Yu-Kemp et al. explore mechanisms that epithelial cells use to assemble supramolecular actomyosin structures at E-Cadherin–based cell–cell junctions. They find that individual actin assembly pathways are not essential. Instead, microscopy and pharmacologic inhibition suggest that micron-scale supramolecular myosin arrays help bundle actin at mature junctions.
Abstract
Epithelial cells assemble specialized actomyosin structures at E-Cadherin–based cell–cell junctions, and the force exerted drives cell shape change during morphogenesis. The mechanisms that build this supramolecular actomyosin structure remain unclear. We used ZO-knockdown MDCK cells, which assemble a robust, polarized, and highly organized actomyosin cytoskeleton at the zonula adherens, combining genetic and pharmacologic approaches with superresolution microscopy to define molecular machines required. To our surprise, inhibiting individual actin assembly pathways (Arp2/3, formins, or Ena/VASP) did not prevent or delay assembly of this polarized actomyosin structure. Instead, as junctions matured, micron-scale supramolecular myosin arrays assembled, with aligned stacks of myosin filaments adjacent to the apical membrane, overlying disorganized actin filaments. This suggested that myosin arrays might bundle actin at mature junctions. Consistent with this idea, inhibiting ROCK or myosin ATPase disrupted myosin localization/organization and prevented actin bundling and polarization. We obtained similar results in Caco-2 cells. These results suggest a novel role for myosin self-assembly, helping drive actin organization to facilitate cell shape change.
Introduction
E-Cadherin (Ecad)–based adherens junctions (AJs) play a pivotal role in maintaining epithelial tissue homeostasis by mediating cell–cell adhesion and anchoring the actomyosin cytoskeleton (Lecuit and Yap, 2015). During morphogenesis, AJs ensure tissue integrity when cells change shape, divide, and move, events involving force exerted on AJs, which must be remodeled to accommodate tissue-wide mechanical forces. The relationship between AJs and the actomyosin cytoskeleton is one of reciprocal reinforcement (Michael and Yap, 2013). Apical AJs polarize actin, and connections to actin stabilize AJs at the plasma membrane. This focused attention on the complex supramolecular actomyosin structures assembled at AJs.
One key issue in the field is to define the mechanisms cells use to assemble and polarize this complex structure. Significant progress has been made in examining polarization of AJs and the junctional cytoskeleton. Drosophila embryogenesis provides an important model, as 6,000 cells simultaneously form and polarize their AJs during cellularization (Schmidt and Grosshans, 2018). Gastrulation then begins within minutes and requires intricate cross-talk between AJs and the cytoskeleton, allowing dramatic cell shape changes while maintaining tissue integrity. However, the complexity of the in vivo system provides major challenges. For example, myosin, its activator Rho (e.g., Crawford et al., 1998; Royou et al., 2004; Xue and Sokac, 2016), Arp2/3, and the formin Diaphanous (Afshar et al., 2000; Stevenson et al., 2002; Zallen et al., 2002) are required for cellularization. Cultured mammalian cells provide a simpler place to examine how the actomyosin cytoskeleton assembles at AJs, with pharmacologic tools allowing disruption of protein function in a temporally controlled way.
Often the junctional cytoskeleton is polarized, with Ecad and actin enriched at the apical end of lateral cell borders, in a structure known as the zonula adherens (ZA). Scientists have taken several approaches to explore molecular mechanisms involved in ZA assembly and maintenance. One approach was to explore roles of actin and its regulators. Actin filament assembly involves nucleation, elongation, and bundling. The Arp2/3 complex nucleates new filaments from the sides of existing ones, creating branched networks, while formin family members nucleate new unbranched filaments (Buracco et al., 2019; Pollard, 2016). Formins and Ena/vasodilator-stimulated phosphoprotein (VASP) proteins promote filament elongation. Blocking actin polymerization using cytochalasin or latrunculin can block de novo AJ assembly (Ivanov et al., 2005), but mature AJs are more resistant (Ivanov et al., 2004; Tang and Brieher, 2012). The Arp2/3 complex coimmunoprecipitates with Ecad (Kovacs et al., 2002) and is enriched at AJs in many epithelial cell types (Yamada and Nelson, 2007). RNAi knockdown (KD; Verma et al., 2012) or use of pharmacologic inhibitors (Kovacs et al., 2011; Tang and Brieher, 2012) revealed a role for Arp2/3 in maintaining actin levels at AJs, but inhibiting Arp2/3 reduced but did not eliminate junctional actin. Multiple Arp2/3 activators, including cortactin (Han et al., 2014; Helwani et al., 2004), N-WASP, WAVE, and WIRE (Wiskott-Aldrich syndrome protein, WASP-family verprolin-homologous protein, and WIP-related; Verma et al., 2004; Kovacs et al., 2011; Verma et al., 2012) also are enriched at AJs and interact with Ecad. However, once again loss-of-function reduced but did not eliminate junctional actin in established monolayers, suggesting that Arp2/3 acts in parallel with other mechanisms to assemble junctional actin. Scientists also explored formins (Carramusa et al., 2007; Kobielak et al., 2004; Nishimura et al., 2016; Rao and Zaidel-Bar, 2016; Sahai and Marshall, 2002). Individual KD of DAAM1 or Dia1 reduced but did not eliminate junctional actin. Finally, scientists explored Ena/VASP proteins. While Ena/VASP proteins are enriched at AJs (e.g., Oldenburg et al., 2015; Vasioukhin et al., 2000), KD or sequestration away from AJs reduced but did not eliminate junctional actin (Scott et al., 2006; Yu-Kemp et al., 2017). Taken together, these data suggest that multiple parallel mechanisms drive assembly and maintenance of the junctional actin cytoskeleton.
Scientists also explored roles of nonmuscle myosin II (hereafter referred to as myosin). Junctional contractility at the ZA is driven by myosin and regulated by complex feedback loops between actin, myosin, and Ecad. Myosin monomers assemble into bipolar filaments, and their ATPase-powered motor activity produces contractile force on the AJ-associated actin cytoskeleton (Agarwal and Zaidel-Bar, 2019; Vicente-Manzanares et al., 2009). Myosin filament assembly requires phosphorylation via Rho kinase (ROCK) or myosin light chain kinase (MLCK). Myosin also cross-links actin, which, coupled with its motor activity, allows myosin to organize actin. Inhibiting myosin motor activity or myosin activation reduced, but did not eliminate, actin assembly at AJs (Leerberg et al., 2014; Sahai and Marshall, 2002; Shewan et al., 2005; Zhang et al., 2005). Myosin-generated contractility can stimulate actin assembly (Leerberg et al., 2014), and myosin contractility can stabilize AJs by pulling on α-catenin and shifting it to an open state, strengthening AJ–actin connections (Ozawa, 2018).
Advanced imaging revealed new insights into supramolecular myosin organization. Two prescient papers from the 1990s explored myosin organization in migrating fibroblast lamella, where myosin filaments stacked on one another in arrays involving dozens of filaments (Svitkina et al., 1997; Verkhovsky et al., 1995). Structured illumination microscopy (SIM) allowed direct observation of myosin stacks and associated actin filaments in living cells (Fig. 1 A; Beach et al., 2017; Burnette et al., 2014; Fenix et al., 2016; Hu et al., 2017). Individual myosin filaments organized into stacks, with stacks oriented perpendicular to peripheral stress fibers, thus linking actin filaments (Fig. 1 A), and aligned myosin stacks formed 2D arrays tightly apposed to the plasma membrane. Inhibiting myosin activation or myosin motor activity reduced stack assembly. Hu and Bershadsky made an interesting prediction: “Since the interaction of myosin filaments associated with different actin bundles creates forces attracting these bundles toward each other, the organization of myosin filaments into stacks is a plausible mechanism for the formation of the densely packed arrays of parallel actin bundles often seen in polarized fibroblast-type cells” (Hu et al., 2017). Advances in imaging also provided insights into actin and myosin organization at the ZA, revealing two zones of actin at the bicellular border ZA of endothelial cells (Efimova and Svitkina, 2018): a central zone of branched actin immediately adjacent to Ecad, colocalizing with Arp2/3, and more lateral bundled actin filaments decorated by myosin, forming an actin belt along each bicellular border. Superresolution microscopy supports a similar organization of actin and myosin at bicellular junctions in other epithelial cells (Choi et al., 2016; Heuzé et al., 2019; Kovacs et al., 2011). At tricellular junctions, bundled bicellular border actin filaments may anchor end-on in cadherin–catenin complexes (Choi et al., 2016; Efimova and Svitkina, 2018).
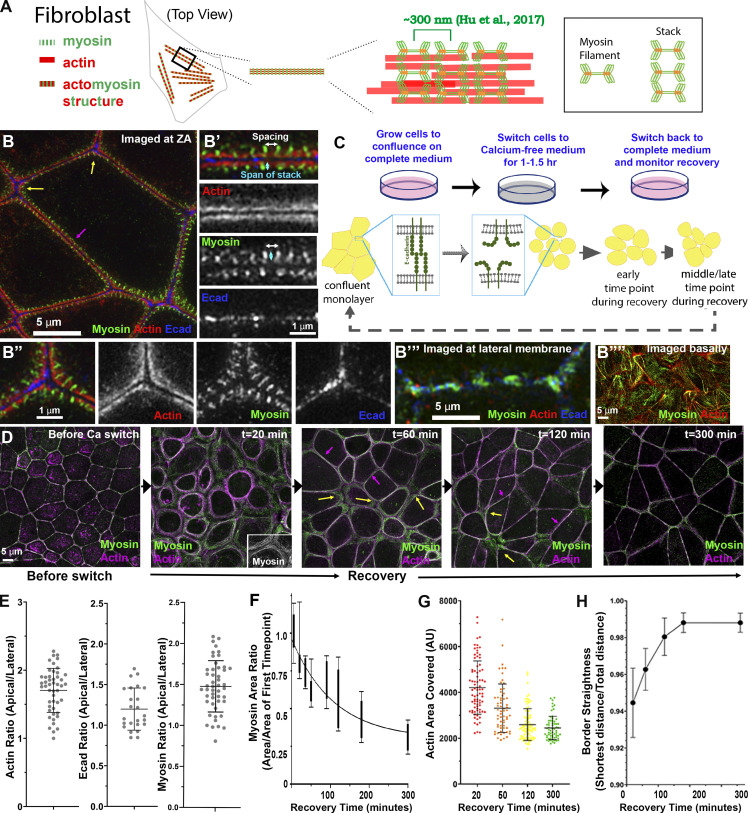
Figure 1.
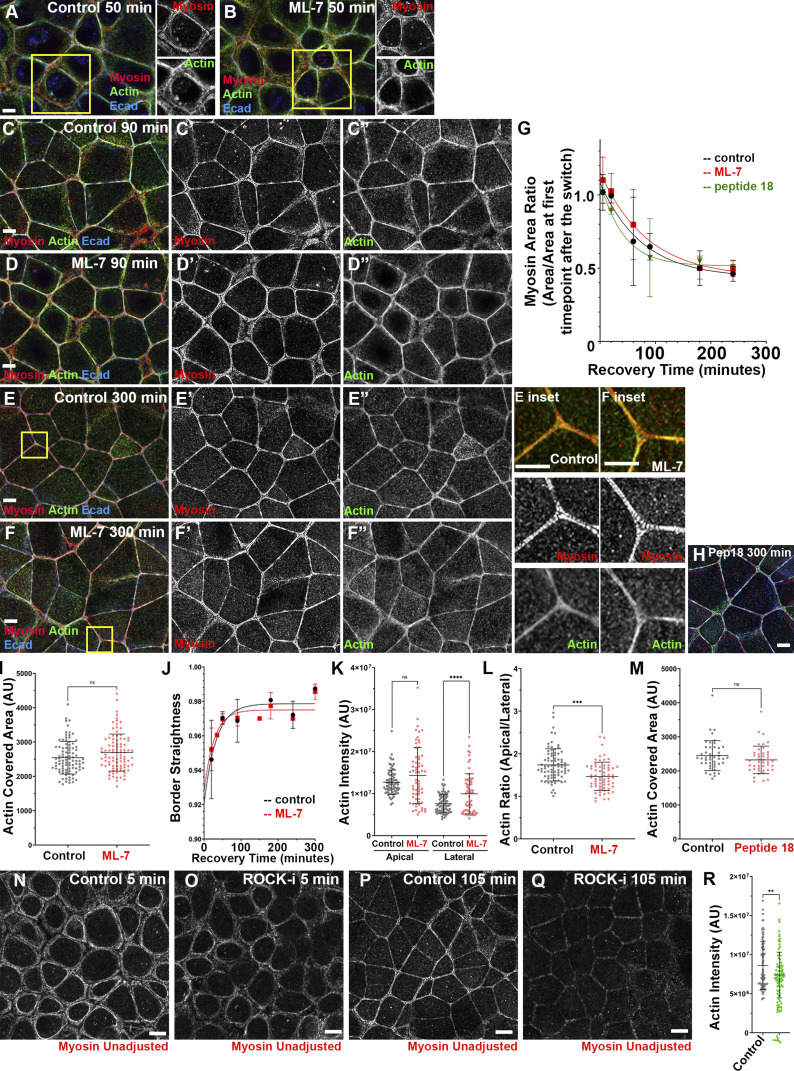
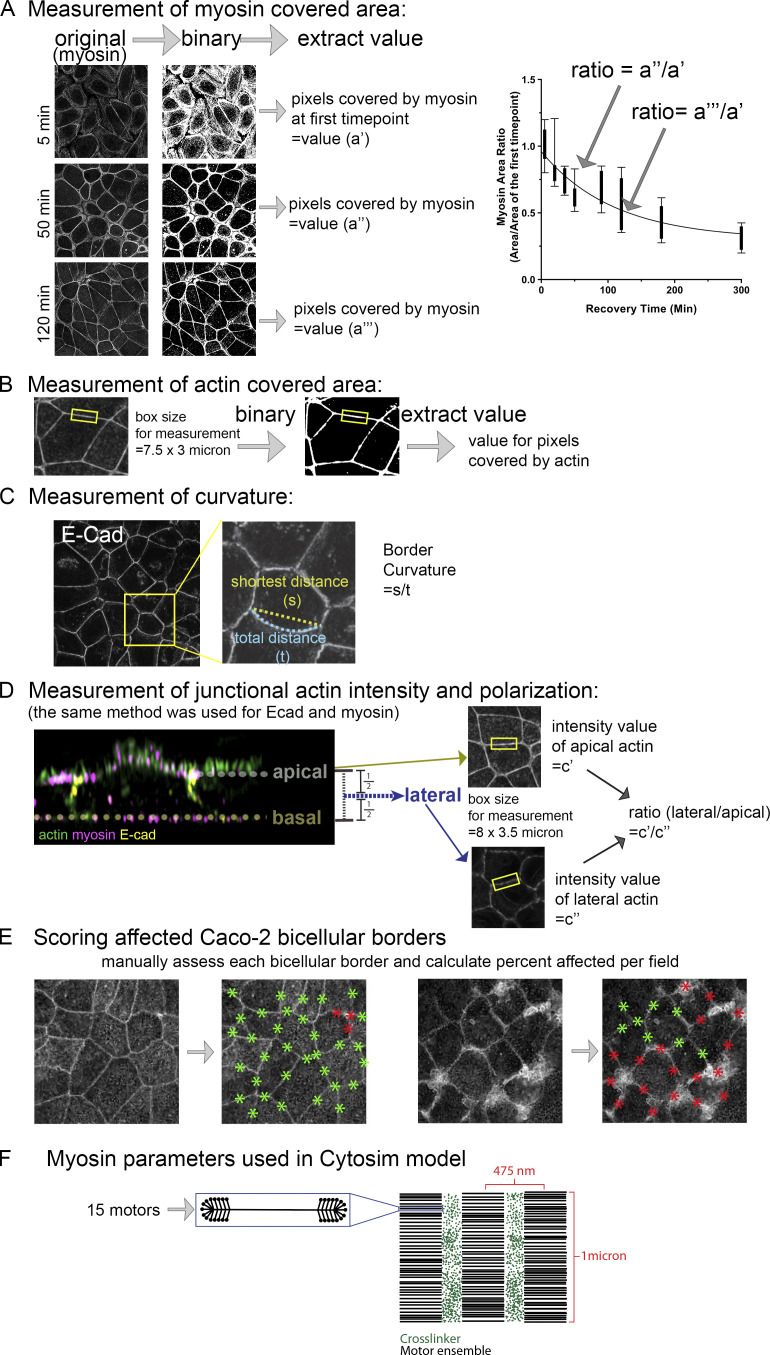
ZO KD MDCK cells as a model to study formation of ZA actomyosin structures. (A) Schematic of myosin filament stacks in fibroblasts. (B) ZO KD MDCK cells. ZA at bicellular borders (B′) and tricellular junctions (B″). Actin is bundled and myosin organized into a sarcomeric pattern. (B″′) Lateral membrane. (B″″) Basal stress fibers. (C) Schematic diagram of calcium switch. (D) Representative images of actin and myosin at the apical surface as junctions recover. (E) Ratio of apical to lateral signals of actin, Ecad, or myosin at the end of recovery. (F–H) Quantification. Changes of area covered by myosin (F), ZA actin bundling (G), and border curvature (H) during junction maturation. Error bars represent mean ± SD. Magenta arrows, bicellular borders; yellow arrows, tri-/multicellular junctions. Scale bars = 5 µm. For all figures, unless indicated, top-view images are apical MIPs. In E, n = individual borders; actin = 48, Ecad = 24, myosin = 48. In F, numbers for each time point are in Table S1. In H, representative of three experiments with one to three fields of cells/experiment/time point, with seven to nine borders quantified/field.
Zonula occludens protein-1 (ZO-1)/ZO-2-KD MDCK cells (hereafter ZO-KD cells) assemble an “Albert’s textbook” ZA (Choi et al., 2016; Fanning et al., 2012), with a robust actomyosin array much like the ZA sarcomeric array of cochlear hair cells (Ebrahim et al., 2013). To define mechanisms by which cells assemble and position the contractile cytoskeleton at the ZA as adhesion is established, we used the calcium switch assay to visualize reassembly of the supramolecular actomyosin array at the ZA in this model epithelial cell line via superresolution microscopy, to determine the ZA organization/assembly pathway, and combined this with genetic and pharmacologic approaches to define the molecular machines and mechanisms involved.
Results
A model system to study actomyosin assembly at the ZA
ZO-KD MDCK cells (Choi et al., 2016; Fanning et al., 2012) assemble a textbook-like ZA with a highly organized actomyosin array positioned at the apical end of lateral cell borders (Fig. 1 B). SIM superresolution imaging revealed tightly bundled actin cables along bicellular junctions (Fig. 1 B, magenta arrow; Fig. 1 B′), decorated by a sarcomeric array of myosin (Fig. 1 B′; white arrow = sarcomeric spacing), underlain by puncta of Ecad (Choi et al., 2016; Fig. 1 B′). There are elevated levels of Ecad and increased spacing between actomyosin arrays at tricellular junctions (Fig. 1 B, yellow arrows; Fig. 1 B′′), where molecular tension is exerted on Ecad–catenin complexes (Choi et al., 2016). Ecad is enriched at the apical ZA (Fig. S1 A, red arrows; quantified in Fig. 1 E) and also present at lower levels all along lateral cell borders (Fig. S1 A, green arrows), where it associates with a distinct actin population that is less bundled and had little associated myosin (Fig. 1 B′′′). Actin and myosin (visualized using antibodies to myosin-2B) are enriched at the apical ZA (Fig. 1 E) and are also found in basal stress fibers (Fig. 1 B′′′′). Myosin-2A showed a similar sarcomeric localization at the ZA (Fig. S1 R).
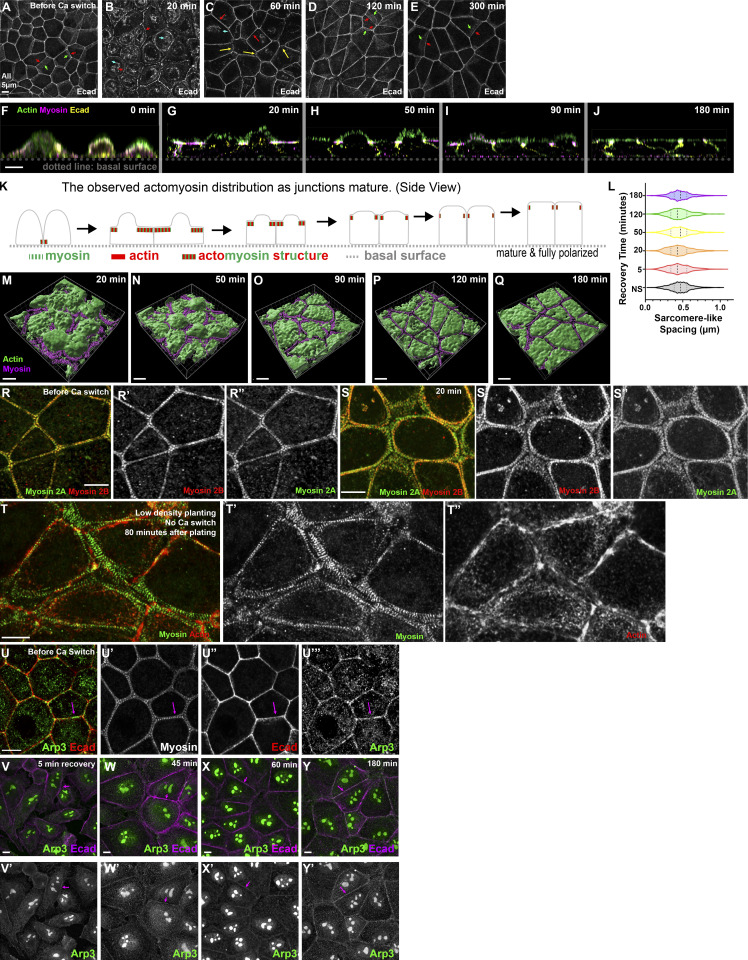
Figure S1.
Ecad localization, 3D cell shapes, sarcomere spacing, myosin 2A localization, arrays without Ca switch, and Arp3 localization before and during recovery. (A–E) The localization of Ecad throughout Ca recovery. Ecad is internalized right after the Ca switch (cyan arrows). As the recovery proceeded, Ecad regained its continuous distribution at the ZA, and cell borders straightened. Arrows: red, apical ZA; green, lateral membrane; yellow, tricellular junctions. (F–J) Cross-section view of monolayer as junctions mature. The apical membrane flattened as cells polarized. Myosin is enriched in arrays underlying the apical membrane, which narrow until it is enriched at the ZA. (K) Diagram illustrating actomyosin localization along the z axis as junctions mature. (L) Quantification of the myosin sarcomere-like spacing at different recovery time points. The spacing remained ∼0.45 µm throughout junction maturation, indicating that the age of the junction did not modulate myosin spacing; n = 650 for each time point. NS = nonswitched. (M–Q) 3D surface view of cells at different recovery time points. These images reveal how the apical surface of the cell changed as junctions and myosin structure matured. (R and S) Both myosin-2A and -2B localize to the sarcomeric array at the ZA (R) and in the extended myosin stacks seen during recovery (S). (T) Cells plated without Ca switch, to observe the ZA form de novo. These cells also formed similar myosin arrays during junction establishment. (U–Y) The Arp2/3 complex localizes to the ZA in confluent ZO KD MDCK cells but returns there only slowly after Ca switch, while VASP localizes to cell borders with enrichment at tricellular junctions. (U) Arp3 at AJs in a confluent, mature monolayer. (V–Y) Arp3 is only weakly localized to junctions during early stages of recovery.
To study AJ reassembly, we used the calcium (Ca) switch assay (Gumbiner and Simons, 1986), which relies on the calcium dependence of classic cadherin structure/function (Shapiro et al., 1995; Takeichi, 1988). Before perturbation, cells were columnar, with a slightly domed apical microvillar surface. We removed Ca for 1.5 h, disrupting cell–cell adhesion, and cells went from columnar to rounded (Fig. 1 D, 0 min to 20 min; Fig. S1 F). Ecad was endocytosed, accumulating in an apparent vesicular compartment (Fig. S1 B, cyan arrows), and cells unzipped along their lateral borders (magnesium was not chelated, so cell–ECM adhesion was not disrupted). When Ca was added back, Ecad returned to the bases of the cells where contact was preserved (Fig. S1 B, red arrows); at this stage, cells were highly rounded (Fig. S1, F and M). As AJs reassembled over several hours (Fig. 1 D), cells zipped together, with Ecad going from punctate to more continuous at reestablished AJs (Fig. S1 B vs. Fig. S1, C and D; red arrows). Over the next 2–3 h, borders slowly straightened; bicellular borders straightened first (Fig. 1 D, magenta arrows), with tricellular and short multicellular junctions the last sites of reassembly (Figs. 1 D and S1 C, yellow arrows), until columnar architecture was restored (Fig. S1, F–Q), with Ecad enriched at the ZA (Fig. S1 E, red vs. green arrows). We adopted three methods to quantify the rate of junctional actomyosin reassembly. First, we quantified the area occupied by myosin over the entire field. As AJs reassembled, myosin arrays at junctions narrowed, and thus the area occupied by myosin decreased until it reached that seen before Ca withdrawal (Fig. 1 F). Similarly, we quantified increased actin bundling into the tight array seen before perturbation, by selecting apical regions of bicellular junctions, binarizing images, and calculating the area occupied by actin (Fig. 1 G). Finally, we quantified junctional straightening as AJs reassembled (Fig. 1 H). These data were the baseline for our subsequent perturbations.
Junctional reassembly involves generation of very large-scale myosin arrays
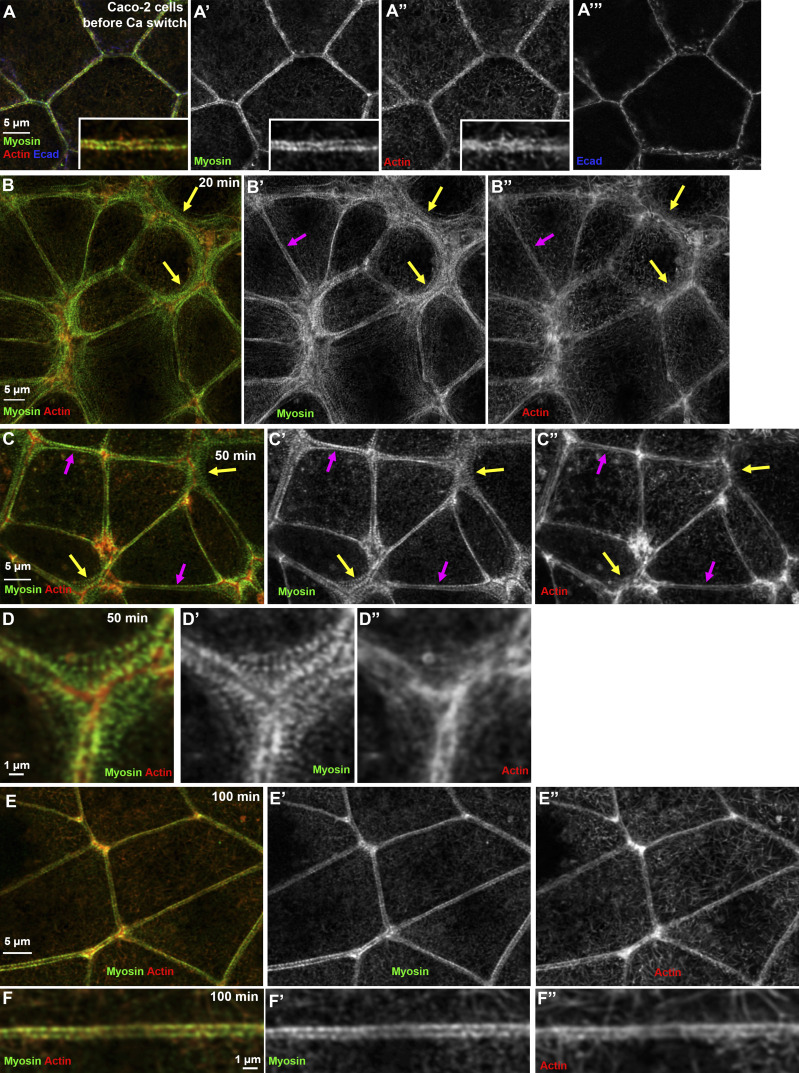
We next examined AJ and cytoskeletal proteins at high resolution as AJs reassembled, using Zeiss Airyscan or Nikon SIM imaging. Myosin and actin localization proved quite surprising. Before perturbation, myosin localized to the tight ZA sarcomeric array (Fig. 1, B–B′′) and to basal stress fibers (Fig. 1 B′′′′). As cells rounded up after Ca removal, myosin initially associated with actin “arcs” aligned along reforming lateral cell borders (Fig. 2, A–A′′′; closeup in Fig. 2 B), similar to those seen in keratinocytes during Ca recovery (Zhang et al., 2005). Strikingly, however, within 20 min of recovery, extensive stacks of myosin filaments assembled around the cell periphery (Fig. 1 D, 20 min; Fig. 2 C), similar to but more extensive than the myosin stacks seen in migrating fibroblasts (Beach et al., 2017; Burnette et al., 2014; Fenix et al., 2016; Hu et al., 2017; Verkhovsky et al., 1995). These myosin stacks rapidly returned to their mature form at many bicellular borders (Fig. 1 D, 60 min; Fig. 2 C, magenta arrows) but remained extensive at a subset of tricellular and multicellular borders as recovery proceeded (Fig. 1 D, 60 min; Fig. 2 C, yellow arrows).
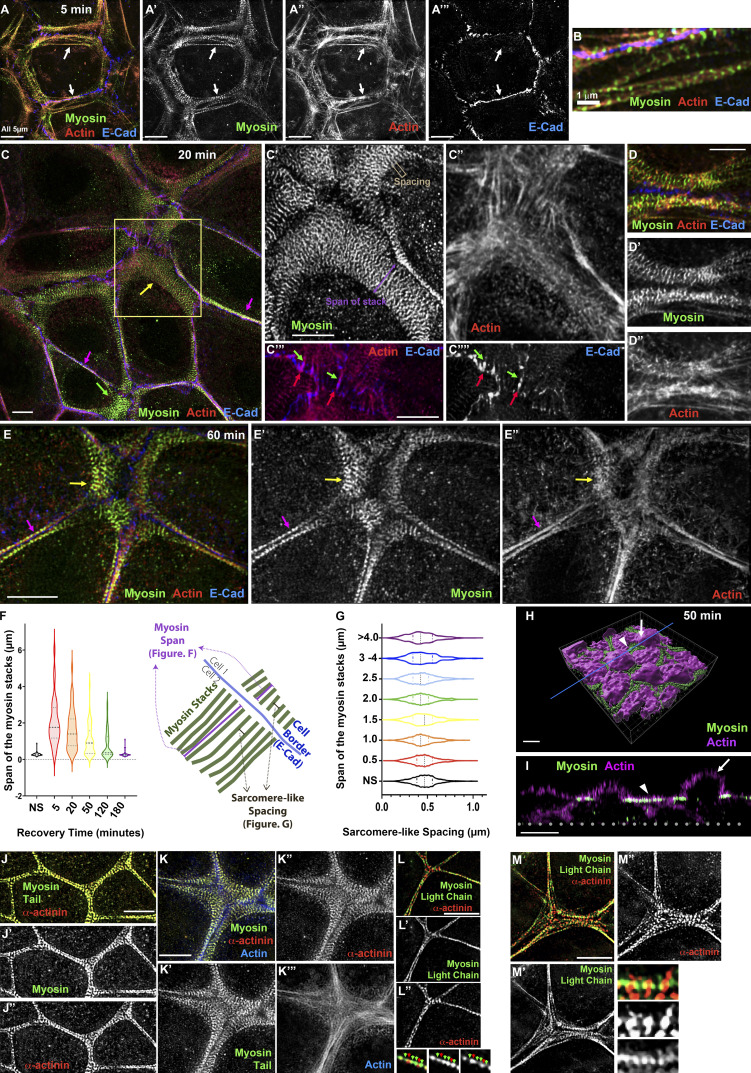
Figure 2.
Extensive stacks of myosin precede the formation of bundled actin as Ecad–based AJs assemble. (A–E) Images collected at different time points of Ca recovery reveal that an extensive myosin array forms during midrecovery and localizes with a less well-organized actin network. (F and G) Quantification. Despite changes in the span of the myosin stacks at different time points (F), spacing between myosin stacks remains similar (G). (H) 3D surface rendering of a cell in midrecovery. (I) Cross-section view at the line indicated in H. The extensive myosin stacks localize to a restricted Z plane underlying the apical plasma membrane (arrowheads), between the domed microvillar caps (arrows). In F, n = (nonswitched [NS] = 75, 5 min = 34, 20 min = 44, 50 min = 63, 120 min = 51, 180 min = 43). In G, n = 550 for each time point.
The micron-scale supramolecular myosin arrays assembled during recovery were quite striking. In unperturbed cells, the sarcomere-like myosin arrays decorating the bundled actin filaments spanned ∼200–300 nm in the direction perpendicular to the membrane (Fig. 1 B′, cyan arrow; Fig. 2 F, nonswitched [NS]). In contrast, mid-assembly, stacks could exceed many times that span, extending to 4–6 µm (Fig. 2, C and C′; purple arrow; Fig. 2 F; visualized using antibodies to myosin-2B; myosin-2A also localized to these stacks; Fig. S1 S). In unperturbed cells, myosin at the ZA had a spacing between the short myosin filament stacks of ∼400 nm (Fig. 1 B′, white arrow; Choi et al., 2016); the antibody detects myosin heavy chain’s tail), similar to spacing in stress fibers. Strikingly, spacing between stacks remained similar throughout assembly (Fig. 2 C′, bracket), regardless of the span of the stacks (Fig. 2 G) or age of the junction (Fig. S1 L). Actin underlying these myosin stacks was less organized than myosin, often forming a meshwork (Fig. 2 C′′). There also were actin structures associated with Ecad at reforming junctions, with short robust actin filaments (Fig. 2, C′′′ and C′′′′, red arrows) terminating in interdigitating cadherin-zippers (Fig. 2, C′′′ and C′′′′, green arrows), like those in keratinocytes or endothelial cells (e.g., Huveneers et al., 2012; Vasioukhin et al., 2000). At times, remnant actin “arcs” parallel to the lateral membrane were seen underlying the myosin stacks (Fig. 2, D–D′′), similar to those seen earlier in recovery (Fig. 2, A–A′′). Myosin arrays at bicellular and some tricellular junctions rapidly reduced their span, but at a subset of tricellular and multicellular junctions, extensive myosin stacks remained, and actin in these regions remained less well organized (Fig. 2 E). These were the last places to resume the bundled actin architecture seen before the Ca switch (Fig. 1 D, 120 min, yellow arrows).
We next examined myosin stack positioning along the z axis. Strikingly, myosin stacks occupied an ∼0.5-µm region in the Z plane, close to and underlying a flattened region of the plasma membrane “apical” to lateral borders (Fig. 2, H–I′, arrowheads; Fig. S1 M) and separate from the domed center (Fig. 2, H–I, arrows). The domed membrane flattened as AJs matured, but residual myosin stacks continued to occupy flattened regions underlying the apical membrane throughout the process (Fig. S1, K and M–Q). Together, these data reveal that very extensive myosin stacks form during ZA reassembly, often before there is any apparent organization of underlying actin, and these stacks narrow as actin is bundled into tight polarized sarcomeric structures, reforming the ZA. We then explored whether these arrays were specific to cells undergoing a Ca switch, by replating cells in medium with normal Ca levels and watching the ZA form de novo. These cells formed similar myosin arrays during AJ establishment (Fig. S1 T). Finally, we explored whether the actin cross-linker α-actinin, known to localize to both stress fibers and myosin arrays in fibroblasts (Hu et al., 2017; Verkhovsky et al., 1995), localized to the ZA arrays in unperturbed cells or the myosin arrays seen during recovery. α-Actinin-4 localized to both structures (Fig. 2, J–M). Intriguingly, it colocalized with antibodies to the myosin tail (Fig. 2, J and K) and alternated in localization with a GFP-tagged myosin regulatory light chain, which is localized to the myosin head (Fig. 2, L and M).
Neither Arp2/3 activity nor formin activity is essential to assemble specialized actomyosin structures at the ZA, although they do enhance cortical actin
We next used functional assays to define mechanisms driving assembly of specialized ZA actomyosin structures. We tested two broad mechanistic hypotheses: (a) ZA actomyosin assembly is driven by actin filament nucleation and elongation, and (b) ZA actomyosin assembly is driven by myosin’s motor activity, gathering and bundling preexisting actin filaments. We initially favored a role for the Arp2/3 complex, as it can coimmunoprecipitate with Ecad, localizes to AJs, can stimulate actin assembly at Ecad-based contacts, and along with its regulators, helps stimulate junctional actin assembly in confluent monolayers (Efimova and Svitkina, 2018; Kovacs et al., 2011; Tang and Brieher, 2012; Verma et al., 2012). We first examined Arp2/3 localization during junction recovery. As in other cell types, the Arp2/3 complex was enriched at the ZA in unperturbed confluent cells (Fig. S1 U, arrow). However, enrichment was weak until relatively late in AJ reassembly (Fig. S1, V–Y).
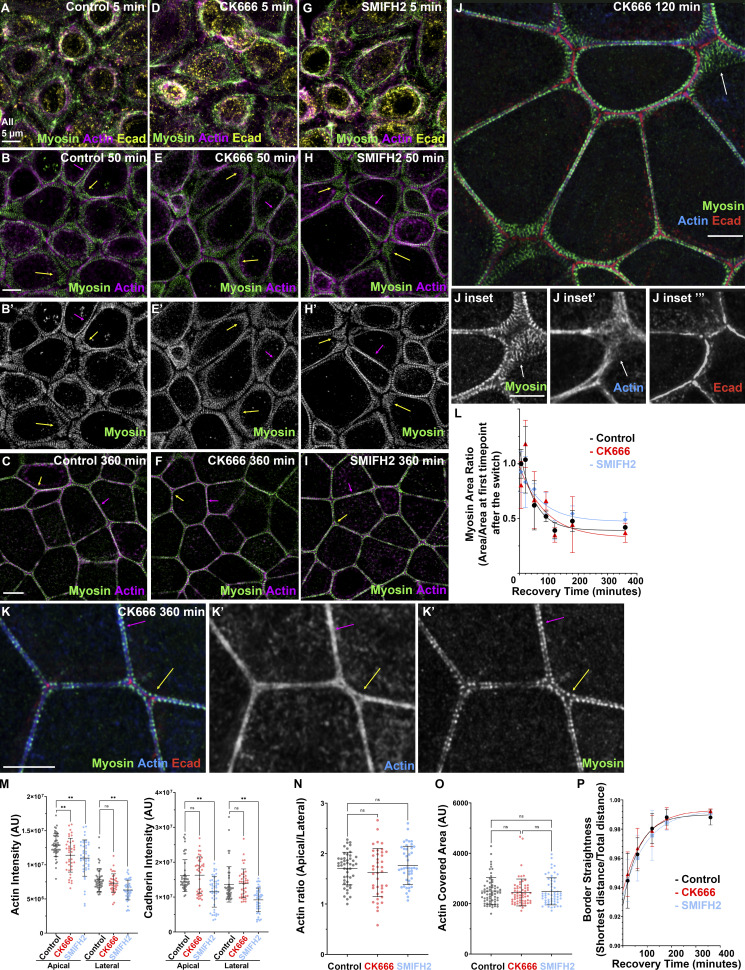
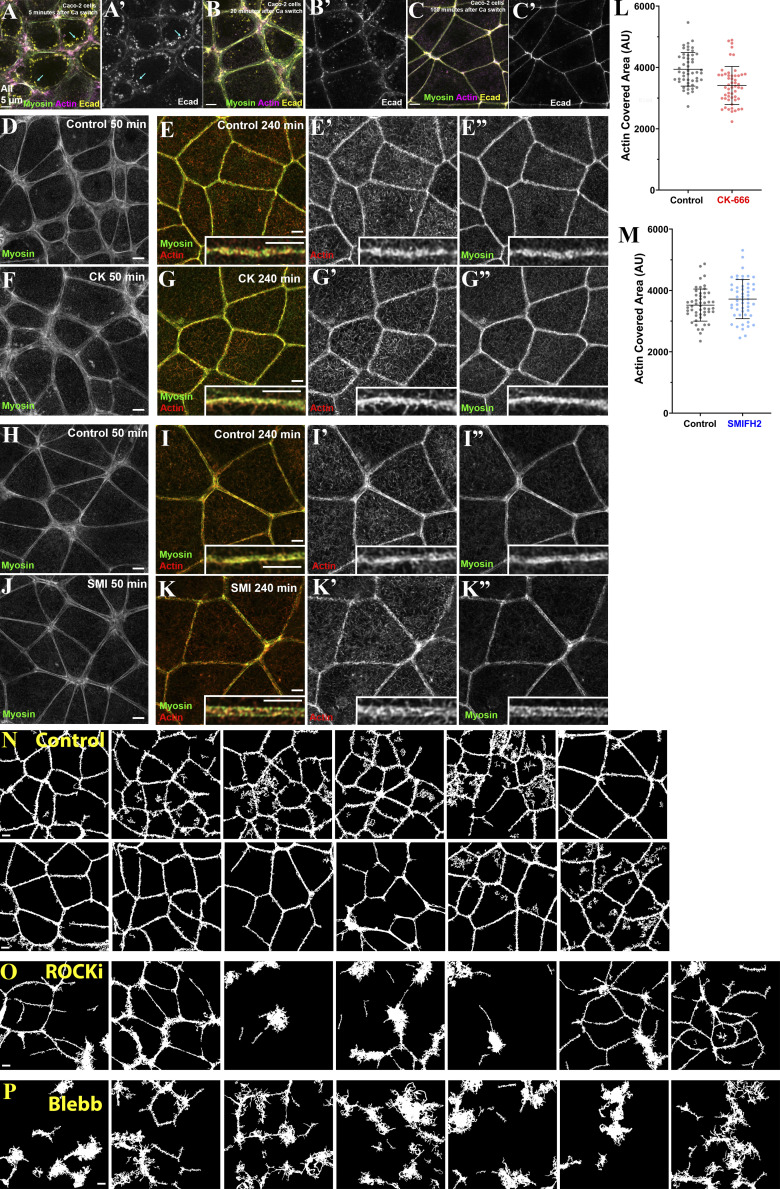
To assess Arp2/3′s role, we used the well-characterized inhibitor CK666 (Hetrick et al., 2013; Nolen et al., 2009), after verifying its activity by assessing its ability to block cell spreading on the substrate (Fig. S2, A–E). We performed the Ca switch in the presence or absence of CK666. To our surprise, assembly of the tight ZA actomyosin array was qualitatively unaltered by the inhibitor (Fig. 3, A–C vs. Fig. 3, D–F). Cells rounded up at early time points (Fig. 3 A vs. Fig. 3 D), and then expansive myosin stacks formed as junctions were reestablished (Fig. 3 B vs. Fig. 3 E, yellow arrows). These stacks slowly reduced in span, first at bicellular junctions (Fig. 3 B vs. Fig. 3 E, magenta arrows) and finally at tricellular junctions, until the ZA sarcomeric actomyosin array was reestablished (Fig. 3 C vs. Fig. 3 F). Superresolution imaging confirmed that in places with extensive myosin stacks, actin was present as a disordered array of filaments (Fig. 3, J and J inset, arrows), as in controls. In CK666-treated cells, tightly bundled actin and narrowed stacks of sarcomeric myosin were restored at the ZA at both bicellular and tricellular junctions (Fig. 3 K, magenta and yellow arrows). To assess whether junctional reestablishment was delayed after Arp2/3 inhibition, we used the reduction in span of myosin stacks to quantitatively assess these events (as in Fig. 1 F). While there was experiment-to-experiment variability, there was no substantial delay of myosin stack narrowing after Arp2/3 inhibition (Fig. 3 L). Intriguingly, Arp2/3 inhibition reduced total apical actin levels, with less effect on lateral actin (Fig. 3 M), but apical actin enrichment at the ZA was unaffected (Fig. 3 N). We assessed ZA actin bundling (as in Fig. 1 G). At the completion of ZA reassembly, actin was equally bundled after Arp2/3 inhibition (Fig. 3 O). Finally, border straightening occurring as the ZA array assembled was also unaltered (Fig. 3 P). Together, these data suggest that Arp2/3 activity is not essential for timely and accurate assembly of specialized ZA actomyosin structures, even though it promotes overall cortical actin levels in this cell type.
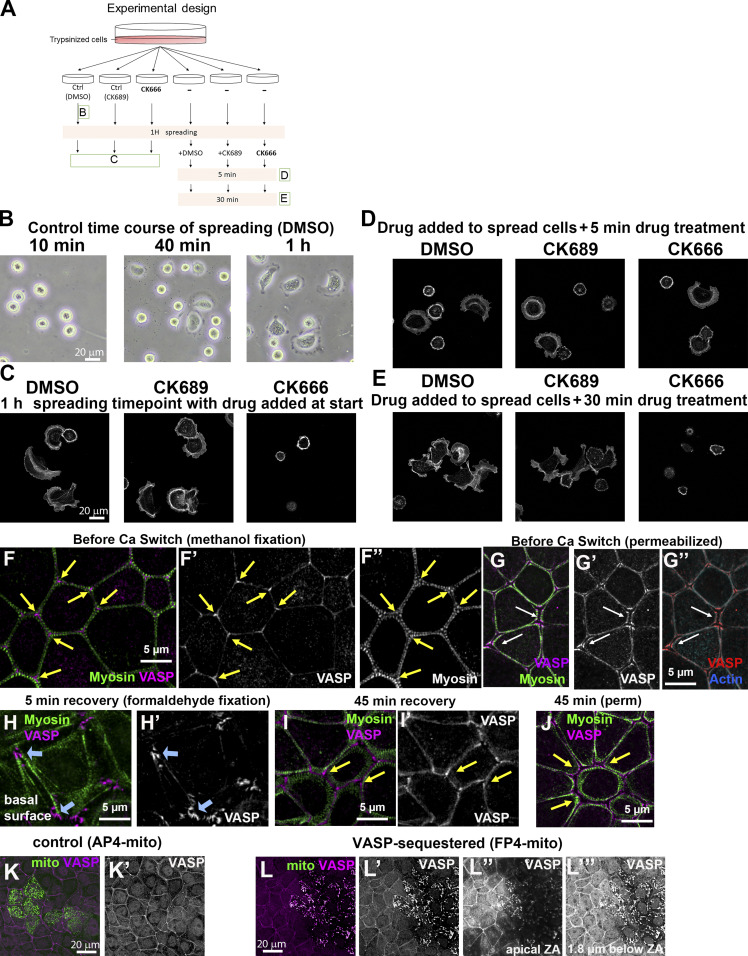
Figure S2.
Verification of the activity of CK666 for inhibiting Arp2/3 function and of AP4/FP4 constructs for sequestering Ena/VASP to mitochondria. (A–E) Verification of CK666 function. (A) Schematic diagram of the experiment. Ctrl, control. (B) Bright-field image of control cells at different spreading time points. More Arp2/3-dependent lamellipodia structures were formed the longer the cells were plated on extracellular matrix. (C–E) Phalloidin staining. (C) CK666 inhibited Arp2/3-dependent cell spreading, whereas the control (DMSO or CK689 [the inactive molecule]) did not. (D and E) Arp2/3-dependent lamellipodia disappeared in the presence of CK666. (F and G) In confluent monolayers, VASP localizes to bicellular borders and is enriched in tricellular junctions. (H) At early stages of recovery, VASP also localizes to basal focal adhesions. (I and J) As Ecad junctions start to form, VASP is detected at cell borders and tricellular junctions. (K and L) Verification of VASP-sequestering construct. (K) In cells transfected with the control construct (AP4mito), VASP remained enriched at the cortex. (L) In FP4mito-transfected cells, VASP localization to apical junctions was strongly reduced or lost, and it relocalized to internal structures we presume are mitochondria. VASP loss at cell junctions was apparent both apically (L″) and more basally (L″′).
Figure 3.
Neither Arp2/3 activity nor formin activity is essential to assemble specialized actomyosin structures at the ZA, although they do enhance cortical actin. (A–K) Recovery from Ca switch in control, CK666 (Arp2/3 inhibitor)-treated, or SMIFH2 (formin inhibitor)-treated cells. (L–P) Quantification. Narrowing of myosin stacks (L), actin bundling (O), and cell border straightening (P) remain similar among all three conditions. However, inhibition of actin nucleators reduced apical actin levels (M, left), but parallel changes in lateral actin mean apical polarization of actin is not altered (N). In L, numbers for each time point are in Table S1. n = individual borders (M–O); control = 48 (M and N), CK666 = 40 (M and N), SMIFH2 = 44 (M and N), control = 63 (O), CK666 = 63 (O), SMIFH2 = 58 (O). In P, representative of three experiments with one to three fields of cells/experiment/time point, with seven to nine borders quantified/field. Statistical analysis performed using one-way ANOVA tests and post hoc Tukey tests (M–O). Error bars represent mean ± SD. **, P < 0.01.
Formins have also been implicated in AJ assembly/maintenance (Carramusa et al., 2007; Kobielak et al., 2004; Nishimura et al., 2016; Rao and Zaidel-Bar, 2016; Sahai and Marshall, 2002). Mammals have many formins, rendering KD approaches challenging. We thus used a formin FH2 domain inhibitor, SMIFH2 (Rizvi et al., 2009), at a concentration used by others (50 µM), in the Ca switch to determine its effect on actomyosin architecture. As with Arp2/3 inhibition, we saw no substantial qualitative (Fig. 3, A–C; vs. Fig. 3, G–I) or quantitative (Fig. 3, L–P) differences in assembling specialized actomyosin structures at the ZA. Formin inhibition did lower levels of both apical and lateral actin (Fig. 3 M), as others saw (e.g., Rao and Zaidel-Bar, 2016), but did not alter its apical polarization at the ZA (Fig. 3 N). We also noted what appeared to be cell toxicity at later time points, in which cells were lost in small regions of the field (Fig. S3, A–F); this may be due to off-target effects. It is important to note that the specificity of this inhibitor has come into question, with evidence that it can also inhibit myosin-2 (Nishimura et al., 2021). However, since SMIFH2 treatment did not alter ZA actomyosin assembly, and myosin inhibition did (see below), we do not think those particular off-target effects alter our conclusions. Thus, with those caveats, there was no requirement for formin activity in assembling specialized ZA actomyosin structures.
Figure S3.
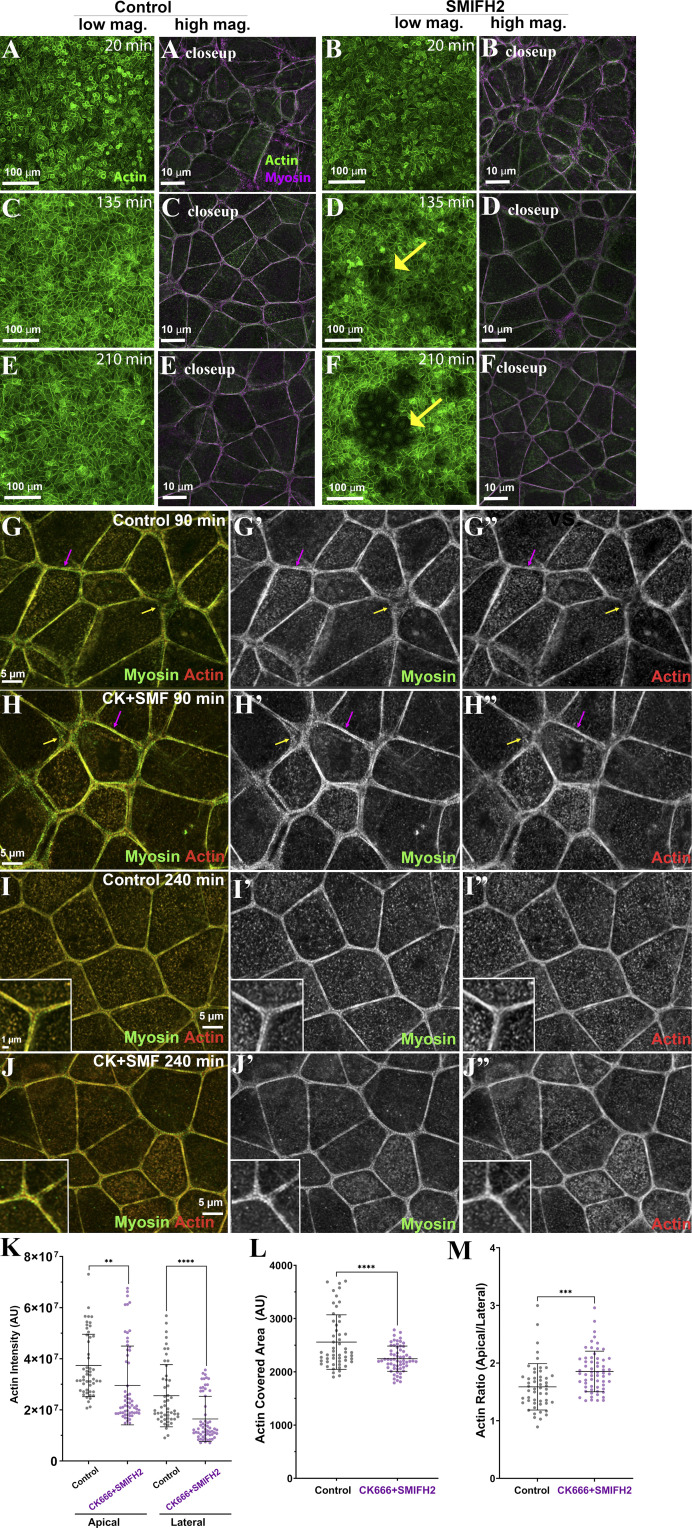
Level of toxicity of SMIFH2 and tests of the effect of combined inhibition of the Arp2/3 complex and formins. (A–F) Images demonstrating the effect of SMIFH2 on a ZO KD MDCK monolayer. When the cells recovered from Ca switched in the presence of SMIFH2, we observed regions in the monolayer with reduced actin signal (yellow arrows) and abnormal cell shape. The longer the cells were incubated with SMIFH2, the more noticeable these “defects” were in a monolayer. mag., magnification. (G–M) Combined treatment with Arp2/3 and formin inhibitors does not prevent or delay the assembly of specialized actomyosin structures at the ZA. (G and H) After 90 min, a subset of bicellular borders have already assembled organized ZA actin and myosin (magenta arrows), while many tricellular junctions remain less organized (yellow arrows). (I and J) Combined inhibition of Arp2/3 and formins does not prevent the tight bundling of actin or assembly of sarcomeric myosin at the ZA (inset = closeup). (K–M) Quantification. Inhibiting both actin nucleators reduced apical and lateral actin levels (K). However, it did not prevent actin bundling (L) or apical polarization of actin (M). In K, M, and L, n = individual borders; control = 51, CK666 + SMIFH2 = 61 (K and M); control = 53, CK666 + SMIFH2 = 62 (L). Statistical analysis was performed using unpaired two-way t tests (K–M). Error bars represent mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01.
One possibility was that these two actin nucleators acted in parallel in junctional assembly. To test this, we treated cells simultaneously with CK666 plus SMIFH2 during recovery from the Ca switch. Intriguingly, this did not prevent ZA actomyosin assembly, nor was there an apparent delay in the process. Cells treated with CK666 plus SMIFH2 reassembled a sarcomeric myosin array with tightly bundled actin (Fig. S3 I vs. Fig. S3 J, insets), with timing that was parallel to the controls (Fig. S3, G–J; representative of three experiments). As in the individual inhibitor treatments, overall junctional and lateral actin intensity was reduced (Fig. S3 K). However, quantification confirmed that neither tight bundling of actin at the ZA (Fig. S3 L) nor apical polarization of actin at the ZA were reduced (Fig. S3 M). We did note some cell toxicity, similar to that seen after SMIFH2 treatment alone. These data suggest our cells can reassemble the ZA without either Arp2/3 or formin activity.
Sequestering Ena/VASP proteins does not block or slow assembly of specialized actomyosin structures at the ZA
Ena/VASP proteins bind to the growing ends of actin filaments to antagonize capping and stimulate monomer addition (Bear and Gertler, 2009). In some cell types, Ena/VASP proteins are implicated in AJ assembly/maintenance (Scott et al., 2006; Yu-Kemp et al., 2017). We first examined VASP localization in our cell line. In confluent, unperturbed cells, VASP was enriched at the ZA, with special enrichment at tricellular junctions (Fig. S2 F, arrows), matching Ena localization in Drosophila tissues (Gates et al., 2007; Rauskolb et al., 2019). A different fixation technique increased junctional signal, and, because AJs separated a bit, revealed that VASP localizes slightly membrane proximal to the actomyosin array (Fig. S2 G, arrows). We then explored how VASP localization changes during junctional reassembly. Right after Ca switch, VASP primarily localized basally to the ends of stress fibers (Fig. S2 H, arrows). Midway through recovery, VASP localized between the expanded myosin stacks, with some enrichment at tri-/multicellular junctions (Fig. S2, I and J). This was consistent with a potential role in actomyosin assembly at the ZA.
The presence of three Ena/VASP family members complicates KD, but a clever approach allows sequestration of all three at mitochondria, thus inactivating them. FP4mito fuses a sequence that inserts in the mitochondrial outer membrane with sequences encoding high-affinity Ena/VASP binding sites, while in the control construct, AP4, the Ena/VASP sites are mutated (Bear et al., 2000). We confirmed that FP4mito sequestered VASP at mitochondria in our cell line, while AP4 did not (Fig. S2 K vs. Fig. S2 L); VASP was no longer detectable at apical junctions, and enhancing the signal more basally suggested that most or all VASP was lost from the cortex (Fig. S2 L).
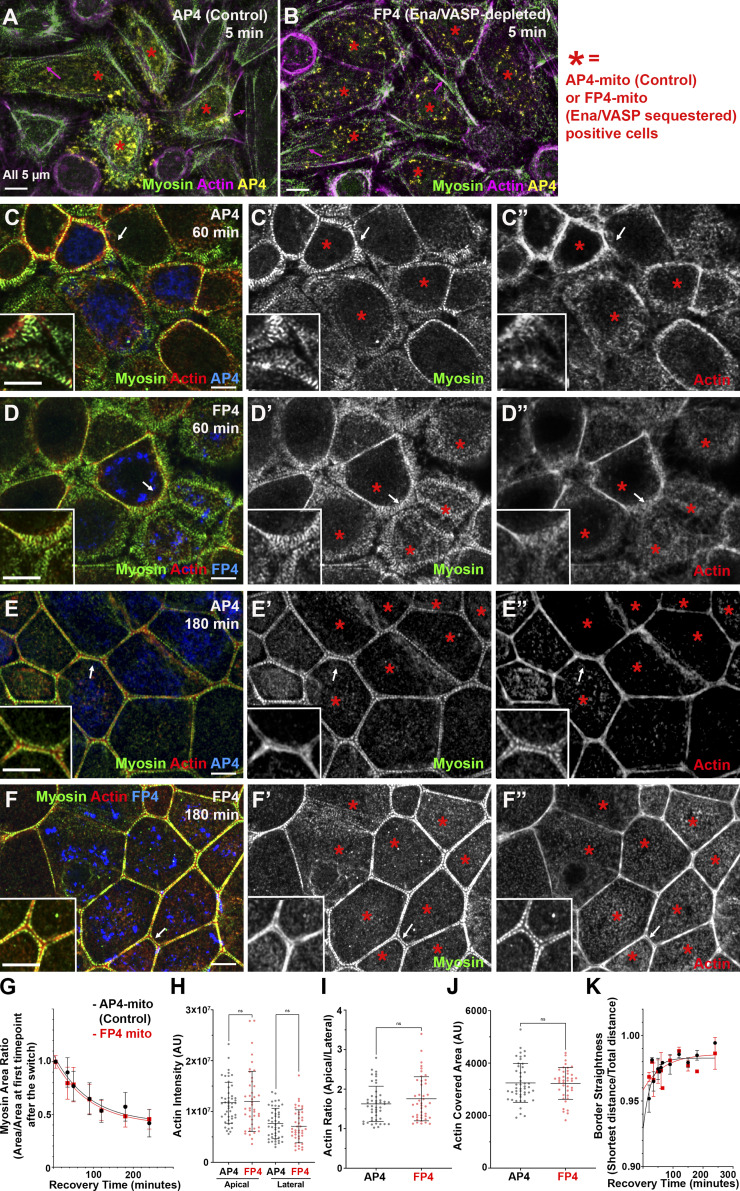
We thus used FP4mito to test whether Ena/VASP proteins play an important role in assembling specialized ZA actomyosin structures, transfecting cells with FP4mito, or AP4mito as a control, and subjecting them to Ca switch (Fig. 4). Transfection efficiency was not 100%, so we focused on regions where several FP4mito- or AP4mito-expressing cells (indicated by asterisks) were adjacent. Surprisingly, sequestering Ena/VASP proteins did not affect ZA actomyosin array assembly, as this proceeded in a similar way in FP4mito and control AP4mito cells. Cells rounded up after Ca withdrawal, with myosin and actin predominantly in basal stress fibers (Fig. 4 A vs. Fig. 4 B, arrows). Midway through recovery, broad myosin stacks were seen in both cell populations (Fig. 4 C vs. Fig. 4 D, arrows, insets), with narrowing occurring first at a subset of bicellular junctions. The myosin stacks were qualitatively similar to those seen in untransfected cells. Finally, expressing FP4mito did not alter the final ZA sarcomeric myosin array decorating tightly bundled actin filaments (Fig. 4 E vs. Fig. 4 F, arrows, insets). Quantitative analysis of myosin stack narrowing revealed no delay after FP4 transfection relative to AP4 (Fig. 4 G). In contrast to inhibiting Arp2/3 or formins, sequestering Ena/VASP did not reduce overall cortical actin levels (Fig. 4 H). Actin apical polarization was unaffected (Fig. 4 I), as was actin bundling at the ZA (Fig. 4 J). Finally, Ena/VASP sequestration did not slow junction straightening (Fig. 4 K). Thus Ena/VASP proteins do not appear to be essential for assembling the highly ordered ZA actomyosin array.
Figure 4.
Ena/VASP proteins are not essential for assembling specialized actomyosin structures at the ZA. (A–F) Representative images of different time points during recovery. Control (AP4) or VASP-sequestered (FP4). *, FP4mito- or AP4mito-expressing cells. (G–K) Quantification. There was no difference between control and VASP-sequestered cells in narrowing of myosin arrays (G), actin intensity (H), apical actin polarization (I), actin bundling (J), or junction straightening (K). White arrows, locations of the zoom-in insets. In G, numbers for each time point are in Table S1. n = individual borders (H–J); AP4 = 47 (H and I), FP4 = 43 (H and I), AP4 = 47 (J), FP4 = 40 (J). mito., mitochondria. In K, representative of nine experiments with two to six borders quantified/experiment/time point. Statistical analysis performed using unpaired two-way t tests (H–J). Error bars represent mean ± SD.
Modeling suggests that actomyosin arrays can help gather actin filaments at a border
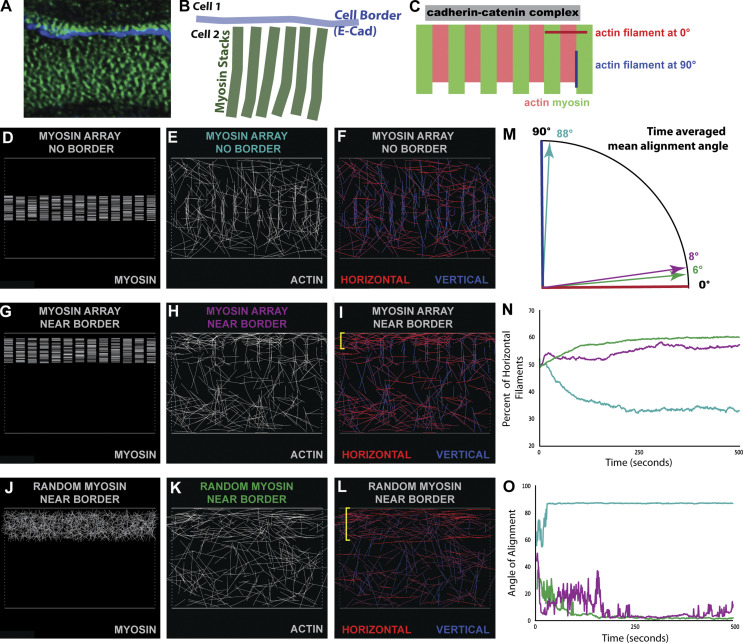
The data above suggest that individual actin polymerization machines are not essential for assembling the highly organized ZA actomyosin structures. An alternate hypothesis was that ZA actomyosin assembly is driven by myosin motor activity, gathering and bundling preexisting actin filaments. To explore whether myosin might have this ability, we used a published molecular model of fully discretized nonmuscle myosin II motor filaments (Cortes et al., 2020; see Materials and methods for details) to examine how myosin arrays altered organization of actin filaments. We made this model 2D, reflecting the actomyosin arrays we saw tightly apposed to the plasma membrane apical to the reforming Ecad-based junctions (Fig. 5, A and B; from data in Fig. 2, C, H, and I). If one looks down on these arrays from above (apical), the array extends toward a boundary, where it encounters the Ecad–catenin complexes (Fig. 5, B and C). We thus first contrasted two scenarios: an actomyosin array in the midst of disordered actin filaments, vs. one next to a “boundary,” which in our cells would be the adjacent Ecad–catenin complexes. We created an array of stacked myosin minifilaments that was not adjacent to a boundary (Fig. 5 D), added a set of randomly oriented actin filaments, and performed 10 simulations. This myosin array effectively moved actin filaments bidirectionally out of its space, with filaments mostly moving vertically along the horizontal motor stacks (perpendicular to the boundary) with a time-averaged mean alignment angle close to 88° (Fig. 5, E, F, and M). This was apparent in quantification of both mean angle over 300 simulated seconds of runtime (Fig. 5 N; averages of 10 simulations) and the percentage of filaments in horizontal orientation, which dropped rapidly to <40% (Fig. 5 O) as the mean angle of alignment rapidly approached 90° (Fig. 5 N). This was the opposite of what is seen in our cell line. However, if the same array was apposed to a boundary (Fig. 5 G), the results were quite different. Once again, the myosin array moved filaments out of its space, but on one side, as they approached the boundary, they forcibly reoriented, becoming aligned parallel to the boundary (horizontal) at angles close to 0° (Fig. 5, H and I). The mean angle over 300 s of runtime for these simulations was ∼8° (Fig. 5 M); for these simulations, the percentage of horizontal filaments increased over time up to ∼60% (Fig. 5 N) as the mean angle of alignment rapidly dropped (Fig. 5 O). The fluctuations seen in the plotted data (Fig. 5 O) reflect the fact that the motor array continually orients filaments vertically as it shuttles them either upward to the boundary or downward into empty space. Finally, we asked if the myosin needed to be in an ordered array, by simulating a zone of randomly oriented myosin filaments adjacent to a boundary (Fig. 5 J). To our surprise, the zone of disordered myosin filaments was equally able to orient filaments parallel to the boundary (Fig. 5, K and L; quantified in Fig. 5, M–O) and did so somewhat more rapidly (Fig. 5). These data suggest that the boundary provided a key element in enabling a myosin array to orient filaments, regardless of the organization of the myosin motor array. One possible advantage of the ordered array (Fig. 5, G–I) over the random array (Fig. 5, J–L) is that actin filaments are bundled in a tighter region (Fig. 5 I vs. Fig. 5 L), which is approximately half the overall width (blue brackets) when myosin filaments are oriented in horizontal stacks.
Figure 5.
Modeling effects of myosin arrays on actin organization. (A and B) Image and diagram of myosin array from above. (C) Schematic of an ordered array with cadherin–catenin complexes as a boundary, illustrating extremes of actin alignment. (D, G, and J) Myosin organization in the three conditions. (E, F, H, I, K, and L) Representative images of actin filaments at the end of run. Filaments in F, I, and L are colorized with regard to alignment. Brackets in I and L indicate regions of maximal alignment. (M) Time-averaged mean alignment angle of actin filaments, averaged over 10 simulations. (N) Percentage of filaments that are horizontal over time. Averaged value for 10 simulations at each simulated time point. In O, mean alignment angle of actin filaments over time.
Proper assembly of tightly bundled actin at the ZA requires ROCK activity but not MLCK
With these modeling data in hand, we tested our second hypothesis: The expansive myosin stacks assembled as junctions reformed helped align actin for bundling. We first examined kinases that phosphorylate and activate myosin: ROCK and MLCK. We used well-characterized, specific inhibitors of each to explore roles of ROCK and MLCK.
To test if MLCK activation of myosin is essential to drive ZA actomyosin assembly, we incubated cells with ML-7, which inhibits MLCK’s catalytic activity (Saitoh et al., 1987), during Ca switch recovery. After ML-7 treatment, myosin still localized to cell borders. There was no noticeable delay in myosin stack narrowing or junctional actin bundling as junctions matured, compared with control (Fig. S4, A–F). Quantification confirmed that ML-7–treated cells had similar rates of myosin stack narrowing (Fig. S4 G) and ZA actin bundling (Fig. S4 I). Borders straightened at the same rate as in controls (Fig. S4 J). ML-7–treated cells had somewhat elevated actin levels laterally, thus reducing apical enrichment (Fig. S4, K and L). We did less extensive experiments with a second MLCK inhibitor, peptide-18 (Lukas et al., 1999), and saw a similar lack of effect (Fig. S4 H; quantified in Fig. S4, G and M). These data suggest that MLCK signaling is not essential for assembling actomyosin structures at the ZA.
Figure S4.
Inhibiting MLCK using ML-7 or peptide-18 does not alter myosin localization during recovery or prevent or delay assembly of the final ZA actomyosin structure. (A–F) Representative images showing cells in control vs. ML-7–treated cells at different time points during Ca recovery. By the last time point, cells assembled bundled F-actin decorated by sarcomeric myosin in both control and drug-treated conditions (E vs. F; H). (G and I–M) Quantification. Myosin maturation (G), actin bundling at the ZA (I and M), and border straightening (J) were similar in control and drug-treated conditions. After ML-7 treatment, there was some elevation of lateral actin and thus apical actin polarization was reduced (L). (N–Q) Representative images showing the difference in apical myosin signal in control (N and P) vs. ROCK-inhibited (O and Q) conditions. Myosin signal at the junction is reduced when ROCK is inhibited. (R) Quantification. Levels of apical actin were reduced after ROCK inhibition. In G, numbers for each time point are in Table S1. In I, K–M, and R, n = individual borders; control and ML7 = 94 (I); control = 80, ML7 = 63 (K and L); control = 45, peptide 18 = 43 (M); and control = 95, Y = 85 (R). In J, representative of five experiments with two fields of cells/experiment/time point, with seven to nine borders quantified/field. Statistical analysis was performed with unpaired two-way t tests (M) or one-way ANOVA tests and post hoc Tukey tests (I, K, L, and R). Error bars represent mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01. Boxes indicate areas magnified at right.
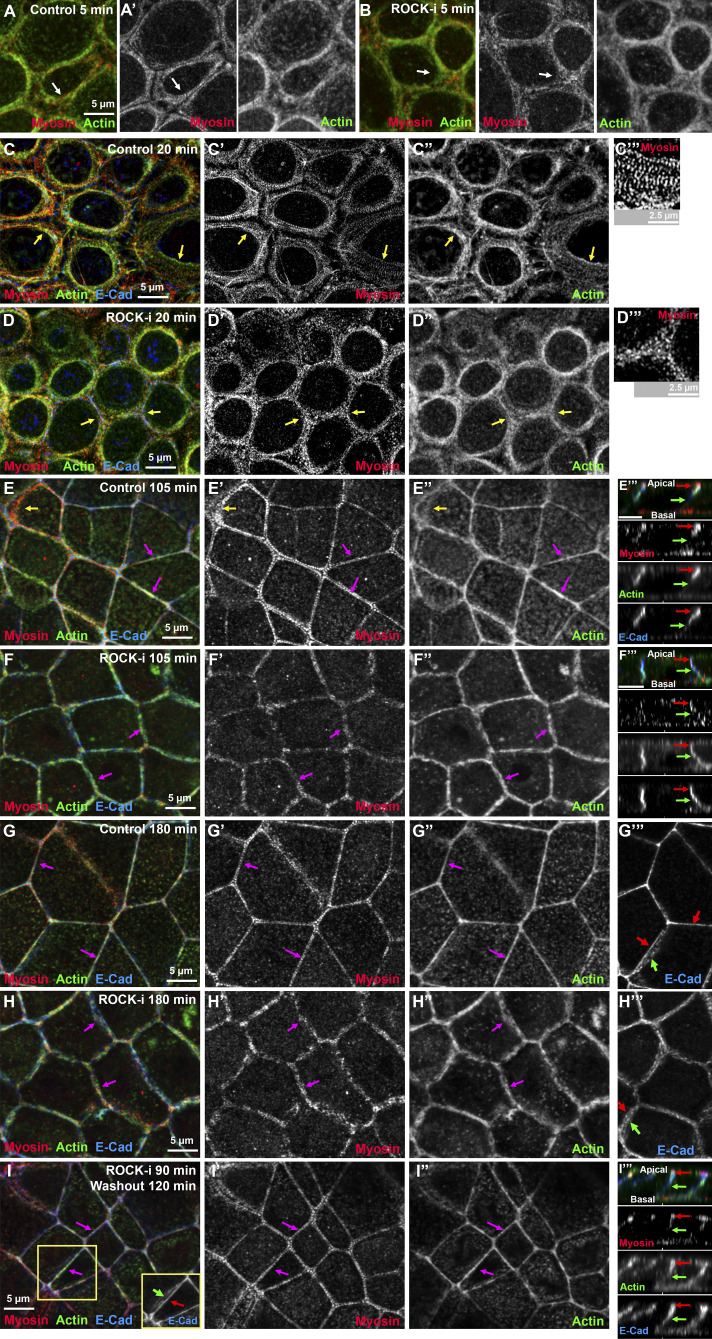
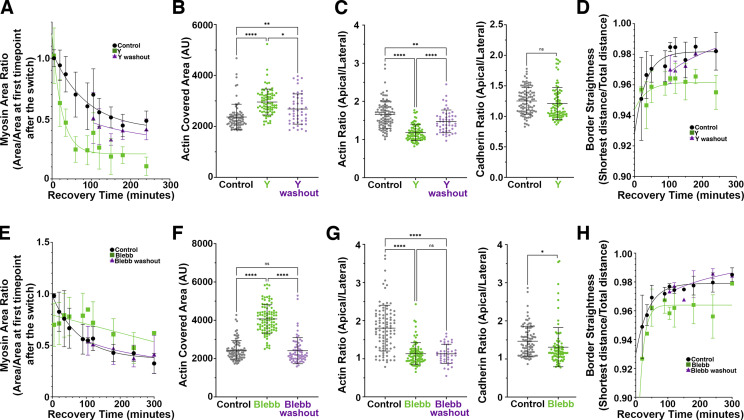
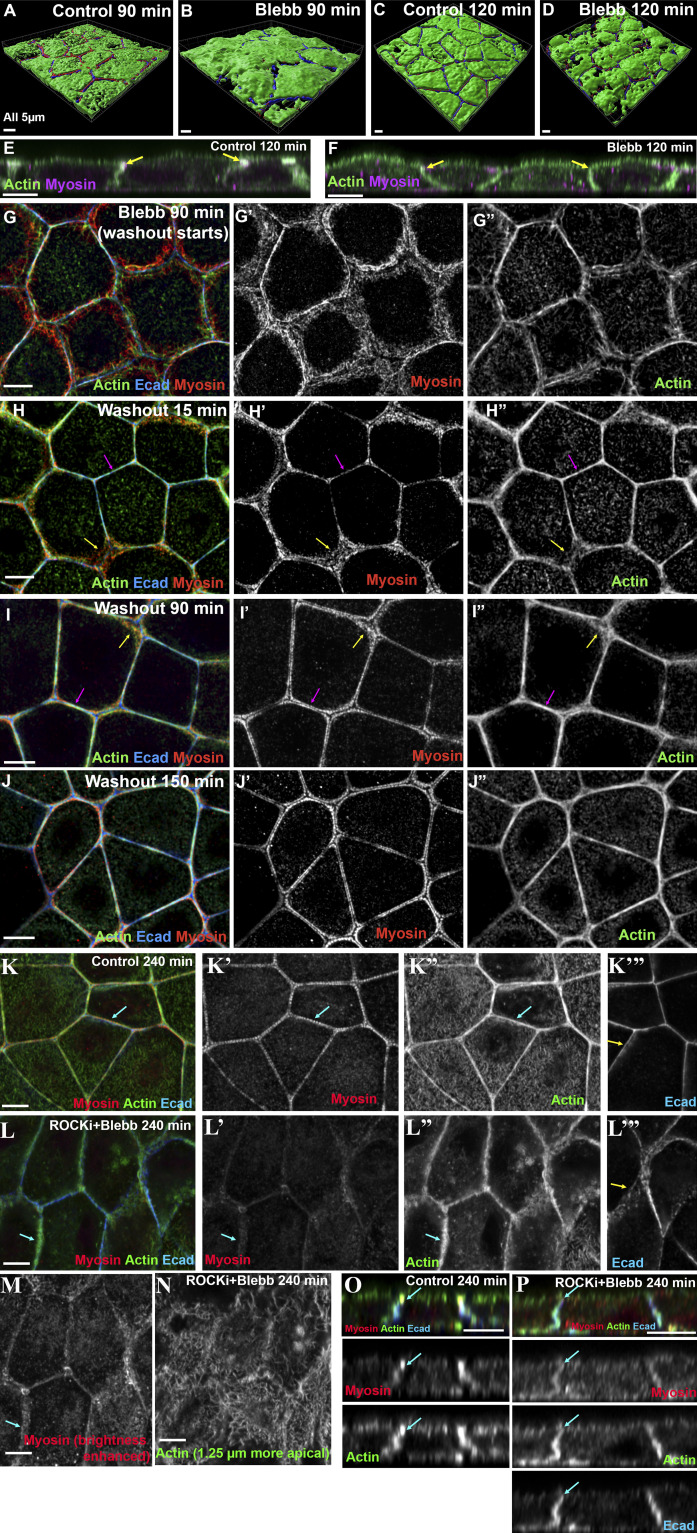
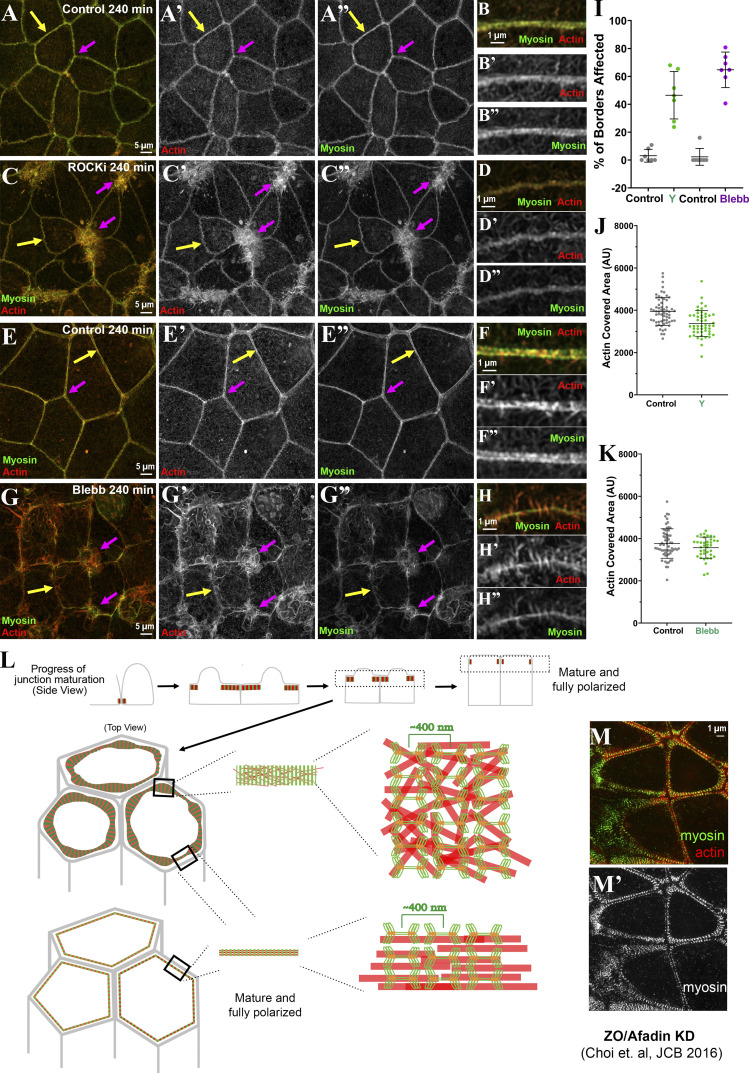
We next tested whether ROCK has a role, using a well-characterized ROCK inhibitor, Y-27632 (Ishizaki et al., 2000), during recovery. The result was quite different. ROCK inhibition reduced overall cortical myosin levels (Fig. S4 N vs. Fig. S4 O; and Fig. S4 P vs. Fig. S4 Q) and disrupted the organized myosin stacks seen in controls—this was seen even at our earliest time points (Fig. 6 A vs. Fig. 6 B, arrows) and was reflected in the reduced area covered by myosin through most of the time course (Fig. 7 A). In controls, extensive myosin stacks appeared at tricellular and multicellular junctions by 20 min of recovery (Fig. 6 C, arrows; Fig. 6 C′′′, magnification of the sarcomere-like array). In contrast, after ROCK inhibition, while some myosin accumulated overlapping cortical actin near junctions (Fig. 6 D, arrows), organized myosin stacks were absent. In controls, myosin stacks at the ZA narrowed by 105 min, with a tight sarcomeric array and bundled actin at many bicellular borders (Fig. 6 E, magenta arrows), although a subset of tricellular junctions retained more extensive myosin stacks and less organized actin (Fig. 6 E, yellow arrows). By 180 min, controls returned to the highly organized bundled actin and sarcomeric myosin seen before perturbation (Fig. 6 G). In contrast, while actin accumulated at AJs in ROCK inhibitor–treated cells, it did not form the tight bundled arrays seen in controls (Fig. 6 E vs. Fig. 6 F; Fig. 6 G vs. Fig. 6 H, magenta arrows; quantified in Fig. 7 B), even at the latest time points (Fig. 6 H). Borders did not become straight, even at time points where recovery was complete in controls (Fig. 6 G vs. Fig. 6 H, arrows; quantified in Fig. 7 D). ROCK inhibition also reduced apical polarization of actin (Fig. 6 E′′′ vs. Fig. 6 F′′′, apical [red arrows] vs. lateral [green arrows]; Fig. 6 G′′′ vs. Fig. 6 H′′′; actin quantified in Fig. 7 C), and reduced overall levels of apical actin (Fig. S4 R). Interestingly, effects of ROCK inhibition were reversible, with tight sarcomeric myosin and bundled actin reforming after inhibitor washout (Fig. 6 I, magenta arrows; quantified in Fig. 7, A and B) and apical polarization reestablished (Fig. 6 I, inset, Fig. 6 I′′′, and Fig. 7 C). Thus myosin activation by ROCK is essential for assembly of sarcomeric myosin arrays and tightly bundled actin at the ZA.
Figure 6.
Inhibiting ROCK reduces myosin recruitment and organization at cell borders, leaving less bundled F-actin and less actin polarization at the ZA. (A–I) Representative images, different time points during recovery. Control vs. ROCK-inhibited (ROCK-i). ROCK inhibition reduces myosin at cell borders (Fig. S4, N–Q), but when its signal is intensified, it becomes clear that myosin stacks are disrupted (C′ vs. D′, arrows; C″′ vs. D″′), and at later time points, sarcomeric organization at the ZA is lost (E′ vs. F′; G′ vs. H′). Actin is less bundled than control (E″ vs. F″; G″ vs. H″). Apical polarization of Ecad, myosin, and actin to the ZA is reduced after ROCK-i (E″′ vs. F″′; G″′ vs. H″′, red vs. green arrows). (I–I″′) After inhibitor washout, ZA actomyosin architecture is restored.
Figure 7.
Myosin activation and motor activity are important for actin bundling and polarization at the ZA. Quantification. (A–D) ROCK inhibition. (E–H) Myosin ATPase inhibition. Inhibiting ROCK reduces junctional myosin (A), while blebbistatin reduces narrowing of myosin stacks (E). Both inhibitors reduce actin bundling at the ZA (B and F), apical actin polarization (C and G, left), and border straightening (D and H). In A and E, numbers for each time point are in Table S1. n = individual borders; control = 80, Y = 80, Y washout = 48 (B); control = 95, Y = 85, Y washout = 49 (C, right); control = 95, Y = 85 (C left); control = 93, blebb = 97, blebb washout = 69 (F); control = 95, blebb = 89, blebb washout =42 (G, right); and control = 95, blebb = 89 (G, left). In D, representative of seven (control, Y) or three (washout) experiments with two fields of cells/experiment/time point, with seven to nine borders quantified/field. In H, representative of five (control, blebb) or two (washout) experiments with two fields of cells/experiment/time point, with seven to nine borders quantified/field. Statistical analysis was performed using one-way ANOVA tests and post hoc Tukey tests (B, C, F, and G). Error bars represent mean ± SD. ****, P < 0.0001; **, P < 0.01; *, P < 0.05.
Proper assembly of tightly bundled actin at the ZA requires myosin motor activity
We next explored the role of myosin motor activity, which can play an important role in actin remodeling. We used the inhibitor blebbistatin, which inhibits myosin motor activity without abolishing its ability to bind actin (Kovács et al., 2004). We repeated the Ca switch in the presence of this inhibitor. The results were interesting and surprising.
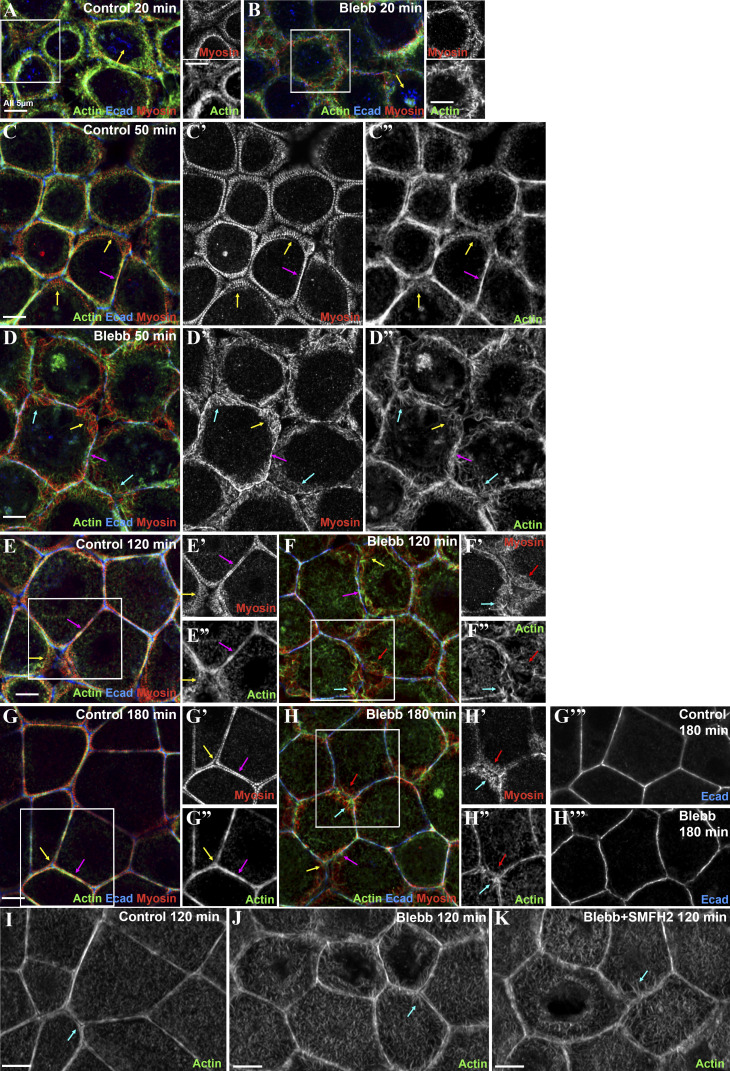
As expected, myosin was disrupted even at the earliest time points (Fig. 8 A vs. Fig. 8 B). Assembly of the extended myosin stacks seen in the controls (Fig. 8 C′, yellow arrows) was prevented by blebbistatin. Instead, myosin accumulated in a punctate pattern overlapping cortical actin (Fig, 7, D′ and D′′, yellow arrows), with some enrichment near Ecad-based AJs (Fig. 8, D and D′, magenta arrows). Cortical myosin remained disorganized even after extended recovery in the presence of blebbistatin (Fig. 8, F′ and H′, red arrows), by which point control cells had resumed tight sarcomeric myosin arrays and bundled actin filaments and both bicellular and tricellular junctions (Fig. 8, G–G′′, magenta and yellow arrows). Not surprisingly, blebbistatin treatment reduced the narrowing of myosin stacks seen in controls (Fig. 8 H; quantified in Fig. 7 E). However, cells retained the ability to regain a more columnar architecture as lateral borders zipped up, although apical flattening was reduced (Fig. S5, A–F).
Figure 8.
Blebbistatin (myosin ATPase inhibitor) treatment disrupted assembly of myosin stacks and altered F-actin structures at the ZA. (A–H) Representative images of different time points during recovery. Control vs. blebbistatin (blebb). At middle time points, assembly of expansive stacks of myosin is lost (C′ vs. D′, arrows), and later in recovery, the tight sarcomeric ZA array of myosin is lost (E′ vs. F′; G′ vs. H′). Actin bundling at the ZA is disrupted, and instead actin has a spiky appearance (D″, F″, and H″) Also see quantifications in Fig. 7, E–H. (I–K) Formin inhibition does not prevent stimulation by blebbistatin of spiky actin at the ZA (arrows). Boxes indicated areas magnified at right.
Figure S5.
3D cell shapes after blebbistatin treatment, blebbistatin washout restores the ZA, and dual inhibition of both ROCK and the myosin ATPase. (A–D) 3D images of cell surface in control vs. blebbistatin-treated cells. The apical surface of the control cells becomes flatter as junctions mature (A vs. C; also in Fig. S1, M–Q). In contrast, blebbistatin-treated cells were not able to flatten the apical surface, even at the later recovery time points (B and D). (E and F) Cross-section views. After blebbistatin treatment, cells still retained the ability to regain a more columnar architecture as lateral borders zipped up, but actin and myosin were not apically polarized (E vs. F, arrows). (G–J) Representative images showing the recovery of ZA actomyosin structures after blebbistatin washout. Within 15 min after washout, actin and myosin already began to focus at bicellular borders (H′ and H″, magenta arrows). Myosin at the tricellular borders was slower to recover (H and I, yellow arrows), but by 150 min (J), ZA actomyosin structures returned to those seen in the control mature monolayer. (K–P) Combined inhibition of ROCK and blebbistatin does not prevent Ecad-based adhesion but does disrupt ZA assembly and Ecad polarization. (K and L) Combined treatment reduces junctional myosin, actin bundling at the ZA, and Ecad apical polarization (K″′ vs. L″′). (M) Dual treatment with myosin signal enhanced. (N) After dual treatment, spiky actin covers the apical surface. (O and P) Cross sections. Ecad-based junctions zip up, but apical enrichment of actin and Ecad is reduced (arrows).
After blebbistatin treatment, Ecad, which was endocytosed after Ca removal (Fig. 8, A and B, arrows), still returned to cell borders, where it associated with a junctional population of actin. However, junctional actin architecture was dramatically altered by blebbistatin treatment. This began early in recovery (Fig. 8 B) and continued to the end of the process (Fig. 8 H). In controls at the recovery midpoint, extended myosin stacks (Fig. 8 C′, yellow arrows) overlaid a disordered meshwork of actin filaments (Fig. 8 C′′, yellow arrows), and as myosin stacks narrowed, beginning at bicellular borders (Fig. 8 E′, magenta arrow), actin filaments became increasingly concentrated at nascent AJs, leading to assembly of bundled arrays (Fig. 8, E′′ and G′′, arrows). In contrast, in blebbistatin-treated cells, spiky actin filaments emanated from cell junctions. These spiky filaments were already evident within 5 min of recovery (Fig. 8 B, inset), became prominent at middle stages (Fig. 8 D′′, cyan arrows), and remained present even late in recovery (Fig. 8 H′′, cyan arrow). Quantification of junctional actin confirmed the failure to tightly bundle actin and its replacement by a disordered apical actin array (Fig. 7 F). Blebbistatin also prevented actin polarization to the ZA (Fig. 7 G), and cell border straightening was substantially reduced (Fig. 8 H; quantified in Fig. 7 H), suggesting that inability to assemble ZA sarcomeric actomyosin may reduce border contractility.
Strikingly, all these effects were rapidly reversed after blebbistatin washout (which began at 90 min of recovery). Within 15 min, some bicellular borders straightened, and the highly spiky actin at junctions was reduced (Fig. S5 G vs. Fig. S5 H). Within 90 min, most borders had assembled sarcomeric myosin and tightly bundled actin (Fig. S5 I), and by 150 min, cells resembled controls (Fig. S5 J), with bundled actin and myosin (quantified in Fig. 7, E and F) at the ZA and straightened cell borders (quantified in Fig. 7 H). Consistent with a role for myosin in actin bundling, as recovery proceeded after washout, there was a correlation between borders where myosin stacks narrowed and those where actin was bundled (Fig. S5, H and I, magenta arrows), while borders where myosin remained disorganized retained disorganized cortical actin (Fig. S5, H and I, yellow arrows). We completed this analysis by exploring the mechanism involved in generating the spiky actin structures seen after blebbistatin treatment. These long linear structures resemble those generated by elevating formin activity, but inhibiting formins using SMIFH2 did not block their formation (Fig. 8 J vs. Fig. 8 K). Together, these data support the hypothesis that myosin motor activity is critical for assembling the ZA supramolecular contractile actomyosin array and reveal that inhibiting myosin motor activity triggers striking changes in actin architecture at nascent junctions. Recent work revealed a similar effect of myosin inhibition on brush border microvilli; the authors suggested that myosin contractility stimulates actin network turnover (Chinowsky et al., 2020).
Finally, we explored the effects of inhibiting both myosin activation and motor activity, treating cells with both the ROCK inhibitor and blebbistatin. The effect on myosin assembly was similar to that seen after ROCK inhibition alone, with strongly reduced cortical myosin and total loss of the myosin arrays (Fig. S5 K′ vs. Fig. S5, L′ and M, arrows). At the level of the apical junctions, the effect on actin organization also resembled that of ROCK inhibition alone—actin accumulated in broad disorganized cortical arrays (Fig. S5 L′′, arrow) rather than the tightly bundled actin seen in controls (Fig. S5 K′′, arrow). The spiky actin protrusions seen at the level of apical junctions after blebbistatin treatment were absent (Fig. S5 L′′). However, when we focused on the apical surface of the cells, spiky protrusions were seen there (Fig. S5 N), similar to those seen at the level of the AJs in cells treated with blebbistatin alone (e.g., Fig. 8 J). Perhaps most interesting, combining ROCK inhibition and blebbistatin did not prevent cells from reestablishing Ecad-based junctions and zipping up, but apical enrichment of Ecad (Fig. S5 K′′′ vs. Fig. S5 L′′′, arrows) and actin (Fig. S5 O vs. Fig. S5 P, arrows) at the ZA were strongly reduced or abolished. Thus, in this cell line, myosin activation and motor activity are not essential for reestablishment of cell adhesion.
Caco-2 cells also assemble myosin arrays during recovery
Thus, in ZO-KD MDCK cells, large-scale myosin arrays assemble during junctional reassembly, and recovery of the tightly bundled actin at the ZA is independent of Arp2/3, formins, and Ena/VASP proteins but dependent on myosin activity. However, the ZA in ZO-KD MDCK cells is exceptionally well-organized, likely due to the elevated ROCK activity triggered by ZO-KD (Choi et al., 2016). Cultured cells vary widely in the organization of actin and myosin at the ZA and in border contractility. We thus looked for a “nonmutant” cell line with a reasonably robust ZA actomyosin cytoskeleton. The human intestinal cell line Caco-2 met these criteria (e.g., Cavanaugh et al., 2020; Kovacs et al., 2011). At confluence, cell borders are reasonably straight, Ecad is enriched apically and a sarcomeric array of myosin can be seen at the apical ZA (Fig. 9 A). Actin is strongly concentrated at the ZA (Fig. 9 A; Kovacs et al., 2011), although it does not appear as tightly bundled as in ZO-KD MDCK cells. Because the cells are flatter than the ZO-KD MDCK cells, the apical microvilli are often seen in the apical slice including the ZA.
Figure 9.
Caco-2 cells also assemble extensive myosin stacks during ZA assembly. (A–F) Images at different time points of Ca recovery reveal that an extensive myosin array forms during midrecovery and localizes with a less well-organized actin network. (A) Before switch the ZA has bundled actin and sarcomeric myosin. (B) Myosin arrays overlaying disorganized actin early in recovery (yellow arrows). A subset of bicellular borders tighten up first (magenta arrows). (C and D) Tricellular and short multicellular junctions tighten up last (C, yellow arrows; D). (E and F) At the end of recovery, ZA assembly is complete.
We then performed a Ca switch, as we had done with ZO-KD MDCK cells. Ecad was initially internalized into a vesicular compartment (Fig. S6 A) and then began to return to cell junctions (Fig. S6 B). Strikingly, as cells begin to recover, we saw extensive myosin arrays at both bicellular (Fig. 9 B, yellow arrows) and tricellular junctions (Fig. 9 C, yellow arrows; Fig. 9 D), similar to but not quite as organized as those in the ZO-KD MDCK cells. These often appeared in regions where the underlying actin was not well organized (Fig. 9, B and C, yellow arrows; Fig. 8 D). As we saw in the ZO-KD MDCK cells, tightening occurred first at a subset of bicellular junctions (Fig. 9, B and C, magenta arrows) and was slowest at tricellular and short multicellular junctions. However, within 100–240 min, the cells had restored their original architecture, with polarized Ecad (Fig. S6 C), and sarcomeric myosin and actin strongly enriched at the ZA (Fig. 9, E and F).
Figure S6.
Inhibiting the Arp2/3 complex or formins does not prevent or delay ZA assembly in Caco-2 cells, and use of binarization of images to assess the effects of Rock inhibition or blebbistatin on cortical actin. (A–C) Ecad relocalization to the ZA in Caco-2 cells during recovery from Ca switch. (A) Ecad was internalized right after the Ca switch (cyan arrows). (B and C) As the recovery proceeded, Ecad regained its continuous distribution at the ZA, as the actomyosin cytoskeleton tightened. (D–M) Inhibiting the Arp2/3 complex or formins does not prevent reassembly of specialized actomyosin structures at the ZA in Caco-2 cells. (D–K) Recovery from Ca switch in control, CK666 (Arp2/3 inhibitor)-treated, or SMIFH2 (formin inhibitor)-treated cells. Controls were performed separately for each treatment. (D, F, H, and J) Midrecovery, with myosin arrays remaining at tricellular and short multicellular junctions. (E, G, I, and K) Recovery was completed in a similar time frame, and actin and myosin organization at the final ZA was unaltered by the treatments. (L and M) Quantification. Actin bundling remains similar among all three conditions. (N–P) Blebbistatin treatment reduced actin assembly at bicellular borders in Caco-2 cells. Apical MIPs of actin were equally thresholded using Fiji’s default algorithm and then binarized, to show actin remaining after equivalent selection for areas of contiguous particles. ROCK-i treatment reduced actin assembly at some bicellular borders, but many contiguous bicellular borders remained after selection. Blebbistatin treatment reduced actin more dramatically, leaving few contiguous bicellular borders after selection. Large areas of disorganized actin at disrupted tricellular borders were seen in both ROCK-i– and blebbistatin-treated cells. Three experiments are shown. In L and M, n = individual borders; control = 51, CK666 = 53 (L); control = 51, SMIFH2 = 52 (M). Error bars represent mean ± SD.
Caco-2 cell ZA reassembly is not prevented by actin nucleation inhibitors but is altered by myosin inhibition
In our final set of experiments, we explored whether Caco-2 cells shared similar insensitivity to inhibition of actin nucleation but sensitivity to myosin inhibition. We first tested sensitivity to Arp-2/3 or formin inhibition, using the inhibitors CK666 and SMIFH2. Neither substantially altered the time course of ZA reassembly (Fig. S6, D and E vs. Fig. S6, F and G; Fig. S6, H and I vs. Fig. S6, J and K; images are representative of three experiments). By the point when control cells had fully assembled the ZA, the organization of actin and myosin at AJs was similar in control and inhibitor-treated cells (Fig. S6, E′–E′′ vs. Fig. S6, G′–G′′′′; Fig. S6, E vs. G insets; Fig. S6, I′ and I′′ vs. Fig. S6, K′ and K′′; and Fig. S6 I vs. Fig. S6 K insets). We quantified this as we had above, measuring tightening of borders by the 240-min time point. Neither treatment reduced border tightening (Fig. S6, L and M); in fact, CK666-treated borders appeared slightly tighter (Fig. S6 L), perhaps due to a reduction in overall F-actin assembly.
We next explored sensitivity of Caco-2 cells to myosin inhibition using the ROCK inhibitor Y-27632 or the myosin ATPase inhibitor blebbistatin. Both had striking effects on ZA reassembly (Fig. 10, A–K), but these were distinct from the effects in ZO-KD MDCK cells. Treatment with the ROCK inhibitor prevented ZA reassembly at many tricellular and short bicellular junctions. At these locations, actin remained highly disorganized and protrusive (Fig. 10 A vs. Fig. 10 C, magenta arrows; and Fig. S6 P). Bicellular borders were less affected (Fig. 10 A vs. Fig. 10 C, yellow arrow; and Fig. 10 B vs. Fig. 10 D). We first quantified the fraction of bicellular borders ending in one or two affected tricellular junctions. 46% of bicellular borders were affected by ROCK inhibition, while in the control only 2–3% were altered. To assess the effect in bicellular border actin assembly, we examined tightening of actin as we had done above; ROCK inhibition did not prevent border tightening (Fig. 10 C, yellow arrows; quantified in Fig. 10 J). Blebbistatin had a qualitatively similar but even more dramatic/penetrant effect at tricellular junctions (Fig. 10 E vs. Fig. 10 G, magenta arrows), with ∼64% of borders affected (Fig. 10 I). In addition, spiky actin protrusions were present at bicellular borders (Fig. 10 E vs. Fig. 10 G, yellow arrow; Fig. 10 F vs. Fig. 10 H), as we had seen in ZO-KD MDCK cells. While bicellular border tightening was not dramatically altered (Fig. 10 K), when we binarized images equivalently, in blebbistatin-treated cells, actin at many bicellular borders was revealed to be weaker (Fig. S6 N vs. Fig. S6 P); this was less apparent after ROCK inhibition (Fig. S6 O). Thus, in this second cell line, myosin activity is also critical for correctly assembling the ZA supramolecular contractile actomyosin array.
Figure 10.
Inhibiting myosin activation or motor activity disrupts Caco-2 ZA reassembly. (A–D) ROCK inhibition leads to failure to reassemble the ZA at many tricellular and short multicellular junctions (A vs. C, magenta arrows), while bicellular borders are less affected (A vs. C, yellow arrows; B vs. D). (E–H) Blebbistatin disrupts ZA reassembly at most tricellular and short multicellular junctions (E vs. G, magenta arrows), while actin at bicellular borders is more spiky (E vs. G, yellow arrows; F vs. H). (I–K) Quantification, affected bicellular borders (I), and tightening of actin at bicellular borders (J and K). (L) Summary diagram illustrating the model. (M) Broad actin stacks assemble when Afadin is knocked down in ZO-KD MDCK cells. In I, n = 7 fields of cells assessed; in J and K, n = individual borders; control = 62, Y = 50 (J); control = 62, blebb = 38 (K). Error bars represent mean ± SD.
Discussion
The assembly of polarized supramolecular actomyosin structures at the ZA underlies the remarkable ability of cells to change shape and move during embryonic development and tissue homeostasis. Despite decades of work, key questions remain about the respective contributions of actin and myosin regulators in this process. We used ZO-KD MDCK cells, which assemble a textbook ZA, to define underlying mechanisms. Our data reveal that myosin activity plays a key role in driving actin organization at cell–cell contacts, via self-organization of extensive micron-scale stacks of myosin filaments underlying the plasma membrane apical to forming cell junctions, which then go through a process of compaction, driving bundling of actin filaments as cell–cell AJs mature (Fig. 10 L).
No single actin polymerization machine is essential for assembling supramolecular actomyosin structures at the ZA
Many previous studies explored roles of actin nucleation and continued polymerization in building and maintaining actin structure at AJs. We initially hypothesized that one or more of the machines driving actin polymerization would be essential. Consistent with earlier work (see Introduction), inhibiting Arp2/3 or formins reduced actin levels at cell junctions. However, our data suggest that, at least in the two cell types we examined, individual activities of Arp2/3, formins, or Ena/VASP proteins are not essential for assembling supramolecular actomyosin structures at the ZA, with sarcomeric myosin arrays and tightly bundled actin. Previous work is consistent with the idea that this is true in a broader array of cell types, as most previous perturbations reduced junctional actin by 40–60% but did not eliminate it. We also considered the possibility that the two actin nucleators acted semiredundantly, but ZA assembly was not prevented by simultaneous Arp2/3 and formin inhibition. However, we did not test combinations of these with inhibiting Ena/VASP proteins. One possibility is that multiple actin polymerization machines act in concert with one another, and simultaneous perturbation of most or all would be needed to eliminate junctional actin assembly. This hypothesis will be important to test. It will also be important to test roles of actin-binding proteins, which may work with myosin to organize and bundle actin filaments.
Myosin activation and motor activity play key roles in ZA assembly
Our data suggest that myosin plays a key role in assembling ordered actin to create tight actin bundles at the ZA. This required both myosin activation and myosin motor activity. Myosin recruitment to cell borders was regulated by ROCK, which activates and allows myosin to interact with its binding partners. Myosin motor activity was not required for it to localize to AJs, but sarcomeric organization required myosin motor function. After blebbistatin treatment, myosin’s ability to cross-link actin filaments may remain intact, suggesting this activity is not sufficient for myosin to drive junctional actin bundling. Myosin’s ability to integrate existing actin filaments into larger actomyosin bundles has been described in studies of stress fibers in lamella (e.g., Anderson et al., 2008; Machesky and Hall, 1997; Nemethova et al., 2008), where actin polymerization is slow compared with lamellipodia (Glacy, 1983; Wang, 1984; Zicha et al., 2003). In the Caco-2 cells, the effects of myosin inhibition were even more dramatic, especially at tricellular junctions, the site where work from many laboratories suggests tension exerted on Ecad–catenin complexes may be maximal.
It’s important to note that inhibiting neither myosin activation nor motor activity prevented reassembly of Ecad-based AJs, as cells zipped up along their lateral borders. Ecad was associated with junctional actin; however, this actin was differently organized, and apical polarization of Ecad and actin was reduced. Similar alterations were seen after blebbistatin treatment of keratinocytes (Zhang et al., 2005). This suggests that multiple mechanisms drive actin assembly at AJs, consistent with recent analysis suggesting that actin at the ZA includes both a central branched actin network and adjacent zones of linear bundled actin (Efimova and Svitkina, 2018). The actin associated with interdigitated Ecad complexes seen at midpoints of junctional reassembly may be generated by one of these mechanisms; perhaps it is Ena/VASP driven, as was suggested in keratinocytes. We also were intrigued by the “spiky” nature of junctional actin in blebbistatin-treated cells; this was even more dramatic in myosin inhibitor–treated Caco-2 cells. Perhaps myosin motor activity, or the actin bundling it promotes, inhibits other actin assembly pathways at AJs.
A role for self-organized micron-scale stacks of myosin filaments
Perhaps our biggest surprise was to see self-assembly of micron-scale stacks of myosin filaments with a sarcomeric organization quite early during junctional reassembly (Fig. 10 L). These arrays were similar to but more extensive than myosin filament stacks in migrating fibroblasts (Beach et al., 2017; Burnette et al., 2014; Fenix et al., 2016; Hu et al., 2017; Verkhovsky et al., 1995). It was especially intriguing that myosin organized into this large-scale sarcomere-like pattern at locations where actin remained much less well organized, suggesting that myosin supramolecular organization does not require preorganized actin. This echoes seminal observations by the Borisy group, who noted that in fibroblasts, “[s]ome myosin spots and ribbons were found in the zone of diffuse actin distribution, suggesting the formation of myosin spots may precede the organization of actin filament bundles” (Verkhovsky et al., 1995).
The mechanisms mediating myosin stack assembly and lateral interactions remain unclear. Actin- and myosin-binding partners might play roles in myosin supramolecular organization. Verkhovsky et al. (1995) suggested a role for α-actinin, as it is observed in alternating A- and I-bands of muscle sarcomeres. Consistent with this, α-actinin-4 KD disrupted myosin stack assembly on stress fibers in REF52 cells. KD also suggested roles for Cofilin 1 and the formin Fmnl3 (Hu et al., 2017). Given our localization of α-actinin to the myosin arrays in our cells, it would be interesting in the future to knock it down in our cell type. Another possible myosin-binding partner that might regulate assembly is myosin-18, which can coassemble with other myosin isoforms to regulate their biophysical properties and localization. It was proposed that myosin-18 can anchor mixed filaments to the plasma membrane via binding to PDZ ligand–containing proteins (Billington et al., 2015). Heterotypic filaments of myosin-18 and myosin-2 exist both in vitro and in stress fibers of several cell types (Billington et al., 2015; Jiu et al., 2019). Myosin-18B promoted formation of myosin-2 stacks and higher-order structure in contractile stress fibers (Jiu et al., 2019). Perhaps myosin-18 isoforms are present in our cells, helping anchor myosin stacks to the membrane and assisting in stack formation. Myosin-18 KD would provide an interesting future test of this possibility.
Mammals have three myosin-2 isoforms: Myo2A, Myo2B, and Myo2C. Intriguingly, Myo2B localizes to the central branched actin region, while Myo2A localizes along the adjacent linear actin bundles (Heuzé et al., 2019). Myo2A and Myo2B KD also suggests differential roles: e.g., Myo2A and Myo2B have distinct localizations and functions at Ecad-mediated AJs in parental MDCK cells, a cell line that does not form sarcomere-like actomyosin bundles at AJs (Smutny et al., 2010; Ozawa, 2018; Heuzé et al., 2019). Myo2A plays a more central role in vivo: myo2A knockout mice have defects in cell adhesion by embryonic day 6.5 and die by embryonic day 7.5 (Conti et al., 2004), while myo2B knockout mice die perinatally with heart and brain defects (Tullio et al., 1997). Different isoforms also have distinct dynamic properties (Vicente-Manzanares et al., 2009). Myo2A has the highest rate of ATP hydrolysis and thus can move actin more rapidly, while Myo2B has the highest duty ratio, allowing it to exert force on actin longer (Melli et al., 2018; Sellers and Heissler, 2019). Despite their different biophysical properties, the three isoforms have both unique and redundant localizations and functions (Bao et al., 2005; Conti et al., 2004; Sandquist and Means, 2008; Beach et al., 2014) and can coassemble into heterotypic filaments (Beach et al., 2014). It will be interesting to explore roles of different myosin isoforms throughout junction maturation, during formation of both micron-scale stacks of myosin filaments and the final robust sarcomeric actomyosin structure. We focused on localizing Myo2B, but also detected Myo2A in both large myosin arrays and in the final ZA. It will be interesting to determine whether heterotypic myosin filaments form in our cell type, and whether the composition and ratios of heterotypic filaments change over time as actin filaments become bundled, supporting the sorting mechanism proposed by Beach et al. (2014).
One final issue of interest is the role of Ecad and associated linker proteins in directing ZA assembly. In previous work, we explored the role of the junction-actin cross-linker Afadin in ZO-KD MDCK cells. Afadin KD had multiple effects in this cell type (Choi et al., 2016). Cell border contractility homeostasis was disrupted, with some borders hyperconstricted and others hyperextended. Most intriguing with regard to our results here was that effects were most striking at tricellular and short multicellular junctions, the same places where ZA reassembly was slowest. At these “weak points,” we saw expansion of the “sarcomere-like” array seen in ZO-KD MDCK cells into more extensive myosin stacks (Fig. 10 M). Thus Afadin KD cells resembled those seen here during mid-assembly. These data suggest that Afadin helps maintain the tight bundling of actin at the ZA, and in its absence, extensive myosin arrays assemble at tricellular junctions. It will be interesting to explore mechanisms by which Afadin acts to regulate actomyosin assembly and homeostasis.
Materials and methods
Cell lines, reagents, and antibodies
MDCK ZO-1/ZO-2-KD (ZO KD, clone 3B3; Fanning et al., 2012) and Ecad-GFP CRISPR Caco-2 (Cavanaugh et al., 2020) cells were cultured in complete medium (high-glucose DMEM [Corning] supplemented with 10% FBS and 20 mM Hepes [Corning]) and maintained at 37°C with 5% CO2. For imaging, cells were plated on Matrigel (356231; Corning)-coated coverslips. For transfection of GFP-mito-AP4/FP4 constructs (a gift from Dr. Stephanie Gupton, University of North Carolina at Chapel Hill; Bear et al., 2000), the PolyJet Transfection Reagent (SignaGen Laboratories) was used per manufacturer’s instructions. Calcium switch buffer (CSB) was prepared by adding 2 mM magnesium dichloride to magnesium-calcium–free PBS (Corning). Antibodies and concentrations used for immunostaining are as follows: rabbit anti-nonmuscle myosin IIB-targeted at myosin tail (909902; BioLegend, 1:300), rat anti-Ecad (sc59778; Santa Cruz, clone DECMA-1, 1:300), mouse anti-VASP (VM2771; ECM Biosciences, 1:150), mouse anti-ARP3 (A5979; Sigma-Aldrich, 1:100), mouse anti-nonmuscle myosin IIA (ab55456; Abcam, 1:100), and mouse anti-α-actinin-4 (sc393495; Santa Cruz, 1:100). Secondary antibodies were purchased from Thermo Fisher Scientific. Actin staining using FITC-phalloidin or Alexa Fluor 647–phalloidin (Invitrogen) was done according to manufacturer’s instructions.
Calcium switch and immunostaining
Calcium switch experiments have been used in multiple epithelial cell types for more than three decades (Angst et al., 1990; Gumbiner and Simons, 1986; Heimark et al., 1990; Rajasekaran et al., 1996). We used previously characterized methods with minor modifications, as follows. MDCK ZO-1/ZO-2 KD and Ecad-GFP CRISPR Caco-2 cells were grown 48 and 96 h after confluency, respectively, before going through calcium switch. Cells were washed once with CSB, incubated in CSB for 1.5 h (MDCK ZO-KD) or 2 h (Caco-2), and then switched back to complete medium, which we refer to as the recovery period. Samples were collected and immunostained at different recovery time points. For drug treatment, inhibitors were added and incubated with cells during the recovery period. Concentrations for inhibitors are as follows: 100 µM CK666 (Sigma-Aldrich), 50 µM SMIFH2 (Sigma-Aldrich), 100 µM Y-27632 (Sigma-Aldrich), and 10 µM blebbistatin (Sigma-Aldrich). For Y-27632 or blebbistatin washout experiments, complete medium without drug was used to replace the drug-containing medium 90 min into the recovery period.
Formaldehyde fixation was used for most experiments, except those in Fig. S1 U and Fig. S2, F, G, and J. For formaldehyde fixation, 1% of formaldehyde was used to fix cells. Cells were then quenched and permeabilized with 0.5% Triton X-100 and incubated with primary and secondary antibodies. For Fig. S2, G and J, cells were permeabilized before fixation, using a method described in Yu-Kemp et al. (2017). For Fig. S1 U and Fig. S2 F, cells were fixed with ice-cold methanol for 5 min and then permeabilized with 0.1% Triton X-100. Cells were incubated with primary antibodies in PBS at RT for 1.5 h, and then with secondary antibodies in PBS for 4 h or overnight at 4°C. For imaging, cells were mounted in Prolong Diamond anti-fade mountant (Fisher).
Imaging and image processing
Images were collected via Airyscan or SIM imaging. Airyscan imaging was performed on a Zeiss LSM880 system, with a 63× 1.4-NA oil objective. Z collection was done with intervals at 0.18 µm. Raw images were processed using the 3D Airyscan processing function in Zen software (Carl Zeiss). SIM images were collected using a Nikon N-SIM and SR APO TIRF with a 100× 1.49-NA oil objective with 3D-SIM function. Images were processed with stack reconstruction using NIS-Elements AR v4.51 software 2016 (Nikon). Imaging parameters across conditions were kept identical for each set of experiments, to allow comparison of intensity between control and drug-treated conditions. All x–y (top view) images in the figures are maximum-intensity projections (MIPs) of the apical region of the cell, unless otherwise noted. 3D images were generated using Imaris v9.6 (Bitplane) with the surface function. To visualize myosin and Ecad signals at the border in the surface view, the actin signal at the borders was selected with surface gain size of 0.3–0.5 µm and then deducted from the total actin surface area. Photoshop CS6 (Adobe) was used to adjust brightness and contrast and adjust input levels so that the signal spanned the entire grayscale output.
Quantification
Quantifications were done using Fiji (National Institutes of Health) or Matlab. Individual cells or borders for measuring signal intensity or border curvature were manually selected based on the 3 × 3 grid in Fiji/ImageJ. Our approaches are illustrated diagrammatically in Fig. S7. For measuring the myosin-covered area, the myosin signal was binarized, and the value of pixels that are covered by myosin were extracted using Matlab. For measuring myosin sarcomere spacing, a straight line was drawn at the cell–cell border in Fiji, the intensity value was extracted, and the mean interval between maxima, which indicates sarcomere spacing, was calculated in Matlab using the findpeaks function. For measuring border curvature, the length of a line drawn fully tracing the cell border from one tricellular junction to the next was compared with the length of a straight line drawn directly from one tricellular junction to another. Only borders with a straight-line length of 150–450 pixels (6.4–19.2 µm) were measured. For measuring actin and Ecad intensity, borders were surrounded by an 8 × 3.5–µm box, and the intensity values were measured at the apical z-slice and the lateral z-slice, with lateral defined as halfway between the apical and basal z-slices. For measuring the actin-covered area, borders were surrounded by a 7.5 × 3-µm box. Each box was binarized, and the value of the pixels corresponding to the actin-covered area were extracted using the histogram tool. For the Caco-2 actin-covered area, border measurements, only unaffected borders were included. For visualizing Caco-2 border continuity, MIP images were binarized, and the analyze particles tool was used to exclude apical microvilli signal and other separated particles (size, 0.0 to infinity; circularity, 0.00–0.01). All statistical analyses were performed using Prism 8 (GraphPad). All data were analyzed using two-way unpaired t tests or ordinary one-way ANOVA tests with post hoc Tukey tests. Data distribution was assumed to be normal but not formally tested. Fitted curves for myosin area ratio or border straightness were calculated using nonlinear fit in Prism. Scatter plots present all the quantified data points, with mean ± SD.
Figure S7.
Visual illustration of quantification methods used. Details are in Materials and methods section Quantification.
Modeling
We used a customized build of Cytosim (Belmonte et al., 2017; Nedelec and Foethke, 2007), an agent-based stochastic physics simulator of cytoskeletal processes, that incorporates code for simulating nonmuscle myosin II motors in large bipolar ensembles (Cortes et al., 2020). We constructed a 2D space that was 12 microns wide by 8 microns tall and was periodic along its x axis, such that components near the left or right edges could interact with each other. The top and bottom edges were set to be rigid confinement boundaries beyond which components could not extend. In our simulations, the top boundary represented the cadherin complex at cell junctions when looking apically down on cells (Fig. 5, A–C), while the bottom boundary served only to limit the area of simulated space (speeding up computation) and represented a distance beyond which actin-like filaments are too far to interact with simulated motor stacks.
Motor ensembles were simulated as 350-nm-long filaments with 15 motors on either side in a bipolar arrangement (Fig. S7 F). Motors were coupled to motor filament backbones by 30-nm-long springs with a stiffness of 150 pN/nm. Actin filaments were simulated as 1 ± 0.6 microns in length and a high rigidity approximately fivefold higher than the rigidity reported for individual actin filaments (Gittes et al., 1993). A nonmotoring cross-linker, akin to α-actinin, was simulated as a single actin-binder with a binding rate of 2 s−1, an unbinding rate of 0.2 s−1, and a binding range of 40 nm.
Three different motor ensemble arrangements were tested for actin alignment capabilities. First, we tested an arrangement where motor ensembles were set in stacks near the surface. In this setup, 80 motor ensembles were seeded aligned horizontally along a 1-micron-tall strip (along the y axis) every 475 nm (Fig. S7 F). 13 such strips were generated within 300 nm of the top surface boundary of the simulation space. Motor ensembles were seeded with affixed anchors that kept them in horizontal orientation and fixed in space. 2,000 actin-binding components were seeded in the spaces between motor stacks, generating an alternating pattern of motors and cross-linkers (Fig. S7 F) as seen in vivo. 300 actin filaments were then seeded in isotropy throughout the simulation space, and the simulations were allowed to evolve for 500 s with 1-ms time steps. 10 simulations were run for each condition.
Second, we tested an arrangement where the same motor ensemble stacks were seeded but in the center of the simulation space. Here, all components were arranged as before, but the motor stacks were placed in the middle of the space, ∼3.5 microns away from the top boundary. Once again, simulations were evolved for 500 s with 1-ms time steps. Finally, we tested an isotropic arrangement where 1,040 motor ensembles were seeded in a rectangular area 12 microns wide by 2 microns tall, right up on the top boundary of the space. Motor ensembles were allowed to orient in any direction within their confined space and were affixed in space as before. Cross-linkers were seeded interdispersed with the motor ensembles and affixed in space, and actin filaments were seeded as before and evolved for 500 s of simulated time. The line graphs are the averaged value for 10 simulations at each simulated time point.
Online supplemental material
Fig. S1 illustrates Ecad localization, 3D cell shapes, sarcomere spacing, myosin-2A localization, arrays without Ca switch, and Arp3 localization before and during recovery. Fig. S2 shows tests for effectiveness of CK666 and FP4mito, and VASP localization before and during recovery. Fig. S3 reveals level of toxicity of SMIFH2 and tests of the effect of combined inhibition of the Arp2/3 complex and formins. Fig. S4 reveals that MLCK does not alter or delay recovery from Ca switch and illustrates the effect of ROCK inhibition on cortical myosin levels. Fig. S5 illustrates 3D cell shapes after blebbistatin treatment, reveals that blebbistatin washout restores the ZA, and illustrates effects of dual inhibition of both ROCK and the myosin ATPase. Fig. S6 reveals that inhibiting the Arp2/3 complex or formins does not prevent or delay ZA assembly in Caco-2 cells, and uses binarization of images to assess the effects of ROCK inhibition or blebbistatin on cortical actin. Fig. S7 provides visual illustration of the quantification methods used. Table S1 lists the number of experiments (n) for each time point in Figs. 1, 3, 4, 7, and S4.
Supplementary Material
lists the number of experiments (n) for each time point in Figs. 1, 3, 4, 7, and S4.
Acknowledgments
We thank Wangsun Choi for advice getting started with the ZO KD MDCK cells; Josh Lawrimore for advice on data analysis and MATLAB codes; Stephanie Gupton for sharing reagents; Alpha Yap for suggesting Caco-2 cells as an alternate cell line; Yvonne Beckham, Kate Cavanaugh, and Margaret Gardel for supplying Caco-2 cells and information on culture; Tony Perdue of the Biology Imaging Center for advice and support; Wangsun Choi, Jordan Beach, Richard Cheney, Steve Rogers, Anja Schmidt, and other Peifer laboratory members for helpful advice and comments; Bob Duronio for advice and support throughout; and the editor and reviewers for helpful suggestions.
This work was supported by National Institutes of Health R35 GM118096 to M. Peifer. H-C. Yu-Kemp and D.B. Cortes were supported in part by National Institutes of Health T32 CA009156. Work in the Maddox laboratory is supported by National Institutes of Health R01 GM102390 and National Science Foundation 1616661.
The authors declare no competing financial interests.
Author contributions: This project was initiated and designed by H-C. Yu-Kemp, with input from M. Peifer. H-C. Yu-Kemp led the experimental team, designed and set up all the protocols for the study, and together with R.A. Szymanski, N.C. Gadda, and M.L. Lilich, carried out all the MDCK experiments. R.A. Szymanski designed and carried out all the Caco-2 experiments, using protocols from H-C. Yu-Kemp and with input from H-C. Yu-Kemp and M. Peifer. H-C. Yu-Kemp and R.A. Szymanski designed and implemented the quantification approaches. D.B. Cortes designed and implemented the modeling experiments, with input from A.S. Maddox and M. Peifer. H-C. Yu-Kemp, R.A. Szymanski, D.B. Cortes, and M. Peifer wrote the manuscript with input from the other authors.
References
- Afshar, K., Stuart B., and Wasserman S.A.. 2000. Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development. 127:1887–1897. 10.1242/dev.127.9.1887 [DOI] [PubMed] [Google Scholar]
- Agarwal, P., and Zaidel-Bar R.. 2019. Diverse roles of non-muscle myosin II contractility in 3D cell migration. Essays Biochem. 63:497–508. 10.1042/EBC20190026 [DOI] [PubMed] [Google Scholar]
- Anderson, T.W., Vaughan A.N., and Cramer L.P.. 2008. Retrograde flow and myosin II activity within the leading cell edge deliver F-actin to the lamella to seed the formation of graded polarity actomyosin II filament bundles in migrating fibroblasts. Mol. Biol. Cell. 19:5006–5018. 10.1091/mbc.e08-01-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angst, B.D., Nilles L.A., and Green K.J.. 1990. Desmoplakin II expression is not restricted to stratified epithelia. J. Cell Sci. 97:247–257. 10.1242/jcs.97.2.247 [DOI] [PubMed] [Google Scholar]
- Bao, J., Jana S.S., and Adelstein R.S.. 2005. Vertebrate nonmuscle myosin II isoforms rescue small interfering RNA-induced defects in COS-7 cell cytokinesis. J. Biol. Chem. 280:19594–19599. 10.1074/jbc.M501573200 [DOI] [PubMed] [Google Scholar]
- Beach, J.R., Shao L., Remmert K., Li D., Betzig E., and Hammer J.A. III. 2014. Nonmuscle myosin II isoforms coassemble in living cells. Curr. Biol. 24:1160–1166. 10.1016/j.cub.2014.03.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach, J.R., Bruun K.S., Shao L., Li D., Swider Z., Remmert K., Zhang Y., Conti M.A., Adelstein R.S., Rusan N.M., et al. 2017. Actin dynamics and competition for myosin monomer govern the sequential amplification of myosin filaments. Nat. Cell Biol. 19:85–93. 10.1038/ncb3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear, J.E., and Gertler F.B.. 2009. Ena/VASP: towards resolving a pointed controversy at the barbed end. J. Cell Sci. 122:1947–1953. 10.1242/jcs.038125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear, J.E., Loureiro J.J., Libova I., Fässler R., Wehland J., and Gertler F.B.. 2000. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 101:717–728. 10.1016/S0092-8674(00)80884-3 [DOI] [PubMed] [Google Scholar]
- Belmonte, J.M., Leptin M., and Nédélec F.. 2017. A theory that predicts behaviors of disordered cytoskeletal networks. Mol. Syst. Biol. 13:941. 10.15252/msb.20177796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington, N., Beach J.R., Heissler S.M., Remmert K., Guzik-Lendrum S., Nagy A., Takagi Y., Shao L., Li D., Yang Y., et al. 2015. Myosin 18A coassembles with nonmuscle myosin 2 to form mixed bipolar filaments. Curr. Biol. 25:942–948. 10.1016/j.cub.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buracco, S., Claydon S., and Insall R.. 2019. Control of actin dynamics during cell motility. F1000 Res. 8:1977. 10.12688/f1000research.18669.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette, D.T., Shao L., Ott C., Pasapera A.M., Fischer R.S., Baird M.A., Der Loughian C., Delanoe-Ayari H., Paszek M.J., Davidson M.W., et al. 2014. A contractile and counterbalancing adhesion system controls the 3D shape of crawling cells. J. Cell Biol. 205:83–96. 10.1083/jcb.201311104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carramusa, L., Ballestrem C., Zilberman Y., and Bershadsky A.D.. 2007. Mammalian diaphanous-related formin Dia1 controls the organization of E-cadherin-mediated cell-cell junctions. J. Cell Sci. 120:3870–3882. 10.1242/jcs.014365 [DOI] [PubMed] [Google Scholar]
- Cavanaugh, K.E., Staddon M.F., Munro E., Banerjee S., and Gardel M.L.. 2020. RhoA Mediates Epithelial Cell Shape Changes via Mechanosensitive Endocytosis. Dev. Cell. 52:152–166.e5. 10.1016/j.devcel.2019.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinowsky, C.R., Pinette J.A., Meenderink L.M., Lau K.S., and Tyska M.J.. 2020. Nonmuscle myosin-2 contractility-dependent actin turnover limits the length of epithelial microvilli. Mol. Biol. Cell. 31:2803–2815. 10.1091/mbc.E20-09-0582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W., Acharya B.R., Peyret G., Fardin M.A., Mège R.M., Ladoux B., Yap A.S., Fanning A.S., and Peifer M.. 2016. Remodeling the zonula adherens in response to tension and the role of afadin in this response. J. Cell Biol. 213:243–260. 10.1083/jcb.201506115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti, M.A., Even-Ram S., Liu C., Yamada K.M., and Adelstein R.S.. 2004. Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II-A in mice. J. Biol. Chem. 279:41263–41266. 10.1074/jbc.C400352200 [DOI] [PubMed] [Google Scholar]
- Cortes, D.B., Gordon M., Nédélec F., and Maddox A.S.. 2020. Bond Type and Discretization of Nonmuscle Myosin II Are Critical for Simulated Contractile Dynamics. Biophys. J. 118:2703–2717. 10.1016/j.bpj.2020.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, J.M., Harden N., Leung T., Lim L., and Kiehart D.P.. 1998. Cellularization in Drosophila melanogaster is disrupted by the inhibition of rho activity and the activation of Cdc42 function. Dev. Biol. 204:151–164. 10.1006/dbio.1998.9061 [DOI] [PubMed] [Google Scholar]
- Ebrahim, S., Fujita T., Millis B.A., Kozin E., Ma X., Kawamoto S., Baird M.A., Davidson M., Yonemura S., Hisa Y., et al. 2013. NMII forms a contractile transcellular sarcomeric network to regulate apical cell junctions and tissue geometry. Curr. Biol. 23:731–736. 10.1016/j.cub.2013.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimova, N., and Svitkina T.M.. 2018. Branched actin networks push against each other at adherens junctions to maintain cell-cell adhesion. J. Cell Biol. 217:1827–1845. 10.1083/jcb.201708103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning, A.S., Van Itallie C.M., and Anderson J.M.. 2012. Zonula occludens-1 and -2 regulate apical cell structure and the zonula adherens cytoskeleton in polarized epithelia. Mol. Biol. Cell. 23:577–590. 10.1091/mbc.e11-09-0791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenix, A.M., Taneja N., Buttler C.A., Lewis J., Van Engelenburg S.B., Ohi R., and Burnette D.T.. 2016. Expansion and concatenation of non-muscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol. Biol. Cell. 27:1465–1478. 10.1091/mbc.E15-10-0725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates, J., Mahaffey J.P., Rogers S.L., Emerson M., Rogers E.M., Sottile S.L., Van Vactor D., Gertler F.B., and Peifer M.. 2007. Enabled plays key roles in embryonic epithelial morphogenesis in Drosophila. Development. 134:2027–2039. 10.1242/dev.02849 [DOI] [PubMed] [Google Scholar]
- Gittes, F., Mickey B., Nettleton J., and Howard J.. 1993. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 120:923–934. 10.1083/jcb.120.4.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glacy, S.D. 1983. Subcellular distribution of rhodamine-actin microinjected into living fibroblastic cells. J. Cell Biol. 97:1207–1213. 10.1083/jcb.97.4.1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner, B., and Simons K.. 1986. A functional assay for proteins involved in establishing an epithelial occluding barrier: identification of a uvomorulin-like polypeptide. J. Cell Biol. 102:457–468. 10.1083/jcb.102.2.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, S.P., Gambin Y., Gomez G.A., Verma S., Giles N., Michael M., Wu S.K., Guo Z., Johnston W., Sierecki E., et al. 2014. Cortactin scaffolds Arp2/3 and WAVE2 at the epithelial zonula adherens. J. Biol. Chem. 289:7764–7775. 10.1074/jbc.M113.544478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimark, R.L., Degner M., and Schwartz S.M.. 1990. Identification of a Ca2(+)-dependent cell-cell adhesion molecule in endothelial cells. J. Cell Biol. 110:1745–1756. 10.1083/jcb.110.5.1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwani, F.M., Kovacs E.M., Paterson A.D., Verma S., Ali R.G., Fanning A.S., Weed S.A., and Yap A.S.. 2004. Cortactin is necessary for E-cadherin-mediated contact formation and actin reorganization. J. Cell Biol. 164:899–910. 10.1083/jcb.200309034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick, B., Han M.S., Helgeson L.A., and Nolen B.J.. 2013. Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem. Biol. 20:701–712. 10.1016/j.chembiol.2013.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuzé, M.L., Sankara Narayana G.H.N., D’Alessandro J., Cellerin V., Dang T., Williams D.S., Van Hest J.C., Marcq P., Mège R.M., and Ladoux B.. 2019. Myosin II isoforms play distinct roles in adherens junction biogenesis. eLife. 8:e46599. 10.7554/eLife.46599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, S., Dasbiswas K., Guo Z., Tee Y.H., Thiagarajan V., Hersen P., Chew T.L., Safran S.A., Zaidel-Bar R., and Bershadsky A.D.. 2017. Long-range self-organization of cytoskeletal myosin II filament stacks. Nat. Cell Biol. 19:133–141. 10.1038/ncb3466 [DOI] [PubMed] [Google Scholar]
- Huveneers, S., Oldenburg J., Spanjaard E., van der Krogt G., Grigoriev I., Akhmanova A., Rehmann H., and de Rooij J.. 2012. Vinculin associates with endothelial VE-cadherin junctions to control force-dependent remodeling. J. Cell Biol. 196:641–652. 10.1083/jcb.201108120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki, T., Uehata M., Tamechika I., Keel J., Nonomura K., Maekawa M., and Narumiya S.. 2000. Pharmacological properties of Y-27632, a specific inhibitor of rho-associated kinases. Mol. Pharmacol. 57:976–983. [PubMed] [Google Scholar]
- Ivanov, A.I., McCall I.C., Parkos C.A., and Nusrat A.. 2004. Role for actin filament turnover and a myosin II motor in cytoskeleton-driven disassembly of the epithelial apical junctional complex. Mol. Biol. Cell. 15:2639–2651. 10.1091/mbc.e04-02-0163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, A.I., Hunt D., Utech M., Nusrat A., and Parkos C.A.. 2005. Differential roles for actin polymerization and a myosin II motor in assembly of the epithelial apical junctional complex. Mol. Biol. Cell. 16:2636–2650. 10.1091/mbc.e05-01-0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiu, Y., Kumari R., Fenix A.M., Schaible N., Liu X., Varjosalo M., Krishnan R., Burnette D.T., and Lappalainen P.. 2019. Myosin-18B Promotes the Assembly of Myosin II Stacks for Maturation of Contractile Actomyosin Bundles. Curr. Biol. 29:81–92.e5. 10.1016/j.cub.2018.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak, A., Pasolli H.A., and Fuchs E.. 2004. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 6:21–30. 10.1038/ncb1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs, E.M., Goodwin M., Ali R.G., Paterson A.D., and Yap A.S.. 2002. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr. Biol. 12:379–382. 10.1016/S0960-9822(02)00661-9 [DOI] [PubMed] [Google Scholar]
- Kovács, M., Tóth J., Hetényi C., Málnási-Csizmadia A., and Sellers J.R.. 2004. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 279:35557–35563. 10.1074/jbc.M405319200 [DOI] [PubMed] [Google Scholar]
- Kovacs, E.M., Verma S., Ali R.G., Ratheesh A., Hamilton N.A., Akhmanova A., and Yap A.S.. 2011. N-WASP regulates the epithelial junctional actin cytoskeleton through a non-canonical post-nucleation pathway. Nat. Cell Biol. 13:934–943. 10.1038/ncb2290 [DOI] [PubMed] [Google Scholar]
- Lecuit, T., and Yap A.S.. 2015. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat. Cell Biol. 17:533–539. 10.1038/ncb3136 [DOI] [PubMed] [Google Scholar]
- Leerberg, J.M., Gomez G.A., Verma S., Moussa E.J., Wu S.K., Priya R., Hoffman B.D., Grashoff C., Schwartz M.A., and Yap A.S.. 2014. Tension-sensitive actin assembly supports contractility at the epithelial zonula adherens. Curr. Biol. 24:1689–1699. 10.1016/j.cub.2014.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas, T.J., Mirzoeva S., Slomczynska U., and Watterson D.M.. 1999. Identification of novel classes of protein kinase inhibitors using combinatorial peptide chemistry based on functional genomics knowledge. J. Med. Chem. 42:910–919. 10.1021/jm980573a [DOI] [PubMed] [Google Scholar]
- Machesky, L.M., and Hall A.. 1997. Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J. Cell Biol. 138:913–926. 10.1083/jcb.138.4.913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melli, L., Billington N., Sun S.A., Bird J.E., Nagy A., Friedman T.B., Takagi Y., and Sellers J.R.. 2018. Bipolar filaments of human nonmuscle myosin 2-A and 2-B have distinct motile and mechanical properties. eLife. 7:e32871. 10.7554/eLife.32871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, M., and Yap A.S.. 2013. The regulation and functional impact of actin assembly at cadherin cell-cell adhesions. Semin. Cell Dev. Biol. 24:298–307. 10.1016/j.semcdb.2012.12.004 [DOI] [PubMed] [Google Scholar]
- Nedelec, F., and Foethke D.. 2007. Collective Langevin dynamics of flexible cytoskeletal fibers. N. J. Phys. 9:427. 10.1088/1367-2630/9/11/427 [DOI] [Google Scholar]
- Nemethova, M., Auinger S., and Small J.V.. 2008. Building the actin cytoskeleton: filopodia contribute to the construction of contractile bundles in the lamella. J. Cell Biol. 180:1233–1244. 10.1083/jcb.200709134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, T., Ito S., Saito H., Hiver S., Shigetomi K., Ikenouchi J., and Takeichi M.. 2016. DAAM1 stabilizes epithelial junctions by restraining WAVE complex-dependent lateral membrane motility. J. Cell Biol. 215:559–573. 10.1083/jcb.201603107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, Y., Shi S., Zhang F., Liu R., Takagi Y., Bershadsky A.D., Viasnoff V., and Sellers J.R.. 2021. The formin inhibitor SMIFH2 inhibits members of the myosin superfamily. J. Cell Sci. 134:jcs253708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen, B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., and Pollard T.D.. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034. 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg, J., van der Krogt G., Twiss F., Bongaarts A., Habani Y., Slotman J.A., Houtsmuller A., Huveneers S., and de Rooij J.. 2015. VASP, zyxin and TES are tension-dependent members of Focal Adherens Junctions independent of the α-catenin-vinculin module. Sci. Rep. 5:17225. 10.1038/srep17225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa, M. 2018. Nonmuscle myosin IIA is involved in recruitment of apical junction components through activation of α-catenin. Biol. Open. 7:bio031369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D. 2016. Actin and Actin-Binding Proteins. Cold Spring Harb. Perspect. Biol. 8:a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran, A.K., Hojo M., Huima T., and Rodriguez-Boulan E.. 1996. Catenins and zonula occludens-1 form a complex during early stages in the assembly of tight junctions. J. Cell Biol. 132:451–463. 10.1083/jcb.132.3.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, M.V., and Zaidel-Bar R.. 2016. Formin-mediated actin polymerization at cell-cell junctions stabilizes E-cadherin and maintains monolayer integrity during wound repair. Mol. Biol. Cell. 27:2844–2856. 10.1091/mbc.e16-06-0429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb, C., Cervantes E., Madere F., and Irvine K.D.. 2019. Organization and function of tension-dependent complexes at adherens junctions. J. Cell Sci. 132:jcs.224063. 10.1242/jcs.224063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi, S.A., Neidt E.M., Cui J., Feiger Z., Skau C.T., Gardel M.L., Kozmin S.A., and Kovar D.R.. 2009. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16:1158–1168. 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou, A., Field C., Sisson J.C., Sullivan W., and Karess R.. 2004. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol. Biol. Cell. 15:838–850. 10.1091/mbc.e03-06-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai, E., and Marshall C.J.. 2002. ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat. Cell Biol. 4:408–415. 10.1038/ncb796 [DOI] [PubMed] [Google Scholar]
- Saitoh, M., Ishikawa T., Matsushima S., Naka M., and Hidaka H.. 1987. Selective inhibition of catalytic activity of smooth muscle myosin light chain kinase. J. Biol. Chem. 262:7796–7801. 10.1016/S0021-9258(18)47638-7 [DOI] [PubMed] [Google Scholar]
- Sandquist, J.C., and Means A.R.. 2008. The C-terminal tail region of nonmuscle myosin II directs isoform-specific distribution in migrating cells. Mol. Biol. Cell. 19:5156–5167. 10.1091/mbc.e08-05-0533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A., and Grosshans J.. 2018. Dynamics of cortical domains in early Drosophila development. J. Cell Sci. 131:jcs212795. 10.1242/jcs.212795 [DOI] [PubMed] [Google Scholar]
- Scott, J.A., Shewan A.M., den Elzen N.R., Loureiro J.J., Gertler F.B., and Yap A.S.. 2006. Ena/VASP proteins can regulate distinct modes of actin organization at cadherin-adhesive contacts. Mol. Biol. Cell. 17:1085–1095. 10.1091/mbc.e05-07-0644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers, J.R., and Heissler S.M.. 2019. Nonmuscle myosin-2 isoforms. Curr. Biol. 29:R275–R278. 10.1016/j.cub.2019.03.022 [DOI] [PubMed] [Google Scholar]
- Shapiro, L., Fannon A.M., Kwong P.D., Thompson A., Lehmann M.S., Grübel G., Legrand J.F., Als-Nielsen J., Colman D.R., and Hendrickson W.A.. 1995. Structural basis of cell-cell adhesion by cadherins. Nature. 374:327–337. 10.1038/374327a0 [DOI] [PubMed] [Google Scholar]
- Shewan, A.M., Maddugoda M., Kraemer A., Stehbens S.J., Verma S., Kovacs E.M., and Yap A.S.. 2005. Myosin 2 is a key Rho kinase target necessary for the local concentration of E-cadherin at cell-cell contacts. Mol. Biol. Cell. 16:4531–4542. 10.1091/mbc.e05-04-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smutny, M., Cox H.L., Leerberg J.M., Kovacs E.M., Conti M.A., Ferguson C., Hamilton N.A., Parton R.G., Adelstein R.S., and Yap A.S.. 2010. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat. Cell Biol. 12:696–702. 10.1038/ncb2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, V., Hudson A., Cooley L., and Theurkauf W.E.. 2002. Arp2/3-dependent pseudocleavage furrow assembly in syncytial Drosophila embryos. Curr. Biol. 12:705–711. 10.1016/S0960-9822(02)00807-2 [DOI] [PubMed] [Google Scholar]
- Svitkina, T.M., Verkhovsky A.B., McQuade K.M., and Borisy G.G.. 1997. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 139:397–415. 10.1083/jcb.139.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi, M. 1988. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development. 102:639–655. 10.1242/dev.102.4.639 [DOI] [PubMed] [Google Scholar]
- Tang, V.W., and Brieher W.M.. 2012. α-Actinin-4/FSGS1 is required for Arp2/3-dependent actin assembly at the adherens junction. J. Cell Biol. 196:115–130. 10.1083/jcb.201103116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullio, A.N., Accili D., Ferrans V.J., Yu Z.X., Takeda K., Grinberg A., Westphal H., Preston Y.A., and Adelstein R.S.. 1997. Nonmuscle myosin II-B is required for normal development of the mouse heart. Proc. Natl. Acad. Sci. USA. 94:12407–12412. 10.1073/pnas.94.23.12407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin, V., Bauer C., Yin M., and Fuchs E.. 2000. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 100:209–219. 10.1016/S0092-8674(00)81559-7 [DOI] [PubMed] [Google Scholar]
- Verkhovsky, A.B., Svitkina T.M., and Borisy G.G.. 1995. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J. Cell Biol. 131:989–1002. 10.1083/jcb.131.4.989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, S., Shewan A.M., Scott J.A., Helwani F.M., den Elzen N.R., Miki H., Takenawa T., and Yap A.S.. 2004. Arp2/3 activity is necessary for efficient formation of E-cadherin adhesive contacts. J. Biol. Chem. 279:34062–34070. 10.1074/jbc.M404814200 [DOI] [PubMed] [Google Scholar]
- Verma, S., Han S.P., Michael M., Gomez G.A., Yang Z., Teasdale R.D., Ratheesh A., Kovacs E.M., Ali R.G., and Yap A.S.. 2012. A WAVE2-Arp2/3 actin nucleator apparatus supports junctional tension at the epithelial zonula adherens. Mol. Biol. Cell. 23:4601–4610. 10.1091/mbc.e12-08-0574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares, M., Ma X., Adelstein R.S., and Horwitz A.R.. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790. 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.L. 1984. Reorganization of actin filament bundles in living fibroblasts. J. Cell Biol. 99:1478–1485. 10.1083/jcb.99.4.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, Z., and Sokac A.M.. 2016. -Back-to-back mechanisms drive actomyosin ring closure during Drosophila embryo cleavage. J. Cell Biol. 215:335–344. 10.1083/jcb.201608025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, S., and Nelson W.J.. 2007. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J. Cell Biol. 178:517–527. 10.1083/jcb.200701058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Kemp, H.C., Kemp J.P. Jr., and Brieher W.M.. 2017. CRMP-1 enhances EVL-mediated actin elongation to build lamellipodia and the actin cortex. J. Cell Biol. 216:2463–2479. 10.1083/jcb.201606084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen, J.A., Cohen Y., Hudson A.M., Cooley L., Wieschaus E., and Schejter E.D.. 2002. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 156:689–701. 10.1083/jcb.200109057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Betson M., Erasmus J., Zeikos K., Bailly M., Cramer L.P., and Braga V.M.. 2005. Actin at cell-cell junctions is composed of two dynamic and functional populations. J. Cell Sci. 118:5549–5562. 10.1242/jcs.02639 [DOI] [PubMed] [Google Scholar]
- Zicha, D., Dobbie I.M., Holt M.R., Monypenny J., Soong D.Y., Gray C., and Dunn G.A.. 2003. Rapid actin transport during cell protrusion. Science. 300:142–145. 10.1126/science.1082026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the number of experiments (n) for each time point in Figs. 1, 3, 4, 7, and S4.