Summary
Bacterial bioluminescence is widely used to study the spatiotemporal dynamics of bacterial populations and gene expression in vivo at a population level but cannot easily be used to study bacterial activity at the level of individual cells. In this study, we describe the development of a new library of mini‐Tn7‐lux and lux::eyfp reporter constructs that provide a wide range of lux expression levels, and which combine the advantages of both bacterial bioluminescence and fluorescent proteins to bridge the gap between macro‐ and micro‐scale imaging techniques. We demonstrate that a dual bioluminescence‐fluorescence approach using the lux operon and eYFP can be used to monitor bacterial movement in plants both macro‐ and microscopically and demonstrate that Pseudomonas syringae pv phaseolicola can colonize the leaf vascular system and systemically infect leaves of common bean (Phaseolus vulgaris). We also show that bacterial bioluminescence can be used to study the impact of plant immune responses on bacterial multiplication, viability and spread within plant tissues. The constructs and approach described in this study can be used to study the spatiotemporal dynamics of bacterial colonization and to link population dynamics and cellular interactions in a wide range of biological contexts.
Introduction
In recent years, advances in DNA sequencing technology and molecular biology techniques have provided important new insights into the composition and functions of host‐associated bacteria for both plants (Bulgarelli et al., 2012; Rybakova et al., 2017) and animals (Turnbaugh et al., 2008; Le Chatelier et al., 2013). However, although these methods are of pivotal importance in furthering our understanding of the biological processes governing microbial communities and their functions, they often provide limited insight into the spatio‐temporal aspects of host–microbe and microbe–microbe interactions. Such methods typically aggregate information across spatially distributed populations, with information being collected for a limited number of samples and timepoints (Prosser, 2010; Fierer and Ladau, 2012). However, the output of an interaction between a bacterial pathogen and its host (animal or plant) greatly depends on the type of tissue that is colonized, the local microenvironment (spatial relationship) and the movement of the pathogen over time within the host (spatiotemporal relationship) (Lamichhane et al., 2015). The homogenisation of a limited number of biological samples to isolate macromolecules or to enumerate microbial cells can obscure variation in microbial physiology at the macro‐ and micro‐scale. For this reason, visualizing and studying bacteria in their environment continue to be important (Propheter and Hooper, 2015) and considerable effort has been invested into developing tools to allow visualization of bacteria in the host environment (Seleem et al., 2008; Monteiro et al., 2012).
For example, the spatial relationships of microbe–microbe interactions can be studied using fluorescent in situ hybridization (FISH) coupled with confocal laser scanning microscopy (CLSM) (Earle et al., 2015; Soldan et al., 2019). The main advantage of FISH is the observation of spatial relationships between multiple members of a microbial community without necessarily knowing a priori the composition of the community itself. However, FISH requires fixed samples, thus preventing the study of real‐time interactions. For this reason, bacteria tagged with fluorescent proteins (e.g. YFP, GFP) have been extensively used to visualize the spatial and temporal relationships of host–microbe interactions for both detrimental (Godfrey et al., 2010; Cerutti et al., 2017; Donati et al., 2018; Planas‐Marquès et al., 2020) and beneficial (Compant et al., 2005; Fan et al., 2011; Mitter et al., 2017) relationships.
However, while fluorescent proteins allow researchers to investigate bacterial colonization at micro‐scale resolution, they are not suitable for macro‐scale resolution studies, such as analyses of systemic infection. This is partially due to the relatively weak fluorescence of labelled bacteria and the autofluorescence of the host (Wang et al., 2007). This can result in a labour‐intensive process of collecting a series of host samples that can be visualized with fluorescence microscopy in an attempt to localize labelled bacteria without knowing a priori their location. Light‐sheet microscopy, such as the ultramicroscope from La Vision Bio Tec, allows imaging of tissue samples up to centimetres in size (Reynaud et al., 2015), but even with this technology, imaging a whole plant organ, such as a leaf, would not be feasible for many plant species. Moreover, this specialized equipment is not commonly accessible to many laboratories. Wang et al. (2007) proposed that a brighter green‐fluorescent protein variant, called GFPuv, encoded in a broad‐host‐range high‐copy number plasmid could allow monitoring of bacterial disease at whole‐plant level using UV light. However, with a few notable exceptions (Rodríguez‐Moreno et al., 2009; Fujie et al., 2010), this approach has not been widely adopted for macro‐scale imaging. In practise, this is commonly due to limitations in sensitivity and difficulties in imaging acquisition and analysis.
An alternative approach is to use bacteria labelled with the lux operon. Bacterial bioluminescence (Huang et al., 2006; Rico et al., 2010; Brodl et al., 2018; Fleiss and Sarkisyan, 2019) offers several advantages over fluorescent proteins for macro‐scale imaging. Bioluminescence has generally little or no background, excitation light is not needed and emission of light depends on bacterial metabolism, so only active cells are visible (Gregor et al., 2018; Planas‐Marquès et al., 2020). Consequently, bacterial bioluminescence has become a valuable tool to monitor bacterial localization in plants (Fukui et al., 1996; Bogs et al., 1998; Seleem et al., 2008; Xu et al., 2010) and animals (Yu et al., 2004; Burkatovskaya et al., 2006; Mortin et al., 2007; Seleem et al., 2008) at a macroscopic scale. Advanced imaging technologies, such as electron multiplying CCD (EMCDD) cameras, can be used to detect bioluminescence at a single‐cell level, as primarily demonstrated in mammalian cell systems to date (Asai et al., 2008; Iwano et al., 2018). However, this equipment is not widely available within the scientific community. Therefore at present, bioluminescence is most often used for temporal analyses of cell growth, viability and gene expression in bacterial populations using luminometers and macroscopic imaging using standard CCD cameras.
In an attempt to solve the low signal intensity limitation, Gregor et al. (2018) recently developed an enhanced lux operon, called ilux, which could allow single‐cell imaging of Escherichia coli. The ilux operon consists of a series of random mutations within the lux operon and an frp gene from Vibrio campbellii inserted at the end of luxE to maximize brightness (Gregor et al., 2018). The frp gene codes for an FMN reductase responsible for generating FMNH2 which is then oxidized in the process of light production (Gregor et al., 2018). In E. coli, the additional FMN reductase caused a 2.3‐fold increase in brightness, suggesting that FMNH2 generated by the endogenous FMN reductase in E. coli was limiting for lux activity under the conditions tested (Gregor et al., 2018). However, this has not yet been tested in a wide range of bacteria and under a range of environmental conditions.
When using either fluorescent proteins or the lux operon as a bioreporter, the system used to tag bacteria has to be chosen carefully. Ideally, the marker genes should integrate into the bacterial chromosome in a single copy and in a neutral position to avoid disrupting endogenous gene functions (Lambertsen et al., 2004). These criteria are satisfied by the bacterial transposon Tn7 (Craig, 1991) which can be used to insert cloned DNA at high efficiency into a specific intergenic site attTn7, present in the chromosome of many Proteobacteria (Koch et al., 2001).
This study aimed to develop a novel library of mini‐Tn7 delivery plasmids combining the benefits of both bacterial luminescence and fluorescence for macro‐ and micro‐scale resolution imaging, which can be used to chromosomally tag Proteobacteria. We assessed both ilux (Gregor et al., 2018) and modified lux operons (luxCDABE and luxCDABE‐frp, this study) in different bacterial strains to select the brightest constructs before developing the mini‐Tn7 vector library. We demonstrate that these constructs can be used to visualize both local and systemic host–microbe interactions between Phaseolus vulgaris and the bacterial plant pathogen Pseudomonas syringae pv. phaseolicola (Pph) and show that Pph can move systemically through bean leaves by colonizing the vascular system. While we have demonstrated the applicability of these constructs to study spatio‐temporal relationships in plant–microbe interactions, this toolbox will be instrumental to study a wide range of host–microbe, microbe–microbe and microbe–environment interactions in situ.
Results and discussion
Comparison between lux, ilux and lux frp operons
To select the brightest bioluminescence operon and achieve a low detection threshold when visualizing bacteria in the environment, we tested whether ilux (Gregor et al., 2018) was brighter than lux in bacteria other than E. coli. We developed a low copy‐number plasmid, called pRSJ‐pnptII::ilux (Supporting Information Table S1) derived from the reporter plasmid pIJ11282 (Frederix et al., 2014), in which lux was replaced with ilux. Additionally, to test the effect of frp on lux‐mediated bioluminescence, we included frp from pGEX(−) (Gregor et al., 2018) after luxE in pIJ11282, generating pRSJ‐pnptII::lux‐frp (Supporting Information Table S1). The three plasmids pIJ11282, pRSJ‐pnptII::ilux and pRSJ‐pnptII:lux‐frp have the same backbone and promoter.
pRSJ‐pnptII::ilux did not yield brighter bioluminescence than pIJ11282 in Acinetobacter baylyi strain ADP1, Pseudomonas fluorescens NZ011 and Pseudomonas syringae pv. phaseolicola (Pph) strain 1302A (Supporting Information Fig. S1A). On the contrary, in P. fluorescens NZ011, pRSJ‐pnptII::ilux was approximately 20‐fold less bright than pIJ11282 in both M9 minimal medium and King's B (KB) medium (Supporting Information Fig. 1A). We speculate that the mutations introduced into the ilux operon were optimized for light production under the conditions tested in E. coli and similar results are not guaranteed for other bacterial strains. However, pRSJ‐pnptII:lux‐frp was two‐fold brighter than pIJ11282 in P. fluorescens NZ011 without affecting bacterial growth (Supporting Information Fig. S1B), indicating that, depending on the bacterial strain, an additional FMN reductase can increase the amount of light produced. FMN reductases oxidize NADPH to NADP+ in order to reduce FMN to FMNH2. To test for the activity of the FMN reductase encoded by the frp gene, we measured the NADP+ concentration of cell extracts of P. fluorescens NZ011 harbouring pIJ11282 or pRSJ‐pnptII:lux‐frp. We observed a higher concentration of NADP+ in bacterial cells expressing the frp gene, which supports the conclusion that the increased bioluminescence of P. fluorescens NZ011 (pRSJ‐pnptII:lux‐frp) relative to P. fluorescens NZ011 (pIJ11282) is due to increased FMN reductase activity (Supporting InformationFig. S2).
Generation of an improved mini‐Tn7 vector library for bioluminescence and combined bioluminescence‐fluorescence expression
To combine the benefit of using bacterial bioluminescence for macro‐scale localization with the advantages of fluorescent proteins for micro‐scale localization, we developed a library of mini‐Tn7 plasmids containing both a bioluminescent operon and eYFP (Supporting Information Fig. S3). eYFP was chosen over other fluorescent proteins for its higher brightness over eGFP (Shaner et al., 2005) and for its peak emission spectra (527 nm), which would easily allow visualization in an environment with red autofluorescence (e.g. leaves). To provide a wide range of expression levels of the bioluminescent operon, which would allow researchers to modularly choose the best expression level according to their experimental needs, we designed the library to have five different constitutive promoters driving lux transcription. Promoters were selected to be highly conserved, constitutive and to provide a range of expression levels. For these reasons, we chose the recA promoter (Giliberti et al., 2006), named p OXB20 and its derivatives p OXB16, p OXB13 and p OXB11 (Oxford Genetics Limited, https://www.oxgene.com/Products; Lynn et al., 2015; Presnell et al., 2019), and the promoter p nptII (Wright and Beattie, 2004). We also included two different constitutive promoters driving the expression of eYFP, namely p lac and p A1/04/03 (Koch et al., 2001). p lac is a common and conserved constitutive promoter, while p A1/04/03 has been widely adopted for gene expression in Pseudomonas (Godfrey et al., 2010)..
To generate the mini‐Tn7 vector library, we used the two cassettes with the highest light production in the tested bacterial strains, which were the lux and the modified lux and frp operons (Supporting Information Fig. S1). Initially, we used pUC18‐mini‐Tn7T‐Gm‐lux (Choi et al., 2005) as a backbone. Plasmid pUC18‐mini‐Tn7T‐Gm‐lux was developed to easily insert a promoter of interest into a multiple cloning site (MCS) upstream of luxC. However, this backbone has a different ribosome binding site (RBS) upstream of luxC than the RBS found upstream of luxC in pIJ11282 (Frederix et al., 2014). The latter RBS has been predicted to increase the translation initiation rate more than fourfold (14243 arbitrary units compared to 3371 calculated with the RBS calculator; Farasat et al., 2014; Ng et al., 2015). Additionally, the upstream sequence of luxC, encompassing the MCS in pUC18‐mini‐Tn7T‐Gm‐lux was found to negatively affect transcription (Glassing and Lewis, 2015) (Supporting Information Fig. S4A). We therefore constructed a second plasmid (pRS‐pOXB20::lux‐pA1/04/03::eYFP) containing the RBS from pIJ11282 without the long MCS of pUC18‐mini‐Tn7T‐Gm‐lux (Supporting Information Fig. S4B).
We tested both constructs in Pph 1302A and found that the promoter with the alternate RBS and without the MCS of pUC18‐mini‐Tn7T‐Gm‐lux (p OXB20) resulted in four to seven times more luminescence compared to the original promoter (p OXB20(1)) in both KB and M9 media, with no consequences for bacterial growth (Supporting Information Fig. S5A and B).
As we found these backbones to be an improvement over constructs based on pUC18‐mini‐Tn7T‐Gm‐lux, we also included constructs with the lux operon by itself in our library, without eYFP (Table 1; Supporting Information Table S1). The five different constitutive promoters used encompass a 15‐fold range between the weakest promoter p OXB11 and the strongest promoter p nptII in Pph 1302A (Fig. 1; Supporting Information Fig. S6A). We observed a small but significant decrease in bacterial growth both in vitro and in planta when comparing Pph 1302A with Pph 1302A pnptII::lux‐pA1/04/03::eYFP inoculated into the susceptible host plant P. vulgaris cultivar Canadian Wonder (Supporting InformationFig. 6B and Fig. 7A and B). However, we did not detect any difference between the growth of Pph 1302A and Pph 1302A pOXB16::lux‐pA1/04/03::eYFP (intermediate expression) or Pph 1302A pOXB11::lux‐pA1/04/03::eYFP (low expression) in planta (Supporting Information Fig. S7B).
Table 1.
Mini‐Tn7 vector library constructed in this study. a
| Lux | Lux and eYFP | Lux and eYFP |
|---|---|---|
| pRS‐pnptII::lux | pRS‐pnptII::lux‐pA1/04/03::eYFP | pRS‐pnptII::lux‐plac::eYFP |
| pRS‐pOXB20::lux | pRS‐pOXB20::lux‐pA1/04/03::eYFP | pRS‐pOXB20::lux‐plac::eYFP |
| pRS‐pOXB16::lux | pRS‐pOXB16::lux‐pA1/04/03::eYFP | pRS‐pOXB16::lux‐plac::eYFP |
| pRS‐pOXB13::lux | pRS‐pOXB13::lux‐pA1/04/03::eYFP | pRS‐pOXB13::lux‐plac::eYFP |
| pRS‐pOXB11::lux | pRS‐pOXB11::lux‐pA1/04/03::eYFP | pRS‐pOXB11::lux‐plac::eYFP |
| pRS‐pnptII::lux‐frp | pRS‐pnptII::lux‐frp‐pA1/04/03::eYFP | pRS‐pnptII::lux‐frp‐plac::eYFP |
| pRS‐pOXB20::lux‐frp | pRS‐pOXB20::lux‐frp‐pA1/04/03::eYFP | pRS‐pOXB20::lux‐frp‐plac::eYFP |
For more details, see the Supporting Information Table S1.
Fig. 1.
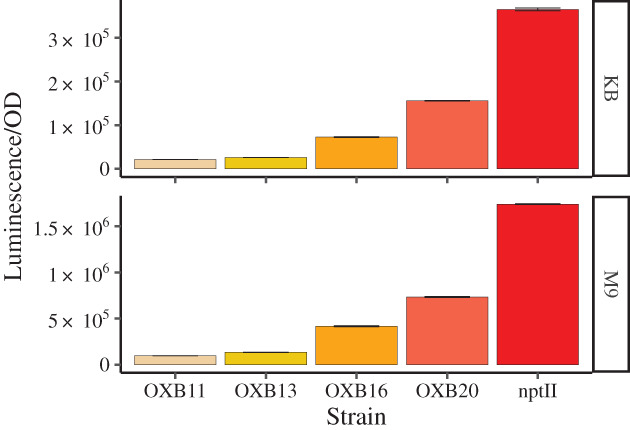
A library of mini‐Tn7 constructs with different promoters upstream of theluxoperon provides a wide dynamic range of luminescence when introduced intoPseudomonas syringaepv. phaseolicola1302A (Pph1302A). Luminescence values were detected at 4 h. Time series data are presented in the Supporting Information Fig. S6.OXB11:Pph1302A pOXB11::lux‐pA1/04/03::eYFP;OXB13:Pph1302A pOXB13::lux‐pA1/04/03::eYFP;OXB16:Pph1302A pOXB16::lux‐pA1/04/03::eYFP;OXB20:Pph1302A pOXB20::lux‐pA1/04/03::eYFP;nptII:Pph1302A pnptII::lux‐pA1/04/03:::eYFP. KB (King's B medium), M9 (M9 minimal medium). OD = optical density at 600 nm. Error bar ± SE.n = 3. [Color figure can be viewed at wileyonlinelibrary.com]
Access to a range of promoters will enable researchers to choose an expression level of the lux cassette that is suitable for their experimental conditions, balancing both signal strength and any fitness cost observed (e.g. purpose of the experiment, equipment used to detect luminescence). While we would expect the constructs to give different expression levels in different bacterial strains, the constructs should maintain a wide expression range as the OXB promoters (Oxford Genetics Limited, UK) were all derived from the constitutive and conserved recA promoter (Weisemann and Weinstock, 1991).
Pseudomonas syringae pv. phaseolicola colonizes the vascular system of Phaseolus vulgaris leaves
We applied a combined luminescence‐fluorescence approach to investigate whether Pph systemically infects P. vulgaris leaves. Pph, the causal agent of halo blight of bean, is known to be a seed‐borne pathogen and to be able to cause systemic symptoms and spread within infected plants (Taylor et al., 1979), but to date, there is limited knowledge of the routes and mechanisms used by Pph to spread systemically within host plants.
We tagged Pph RJ3 (Jackson et al., 2000) using pRS pnptII::lux‐pA1/04/03::eYFP (Table 1, Supporting Information Table S1) as p nptII was the strongest promoter (Fig. 1) and pA1/04/03 has previously been successfully used in Pph (Godfrey et al., 2010). Pph RJ3 differs from Pph 1302A as it lacks a > 40 Kb genomic island that contains the gene avrPphB, which encodes a secreted effector protein that elicits effector triggered immunity (ETI) in plants carrying the resistance gene R3 (Jackson et al., 2000). Due to the loss of avrPphB, Pph RJ3 can successfully colonize P. vulgaris cultivar Tendergreen (TG) without triggering a plant immune response known as the hypersensitive response (HR), which acts to restrict bacterial growth. In experiments with bacteria incubated in agar plates, we calculated that the detection limit for tagged cells, in terms of number of cells detectable per unit area, was approximately 100 CFU mm−2 (Supporting Information Fig. S8) and observed a high correlation (0.99) between luminescence and CFU, as also reported by Fan et al. (2008).
We used bacterial bioluminescence to macroscopically observe Pph RJ3 in P. vulgaris cultivar TG leaves. We detected a clear co‐localization of Pph RJ3 with the leaf vasculature within 3 days after syringe infiltration (Fig. 2A). We then used this information to select and collect leaf samples (vasculature cross sections) from leaves infected with Pph RJ3 to be visualized with confocal microscopy. This confirmed that Pph RJ3 was localized inside the leaf vasculature (Fig. 2B and C). Both data types (bioluminescence and fluorescence) showed the ability of Pph RJ3 to move from the infection site.
Fig. 2.
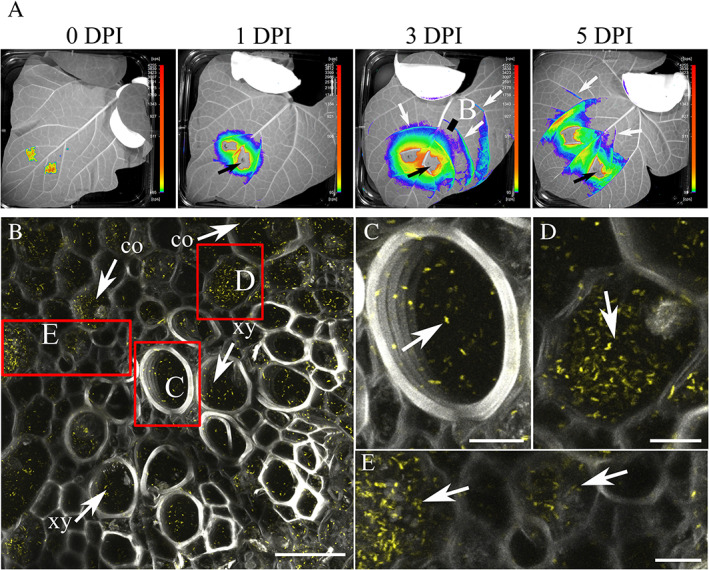
Pseudomonas syringae pv. phaseolicola RJ3 (Pph RJ3) colonizes the leaf vasculature of P. vulgaris cultivar TG.
A. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP was syringe‐infiltrated in localized areas on the abaxial surface of Phaseolus vulgaris cv. Tendergreen leaves. Images were taken after 0, 1, 3, 5 DPI with the nightOWL LB 983 at 10‐min exposure. The black line indicates cross sections used for confocal microscopy imaging (B). The white arrows indicate co‐localization of Pph RJ3 pnptII::lux‐pA1/04/03::eYFP with the leaf vasculature. The black arrows indicate regions of the leaves that have cts values higher than the maximum detection limit.
B. Maximum projections of the leaf vasculature cross section. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow). Xylem autofluorescence (bright grey). Collenchyma autofluorescence (dark grey). co; collenchyma. xy; xylem. Red squares indicate areas shown at higher magnification in panels C–E. Scale bar: 40 μm.
C. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow; white arrow). Xylem autofluorescence (grey).
D. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow; white arrow). Collenchyma autofluorescence (dark grey).
E. Zoom of panel B. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow; white arrow). Collenchyma autofluorescence (dark grey). Scale bar: 10 μm. [Color figure can be viewed at wileyonlinelibrary.com]
We were able to detect Pph RJ3 cells within the xylem vessels (Fig. 2B and C), and in the collenchyma (Fig. 2B, D, and E). Pph RJ3 appeared to accumulate to higher densities in the collenchymatic cells of the leaf vascular system than in the xylem (Fig. 2D and E; Supporting Information Video S1). Pph RJ3 movement in the xylem was more rapid than movement observed in collenchymatic cells (Supporting Information Video S2), suggesting that Pph RJ3 moves within, but does not produce biofilms inside the xylem. We confirmed that Pph RJ3 can colonize the vascular system of intact leaves following epiphytic colonization by spray‐inoculating Pph RJ3 onto P. vulgaris leaves (Supporting Information Fig. S9). Images of negative controls show that there is little or no background signal in the absence of tagged bacteria (Supporting Information Fig. S10).
Pph strains have been reported to show substantial variation in their ability to cause systemic symptoms in host plants, while host cultivars have been reported to vary in their susceptibility to systemic symptoms. This has been attributed in some studies to systemic movement of the toxin, phaseolotoxin, within host plants (Mitchell and Bieleski, 1977). Our study confirms that the pathogen itself can move systemically within host tissues, although under our experimental conditions Pph RJ3 was not an aggressive systemic pathogen, consistent with the absence of detectable bacterial biofilms in the xylem and wilting symptoms, which are often the manifestation of systemic bacterial infections (Bae et al., 2015). The constructs and methods established in this study will provide valuable tools to investigate the underlying causes of variation in systemic infection and will enable researchers to better understand the biology and epidemiology of this disease.
Bacterial bioluminescence can be used to monitor the effect of plant immune responses on bacterial growth and viability in planta
Bacterial bioluminescence has been proposed as a high‐throughput method to detect in planta growth of bacterial pathogens (Fan et al., 2008). Fan et al. (2008) showed that Pseudomonas syringae tagged with luxCDABE can be used as a quantitative assay to monitor bacterial growth and that CFU is correlated with bacterial bioluminescence. Moreover, luminescence allows the detection of bacterial growth without performing tissue extraction, serial dilutions, plating, and scoring, thus reducing the time needed for the experiment. We therefore examined whether the luminescent constructs developed in this study could also be used to rapidly and sensitively monitor the effect of different plant immune responses on bacterial growth and viability in planta. As expression of high levels of lux activity could have a detrimental effect on bacterial fitness in the plant environment, we examined whether a relatively weak promoter driving lux expression could be used for this purpose.
We tagged Pph 1302A ΔhrpA (Supporting Information Table S2), Pph 1302A ΔxerC (Lovell et al., 2009) and Pph 1302A ΔavrPphB (Supporting Information Table S2) using pRS‐pOXB13::lux (Table 1; Supporting Information Table S1). Pph 1302A ΔxerC (Lovell et al., 2009) lacks a recombinase (XerC) that is able to excise the genomic island containing avrPphB, thus it stably triggers ETI in P. vulgaris cultivar TG. On the contrary, Pph 1302A is able to excise the island and overcome resistance. Pph 1302A ΔhrpA (this study; Supporting Information Table S2) cannot assemble a functioning type III secretion system (T3SS), thus it cannot secrete effectors to counteract plant defences. Therefore, its growth in planta is suppressed by PAMP‐Triggered Immunity (PTI) (Jones and Dangl, 2006). Pph 1302A ΔPphB (this study; Supporting Information Table S2) has a functional T3SS, but lacks the avr gene avrPphB, thus it can suppress PTI and colonize TG leaves. This compatible interaction is named effector‐triggered susceptibility (ETS) (Jones and Dangl, 2006).
A 0 days post inoculation (DPI) we could not detect any difference between strains for both luminescence value (cts) [ANOVA, F(2,14) = 0.0616,P = 0.94] and infected area (mm2) [ANOVA, F(2,14) = 0.18, P = 0.83] (Fig. 3A–C). At 2 DPI, both luminescence signal (cts) [ANOVA, F(2,14) = 35.01, P < 0.0001] and infected area (mm2) [ANOVA, F(2,14) = 19.21, P < 0.0001] for the different strains displayed significant differences using a two way ANOVA, as well as pairwise comparisons between strains (Fig. 3A–C), indicating that both bacterial bioluminescence and infected area can be used to detect the effect of different plant immune responses on bacterial viability and growth, with significantly lower bioluminescence in the context of PTI and ETI.
Fig. 3.
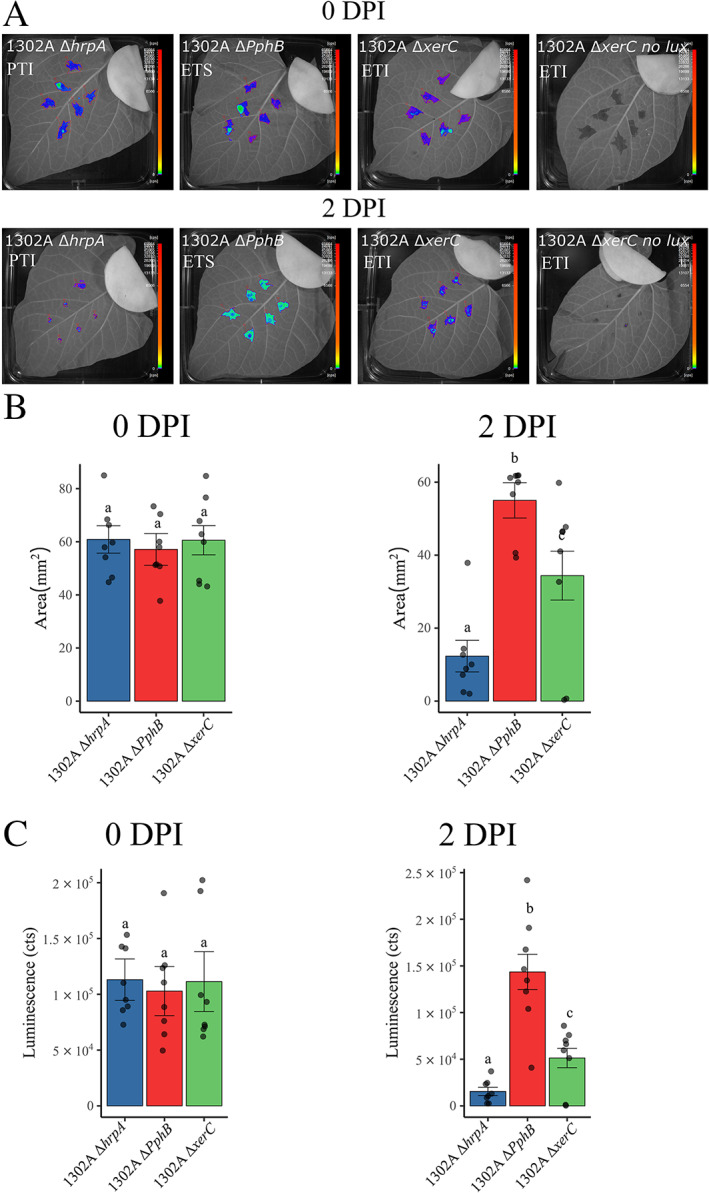
Bacterial bioluminescence can be used to study the effect of plant immune responses on bacterial growth and viability. Phaseolus vulgaris cultivar Tendergreen leaves were syringe infiltrated with 106 CFU ml−1 of Pseudomonas syringae pv. phaseolicola 1302A (Pph 1302A) ΔhrpA, ΔPphB and ΔxerC tagged with pOXB13::lux.
A. After infiltration, leaves were kept in the dark for 10 min and then imaged with the nightOWL LB 983 at 2 min exposure.
B. After 2 days post inoculation, leaves were detached from the plants, kept in the dark for 10 min and imaged with the nightOWL LB 983 at 5 min exposure. 1302A ΔPphB : Pph 1302A ΔPphB pOXB13::lux; 1302A ΔxerC : Pph1302A ΔxerC pOXB13::lux; 1302A ΔhrpA : Pph 1302A ΔhrpA pOXB13::lux; 1302A ΔxerC no lux: Pph 1302A ΔxerC. Luminescence and area values are reported for the eight biological replicates. Technical replicates (spots within the same leaf) were averaged. Error bar ± SE. n = 8. Significant differences (Student's T‐test, P < 0.05) are indicated by letters. [Color figure can be viewed at wileyonlinelibrary.com]
Colony count analyses performed at 2 DPI agreed with luminescence data [ANOVA, F(2,14) = 56.73, P < 0.0001] identifying a statistically significant effect of strain. However, pairwise comparisons failed to distinguish between plants infected with Pph 1302A ΔxerC and Pph 1302A ΔPphB [t(14) = 1.6, P = 0.06] (Supporting Information Fig. 11A and B). The higher variability of colony count data compared to bioluminescence data (Fig. 3A–C; Supporting Information Fig. S11B) may be associated with variability introduced during tissue extraction, serial dilution and plating that are avoided in bioluminescence detection. Therefore, the bioluminescent reporter constructs developed in this study could be used to effectively monitor the effect of plant immune responses on bacterial growth, spread and viability in interactions between Pph and P. vulgaris.
Conclusion
Dual bioluminescence and fluorescence reporter constructs have previously been described for both gram‐negative and gram‐positive bacteria and used to study transcription, metabolic activity and host colonization (Unge et al., 1999; Unge and Jansson, 2001; Perehinec et al., 2007; Benedetti et al., 2012; Kim et al., 2018). Here, we describe the development of a flexible library of mini‐Tn7 constructs that can be used to tag Gram‐negative bacteria with a combination of lux, lux‐frp and lux‐eYFP operons with a wide selection of promoter strengths to drive lux expression. The strongest promoter used (p nptII ), together with a modified RBS and removal of the MCS, gives a fourfold to sevenfold increase in luminescence over a previously described mini‐Tn7‐lux construct. We also show that introducing an additional FMN reductase can positively increase bioluminescence in some bacterial strains and that the relative performance of lux and ilux depends on the bacterial strain used.
We have demonstrated the application of these reporter constructs by showing that Pph is able to systemically colonize the leaves of P. vulgaris. By combining bioluminescence and fluorescence, the process of collecting samples to be imaged with confocal microscopy could be greatly accelerated. Interestingly, Pph did not form biofilms within the xylem vessels, where they were observed to be highly mobile. Instead Pph preferentially colonized collenchymatic cells. While previous studies have shown that Pph causes systemic symptoms in P. vulgaris (Zaiter and Coyne, 1984), the question of whether, and how Pph is able to move systemically through host plants has not been fully addressed. Having shown that Pph can colonize the leaf vascular system it will be of interest to elucidate the molecular mechanisms underpinning vascular colonization by Pph and to investigate the contribution of vascular colonization to the development of systemic symptoms and to seed‐borne transmission of Pph.
Our results also show that bacterial bioluminescence can be used to rapidly detect the effect of different plant immune responses on bacterial viability and growth and to monitor the effectiveness of plant immune responses in restricting systemic colonization of plant tissues. Importantly, bioluminescence is a sensitive indicator of bacterial viability and metabolic activity as well as localisation, while the relatively high stability of fluorescent proteins means that they can be used to detect both living and metabolically inactive or dead cells. Thus, it is of interest to note that while ETI and PTI could be observed to restrict the movement and population levels of Pph in infected tissue, we were still able to detect a substantial number of bioluminescent cells 2 days, and even 20 days after infection (Fig. 3A; data not shown), showing that neither PTI nor ETI eliminated invading bacteria. Bioluminescent imaging may be of particular value in studying diseases such as bacterial leaf streak of wheat caused by Xanthomonas translucens pv. undulosa, or bacterial blight of rice caused by Xanthomonas oryzae pv. oryzae, in which resistance is typically assessed by monitoring lesion length or area, allowing rapid comparative imaging of bacterial spread within host plants even before lesions develop (Kandel et al., 2012; Oliva et al., 2019; Sapkota et al., 2020). We have already confirmed that the constructs described here can be used to transform and produce bioluminescence in Xanthomonas (data not shown).
Mini‐Tn7 constructs have previously been used to transform a wide range of Proteobacteria by exploiting the presence of a conserved attTn7 site downstream of the glmS gene, in which the effects of transposon insertion have generally been found to be relatively neutral (Choi et al., 2005; Choi and Schweizer, 2006). More recently, researchers have shown that the range of bacteria that can be tagged using this system can be further expanded through the introduction of an artificial attTn7 site, which can also be used to extend the use of this system to bacteria in which insertion into the pre‐existing attTn7 site has been found to not be neutral (Figueroa‐Cuilan et al., 2016). We therefore believe that the constructs and approaches described in this study will be a valuable resource for researchers who are interested in tracking both the spatio‐temporal and physiological dynamics of bacterial populations in a wide range of biological contexts.
Experimental procedures
Bacterial strains, media and growth conditions
E. coli DH5α strains carrying the plasmids listed in the Supporting Information Table S1 were inoculated from frozen glycerol stock onto Luria Bertani (LB) (Sambrook et al., 1989) agar (1.5% agar) and grown for 24 h at 37°C. Single colonies were used to inoculate 10 ml LB cultures, which were incubated at 37°C with shaking at 200 r.p.m. Pseudomonas and Acinetobacter strains were routinely grown in the same manner but incubated at 28°C. Antibiotics were included where relevant at the following concentrations: gentamicin (10 μg ml−1), tetracycline (10 μg ml−1), kanamycin (20 μg ml−1), carbenicillin (50 μg ml−1), nitrofurantoin (25 μg ml−1).
Plant growth conditions
Phaseolus vulgaris cultivars Tendergreen (TG) and Canadian Wonder were grown at 20–22°C, 70% humidity and with artificial light maintained for 8 h periods within the 24‐h cycle. Two‐week old plants (first true leaves fully developed) were used for all experiments.
Cloning, restriction enzyme digestion and polymerase chain reaction
Cloning was performed using Gibson assembly (Gibson et al., 2009) and constructs heat‐shock transformed into E. coli strain DH5α. Transformants harbouring the lux operon were screened for luminescence using a nightOWL LB 983 in vivo imaging system (Berthold Technologies, Germany). For constructs combining luminescence and fluorescence, transformants were screened for both luminescence and fluorescence using a nightOWL LB 983 in vivo imaging system (Berthold Technologies) and a FastGene blue light LED illuminator (Geneflow Ltd, UK) respectively. PCR was conducted according to the manufacturer's instructions using Q5 high‐fidelity DNA polymerase (Thermo Fisher Scientific) unless otherwise specified. Colony‐PCR was performed according to the manufacturer's instructions using GoTaq Green Master Mix (Promega Corporation, USA). DNA Sanger sequencing was performed by Source Bioscience (UK). Restriction enzyme digestions were performed according to the manufacturer's instructions (Thermo Fisher Scientific).
Generation of a low copy‐number plasmid harbouring ilux and lux constructs
pRSJ‐pnptII::ilux was derived from pIJ11282 (Frederix et al., 2014) and pGEX(−) (Gregor et al., 2018). Briefly, pIJ11282 was digested with PstI and SnaBI to remove the lux operon. ilux was amplified with primers OXRS_F1 and OXRS_R1 (Supporting Information Table S3) from pGEX(−). The PCR product was ligated into PstI/SnaBI‐digested pIJ11282 and transformed into E. coli. Potential clones were selected in LB agar supplemented with 10 μg ml−1 tetracycline. DNA sequencing was performed with primers listed in the Supporting Information Table S3.
pRSJ‐pnptII::lux‐frp was generated by ligating frp from pGEX(−) into pIJ11282. Briefly, frp was amplified with primer OXRS_F2 and OXRS_R2 (Supporting Information Table S3) from pGEX(−) and ligated into PstI‐digested pIJ11282. Transformants were selected on LB agar supplemented with 10 μg ml−1 tetracycline. DNA sequencing was performed using primer OXRS_S1 (Supporting Information Table S5).
Generation of a mini‐Tn7 vector library for lux expression
pRS‐pOXB20(1)::lux was generated by ligating the OXB20 promoter (Oxford Genetics Limited, UK) synthesized by Eurofins Genomics into SmaI‐digested pUC18‐mini‐Tn7T‐Gm‐lux (Choi et al., 2005). To make pUC18‐mini‐Tn7T‐Gm‐lux compatible for conjugation, oriT was PCR amplified with primers OXRS_FT and OXRS_RT (Supporting Information Table S3) from pK18mobsacB (Schäfer et al., 1994) and ligated into EheI‐digested pUC18‐mini‐Tn7T‐Gm‐lux. Potential clones were selected on LB agar supplemented with 10 μg ml−1 gentamicin. DNA sequencing was performed using primer OX26 (Supporting Informationy Table S4).
pRS‐pnptII::lux was generated by ligating lux amplified with primers OXRS_F3 and OXRS_R3 (Supporting Information Table S3) from pIJ11282 into the pUC18T backbone amplified with primers OXRS_F4 and OXRS_R4 (Supporting Information Table S3) from pUC18T‐mini‐Tn7T‐Gm‐dsRedExpress (Choi et al., 2005). Transformants were selected on LB agar supplemented with 10 μg ml−1 gentamicin. DNA sequencing was performed using primers listed in the Supporting Information Table S6.
pRS‐pOXB20::lux, pRS‐pOXB16::lux, pRS‐pOXB13::lux and pRS‐pOXB11::lux plasmids were generated by ligating OXB20, OXB16, OXB13 and OXB11 promoters (Oxford Genetics Limited, UK) into PdiI (NaeI)/SnaBI‐digested pRS‐pnptII::lux. Transformants were selected on LB agar supplemented with 10 μg ml−1 gentamicin. DNA sequencing was performed using primer OXB26 (Supporting Information Table S4).
For constructs pRS‐pOXB20::lux and pRS‐pnptII::lux, we additionally included frp downstream of the lux operon. Briefly, frp was amplified from pGEX(−) with primers OXRS_F5 and OXRS_R5 (Supporting Information Table S3) and ligated into MscI‐digested pRS‐pnptII::lux and MscI‐digested pRS‐pOXB20::lux. DNA sequencing was performed using primer OXRS_S1 (Supporting Information Table S5).
Generation of a mini‐Tn7 vector library for combined bioluminescence and fluorescence expression
pA1/04/03::eYFP amplified with OXRS_F6 and OXRS_R6 (Supporting Information Table S3) from miniTn7(Gm)PA1/04/03‐eyfp‐a (Klausen et al., 2003), was ligated into MscI‐digested pRS‐pnptII::lux, pRS‐pOXB20::lux, pRS‐pOXB16::lux, pRS‐pOXB13::lux and pRS‐pOXB11::lux plasmids. pA1/04/03::eYFP amplified with OXRS_F7 and OXRS_R7 (Supporting Information Table S3) from miniTn7(Gm)PA1/04/03‐eyfp‐a (Klausen et al., 2003), was ligated into MscI‐digested pRS‐pnptII::lux‐frp and pRS‐pOXB20::lux‐frp. Transformants were selected on LB agar supplemented with 10 μg ml−1 gentamicin. For plasmids pRS‐pnptII::lux‐pA1/04/03::eYFP, pRS‐pOXB20::lux‐pA1/04/03::eYFP, pRS‐pOXB16::lux‐pA1/04/03::eYFP, pRS‐pOXB13::lux‐pA1/04/03::eYFP and pRS‐pOXB11::lux‐pA1/04/03::eYFP, DNA sequencing was performed using primer OXRS_S1 (Supporting Information Table S5). For plasmids pRS‐pnptII::lux‐frp‐pA1/04/03::eYFP and pRS‐pOXB20::lux‐frp‐pA1/04/03::eYFP DNA sequencing was performed using primer OXRS_S2 (Supporting Information Table S5).
The same cloning procedure, primers and PCR conditions used to ligate pA1/04/03::eYFP into pRS‐pnptII::lux, pRS‐pOXB20::lux, pRS‐pOXB16::lux, pRS‐pOXB13::lux, pRS‐pOXB11::lux, pRS‐pnptII::lux‐frp and pRS‐pOXB20::lux‐frp plasmids were used to amplify plac::eYFP from pRS plac::eYFP (this study) and ligate it into MscI‐digested pRS‐pnptII::lux, pRS‐pOXB20::lux, pRS‐pOXB16::lux, pRS‐pOXB13::lux, pRS‐pOXB11::lux, pRS‐pnptII::lux‐frp and pRS‐pOXB20::lux‐frp.
pRS‐pOXB20(1)::lux‐pA1/04/03::eYFP was generated by ligating pA1/04/03::eYFP amplified with primers OXRS_F8 and OXRS_R8 from miniTn7(Gm)PA1/04/03‐eyfp‐a (Klausen et al., 2003) into StuI‐digested pRS‐pOXB20(1)::lux. Potential clones were selected on LB agar supplemented with 10 μg ml−1 gentamicin.
Construction of Pph 1302A PphB and hrpA knock‐out mutants
To generate the Pph 1302A avrPphB knock‐out mutant (Pph 1302A PphB‐), 1 kb genomic regions flanking PphB were PCR amplified with primers OXRS_F9 and OXRS_R9 (left end) and primers OXRS_F10 and OXRS_R10 (right end) (Supporting Information Table S3). Primer OXRS_R9 was designed to include the starting codon of avrPphB while primer OXRS_R10 included the stop codon of avrPphB. PCR products were ligated into pK18mobsacB (Schäfer et al., 1994) amplified with primers OXRS_F11 and OXRS_R11 (Supporting Information Table S3). Transformants were selected in LB agar supplemented with 20 μg ml−1 kanamycin. Colony‐PCR was performed with primers OXRS_F12 and OXRS_R12 (Supporting Information Table S3) to validate the assembly. The plasmid was conjugated into Pph 1302A, as described below, and transformants were plated onto LB plates supplemented with kanamycin and nitrofurantoin. Transformants were the result of plasmid integration (a single recombination event). To allow allelic exchange, four colonies were cultured overnight in LB and 20 μl plated onto LB (1.5% agar) supplemented with 10% sucrose. Colony PCR with primers OXRS_F9 and OXRS_R10 (Supporting Information Table S3) was used to confirm the deletion of avrPphB from the Pph 1302A genome.
A similar procedure was used to generate Pph 1302A ΔhrpA. Briefly, hrpA upstream and downstream regions were amplified from the Pph 1302A genome with primers OXRS_F13, OXRS_R13 and OXRS_F14, OXRS_R14 (Supporting Information Table S3) and ligated into pK18mobsacB amplified with primers OXRS_F11 and OXRS_R11 (Supporting Information Table S3). Conjugation and selections of knock‐out mutants were carried out as described for Pph 1302A ΔPphB.
Mini‐Tn7 delivery by four‐parental mating conjugation
Pph 1302A strains were tagged by four‐parental mating conjugation (Choi and Schweizer, 2006) with minor modifications. Details of the protocol used can be found in the Supporting Information File S1. Pph 1302A transformants were screened for luminescence (if tagged with lux constructs) and luminescence and fluorescence (if tagged with lux and eYFP constructs) as reported for E. coli. The correct insertion of the mini‐Tn7 delivery construct was verified by colony‐PCR using primers OXRS_F15 and OXRS_R15 (Supporting Information Table S3). Primer OXRS_F15 anneals in the glmS gene of Pph and primer OXRS_R15 binds in the Tn7R (right end of the Tn7 transposon).
Three‐parental mating conjugation
Pseudomonas and Acinetobacter strains (Supporting Information Table S2) were inoculated from frozen glycerol stock onto Luria Bertani (LB) (Sambrook et al., 1989) agar (1.5% agar) and grown for 24 h at 28°C. Single colonies were used to inoculate 10 ml LB cultures, which were incubated at 28°C with shaking at 200 rpm. Bacterial cultures were then used for three‐parental mating conjugation as described for four‐parental mating conjugation (Supporting Information File S1), using the helper plasmid pRK2013 (Knauf and Nester, 1982).
Plate reader assay
Bacterial cultures were inoculated from frozen glycerol stock onto Luria Bertani (LB) (Sambrook et al., 1989) agar (1.5% agar) and grown for 24 h at 28°C. Single colonies were used to inoculate 10 ml LB cultures, which were incubated at 28°C with shaking at 200 rpm. After 24 h, 500 μl pre‐culture was used to set up an overnight 10 ml LB culture. Cultures were centrifuged at 4000g for 6 min and resuspended in 10 ml 10 mM MgCl2. The 15 μl of resuspended bacterial cultures were added to 135 μl King's B medium (KB) (King et al., 1954) or M9 medium (20% glucose) (CSH protocols) placed into a 96‐well assay black plate (Corning incorporated, USA). Luminescence and OD600 were measured using a Infinite M200 plate reader (Tecan Group Ltd, Switzerland) over 20 h. The temperature was set at 28°C and measurements were taken every 30 min.
NADP+ assay
Bacterial cultures were inoculated from frozen glycerol stock onto Luria Bertani (LB) (Sambrook et al., 1989) agar (1.5% agar) supplemented with 10 μg ml−1 tetracycline and grown for 24 h at 28°C. Single colonies were used to inoculate 10 ml LB cultures supplemented with 10 μg ml−1 tetracycline, which were incubated at 28°C with shaking at 200 rpm. After 24 h, 200 μl pre‐culture was used to set up 200 ml LB cultures supplemented with 10 μg ml−1 tetracycline which were incubated at 28°C with shaking at 200 rpm for 6 h. Cultures were processed according to the manufacturer's instructions (NADP/NADPH assay ab176724, Abcam, UK) at a final OD600 of 3,3.
Determining the effect of bacterial bioluminescence expression on bacterial growth in planta
Pph strains were inoculated from frozen glycerol stock onto Luria Bertani (LB) (Sambrook et al., 1989) agar (1.5% agar) and grown for 24 h at 28°C. Single colonies were used to inoculate 10 ml LB cultures, which were incubated at 28°C with shaking at 200 rpm. After 24 h, 500 μl pre‐culture was used to set up an overnight 10 ml LB culture. Cultures were centrifuged at 4000g for 6 min and resuspended in 10 ml 10 mM MgCl2 (5 × 106 CFU ml−1). Four plants per bacterial culture were used for the experiment. Briefly, the first two fully developed leaves of 12 two‐weeks old plants (P. vulgaris cultivar Canadian Wonder) were syringe‐infiltrated with Pph 1302A pnptII::lux pA1/04/03, Pph 1302A pOXB16::lux pA1/04/03, Pph 1302A pOXB11::lux pA1/04/03 and Pph 1302A (5 × 106 CFU ml−1) (Supporting Information Table S2). Each leaf was infiltrated four times in four small areas (50–80 mm2). The four spots within the same leaf were considered technical replicates and were averaged for the calculation of the CFU mm−2. At 2 DPI (Days Post Inoculation) four leaf discs per leaf (four leaves total per bacterial culture) were detached from the four inoculated spots with a 1 cm2 cork and pooled together. Samples were ground with a TissueLyser (Qiagen) in 1 ml 10 mM MgCl2, and serial dilutions (20 μl drops) were spotted onto LB agar supplemented with nitrofurantoin 25 μg ml−1. Plates were incubated at 28°C for 3 days. At 5 DPI, the same process described above was repeated.
Determining the detection limit of Pph RJ3 pnptII ::lux pA1 /04/03::eYFP
Bacterial cultures were prepared as described above. Serial dilutions (100/1000 μl) were performed in 10 mM MgCl2 and 100 μl aliquots were spotted onto LB (1.5% agar) square plates. Plates were incubated at 28°C for 1 h before imaging with the nightOWL LB 983 in vivo imaging system (Berthold Technologies) at 20 min exposure. Luminescence values and area (mm2) of each spot were measured for the 10−1, 10−2, 10−3, and 10−4 dilutions. Plates were then incubated at 28°C for 3 days to allow bacterial growth and colony count to be performed.
Combined luminescence‐fluorescence approach to monitor Pph dispersal in planta
Syringe‐infiltration
Pph RJ3 pnptII::lux pA1/04/03::eYFP (Supporting Information Table S1) was inoculated from frozen glycerol stock onto Luria Bertani (LB) (Sambrook et al., 1989) agar (1.5% agar) and grown for 24 h at 28°C. Single colonies were used to inoculate 10 ml LB cultures, which were incubated at 28°C with shaking at 200 rpm. After 24 h, 500 μl pre‐culture was used to set up an overnight 10 ml LB culture. Cultures were centrifuged at 4000g for 6 min and resuspended in 20 ml 10 mM MgCl2. The bacterial suspension (5 × 106 CFU ml−1) was syringe‐infiltrated into abaxial areas of the first true leaves of TG plants without causing any damage to the leaf mesophyll. To minimize damage to the leaf while infiltrating, we modified a 10 ml syringe by attaching to it a P20 pipette tip. We removed the last 1 cm of the P20 pipette tip and attached to it a rubber O‐ring to ensure a soft contact between the tip and the leaf mesophyll. Plants were incubated for 0,1,3 and 5 days at 22°C and artificial light was maintained for 16 h within the 24‐h cycle.
At designated times (0, 1, 3, and 5 DPI), leaves were detached and placed on square Petri dishes. Leaf petioles were wrapped in water‐soaked cotton discs to allow plant transpiration. Leaves were incubated in the dark for 10 min before imaging with the nightOWL LB 983 in vivo imaging system (Berthold Technologies) at 10 min exposure. Bacterial bioluminescence was used to visualize Pph localisation and to identify portions of leaves to be imaged with confocal laser scanning microscopy. Leaf vasculatures showing Pph colonization were cross‐sectioned by hand‐sectioning (50–80 μm) without embedding. Samples were imaged with a confocal laser scanning microscope Zeiss LSM 880 (Carl Zeiss, Germany). eYFP was excited at 514 nm (5.50%) and detected in the range of 520–610 nm; Phenolic compounds were excited at 405 nm (1.10%) and detected in the range of 200–450 nm to visualize xylem structure and collenchyma. Confocal stacks were acquired with a Z‐step of 50–80 μm using the objectives C‐Apochromat 40x/1.2 W Korr FCS M27.
Spray‐inoculation
Pph RJ3 pnptII::lux pA1/04/03::eYFP (Supporting Information Table S1) was incubated prior to inoculation as described above. The bacterial suspension (5 × 107 CFU ml−1) was deposited on restricted abaxial areas of the first true leaves of TG plants by spraying the resuspended bacterial culture through a round aperture (about 1 cm2) cut into a folded aluminium foil. When spraying, close contact between leaves and aluminium foil ensured that there was no contamination of adjacent areas of the leaf. Four inoculated plants were placed inside a closed transparent box (30 × 20 × 30 cm) with two Oasis flower foams (Oasis Floral Products, UK) previously soaked in water. Plants were incubated for 3 days at 22°C and artificial light was maintained for 16 h within the 24‐h cycle. Negative controls were represented by leaves inoculated solely with 10 mM MgCl2.
After 3 days, leaves were detached and placed on square Petri dishes. Leaf petioles were wrapped in water‐soaked cotton discs to allow plant transpiration. Leaves were incubated in the dark for 10 min before imaging with the nightOWL LB 983 in vivo imaging system (Berthold Technologies) at 20 min exposure. Bacterial bioluminescence was used to visualize Pph localisation and to identify portions of leaves to be imaged with confocal laser scanning microscopy. Leaf vasculatures showing Pph colonization were cross‐sectioned by hand‐sectioning (50–80 μm) or imaged as part of leaf discs (50 mm2). Samples were imaged with a confocal laser scanning microscope Zeiss LSM 880 (Carl Zeiss).
eYFP was excited at 514 nm (5.50%) and detected in the range of 520–610 nm; phenolic compounds were excited at 405 nm (1.10%) and detected in the range of 200–450 nm to visualize xylem structure; and chlorophyll was excited at 514 nm (5.50%) and detected in the range of 650–720 nm.
Confocal stacks were acquired with a Z‐step of 50–80 μm (for cross sections) or 10–30 μm (for leaf discs) using the objectives LD LCI Plan‐Apochromat 25x/0.8 Imm Korr DIC M27. Leaf volume‐rendering and three‐dimensional models of the confocal stacks were created with the software Imaris 8 (Bitplane AG, Zürich, Switzerland).
Bacterial bioluminescence assays to detect the effect of plant immune responses on bacterial growth and viability
Pph 1302A ΔhrpA (this study), Pph 1302A ΔPphB (this study) and Pph 1302A ΔxerC (Lovell et al., 2009) were tagged using pRS‐pOXB13::lux (Table 1; Supporting Information Table S1) as described above. Bacterial cultures were inoculated from frozen glycerol stock onto Luria Bertani (LB) (Sambrook et al., 1989) agar (1.5% agar) and grown for 24 h at 28°C. Single colonies were used to inoculate 10 ml LB cultures, which were incubated at 28°C with shaking at 200 rpm. After 24 h, 500 μl pre‐culture was used to set up an overnight 10 ml LB culture. Cultures were centrifuged at 4000g for 6 min and resuspended in 10 ml 10 mM MgCl2 (106 CFU ml−1). Eight plants per bacterial culture were used for the experiment.
The optimal number of replicates were calculated with a jacknife resampling (Hanna, 1989) derived approach before performing the experiment. Briefly, the first two fully developed leaves of 12 two‐week‐old plants (P. vulgaris cultivar TG) were syringe‐infiltrated with Pph 1302A ΔPphB pOXB13::lux (106 CFU ml−1) (Supporting Information Table S1). Each leaf was infiltrated six times in six small areas (50–80 mm2). The six spots within the same leaf were considered technical replicates and were averaged for the calculation of the optimal sample size. Leaves were detached and incubated in the dark for 10 min before imaging with the nightOWL LB 983 in vivo imaging system (Berthold Technologies) at 2‐min exposure. Luminescence values were recorded and used for the calculation of the optimal sample size. A custom R script (Supporting Information File S2) was used to randomly sample 1000 thousand times the generated data (12 values, in this case) with a jackknife approach. Standard deviation (SD) for each 1000 resampling per each sample size (from 12 to 1) was used to assess the difference in SD between sample sizes (12‐n), where n is the sample size. The process was repeated 300 times and statistically significance differences of differences in SD between sample sizes were assessed with Benjamin–Hochberg (BH) post hoc test. Eight was assessed to be the optimal sample size as the reduction in SD from sample size 8 to sample size 9 was not statistically significant.
Subsequently, 2‐week‐old TG plants were syringe‐infiltrated with pOXB13::lux expressing Pph 1302A ΔhrpA, Pph 1302A ΔPphB and Pph 1302A ΔxerC bacterial cultures (Supporting Information Table S2). The experiment was conducted according to a randomized complete block design (RCBD) (Liu and Berger, 2014) in which the block corresponded to the inoculation time. This was done to account for the passage of time from the first plant inoculations to the last plant inoculations (about 4 h). At time 1, three plants (two leaves per plant) were syringe‐infiltrated with the corresponding bacterial cultures. One leaf per plant was detached and incubated in the dark for 10 min before imaging with the nightOWL LB 983 in vivo imaging system (Berthold Technologies) at 2‐min exposure. Luminescence and area of each spot within leaves were recorded. Plants were then incubated for 2 days at 22°C and artificial light was maintained for 16 h within the 24‐h cycle. The same procedure was repeated for subsequent time points. Within each block (time point and tray) plants were randomized. The same bacterial cultures were used for all time points. Negative controls were represented by plants infiltrated with 10 mM MgCl2 and infiltrated with the non‐tagged Pph 1302A ΔxerC bacterial culture to detect any bioluminescence associated with induction of plant immune responses (Bennett et al., 2005).
After 2 DPI, luminescence and the area where luminescence is detected were measured as described previously, for the remaining leaves. Exposure time was set at 5 min. Immediately after measuring bioluminescence signal, leaves were used to perform colony counts. Briefly, six leaf discs per leaf were detached from the six inoculated spots with a 1‐cm2 cork and pooled together. Samples were ground with a TissueLyser (Qiagen) in 1 ml 10 mM MgCl2, and serial dilutions (20 μl drops) were spotted onto LB agar supplemented with gentamicin 10 μg ml−1 and nitrofurantoin 25 μg ml−1. Plates were incubated at 28°C for 3 days.
Data analysis and statistics
Processing of data was performed using R (RDC Team (R Development Core Team), 2006) version 3.5.3 and ggplot2 (Wickham, 2016). Requirements of linear models, namely normal distribution of the residuals and homogeneity of variance were assessed using diagnostic plots (Harrison et al., 2018). When diagnostic plots were not clearly interpretable, we used Leven's test to assess the homogeneity of variance. Independence of the observations was guaranteed by the experimental design.
Author contributions
R.S., N.S., and G.M.P. designed the study and the experiments. R.S. performed the experiments and analysed the data. M.B.P. performed the NADP+ assay and discussed the experiments with R.S, I.B. and W.H. contributed to the design for pRS pnptII::ilux and I.B. also contributed to the design of the mini‐Tn7 library. R.S. and G.M.P. wrote the manuscript.
Supporting information
Supplementary Fig. 1 Expression of FMN reductase (frp) increases luminescence in P. fluorescens NZ011 when combined with the lux operon. Acinetobacter strains: A. baylyi ADP1 (pIJ11282), A. baylyi ADP1 (pRSJ‐pnptII::ilux), A. baylyi ADP1 (pRSJ‐pnptII::lux‐frp). Pseudomonas fluorescens strains: P. fluorescens NZ011 (pIJ11282), P. fluorescens NZ011 (pRSJ‐pnptII::ilux), P. fluorescens NZ011 (pRSJ‐pnptII::lux‐frp). Pseudomonas syringae pv. phaseolicola (Pph) strains: Pph 1302A (pIJ11282), Pph 1302A (pRSJ‐pnptII::ilux), Pph 1302A (pRSJ‐pnptII::lux‐frp). Plasmids pIJ11282, pRSJ‐pnptII::ilux and pRSJ‐pnptII::lux‐frp have the same backbone (pIJ11282) and promoter (pnptII). A: Normalized luminescence of reporter strains in KB (King's B medium) and M9 (M9 minimal medium). B: Growth of reporter strains in KB and M9. OD = optical density at 600 nm. Error bar, +/− SE. n = 3.
Supplementary Fig. 2. Expression of frp in P. fluorescens NZ011 (pRSJ‐pnptII:lux‐frp) results in an increase in NADP+ concentration. Lux: P. fluorescens NZ011 (pIJ11282). Lux‐frp: P. fluorescens NZ011 (pRSJ‐pnptII:lux‐frp). NADP+ concentration refers to 25 ul of bacterial culture, for which a 10‐fold dilution was measured as having an optical density (OD600) of 0.33. Error bar +/− SE. Significant differences (t‐test, p < 0.05) are indicated by asterisks. n = 6.
Supplementary Fig. 3. The lux‐eYFP operon constructed in this study. A: Schematic representation of the lux‐eYFP construct made in this study. p1: pnptII, pOXB20, pOXB16, pOXB13, pOXB11. p2: pA1/04/03, plac. The red ‘T’ represents the T0 terminator. B: Pseudomonas syringae pv. phaseolicola RJ3 (Pph RJ3) pnptII::lux‐pA1/04/03::eYFP imaged with the Typhoon scanner (Amersham/GE Healthcare, UK) using Cy3 settings. eYFP expression is detected. C: Pph RJ3 pnptII::lux pA1/04/03::eYFP imaged with the nightOWL LB 983 at 10 s exposure. Bioluminescence is detected.
Supplementary Fig. 4. A: Schematic representation of the pRS‐pOXB20(1)::lux plasmid. pRS‐pOXB20(1)::lux is derived from pUC18‐mini‐Tn7T‐Gm‐lux. MCS: Long multiple cloning site (490 bp) of pUC18‐mini‐Tn7T‐Gm‐lux plasmid. RBS1: Ribosome Binding site of plasmid pUC18‐mini‐Tn7T‐Gm‐lux. B: Schematic representation of the pRS‐pOXB20::lux plasmid. RBS2: Ribosome‐binding site of plasmid pIJ11282. The RBS strength was calculated with the RBS calculator (Farasat et al., 2014; Ng et al., 2015) considering a total DNA length of 45 bp from the starting codon of luxC.
Supplementary Fig. 5. pRS‐pOXB20::lux generates increased luminescence compared to pRS‐pOXB20(1)::lux in Pseudomonas syringae pv. phaseolicola 1302A. Plasmid pRS‐pOXB20(1)::lux was derived from the existing mini‐Tn7 plasmid pUC18‐mini‐Tn7T‐Gm‐lux (Choi et al., 2005) by inserting oriT and ligating the OXB20 promoter upstream of luxC. Plasmid pRS‐pOXB20::lux has a stronger RBS (derived from pIJ11282) upstream of luxC compared to pUC18‐mini‐Tn7T‐Gm‐lux. A: Normalized luminescence of Pph reporter strains in KB (King's B medium) and M9 (M9 minimal medium). B: Growth of reporter strains in KB and M9. OD = optical density at 600 nm. Error bar +/− SE. n = 3
Supplementary Fig. 6. Pseudomonas syringae pv. phaseolicola 1302A (Pph 1302A) reporter strains exhibit growth curves similar to Pph 1302A in both rich and minimal medium while providing a wide range of luminescence. Strains: Pph 1302A, Pph 1302A pOXB11::lux‐pA1/04/03::eYFP, Pph 1302A pOXB13::lux‐pA1/04/03::eYFP, Pph 1302A pOXB16::lux‐pA1/04/03::eYFP, Pph 1302A pOXB20::lux‐pA1/04/03::eYFP and Pph 1302A pnptII::lux‐pA1/04/03::eYFP. A: Normalized luminescence of Pph reporter strains in KB (King's B medium) and M9 (M9 minimal medium) B: Growth of Pph 1302A reporter strains compared to Pph 1302A in KB and M9. OD = optical density at 600 nm.. Error bar +/− SE. n = 3.
Supplementary Fig. 7. Strong expression of the lux cassette has a small but significant effect on Pph 1302A bacterial growth both in vitro and in planta. A: OD values at 3 h of data presented in Supplementary Fig. 6. nptII: Pph 1302A pnptII::lux‐pA1/04/03:::eYFP; OXB20: Pph 1302A pOXB20::lux‐pA1/04/03::eYFP; OXB16: Pph 1302A pOXB16::lux‐pA1/04/03::eYFP; OXB13: Pph 1302A pOXB13::lux‐pA1/04/03::eYFP; OXB11: Pph 1302A pOXB11::lux‐pA1/04/03::eYFP; 1302A: Pph 1302A. Error bar +/− SE. Significant differences (Tukey HSD, p < 0.05) are indicated by letters. n = 3. B: In planta growth of Pph 1302A reporter strains. TG leaves were syringe infiltrated with Pph 1302A reporter strains at 5x106 CFU ml−1 in 10 mM MgCl2. nptII: Pph 1302A pnptII::lux‐pA1/04/03:::eYFP; OXB16: Pph 1302A pOXB16::lux‐pA1/04/03::eYFP; OXB11: Pph 1302A pOXB11::lux‐pA1/04/03::eYFP; 1302A: Pph 1302A. Error bar +/− SE. Significant differences (Tukey HSD, p < 0.05) are indicated by letters. n = 4.
Supplementary Fig. 8. Detection limit for bioluminescent Pseudomonas syringae pv. phaseolicola RJ3 (Pph RJ3) in agar plates. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP was spotted in serial dilution on an LB agar plate. Images were taken after 1 h using the nightOWL LB 983 at 20 min exposure. Approximately 100 cells/mm2 can be detected. Colour scales have different minimum values to avoid oversaturation. CFU/mm2 was estimated by colony counting. SE for CFU and SE for luminescence constitute on average 27.7% and 2.66% of the means respectively. n = 3.
Supplementary Fig. 9. Pseudomonas syringae pv. phaseolicola RJ3 (Pph RJ3) colonizes the leaf vasculature of P. vulgaris cultivar TG following spray inoculation onto the leaf surface. A, B: Pph RJ3 pnptII::lux‐pA1/04/03::eYFP was sprayed in a localized area on the abaxial surface of Phaseolus vulgaris cv. Tendergreen leaves through an aperture in an aluminium foil (white dotted line). Images were taken after 2 DPI with the nightOWL LB 983 at 20 min exposure. Red circle indicates leaf discs used for confocal microscopy imaging (C). Red lines indicates cross sections used for confocal microscopy imaging (D,E). C: Maximum projections of the leaf vasculature. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow; white arrow). Chlorophyll (red; black arrow). D: image of vasculature cross section. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow). Xylem autofluorescence (grey). Red circular line indicates area zoomed in panel F. E: Maximum projection of vasculature cross section. Xylem autofluorescence (grey). Red circular line indicates area modelled in panel G. F: zoom of panel D. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow; white arrow). Xylem autofluorescence (grey). G: Three‐dimensional model of panel E (Imaris 9 Bitplane AG, Zürich, Switzerland). Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow; white arrow). H: Three‐dimensional model of panel C (Imaris 9 Bitplane AG, Zürich, Switzerland). Co‐localization of chlorophyll signal (red) and eYFP (yellow), indicates endophytic localization of Pph RJ3 pnptII::lux‐pA1/04/03::eYFP in the leaf vasculature. Scale bars: 40 μm.
Supplementary Fig. 10. Phaseolus vulgaris cultivar Tendergreen leaves do not display background bioluminescence or fluorescence under conditions used to image luminescence or eYFP. A: nightOWL LB 983 image. The image is automatically generated by the nightOWL LB 983 by overlaying the image acquired with CCD camera with a photo of the sample. Exposure time 20 min. The leaf was incubated for 10 min in darkness before imaging. Black arrows indicate tissue used for confocal microscopy imaging. B: details of the Phaseolus vulgaris cultivar TG vascular system. In non‐inoculated plants, eYFP signal is not detectable. Chlorophyll (red), eYFP (yellow). C: details of xylem vessels of non‐inoculated plants. eYFP signal is not detectable. Xylem (grey), eYFP (yellow). Settings used were the same as for inoculated plants. Scale bars: 40 μm.
Supplementary Fig. 11. The colony count approach is able to detect the effect of PTI but not ETI on the growth of Pseudomonas syringae pv. phaseolicola in the early stages of infection (2 DPI). P. vulgaris cultivar Tendergreen (TG) leaves were syringe infiltrated with Pph 1302A ΔxerC, ΔhrpA and ΔPphB at 106 CFU ml−1 in 10 mM MgCl2. A: Details of the symptoms caused by Pph 1302A knock‐out mutants in TG leaves at 2 days post inoculation (DPI). The hypersensitive response (HR) is evident on leaves inoculated with Pph 1302A ΔxerC. B: Colony count data for inoculated leaves at 2 DPI. Error bar +/− SE. Significant differences (Student's T‐test, p < 0.05) are indicated by letters. n = 8.
Supplementary Video 1 Pph RJ3‐eYFP colonizes the collenchyma of the Phaseolus vulgaris cultivar Tendergreen (TG) leaf vasculature. Pph RJ3‐eYFP (Godfrey et al. 2010) was syringe‐infiltrated at 106 CFU ml−1 into the first true leaves of TG plants. Leaf disks were detached at 3 days post inoculation and imaged with a TCS Leica SP5 confocal microscope in a xyz series (objective used: HCX PL APO CS 20.0x0.70 IMM UV). eYFP was excited with the 488 nm wave length laser. Cyan (Pph RJ3 cells); Red (Chlorophyll autofluorescence); Grey (bright field).
Supplementary Video 2 Pph RJ3‐eYFP (Godfrey et al., 2010) moves systemically inside the vasculature of Phaseolus vulgaris cultivar Tendergreen (TG). Pph RJ3‐eYFP was syringe‐infiltrated at 106 CFU ml−1 into the first true leaves of TG plants. Leaf disks were detached at 1 day post inoculation and imaged with the a TCS Leica SP5 confocal microscope in a xyz series (objective used: HC PL APO CS2 63.0x1.20 WATER UV). eYFP was excited with the 488 nm wave length laser. Cyan (Pph RJ3 cells); Red (Chlorophyll autofluorescence); Grey (bright field).
Supplementary Table 1 Plasmids used in this study.
Supplementary Table 2. List of strains used
Supplementary Table 3. List of primers used for cloning
Supplementary Table 5. List of primers used to sequence eYFP and frp.
Supplementary Table 6. List of primers used to sequence the lux operon
Appendix S1. Supporting Information.
Appendix S2. Supporting Information.
Acknowledgements
The authors thank Philip Poole (Department of Plant Sciences, University of Oxford) for access to the nightOWL LB 983 and the gift of pIJ11282 and thank the Micron Advanced Bioimaging Unit (University of Oxford) for providing access to the software IMARIS. The authors warmly thank Vinoy Ramachandran for providing training in use of the nightOWL LB 983 and Dawn Arnold and Helen Neale (University of Bristol) for providing P. syringae pv. phaseolicola strains and useful discussions. The authors wish to thank all the members of Gail Preston and Renier van der Hoorn's research groups for support and discussion. The authors particularly thank Phillippe V. Jutras for advice regarding the figures. R.S. is supported by Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/M011224/1, the Oxford Interdisciplinary Bioscience Doctoral Training Partnership (Doctoral Training Centre, University of Oxford) and the Ermenegildo Zegna's founder scholarship. This project was also supported by BBSRC grant BB/R009236/1 awarded to G.M.P.
Data Availability Statement
The research materials supporting this publication have been deposited in the Oxford Research Archive (ORA) as https://doi.org/10.5287/bodleian:y0r8ErQja and https://doi.org/10.5287/bodleian:KOeJ8agdn. If you wish to access the biological material that is described in this study please contact gail.preston@plants.ox.ac.uk.
References
- Asai, S. , Takamura, K. , Suzuki, H. , and Setou, M. (2008) Single‐cell imaging of c‐fos expression in rat primary hippocampal cells using a luminescence microscope. Neurosci Lett 434: 289–292. [DOI] [PubMed] [Google Scholar]
- Bae, C. , Han, S.W. , Song, Y.‐R. , Kim, B.‐Y. , Lee, H.‐J. , Lee, J.‐M. , et al. (2015) Infection processes of xylem‐colonizing pathogenic bacteria: possible explanations for the scarcity of qualitative disease resistance genes against them in crops. Theor Appl Genet 128: 1219–1229. [DOI] [PubMed] [Google Scholar]
- Benedetti, I.M. , de Lorenzo, V. , and Silva‐Rocha, R. (2012) Quantitative, non‐disruptive monitoring of transcription in single cells with a broad‐host range GFP‐luxCDABE dual reporter system. PLOS ONE 7: e52000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M. , Mehta, M. , and Grant, M. (2005) Biophoton imaging: a nondestructive method for assaying R gene responses. MPMI 18: 95–102. [DOI] [PubMed] [Google Scholar]
- Bogs, J. , Bruchmüller, I. , Erbar, C. , and Geider, K. (1998) Colonization of host plants by the fire blight pathogen Erwinia amylovora marked with genes for bioluminescence and fluorescence. Phytopathology™ 88: 416–421. [DOI] [PubMed] [Google Scholar]
- Brodl, E. , Winkler, A. , and Macheroux, P. (2018) Molecular mechanisms of bacterial bioluminescence. Comput Struct Biotechnol J 16: 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli, D. , Rott, M. , Schlaeppi, K. , Ver Loren van Themaat, E. , Ahmadinejad, N. , Assenza, F. , et al. (2012) Revealing structure and assembly cues for Arabidopsis root‐inhabiting bacterial microbiota. Nature 488: 91–95. [DOI] [PubMed] [Google Scholar]
- Burkatovskaya, M. , Tegos, G.P. , Swietlik, E. , Demidova, T.N. , P Castano, A. , and Hamblin, M.R. (2006) Use of chitosan bandage to prevent fatal infections developing from highly contaminated wounds in mice. Biomaterials 27: 4157–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti, A. , Jauneau, A. , Auriac, M.‐C. , Lauber, E. , Martinez, Y. , Chiarenza, S. , et al. (2017) Immunity at cauliflower Hydathodes controls systemic infection by Xanthomonas campestris pv campestris . Plant Physiol 174: 700–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K.‐H. , Gaynor, J.B. , White, K.G. , Lopez, C. , Bosio, C.M. , Karkhoff‐Schweizer, R.R. , and Schweizer, H.P. (2005) A Tn 7 ‐based broad‐range bacterial cloning and expression system. Nat Methods 2: 443–448. [DOI] [PubMed] [Google Scholar]
- Choi, K.‐H. , and Schweizer, H.P. (2006) Mini‐Tn 7 insertion in bacteria with single att Tn 7 sites: example Pseudomonas aeruginosa . Nature Protoc 1: 153–161. [DOI] [PubMed] [Google Scholar]
- Compant, S. , Reiter, B. , Sessitsch, A. , Nowak, J. , Clément, C. , and Ait Barka, E. (2005) Endophytic colonization of Vitis vinifera L. by plant growth‐promoting bacterium Burkholderia sp. strain PsJN. Appl Environ Microbiol 71: 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, N.L. (1991) Tn7: a target site‐specific transposon. Mol Microbiol 5: 2569–2573. [DOI] [PubMed] [Google Scholar]
- Donati, I. , Cellini, A. , Buriani, G. , Mauri, S. , Kay, C. , Tacconi, G. , and Spinelli, F. (2018) Pathways of flower infection and pollen‐mediated dispersion of Pseudomonas syringae pv. actinidiae, the causal agent of kiwifruit bacterial canker. Hortic Res 5: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle, K.A. , Billings, G. , Sigal, M. , Lichtman, J.S. , Hansson, G.C. , Elias, J.E. , et al. (2015) Quantitative imaging of gut microbiota spatial organization. Cell Host Microb 18: 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, B. , Chen, X.H. , Budiharjo, A. , Bleiss, W. , Vater, J. , and Borriss, R. (2011) Efficient colonization of plant roots by the plant growth promoting bacterium Bacillus amyloliquefaciens FZB42, engineered to express green fluorescent protein. J Biotechnol 151: 303–311. [DOI] [PubMed] [Google Scholar]
- Fan, J. , Crooks, C. , and Lamb, C. (2008) High‐throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J 53: 393–399. [DOI] [PubMed] [Google Scholar]
- Farasat, I. , Kushwaha, M. , Collens, J. , Easterbrook, M. , Guido, M. , and Salis, H.M. (2014) Efficient search, mapping, and optimization of multi‐protein genetic systems in diverse bacteria. Mol Syst Biol 10: 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer, N. , and Ladau, J. (2012) Predicting microbial distributions in space and time. Nat Methods 9: 549–551. [DOI] [PubMed] [Google Scholar]
- Figueroa‐Cuilan, W. , Daniel, J.J. , Howell, M. , Sulaiman, A. , and Brown, P.J.B. (2016) Mini‐Tn7 insertion in an artificial attTn7 site enables depletion of the essential master regulator CtrA in the Phytopathogen Agrobacterium tumefaciens . Appl Environ Microbiol 82: 5015–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleiss, A. , and Sarkisyan, K.S. (2019) A brief review of bioluminescent systems (2019). Curr Genet 65: 877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederix, M. , Edwards, A. , Swiderska, A. , Stanger, A. , Karunakaran, R. , Williams, A. , et al. (2014) Mutation of praR in Rhizobium leguminosarum enhances root biofilms, improving nodulation competitiveness by increased expression of attachment proteins. Mol Microbiol 93: 464–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujie, M. , Takamoto, H. , Kawasaki, T. , Fujiwara, A. , and Yamada, T. (2010) Monitoring growth and movement of Ralstonia solanacearum cells harboring plasmid pRSS12 derived from bacteriophage ϕRSS1. J Biosci Bioeng 109: 153–158. [DOI] [PubMed] [Google Scholar]
- Fukui, R. , Fukui, H. , McElhaney, R. , Nelson, S.C. , and Alvarez, A.M. (1996) Relationship between symptom development and actual sites of infection in leaves of Anthurium inoculated with a bioluminescent strain of Xanthomonas campestris pv. dieffenbachiae . Appl Environ Microbiol 62: 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, D.G. , Young, L. , Chuang, R.‐Y. , Venter, J.C. , Hutchison, C.A. , and Smith, H.O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Giliberti, G. , Baccigalupi, L. , Cordone, A. , Ricca, E. , and De Felice, M. (2006) Transcriptional analysis of the recA gene of Streptococcus thermophilus . Microb Cell Factor 5: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassing, A. , and Lewis, T.A. (2015) An improved Tn7‐lux reporter for broad host range, chromosomally‐integrated promoter fusions in gram‐negative bacteria. J Microbiol Methods 118: 75–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey, S.a.C. , Mansfield, J.W. , Corry, D.S. , Lovell, H.C. , Jackson, R.W. , and Arnold, D.L. (2010) Confocal imaging of Pseudomonas syringae pv. phaseolicola colony development in bean reveals reduced multiplication of strains containing the genomic Island PPHGI‐1. Mol Plant Microbe Interact 23: 1294–1302. [DOI] [PubMed] [Google Scholar]
- Gregor, C. , Gwosch, K.C. , Sahl, S.J. , and Hell, S.W. (2018) Strongly enhanced bacterial bioluminescence with the ilux operon for single‐cell imaging. Proc Natl Acad Sci USA 115: 962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, S.R. (1989) Confidence limits for air quality model evaluations, as estimated by bootstrap and jackknife resampling methods. Atmos Environ (1967) 23: 1385–1398. [Google Scholar]
- Harrison, X.A. , Donaldson, L. , Correa‐Cano, M.E. , Evans, J. , Fisher, D.N. , Goodwin, C.E.D. , et al. (2018) A brief introduction to mixed effects modelling and multi‐model inference in ecology. PeerJ 6: e4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W.E. , Huang, L. , Preston, G.M. , Naylor, M. , Carr, J.P. , Li, Y. , et al. (2006) Quantitative in situ assay of salicylic acid in tobacco leaves using a genetically modified biosensor strain of Acinetobacter sp. ADP1. Plant Journal 46: 1073–1083. [DOI] [PubMed] [Google Scholar]
- Iwano, S. , Sugiyama, M. , Hama, H. , Watakabe, A. , Hasegawa, N. , Kuchimaru, T. , et al. (2018) Single‐cell bioluminescence imaging of deep tissue in freely moving animals. Science 359: 935–939. [DOI] [PubMed] [Google Scholar]
- Jackson, R.W. , Mansfield, J.W. , Arnold, D.L. , Sesma, A. , Paynter, C.D. , Murillo, J. , et al. (2000) Excision from tRNA genes of a large chromosomal region, carrying avrPphB, associated with race change in the bean pathogen, Pseudomonas syringae pv. phaseolicola . Mol Microbiol 38: 186–197. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. , and Dangl, J.L. (2006) The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kandel, Y.R. , Glover, K.D. , Tande, C.A. , and Osborne, L.E. (2012) Evaluation of spring wheat germplasm for resistance to bacterial leaf streak caused by Xanthomonas campestris pv. translucens . Plant Disease 96: 1743–1748. [DOI] [PubMed] [Google Scholar]
- Kim, Y.‐H. , Park, P.‐G. , Seo, S.‐H. , Hong, K.‐J. , and Youn, H. (2018) Development of dual reporter imaging system for Francisella tularensis to monitor the spatio‐temporal pathogenesis and vaccine efficacy. Clin Exp Vaccine Res 7: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, E.O. , Ward, M.K. , and Raney, D.E. (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44: 301–307. [PubMed] [Google Scholar]
- Klausen, M. , Heydorn, A. , Ragas, P. , Lambertsen, L. , Aaes‐Jørgensen, A. , Molin, S. , and Tolker‐Nielsen, T. (2003) Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48: 1511–1524. [DOI] [PubMed] [Google Scholar]
- Knauf, V.C. , and Nester, E.W. (1982) Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8: 45–54. [DOI] [PubMed] [Google Scholar]
- Koch, B. , Jensen, L.E. , and Nybroe, O. (2001) A panel of Tn7‐based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into gram‐negative bacteria at a neutral chromosomal site. J Microbiol Methods 45: 187–195. [DOI] [PubMed] [Google Scholar]
- Lambertsen, L. , Sternberg, C. , and Molin, S. (2004) Mini‐Tn7 transposons for site‐specific tagging of bacteria with fluorescent proteins. Environ Microbiol 6: 726–732. [DOI] [PubMed] [Google Scholar]
- Lamichhane, J.R. , Messéan, A. , and Morris, C.E. (2015) Insights into epidemiology and control of diseases of annual plants caused by the Pseudomonas syringae species complex. J Gen Plant Pathol 81: 331–350. [Google Scholar]
- Le Chatelier, E. , Nielsen, T. , Qin, J. , Prifti, E. , Hildebrand, F. , Falony, G. , et al. (2013) Richness of human gut microbiome correlates with metabolic markers. Nature 500: 541–546. [DOI] [PubMed] [Google Scholar]
- Liu, L. , and Berger, V.W. (2014) Randomized block design: nonparametric analyses. In Wiley StatsRef: Statistics Reference Online, Atlanta, GA: American Cancer Society. [Google Scholar]
- Lovell, H.C. , Mansfield, J.W. , Godfrey, S.A.C. , Jackson, R.W. , Hancock, J.T. , and Arnold, D.L. (2009) Bacterial evolution by Genomic Island transfer occurs via DNA transformation in planta. Curr Biol 19: 1586–1590. [DOI] [PubMed] [Google Scholar]
- Lynn, G.M. , Laga, R. , Darrah, P.A. , Ishizuka, A.S. , Balaci, A.J. , Dulcey, A.E. , et al. (2015) In vivo characterization of the physicochemical properties of polymer‐linked TLR agonists that enhance vaccine immunogenicity. Nat Biotechnol 33: 1201–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, R.E. , and Bieleski, R.L. (1977) Involvement of Phaseolotoxin in halo blight of beans: transport and conversion to functional toxin. Plant Physiol 60: 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitter, B. , Pfaffenbichler, N. , Flavell, R. , Compant, S. , Antonielli, L. , Petric, A. , et al. (2017) A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front Microbiol 8: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro, F. , Genin, S. , van Dijk, I. , and Valls, M. (2012) A luminescent reporter evidences active expression of Ralstonia solanacearum type III secretion system genes throughout plant infection. Microbiology (Reading, Engl) 158: 2107–2116. [DOI] [PubMed] [Google Scholar]
- Mortin, L.I. , Li, T. , Praagh, A.D.G.V. , Zhang, S. , Zhang, X.‐X. , and Alder, J.D. (2007) Rapid bactericidal activity of Daptomycin against methicillin‐resistant and methicillin‐susceptible Staphylococcus aureus peritonitis in mice as measured with bioluminescent bacteria. Antimicrob Agents Chemother 51: 1787–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, C.Y. , Farasat, I. , Maranas, C.D. , and Salis, H.M. (2015) Rational design of a synthetic Entner–Doudoroff pathway for improved and controllable NADPH regeneration. Metabol Eng 29: 86–96. [DOI] [PubMed] [Google Scholar]
- Oliva, R. , Ji, C. , Atienza‐Grande, G. , Huguet‐Tapia, J.C. , Perez‐Quintero, A. , Li, T. , et al. (2019) Broad‐spectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol 37: 1344–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perehinec, T.M. , Qazi, S.N. , Gaddipati, S.R. , Salisbury, V. , Rees, C.E. , and Hill, P.J. (2007) Construction and evaluation of multisite recombinatorial (gateway) cloning vectors for gram‐positive bacteria. BMC Mol Biol 8: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas‐Marquès, M. , Kressin, J.P. , Kashyap, A. , Panthee, D.R. , Louws, F.J. , Coll, N.S. , and Valls, M. (2020) Four bottlenecks restrict colonization and invasion by the pathogen Ralstonia solanacearum in resistant tomato. J Exp Bot 71: 2157–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnell, K.V. , Flexer‐Harrison, M. , and Alper, H.S. (2019) Design and synthesis of synthetic UP elements for modulation of gene expression in Escherichia coli . Synth Syst Biotechnol 4: 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Propheter, D.C. , and Hooper, L.V. (2015) Bacteria come into focus: new tools for visualizing the microbiota. Cell Host Microbe 18: 392–394. [DOI] [PubMed] [Google Scholar]
- Prosser, J.I. (2010) Replicate or lie. Environ Microbiol 12: 1806–1810. [DOI] [PubMed] [Google Scholar]
- RDC Team (R Development Core Team) (2006) R: A Language and Environment for Statistical Computing, Vienna, Austria: R foundation for Statistical Computing. [Google Scholar]
- Reynaud, E.G. , Peychl, J. , Huisken, J. , and Tomancak, P. (2015) Guide to light‐sheet microscopy for adventurous biologists. Nat Methods 12: 30–34. [DOI] [PubMed] [Google Scholar]
- Rico, A. , Bennett, M.H. , Forcat, S. , Huang, W.E. , and Preston, G.M. (2010) Agroinfiltration reduces ABA levels and suppresses Pseudomonas syringae‐elicited salicylic acid production in Nicotiana tabacum . PLoS ONE 5: e8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Moreno, L. , Jiménez, A.J. , and Ramos, C. (2009) Endopathogenic lifestyle of Pseudomonas savastanoi pv. savastanoi in olive knots. Microb Biotechnol 2: 476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakova, D. , Mancinelli, R. , Wikström, M. , Birch‐Jensen, A.‐S. , Postma, J. , Ehlers, R.‐U. , et al. (2017) The structure of the Brassica napus seed microbiome is cultivar‐dependent and affects the interactions of symbionts and pathogens. Microbiome 5: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch, E.F. , and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, New York, NY: Cold Spring Harbor. [Google Scholar]
- Sapkota, S. , Mergoum, M. , and Liu, Z. (2020) The translucens group of Xanthomonas translucens: complicated and important pathogens causing bacterial leaf streak on cereals. Mol Plant Pathol 21: 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, A. , Tauch, A. , Jäger, W. , Kalinowski, J. , Thierbach, G. , and Pühler, A. (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145: 69–73. [DOI] [PubMed] [Google Scholar]
- Seleem, M.N. , Ali, M. , Boyle, S.M. , and Sriranganathan, N. (2008) Reporter genes for real‐time in vivo monitoring of Ochrobactrum anthropi infection. FEMS Microb Lett 286: 124–129. [DOI] [PubMed] [Google Scholar]
- Shaner, N.C. , Steinbach, P.A. , and Tsien, R.Y. (2005) A guide to choosing fluorescent proteins. Nat Methods 2: 905–909. [DOI] [PubMed] [Google Scholar]
- Soldan, R. , Mapelli, F. , Crotti, E. , Schnell, S. , Daffonchio, D. , Marasco, R. , et al. (2019) Bacterial endophytes of mangrove propagules elicit early establishment of the natural host and promote growth of cereal crops under salt stress. Microbiol Res 223–225: 33–43. [DOI] [PubMed] [Google Scholar]
- Taylor, J.D. , Dudley, C.L. , and Presly, L. (1979) Studies of halo‐blight seed infection and disease transmission in dwarf beans. Ann Appl Biol 93: 267–277. [Google Scholar]
- Turnbaugh, P.J. , Bäckhed, F. , Fulton, L. , and Gordon, J.I. (2008) Diet‐induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unge, A. , and Jansson, J. (2001) Monitoring population size, activity, and distribution of gfp‐luxAB‐tagged Pseudomonas fluorescens SBW25 during colonization of wheat. Microb Ecol 41: 290–300. [DOI] [PubMed] [Google Scholar]
- Unge, A. , Tombolini, R. , Mølbak, L. , and Jansson, J.K. (1999) Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a Dualgfp‐luxAB marker system. Appl Environ Microbiol 65: 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. , Kang, L. , Anand, A. , Lazarovits, G. , and Mysore, K.S. (2007) Monitoring in planta bacterial infection at both cellular and whole‐plant levels using the green fluorescent protein variant GFPuv. New Phytologist 174: 212–223. [DOI] [PubMed] [Google Scholar]
- Weisemann, J.M. , and Weinstock, G.M. (1991) The promoter of the recA gene of Escherichia coli . Biochimie 73: 457–470. [DOI] [PubMed] [Google Scholar]
- Wickham, H. (2016) ggplot2: Elegant Graphics for Data Analysis. Springer‐Verlag: New York.
- Wright, C.A. , and Beattie, G.A. (2004) Bacterial species specificity in proU Osmoinducibility and nptII and lacZ expression. MMB 8: 201–208. [DOI] [PubMed] [Google Scholar]
- Xu, X. , Miller, S.A. , Baysal‐Gurel, F. , Gartemann, K.‐H. , Eichenlaub, R. , and Rajashekara, G. (2010) Bioluminescence imaging of Clavibacter michiganensis subsp. michiganensis infection of tomato seeds and plants. Appl Environ Microbiol 76: 3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.A. , Shabahang, S. , Timiryasova, T.M. , Zhang, Q. , Beltz, R. , Gentschev, I. , et al. (2004) Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light‐emitting proteins. Nat Biotechnol 22: 313–320. [DOI] [PubMed] [Google Scholar]
- Zaiter, H.Z. , and Coyne, D.P (1984). Testing inoculation methods and sources of resistance to the halo blight bacterium (Pseudomonas syringae pv. phaseolicola) in Phaseolus vulgaris. Euphytica, 33, 133–141. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1 Expression of FMN reductase (frp) increases luminescence in P. fluorescens NZ011 when combined with the lux operon. Acinetobacter strains: A. baylyi ADP1 (pIJ11282), A. baylyi ADP1 (pRSJ‐pnptII::ilux), A. baylyi ADP1 (pRSJ‐pnptII::lux‐frp). Pseudomonas fluorescens strains: P. fluorescens NZ011 (pIJ11282), P. fluorescens NZ011 (pRSJ‐pnptII::ilux), P. fluorescens NZ011 (pRSJ‐pnptII::lux‐frp). Pseudomonas syringae pv. phaseolicola (Pph) strains: Pph 1302A (pIJ11282), Pph 1302A (pRSJ‐pnptII::ilux), Pph 1302A (pRSJ‐pnptII::lux‐frp). Plasmids pIJ11282, pRSJ‐pnptII::ilux and pRSJ‐pnptII::lux‐frp have the same backbone (pIJ11282) and promoter (pnptII). A: Normalized luminescence of reporter strains in KB (King's B medium) and M9 (M9 minimal medium). B: Growth of reporter strains in KB and M9. OD = optical density at 600 nm. Error bar, +/− SE. n = 3.
Supplementary Fig. 2. Expression of frp in P. fluorescens NZ011 (pRSJ‐pnptII:lux‐frp) results in an increase in NADP+ concentration. Lux: P. fluorescens NZ011 (pIJ11282). Lux‐frp: P. fluorescens NZ011 (pRSJ‐pnptII:lux‐frp). NADP+ concentration refers to 25 ul of bacterial culture, for which a 10‐fold dilution was measured as having an optical density (OD600) of 0.33. Error bar +/− SE. Significant differences (t‐test, p < 0.05) are indicated by asterisks. n = 6.
Supplementary Fig. 3. The lux‐eYFP operon constructed in this study. A: Schematic representation of the lux‐eYFP construct made in this study. p1: pnptII, pOXB20, pOXB16, pOXB13, pOXB11. p2: pA1/04/03, plac. The red ‘T’ represents the T0 terminator. B: Pseudomonas syringae pv. phaseolicola RJ3 (Pph RJ3) pnptII::lux‐pA1/04/03::eYFP imaged with the Typhoon scanner (Amersham/GE Healthcare, UK) using Cy3 settings. eYFP expression is detected. C: Pph RJ3 pnptII::lux pA1/04/03::eYFP imaged with the nightOWL LB 983 at 10 s exposure. Bioluminescence is detected.
Supplementary Fig. 4. A: Schematic representation of the pRS‐pOXB20(1)::lux plasmid. pRS‐pOXB20(1)::lux is derived from pUC18‐mini‐Tn7T‐Gm‐lux. MCS: Long multiple cloning site (490 bp) of pUC18‐mini‐Tn7T‐Gm‐lux plasmid. RBS1: Ribosome Binding site of plasmid pUC18‐mini‐Tn7T‐Gm‐lux. B: Schematic representation of the pRS‐pOXB20::lux plasmid. RBS2: Ribosome‐binding site of plasmid pIJ11282. The RBS strength was calculated with the RBS calculator (Farasat et al., 2014; Ng et al., 2015) considering a total DNA length of 45 bp from the starting codon of luxC.
Supplementary Fig. 5. pRS‐pOXB20::lux generates increased luminescence compared to pRS‐pOXB20(1)::lux in Pseudomonas syringae pv. phaseolicola 1302A. Plasmid pRS‐pOXB20(1)::lux was derived from the existing mini‐Tn7 plasmid pUC18‐mini‐Tn7T‐Gm‐lux (Choi et al., 2005) by inserting oriT and ligating the OXB20 promoter upstream of luxC. Plasmid pRS‐pOXB20::lux has a stronger RBS (derived from pIJ11282) upstream of luxC compared to pUC18‐mini‐Tn7T‐Gm‐lux. A: Normalized luminescence of Pph reporter strains in KB (King's B medium) and M9 (M9 minimal medium). B: Growth of reporter strains in KB and M9. OD = optical density at 600 nm. Error bar +/− SE. n = 3
Supplementary Fig. 6. Pseudomonas syringae pv. phaseolicola 1302A (Pph 1302A) reporter strains exhibit growth curves similar to Pph 1302A in both rich and minimal medium while providing a wide range of luminescence. Strains: Pph 1302A, Pph 1302A pOXB11::lux‐pA1/04/03::eYFP, Pph 1302A pOXB13::lux‐pA1/04/03::eYFP, Pph 1302A pOXB16::lux‐pA1/04/03::eYFP, Pph 1302A pOXB20::lux‐pA1/04/03::eYFP and Pph 1302A pnptII::lux‐pA1/04/03::eYFP. A: Normalized luminescence of Pph reporter strains in KB (King's B medium) and M9 (M9 minimal medium) B: Growth of Pph 1302A reporter strains compared to Pph 1302A in KB and M9. OD = optical density at 600 nm.. Error bar +/− SE. n = 3.
Supplementary Fig. 7. Strong expression of the lux cassette has a small but significant effect on Pph 1302A bacterial growth both in vitro and in planta. A: OD values at 3 h of data presented in Supplementary Fig. 6. nptII: Pph 1302A pnptII::lux‐pA1/04/03:::eYFP; OXB20: Pph 1302A pOXB20::lux‐pA1/04/03::eYFP; OXB16: Pph 1302A pOXB16::lux‐pA1/04/03::eYFP; OXB13: Pph 1302A pOXB13::lux‐pA1/04/03::eYFP; OXB11: Pph 1302A pOXB11::lux‐pA1/04/03::eYFP; 1302A: Pph 1302A. Error bar +/− SE. Significant differences (Tukey HSD, p < 0.05) are indicated by letters. n = 3. B: In planta growth of Pph 1302A reporter strains. TG leaves were syringe infiltrated with Pph 1302A reporter strains at 5x106 CFU ml−1 in 10 mM MgCl2. nptII: Pph 1302A pnptII::lux‐pA1/04/03:::eYFP; OXB16: Pph 1302A pOXB16::lux‐pA1/04/03::eYFP; OXB11: Pph 1302A pOXB11::lux‐pA1/04/03::eYFP; 1302A: Pph 1302A. Error bar +/− SE. Significant differences (Tukey HSD, p < 0.05) are indicated by letters. n = 4.
Supplementary Fig. 8. Detection limit for bioluminescent Pseudomonas syringae pv. phaseolicola RJ3 (Pph RJ3) in agar plates. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP was spotted in serial dilution on an LB agar plate. Images were taken after 1 h using the nightOWL LB 983 at 20 min exposure. Approximately 100 cells/mm2 can be detected. Colour scales have different minimum values to avoid oversaturation. CFU/mm2 was estimated by colony counting. SE for CFU and SE for luminescence constitute on average 27.7% and 2.66% of the means respectively. n = 3.
Supplementary Fig. 9. Pseudomonas syringae pv. phaseolicola RJ3 (Pph RJ3) colonizes the leaf vasculature of P. vulgaris cultivar TG following spray inoculation onto the leaf surface. A, B: Pph RJ3 pnptII::lux‐pA1/04/03::eYFP was sprayed in a localized area on the abaxial surface of Phaseolus vulgaris cv. Tendergreen leaves through an aperture in an aluminium foil (white dotted line). Images were taken after 2 DPI with the nightOWL LB 983 at 20 min exposure. Red circle indicates leaf discs used for confocal microscopy imaging (C). Red lines indicates cross sections used for confocal microscopy imaging (D,E). C: Maximum projections of the leaf vasculature. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow; white arrow). Chlorophyll (red; black arrow). D: image of vasculature cross section. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow). Xylem autofluorescence (grey). Red circular line indicates area zoomed in panel F. E: Maximum projection of vasculature cross section. Xylem autofluorescence (grey). Red circular line indicates area modelled in panel G. F: zoom of panel D. Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow; white arrow). Xylem autofluorescence (grey). G: Three‐dimensional model of panel E (Imaris 9 Bitplane AG, Zürich, Switzerland). Pph RJ3 pnptII::lux‐pA1/04/03::eYFP colonies (yellow; white arrow). H: Three‐dimensional model of panel C (Imaris 9 Bitplane AG, Zürich, Switzerland). Co‐localization of chlorophyll signal (red) and eYFP (yellow), indicates endophytic localization of Pph RJ3 pnptII::lux‐pA1/04/03::eYFP in the leaf vasculature. Scale bars: 40 μm.
Supplementary Fig. 10. Phaseolus vulgaris cultivar Tendergreen leaves do not display background bioluminescence or fluorescence under conditions used to image luminescence or eYFP. A: nightOWL LB 983 image. The image is automatically generated by the nightOWL LB 983 by overlaying the image acquired with CCD camera with a photo of the sample. Exposure time 20 min. The leaf was incubated for 10 min in darkness before imaging. Black arrows indicate tissue used for confocal microscopy imaging. B: details of the Phaseolus vulgaris cultivar TG vascular system. In non‐inoculated plants, eYFP signal is not detectable. Chlorophyll (red), eYFP (yellow). C: details of xylem vessels of non‐inoculated plants. eYFP signal is not detectable. Xylem (grey), eYFP (yellow). Settings used were the same as for inoculated plants. Scale bars: 40 μm.
Supplementary Fig. 11. The colony count approach is able to detect the effect of PTI but not ETI on the growth of Pseudomonas syringae pv. phaseolicola in the early stages of infection (2 DPI). P. vulgaris cultivar Tendergreen (TG) leaves were syringe infiltrated with Pph 1302A ΔxerC, ΔhrpA and ΔPphB at 106 CFU ml−1 in 10 mM MgCl2. A: Details of the symptoms caused by Pph 1302A knock‐out mutants in TG leaves at 2 days post inoculation (DPI). The hypersensitive response (HR) is evident on leaves inoculated with Pph 1302A ΔxerC. B: Colony count data for inoculated leaves at 2 DPI. Error bar +/− SE. Significant differences (Student's T‐test, p < 0.05) are indicated by letters. n = 8.
Supplementary Video 1 Pph RJ3‐eYFP colonizes the collenchyma of the Phaseolus vulgaris cultivar Tendergreen (TG) leaf vasculature. Pph RJ3‐eYFP (Godfrey et al. 2010) was syringe‐infiltrated at 106 CFU ml−1 into the first true leaves of TG plants. Leaf disks were detached at 3 days post inoculation and imaged with a TCS Leica SP5 confocal microscope in a xyz series (objective used: HCX PL APO CS 20.0x0.70 IMM UV). eYFP was excited with the 488 nm wave length laser. Cyan (Pph RJ3 cells); Red (Chlorophyll autofluorescence); Grey (bright field).
Supplementary Video 2 Pph RJ3‐eYFP (Godfrey et al., 2010) moves systemically inside the vasculature of Phaseolus vulgaris cultivar Tendergreen (TG). Pph RJ3‐eYFP was syringe‐infiltrated at 106 CFU ml−1 into the first true leaves of TG plants. Leaf disks were detached at 1 day post inoculation and imaged with the a TCS Leica SP5 confocal microscope in a xyz series (objective used: HC PL APO CS2 63.0x1.20 WATER UV). eYFP was excited with the 488 nm wave length laser. Cyan (Pph RJ3 cells); Red (Chlorophyll autofluorescence); Grey (bright field).
Supplementary Table 1 Plasmids used in this study.
Supplementary Table 2. List of strains used
Supplementary Table 3. List of primers used for cloning
Supplementary Table 5. List of primers used to sequence eYFP and frp.
Supplementary Table 6. List of primers used to sequence the lux operon
Appendix S1. Supporting Information.
Appendix S2. Supporting Information.
Data Availability Statement
The research materials supporting this publication have been deposited in the Oxford Research Archive (ORA) as https://doi.org/10.5287/bodleian:y0r8ErQja and https://doi.org/10.5287/bodleian:KOeJ8agdn. If you wish to access the biological material that is described in this study please contact gail.preston@plants.ox.ac.uk.


