused by the saturation or full cleavage of the sensors. In comparison, with the same treatment, the inactive DNAzyme sensor displayed a minor increase inside cells (Figure 3b), ruling out nonspecific degradation or activation of the sensors during detection.
Figure 3.
Li+ fluorescence imaging in HeLa cells. (a, b) Fluorescence imaging of HeLa cells treated with different Li+ concentrations with active (a) or inactive (b) DNAzyme sensors. Scale bar: 20 μm. (c) Plot graph shows the quantification of fluorescence intensity from HeLa cells. Data are shown as mean ± SD. Two-tailed Mann–Whitney test: For the active sensor, ****p = 3.392 × 10–6 < 0.0001 between 0 mM and 1 mM groups, ****p = 4.143 × 10–6 < 0.0001 between 1 mM and 3 mM groups, ns p = 0.2169 > 0.05 between 3 mM and 5 mM groups; for the inactive sensor, ns p = 0.4610 > 0.05 between 0 mM and 1 mM groups, ns p = 0.5741 > 0.05 between 1 mM and 3 mM groups, ns p = 0.7437 > 0.05 between 3 mM and 5 mM groups. n = 15 for each group by quantifying fluorescence intensity of five cells from three figures.
To assess the generalizability of the 20-4 DNAzyme sensor for imaging Li+ in other cell models, we tested the Li+ sensor in PC12 NS1, a widely used neuronal-like cell line derived from rat pheochromocytoma cells. Both progenitor-like PC12 and NGF-differentiated PC12-derived neurons were transfected with 20-4 DNAzyme and then incubated with 1 mM Li+ as a typical concentration within the therapeutic window (Supporting Information, Figures S17 and S18). Signaling was observed in the 1 mM Li+ group. These data confirmed the ability of the DNAzyme-based sensors to achieve Li+ detection at a low concentration range in living cells.
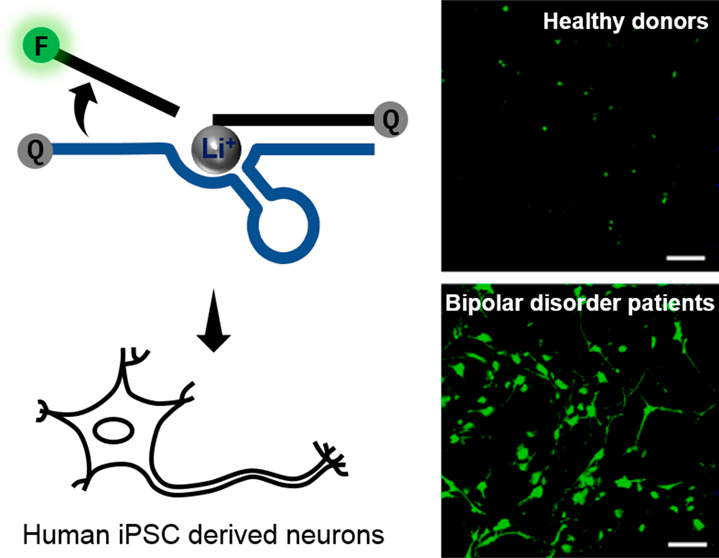
Li+ Enrichment in BD Patient-Derived Neurons
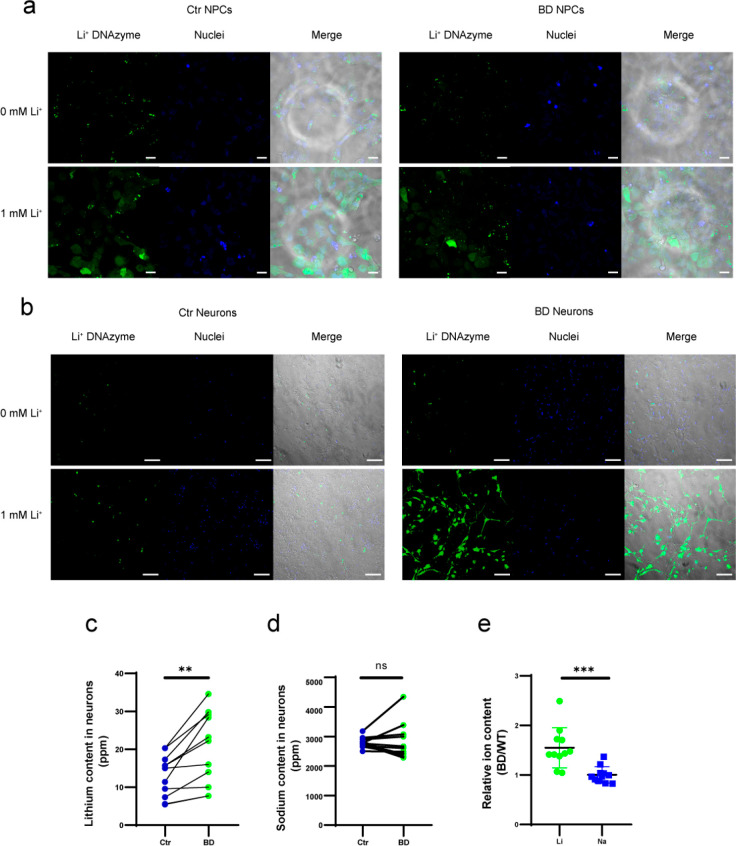
As a proof of concept for applying our sensor to study the therapeutic response in BD, we investigated the Li+ concentration in a human-induced pluripotent stem cell (iPSC)-based model, a powerful tool for investigating the pathogenesis of neuropsychological disorders. Neuronal cells generated from BD patient-derived iPSCs maintain the disease-related genetic background of the patient and disease-related phenotypes, which can be identified by stepwise differentiation to cell types of interest.58 It was reported that the neurons from BD patient iPSCs have different responses to lithium when compared with healthy controls, indicating the usefulness of iPSC-derived neurons as models of BD for lithium responsiveness studies.59 We collected and reprogrammed fibroblasts from patients who were diagnosed with bipolar I disorder as well as from healthy donors.60−62 To compare intracellular Li+ fluorescent signals upon lithium treatment at early differentiation stages, the active or inactive DNAzyme sensors were transfected into iPSC-derived neural progenitor cells (NPCs) (Figure 4a). Increased fluorescence signals from active Li+ sensors were observed when 1 mM Li+ was added to cell media (Figure 4a, Supporting Information, Figure S20). In contrast, the sensor signal intensity was not elevated in the inactive sensor group (Supporting Information, Figures S19 and S20). No significant difference was observed between NPCs from BD patients and healthy controls under the same conditions when the signal outputs from the BD and Ctrl group treated with 1 mM Li+ were compared. This indicated that the uptake of extracellular Li+ was comparable between NPCs from BD patients and healthy individuals during the early stages of differentiation.
Figure 4.
Intracellular Li+ accumulation in neural progenitor cells and iPSC-derived neurons. Intracellular Li+ confocal imaging was performed in NPCs (a, scale bar 20 μm) and neurons (b, scale bar 100 μm) from both healthy donors (left) and BD patients (right). ICP-MS measurements of Li (c) and Na (d) in neurons are shown; the connected dots are from the same differentiation batch. n = 11, **p = 0.0016 < 0.01 (Li) and ns p = 0.8658 > 0.05 (Na) using two-tailed paired t test. (e) BD over healthy ratios of Li and Na were calculated using the ICP results from the same differentiation batch, ***p = 0.0005 < 0.001 with a two-tailed unpaired t test.
Published methods have shown that differentiated neurons from BD patients perform differently from healthy human -derived neurons, possibly reflecting BD symptoms, and are useful as a disease model to provide insight into lithium-based treatments.63,64 Therefore, we further differentiated NPCs into neurons to study differential Li+ dynamics in mature neurons (Figure 4b, Supporting Information, Figures S21 and S22). Following the 30-day differentiation period, we obtained differentiated neurons from both patients and healthy controls. Interestingly, with the 20-4 DNAzyme Li+ sensor, BD-derived neurons showed a higher Li+ signal than control neurons when treated with 1 mM (Figure 4b, Supporting Information, Figure S22) or 3 mM (Supporting Information, Figure S23B) Li+, which was completely different from the observation with NPCs at early differentiation stages (Figure 4a, Supporting Information, Figure S23A). To exclude the possibility that the signal difference was caused by distinct sensor delivery efficiency between BD and control neurons, a delivery control DNA containing fluorophores without any quencher was transfected into neurons, showing that the transfection efficiencies were similar (Supporting Information, Figure S24). To confirm our findings, Li+ concentrations in those neurons were also measured using inductively coupled plasma mass spectrometry (ICP-MS) for absolute amounts of elements Li and Na, in which Na was used as the reference element for comparison.65 The ICP-MS analysis showed a similar result as what we observed by the DNAzyme-based Li+ sensor. From the ICP-MS data, BD patient-derived neurons showed higher intracellular Li concentrations than control neurons (BD:20.25 ± 9.20 ppm, Ctrl: 13.06 ± 5.24 ppm; ppm, mg/kg sample) with no significant differences in the amount of Na (BD: 2913.73 ± 604.78 ppm; Ctrl: 2887.46 ± 173.77 ppm) (Figure 4c–e). These data further confirmed the performance of our Li+ DNAzyme sensors within the therapeutic window and demonstrated that BD neurons accumulate more Li+ than control cortical neurons when incubated with Li+ in therapeutic ranges, even though the NPCs from BD and Ctrl have similar intracellular Li+ concentrations (Supporting Information, Figures S20 and S25).
In this work, we have developed a Li+-specific DNAzyme with high selectivity toward Li+ over other competing metal ions, including other chemically similar and biologically abundant monovalent metal ions, such as Na+ and K+. Most types of previously published chemical Li+ sensors seldom achieved this high selectivity of Li+ imaging.20−24 Taking advantage of this selectivity, we have developed a catalytic beacon fluorescent sensor for intracellular Li+ sensing in multiple types of cells including human cell lines (HeLa) and human iPSC-derived neurons and neuronal progenitor cells. The high selectivity together with the inactive DNAzyme negative control ensured that the signals from cell imaging reflect the Li+ concentration in cells, instead of artifacts from sensor activation by other biological metal ions, nucleases, and strand dehybridization.
In addition, constituted by DNA, a fluorophore, and quenchers, DNAzyme-based sensors are intrinsically biocompatible. This DNAzyme-based Li+ sensor is soluble and functional in the aqueous environment with minimum cell toxicity, which is a key advantage over most published Li+ sensors. Through careful in vitro selection and reselection to allow the DNAzyme sequence to evolve, our DNAzyme-based Li+ sensor is effective under physiological ionic strength, pH, and temperature, which fulfills the biocompatible requirement for live cell applications.
More importantly, detection of Li+ in living cells has been achieved within a biomedically relevant concentration range in this work, with 0 mM for no treatment, 1 mM for a representative level of effective lithium treatment (0.6–1.2 mM), and 3 mM for the toxic effect concentration. Detecting Li+ in this low biomedical concentration window is difficult for previously reported Li+ sensors, especially with the above considerations of selectivity and biocompatibility of the sensor. Our DNAzyme-based Li+ sensor provides a good detection range covering the biomedically informative concentration range (0–2 mM) that will be fine-tuned with lithium biomedical and toxicity studies.7,8
It is interesting to observe that the sensor could detect Li+ in living cells when 1 mM Li+ was added to the cell media, which is even lower than the limit of detection of the sensor (1.1 mM) obtained in the test tube (see Figure 2d). This difference suggests that the Li+ concentration in cells may not be the same as that in the extracellular culture media. Li+ accumulation in hippocampus was observed in both mouse models and human patients, indicating a higher level of Li+ in brain cells and tissues compared with serum. Enrichments of Li+ in cultured glioma cells and yeast are also observed, resulting in a higher intracellular Li+ level than the extracellular media. Therefore, the reason we were able to observe a fluorescence signal inside the cells when 1 mM Li+ was added to the cell media may have been due to accumulation of Li+ to levels higher than 1 mM. To support this explanation, we used ICP-MS to measure the Li concentrations in the cells and found 20.25 and 13.06 ppm Li from BD patient and healthy people neuron samples, which correspond to ∼3.145 mM and 2.028 mM of Li+, respectively.
Interestingly, when we applied the sensor at different stages of neuron differentiation in both BD patients and healthy donors, we observed a differential Li+ accumulation between the two groups. This difference was further confirmed by ICP analysis (Figure 4c–e, Supporting Information, Figure S23), suggesting that BD neurons accumulated more Li+ than healthy neurons when Li+ is provided in the culture environment. Our observation of this differential Li+ accumulation is consistent with a published study on differential responses to Li+ in hyperexcitable neurons.59 In that research, Yao and co-workers generated iPSC-derived neurons from BD patients and healthy controls and characterized hyperactive action-potential firing in differentiated BD neurons by patch-clamp recording and somatic Ca2+ imaging assays. This hyperexcitability phenotype of BD neurons was selectively reversed by adding Li+, while the action-potential firing from healthy normal neurons was not affected. Connecting this observation with our imaging data, it is logical to expect that the rescue of the action-potential firing in BD neurons is corrected by differential Li+ enrichment in BD cells, in addition to other transcriptome changes.
The observed differential Li+ accumulation between BD patients and healthy controls is only observed in differentiated neurons and not at the NPC stages. In addition, from ICP-MS, we observed a slight increase of Na in BD patient NPCs over the healthy controls but not in neurons (see Figure 4d, Supporting Information, Figure S25). These results suggest that BD patient-derived neural cells have overall higher Li+ and Na+ enrichment compared with the cells derived from healthy controls, and different ion types are preferred at different stages of neuron differentiation. This intriguing preference between Li+ and Na+ in cellular accumulation is also consistent with the knowledge that Li+ and Na+ are being transported by, and maybe competing for, the same system when entering cells.66−69 Other biorelevant metal ions and ion channels may also play roles in this differential accumulation process, considering the differential expressions of membrane ion channels during neuron differentiation and between neurons and NPCs. Further comparison and analysis of ion channels and the downstream transcriptome during neuronal differentiation would be beneficial in identifying potential transporters and targets of Li+. Once verified, these studies of Li+ transportation and function will provide mechanistic understandings of lithium therapy.
Conclusion
In conclusion, we have developed a Li+-specific DNAzyme, with high selectivity toward Li+ over other competing metal ions, including Na+ and K+, the two major monovalent metal ions in biological systems. Taking advantage of this selectivity, we have developed a catalytic beacon fluorescent sensor for intracellular Li+ sensing in multiple cell lines including human cell lines (HeLa) and human iPSC-derived neurons and neuronal progenitor cells. Interestingly, when we applied the sensor at different stages of iPSC differentiation from both BD patients and healthy donors, we observed a different Li+ accumulation pattern between BD and controls in differentiated neurons but not at progenitor stages. This difference was further confirmed by ICP microanalysis, suggesting that BD neurons can accumulate more Li+ than healthy neurons and that a different Li+ regulation system may exist in these cells. These new insights demonstrate that our Li+ sensor can be a powerful tool for future investigations of Li+ dynamics in many biological systems to better understand the effects of lithium in treating diseases like BD and TBI.
Methods
Safety Statement
No unexpected or unusually high safety hazards were encountered.
Template Preparation
All DNA sequences were purchased from Integrated DNA Technologies (IDT) with standard desalting. Selection was completed with a pool with random regions of 35 (N35) nucleobases. The full N35 pool was purified via denaturing PAGE (10% acrylamide, 8 M urea, 90 mM Tris, 90 mM boric acid, 2.75 mM Na2EDTA), extraction in Buffer A (0.3 M NaCl, 0.001 M Na2EDTA, 0.015 M N(Et)4OH, 0.02 MOPs pH 7.4), and was desalted using Waters Sep-Pak columns prior to use. The DNA sequences are summarized in Supporting Information, Table S1.
Pool Generation
The partial counter sequence of the N35 pool was ordered, and the pool was generated via asymmetric PCR, due to IDT synthesis limitations. To generate the N35 of 1× standard Taq buffer, 6 U/100 μL Taq polymerase, 0.2 mM of each dNTP, 1 μM P3-rG, 100 nM IDT template (N35), 1 μL of 32P-labeled α-ATP were mixed and underwent the follow temperature treatment: 3 min at 95 °C, repeated 14 (30 s at 95 °C, 1.25 min 60 °C, 1.25 min 72 °C), 10 min at 72 °C, then cooled to 4 °C. The 3 mL of DNA product was then desalted using ethanol precipitation in ∼65% EtOH, ∼0.25 NaOAc pH 5.2 chilled at −80 °C for at least 2 h, before the samples underwent centrifugation at −11 °C at 15 000 rpm for 30 min and then washed with 70% EtOH and spun down for an additional 20 min at −11 °C at 15 000 rpm. Samples were dried by a vacuum. Dried samples were resuspended in Millipore water and Stop Solution A (9.2 M urea, 1/5 TB) and purified on a 10% acrylamide denaturing PAGE gel. The purified sample was excised from the gel, the extricated gel was crushed, and the sample DNA was extracted in Extraction Buffer B (0.1 M LiCl, 0.001 M H2EDTA, 0.015 M LiOH, 0.02 MOPs pH 7.4) using a quick extraction technique. Extracted DNA was desalted using EtOH precipitation with approximately 70% EtOH, and 0.08 M LiCl, and chilled at −80 °C for 2 h or more. Samples were spun down at −6 °C at 15 000 rpm for 30 min. The supernatant was removed, and the sample was washed with chilled (−20 °C) 70% EtOH for an additional 20 min at −6 °C at 15 000 rpm. Samples were dried by a vacuum.
Selection in Li+-Containing Buffer
For positive selection, samples were resuspended in Milli-Q water, and then the reaction was initiated by the addition of 2× positive selection buffer (0.2 M LiCl, 0.001 M EDTA, 0.015 M LiOH, 0.02 M MOPs, pH 7.4). Samples were incubated at room temperature for 2 h before the addition of Stop Solution B (9.2 M Urea, 0.001 M H2EDTA, 0.015 LiOH, 180 mM Tris, 180 mM boric acid). Samples were purified using denaturing PAGE, from which the cleaved sequence was extracted using Extraction Buffer A and desalted using precipitation with 0.3 M NaOAc for the selection.
PCR Amplification
To regenerate the cleaved N35 pool, there is a two-step PCR process. PCR 1 extends the cleaved DNA sequence with P2. Subsequently, PCR 2 fully restores the extended sequence to the initial full-length DNA pool sequence and reincorporates the RNA active site. PCR 1 uses 1× standard Taq buffer, 6 U/100 μL Taq polymerase, 0.2 mM dNTP, 0.6 μM P1-iSp3, 1 μM P2, 50 μL of the 150 μL of redissolved desalted Positive Selection product, and 1 μL of 32P-labeled α-ATP. The PCR 1 mixture undergoes the following heat treatment: 3 min at 95 °C, repeated 14–22 (30 s at 95 °C, 1.25 min 60 °C, 1.25 min 72 °C), 10 min at 72 °C, and then the mixture is cooled to 4 °C. PCR 2 uses 1× standard Taq buffer, U Taq, mM dNTP. 1 μM P1-iSp3, 1 μM P3-rA, 5 μL of PCR 1 product, and 1 μL of 32P-labeled α-ATP were mixed and underwent the follow temperature treatment: 3 min at 95 °C, repeated 14–18 (30 s at 95 °C, 1.25 min 52 °C, 1.25 min 72 °C), 10 min at 72 °C, and then cooled to 4 °C. The remainder of PCR 1 is stored at −20 °C. All of the PCR 2 products are PAGE purified. The regenerated pool was extracted into Extraction Buffer B and was desalted using ethanol precipitation with 0.08 M LiCl as a counterion.
Cloning
Round 8 of Selection B was PCR amplified using primers P1 and P3-Opt. The PCR product was PAGE purified, desalted, and incorporated into a plasmid using Invitrogen TOPO TA cloning kit for sequencing and grown in Transform One Shot TOP10 competent cells according to the provided protocol. TOP10 competent cells were plated under sterile conditions and grown overnight. Under sterile conditions, 100 colonies were picked and grown in 4–6 mL of LB. Plasmids were extracted using Qiagen Miniprep.
Sequencing
100 plasmid sequences were submitted for sequencing to ACGT, Inc. Sequencing results were analyzed and aligned by ClustalW2 from the EBI web server.
Gel-Based Activity Assays
Activity assays were carried out to determine the best round of selection and selection conditions to submit for sequencing, to characterize individual sequences. All activity assay experiments were carried out with 32P-labeled DNA, labeled internally for all PCR-generated sequences, dissolved in Milli-Q H2O. DNA was mixed with 2× reaction buffer to initiate the reaction. Selectivity of the sensor for Li+ over other metal ions was tested in the presence of 13 different metal salts. Sensor response to monovalent competing cations (Na+, K+, Rb+, Cs+, NH4+) was tested at 200 mM, while divalent (Mg2+, Ca2+, Sr2+, Ba2+, Mn2+, Co2+, Hg2+, Pb2+) and trivalent (Fe3+, In3+) metal ions were tested at 4 mM and 0.4 mM respectively.
At predetermined time points, 2 μL aliquots of the reaction mixture were removed and added to 20 μL of Stop Solution C (9.2 M urea, 0.001 M H2EDTA, 0.015 M LiOH, 180 mM Tris, 180 mM boric acid, 0.05% xylene cyanol, 0.05% bromophenol blue). Samples were run on 10% dPAGE gels, gels were wrapped in cellophane and exposed to a phosphoimage film, and the image was captured using a Molecular Dynamics Storm 430 Phosphorimager (from Amersham Biosciences). The total percent of DNA cleaved at each time point was quantified using Image Quant (Molecular Dynamics) Software, and each image was adjusted for background. Because of the exceeding slow reaction rate for both the DNA pool and DNAzymes ultimately obtained, the rate was determined using eq 1 in which [P]t is the cleavage at time t, t is time, [P]0 is initial cleavage, and kobs is the pseudo-first-order rate constant or eq 2 when the reaction was run for more than 12 h.
| 1 |
| 2 |
In eq 2 [P]t is cleavage at time t, t is time, [P]∞ is the final cleavage ratio, and kobs is the rate constant.
Reselection
Reselection samples were resuspended in Milli-Q water, and then the reaction was initiated by the addition of 2× positive selection buffer (0.07 M LiCl, 0.001 M EDTA, 0.02 M MOPs pH 7.4). Samples were incubated at room temperature for the allotted incubation time in Table S3 before the addition of Stop Solution B (9.2 M urea, 0.001 M H2EDTA, 0.015 LiOH, 180 mM Tris, 180 mM boric acid). Samples were purified using denaturing PAGE, from which the cleaved sequence was extracted using Buffer C and desalted using ethanol precipitation 0.08 M LiCl.
In Vitro Fluorescence Activity Assay
The enzyme–substrate complex was formed by annealing a mixture of 20-4 enzyme and 20-4 substrate strands for sensor in buffer C with the ratio of 1.6:1. Concentrated Li+ or other metal solutions in buffer C were mixed with annealed complex before recording the fluorescence change. The final concentration of the sensor complex was calculated to be 50 nM. The fluorescence change was continuously monitored by a FluoroMax-2 fluorometer (Horiba Jobin Yvon, Inc., Edison, NJ) every 30 min for at least 6 h. The excitation wavelength was 488 nm, and the emission was monitored at 520 nm.
Cell Line Culture and Transfection
A HeLa cell line was directly purchased from ATCC and cultured in Dulbecco’s modification of Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 U/mL streptomycin, in 25 cm2 culture flasks at 37 °C in a humidified 5% CO2 incubator. The PC12 NS1 cell line was cultured in DMEM F12-K supplemented with 2.5% FBS, 15% horse serum (Cat. No. 16050130) and 100 U/mL penicillin and 100 U/mL streptomycin. To induce PC12 neuronal differentiation, differentiation media (DMEM F12K, 0.25% horse serum, 0.025% FBS and penicillin, streptomycin) supplemented with 50 μg/mL recombinant human β-NGF (PeproTech, Cat. No. 450-01) was prepared and introduced to culture the cells for 24–48 h. Before imaging, cells were plated in 35 mm glass-bottom dishes (MatTek, Cat: P35GC-1.0-14-C) or glass-bottom eight-well chamber slides (ibidi, Cat. No. 80806) and grown to 50–70% confluence.
The DNAzyme sensor was added into the HeLa cell culture Petri dish together with Lipofectamine 3000 transfection reagent (Invitrogen, L3000001). The working DNAzyme sensor concentration is estimated to be 200 nM final concentration. The DNAzyme sensor was added into the PC12 cell culture Petri dish together with Lipofectamine MessengerMAX transfection reagent (Invitrogen, LMRNA001) after comparison with other commercialized cellular delivery reagents. The working DNAzyme sensor concentration is estimated to be 800 nM final concentration. After 4 h transfection, cells were washed thoroughly with PBS to remove the excess amount of DNAzyme in the medium.
Human iPSC Generation
The iPSCs cells were generated by Dr. O’Shea at University of Michigan from the fibroblasts of diagnosed Bipolar 1 Disorder patients or healthy unaffected controls as previous70,71 with the following modifications. Patient fibroblasts were expanded and reprogrammed to pluripotency by transfecting them with episomal expression vectors containing the reprogramming factors: SOX2, OCT4, KLF4, L-MYC, and LIN28 (Epi5 Reprogramming Kit, ThermoFisher). Clonal lines were selected and plated onto Matrigel (Corning) in TesR-E8 (Stemcell Technologies), tested for karyotype, lineage differentiation capacity, for mycoplasma, and to ensure that the episomal vectors were no longer present in the cells.
NPC Differentiation from iPSCs
iPSCs were cultured on Matrigel in TesR-E8 for 4 days. Cell clusters were lifted from the plates using L7 Passaging Solution (Lonza), and suspended in EB medium (DMEM/F12, 20% KnockOut Serum Replacement (Gibco), 1% NEAA, 2 mM GlutaMAX, 0.1 mM β-mercaptoethanol, 2 μM dorsomorphin (R&D Systems) and 5 μM SB431542 (R&D Systems)) for 4 days, with 10 μM ROCK inhibitor Y-27632 (Tocris) for the first day. Then medium was replaced with neural induction medium consisting of DMEM/F12, 1X N2 supplement (Gibco), 1% NEAA, and 2 mM GlutaMAX for another 3 days. The neurospheres were plated onto 100 μg/mL poly-l-ornithine (Sigma)- and 10 ng/mL laminin (Corning)-coated culture dishes. Rosettes were grown and picked after 7 days, dissociated with Accutase (Corning) into single NPCs, and reseeded onto Matrigel-coated plates in NPC maintaining medium (Neurobasal Medium (Gibco), 1× B27 minus vitamin A supplement (Gibco), 2 mM GlutaMAX, 1% NEAA, 1× penicillin/streptomycin and 20 ng/mL FGF2).
Neuronal Differentiation from NPCs
NPCs were reseeded on 20 ng/mL laminin and poly-d-lysine-coated plates in NPC culture medium for 1 day. Then, half-medium change with neuronal differentiation medium composed of BrainPhys medium (Stem Cell, 5792), 1× SM1 supplement (Stem Cell, 5711), 1× N2 supplement, 20 ng/mL BDNF (Peprotech, 450-02-10), 20 ng/mL GDNF (Peprotech, 450-10-10), and 200 nM l-ascorbic acid (Sigma, A92902). The cells were fed by half-change of the medium every 3 days. For the first culture change only, 200 nM compound E (Calbiochem, 565790) was added. In the first 2 weeks of differentiation, 1 mM dibutyryl cyclic AMP (dbcAMP, Cayman, 14408) was added. Once a week, 1 μg/mL laminin was added when doing the half-change. Thereafter, the cells were fed by performing half-media change until Day 28 for future experiments.
Human Neural Cell Delivery
The DNAzyme sensor was added into the cell culture Petri dish together with TurboFect transfection reagent (ThermoFisher Scientific, R0534) after comparison with other commercialized cellular delivery reagents. The working DNAzyme sensor concentration is estimated to be 400 nM final concentration. After 4 h transfection, cells were washed thoroughly with PBS to remove excess amounts of DNAzyme in the medium.
Intracellular Li+ Imaging
After transfection and washing steps with DPBS, HeLa or human neuron cells were immersed in serum-free culture media (DMEM for HeLa, DMEM F12K for PC12 and PC12-induced neurons, Neurobasal for NPC and BrainPhys for neurons) with different final concentrations of Li+ for cell treatment (diluted from 1 M LiCl stock in nuclease-free water). Another 6 h incubation was given to allow exogenous Li+ delivery into cells and trigger the reaction of the activated DNAzyme sensor. Fluorescent images were taken immediately after incubation.
Images were obtained using either Zeiss LSM 710 NLO or Zeiss LSM 880 confocal microscope at 20× and 63× magnification. Fluorescence emission of Hoechst 33258 was measured over 415–475 nm ranges, with excitation at 405 nm. Fluorescence emission of LysoTracker was obtained by exciting at 561 nm and measuring over 570–735 nm. Fluorescence emission of FAM or Alexa488 was obtained by exciting at 488 nm and measuring over 497–550 nm. The pinhole and gain settings were kept constant throughout the whole imaging process.
ICP Measurement of Elements Li and Na
The neurons were differentiated for more than 24 days as described previously. At day 27–30, the cell culture medium was replaced with differentiation media supplemented with 1 mM LiCl. After 8 h incubation, the cells were collected by softly lifting. The cell pellets were then weighed and sent to Microanalysis Lab of University of Illinois at Urbana-Champaign. In brief, the cell pellets were digested with 0.5 mL of nitric acid and were diluted to 25 mL final volume before.
Acknowledgments
The work described in this manuscript is supported by the U.S. National Institutes of Health (R21MH110975 and R35GM141931). Derivation and differentiation of human iPSC lines were supported by 5U19MH106434-05. We would like to thank Dr. Seyed Torabi for his pioneering work with monovalent-selective DNAzymes and guidance with in vitro selection. We wish to thank Gang Xiao and Karen Zhang for their assistance in fluorescence activity assays and Anne B. Farrell for proofreading the manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.1c00843.
Additional experimental materials, figures, and tables including in vitro selection schemes, DNAzyme characterizations, sensor performance in PC12 cells and human cells, and ICP-MS experiments (PDF)
Author Contributions
# C.E.M., Z.Y., and W.G. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Ferrari A. J.; Baxter A. J.; Whiteford H. A. A systematic review of the global distribution and availability of prevalence data for bipolar disorder. J. Affective Disord. 2011, 134 (1), 1–13. 10.1016/j.jad.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Bessonova L.; Ogden K.; Doane M. J.; O’Sullivan A. K.; Tohen M. The Economic Burden of Bipolar Disorder in the United States: A Systematic Literature Review. Clinicoecon Outcomes Res. 2020, 12, 481–497. 10.2147/CEOR.S259338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui T. P.; Kandola A.; Shen L.; Lewis G.; Osborn D. P. J.; Geddes J. R.; Hayes J. F. A systematic review and meta-analysis of clinical predictors of lithium response in bipolar disorder. Acta Psychiatr. Scand. 2019, 140 (2), 94–115. 10.1111/acps.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baastrup P. C.; Schou M. Lithium as a prophylactic agents. Its effect against recurrent depressions and manic-depressive psychosis. Arch. Gen. Psychiatry 1967, 16 (2), 162–72. 10.1001/archpsyc.1967.01730200030005. [DOI] [PubMed] [Google Scholar]
- Volkmann C.; Bschor T.; Kohler S. Lithium Treatment Over the Lifespan in Bipolar Disorders. Front Psychiatry 2020, 11, 377. 10.3389/fpsyt.2020.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S. S. Lithium: A Novel Therapeutic Drug for Traumatic Brain Injury. J. Alzheimers Dis. Parkinsonism 2017, 7 (3), 327. 10.4172/2161-0460.1000327. [DOI] [Google Scholar]
- Geddes J. R.; Burgess S.; Hawton K.; Jamison K.; Goodwin G. M. Long-term lithium therapy for bipolar disorder: systematic review and meta-analysis of randomized controlled trials. Am. J. Psychiatry 2004, 161 (2), 217–22. 10.1176/appi.ajp.161.2.217. [DOI] [PubMed] [Google Scholar]
- McKnight R. F.; Adida M.; Budge K.; Stockton S.; Goodwin G. M.; Geddes J. R. Lithium toxicity profile: a systematic review and meta-analysis. Lancet 2012, 379 (9817), 721–728. 10.1016/S0140-6736(11)61516-X. [DOI] [PubMed] [Google Scholar]
- Tondo L.; Abramowicz M.; Alda M.; Bauer M.; Bocchetta A.; Bolzani L.; Calkin C. V.; Chillotti C.; Hidalgo-Mazzei D.; Manchia M.; Müller-Oerlinghausen B.; Murru A.; Perugi G.; Pinna M.; Quaranta G.; Reginaldi D.; Reif A.; Ritter P.; Rybakowski J. K.; Saiger D.; Sani G.; Selle V.; Stamm T.; Vázquez G. H.; Veeh J.; Vieta E.; Baldessarini R. J. Long-term lithium treatment in bipolar disorder: effects on glomerular filtration rate and other metabolic parameters. Int. J. Bipolar Disord. 2017, 5 (1), 27. 10.1186/s40345-017-0096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurteaux C.; Ripoll C.; Ouznadji S. d.; Ouznadji H.; Wissocq J.-C.; Thellier M. Lithium transport in the mouse brain. Brain Res. 1991, 547 (1), 123–129. 10.1016/0006-8993(91)90582-G. [DOI] [PubMed] [Google Scholar]
- Ríos C.; Cuzmán-Méndez R. Determination of lithium in rat brain regions and synaptosomes by graphite furnace atomic absorption spectrophotometry. J. Pharmacol. Methods 1990, 24 (4), 327–332. 10.1016/0160-5402(90)90016-E. [DOI] [PubMed] [Google Scholar]
- Smith F. E.; Thelwall P. E.; Necus J.; Flowers C. J.; Blamire A. M.; Cousins D. A. 3D 7Li magnetic resonance imaging of brain lithium distribution in bipolar disorder. Mol. Psychiatry 2018, 23 (11), 2184–2191. 10.1038/s41380-018-0016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell M. B.; Berlin J. R.; Lederer W. J. Intracellular calcium in cardiac myocytes: calcium transients measured using fluorescence imaging. Soc. Gen. Physiol. Ser. 1987, 42, 201–214. [PubMed] [Google Scholar]
- Carter K. P.; Young A. M.; Palmer A. E. Fluorescent Sensors for Measuring Metal Ions in Living Systems. Chem. Rev. 2014, 114 (8), 4564–4601. 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron A. T.; Ramos-Torres K. M.; Cotruvo J. A.; Chang C. J. Recognition- and Reactivity-Based Fluorescent Probes for Studying Transition Metal Signaling in Living Systems. Acc. Chem. Res. 2015, 48 (8), 2434–2442. 10.1021/acs.accounts.5b00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotruvo J. A. Jr.; Aron A. T.; Ramos-Torres K. M.; Chang C. J. Synthetic fluorescent probes for studying copper in biological systems. Chem. Soc. Rev. 2015, 44 (13), 4400–4414. 10.1039/C4CS00346B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner S. V.; Arslan H.; Sunbul M.; Yin J.; He C. Dynamic Copper(I) Imaging in Mammalian Cells with a Genetically Encoded Fluorescent Copper(I) Sensor. J. Am. Chem. Soc. 2010, 132 (8), 2567–2569. 10.1021/ja9097324. [DOI] [PubMed] [Google Scholar]
- Chang C. J.; Jaworski J.; Nolan E. M.; Sheng M.; Lippard S. J. A tautomeric zinc sensor for ratiometric fluorescence imaging: Application to nitric oxide-induced release of intracellular zinc. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (5), 1129. 10.1073/pnas.0308079100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. J.; zum Alten Borgloh S. M.; Astori S.; Yang Y.; Bausen M.; Kügler S.; Palmer A. E.; Tsien R. Y.; Sprengel R.; Kerr J. N. D.; Denk W.; Hasan M. T. Single-spike detection in vitro and in vivo with a genetic Ca2+ sensor. Nat. Methods 2008, 5 (9), 797–804. 10.1038/nmeth.1242. [DOI] [PubMed] [Google Scholar]
- Caballero A.; Tormos R.; Espinosa A.; Velasco M. D.; Tárraga A.; Miranda M. A.; Molina P. Selective Fluorescence Sensing of Li+ in an Aqueous Environment by a Ferrocene–Anthracene-Linked Dyad. Org. Lett. 2004, 6 (24), 4599–4602. 10.1021/ol047972m. [DOI] [PubMed] [Google Scholar]
- Stubing D. B.; Heng S.; Abell A. D. Crowned spiropyran fluoroionophores with a carboxyl moiety for the selective detection of lithium ions. Org. Biomol. Chem. 2016, 14 (15), 3752–3757. 10.1039/C6OB00468G. [DOI] [PubMed] [Google Scholar]
- Citterio D.; Takeda J.; Kosugi M.; Hisamoto H.; Sasaki S.-i.; Komatsu H.; Suzuki K. pH-Independent Fluorescent Chemosensor for Highly Selective Lithium Ion Sensing. Anal. Chem. 2007, 79 (3), 1237–1242. 10.1021/ac061674g. [DOI] [PubMed] [Google Scholar]
- Kamenica M.; Kothur R. R.; Willows A.; Patel B. A.; Cragg P. J. Lithium Ion Sensors. Sensors 2017, 17 (10), 2430. 10.3390/s17102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnlaugsson T.; Bichell B.; Nolan C. Fluorescent PET chemosensors for lithium. Tetrahedron 2004, 60 (27), 5799–5806. 10.1016/j.tet.2004.04.082. [DOI] [Google Scholar]
- Yan X.; Le X. C. Fluorescence imaging of Cu(I) in endoplasmic reticulum of live cells and tissue. Sci. China: Chem. 2019, 62 (7), 887–888. 10.1007/s11426-019-9500-6. [DOI] [Google Scholar]
- Pei J. V.; Heng S.; De Ieso M. L.; Sylvia G.; Kourghi M.; Nourmohammadi S.; Abell A. D.; Yool A. J. Development of a Photoswitchable Lithium-Sensitive Probe to Analyze Nonselective Cation Channel Activity in Migrating Cancer Cells. Mol. Pharmacol. 2019, 95 (5), 573. 10.1124/mol.118.115428. [DOI] [PubMed] [Google Scholar]
- McConnell E. M.; Cozma I.; Mou Q.; Brennan J. D.; Lu Y.; Li Y. Biosensing with DNAzymes. Chem. Soc. Rev. 2021, 50, 8954. 10.1039/D1CS00240F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker R. R.; Joyce G. F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1 (4), 223–9. 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Li Y.; Breaker R. R. Deoxyribozymes: New players in the ancient game of biocatalysis. Curr. Opin. Struct. Biol. 1999, 9 (3), 315–323. 10.1016/S0959-440X(99)80042-6. [DOI] [PubMed] [Google Scholar]
- Lyu M.; Kong L.; Yang Z.; Wu Y.; McGhee C. E.; Lu Y. PNA-Assisted DNAzymes to Cleave Double-Stranded DNA for Genetic Engineering with High Sequence Fidelity. J. Am. Chem. Soc. 2021, 143 (26), 9724–9728. 10.1021/jacs.1c03129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake R. J.; Yang Z.; Zhang J.; Lu Y. DNAzymes as Activity-Based Sensors for Metal Ions: Recent Applications, Demonstrated Advantages, Current Challenges, and Future Directions. Acc. Chem. Res. 2019, 52 (12), 3275–3286. 10.1021/acs.accounts.9b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y.; Lu Y. DNA as Sensors and Imaging Agents for Metal Ions. Inorg. Chem. 2014, 53 (4), 1925–1942. 10.1021/ic4019103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Lu Y. Improving Fluorescent DNAzyme Biosensors by Combining Inter- and Intramolecular Quenchers. Anal. Chem. 2003, 75 (23), 6666–6672. 10.1021/ac034924r. [DOI] [PubMed] [Google Scholar]
- Hwang K.; Wu P.; Kim T.; Lei L.; Tian S.; Wang Y.; Lu Y. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew. Chem., Int. Ed. 2014, 53 (50), 13798–802. 10.1002/anie.201408333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Yang Z.; Lu Y. Photocaged functional nucleic acids for spatiotemporal imaging in biology. Curr. Opin. Chem. Biol. 2020, 57, 95–104. 10.1016/j.cbpa.2020.05.003. [DOI] [PubMed] [Google Scholar]
- Zhou W.; Saran R.; Liu J. Metal Sensing by DNA. Chem. Rev. 2017, 117 (12), 8272–8325. 10.1021/acs.chemrev.7b00063. [DOI] [PubMed] [Google Scholar]
- McGhee C. E.; Lake R. J.; Lu Y. Preparation of MetalloDNAzymes. Artificial Metalloenzymes and MetalloDNAzymes in Catalysis 2018, 41–68. 10.1002/9783527804085.ch2. [DOI] [Google Scholar]
- Wu P.; Hwang K.; Lan T.; Lu Y. A DNAzyme-Gold Nanoparticle Probe for Uranyl Ion in Living Cells. J. Am. Chem. Soc. 2013, 135 (14), 5254–5257. 10.1021/ja400150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihms H. E.; Lu Y.. In Vitro Selection of Metal Ion-Selective DNAzymes. In Ribozymes: Methods and Protocols; Hartig J. S., Ed.; Humana Press: Totowa, NJ, 2012; pp 297–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnapen D. J. F.; Sen D. A deoxyribozyme that harnesses light to repair thymine dimers in DNA. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (1), 65. 10.1073/pnas.0305943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D.; Rothenbroker M.; Li Y. DNAzymes: Selected for Applications. Small. Methods 2018, 2 (3), 1700319. 10.1002/smtd.201700319. [DOI] [Google Scholar]
- Geyer C. R.; Sen D. Evidence for the metal-cofactor independence of an RNA phosphodiester-cleaving DNA enzyme. Chem. Biol. 1997, 4 (8), 579–593. 10.1016/S1074-5521(97)90244-1. [DOI] [PubMed] [Google Scholar]
- Cruz R. P. G.; Withers J. B.; Li Y. Dinucleotide Junction Cleavage Versatility of 8–17 Deoxyribozyme. Chem. Biol. 2004, 11 (1), 57–67. 10.1016/j.chembiol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Liu J.; Cao Z.; Lu Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009, 109 (5), 1948–1998. 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian R. C.; Zhou Z. R.; Guo W.; Wu Y.; Yang Z.; Lu Y. Cell Surface Engineering Using DNAzymes: Metal Ion Mediated Control of Cell-Cell Interactions. J. Am. Chem. Soc. 2021, 143 (15), 5737–5744. 10.1021/jacs.1c00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Brown A. K.; Meng X.; Cropek D. M.; Istok J. D.; Watson D. B.; Lu Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (7), 2056. 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Huang H.; Xu N.; Yin Q. Functionalization of cationic poly(p-phenylene ethynylene) with dendritic polyethylene enables efficient DNAzyme delivery for imaging Pb2+ in living cells. J. Mater. Chem. B 2014, 2 (30), 4935–4942. 10.1039/C4TB00680A. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Loh K. Y.; Chu Y.-T.; Feng R.; Satyavolu N. S. R.; Xiong M.; Nakamata Huynh S. M.; Hwang K.; Li L.; Xing H.; Zhang X.; Chemla Y. R.; Gruebele M.; Lu Y. Optical Control of Metal Ion Probes in Cells and Zebrafish Using Highly Selective DNAzymes Conjugated to Upconversion Nanoparticles. J. Am. Chem. Soc. 2018, 140 (50), 17656–17665. 10.1021/jacs.8b09867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Zheng W.; Kwon A. H.; Lu Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000, 28 (2), 481–488. 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro S. W.; Joyce G. F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. U. S. A. 1997, 94 (9), 4262–4266. 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.; Yang Z.; Lake R. J.; Zheng C.; Lu Y. Enzyme-Mediated Endogenous and Bioorthogonal Control of a DNAzyme Fluorescent Sensor for Imaging Metal Ions in Living Cells. Angew. Chem., Int. Ed. 2019, 58 (47), 17061–17067. 10.1002/anie.201910343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong M.; Yang Z.; Lake R. J.; Li J.; Hong S.; Fan H.; Zhang X.-B.; Lu Y. DNAzyme-Mediated Genetically Encoded Sensors for Ratiometric Imaging of Metal Ions in Living Cells. Angew. Chem., Int. Ed. 2020, 59 (5), 1891–1896. 10.1002/anie.201912514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi S.-F.; Wu P.; McGhee C. E.; Chen L.; Hwang K.; Zheng N.; Cheng J.; Lu Y. In vitro selection of a sodium-specific DNAzyme and its application in intracellular sensing. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (19), 5903. 10.1073/pnas.1420361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y.; Zhang J.; Yang Z.; Mou Q.; Ma Y.; Xiong Y.; Lu Y. Functional DNA Regulated CRISPR-Cas12a Sensors for Point-of-Care Diagnostics of Non-Nucleic-Acid Targets. J. Am. Chem. Soc. 2020, 142 (1), 207–213. 10.1021/jacs.9b09211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K. K.; Lowe J. E. W.; Aboud A. A.; Neely M. D.; Redha R.; Bauer J. A.; Odak M.; Weaver C. D.; Meiler J.; Aschner M.; Bowman A. B. Cellular manganese content is developmentally regulated in human dopaminergic neurons. Sci. Rep. 2015, 4 (1), 6801. 10.1038/srep06801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Lu Y.. Fluorescent DNAzyme Biosensors for Metal Ions Based on Catalytic Molecular Beacons. In Fluorescent Energy Transfer Nucleic Acid Probes: Designs and Protocols; Didenko V. V., Ed.; Humana Press: Totowa, NJ, 2006; pp 275–288. [DOI] [PubMed] [Google Scholar]
- Li J.; Lu Y. A Highly Sensitive and Selective Catalytic DNA Biosensor for Lead Ions. J. Am. Chem. Soc. 2000, 122 (42), 10466–10467. 10.1021/ja0021316. [DOI] [Google Scholar]
- O’Shea K. S.; McInnis M. G. Neurodevelopmental origins of bipolar disorder: iPSC models. Mol. Cell. Neurosci. 2016, 73, 63–83. 10.1016/j.mcn.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Mertens J.; Wang Q.-W.; Kim Y.; Yu D. X.; Pham S.; Yang B.; Zheng Y.; Diffenderfer K. E.; Zhang J.; Soltani S.; Eames T.; Schafer S. T.; Boyer L.; Marchetto M. C.; Nurnberger J. I.; Calabrese J. R.; Oedegaard K. J.; McCarthy M. J.; Zandi P. P.; Alda M.; Nievergelt C. M.; Mi S.; Brennand K. J.; Kelsoe J. R.; Gage F. H.; Yao J. The Pharmacogenomics of Bipolar Disorder, S., Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 2015, 527 (7576), 95–99. 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof A. M.; Keros S.; Tyson J. A.; Ying S.-W.; Ganat Y. M.; Merkle F. T.; Liu B.; Goulburn A.; Stanley E. G.; Elefanty A. G.; Widmer H. R.; Eggan K.; Goldstein P. A.; Anderson S. A.; Studer L. Directed Differentiation and Functional Maturation of Cortical Interneurons from Human Embryonic Stem Cells. Cell Stem Cell 2013, 12 (5), 559–572. 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K.; Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126 (4), 663–676. 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Chen H. M.; DeLong C. J.; Bame M.; Rajapakse I.; Herron T. J.; McInnis M. G.; O’Shea K. S. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl. Psychiatry 2014, 4 (3), e375–e375. 10.1038/tp.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. L.; Shamah S. M.; Sun A. X.; Waldman I. D.; Haggarty S. J.; Perlis R. H. Label-free, live optical imaging of reprogrammed bipolar disorder patient-derived cells reveals a functional correlate of lithium responsiveness. Transl. Psychiatry 2014, 4 (8), e428–e428. 10.1038/tp.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. H.; Reimers M.; Maher B.; Williamson V.; McMichael O.; McClay J. L.; van den Oord E. J. C. G.; Riley B. P.; Kendler K. S.; Vladimirov V. I. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr. Res. 2010, 124 (1), 183–191. 10.1016/j.schres.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richelson E. Lithium ion entry through the sodium channel of cultured mouse neuroblastoma cells: a biochemical study. Science 1977, 196 (4293), 1001–2. 10.1126/science.860126. [DOI] [PubMed] [Google Scholar]
- Inoue K.; Zhuang L.; Maddox D. M.; Smith S. B.; Ganapathy V. Human sodium-coupled citrate transporter, the orthologue of Drosophila Indy, as a novel target for lithium action. Biochem. J. 2003, 374 (1), 21–26. 10.1042/bj20030827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwai Y.; Arima R.; Takatsu C.; Furuta R.; Kawasaki T.; Nabekura T. Sodium-phosphate cotransporter mediates reabsorption of lithium in rat kidney. Pharmacol. Res. 2014, 87, 94–8. 10.1016/j.phrs.2014.06.012. [DOI] [PubMed] [Google Scholar]
- Luo H.; Gauthier M.; Tan X.; Landry C.; Poupon J.; Dehouck M. P.; Gosselet F.; Perriere N.; Bellivier F.; Cisternino S.; Decleves X. Sodium Transporters Are Involved in Lithium Influx in Brain Endothelial Cells. Mol. Pharmaceutics 2018, 15 (7), 2528–2538. 10.1021/acs.molpharmaceut.8b00018. [DOI] [PubMed] [Google Scholar]
- Gopal E.; Babu E.; Ramachandran S.; Bhutia Y. D.; Prasad P. D.; Ganapathy V. Species-specific influence of lithium on the activity of SLC13A5 (NaCT): lithium-induced activation is specific for the transporter in primates. J. Pharmacol. Exp. Ther. 2015, 353 (1), 17–26. 10.1124/jpet.114.221523. [DOI] [PubMed] [Google Scholar]
- Bame M.; McInnis M. G.; O’Shea K. S. MicroRNA Alterations in Induced Pluripotent Stem Cell-Derived Neurons from Bipolar Disorder Patients: Pathways Involved in Neuronal Differentiation, Axon Guidance, and Plasticity. Stem Cells Dev. 2020, 29 (17), 1145–1159. 10.1089/scd.2020.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. M.; DeLong C. J.; Bame M.; Rajapakse I.; Herron T. J.; McInnis M. G.; O’Shea K. S. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl. Psychiatry 2014, 4, e375. 10.1038/tp.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.