Abstract
Simple Summary
Cryptosporidium is a parasite that infects humans and a broad range of animals. There are few diagnostic features that can be used to identify and differentiate between species and therefore DNA-based detection and genetic typing methods are required. This is important as some species are transmitted from animals to humans (a process called zoonotic transmission) and understanding this is central to control. These DNA-based tools have greatly facilitated our current understanding of which of the many Cryptosporidium species and genotypes that have been identified are capable of infecting humans. More studies need to be conducted in areas where the potential for zoonotic transmission is greatest and recently developed genetic tools should be applied more extensively to further our understanding of the zoonotic transmission of this important parasite.
Abstract
The enteric parasite, Cryptosporidium is a major cause of diarrhoeal illness in humans and animals worldwide. No effective therapeutics or vaccines are available and therefore control is dependent on understanding transmission dynamics. The development of molecular detection and typing tools has resulted in the identification of a large number of cryptic species and genotypes and facilitated our understanding of their potential for zoonotic transmission. Of the 44 recognised Cryptosporidium species and >120 genotypes, 19 species, and four genotypes have been reported in humans with C. hominis, C. parvum, C. meleagridis, C. canis and C. felis being the most prevalent. The development of typing tools that are still lacking some zoonotic species and genotypes and more extensive molecular epidemiological studies in countries where the potential for transmission is highest are required to further our understanding of this important zoonotic pathogen. Similarly, whole-genome sequencing (WGS) and amplicon next-generation sequencing (NGS) are important for more accurately tracking transmission and understanding the mechanisms behind host specificity.
Keywords: Cryptosporidium, zoonotic, transmission, molecular tools
1. Introduction
Cryptosporidium species are enteric parasites with a global distribution and a wide range of hosts. Transmission is by the faecal–oral route via contaminated water, food or direct contact with humans and animals [1]. Although first described in 1907 by Tyzzer [2], Cryptosporidium did not come to prominence until the early 1980s, when it was identified as a cause of severe protracted diarrhoea and death in HIV+/AIDS patients [3]. It is now recognised as a major pathogen in children and immunocompromised adults [4,5] and after rotavirus is the most important diarrheal pathogen in young children [6,7]. In 1993, Cryptosporidium was responsible for a large waterborne outbreak affecting over 400,000 residents of Milwaukee, Wisconsin [8] and although still frequently under-reported, is a well-known and major cause of both waterborne and foodborne outbreaks of gastroenteritis globally [9,10,11]. This is due in part to the resistance of the environmental stage, the oocyst, to disinfectants including chlorine treatment of both drinking and recreational water [12,13].
The parasite has a complex life cycle that initiates upon ingestion and excystation of oocysts and involves both asexual and sexual phases, which culminate in the shedding of infectious thick-walled oocysts in faeces [14]. In humans, in addition to watery diarrhoea, cryptosporidiosis can cause abdominal pain, vomiting, headaches, joint pain, malnutrition, failure to thrive and cognitive deficits and has been linked with colon cancer [15,16,17,18,19,20]. Although usually self-limiting in immunocompetent hosts, cryptosporidiosis can become chronic, persisting for 2 years or more [5,17]. In neonatal livestock, cryptosporidiosis can cause profuse diarrhoea, weight loss and death [11,21,22,23], resulting in significant production losses [22,24,25,26]. The only US Food and Drug Administration approved drug, nitazoxanide, is ineffective in the most affected populations (children and immunocompromised individuals) and there is no FDA-approved vaccine [27,28,29,30]. In animals, nitazoxanide is also largely ineffective and while Halocur® (halofuginone lactate) has been licensed in some countries as a prophylactic, its effectiveness is variable and it cannot be given to animals that already have diarrhoea [11,22].
Molecular typing of Cryptosporidium has advanced significantly over the last few decades and is particularly important for this parasite as many species are morphologically identical [31]. Identification to species level is most commonly conducted using the 18S ribosomal RNA locus due to the existence of both hypervariable and conserved sequences [31,32]. Other loci examined include actin, heat shock protein 70 (hsp70) and the Cryptosporidium oocyst wall protein gene (COWP) [31]. Currently, at least 44 species of Cryptosporidium and >120 genotypes have been identified, with C. hominis and C. parvum being the most important species infecting humans [33].
Contact with animals has been identified as a risk factor in many studies [34,35,36,37]; therefore, zoonotic transmission plays a major role in the epidemiology of cryptosporidiosis [1,23,38,39,40,41]. In order to track transmission, multi-locus sequence typing (MLST) tools have been developed, including analysis of the highly polymorphic glycoprotein 60 (gp60) gene, the most commonly used locus [31]. The gp60-based typing system is based on a combination of tandem serine-coding trinucleotide repeats (TCA, TCG and TCT) and extensive sequence divergence in non-repeat regions [31,42,43]. The nomenclature system [43], starts with a Roman numeral and lower-case letter for each Cryptosporidium and genotype (e.g., C. hominis, Ia, Ib, etc., C. parvum IIa, IIb, etc.), followed by uppercase letters denoting numbers of repeats. For example, C. hominis subtype IfA19G1R5 has 19 TCA (A) repeats, 1 TCG repeat (G) repeat and 5 AAGAAGGCAAAGAAG repeats (R). For some divergent species and genotypes, including C. ubiquitum, C. felis, C. canis, C. ryanae, C. xiaoi, C. bovis and Cryptosporidium chipmunk genotype I and skunk genotype, whole-genome sequencing (WGS) has been required to identify the gp60 loci in order to develop typing systems [44,45,46,47,48,49,50]. The trinucleotide repeats (TCA, TCG, TCT) are absent from gp60 genes in C. ubiquitum, C. felis, C. canis, C. ryanae, C. bovis and C. xiaoi [44,46,47,48,49,50].
This review aims to summarise the currently available data on the zoonotic transmission of Cryptosporidium species and genotypes as well as outlining future studies that are required to better understand the transmission dynamics of this ubiquitous parasite. This review could provide a valuable reference for One Health scientists and help guide future research.
2. Zoonotic Cryptosporidium Species and Genotypes
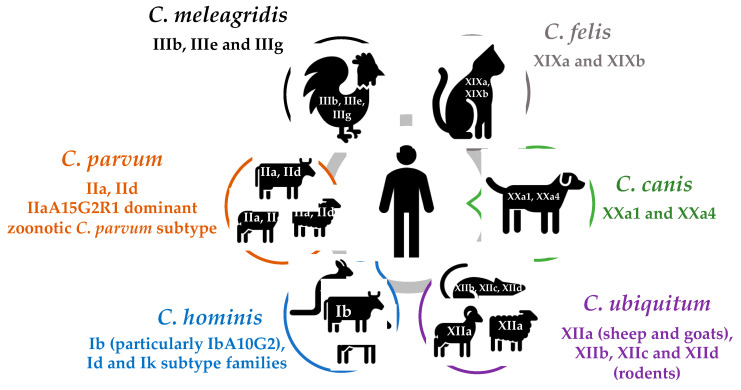
In total, 19 species (including C. hominis and C. parvum) and 4 genotypes have been reported in humans including C. meleagridis, C. canis, C. felis, C. ubiquitum, C. cuniculus, C. viatorum, C. muris, C. andersoni, C. erinacei, C. tyzzeri, C. bovis, C. suis, C. scrofarum, C. occultus, C. xiaoi, C. fayeri, C. ditrichi, Cryptosporidium chipmunk genotype I, mink genotype, skunk genotype and horse genotype [1,33] (Table 1). Where possible, subtyping analysis at the gp60 locus has been used to document gp60 subtypes common to humans and animals, to support zoonotic transmission (Table 2). Cryptosporidium hominis and C. parvum are responsible for ~95% of human infections, followed by C. meleagridis, C. felis, C. canis and C. ubiquitum [1,38] (Figure 1). Some species such as C. meleagridis, C. canis, C. felis, C. viatorum and C. muris are more commonly identified in humans in developing countries, whereas others such as C. ubiquitum, C. cuniculus and chipmunk genotype I are predominantly seen in developed countries [1].
Table 1.
Cryptosporidium species and genotypes in humans in order of frequency.
| Species Name | Main Reservoir Hosts | Reports in Humans |
|---|---|---|
| C. hominis | Non-human primates, donkeys | Most common species in humans |
| C. parvum | Ruminants, wildlife | Second most common species in humans |
| C. meleagridis | Birds | Third most commonly reported species in humans |
| C. felis | Cats | Commonly reported [40] |
| C. canis (previously canine genotype) | Dog | Commonly reported [40] |
| C. ubiquitum (previously cervine genotype) | Ruminants, rodents, carnivores, non-human primates |
Commonly reported [31,44,51,52] |
| C. cuniculus (previously rabbit genotype) | Rabbits | Many reports in humans [51,52,53,54,55,56,57,58,59,60,61] |
| C. viatorum | Rodents | [34,51,62,63,64,65,66,67,68,69,70,71,72,73,74] |
| Chipmunk genotype I | Rodents | [37,45,51,60,71,72,75,76,77] |
| C. muris | Rodents | [52,60,78,79,80,81,82,83,84,85,86,87,88,89] |
| C. andersoni (previously C. muris-like genotype) | Cattle | [65,81,90,91,92,93,94,95,96,97] |
| C. suis (previously pig genotype I) | Pigs, wild boars | [79,96,98,99,100,101,102,103] |
| Horse genotype | Horses | [51,58,82,104,105] |
| C. erinacei (previously hedgehog genotype) | Hedgehogs, horses | [51,53,54,106,107] |
| C. bovis (previously bovine B genotype) | Cattle | [78,108,109,110] |
| Skunk genotype | Skunk | [37,82,105,111,112] |
| C. tyzzeri (previously mouse genotype I) | Rodents | [43,54,113] |
| C. occultus (previously C. suis-like genotype and C. parvum VF383) | Rodents | Refs. [114,115] Unpublished report in a human in GenBank (HQ822146) |
| C. ditrichi | Rodents (mainly mice) | [51,116] |
| Mink genotype | Mink | [117,118] |
| C. fayeri (previously marsupial genotype I) | Marsupials | [119,120] |
| C. xiaoi (previously C. bovis-like genotype) | Sheep, goats | [34] |
| C. scrofarum (previously pig genotype II) | Pigs, wild boar | [121] |
Table 2.
Zoonotic gp60 subtypes common to humans and animals (excluding non-human primates).
| Species Name | gp60 Subtypes | References |
|---|---|---|
| C. hominis | IbA9G3, IbA13G3, IbA14G2, IbA10G2, IdA15G1, IbA10G2R2 and Ik subtype family | [22,125,127,131,132,133,134,139,221] |
| C. parvum | Many subtypes but mainly the IIa (particularly IIaA15G2R1) and IId subtype families | [11,22,23,31,117,140] |
| C. meleagridis | IIIbA21G1R1b, IIIbA22G1R1cIIIbA23G1R1b, IIIbA23G1R1c, IIIbA24G1R1, IIIbA26G1R1b, IIIeA17G2R1, IIIeA19G2R1, IIIeA21G2R1, IIIeA21G2R1 and IIIgA31G3R1 | [51,52,79,181,182,183,184] |
| C. felis | XIXa and XIXb | [40,46,186,192] |
| C. canis | XXa1 and XXa4 | [40,49] |
| C. ubiquitum | XIIa, XIIb, XIIc and XIId | [23,44,51,52] |
| C. cuniculus | VaA18, VbA19, VbA22 to Vb26, Vb28, VbA29 and VbA31 to VbA33 | [51,52,53,54,55,56,58,210,211] |
| C. viatorum | XVaA3g, XVaA3h and XVcA2G1 | [65,66,114,207,209] |
| Chipmunk genotype I | XIVaA18G2T2 | [37,45,51,60,71,72,75,76,77] |
| C. muris | Gastric Cryptosporidium species do not appear to have the gp60 gene | |
| C. andersoni | ||
| C. suis | A gp60 typing tool has not yet been developed | |
| Horse genotype | VIbA13 and VIcA16 | [51,58,82,104,105] |
| C. erinacei | XIIIaA20R10, XIIIaA23R12, XIIIaA24R9, XIIIaA24R10, XIIIaA25R10 XIIIaA25R11, XIIIaA26R9 and XIIIaA26R10 (humans only) | [53,54] |
| C. bovis | No gp60 sequences available from humans | |
| Skunk genotype | XVIbA16G2b and XVIcA22 (humans only but XVIbA16G2a in wildlife) | [111,224] |
| C. tyzzeri | IXaA8 and IXbA6 | [43,113] |
| C. occultus | A gp60 typing tool has not yet been developed | |
| C. ditrichi | A gp60 typing tool has not yet been developed | |
| Mink genotype | No gp60 sequences available from humans | |
| C. fayeri | IVaA9G4T1R1, IVgA10G1T1R1 | [119,120] |
| C. xiaoi | No gp60 sequences available from humans | |
| C. scrofarum | A gp60 typing tool has not yet been developed |
Figure 1.
Zoonotic transmission of the six most common species of Cryptosporidium in humans.
2.1. Cryptosporidium hominis
Cryptosporidium hominis is the dominant species in humans in many industrialised countries and in developing countries, whereas, in the Middle East, European countries and New Zealand, C. parvum occurs at similar rates to C. hominis [1], indicating that zoonotic transmission is more prevalent in countries with intensive farming [122]. Although largely anthropologically transmitted, there have been numerous reports of C. hominis in animals (including non-human primates, cattle, sheep, goats, horses, donkeys, Bactrian camels, birds, marsupials, a dugong, badger, dingo, foxes, flying fox, rodents, and fish) and experimental infections has been established in calves, lambs, piglets, gerbils, and mice [123,124,125]. In human infectivity trials, the 50% infectious dose (ID50) was as low as 10 C. hominis oocysts [126], however, in animal models, much larger numbers of oocysts were required to achieve infections, suggesting that higher doses are required to cause infection in animals [125].
Cryptosporidium hominis has been detected at a relatively low frequency in livestock [22,23,125], although in one study, 80% of Cryptosporidium positives in calves in New Zealand were identified as C. hominis [127] and C. hominis was the only Cryptosporidium sp. detected in goats in South Korea [128]. These studies demonstrate that cattle, sheep and goats can potentially serve as animal reservoirs for infections with C. hominis [22,23,125]. In Australia, in addition to cattle and sheep, C. hominis has been detected in deer, dingoes and marsupials inhabiting drinking water catchments [129,130,131]. This likely resulted from human spill-back either via direct contact or via sewage contamination of pastures or drinking water.
The C. hominis subtypes identified at the gp60 locus in non-human hosts has been extensively reviewed [125]. Briefly, the C. hominis IbA10G2 subtype dominates in livestock that are positive for C. hominis [22,125] and has also been detected in hedgehogs and mice in Europe [132,133,134]. As IbA10G2 is one of the most prevalent subtypes in humans in many countries and is also a major cause of waterborne outbreaks [1,11,38], this further supports a human origin for C. hominis in animals. In equines and particularly donkeys and horses, the Ik C. hominis subtype family (which is relatively rare in humans) is endemic and a reservoir of infection for humans [135,136,137,138]. Studies in Australia also suggest that C. hominis may become endemic in marsupials [131,139,140]. Continued human encroachment into wildlife areas may result in C. hominis becoming increasingly endemic in livestock and wildlife populations escalating the risk of zoonotic C. hominis transmission.
2.2. Cryptosporidium parvum and Other Livestock-Associated Species
Cryptosporidium parvum has a very wide host range infecting ungulates, wildlife (including carnivores, rodents, non-human primates, marine mammals and fish) and is the most important zoonotic species in humans, particularly in rural areas with frequent contact with livestock [1,11,22,23,124,141,142]. Calves, sheep and goats are major contributors to zoonotic C. parvum transmission with significant differences in prevalence [11,22,23,143]. In cattle, the dominant species are C. parvum, C. bovis, C. ryanae and C. andersoni, although other species (C. hominis, C. felis, C. suis, C. scrofarum, C. meleagridis, C. tyzzeri, C. serpent is, a novel genotype and C. occultus) have been reported [11,22]. In sheep and goats, C. parvum, C. ubiquitum and C. xiaoi are the predominant species, but a range of other species including C. bovis, C. hominis, C. andersoni, C. suis, C. muris, C. fayeri, C. baileyi, C. ryanae, C. scrofarum, C. canis, C. occultus, Cryptosporidium sheep genotype I and rat genotype II have occasionally been reported [11,22,23]. In pre-weaned calves, C. parvum is the dominant species in most studies [22], with the exception of China where C. bovis mostly dominates [144]. There does not appear to be an age-related prevalence of C. parvum in sheep and goats and while C. parvum dominates in these hosts in Europe and Australia, in other regions including the Americas, Asia, C. xiaoi and C. ubiquitum dominate in sheep and goats [23].
Over 20 C. parvum gp60 subtype families have been identified with geographic variation in subtype distribution as well as host adaptation demonstrated [23,31]. For example, the three dominant C. parvum gp60 subtype families in humans are IIa, IId and IIc, and of these, IIc appears to be almost exclusively anthropologically transmitted whereas IIa and IId are zoonotic [1,33,145]. WGS also supports this and it has been suggested that the C. parvum IIc subtype should be considered a separate subspecies (Cryptosporidium parvum anthroponosum), while the zoonotically transmitted IIa and IId subtypes referred to as Cryptosporidium parvum parvum [146]. In cattle, IIa is the dominant C. parvum subtype family in most countries and particularly in Europe and contact with calves has been identified as a risk factor for infection with the IIa subtypes [34]. In other countries, however, including China, Egypt, Malaysia, Romania and Sweden, the IId subtype dominates in cattle [31,144]. Similarly in sheep and goats, IIa dominates in European countries but in Africa, Asia and Australia, IId is the dominant C. parvum subtype family [23,144]. Subtyping studies have confirmed the identification of identical IIa and IId subtypes in humans and animals indicating zoonotic transmission [11,22,23,31,117,139]. For example, the relatively rare IIaA19G1R1 was identified in children, goat kids, and lambs during an outbreak of cryptosporidiosis at a holiday farm in Norway [147]. Similarly, subtypes IIaA15G2R1, IIaA18G2R1 and IIaA19G1R1 were responsible for several outbreaks in the UK, with the same subtypes identified from livestock on the same farms where the outbreaks occurred [38]. Of these, IIaA15G2R1 is the dominant C. parvum subtype in humans and cattle in industrialised countries and MLST analysis indicates that genetic recombination is the driving force behind its emergence as a dominant and hyper-transmissible subtype [148].
Cryptosporidium bovis is prevalent in cattle and other bovid species, while C. xiaoi is a related species in sheep and goats worldwide [11,22,23]. A gp60 typing system has been established for C. bovis based on WGS sequences. Sixty-eight subtypes in six subtype families (XXVIa to XXVIf) have been identified, with apparent genetic recombination among subtype families [50]. There have been several reports of C. bovis in humans (Table 1). It was first reported in a dairy farm worker (and dairy calves) in Bengal, India in 2010 [108] and again in 2012 in a study in Australia, which collected and screened faecal samples from diarrheic calves in rural NSW and farmers from these farms [109]. In that study, C. bovis was identified in a 3-year-old child and a 23-year-old adult, from separate farms and both were asymptomatic [109]. Both individuals drank raw milk and had frequent contact with calves [109]. In another study, a mixed C. parvum and C. bovis infection was identified in a diarrhoeic child (<6 years) (along with mixed infections in cattle) from Ismailia province in Egypt [110]. A gp60 typing system has recently been established for C. xiaoi based on WGS, with 12 subtype families, (XXIIIa to XXIIIl) and high subtype diversity identified [48]. It has previously been reported in two HIV/AIDS patients from Ethiopia based on 18S sequences only [34]. Both C. bovis and C. xiaoi appear to have narrow host ranges but further research is required to understand the zoonotic potential of these common livestock species.
Like C. parvum, C. ubiquitum (previously cervine genotype) also has a wide host range and is commonly detected in both domestic and wild ruminants, rodents, carnivores, primates, and humans, particularly in industrialised countries [44,51,52,139,149,150]. Both gp60 and MLST tools have been developed for C. ubiquitum [44,150]. At the gp60 locus, C. ubiquitum lacks the TCA, TCG and TCT repeats and to date, eight subtype families (XIIa–XIIh) have been identified [44], yet the transmission routes between animals and humans are not well understood. In the UK, XIIa is the dominant C. ubiquitum subtype in humans, which is also the main subtype in small ruminants [23,44]. In contrast, human C. ubiquitum infections in the US are caused by XIIb, XIIc and XIId subtype families which are common in rodents [44]. Subtype families XIIe and XIIf are also found in rodents but at present have only been identified in the Slovak Republic [44]. In France and Sweden, XIIb and XIId subtype families have been reported in humans [51,52]. MLST analysis has also re-affirmed that XIIa is ruminant-adapted and XIIb, XIIc and XIId subtype families are rodent-adapted [150].
Cryptosporidium andersoni, a gastric species, was originally thought to be C. muris due to its morphological similarity, until it was established as a separate species [151]. It is commonly reported in ruminants, particularly adult cattle and other bovids and is also common in sheep and goats, mainly in China [23] and has been reported in deer [152] and rodents, particularly hamsters [153,154,155,156]. As gastric Cryptosporidium species do not appear to have the gp60 gene, a gp60 typing tool has not been established for C. andersoni. However, an MLST tool has been developed and a mostly epidemic population structure has been identified [157,158,159]. There have been numerous reports in humans, but the extent of zoonotic transmission remains to be determined (Table 1).
Cryptosporidium suis and C. scrofarum are the dominant species infecting pigs (and wild boars) and mostly cause subclinical infections (although C. parvum, C. muris, C. tyzzeri, C. felis, C. hominis, C. andersoni and C. meleagridis can also infect pigs) [160,161,162]. Cryptosporidium suis is mainly found in pre-weaned animals and C. scrofarum in post-weaned pigs. Neither C. suis nor C. scrofarum are commonly identified in humans. There have been a few reports of C. suis in HIV-positive individuals in China, Peru and Thailand [79,92,101,102,103], as well as children in Cambodia [99] and symptomatic individuals in the UK and Madagascar [96,100]. There has only been one report of C. scrofarum in a symptomatic immunocompetent 29-year-old male co-infected with Giardia [121]. A gp60 typing tool has not yet been developed for C. suis or C. scrofarum and need to be developed to explore the zoonotic potential of C. suis and C. scrofarum in pigs and transmission routes via water and sewage.
The horse genotype is commonly reported in donkeys and horses [135,137,138]. Typing of the horse genotype at the gp60 locus has identified subtype families VIa, VIb and VIc [31,51]. Subtype VIaA15G4 is one of the most common subtypes identified in horses and donkeys, with subtype VIbA13 and VIaA14G4 identified in a hedgehog and calf respectively [135]. The horse genotype (subtype VIcA16) has been identified in a traveller to Sweden [51], an 18-year-old symptomatic patient from the US (subtype VIbA13) [104], a symptomatic 30-year-old immunocompetent woman from the rural UK (subtype VIbA13) [105] and in two adult females from the UK [82].
2.3. C. meleagridis
Avian cryptosporidiosis was first described in 1929 [163], but the first avian species, Cryptosporidium meleagridis was not named until 1955 in turkeys [164]. The parasite has a broad host range and is commonly reported in wild birds but less so in poultry [165,166]. It has also been reported in foxes, minks, cattle, wallabies, gorillas, and dogs [167,168,169,170,171,172]. Experimental transmission studies have also confirmed its infectivity for calves, pigs, rabbits, rats and mice and humans [173,174]. It is the third most common Cryptosporidium species in humans [1,175] and has also been identified in colon adenocarcinoma tissue in an immunocompetent Polish patient [176]. Phylogenetic analysis suggests that C. meleagridis may originally have been a mammalian parasite that secondarily became established in birds [177]. In children and HIV+ individuals in developing countries, C. meleagridis is frequently the dominant Cryptosporidium species [1,5,178], whereas, in developed countries, C. meleagridis is usually responsible for ~1–4% of human infections [38,66,174,179]. Despite being so commonly identified in humans, the extent of zoonotic transmission is not well understood. One study in an organic farm in Sweden identified identical C. meleagridis 70 kDa Heat Shock Protein (hsp70) gene sequences in samples from one human, three chickens and one hen, suggesting zoonotic transmission [180].
Typing at the gp60 locus indicates that humans are susceptible to most C. meleagridis gp60 subtypes, as of the 10 known subtype families (IIIa to IIIi), eight (IIIa to IIIh) (and 30 subtypes) have been identified in humans with IIIb among the most common [31,181,182,183,184]. Relatively few subtyping studies have been conducted in birds but both MLST and gp60 studies have provided some support for zoonotic transmission. For example, subtype IIIgA31G3R1 has been reported from four poultry and one human from a Swedish farm [181]. MLST analysis of C. meleagridis from humans and birds from Peru did not find evidence of host segregation [182] and the same gp60 subtypes (IIIbA26G1R1b and IIIbA22G1R1c) have been found in children with diarrhoea and in farmed chickens in Hubei province, China, supporting zoonotic transmission [183,184]. Similarly, subtypes IIIeA17G2R1, IIIeA19G2R1, IIIeA21G2R1 and IIIeA22G1R1 has been reported in Swedish patients (and IIIeA21G2R1 in Canadian patients) and in rodents and chickens in Asia [51,52,79,183] (Table 2).
2.4. Companion Animal-Associated Species (C. canis and C. felis)
Companion animals, particularly dogs and cats, have close relationships with their owners contributing to improved mental health and social support but can also contribute to zoonotic disease transmission [185]. There are currently ~470 million pet dogs and 370 million pet cats in the world (www.statista.com/statistics/1044386/dog-and-cat-pet-population-worldwide/accessed on 20 October 2021). Cryptosporidium canis and C. felis are the main species infecting dogs and cats respectively and are among the top five Cryptosporidium species infecting humans [142,186]. There have also been numerous reports of C. parvum and a few reports of C. meleagridis, C. muris, C. andersoni, C. scrofarum, C. ryanae, C. hominis and rat genotype III in cats and dogs, some of which may be due to coprophagy [40,187]. Prevalence rates of C. canis and C. felis vary widely but are commonly below 10% [40,187,188]. Few studies have examined the species of Cryptosporidium in humans and pets living in the same household, but one study identified C. canis in a 32-month-old girl, her 6.5-year-old brother and a dog from the same house in Lima, Peru [189]. Another study reported identical 18S, HSP70 and COWP C. felis sequences from a cat and her 37-year-old immunocompetent owner in Sweden [190]. A case-control study of HIV-infected individuals with and without cryptosporidiosis reported only a weak association between dog ownership and cryptosporidiosis [191].
Cryptosporidium canis and C. felis gp60 loci have recently been characterised and have been shown to lack the serine-coding trinucleotide repeats normally used for typing (similar to C. ubiquitum) [40,46,49,186]. Within C. canis, five gp60 subtype families have been identified (XXa, XXb, XXc, XXd and XXe), with subtypes XXa1 and XXa4 detected in both humans and dogs [49]. The previously identified household transmission of C. canis between two children and a dog in Lima, Peru [190] was confirmed by gp60 subtyping [49]. Five subtype families have also been identified in C. felis (XIXa, XIXb, XIXc, XIXd and XIXe) [46] and of these, two subtypes (XIXa and XIXb) have been reported in both humans and cats supporting zoonotic transmission, with the remaining three subtypes (XIXc, XIXd and XIXe), possibly transmitted anthropologically [40,46,186,192].
2.5. Wildlife-Associated Species and Genotypes
Due to their wide geographical distribution and abundance and close contact with humans, rodents are considered an important zoonotic reservoir for Cryptosporidium and the global prevalence of Cryptosporidium in rodents has been estimated at ~17% [193]. To date, cryptosporidial infections from five rodent-derived species (C. viatorum, C. muris, C. tyzzeri, C. occultus and C. ditrichi) and one genotype (Cryptosporidium chipmunk genotype I) have been reported in humans (Table 1). Of these, there have only been a few reports of C. tyzzeri, C. occultus and C. ditrichi in humans (Table 1).
Cryptosporidium muris, a gastric species, has a very wide host range including rodents, ruminants, cats and dogs, horses, Bactrian camels, pigs, birds, and non-human primates [22,23,40,162,194,195,196,197,198]. There have been numerous reports in humans, particularly in developing countries (in children and HIV patients) [1,5], (Table 1). As with C. andersoni, an MLST tool has been developed for C. muris [157,158], which revealed that the genetic diversity of C. muris was greater than C. andersoni, possibly reflecting the much smaller host range of C. andersoni and that host clustering was evident, suggesting that some C. muris isolates have co-evolved with their hosts over a long period of time [158]. A C. muris human infectivity trial has been conducted in which six healthy adults were challenged with 105 C. muris oocysts each (isolate RN66) and all became infected (two were symptomatic), confirming the zoonotic potential of C. muris [199].
Typing of C. tyzzeri is at the gp60 locus has identified three subtype families; IXa, IXb and IXc [31,200], including subtype IXbA22R9 from a horse [195]. Cryptosporidium tyzzeri (subtype IXaA6R2) was identified from a child in Kuwait [43] and subtype family IXb has been detected in three human patients in New Zealand [54]. A mixed C. parvum (subtype IIaA13G1R1) and C. tyzzeri (subtypes IXaA8 and IXbA6) infection was identified in a symptomatic 25-year-old female conducting fieldwork trapping wild rodents in the Czech Republic [113]. The same subtypes were identified in the trapped mice supporting zoonotic transmission [113]. Cryptosporidium tyzzeri is genetically very closely related to C. parvum [201], which further supports its potential for zoonotic transmission.
A gp60 typing system has not been established for C. ditrichi or C. occultus. Cryptosporidium ditrichi has been reported in three patients from Sweden, two of which were symptomatic and one had possible contact with mice [51,116]. Although predominantly a rodent species, C. occultus appears to be common in bovids including cattle, yaks (Bos gunniens) and water buffalo (Bubalus bubalis) [202] and was the dominant species detected in Alpacas in China [203]. It has been reported in one human from the UK (HQ822146), two humans from British Columbia [115] and a young child in China [114].
Cryptosporidium viatorum was originally reported in the UK in travellers returning from India and formally described as a human species in 2012 [73]. Since then, C. viatorum have been described in humans from Australia [66], China [114], Columbia [68], Ethiopia [34,64,74], India [63,67], Mozambique [62], Myanmar [65], Nigeria [69,70] and Swedish patients (who had returned from Kenya or Guatemala) [51,71,72]. It has also been detected in urban wastewater in China [204]. Due to the relatively high prevalence of C. viatorum in rats in Australia and China [205,206,207], it is now thought that C. viatorum was originally a rodent species and therefore rodents are likely to be an important reservoir host.
A gp60 typing tool has been established for C. viatorum and to date four subtype families have been identified; XVa, XVb, XVc and XVd [205,206,207,208]. Subtyping supports the potential zoonotic transmission between rodents and humans. For example, human-derived C. viatorum isolates have been subtyped to date as XVa3a to XVa3h [66,114,209] and XVcA2G1c [65], and subtype XVaA6 was identified in both sewer overflow and wastewater in Shanghai, China [204]. In rodents, XVa (XVaA6, XVaA3g, XVaA3h), XVb (XVbA2G1), XVc (XVcA2G1a, XVcA2G1b) and XVd (XVdA3) subtype families have been identified [205,206,207]. Thus, three subtypes (XVaA3g, XVaA3h and XVcA2G1) are common to humans and rodents and in addition, the XVaA3g subtype identified from wild rats [206] was 100% homologous to an XVaA3g subtype identified in a human patient in Australia [66]. Screening of rodents and rats across wider geographic areas is essential to better understand their role as reservoirs for C. viatorum.
To date, five chipmunk genotypes have been identified in rodents but only one of these, chipmunk genotype 1 has been identified in humans [33]. Chipmunk genotype I was first identified in New York storm water as genotype W17 [209] but was renamed as chipmunk genotype I, when it was detected in rodent faecal samples in 2007 [210]. It infects a range of rodents particularly squirrels, chipmunks, and deer mice and is considered an emerging zoonotic pathogen in humans [45,75], being the third most commonly identified Cryptosporidium sp. in Sweden [75]. A gp60 typing system has been developed for chipmunk genotype I, with one subtype family identified (XIVa) [45]. Chipmunk genotype I was first identified in two humans in 2004 in Wisconsin (subtype XIVaA16G2T1) [77], and in a 41-year-old HIV-positive male in France [60]. In 2013, it was reported in a two-year-old female and 56-year-old male from Sweden [71,72] and typed as XIVaA20G2T1 by Guo et al. [45], who also identified 19 US human isolates and typed 17 as XIVaA14G2T1 (n = 1), XIVaA16G2T1 (n = 1), XIVaA14G2T2 (n = 1), XIVaA16G2T2 (n = 4), XIVaA18G2T1b (n = 2), XIVaA17G2T2 (n = 1), XIVaA19G2T2a (n = 1), XIVaA20G2T2 (n = 2), XIVaA19G2T2b (n = 1), XIVaA15G2T3 (n = 2) and XIVaA17G2T3 (n = 1) [45]. In that study, in the three rodent samples analysed, subtypes XIVaA18G2T1a and XIVaA18G2T2 were detected. It was identified in a human in Nebraska [37] and between 2014 and 2015, chipmunk genotype 1 was identified in five adults (four women and one man) in Sweden and all were typed as XIVaA20G2T1 [51]. Subsequently, it was identified in sixteen sporadic cases, three outbreak-related cases, and one zoonotic case, as well as in two squirrel samples in Sweden between 2018 and 2019, and subtyping of nineteen humans and two squirrels identified subtype XIVaA20G2T1 in all samples, supporting zoonotic transmission [75].
Of the other wildlife species and genotypes identified in humans, C. cuniculus (previously known as rabbit genotype) is a common species in rabbits worldwide [211,212,213] and has also been reported in kangaroos and alpacas in Australia [130,131,214]. It is the only species (other than C. parvum and C. hominis) known to have caused a waterborne outbreak of disease [56,61]. It is more commonly reported in humans in industrialised countries and particularly in the UK (Table 1). Two subtype families (Va and Vb) have been identified [31]. The waterborne outbreak caused by C. cuniculus in the UK was typed as VaA18 [56], however, most C. cuniculus cases typed in humans and rabbits have been caused by subtype family Vb [51,52,53,54,55,58]. In humans, subtypes VaA11, VaA18, VaA19 to VaA22, VaA23, VbA13, VbA15 VbA17, VbA20, VbA22, VbA23, VbA24 to VbA34, VbA36, VbA37 and VbA38 have been reported [51,52,54,58,211,212]. Of these, subtypes VbA19, VbA22 to Vb26, Vb28, VbA29 and VbA31 to VbA33 have been reported in rabbits supporting zoonotic transmission [130,131,140,212,215,216,217,218]. More recently an MLST typing tool has been developed and a clonal population structure identified [219].
Cryptosporidium erinacei (previously hedgehog genotype 1) was first described in European hedgehogs in 2014 [220]. It has also been reported in horses in Algeria and rats in China [206,221] and was detected in cattle and kangaroo faecal samples by next-generation amplicon sequencing in Australia [131]. Cryptosporidium erinacei has been reported in an immunocompetent 26-year-old man from the Czech Republic [107], an immunocompromised patient in France [106], in two cases from Sweden (one locally acquired and one in a traveller from Greece) [51] and in thirteen cases from New Zealand [53,54]. The higher number of cases in New Zealand may be due to the greater abundance of European hedgehogs (Erinaceus europaeus) in New Zealand compared to Europe, but as C. erinacei can infect other hosts, the range and prevalence of this parasite in reservoir hosts remain to be determined.
At the gp60 locus, one subtype family (XIIIa) has been identified [31,220]. Zoonotic transmission has not been demonstrated as the gp60 subtypes identified in humans (XIIIaA20R10, XIIIaA23R12, XIIIaA24R9, XIIIaA24R10, XIIIaA25R10 XIIIaA25R11, XIIIaA26R9 and XIIIaA26R10) [51,54] have not been identified in hedgehogs and horses (XIIIaA19R12, XIIIaA19R13, XIIIaA21R10, XIIIaA21R11, XIIIaA22R9, XIIIaA22R11 [221,222,223]). However as only a small number of isolates have been subtyped, further research is required to better understand its zoonotic potential.
The skunk genotype was first identified in striped skunks (Mephitis mephitis) in the US [177], but has a broad host range and has been described in numerous species of wild mammals [111,223], and is common in surface water in the US and Canada [111]. There have been five reports in humans; an 18-month-old child attending a UK daycare centre [112], a 25-year-old woman from a rural area in the UK [105], an adult female from the UK [82] and a human in Nebraska [37,111], and all except the child attending daycare were symptomatic.
Typing at the gp60 locus has identified four subtype families (XVIa, XVIb, XVIc and XVId) with a high subtype diversity (n = 14) [111,224]. In humans, subtypes XVIbA16G2b and XVIcA22 have been identified, which have not been identified in wildlife. However, a closely related subtype (XVIbA16G2a subtype) has been found in raccoons and storm runoff samples in the US [111].
There have only been two reports of the marsupial derived C. fayeri in humans; a symptomatic 29-year-old immunocompetent female in New South Wales (NSW), Australia in 2010 and a decade later, in 2020, it was identified in a symptomatic 37-year-old immunosuppressed female treated for acute myeloid leukaemia in Western Australia (WA) [119,120]. Eight subtype families have been identified within C. fayeri (Iva–IVh) [31,130,225,226]. In the initial study by Waldron et al. [119], the same C. fayeri subtype (IVaA9G4T1R1) was identified in both the woman and eastern grey kangaroos (Macropus giganteus) inhabiting the main drinking water catchment for Sydney [225] and the woman had regular contact with marsupials supporting zoonotic transmission [119]. In the latter study in WA, C. fayeri subtype IVgA10G1T1R1 was identified in the female [120] and the same subtype had previously been identified in western grey kangaroos (Macropus fuliginosus) in WA and rabbits (Oryctolagus cuniculus) in NSW drinking water catchments [131], again supporting the potential for zoonotic transmission. Marsupials are the dominant animals inhabiting drinking water catchments in Australia and further research is necessary to better understand the zoonotic implications, particularly in addition to C. fayeri and C. macropodum, marsupials can also be infected with C. hominis as discussed above [131].
The Cryptosporidium mink genotype was first identified in minks (Mustela vison) inhabiting a New York City drinking water supply watershed [210] and is a common parasite in minks [227]. Typing at the gp60 locus has identified four subtypes, including XaA5G1, XbA5G1R1, XcA5G1R1 and XdA4G1 [172,227,228]. There have been two reports of the mink genotype in humans [117,118]. However, no gp60 sequences are available and therefore the possibility of zoonotic transmission is unknown.
3. Knowledge Gaps and Future Studies
The prevalence and clinical impact of Cryptosporidium and the potential for zoonotic transmission is highest in developing countries, yet relatively few molecular epidemiological studies have been conducted in these areas [1,33]. Large-scale molecular studies are therefore urgently required to explore the extent of zoonotic transmission [1,33]. Relatively little information is also available on the pathogenicity of some of the less common zoonotic species and genotypes, as most reports do not provide detailed clinical information, and this is another important knowledge gap that needs to be addressed. The majority of gp60 subtyping studies conducted to date have relied on Sanger sequencing, which cannot reliably identify the extent of mixed subtypes. For example, a recent study applied the bioinformatic program TIDE to deconvolute gp60 chromatograms generated using Sanger sequencing and identified previously unrecognised mixed subtype infections and has the advantage of also being able to be applied to retrospective data [229]. Similarly, another bioinformatics tool called “CryptoGenotyper” has been developed to read both 18S and gp60 Sanger sequence data, which can also resolve double peaks in mixed infections and increase the accuracy of sequence identification [230]. Amplicon next-generation sequencing (NGS), which can identify low-abundance sequences in mixed infections, has shown that it can identify additional Cryptosporidium gp60 subtypes not identified by Sanger sequencing in various hosts [141,231]. This has important implications for tracing zoonotic transmission as Sanger sequencing may not detect zoonotic species and subtypes that are present at low abundance and therefore incorrect conclusions regarding zoonotic transmission may be made. Future studies need to examine the extent of intra-sample diversity more closely to better understand zoonotic transmission dynamics.
The application of CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated protein) genome editing technology to Cryptosporidium has been a major advance in the ability to genetically modify the parasite [232]. More recently CRISPR technologies have been applied to the detection of infectious diseases by identifying specific pathogen sequences and then cleaving them in order to produce a readable signal [233]. For example, in the HOLMES (one-HOur Low-cost Multipurpose highly Efficient System) detection platform, if a target DNA is present, a Cas protein called Cas12a forms a complex with the guide CRISPR RNA, which then binds the target DNA and cleaves a non-target ssDNA reporter in the system, resulting in a fluorescent signal [233]. A lateral flow biosensor has been developed which incorporated isothermal amplification of Cryptosporidium DNA with this CRISPR detection method to identify C. parvum sequences belonging to the gp60 IId subtype family (from both humans and cattle), with high sensitivity and specificity [234]. This type of system has the potential to be employed in field situations to rapidly identify zoonotic subtypes and transmission hot-spots for more targeted analysis.
A more complete understanding of the molecular epidemiology of Cryptosporidium and zoonotic transmission dynamics can be gained from WGS studies, which are also increasingly being used to identify gp60 loci in divergent species and genotypes [235]. Currently, WGS data has been generated from the following zoonotic species and genotypes; C. parvum, C. hominis, C. muris, C. meleagridis, C. ubiquitum, C. andersoni, C. tyzzeri, C. cuniculus, C. viatorum, C. felis, C. canis, C. bovis, C. xiaoi, Cryptosporidium skunk genotype and chipmunk genotype I [44,45,46,47,48,49,50,236]. Comparative WGS has identified a variety of invasion-associated proteins including mucin glycoproteins, insulinase-like proteases and MEDLE secretory proteins, which differ between species with narrow and broad host ranges and hence may play a role in host specificity [76,237,238,239]. However, many challenges remain including the development of a universal MLST system that uses polymorphic markers distributed across all chromosomes [236]. Future studies need to conduct WGS on a wider range of species and genotypes and to examine the biological functions of diverse genes in the Cryptosporidium genome to gain a better understanding of the genetic determinants of host specificity.
WGS is expensive and another alternative may be to mine existing raw untargeted (shotgun) sequencing metagenomic data deposited in public databases generated for other purposes (such as the microbiome of humans and animals) [240,241]. For example, one study successfully mined 12 large data sets on the human gut microbiome to examine the population genomics of Blastocystis [242]. As a proof of principle, C. parvum (chromosome 6) was identified and mapped from data sets derived from water concentrates and calf microbiomes, although further bioinformatic pipeline optimisation is required [243]. With the rapid expansion of publicly available metagenomes, this approach offers a cost-effective and unprecedented opportunity to unravel the extent of zoonotic transmission and environmental contamination of Cryptosporidium across wide geographic areas.
4. Conclusions
Our understanding of the extent of genetic diversity and zoonotic potential of Cryptosporidium species and genotypes has been greatly enhanced by the development of molecular detection and typing tools, particularly the gp60 typing tool. The six most common species identified in humans are C. hominis, C. parvum, C. meleagridis, C. canis, C. felis and C. ubiquitum, yet the extent of zoonotic transmission in many of these species is still not clearly understood. Although primarily anthropologically transmitted, C. hominis is increasingly detected in animals, with evidence that it may be endemic in marsupials (IbA10G2 subtype) and equines (Ik subtype family). Cryptosporidium parvum is the most important zoonotic species, with IIa and IId the major zoonotic subtypes. Evidence to date supports the zoonotic transmission of C. meleagridis (IIIbA26G1R1b and IIIbA22G1R1c subtypes), C. canis (XXa1 and XXa4 subtypes), C. felis (XIXa and XIXb subtypes) and C. ubiquitum (XIIa, XIIb, XIIc and XIId subtype families). Cryptosporidium viatorum was originally described as a human species but rodents are now thought to be the major host species with subtypes (XVaA3g, XVaA3h and XVcA2G1) common to both humans and rodents. The rodent-derived chipmunk genotype I is considered an emerging human pathogen in the US and Sweden with evidence supporting the zoonotic transmission (subtype XIVaA20G2T1) between humans and squirrels. Cryptosporidium cuniculus in rabbits is another important zoonotic species, as it was responsible for a waterborne outbreak in the UK, with evidence supporting zoonotic transmission of multiple subtypes. Cryptosporidium erinacei in hedgehogs is a minor zoonotic species in most areas except New Zealand and the limited gp60 subtyping conducted to date has yet to support zoonotic transmission. Similarly, identical skunk genotype gp60 subtypes have not been identified in skunks and humans. Two studies have identified identical C. fayeri subtypes (IVaA9G4T1R1 and IVgA10G1T1R1) in humans and kangaroos in Australia. Gastric species (C. muris and C. andersoni) seem to lack the gp60 gene but gp60 typing tools that are still lacking for C. suis, C. scrofarum, C. ditrichi, C. occultus and the mink genotype are essential to better understand their transmission dynamics. The application of WGS and amplicon NGS in future studies are important for more accurately tracking transmission and understanding the mechanisms behind host specificity.
Author Contributions
Conceptualisation, U.R. and A.Z.; writing—original draft preparation, U.R., A.Z., Y.F. and L.X.; writing—review and editing, U.R., A.Z., Y.F. and L.X.; funding acquisition, Y.F. and L.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Guangdong Major Project of Basic and Applied Basic Research (2020B0301030007) and the 111 Project (D20008), China.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yang X., Guo Y., Xiao L., Feng Y. Molecular epidemiology of human cryptosporidiosis in low- and middle-income countries. Clin. Microbiol. Rev. 2021;34:e00087-19. doi: 10.1128/CMR.00087-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyzzer E.E. A sporozoan found in the peptic glands of the common mouse. Proc. Soc. Exp. Biol. Med. 1907;5:12–13. doi: 10.3181/00379727-5-5. [DOI] [Google Scholar]
- 3.Centers for Disease Control Cryptosporidiosis: Assessment of chemotherapy of males with acquired immune deficiency syndrome (AIDS) MMWR Morb. Mortal. Wkly. Rep. 1982;31:589–592. [PubMed] [Google Scholar]
- 4.Shrivastava A., Kumar S., Smith W.A., Sahu P.S. Revisiting the global problem of cryptosporidiosis and recommendations. Trop. Parasitol. 2017;7:8–17. doi: 10.4103/2229-5070.202290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang R.-J., Li J.-Q., Chen Y.-C., Zhang L.-X., Xiao L.-H. Widespread occurrence of Cryptosporidium infections in patients with HIV/AIDS: Epidemiology, clinical feature, diagnosis, and therapy. Acta Trop. 2018;187:257–263. doi: 10.1016/j.actatropica.2018.08.018. [DOI] [PubMed] [Google Scholar]
- 6.Kotloff K.L., Nataro J.P., Blackwelder W.C., Nasrin D., Farag T.H., Panchalingam S., Wu Y., Sow S.O., Sur D., Breiman R.F., et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 7.Levine M.M., Nasrin D., Ac’acio S., Bassat Q., Powell H., Tennant S.M., Sow S.O., Sur D., Zaidi A.K., Faruque A.S.G., et al. Diarrhoeal disease and subsequent risk of death in infants and children residing in low-income and middle-income countries: Analysis of the GEMS case-control study and 12-month GEMS-1A follow-on study. Lancet Glob. Health. 2020;8:e204–e214. doi: 10.1016/S2214-109X(19)30541-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mac Kenzie W.R., Schell W.L., Blair K.A., Addiss D.G., Peterson D.E., Hoxie N.J., Kazmierczak J.J., Davis J.P. Massive Outbreak of Waterborne Cryptosporidium Infection in Milwaukee, Wisconsin: Recurrence of Illness and Risk of Secondary Transmission. Clin. Infect. Dis. 1995;21:57–62. doi: 10.1093/clinids/21.1.57. [DOI] [PubMed] [Google Scholar]
- 9.Efstratiou A., Ongerth J.E., Karanis P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2011–2016. Water Res. 2017;114:14–22. doi: 10.1016/j.watres.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 10.Ryan U., Hijjawi N., Xiao L. Foodborne cryptosporidiosis. Int. J. Parasitol. 2018;48:1–12. doi: 10.1016/j.ijpara.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Zahedi A., Ryan U. Cryptosporidium—An update with an emphasis on foodborne and waterborne transmission. Res. Vet. Sci. 2020;132:500–512. doi: 10.1016/j.rvsc.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 12.King B., Monis P.T. Critical processes affecting Cryptosporidium oocyst survival in the environment. Pt 3Parasitology. 2007;134:309–323. doi: 10.1017/S0031182006001491. [DOI] [PubMed] [Google Scholar]
- 13.Gharpure R., Perez A., Miller A.D., Wikswo M.E., Silver R., Hlavsa M.C. Cryptosporidiosis outbreaks—United States, 2009–2017. MMWR Morb. Mortal Wkly. Rep. 2019;68:568–572. doi: 10.15585/mmwr.mm6825a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunasekera S., Zahedi A., O’Dea M., King B., Monis P., Thierry B., Carr J.M., Ryan U. Organoids and bioengineered intestinal models: Potential solutions to the Cryptosporidium culturing dilemma. Microorganisms. 2020;8:715. doi: 10.3390/microorganisms8050715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerrant D.I., Lima A., Patrick P.D., Schorling J.B., Moore S., Guerrant R.L. Association of early childhood diarrhea and cryptosporidiosis with impaired physical fitness and cognitive function four-seven years later in a poor urban community in northeast Brazil. Am. J. Trop. Med. Hyg. 1999;61:707–713. doi: 10.4269/ajtmh.1999.61.707. [DOI] [PubMed] [Google Scholar]
- 16.Mondal D., Sack R.B., Haque R., Jr W.A.P., Kirkpatrick B.D. Attribution of malnutrition to cause-specific diarrheal illness: Evidence from a prospective study of preschool children in Mirpur, Dhaka, Bangladesh. Am. J. Trop. Med. Hyg. 2009;80:824–826. doi: 10.4269/ajtmh.2009.80.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter B.L., Chalmers R.M., Davies A.P. Health sequelae of human cryptosporidiosis in industrialised countries: A systematic review. Parasites Vectors. 2020;13:1–14. doi: 10.1186/s13071-020-04308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalantari N., Gorgani-Firouzjaee T., Ghaffari S., Bayani M., Ghaffari T., Chehrazi M. Association between Cryptosporidium infection and cancer: A systematic review and meta-analysis. Parasitol. Int. 2020;74:101979. doi: 10.1016/j.parint.2019.101979. [DOI] [PubMed] [Google Scholar]
- 19.Pinto D.J., Vinayak S. Cryptosporidium: Host-Parasite Interactions and Pathogenesis. Curr. Clin. Microbiol. Rep. 2021;8:62–67. doi: 10.1007/s40588-021-00159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawant M., Baydoun M., Creusy C., Chabé M., Viscogliosi E., Certad G., Benamrouz-Vanneste S. Cryptosporidium and colon cancer: Cause or consequence? Microorganisms. 2020;8:1665. doi: 10.3390/microorganisms8111665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomson S., Hamilton C.A., Hope J.C., Katzer F., Mabbott N.A., Morrison L.J., Innes E.A. Bovine cryptosporidiosis: Impact, host-parasite interaction and control strategies. Vet. Res. 2017;48:1–16. doi: 10.1186/s13567-017-0447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santin M. Cryptosporidium and Giardia in Ruminants. Vet. Clin. N. Am. Food Anim. Pract. 2020;36:223–238. doi: 10.1016/j.cvfa.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Guo Y., Li N., Ryan U., Feng Y., Xiao L. Small ruminants and zoonotic cryptosporidiosis. Parasitol. Res. 2021:1–10. doi: 10.1007/s00436-021-07116-9. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson C., Williams A., Yang R., Ryan U., Carmichael I., Campbell A.J., Gardner G. Greater intensity and frequency of Cryptosporidium and Giardia oocyst shedding beyond the neonatal period is associated with reductions in growth, carcase weight and dressing efficiency in sheep. Vet. Parasitol. 2016;228:42–51. doi: 10.1016/j.vetpar.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Jacobson C., Al-Habsi K., Ryan U., Williams A., Anderson F., Yang R., Abraham S., Miller D. Cryptosporidium infection is associated with reduced growth and diarrhoea in goats beyond weaning. Vet. Parasitol. 2018;260:30–37. doi: 10.1016/j.vetpar.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Shaw H.J., Innes E.A., Morrison L.J., Katzer F., Wells B. Long-term production effects of clinical cryptosporidiosis in neonatal calves. Int. J. Parasitol. 2020;50:371–376. doi: 10.1016/j.ijpara.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sears C.L., Kirkpatrick B.D. Is nitazoxanide an effective treatment for patients with acquired immune deficiency syndrome-related cryptosporidiosis? Nat. Clin. Pract. Gastroenterol. Hepatol. 2007;4:136–137. doi: 10.1038/ncpgasthep0737. [DOI] [PubMed] [Google Scholar]
- 28.Amadi B., Mwiya M., Sianongo S., Payne L., Watuka A., Katubulushi M., Kelly P. High dose prolonged treatment with nitazoxanide is not effective for cryptosporidiosis in HIV positive Zambian children: A randomised controlled trial. BMC Infect. Dis. 2009;9:195. doi: 10.1186/1471-2334-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashigbie P.G., Shepherd S., Steiner K.L., Amadi B., Aziz N., Manjunatha U.H., Spector J.M., Diagana T.T., Kelly P. Use-case scenarios for an anti-Cryptosporidium therapeutic. PLoS Negl. Trop. Dis. 2021;15:e0009057. doi: 10.1371/journal.pntd.0009057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crawford C.K., Kol A. The Mucosal Innate Immune Response to Cryptosporidium parvum, a Global one health issue. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.689401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao L., Feng Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017;8-9:14–32. doi: 10.1016/j.fawpar.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roellig D.M., Xiao L. Cryptosporidium Genotyping for epidemiology tracking. Methods Mol. Biol. 2020;2052:103–116. doi: 10.1007/978-1-4939-9748-0_7. [DOI] [PubMed] [Google Scholar]
- 33.Ryan U.M., Feng Y., Fayer R., Xiao L. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia—A 50 year perspective (1971–2021) Int. J. Parasitol. 2021 doi: 10.1016/j.ijpara.2021.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Adamu H., Petros B., Zhang G., Kassa H., Amer S., Ye J., Feng Y., Xiao L. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl. Trop. Dis. 2014;8:e2831. doi: 10.1371/journal.pntd.0002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouzid M., Kintz E., Hunter P. Risk factors for Cryptosporidium infection in low and middle income countries: A systematic review and meta-analysis. PLoS Negl. Trop. Dis. 2018;12:e0006553. doi: 10.1371/journal.pntd.0006553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan A., Shams S., Khan S., Khan M.I., Khan S., Ali A. Evaluation of prevalence and risk factors associated with Cryptosporidium infection in rural population of district Buner, Pakistan. PLoS ONE. 2019;14:e0209188. doi: 10.1371/journal.pone.0209188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loeck B.K., Pedati C., Iwen P.C., McCutchen E., Roellig D.M., Hlavsa M.C., Fullerton K., Safranek T., Carlson A.V. Genotyping and subtyping Cryptosporidium to identify risk factors and transmission patterns—Nebraska, 2015–2017. MMWR Morb. Mortal. Wkly. Rep. 2020;69:335–338. doi: 10.15585/mmwr.mm6912a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chalmers R.M., Robinson G., Elwin K., Elson R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasites Vectors. 2019;12:1–13. doi: 10.1186/s13071-019-3354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas-Lopez D., Müller L., Vestergaard L.S., Christoffersen M., Andersen A.-M., Jokelainen P., Agerholm J.S., Stensvold C.R. Veterinary students have a higher risk of contracting cryptosporidiosis when calves with high fecal Cryptosporidium loads are used for fetotomy exercises. Appl. Environ. Microbiol. 2020;86:e01250-20. doi: 10.1128/AEM.01250-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Ryan U., Guo Y., Feng Y., Xiao L. Advances in molecular epidemiology of cryptosporidiosis in dogs and cats. Int. J. Parasitol. 2021;51:787–795. doi: 10.1016/j.ijpara.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Smith R.P., Newton K., Rimdap E., Wight A., Robinson G., Chalmers R.M. Review of investigations of premises housing animals that were linked to human outbreaks of cryptosporidiosis in England and Wales between 2009 and 2019. Vet. Rec. 2021;189:e246. doi: 10.1002/vetr.246. [DOI] [PubMed] [Google Scholar]
- 42.Strong W.B., Gut J., Nelson R.G. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of Its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 2000;68:4117–4134. doi: 10.1128/IAI.68.7.4117-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulaiman I.M., Hira P.R., Zhou L., Al-Ali F.M., Al-Shelahi F.A., Shweiki H.M., Iqbal J., Khalid N., Xiao L. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 2005;43:2805–2809. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li N., Xiao L., Alderisio K., Elwin K., Cebelinski E., Chalmers R., Santín M., Fayer R., Kvac M., Ryan U., et al. Subtyping Cryptosporidium ubiquitum, A zoonotic pathogen emerging in humans. Emerg. Infect. Dis. 2014;20:217–224. doi: 10.3201/eid2002.121797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Y., Cebelinski E., Matusevich C., Alderisio K.A., Lebbad M., McEvoy J., Roellig D.M., Yang C., Feng Y., Xiao L. Subtyping novel zoonotic pathogen Cryptosporidium chipmunk genotype I. J. Clin. Microbiol. 2015;53:1648–1654. doi: 10.1128/JCM.03436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rojas-López L., Elwin K., Chalmers R.M., Enemark H.L., Beser J., Troell K. Development of a gp60-subtyping method for Cryptosporidium felis. Parasites Vectors. 2020;13:1–8. doi: 10.1186/s13071-020-3906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X., Huang N., Jiang W., Wang X., Li N., Guo Y., Kváč M., Feng Y., Xiao L. Subtyping Cryptosporidium ryanae: A Common pathogen in bovine animals. Microorganisms. 2020;8:1107. doi: 10.3390/microorganisms8081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan Y., Huang X., Guo S., Yang F., Yang X., Guo Y., Feng Y., Xiao L., Li N. Subtyping Cryptosporidium xiaoi, a common pathogen in sheep and goats. Pathogens. 2021;10:800. doi: 10.3390/pathogens10070800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang W., Roellig D.M., Guo Y., Li N., Feng Y., Xiao L. Development of a subtyping tool for zoonotic pathogen Cryptosporidium canis. J. Clin. Microbiol. 2021;59:1648–1654. doi: 10.1128/JCM.02474-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W., Wan M., Yang F., Li N., Xiao L., Feng Y., Guo Y. Development and Application of a gp60-Based Subtyping Tool for Cryptosporidium bovis. Microorganisms. 2021;9:2067. doi: 10.3390/microorganisms9102067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lebbad M., Winiecka-Krusnell J., Stensvold C., Beser J. High Diversity of Cryptosporidium Species and subtypes identified in cryptosporidiosis acquired in sweden and abroad. Pathogens. 2021;10:523. doi: 10.3390/pathogens10050523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guy R.A., Yanta C.A., Muchaal P.K., Rankin M.A., Thivierge K., Lau R., Boggild A.K. Molecular characterization of Cryptosporidium isolates from humans in Ontario, Canada. Parasites Vectors. 2021;14:1–14. doi: 10.1186/s13071-020-04546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia–R J.C., French N., Pita A., Velathanthiri N., Shrestha R., Hayman D. Local and global genetic diversity of protozoan parasites: Spatial distribution of Cryptosporidium and Giardia genotypes. PLoS Negl. Trop. Dis. 2017;11:e0005736. doi: 10.1371/journal.pntd.0005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia–R J.C., Pita A.B., Velathanthiri N., French N.P., Hayman D.T.S. Species and genotypes causing human cryptosporidiosis in New Zealand. Parasitol. Res. 2020;119:2317–2326. doi: 10.1007/s00436-020-06729-w. [DOI] [PubMed] [Google Scholar]
- 55.Koehler A.V., Whipp M.J., Haydon S.R., Gasser R.B. Cryptosporidium cuniculus—New records in human and kangaroo in Australia. Parasites Vectors. 2014;7:492. doi: 10.1186/s13071-014-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puleston R.L., Mallaghan C.M., Modha D.E., Hunter P., Nguyen-Van-Tam J., Regan C.M., Nichols G.L., Chalmers R.M. The first recorded outbreak of cryptosporidiosis due to Cryptosporidium cuniculus (formerly rabbit genotype), following a water quality incident. J. Water Health. 2014;12:41–50. doi: 10.2166/wh.2013.097. [DOI] [PubMed] [Google Scholar]
- 57.Martínez-Ruiz R., de Lucio A., Fuentes I., Carmena D. Autochthonous Cryptosporidium cuniculus infection in Spain: First report in a symptomatic paediatric patient from Madrid. Enferm. Infecc. Microbiol. Clin. 2016;34:532–534. doi: 10.1016/j.eimc.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 58.Chalmers R.M., Elwin K., Hadfield S.J., Robinson G. Sporadic human cryptosporidiosis caused by Cryptosporidium cuniculus, United Kingdom, 2007–2008. Emerg. Infect. Dis. 2011;17:536–538. doi: 10.3201/eid1703.100410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Molloy S.F., Kirwan P., Asaolu S.O., Holland C.V., Nichols R.A.B., Connelly L., Smith H.V. Identification of a High Diversity of Cryptosporidium Species Genotypes and Subtypes in a Pediatric Population in Nigeria. Am. J. Trop. Med. Hyg. 2010;82:608–613. doi: 10.4269/ajtmh.2010.09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anofel Cryptosporidium National Network Laboratory-based surveillance for Cryptosporidium in France, 2006–2009. Eurosurveilliance. 2010;15:19642. [PubMed] [Google Scholar]
- 61.Chalmers R.M., Robinson G., Elwin K., Hadfield S.J., Xiao L., Ryan U., Modha D., Mallaghan C. Cryptosporidium sp. Rabbit genotype, a newly identified human pathogen. Emerg. Infect. Dis. 2009;15:829–830. doi: 10.3201/eid1505.081419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muadica A.S., Köster P.C., Dashti A., Bailo B., Hernández-de-Mingo M., Balasegaram S., Carmena D. Molecular diversity of Giardia duodenalis, Cryptosporidium spp. and Blastocystis sp. in symptomatic and asymptomatic school children in Zambezia province (Mozambique) Pathogens. 2021;10:255. doi: 10.3390/pathogens10030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sardar S.K., Ghosal A., Saito-Nakano Y., Dutta S., Nozaki T., Ganguly S. Molecular Identification of Cryptosporidium viatorum infection in a patient suffering from unusual cryptosporidiosis in West Bengal, India. Korean J. Parasitol. 2021;59:409–413. doi: 10.3347/kjp.2021.59.4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tarekegn Z.S., Tigabu Y., Dejene H. Cryptosporidium infection in cattle and humans in Ethiopia: A systematic review and meta-analysis. Parasite Epidemiol. Control. 2021;14:e00219. doi: 10.1016/j.parepi.2021.e00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu Y., Gong B., Liu X., Jiang Y., Cao J., Yao L., Li H., Liu A., Shen Y. Identification of uncommon Cryptosporidium viatorum (a novel subtype XVcA2G1c) and Cryptosporidium andersoni as well as common Giardia duodenalis assemblages A and B in humans in Myanmar. Front. Cell. Infect. Microbiol. 2020;10:740. doi: 10.3389/fcimb.2020.614053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Braima K., Zahedi A., Oskam C., Reid S., Pingault N., Xiao L., Ryan U. Retrospective analysis of Cryptosporidium species in Western Australian human populations (2015–2018), and emergence of the C. hominis IfA12G1R5 subtype. Infect. Genet. Evol. 2019;73:306–313. doi: 10.1016/j.meegid.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 67.Mirdha B.R., Khalil S., Paul J., Panda A., Singh Y. Molecular Detection and Identification of Cryptosporidium viatorum in a Human Immunodeficiency Virus-seropositive Patient. J. Glob. Infect. Dis. 2018;10:28–29. doi: 10.4103/jgid.jgid_26_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sánchez A., Munoz M., Gómez N., Tabares J., Segura L., Salazar Á., Restrepo C., Ruíz M., Reyes P., Qian Y., et al. Molecular Epidemiology of Giardia, Blastocystis and Cryptosporidium among Indigenous Children from the Colombian Amazon Basin. Front. Microbiol. 2017;8:248. doi: 10.3389/fmicb.2017.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ukwah B.N., Ezeonu I.M., Ezeonu C.T., Roellig D., Xiao L. Cryptosporidium species and subtypes in diarrheal children and HIV-infected persons in Ebonyi and Nsukka, Nigeria. J. Infect. Dev. Ctries. 2017;11:173–179. doi: 10.3855/jidc.8034. [DOI] [PubMed] [Google Scholar]
- 70.Ayinmode A.B., Zhang H., Dada-Adegbola H.O., Xiao L. Cryptosporidium hominis Subtypes and Enterocytozoon bieneusi Genotypes in HIV-Infected Persons in Ibadan, Nigeria. Zoonoses Public Health. 2014;61:297–303. doi: 10.1111/zph.12072. [DOI] [PubMed] [Google Scholar]
- 71.Lebbad M., Beser J., Insulander M., Karlsson L., Mattsson J.G., Svenungsson B., Axen C. Unusual cryptosporidiosis cases in Swedish patients: Extended molecular characterization of Cryptosporidium viatorum and Cryptosporidium chipmunk genotype I. J. Parasitol. 2013;140:1735–1740. doi: 10.1017/S003118201300084X. [DOI] [PubMed] [Google Scholar]
- 72.Insulander M., Silverlås C., Lebbad M., Karlsson L., Mattsson J.G., Svenungsson B. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol. Infect. 2013;141:1009–1020. doi: 10.1017/S0950268812001665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elwin K., Hadfield S.J., Robinson G., Crouch N.D., Chalmers R.M. Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007–2011. Int. J. Parasitol. 2012;42:675–682. doi: 10.1016/j.ijpara.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 74.De Lucio A., Amor-Aramendía A., Bailo B., Saugar J.M., Anegagrie M., Arroyo A., López-Quintana B., Zewdie D., Ayehubizu Z., Yizengaw E., et al. Prevalence and Genetic Diversity of Giardia duodenalis and Cryptosporidium spp. among School Children in a Rural Area of the Amhara Region, North-West Ethiopia. PLoS ONE. 2016;11:e0159992. doi: 10.1371/journal.pone.0159992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bujila I., Troell K., Fischerström K., Nordahl M., Killander G., Hansen A., Söderlund R., Lebbad M., Beser J. Cryptosporidium chipmunk genotype I—An emerging cause of human cryptosporidiosis in Sweden. Infect. Genet. Evol. 2021;92:104895. doi: 10.1016/j.meegid.2021.104895. [DOI] [PubMed] [Google Scholar]
- 76.Xu Z., Guo Y., Roellig D.M., Feng Y., Xiao L. Comparative analysis reveals conservation in genome organization among intestinal Cryptosporidium species and sequence divergence in potential secreted pathogenesis determinants among major human-infecting species. BMC Genom. 2019;20:406. doi: 10.1186/s12864-019-5788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feltus D.C., Giddings C.W., Schneck B.L., Monson T., Warshauer D., McEvoy J.M. Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. J. Clin. Microbiol. 2006;44:4303–4308. doi: 10.1128/JCM.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Higuera A., Villamizar X., Herrera G., Giraldo J.C., Vasquez-A. L.R., Urbano P., Villalobos O., Tovar C., Ramírez J.D. Molecular detection and genotyping of intestinal protozoa from different biogeographical regions of Colombia. PeerJ. 2020;8:e8554. doi: 10.7717/peerj.8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sannella A.R., Suputtamongkol Y., Wongsawat E., Cacciò S.M. A retrospective molecular study of Cryptosporidium species and genotypes in HIV-infected patients from Thailand. Parasites Vectors. 2019;12:91. doi: 10.1186/s13071-019-3348-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ayinmode A.B., Oliveira B.C.M., Obebe O.O., Dada-Adgebola H.O., Ayede A.I., Widmer G., Dada-Adegbola H. Genotypic Characterization of Cryptosporidium Species in Humans and Peri-Domestic Animals in Ekiti and Oyo States, Nigeria. J. Parasitol. 2018;104:639–644. doi: 10.1645/17-74. [DOI] [PubMed] [Google Scholar]
- 81.Kattula D., Jeyavelu N., Prabhakaran A.D., Premkumar P.S., Velusamy V., Venugopal S., Geetha J.C., Lazarus R.P., Das P., Nithyanandhan K., et al. Natural History of Cryptosporidiosis in a Birth Cohort in Southern India. Clin. Infect. Dis. 2017;64:347–354. doi: 10.1093/cid/ciw730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elwin K., Hadfield S.J., Robinson G., Chalmers R.M. The epidemiology of sporadic human infections with unusual cryptosporidia detected during routine typing in England and Wales, 2000–2008. Epidemiol. Infect. 2011;140:673–683. doi: 10.1017/S0950268811000860. [DOI] [PubMed] [Google Scholar]
- 83.Al-Brikan F.A., Salem H.S., Beeching N., Hilal N. Multilocus genetic analysis of Cryptosporidium isolates from Saudi Arabia. J. Egypt. Soc. Parasitol. 2008;38:645–658. [PubMed] [Google Scholar]
- 84.Gatei W., Kamwati S.K., Mbae C., Waruru A., Hart C.A., Wamae C.N., Revathi G., Mulinge E., Gatika S.M., Waithera T. Cryptosporidiosis: Prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am. J. Trop. Med. Hyg. 2006;75:78–82. doi: 10.4269/ajtmh.2006.75.78. [DOI] [PubMed] [Google Scholar]
- 85.Muthusamy D., Rao S.S., Ramani S., Monica B., Banerjee I., Abraham O.C., Mathai D.C., Primrose B., Muliyil J., Wanke C.A., et al. Multilocus Genotyping of Cryptosporidium sp. Isolates from Human immunodeficiency virus-infected individuals in South India. J. Clin. Microbiol. 2006;44:632–634. doi: 10.1128/JCM.44.2.632-634.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Palmer C.J., Xiao L., Terashima A., Guerra H., Gotuzzo E., Saldias G., Bonilla J.A., Zhou L., Lindquist A., Upton S.J. Cryptosporidium muris, a rodent pathogen, recovered from a human in Peru. Emerg. Infect. Dis. 2003;9:1174–1176. doi: 10.3201/eid0909.030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gatei W., Ashford R.W., Beeching N.J., Kamwati S.K., Greensill J., Hart C.A. Cryptosporidium muris infection in an HIV-infected adult, Kenya. Emerg. Infect. Dis. 2002;8:204–206. doi: 10.3201/eid0802.010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tiangtip R., Jongwutiwes S. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop. Med. Int. Health. 2002;7:357–364. doi: 10.1046/j.1365-3156.2002.00855.x. [DOI] [PubMed] [Google Scholar]
- 89.Katsumata T., Hosea D., Uga S., Kohno S., Ranuh I.G., Yanagi T. Short report: Possible Cryptosporidium muris infection in humans. Am. J. Trop. Med. Hyg. 2000;62:70–72. doi: 10.4269/ajtmh.2000.62.70. [DOI] [PubMed] [Google Scholar]
- 90.Hussain G., Roychoudhury S., Singha B., Paul J. Incidence of Cryptosporidium andersoni in diarrheal patients from southern Assam, India: A molecular approach. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:1023–1032. doi: 10.1007/s10096-016-2887-2. [DOI] [PubMed] [Google Scholar]
- 91.Jiang Y., Ren J., Yuan Z., Liu A., Zhao H., Liu H., Chu L., Pan W., Cao J., Lin Y., et al. Cryptosporidium andersoni as a novel predominant Cryptosporidium species in outpatients with diarrhea in Jiangsu Province, China. BMC Infect. Dis. 2014;14:1–6. doi: 10.1186/s12879-014-0555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu H., Shen Y., Yin J., Yuan Z., Jiang Y., Xu Y., Pan W., Hu Y., Cao J. Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon, Giardia and Cyclospora in diarrheal outpatients in china. BMC Infect. Dis. 2014;14:25. doi: 10.1186/1471-2334-14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Agholi M., Hatam G.M., Motazedian M.H. HIV/AIDS-associated opportunistic protozoal diarrhoea. AIDS Res. Hum. Retrovir. 2013;29:35–41. doi: 10.1089/aid.2012.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Waldron L.S., Dimeski B., Beggs P., Ferrari B., Power M.L. Molecular Epidemiology, Spatiotemporal Analysis, and Ecology of Sporadic Human Cryptosporidiosis in Australia. Appl. Environ. Microbiol. 2011;77:7757–7765. doi: 10.1128/AEM.00615-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morse T.D., Nichols R.A.B., Grimason A.M., Campbell B.M., Tembo K.C., Smith H.V. Incidence of cryptosporidiosis species in paediatric patients in Malawi. Epidemiol. Infect. 2007;135:1307–1315. doi: 10.1017/S0950268806007758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leoni F., Amar C., Nichols G., Pedraza-Díaz S., McLauchlin J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J. Med. Microbiol. 2006;55:703–707. doi: 10.1099/jmm.0.46251-0. [DOI] [PubMed] [Google Scholar]
- 97.Guyot K., Follet-Dumoulin A., Lelièvre E., Sarfati C., Rabodonirina M., Nevez G., Cailliez J.C., Camus D., Dei-Cas E. Molecular Characterization of Cryptosporidium Isolates Obtained from Humans in France. J. Clin. Microbiol. 2001;39:3472–3480. doi: 10.1128/JCM.39.10.3472-3480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu A., Gong B., Liu X., Shen Y., Wu Y., Zhang W., Cao J. A retrospective epidemiological analysis of human Cryptosporidium infection in China during the past three decades (1987–2018) PLoS Negl. Trop. Dis. 2020;14:e0008146. doi: 10.1371/journal.pntd.0008146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moore C.E., Elwin K., Phot N., Seng C., Mao S., Suy K., Kumar V., Nader J., Bousfield R., Perera S., et al. Molecular Characterization of Cryptosporidium Species and Giardia duodenalis from Symptomatic Cambodian Children. PLoS Negl. Trop. Dis. 2016;10:e0004822. doi: 10.1371/journal.pntd.0004822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bodager J.R., Parsons M.B., Wright P.C., Rasambainarivo F., Roellig D., Xiao L., Gillespie T.R. Complex epidemiology and zoonotic potential for Cryptosporidium suis in rural Madagascar. Vet. Parasitol. 2015;207:140–143. doi: 10.1016/j.vetpar.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 101.Wang L., Zhang H.-W., Zhao X., Zhang L., Zhang G., Guo M., Liu L., Feng Y., Xiao L. Zoonotic Cryptosporidium Species and Enterocytozoon bieneusi Genotypes in HIV-positive patients on antiretroviral therapy. J. Clin. Microbiol. 2012;51:557–563. doi: 10.1128/JCM.02758-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cama V.A., Ross J., Crawford S., Kawai V., Chavez-Valdez R., Vargas-Pacherrez D., Vivar A., Ticona E., Ñavincopa M., Williamson J., et al. Differences in Clinical Manifestations among Cryptosporidium Species and Subtypes in HIV-Infected Persons. J. Infect. Dis. 2007;196:684–691. doi: 10.1086/519842. [DOI] [PubMed] [Google Scholar]
- 103.Xiao L., Bern C., Arrowood M., Sulaiman I., Zhou L., Kawai V., Vivar A., Lal A.A., Gilman R.H. Identification of the Cryptosporidium Pig genotype in a human patient. J. Infect. Dis. 2002;185:1846–1848. doi: 10.1086/340841. [DOI] [PubMed] [Google Scholar]
- 104.Xiao L., Hlavsa M.C., Yoder J., Ewers C., Dearen T., Yang W., Nett R., Harris S., Brend S.M., Harris M., et al. Subtype analysis of Cryptosporidium specimens from sporadic cases in Colorado, Idaho, New Mexico, and Iowa in 2007: Widespread Occurrence of One Cryptosporidium hominis subtype and case history of an infection with the Cryptosporidium Horse Genotype. J. Clin. Microbiol. 2009;47:3017–3020. doi: 10.1128/JCM.00226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Robinson G., Elwin K., Chalmers R.M. Unusual Cryptosporidium genotypes in human cases of diarrhea. Emerg. Infect. Dis. 2008;14:1800–1802. doi: 10.3201/eid1411.080239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Costa D., Razakandrainibe R., Sautour M., Valot S., Basmaciyan L., Gargala G., Lemeteil D., Favennec L., Dalle F., Debourgogne A., et al. Human cryptosporidiosis in immunodeficient patients in France (2015–2017) Exp. Parasitol. 2018;192:108–112. doi: 10.1016/j.exppara.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 107.Kvac M., Saková K., Květoňová D., Kicia M., Wesołowska M., McEvoy J., Sak B. Gastroenteritis Caused by the Cryptosporidium Hedgehog Genotype in an Immunocompetent Man. J. Clin. Microbiol. 2014;52:347–349. doi: 10.1128/JCM.02456-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khan S.M., Debnath C., Pramanik A.K., Xiao L., Nozaki T., Ganguly S. Molecular characterization and assessment of zoonotic transmission of Cryptosporidium from dairy cattle in West Bengal, India. Vet. Parasitol. 2010;171:41–47. doi: 10.1016/j.vetpar.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 109.Ng J.S.Y., Eastwood K., Walker B., Durrheim D.N., Massey P.D., Porigneaux P., Kemp R., McKinnon B., Laurie K., Miller D., et al. Evidence of Cryptosporidium transmission between cattle and humans in northern New South Wales. Exp. Parasitol. 2012;130:437–441. doi: 10.1016/j.exppara.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 110.Helmy Y.A., Krücken J., Nöckler K., von Samson-Himmelstjerna G., Zessin K.-H. Molecular epidemiology of Cryptosporidium in livestock animals and humans in the Ismailia province of Egypt. Vet. Parasitol. 2013;193:15–24. doi: 10.1016/j.vetpar.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 111.Yan W., Alderisio K., Roellig D.M., Elwin K., Chalmers R.M., Yang F., Wang Y., Feng Y., Xiao L. Subtype analysis of zoonotic pathogen Cryptosporidium skunk genotype. Infect. Genet. Evol. 2017;55:20–25. doi: 10.1016/j.meegid.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Davies A.P., Campbell B., Evans M.R., Bone A., Roche A., Chalmers R.M. Asymptomatic carriage of protozoan parasites in children in day care centers in the United Kingdom. Pediatr. Infect. Dis. J. 2009;28:838–840. doi: 10.1097/INF.0b013e31819d646d. [DOI] [PubMed] [Google Scholar]
- 113.Rasková V., Kvetonová D., Sak B., McEvoy J., Edwinson A., Stenger B., Kvác M. Human cryptosporidiosis is caused by Cryptosporidium tyzzeri and C. parvum isolates presumably transmitted from wild mice. J. Clin. Microbiol. 2013;51:360–362. doi: 10.1128/JCM.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu N., Liu H., Jiang Y., Yin J., Yuan Z., Shen Y., Cao J. First report of Cryptosporidium viatorum and Cryptosporidium occultus in humans in China, and of the unique novel C. viatorum subtype XVaA3h. BMC Infect. Dis. 2020;20:16. doi: 10.1186/s12879-019-4693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ong C.S.L., Eisler D.L., Alikhani A., Fung V.W.K., Tomblin J., Bowie W.R., Isaac-Renton J.L. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: First report of human infections with a cervine genotype. Emerg. Infect. Dis. 2002;8:263–268. doi: 10.3201/eid0803.010194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Beser J., Bujila I., Wittesjö B., Lebbad M. From mice to men: Three cases of human infection with Cryptosporidium ditrichi. Infect. Genet. Evol. 2020;78:104120. doi: 10.1016/j.meegid.2019.104120. [DOI] [PubMed] [Google Scholar]
- 117.Ng-Hublin J.S.Y., Combs B., MacKenzie B., Ryan U. Human Cryptosporidiosis Diagnosed in Western Australia: A Mixed Infection with Cryptosporidium meleagridis, the Cryptosporidium Mink Genotype, and an Unknown Cryptosporidium Species. J. Clin. Microbiol. 2013;51:2463–2465. doi: 10.1128/JCM.00424-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ebner J., Koehler A., Robertson G., Bradbury R., Jex A., Haydon S., Stevens M.A., Norton R., Joachim A., Gasser R.B. Genetic analysis of Giardia and Cryptosporidium from people in Northern Australia using PCR-based tools. Infect. Genet. Evol. 2015;29:389–395. doi: 10.1016/j.meegid.2015.08.034. [DOI] [PubMed] [Google Scholar]
- 119.Waldron L.S., Cheung-Kwok-Sang C., Power M.L. Wildlife-associated Cryptosporidium fayeri in human, Australia. Emerg. Infect. Dis. 2010;16:2006–2007. doi: 10.3201/eid1612.100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Braima K., Zahedi A., Oskam C., Austen J., Egan S., Reid S., Ryan U. Zoonotic infection by Cryptosporidium fayeri IVgA10G1T1R1 in a Western Australian human. Zoonoses Public Health. 2021;68:358–360. doi: 10.1111/zph.12806. [DOI] [PubMed] [Google Scholar]
- 121.Kváč M., Květoňová D., Sak B., Ditrich O. Cryptosporidium Pig genotype II in immunocompetent man. Emerg. Infect. Dis. 2009;15:982–983. doi: 10.3201/eid1506.071621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Robertson L.J., Johansen H., Kifleyohannes T., Efunshile A.M., Terefe G. Cryptosporidium Infections in Africa—How important is zoonotic transmission? A review of the evidence. Front. Vet. Sci. 2020;7:575881. doi: 10.3389/fvets.2020.575881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barrera J.P., Carmena D., Rodríguez E., Checa R., López A.M., Fidalgo L.E., Gálvez R., Marino V., Fuentes I., Miró G., et al. The red fox (Vulpes vulpes) as a potential natural reservoir of human cryptosporidiosis by Cryptosporidium hominis in Northwest Spain. Transbound. Emerg. Dis. 2020 doi: 10.1111/tbed.13569. [DOI] [PubMed] [Google Scholar]
- 124.Golomazou E., Malandrakis E.E., Panagiotaki P., Karanis P. Cryptosporidium in fish: Implications for aquaculture and beyond. Water Res. 2021;201:117357. doi: 10.1016/j.watres.2021.117357. [DOI] [PubMed] [Google Scholar]
- 125.Widmer G., Köster P.C., Carmena D. Cryptosporidium hominis infections in non-human animal species: Revisiting the concept of host specificity. Int. J. Parasitol. 2020;50:253–262. doi: 10.1016/j.ijpara.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 126.Chappell C.L., Tzipori S., Akiyoshi D.E., Okhuysen P., Tanriverdi S., Langer-Curry R., Widmer G. Cryptosporidium hominis: Experimental challenge of healthy adults. Am. J. Trop. Med. Hyg. 2006;75:851–857. doi: 10.4269/ajtmh.2006.75.851. [DOI] [PubMed] [Google Scholar]
- 127.Abeywardena H., Jex A.R., Nolan M.J., Haydon S.R., Stevens M.A., McAnulty R.W., Gasser R.B. Genetic characterisation of Cryptosporidium and Giardia from dairy calves: Discovery of species/genotypes consistent with those found in humans. Infect. Genet. Evol. 2012;12:1984–1993. doi: 10.1016/j.meegid.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 128.Park J.-H., Guk S.-M., Han E.-T., Shin E.-H., Kim J.-L., Chai J.-Y. Genotype analysis of Cryptosporidium spp. prevalent in a rural village in Hwasun-gun, Republic of Korea. Korean J. Parasitol. 2006;44:27–33. doi: 10.3347/kjp.2006.44.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ng J., Yang R., Whiffin V., Cox P., Ryan U. Identification of zoonotic Cryptosporidium and Giardia genotypes infecting animals in Sydney’s water catchments. Exp. Parasitol. 2011;128:138–144. doi: 10.1016/j.exppara.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 130.Koehler A.V., Haydon S., Jex A.R., Gasser R.B. Cryptosporidium and Giardia taxa in faecal samples from animals in catchments supplying the city of Melbourne with drinking water (2011 to 2015) Parasites Vectors. 2016;9:315. doi: 10.1186/s13071-016-1607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zahedi A., Monis P., Gofton A., Oskam C.L., Ball A., Bath A., Bartkow M., Robertson I., Ryan U. Cryptosporidium species and subtypes in animals inhabiting drinking water catchments in three states across Australia. Water Res. 2018;134:327–340. doi: 10.1016/j.watres.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 132.Krawczyk A.I., Van Leeuwen A.D., Jacobs-Reitsma W., Wijnands L.M., Bouw E., Jahfari S., Hoek A.H.A.M.V., Van Der Giessen J.W.B., Roelfsema J.H., Kroes M., et al. Presence of zoonotic agents in engorged ticks and hedgehog faeces from Erinaceus europaeus in (sub) urban areas. Parasites Vectors. 2015;8:1–6. doi: 10.1186/s13071-015-0814-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Danišová O., Valenčáková A., Stanko M., Luptáková L., Hatalová E., Canády A. Rodents as a reservoir of infection caused by multiple zoonotic species genotypes of C. parvum, C. hominis, C. suis, C. scrofarum, and the first evidence of C. muskrat genotypes I and II of rodents in Europe. Acta Trop. 2017;172:29–35. doi: 10.1016/j.actatropica.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 134.Condlová Š., Horčičková M., Sak B., Květoňová D., Hlásková L., Konecny R., Stanko M., McEvoy J., Kváč M. Cryptosporidium apodemi sp. n. and Cryptosporidium ditrichi sp. n. (Apicomplexa: Cryptosporidiidae) in Apodemus spp. Eur. J. Protistol. 2018;63:1–12. doi: 10.1016/j.ejop.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 135.Jian F., Liu A., Wang R., Zhang S., Qi M., Zhao W., Shi Y., Wang J., Wei J., Zhang L., et al. Common occurrence of Cryptosporidium hominis in horses and donkeys. Infect. Genet. Evol. 2016;43:261–266. doi: 10.1016/j.meegid.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 136.Inácio S.V., Widmer G., De Brito R.L.L., Zucatto A.S., De Aquino M.C.C., Oliveira B.C.M., Nakamura A.A., Neto L.D.S., Carvalho J.G.B., Gomes J.F., et al. First description of Cryptosporidium hominis GP60 genotype IkA20G1 and Cryptosporidium parvum GP60 genotypes IIaA18G3R1 and IIaA15G2R1 in foals in Brazil. Vet. Parasitol. 2017;233:48–51. doi: 10.1016/j.vetpar.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 137.Li F., Su J., Chahan B., Guo Q., Wang T., Yu Z., Guo Y., Li N., Feng Y., Xiao L. Different distribution of Cryptosporidium species between horses and donkeys. Infect. Genet. Evol. 2019;75:103954. doi: 10.1016/j.meegid.2019.103954. [DOI] [PubMed] [Google Scholar]
- 138.Wang W., Zhang Z., Zhang Y., Zhao A., Jing B., Zhang L., Liu P., Qi M., Zhao W. Prevalence and genotypic identification of Cryptosporidium in free-ranging and farm-raised donkeys (Equus asinus asinus) in Xinjiang, China. Parasite. 2020;27:45. doi: 10.1051/parasite/2020042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zahedi A., Monis P., Aucote S., King B., Paparini A., Jian F., Yang R., Oskam C., Ball A., Robertson I., et al. Zoonotic Cryptosporidium species in animals inhabiting sydney water catchments. PLoS ONE. 2016;11:e0168169. doi: 10.1371/journal.pone.0168169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zahedi A., Gofton A.W., Jian F., Paparini A., Oskam C., Ball A., Robertson I., Ryan U. Next Generation Sequencing uncovers within-host differences in the genetic diversity of Cryptosporidium gp60 subtypes. Int. J. Parasitol. 2017;47:601–607. doi: 10.1016/j.ijpara.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 141.Zahedi A., Paparini A., Jian F., Robertson I., Ryan U. Public health significance of zoonotic Cryptosporidium species in wildlife: Critical insights into better drinking water management. Int. J. Parasitol. Parasites Wildl. 2016;5:88–109. doi: 10.1016/j.ijppaw.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Feng Y., Ryan U.M., Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34:997–1011. doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 143.Hatam-Nahavandi K., Ahmadpour E., Carmena D., Spotin A., Bangoura B., Xiao L. Cryptosporidium infections in terrestrial ungulates with focus on livestock: A systematic review and meta-analysis. Parasites Vectors. 2019;12:1–23. doi: 10.1186/s13071-019-3704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Feng Y., Xiao L. Molecular Epidemiology of Cryptosporidiosis in China. Front. Microbiol. 2017;8:1701. doi: 10.3389/fmicb.2017.01701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.King P., Tyler K.M., Hunter P.R. Anthroponotic transmission of Cryptosporidium parvum predominates in countries with poorer sanitation: A systematic review and meta-analysis. Parasites Vectors. 2019;12:16. doi: 10.1186/s13071-018-3263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nader J.L., Mathers T., Ward B.J., Pachebat J., Swain M.T., Robinson G., Chalmers R.M., Hunter P.R., Van Oosterhout C., Tyler K.M. Evolutionary genomics of anthroponosis in Cryptosporidium. Nat. Microbiol. 2019;4:826–836. doi: 10.1038/s41564-019-0377-x. [DOI] [PubMed] [Google Scholar]
- 147.Lange H., Johansen O.H., Vold L., Robertson L.J., Anthonisen I.L., Nygard K. Second outbreak of infection with a rare Cryptosporidium parvum genotype in schoolchildren associated with contact with lambs/goat kids at a holiday farm in Norway. Epidemiol. Infect. 2014;142:2105–2113. doi: 10.1017/S0950268813003002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Feng Y., Torres E., Li N., Wang L., Bowman D., Xiao L. Population genetic characterisation of dominant Cryptosporidium parvum subtype IIaA15G2R1. Int. J. Parasitol. 2013;43:1141–1147. doi: 10.1016/j.ijpara.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 149.Fayer R., Santín M., Macarisin D. Cryptosporidium ubiquitum n. sp. in animals and humans. Vet. Parasitol. 2010;172:23–32. doi: 10.1016/j.vetpar.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 150.Tang Y., Li N., Song M., Roellig D.M., Feng Y., Xiao L. Development of a multilocus sequence typing tool for high-resolution subtyping and genetic structure characterization of Cryptosporidium ubiquitum. Infect. Genet. Evol. 2016;45:256–261. doi: 10.1016/j.meegid.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lindsay D.S., Upton S.J., Owens D.S., Morgan U.M., Mead J.R., Blagburn B.L. Cryptosporidium andersoni n. sp. (Apicomplexa: Cryptosporiidae) from Cattle, Bos taurus. J. Eukaryot. Microbiol. 2000;47:91–95. doi: 10.1111/j.1550-7408.2000.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 152.Huang J., Zhang Z., Zhang Y., Yang Y., Zhao J., Wang R., Jian F., Ning C., Zhang W., Zhang L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in deer in Henan and Jilin, China. Parasites Vectors. 2018;11:239. doi: 10.1186/s13071-018-2813-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen S., Chai Y., Deng L., Liu H., Zhong Z., Fu H., Hu Y., Shen L., Zhou Z., Geng Y., et al. Cryptosporidium spp. in Pet Dwarf Winter White Russian Hamsters (Phodopus sungoris sungoris) in China. J. Parasitol. 2021;107:770–777. doi: 10.1645/20-102. [DOI] [PubMed] [Google Scholar]
- 154.Chen J., Wang W., Lin Y., Sun L., Li N., Guo Y., Kvac M., Ryan U., Feng Y., Xiao L. Genetic characterizations of Cryptosporidium spp. from pet rodents indicate high zoonotic potential of pathogens from chinchillas. One Health. 2021;13:100269. doi: 10.1016/j.onehlt.2021.100269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ježková J., Prediger J., Holubová N., Sak B., Konečný R., Feng Y., Xiao L., Rost M., McEvoy J., Kváč M. Cryptosporidium ratti n. sp. (Apicomplexa: Cryptosporidiidae) and genetic diversity of Cryptosporidium spp. in brown rats (Rattus norvegicus) in the Czech Republic. Parasitology. 2021;148:84–97. doi: 10.1017/S0031182020001833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lv C., Zhang L., Wang R., Jian F., Zhang S., Ning C., Wang H., Feng C., Wang X., Ren X., et al. Cryptosporidium spp. in Wild, Laboratory, and Pet Rodents in China: Prevalence and molecular characterization. Appl. Environ. Microbiol. 2009;75:7692–7699. doi: 10.1128/AEM.01386-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Feng Y., Yang W., Ryan U., Zhang L., Kváč M., Koudela B., Modrý D., Li N., Fayer R., Xiao L. Development of a Multilocus Sequence Tool for Typing Cryptosporidium muris and Cryptosporidium andersoni. J. Clin. Microbiol. 2011;49:34–41. doi: 10.1128/JCM.01329-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang R., Jian F., Zhang L., Ning C., Liu A., Zhao J., Feng Y., Qi M., Wang H., Lv C., et al. Multilocus sequence subtyping and genetic structure of Cryptosporidium muris and Cryptosporidium andersoni. PLoS ONE. 2012;7:e43782. doi: 10.1371/journal.pone.0043782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhao W., Wang R., Zhang W., Liu A., Cao J., Shen Y., Yang F., Zhang L. MLST Subtypes and Population Genetic Structure of Cryptosporidium andersoni from dairy cattle and beef cattle in northeastern china’s heilongjiang province. PLoS ONE. 2014;9:e102006. doi: 10.1371/journal.pone.0102006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kváč M., Hanzlíková D., Sak B., Květoňová D. Prevalence and age-related infection of Cryptosporidium suis, C. muris and Cryptosporidium pig genotype II in pigs on a farm complex in the Czech Republic. Vet. Parasitol. 2009;160:319–322. doi: 10.1016/j.vetpar.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 161.García-Presedo I., Pedraza-Díaz S., González-Warleta M., Mezo M., Gómez-Bautista M., Ortega-Mora L.-M., Castro-Hermida J.A. Presence of Cryptosporidium scrofarum, C. suis and C. parvum subtypes IIaA16G2R1 and IIaA13G1R1 in Eurasian wild boars (Sus scrofa) Vet Parasitol. 2013;196:497–502. doi: 10.1016/j.vetpar.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 162.Ryan U., Fayer R., Xiao L. Cryptosporidium species in humans and animals: Current understanding and research needs. Parasitology. 2014;141:1667–1685. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- 163.Tyzzer E.E. Coccidiosis in gallinaceous birds. Am. J. Epidemiol. 1929;10:269–383. doi: 10.1093/oxfordjournals.aje.a112759. [DOI] [Google Scholar]
- 164.Slavin D. Cryptosporidium Meleagridis (Sp. Nov.) J. Comp. Pathol. Ther. 1955;65:262–266. doi: 10.1016/S0368-1742(55)80025-2. [DOI] [PubMed] [Google Scholar]
- 165.Baroudi D., Khelef D., Goucem R., Adjou K.T., Adamu H., Zhang H., Xiao L. Common occurrence of zoonotic pathogen Cryptosporidium meleagridis in broiler chickens and turkeys in Algeria. Vet. Parasitol. 2013;196:334–340. doi: 10.1016/j.vetpar.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 166.Wang Y., Zhang K., Chen Y., Li X., Zhang L. Cryptosporidium and cryptosporidiosis in wild birds: A One Health perspective. Parasitol. Res. 2021;120:3035–3044. doi: 10.1007/s00436-021-07289-3. [DOI] [PubMed] [Google Scholar]
- 167.Hajdušek O., Ditrich O., Šlapeta J. Molecular identification of Cryptosporidium spp. in animal and human hosts from the Czech Republic. Vet. Parasitol. 2004;122:183–192. doi: 10.1016/j.vetpar.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 168.Sak B., Petrželková K.J., Květoňová D., Mynářová A., Pomajbíková K.J., Modrý D., Cranfield M.R., Mudakikwa A., Kváč M. Diversity of Microsporidia, Cryptosporidium and Giardia in Mountain Gorillas (Gorilla beringei beringei) in Volcanoes National Park, Rwanda. PLoS ONE. 2014;9:e109751. doi: 10.1371/journal.pone.0109751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vermeulen E.T., Ashworth D.L., Eldridge M., Power M. Diversity of Cryptosporidium in brush-tailed rock-wallabies (Petrogale penicillata) managed within a species recovery programme. Int. J. Parasitol. Parasites Wildl. 2015;4:190–196. doi: 10.1016/j.ijppaw.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang W., Wang R., Yang F., Zhang L., Cao J., Zhang X., Ling H., Liu A., Shen Y. Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in northeastern China’s Heilongjiang Province. PLoS ONE. 2013;8:e54857. doi: 10.1371/journal.pone.0054857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang S., Tao W., Liu C., Jiang Y., Wan Q., Li Q., Yang H., Lin Y., Li W. First report of Cryptosporidium canis in foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) and identification of several novel subtype families for Cryptosporidium mink genotype in minks (Mustela vison) in China. Infect. Genet. Evol. 2016;41:21–25. doi: 10.1016/j.meegid.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 172.Yang Z., Zhao W., Wang J., Ren G., Zhang W., Liu A. Molecular detection and genetic characterizations of Cryptosporidium spp. in farmed foxes, minks, and raccoon dogs in northeastern China. Parasitol. Res. 2018;117:169–175. doi: 10.1007/s00436-017-5686-5. [DOI] [PubMed] [Google Scholar]
- 173.Akiyoshi D.E., Dilo J., Pearson C., Chapman S., Tumwine J., Tzipori S. Characterization of Cryptosporidium meleagridis of Human Origin Passaged through Different Host Species. Infect. Immun. 2003;71:1828–1832. doi: 10.1128/IAI.71.4.1828-1832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chappell C.L., Okhuysen P., Tzipori S., Widmer G., Akiyoshi D.E., Langer-Curry R.C. Cryptosporidium meleagridis: Infectivity in Healthy Adult Volunteers. Am. J. Trop. Med. Hyg. 2011;85:238–242. doi: 10.4269/ajtmh.2011.10-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Omolabi K.F., Odeniran P.O., Soliman M.E. A meta-analysis of Cryptosporidium species in humans from southern Africa (2000–2020) J. Parasit. Dis. 2021;18:1–13. doi: 10.1007/s12639-021-01436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kopacz Ż., Kváč M., Karpiński P., Hendrich A.B., Sąsiadek M.M., Leszczyński P., Sak B., McEvoy J., Kicia M. The first evidence of Cryptosporidium meleagridis infection in a colon adenocarcinoma from an immunocompetent patient. Front. Cell Infect. Microbiol. 2019;9:35. doi: 10.3389/fcimb.2019.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xiao L., Sulaiman I.M., Ryan U., Zhou L., Atwill E.R., Tischler M.L., Zhang X., Fayer R., Lal A.A. Host adaptation and host–parasite co-evolution in Cryptosporidium: Implications for taxonomy and public health. Int. J. Parasitol. 2002;32:1773–1785. doi: 10.1016/S0020-7519(02)00197-2. [DOI] [PubMed] [Google Scholar]
- 178.Korpe P.S., Gilchrist C., Burkey C., Taniuchi M., Ahmed E., Madan V., Castillo R., Ahmed S., Arju T., Alam M., et al. Case-Control Study of Cryptosporidium Transmission in Bangladeshi Households. Clin. Infect. Dis. 2019;68:1073–1079. doi: 10.1093/cid/ciy593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ng-Hublin J.S., Combs B., Reid S., Ryan U. Differences in the occurrence and epidemiology of cryptosporidiosis in Aboriginal and non-Aboriginal people in Western Australia (2002–2012) Infect. Genet. Evol. 2017;53:100–106. doi: 10.1016/j.meegid.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 180.Silverlås C., Mattsson J.G., Insulander M., Lebbad M. Zoonotic transmission of Cryptosporidium meleagridis on an organic Swedish farm. Int. J. Parasitol. 2012;42:963–967. doi: 10.1016/j.ijpara.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 181.Stensvold C.R., Beser J., Axén C., Lebbad M. High Applicability of a Novel Method for gp60-Based Subtyping of Cryptosporidium meleagridis. J. Clin. Microbiol. 2014;52:2311–2319. doi: 10.1128/JCM.00598-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang Y., Yang W., Cama V., Wang L., Cabrera L., Ortega Y., Bern C., Feng Y., Gilman R., Xiao L. Population genetics of Cryptosporidium meleagridis in humans and birds: Evidence for cross-species transmission. Int. J. Parasitol. 2014;44:515–521. doi: 10.1016/j.ijpara.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 183.Wang T., Fan Y., Koehler A., Ma G., Li T., Hu M., Gasser R.B. First survey of Cryptosporidium, Giardia and Enterocytozoon in diarrhoeic children from Wuhan, China. Infect. Genet. Evol. 2017;51:127–131. doi: 10.1016/j.meegid.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 184.Liao C., Wang T., Koehler A.V., Fan Y., Hu M., Gasser R.B. Molecular investigation of Cryptosporidium in farmed chickens in Hubei Province, China, identifies ‘zoonotic’ subtypes of C. meleagridis. Parasites Vectors. 2018;11:484. doi: 10.1186/s13071-018-3056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Esch K.J., Petersen C.A. Transmission and epidemiology of zoonotic protozoal diseases of companion animals. Clin. Microbiol. Rev. 2013;26:58–85. doi: 10.1128/CMR.00067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li J., Yang F., Liang R., Guo S., Guo Y., Li N., Feng Y., Xiao L. Subtype Characterization and Zoonotic Potential of Cryptosporidium felis in Cats in Guangdong and Shanghai, China. Pathogens. 2021;10:89. doi: 10.3390/pathogens10020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lucio-Forster A., Griffiths J.K., Cama V.A., Xiao L., Bowman D.D. Minimal zoonotic risk of cryptosporidiosis from pet dogs and cats. Trends Parasitol. 2010;26:174–179. doi: 10.1016/j.pt.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 188.Taghipour A., Olfatifar M., Bahadory S., Godfrey S.S., Abdoli A., Khatami A., Javanmard E., Shahrivar F. The global prevalence of Cryptosporidium infection in dogs: A systematic review and meta-analysis. Vet. Parasitol. 2020;281:109093. doi: 10.1016/j.vetpar.2020.109093. [DOI] [PubMed] [Google Scholar]
- 189.Xiao L., Cama V.A., Cabrera L., Ortega Y., Pearson J., Gilman R.H. Possible Transmission of Cryptosporidium canis among Children and a Dog in a Household. J. Clin. Microbiol. 2007;45:2014–2016. doi: 10.1128/JCM.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Beser J., Toresson L., Eitrem R., Troell K., Winiecka-Krusnell J., Lebbad M. Possible zoonotic transmission of Cryptosporidium felis in a household. Infect. Ecol. Epidemiol. 2015;5:28463. doi: 10.3402/iee.v5.28463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Glaser C.A., Safrin S., Reingold A., Newman T.B. Association between Cryptosporidium infection and animal exposure in HIV-infected individuals. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1998;17:79–82. doi: 10.1097/00042560-199801010-00012. [DOI] [PubMed] [Google Scholar]
- 192.Jiang W., Roellig D.M., Lebbad M., Beser J., Troell K., Guo Y., Li N., Xiao L., Feng Y. Subtype distribution of zoonotic pathogen Cryptosporidium felis in humans and animals in several countries. Emerg. Microbes Infect. 2020;9:2446–2454. doi: 10.1080/22221751.2020.1840312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Taghipour A., Olfatifar M., Foroutan M., Bahadory S., Malih N., Norouzi M. Global prevalence of Cryptosporidium infection in rodents: A systematic review and meta-analysis. Prev. Vet. Med. 2020;182:105119. doi: 10.1016/j.prevetmed.2020.105119. [DOI] [PubMed] [Google Scholar]
- 194.Qi M., Huang L., Wang R., Xiao L., Xu L., Li J., Zhang L. Natural infection of Cryptosporidium muris in ostriches (Struthio camelus) Vet. Parasitol. 2014;205:518–522. doi: 10.1016/j.vetpar.2014.06.035. [DOI] [PubMed] [Google Scholar]
- 195.Wagnerová P., Sak B., McEvoy J., Rost M., Matysiak A.P., Ježková J., Kváč M. Genetic diversity of Cryptosporidium spp. including novel identification of the Cryptosporidium muris and Cryptosporidium tyzzeri in horses in the Czech Republic and Poland. Parasitol. Res. 2015;114:1619–1624. doi: 10.1007/s00436-015-4353-y. [DOI] [PubMed] [Google Scholar]
- 196.Chen L., Hu S., Jiang W., Zhao J., Li N., Guo Y., Liao C., Han Q., Feng Y., Xiao L. Cryptosporidium parvum and Cryptosporidium hominis subtypes in crab-eating macaques. Parasites Vectors. 2019;12:1–9. doi: 10.1186/s13071-019-3604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.García-Livia K., Martín-Alonso A., Foronda P. Diversity of Cryptosporidium spp. in wild rodents from the Canary Islands, Spain. Parasites Vectors. 2020;13:445. doi: 10.1186/s13071-020-04330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang L., Cao L., Zheng S., Chang Y., Zhang K., Zhang S., Zhang L. Molecular identification and biological characterization of Cryptosporidium muris from camels (Camelus bactrianus) in China. Parasites Vectors. 2021;14:1–10. doi: 10.1186/s13071-021-04862-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chappell C.L., Okhuysen P., Tzipori S., Widmer G., Lupo P., Langer-Curry R.C. Cryptosporidium muris: Infectivity and Illness in Healthy Adult Volunteers. Am. J. Trop. Med. Hyg. 2015;92:50–55. doi: 10.4269/ajtmh.14-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Čondlová Š., Horčičková M., Havrdová N., Sak B., Hlásková L., Perec-Matysiak A., Kicia M., McEvoy J., Kváč M. Diversity of Cryptosporidium spp. in Apodemus spp. in Europe. Eur. J. Protistol. 2019;69:1–13. doi: 10.1016/j.ejop.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ren X., Zhao J., Zhang L., Ning C., Jian F., Wang R., Lv C., Wang Q., Arrowood M.J., Xiao L. Cryptosporidium tyzzeri n. sp. (Apicomplexa: Cryptosporidiidae) in domestic mice (Mus musculus) Exp. Parasitol. 2012;130:274–281. doi: 10.1016/j.exppara.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 202.Kváč M., Vlnatá G., Ježková J., Horčičková M., Konečný R., Hlásková L., McEvoy J., Sak B. Cryptosporidium occultus sp. n. (Apicomplexa: Cryptosporidiidae) in rats. Eur. J. Protistol. 2018;63:96–104. doi: 10.1016/j.ejop.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 203.Zhang Q., Li J., Li Z., Xu C., Hou M., Qi M. Molecular identification of Cryptosporidium spp. in alpacas (Vicugna pacos) in China. Int. J. Parasitol. Parasites Wildl. 2020;12:181–184. doi: 10.1016/j.ijppaw.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Huang C., Hu Y., Wang L., Wang Y., Li N., Guo Y., Feng Y., Xiao L. Environmental Transport of Emerging Human-Pathogenic Cryptosporidium Species and Subtypes through Combined Sewer Overflow and Wastewater. Appl. Environ. Microbiol. 2017;83:e00682-17. doi: 10.1128/AEM.00682-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Koehler A.V., Wang T., Haydon S.R., Gasser R.B. Cryptosporidium viatorum from the native Australian swamp rat Rattus lutreolus-An emerging zoonotic pathogen? Int. J. Parasitol. Parasites Wildl. 2018;7:18–26. doi: 10.1016/j.ijppaw.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen Y.-W., Zheng W.-B., Zhang N.-Z., Gui B.-Z., Lv Q.-Y., Yan J.-Q., Zhao Q., Liu G.-H. Identification of Cryptosporidium viatorum XVa subtype family in two wild rat species in China. Parasites Vectors. 2019;12:1–5. doi: 10.1186/s13071-019-3763-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhao W., Zhou H., Huang Y., Xu L., Rao L., Wang S., Wang W., Yi Y., Zhou X., Wu Y., et al. Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: Molecular detection, species/genotype identification and implications for public health. Int. J. Parasitol. Parasites Wildl. 2019;9:317–321. doi: 10.1016/j.ijppaw.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Stensvold C.R., Elwin K., Winiecka-Krusnell J., Chalmers R.M., Xiao L., Lebbad M. Development and Application of a gp60-Based Typing Assay for Cryptosporidium viatorum. J. Clin. Microbiol. 2015;53:1891–1897. doi: 10.1128/JCM.00313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Jiang J., Alderisio K.A., Xiao L. Distribution of Cryptosporidium Genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 2005;71:4446–4454. doi: 10.1128/AEM.71.8.4446-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Feng Y., Alderisio K.A., Yang W., Blancero L.A., Kuhne W.G., Nadareski C.A., Reid M., Xiao L. Cryptosporidium Genotypes in Wildlife from a New York Watershed. Appl. Environ. Microbiol. 2007;73:6475–6483. doi: 10.1128/AEM.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Robinson G., Wright S., Elwin K., Hadfield S.J., Katzer F., Bartley P.M., Hunter P.R., Nath M., Innes E.A., Chalmers R.M. Redescription of Cryptosporidium cuniculus Inman and Takeuchi, 1979 (Apicomplexa: Cryptosporidiidae): Morphology, biology and phylogeny. Int. J. Parasitol. 2010;40:1539–1548. doi: 10.1016/j.ijpara.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 212.Hadfield S.J., Chalmers R.M. Detection and characterization of Cryptosporidium cuniculus by real-time PCR. Parasitol. Res. 2012;111:1385–1390. doi: 10.1007/s00436-012-2874-1. [DOI] [PubMed] [Google Scholar]
- 213.Naguib D., Roellig D., Arafat N., Xiao L. Genetic Characterization of Cryptosporidium cuniculus from Rabbits in Egypt. Pathogens. 2021;10:775. doi: 10.3390/pathogens10060775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Koehler A.V., Rashid M.H., Zhang Y., Vaughan J.L., Gasser R.B., Jabbar A. First cross-sectional, molecular epidemiological survey of Cryptosporidium, Giardia and Enterocytozoon in alpaca (Vicugna pacos) in Australia. Parasites Vectors. 2018;11:498. doi: 10.1186/s13071-018-3055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kaupke A., Kwit E., Chalmers R., Michalski M., Rzeżutka A. An outbreak of massive mortality among farm rabbits associated with Cryptosporidium infection. Res. Vet. Sci. 2014;97:85–87. doi: 10.1016/j.rvsc.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 216.Liu X., Zhou X., Zhong Z., Chen W., Deng J., Niu L., Wang Q., Peng G. New Subtype of Cryptosporidium cuniculus Isolated from Rabbits by Sequencing the Gp60 Gene. J. Parasitol. 2014;100:532–536. doi: 10.1645/13-223.1. [DOI] [PubMed] [Google Scholar]
- 217.Zhang X., Qi M., Jing B., Yu F., Wu Y., Chang Y., Zhao A., Wei Z., Dong H., Zhang L. Molecular Characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Rabbits in Xinjiang, China. J. Eukaryot. Microbiol. 2016;65:854–859. doi: 10.1111/jeu.12629. [DOI] [PubMed] [Google Scholar]
- 218.Zhang W., Shen Y., Wang R., Liu A., Ling H., Li Y., Cao J., Zhang X., Shu J., Zhang L. Cryptosporidium cuniculus and Giardia duodenalis in Rabbits: Genetic Diversity and Possible Zoonotic Transmission. PLoS ONE. 2012;7:e31262. doi: 10.1371/journal.pone.0031262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yang Z., Yang F., Wang J., Cao J., Zhao W., Gong B., Yan J., Zhang W., Liu A., Shen Y. Multilocus sequence typing and population genetic structure of Cryptosporidium cuniculus in rabbits in Heilongjiang Province, China. Infect. Genet. Evol. 2018;64:249–253. doi: 10.1016/j.meegid.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 220.Kváč M., Hofmannová L., Hlásková L., Květoňová D., Vítovec J., McEvoy J., Sak B. Cryptosporidium erinacei n. sp. (Apicomplexa: Cryptosporidiidae) in hedgehogs. Vet. Parasitol. 2010;201:9–17. doi: 10.1016/j.vetpar.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 221.Laatamna A.E., Wagnerová P., Sak B., Květoňová D., Aissi M., Rost M., Kváč M. Equine cryptosporidial infection associated with Cryptosporidium hedgehog genotype in Algeria. Vet. Parasitol. 2013;197:350–353. doi: 10.1016/j.vetpar.2013.04.041. [DOI] [PubMed] [Google Scholar]
- 222.Dyachenko V., Kuhnert Y., Schmaeschke R., Etzold M., Pantchev N., Daugschies A. Occurrence and molecular characterization of Cryptosporidium spp. genotypes in European hedgehogs (Erinaceus europaeus L.) in Germany. Parasitology. 2010;137:205–216. doi: 10.1017/S0031182009991089. [DOI] [PubMed] [Google Scholar]
- 223.Hofmannová L., Hauptman K., Huclová K., Květoňová D., Sak B., Kváč M. Cryptosporidium erinacei and C. parvum in a group of overwintering hedgehogs. Eur. J. Protistol. 2016;56:15–20. doi: 10.1016/j.ejop.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 224.Prediger J., Horčičková M., Hofmannová L., Sak B., Ferrari N., Mazzamuto M.V., Romeo C., Wauters L.A., McEvoy J., Kváč M. Native and introduced squirrels in Italy host different Cryptosporidium spp. Pt AEur. J. Protistol. 2017;61:64–75. doi: 10.1016/j.ejop.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 225.Power M.L., Cheung-Kwok-Sang C., Slade M., Williamson S. Cryptosporidium fayeri: Diversity within the GP60 locus of isolates from different marsupial hosts. Exp. Parasitol. 2009;121:219–223. doi: 10.1016/j.exppara.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 226.Takaki Y., Takami Y., Watanabe T., Nakaya T., Murakoshi F. Molecular identification of Cryptosporidium isolates from ill exotic pet animals in Japan including a new subtype in Cryptosporidium fayeri. Vet. Parasitol. Reg. Stud. Rep. 2020;21:100430. doi: 10.1016/j.vprsr.2020.100430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Qian W., Zhang Y., Jiang Y., Zhao A., Lv C., Qi M. Molecular characterization of Cryptosporidium spp. in minks (Neovison vison), blue foxes (Vulpes lagopus), and raccoon dogs (Nyctereutes procyonoides) in farms from Xinjiang, Northwest China. Parasitol. Res. 2020;119:3923–3927. doi: 10.1007/s00436-020-06909-8. [DOI] [PubMed] [Google Scholar]
- 228.Feng Y., Lal A.A., Li N., Xiao L. Subtypes of Cryptosporidium spp. in mice and other small mammals. Exp. Parasitol. 2011;127:238–242. doi: 10.1016/j.exppara.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 229.Dettwiler I., Troell K., Robinson G., Chalmers R.M., Basso W., Rentería-Solís Z.M., Daugschies A., Mühlethaler K., Dale M.I., Raghavendra J.B., et al. TIDE Analysis of Cryptosporidium Infections by gp60 Typing Reveals Obscured Mixed Infections. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab417. [DOI] [PubMed] [Google Scholar]
- 230.Yanta C.A., Bessonov K., Robinson G., Troell K., Guy R.A. CryptoGenotyper: A new bioinformatics tool for rapid Cryptosporidium identification. Food Waterborne Parasitol. 2021;23:e00115. doi: 10.1016/j.fawpar.2021.e00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Braima K., Zahedi A., Egan S., Austen J., Xiao L., Feng Y., Witham B., Pingault N., Perera S., Oskam C., et al. Molecular analysis of cryptosporidiosis cases in Western Australia in 2019 and 2020 supports the occurrence of two swimming pool associated outbreaks and reveals the emergence of a rare C. hominis IbA12G3 subtype. Infect. Genet. Evol. 2021;92:104859. doi: 10.1016/j.meegid.2021.104859. [DOI] [PubMed] [Google Scholar]
- 232.Vinayak S., Pawlowic M.C., Sateriale A., Brooks C.F., Studstill C.J., Bar-Peled Y., Cipriano M.J., Striepen B. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature. 2015;523:477–480. doi: 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Vangah S.J., Katalani C., Booneh H.A., Hajizade A., Sijercic A., Ahmadian G. CRISPR-Based Diagnosis of Infectious and Noninfectious Diseases. Biol. Proced. Online. 2020;22:22. doi: 10.1186/s12575-020-00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yu F., Zhang K., Wang Y., Li D., Cui Z., Huang J., Zhang S., Li X., Zhang L. CRISPR/Cas12a-based on-site diagnostics of Cryptosporidium parvum IId-subtype-family from human and cattle fecal samples. Parasites Vectors. 2021;14:1–10. doi: 10.1186/s13071-021-04709-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Fan Y., Feng Y., Xiao L. Comparative genomics: How has it advanced our knowledge of cryptosporidiosis epidemiology? Parasitol. Res. 2019;118:3195–3204. doi: 10.1007/s00436-019-06537-x. [DOI] [PubMed] [Google Scholar]
- 236.Baptista R., Cooper G., Kissinger J. Challenges for Cryptosporidium Population Studies. Genes. 2021;12:894. doi: 10.3390/genes12060894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Xu Z., Li N., Guo Y., Feng Y., Xiao L. Comparative genomic analysis of three intestinal species reveals reductions in secreted pathogenesis determinants in bovine-specific and non-pathogenic Cryptosporidium species. Microb. Genom. 2020;6:e000379. doi: 10.1099/mgen.0.000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Su J., Jin C., Wu H., Fei J., Li N., Guo Y., Feng Y., Xiao L. Differential expression of three Cryptosporidium species-specific MEDLE Proteins. Front. Microbiol. 2019;10:1177. doi: 10.3389/fmicb.2019.01177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Guo Y., Tang K., Rowe L.A., Li N., Roellig D.M., Knipe K., Frace M., Yang C., Feng Y., Xiao L. Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum. BMC Genom. 2015;16:1–18. doi: 10.1186/s12864-015-1517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Pallen M.J. Diagnostic metagenomics: Potential applications to bacterial, viral and parasitic infections. Parasitology. 2014;141:1856–1862. doi: 10.1017/S0031182014000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lopes R.J., Merida A.M., Carneiro M. Unleashing the potential of public genomic resources to find parasite genetic data. Trends Parasitol. 2017;33:750–753. doi: 10.1016/j.pt.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 242.Beghini F., Pasolli E., Truong T.D., Putignani L., Cacciò S.M., Segata N. Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 2017;11:2848–2863. doi: 10.1038/ismej.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Franssen F.F.J., Janse I., Janssen D., Caccio S.M., Vatta P., van der Giessen J.W.B., van Passel M.W.J. Mining public metagenomes for environmental surveillance of parasites: A proof of principle. Front. Microbiol. 2021;12:1784. doi: 10.3389/fmicb.2021.622356. [DOI] [PMC free article] [PubMed] [Google Scholar]