Abstract
Enterohemorrhagic Escherichia coli (EHEC) and specifically serotype O157:H7 are a significant cause of hemorrhagic gastrointestinal disease and the hemolytic uremic syndrome. Methods currently used in clinical microbiology labs, such as sorbitol-MacConkey (SMAC) agar, reliably detect only O157:H7. We have evaluated a two-step method that has the potential to identify and isolate all EHEC serotypes, including serotype O157:H7. This method utilizes a chromogenic selective-differential medium for the isolation of E. coli together with an enzyme-linked immunosorbent assay (ELISA) that detects the Shiga-like toxins Stx1 and Stx2. Both are commercially available and usable in a wide range of clinical microbiology laboratories. Compared to a Vero cell cytotoxic assay, SMAC had sensitivities of 23.5% for the identification of all EHEC serotypes and of 50.0% for the identification of O157:H7 alone. The two-step method had sensitivities of 76.5 and 100%, respectively. The ELISA alone had a sensitivity of 82.4% in the detection of Stx1 and Stx2. The specificity was 100% in all cases. Overall, 14 EHEC isolates were obtained: 8 (58%) O157:H7, 2 (14%) O26, 2 (14%) O111:NM, 1 (7%) O103:H2, and 1 (7%) O121:H19. All but one were isolated during the months of May to September. The two-step method was found to be considerably more expensive than SMAC for both positive and negative samples.
Enterohemorrhagic Escherichia coli (EHEC) can cause diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome (HUS) through the production of the Shiga-like toxins Stx1 and Stx2 and other probable virulence factors (1, 29). While >100 serotypes have been found to produce Stx, some serotypes (O26:H11, O103:H2, O111:NM, O103:H21, and especially O157:H7) appear to be the predominant etiologic agents of human disease (15). Clinically amenable methodologies for the detection of all EHEC strains are not widely used, and the overall incidence of EHEC-related disease is therefore unknown. However, studies have found O157:H7 alone to typically be the one-third to one-fourth most common enteric pathogen after Salmonella and Campylobacter spp., and widespread testing for other pathogenic EHEC serotypes could alter this distribution (31). EHEC strains have also been strongly associated with HUS, a disease with significant morbidity and mortality in children and the elderly (22).
EHEC are genetically and phenotypically similar to other, nondiarrheagenic E. coli found in the human gut, making their detection and subsequent isolation from fecal material difficult (6, 9). Nevertheless, the Centers for Disease Control and Prevention and others recommend that at a minimum, bloody stools and stools from HUS patients be cultured for E. coli O157:H7 (10). Many clinical laboratories in the United States and elsewhere now use sorbitol-MacConkey medium (SMAC) agar to identify the slow sorbitol fermentation phenotype (i.e., sorbitol-negative at 24 h) of O157:H7 (12). However, this medium does not detect other, sorbitol-positive EHEC serotypes (24). While many EHEC isolates produce enterohemolysin that can be detected with a specific trypticase sheep blood agar (Unipath GmbH, Wesel, Germany), approximately 10% do not and are missed by this technique (4).
PCR has been used in both single and multiplex formats to detect various EHEC-associated gene sequences including stx1 and stx2, the genes responsible for Shiga-like toxin production (13, 27, 30; D. Swenson, N. Strockbine, L. Kirchoff, L. Barley, B. Swaminathan, and L. Wu, Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998, abstr. P-6, p. 405, 1998). PCR can be both sensitive and specific, but various technical difficulties can result in both false-negative and false-positive results (11). Finally, several immunoassay products are available that can reliably detect O157 lipopolysaccharide, Stx1, and Stx2 (20, 26). However, neither PCR nor immunoassay can recover EHEC isolates for strain confirmation and epidemiology studies.
We report here the results of an evaluation of two commercial products used in tandem that can both detect and isolate O157 and non-O157 EHEC strains. Our intent was to devise a method that is sensitive, specific, and usable in a wide range of clinical laboratory settings. The first product, the Premier EHEC enzyme-linked immunosorbent assay (ELISA; Meridian Diagnostics, Cincinnati, Ohio) detects Stx1 and Stx2 in a batch microwell format. The second product, Rainbow Agar O157 (Biolog, Inc., Hayward, Calif.) is a selective, chromogenic medium that was designed to identify E. coli, and specifically E. coli O157:H7 (5).
(This work was presented in part at the 98th Annual Meeting of the American Society for Microbiology, Atlanta, Ga., May 1998 [T. J. Novicki, J. A. Daly, S. Mottice, and K. C. Carroll, abstr. C257, p. 174, 1998].)
MATERIALS AND METHODS
RCF.
Relative centrifugal force (RCF) was calculated as follows: RCF = 11.17r(n/1,000)2, where r is the radius of the rotor in centimeters and n is the rotor speed in rpm.
Study design.
Over a 12-month period, 488 liquid or semiformed (i.e., taking the shape of the container) stool samples were submitted to three Salt Lake City (Utah) clinical microbiology laboratories: the Associated and Regional University Laboratories (ARUP), the Utah Department of Health (UDOH), and the Primary Children's Medical Center (PCMC). Stool samples came from individuals residing in Utah and southeastern Idaho and were received either as fresh specimens or in Cary-Blair transport medium. Upon receipt, stool samples were initially processed by technologists at the participating clinical microbiology labs. The color and consistency of the stool, any special instructions recorded on the requisition, and the date of culture setup were recorded. An aliquot of each stool was also frozen at −70°C. In addition to a routine stool culture, including the SMAC plate, all stools were inoculated into a MacConkey medium (MAC) broth and onto a Rainbow agar O157 (RA) plate for the study. MAC broths were incubated overnight, and RA plates were incubated for 48 h, after which they were held at 4°C until transport to ARUP for further workup. SMAC plates were also incubated overnight.
SMAC plates were worked up for E. coli O157:H7 by the recipient clinical microbiology laboratories. MAC broths and RA plates were processed at ARUP. Upon arrival, broths were tested for Stx by ELISA, and aliquots of each were frozen at −70°C for batch testing by Vero cell cytotoxicity assay (VCA) (see below). When Stx was detected by ELISA, an attempt was made to isolate EHEC from the corresponding RA plate by using the following protocol. An isolated colony representative of any red, blue, purple, and/or black colonies present on the RA plate was subcultured onto MAC broth for toxin testing and onto sheep blood and MAC plates for identification procedures; all were incubated overnight. (Mucoid gray colonies were later included when it was observed that many O157:H7 isolates had a distinctive watery-gray morphology on RA.) After incubation, MAC broth subcultures were tested for Stx by ELISA; Stx-positive isolates were then identified to species level and serotyped.
Media and chemicals.
Five milliliters of sterile MAC broth in screw-cap tubes was supplied by NEL (Waterville, Maine). RA was prepared according to the manufacturer's directions. Then, 20 ml of molten agar was poured into 100-mm disposable petri dishes (Fisher Scientific, Pittsburgh, Pa.). Prepared plates of MAC agar and Columbia agar with 5% sheep blood were purchased from Remel, Inc. (Lenexa, Kans.). Novobiocin sodium salts were purchased from Sigma (St. Louis, Mo.), dissolved in deionized water at 30 mg/ml, filter sterilized, and frozen at −20°C for up to 1 year. Minimal essential medium (MEM) with 10% calf serum was used as the Vero cell growth medium; MEM with 2% calf serum, 50 U of penicillin per ml, 50 μg of streptomycin per ml, 1 μg of amphotericin B per ml, 40 μg of vancomycin per ml, and 20 μg of gentamicin per ml was used as the Vero cell maintenance medium. MEM and penicillin-streptomycin solution (P&S) (penicillin sodium salt, 5,000 U/ml; streptomycin, 5 mg/ml) were supplied by the ARUP Laboratories Tissue Culture Facility.
RA.
RA is a selective, differential medium that identifies E. coli by chromogenic substrate metabolism. The manufacturer states that RA will identify E. coli as red, blue, or purple colonies and specifically O157:H7 as black or charcoal-colored colonies; other bacteria are said to not grow at all or be nonchromogenic. Novobiocin was added at a concentration of 30 μg/ml at the recommendation of Biolog to increase the selectivity of the medium for E. coli.
Bacterial identification and serotyping.
All bacterial cultures were incubated at 37°C in ambient air. SMAC plates were worked up for E. coli O157:H7 in recipient clinical labs by using accepted biochemical and phenotypic tests. All RA plates were worked up in the research lab of one author (T.J.N.). Stx producers isolated from RA plates were confirmed as EHEC with the Vitek Auto/Microbic GNI card (bioMerieux, Inc., St. Louis, Mo.), the RIM E. coli O157:H7 latex agglutination kit (Remel, Inc.), and VCA. Non-O157 EHEC isolates were sent to UDOH for further serotyping.
Premier EHEC ELISA.
All aspects of ELISA testing were performed according to the manufacturer's instructions by the ARUP Rapid Diagnostics Section. To increase the sensitivity of the toxin test, 10 to 50 μl of each stool sample was inoculated into 5 ml of MAC broth and incubated overnight prior to ELISA testing (23). Aliquots (1 ml) of these broth cultures were also frozen at −70°C for VCA.
VCA.
A modification of the VCA was used as the reference method for Stx detection; all work was performed in the research lab of one author (T.J.N.) (19, 21). Briefly, Vero cell monolayers (Viromed, Minnetonka, Minn.) were prepared by the ARUP Laboratories Tissue Culture Facility in 0.3-ml 96-well microtiter cell culture plates by using standard cell culture techniques. The MEM growth medium was removed from the cells immediately prior to use. Stools were cultured in MAC broth prior to testing as described above. To remove excess bacteria, 1 ml of each broth culture was spun for 5 min at 4°C, 5,000 RCF in a snap cap microcentrifuge tube; 0.5 ml of the supernatant was then decanted into a fresh microcentrifuge tube containing 35 μl of P&S and mixed. The treated supernatant was then immediately centrifuged as described above, diluted to 1:20 and 1:400 in MEM maintenance medium, and placed on the Vero cell monolayers with a final volume of 200 μl. A clinical isolate of O157:H7 EHEC, previously characterized by the UDOH to be a high-titer Stx producer, and the Stx-negative E. coli ATCC strain 25922 were treated in a similar manner and used as positive and negative controls, respectively. The inoculated Vero cell plates were incubated at 35°C in 5% CO2 for 3 days, after which the cells were examined for cytotoxic effects.
Samples found to produce cytotoxic effects were neutralized with an Stx-specific polyclonal antitoxin (Meridian Diagnostics, Cincinnati, Ohio) by mixing 25 μl of the previously P&S-treated Stx-positive broth with 25 μl of antitoxin and then incubating it at 37°C for 1 h. The original P&S-treated broth and the neutralized sample were then placed on Vero cell monolayers at a 1:20 dilution. P&S-treated positive and negative controls as described above were neutralized and set up in a similar manner. The plates were incubated as described above and then read. Cytotoxic effects in the untreated well, accompanied by a loss of cytotoxicity in the antitoxin-neutralized well, were considered to be due to Stx activity.
Discrepancy resolution.
Discrepancies between VCA and ELISA results were resolved in the laboratory of D. W. K. Acheson (Division of Geographic Medicine and Infectious Diseases, New England Medical Center, Boston, Mass.) by PCR amplification of stx gene sequences in stool samples that had been frozen at the time of culture setup. Primer sequences that detect both the stx1 and stx2 genes and thermocycler parameters have been previously described (8). PCR products were detected by electrophoresis in 1.5% agarose-ethidium bromide slab gels.
Statistical analysis.
Positives and negatives are defined in Table 1. A positive SMAC culture required the isolation of an O157 strain; a positive RA culture required isolation of an EHEC strain of any serotype. Formulae for the calculation of sensitivity, specificity, and positive and negative predictive values have been previously published (17).
TABLE 1.
Positive and negatives, defined
| Statistic | Methoda
|
||
|---|---|---|---|
| ELISA | SMAC | ELISA-RA | |
| True positive | E+/Vc+ | Cxs+/Vc+ | E+/Cxr+/Vc+ |
| True negative | E−/Vc− | Cxs−/Vc− | E−/Vc− |
| False positive | E+/Vc−/P− | Cxs+/Vc− | E+/Cxr−/Vc−/P− |
| False negative | E−/Vc+/P+ | Cxs−/Vc+/ER+ | E−/Vc+/P+ or E+/Cxr−/Vc+/P+ |
E, ELISA; Vc, VCA; Cxs is culture on SMAC agar; Cxr is culture on RA; ER, ELISA-RA; P, PCR.
RESULTS
Detection of Stx-positive cultures.
The VCA detected Stx in 17 of the 488 stool samples. Of these, 14 were detected by the ELISA as well. Three additional Stx-positive stool samples were not detected by ELISA; these samples were subsequently found to contain stx gene sequences by PCR and were thus considered to be false-negative Premier EHEC results. (See Table 1 for the definitions of positive and negative results.) The ELISA did not detect any positives that were not also detected by the VCA, and the two methods were in complete agreement on Stx-negative samples as well. The sensitivity, specificity, and predictive values are given in Table 2.
TABLE 2.
Sensitivity, specificity, and predictive values
| Method | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|
| ELISA alonea | 82.4 | 100.0 | 100.0 | 99.4 |
| ELISA-RA | ||||
| All EHECa | 76.5 | 100.0 | 100.0 | 99.2 |
| O157b | 100.0 | 100.0 | 100.0 | 100.0 |
| SMAC | ||||
| All EHECa | 23.5 | 100.0 | 100.0 | 97.3 |
| O157b | 50.0 | 100.0 | 100.0 | 99.2 |
n = 488.
n = 479 (the three PCR-positive–culture-negative samples and the six non-O157:H7 culture-positive samples were deleted from this data set).
Isolation of EHEC from Stx-positive culture.
EHEC strains were detected and isolated from 13 of the 17 VCA-positive samples by the ELISA-RA method. The details of each EHEC isolate are given in Table 3. A 14th VCA-positive sample was identified as positive by ELISA. While EHEC could be detected by ELISA testing of the confluent growth in the primary quadrant of the corresponding RA plate, repeated attempts at isolating the EHEC strain failed. (Simultaneous culturing on cefixime tellurite SMAC agar by the UDOH yielded an EHEC isolate of O26 serotype [culture 253, Table 3].) This sample was thus considered to be a false-negative result by the ELISA-RA method. A mean of three colonies per EHEC-positive sample (range, 1 to 9) was tested by ELISA during the process of EHEC isolation from these 14 samples (data not shown).
TABLE 3.
EHEC isolates
| Culture-isolate | Date collected (mo/day/yr) | Color, consistency of stool | Color on primary RA plate | Identified on SMAC plate | Serotype | Sorbitol fermentera |
|---|---|---|---|---|---|---|
| 212-1 | 7/8/97 | Yellow-bloody, unformed | Gray | Yes | O157:H7 | No |
| 225-2 | 7/2/97 | Yellow-brown, liquid | Gray | No | O157:H7 | No |
| 280-2 | 9/2/97 | Brown, liquid | Cream, red center | No | O111:NMb | Yes |
| 296-1 | 9/5/97 | Bloody, liquid | Gray | Yes | O157:H7 | No |
| 301-1 | 9/19/97 | Brown-red, liquid | Cream | No | O157:H7 | No |
| 438-2 | 9/15/97 | Transport medium | Blue | No | O157:H7 | No |
| 439-1 | 9/15/97 | Transport medium | Black | Yes | O157:H7 | No |
| 305-1 | 9/21/97 | Brown, liquid | Purple | No | O26 | Yes |
| 42-2 | 9/27/97 | Transport medium | Blue | No | O111:NMb | Yes |
| 455-1 | 11/17/97 | Transport medium | Blue-purple | No | O103:H2 | Yes |
| 820-7 | 5/9/98 | Red-brown, liquid | Cream, red center | No | O121:H19 | Yes |
| 817-1 | 5/10/98 | Red-yellow, unformed | Charcoal | Yes | O157:H7 | No |
| 844-1 | 6/20/98 | Bloody, liquid | Gray | No | O157:H7 | No |
| 253c | 8/19/97 | Brown-green, liquid | NDd | No | O26 | No |
As determined by Vitek GNI card.
NM, nonmotile.
Isolated, identified, and serotyped by UDOH.
ND, not determined.
Three additional samples were found to be positive by VCA and PCR but negative by ELISA (see above). No RA plates were available at the time the discrepancies were resolved, and we therefore could not attempt to isolate EHEC strains as part of the ELISA-RA protocol. However, these three were counted as ELISA-RA false-negative results for the statistical analysis presented in Table 2.
Four VCA-positive samples yielded EHEC by the SMAC method: all were of O157:H7 serotype (Table 3). The SMAC method did not detect any EHEC-positive samples that were not also identified by the ELISA-RA method. Statistical analysis of this method is presented in Table 2.
Isolation on RA.
RA is sold as a primary isolation medium that may be used to select and identify E. coli and, specifically, E. coli O157:H7. The non-O157:H7 EHEC strains isolated in this study were chromogenic, as predicted by Biolog, Inc., since all were red, purple, or blue (Table 3). In contrast, only two of the eight (25%) O157:H7 isolates grew as compact black or charcoal-colored colonies as predicted by the manufacturer, the rest being mucoid and light gray in color.
Epidemiology.
Epidemiological data are presented in Table 3. Of 14 EHEC strains isolated in this study (including 1 O26 strain isolated by UDOH), 8 (57%) were of the O157:H7 serotype and 6 (43%) were non-O157 (2 O26, 2 O111:NM, 1 O103:H2, and 1 O121:H19). Bloody stool and diarrhea are two characteristics frequently associated with serotype O157:H7 EHEC (14, 15). In this series, 4 of the 14 EHEC-positive stools had been received in transport medium, and no information was available on their appearance. Of the remaining 10, 3 (30%) were bloody, while another 3 (30%) were noted to have a reddish tinge, which may have indicated the presence of blood. Eight (80%) were liquid, and two (20%) were unformed. Taken together, two (20%) were both bloody and liquid, two (20%) were both red and liquid, one (10%) was bloody and unformed, one (10%) was red and unformed, and four (40%) were non-bloody, but red and liquid.
The overall prevalence of EHEC (i.e., as detected by any culture method or by PCR) was 3.5%; the prevalence of O157:H7 alone was 1.7%. Of the 14 culture-confirmed EHEC-positive samples, 13 (93%) were collected during the months of May through September.
Material costs.
A comparison of material costs is given in Table 4. The cost of the ELISA-RA method is based upon the following factors: one ELISA is performed to screen a stool sample for Stx production, with an average of three isolates screened per sample by ELISA to yield an Stx producer (see above), and two RA plates are needed for primary culture and subculture of the EHEC isolate. Similarly, the cost of SMAC agar per positive sample is based upon the use of two plates for primary culture and subculture of the EHEC isolate. The cost of the other materials, such as serotyping reagents, biochemicals and disposables, has not been included.
TABLE 4.
Comparative material costs
| Parametera | SMAC plate ($) | ELISA ($) | RA plate ($) | ELISA-RA method ($) |
|---|---|---|---|---|
| Cost per item | 0.46 | 9.00 | 1.80 | NA |
| Cost per negative sampleb | 0.46 | 9.00 | 1.80 | 10.80 |
| Cost per positive sample | 0.92c | 9.00d | 3.60c | 39.60d |
Costs do not include positive and negative controls.
Does not include the cost of additional materials for “rule-out” testing.
Two plates were used for primary isolation and subculture.
Based upon an average of four ELISA tests (see the text).
DISCUSSION
SMAC agar is the current standard for the detection of EHEC. While inexpensive and easy to use, this medium lacks sensitivity in that it routinely detects only one serotype, O157:H7. In this study, we have evaluated the use of the Premier EHEC ELISA and RA together to isolate all EHEC serotypes.
The Premier EHEC ELISA was compared to a VCA, considered to be the “gold standard” for the detection of Stx (21). While the Premier EHEC ELISA has been reported to be as sensitive as the VCA in detecting Stx, we did not find this to be the case (20, 23). Three ELISA-negative samples that were VCA positive for Stx, as well as PCR positive for the stx1 and stx2 genes, were identified in this study. While these were low positives by the VCA method (titers of >1:20 and <1:400), a low concentration of Stx alone is an unlikely explanation for the failure of the ELISA, since other low positives in this series were identified by ELISA (data not shown). In spite of this, the ELISA gave an 82.4% sensitivity and a 100% specificity.
The ELISA-RA method was found to be more sensitive than SMAC agar in detecting both O157:H7 and non-O157:H7 EHEC (100 versus 50% for O157:H7 alone; 76.5 versus 23.5% overall). While the overall superiority of the ELISA-RA method over SMAC was not surprising, given that SMAC only reliably detects serotype O157:H7, SMAC proved inferior in the isolation of sorbitol negative O157 as well (24). While there have been reports of sorbitol fermenting O157 EHEC, all of the O157:H7 strains missed by the SMAC method in this study were sorbitol negative (16).
Four Stx-positive samples were not identified by the ELISA-RA method. Three, discussed above, were ELISA negative and both VCA and PCR positive and were thus considered false negatives. A fourth sample was identified as Stx positive by ELISA. While Stx was detected in the confluent growth in the primary quadrant of the RA plate, no EHEC could be found among isolated colonies. This patient had been in the hospital for a period of time for severe colitis before a stool sample was submitted for the study, and it is therefore possible that she had already begun to clear her infection by the time we acquired a stool sample. Such early clearance has been reported by others (32). While this was counted as a false negative, the detection of Stx alone would undoubtedly have helped make the diagnosis.
On average, three colonies from each RA plate were tested by ELISA for Stx production in order to isolate an EHEC strain. While this is considerably less than the 20 or more colonies that may require screening from primary plating media such as MAC or eosin-methylene blue agars, the use of three additional ELISA tests add considerably to the cost of the ELISA-RA method (18).
While the ATCC strain of O157:H7 did form compact, black colonies on RA as predicted by the manufacturer, the majority of patient O157:H7 isolates were large, watery, and gray on primary RA culture. While this morphology was often sufficient to help identify O157:H7 on RA with some precision when the sample was known to be Stx positive, it also resembled other non-EHEC colonies often seen on Stx-negative cultures so as to make this morphology nonspecific for EHEC (data not shown). No evaluation of EHEC isolation from stool with RA has been reported to date, and it is therefore unclear whether these findings are representative of the performance of this medium in general. In contrast, non-O157 EHEC reliably grew as chromogenic colonies in all instances.
Previous reports have found that non-O157:H7 EHEC can account for up to 60% of all EHEC infections (7, 28). The findings of this study are in agreement with those studies, since 43% of our EHEC strains were found to be non-O157 serotypes. Further, E. coli serotype O157:H7 has been found to be most prevalent during the summer months, and we also found that both O157 and non-O157 isolates were most prevalent between the months of May and September, with only one EHEC strain being isolated outside this period (25, 31).
Determining the total cost of a test for comparative purposes can be difficult, since labor, product discounting, and indirect expenses can vary widely by institution. However, comparing material costs alone can prove useful, as the other costs mentioned should be roughly equal for a given institution. In this study, a mean of four ELISA tests and two RA plates were required per positive sample, for an average total material cost of $39.60 for each positive sample tested by the ELISA-RA method. In contrast, each SMAC plate costs only $0.46, for a total of $0.92 per positive sample. Thus, the material required to identify each positive sample by the ELISA-RA method costs, on average, 43 times that of SMAC: a considerable difference. Similarly, each negative sample costs $10.80 to screen by ELISA-RA compared to $0.46 for SMAC, for a 23-fold difference.
Given the low prevalence of EHEC in the population, one must carefully consider whether the additional expense of the ELISA-RA method over culture by SMAC can be justified in the current era of managed health care. This question is made more complex by the fact that there is currently no effective treatment for EHEC-associated colitis, nor is there prophylaxis for the prevention of HUS following EHEC-associated colitis (2). On a strictly cost-based analysis, the use of the ELISA-RA method will be difficult to justify in many laboratories. However, an argument may be made for the use of the more expensive protocol for the following reasons. (i) The ELISA-RA method is superior to SMAC in the detection of all EHEC serotypes including O157:H7. (ii) While no treatment currently exists for the prevention of HUS, meticulous management of HUS is required to keep morbidity and mortality to a minimum (1, 32). (iii) Treatment modalities may become available in the near future that will block progression to HUS or limit the severity of disease: such treatments would make improved testing more relevant (3, 33). (iv) Widespread identification of all EHEC serotypes as a cause of disease will provide much-needed epidemiological data.
Alternatives to an all-or-nothing testing strategy for all EHEC could be considered for the sake of economy. These include only testing stool samples that are grossly bloody, only testing populations during the summer months when the incidence of EHEC infection appears to be greatest, or only testing at-risk (i.e., pediatric and geriatric) populations. However, not all EHEC containing stools are bloody, the seasonal distribution of all EHEC strains is not clearly understood, and EHEC can infect individuals of all ages (14, 25).
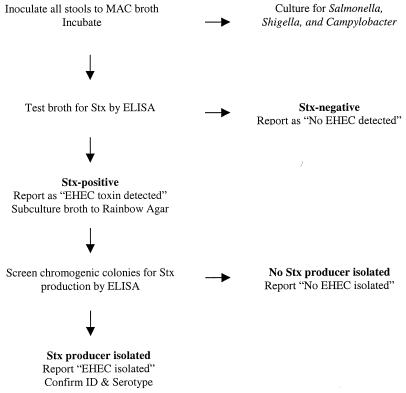
At present, there are no clear answers to this question. One strategy, using a variation of the ELISA-RA method, does not use selective criteria and thus would cover the widest patient population (Fig. 1). Costs for the primary lab could be further reduced by forwarding RA plates that have been inoculated with toxin-positive stool MAC broths to the local public health laboratory for further characterization.
FIG. 1.
Proposed scheme for the detection and isolation of EHEC.
In conclusion, we compared SMAC culture to a novel two-step method that uses ELISA-based Stx testing and a selective-differential chromogenic medium for the detection and isolation of EHEC from human stool samples. Although the predictive values of both methods are high, the ELISA-RA method proved superior to SMAC in isolating both O157:H7 and other EHEC serotypes. While the former method may be rejected on purely cost-specific grounds, an argument can nevertheless be made for its use on the grounds of improved patient care and a better epidemiological understanding of these organisms.
ACKNOWLEDGMENTS
This study was supported in part by Meridian Diagnostics, Inc., and Biolog, Inc.
PCR amplification of stx gene sequences was graciously performed by David W. K. Acheson, Division of Geographic Medicine and Infectious Diseases, New England Medical Center, Boston, Mass. We also thank the technologists of the microbiology and virology laboratories of the ARUP, the PCMC, and the UDOH, William Aldeen and the technologists of the ARUP Rapid Diagnostics Section; and Barry Bochner of Biolog, Inc., for his advice regarding the protocol design and execution.
REFERENCES
- 1.Andreoli S P. Renal manifestations of systemic diseases. Semin Nephrol. 1998;18:270–279. [PubMed] [Google Scholar]
- 2.Anonymous. Clinical approach to initial choice of antimicrobial therapy. In: Gilbert D N, Moellering R C Jr, Sande M A, editors. The Sanford guide to antimicrobial therapy. Vienna, Va: Antimicrobial Therapy, Inc.; 1998. p. 13. [Google Scholar]
- 3.Armstrong G D, Rowe P C, Goodyer P, Orrbine E, Klassen T P, Wells G, MacKenzie A, Lior H, Blanchard C, Auclair F. A phase I study of chemically synthesized verotoxin (Shiga-like) Pk-trisaccharide receptors attached to Chromosorb for preventing hemolytic-uremic syndrome. J Infect Dis. 1995;171:1042–1045. doi: 10.1093/infdis/171.4.1042. [DOI] [PubMed] [Google Scholar]
- 4.Bettelheim K A. Identification of enterohaemorrhagic Escherichia coli by means of their production of enterohaemolysin. J Appl Bacteriol. 1995;79:178–180. doi: 10.1111/j.1365-2672.1995.tb00932.x. [DOI] [PubMed] [Google Scholar]
- 5.Bettelheim K A. Studies of Escherichia coli cultured on Rainbow Agar O157 with particular reference to enterohemorrhagic Escherichia coli (EHEC) Microbiol Immunol. 1998;42:265–269. doi: 10.1111/j.1348-0421.1998.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 6.Beutin L, Zimmermann S, Gleier K. Rapid detection and isolation of Shiga-like toxin (Verocytotoxin)-producing Escherichia coli by direct testing of individual enterohemolytic colonies from washed sheep blood agar plates in the VTEC-RPLA assay. J Clin Microbiol. 1996;34:2812–2814. doi: 10.1128/jcm.34.11.2812-2814.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bokete T N, O'Callahan C M, Clausen C R, Tang N M, Tran N, Moseley S L, Fritsche T R, Tarr P I. Shiga-like toxin-producing Escherichia coli in Seattle children: a prospective study. Gastroenterology. 1993;105:1724–1731. doi: 10.1016/0016-5085(93)91069-t. [DOI] [PubMed] [Google Scholar]
- 8.Brian M J, Frosolono M, Murray B E, Miranda A, Lopez E L, Gomez H F, Cleary T G. Polymerase chain reaction for diagnosis of enterohemorrhagic Escherichia coli infection and hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1801–1806. doi: 10.1128/jcm.30.7.1801-1806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown J E, Echeverria P, Taylor D N, Seriwatana J, Vanapruks V, Lexomboon U, Neill R J, Newland J W. Determination by DNA hybridization of Shiga-like toxin-producing Escherichia coli in children with diarrhea in Thailand. J Clin Microbiol. 1989;27:291–294. doi: 10.1128/jcm.27.2.291-294.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis M, Osaki C, Gordon D, Hinds M W, Motram K, et al. Update: multistate outbreak of Escherichia coli O157:H7 infections from hamburgers - western United States, 1992–1993. Morbid Mortal Weekly Rep. 1993;42:257–263. [PubMed] [Google Scholar]
- 11.Dragon E A, Spadoro J P, Madej R. Quality control of polymerase chain reaction. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. p. 163. [Google Scholar]
- 12.Farmer J J, III, Davis B R. H7 antiserum-sorbitol fermentation medium: a single-tube screening method for detecting Escherichia coli O157:H7 associated with hemorrhagic colitis. J Clin Microbiol. 1985;22:620–625. doi: 10.1128/jcm.22.4.620-625.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gannon V P J, Rashed M, King R K, Golsteyn Thomas E J. Detection and characterization of the eae gene of Shiga-like toxin-producing Escherichia coli by using polymerase chain reaction. J Clin Microbiol. 1993;31:1268–1274. doi: 10.1128/jcm.31.5.1268-1274.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin P M, Ostroff S M, Tauxe R V, Greene K D, Wells J G, Lewis J H, Blake P A. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. An Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- 15.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 16.Gunzer F, Bohm H, Russmann H, Bitzan M, Aleksic S, Karch H. Molecular detection of sorbitol-fermenting Escherichia coli O157 in patients with hemolytic uremic syndrome. J Clin Microbiol. 1992;30:1807–1810. doi: 10.1128/jcm.30.7.1807-1810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermann J E. Immunoassays for the diagnosis of infectious diseases. In: Murray P R, Baron E J, Phaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. p. 111. [Google Scholar]
- 18.Karmali A K. Laboratory diagnosis of Verotoxin-producing Escherichia coli infections. Clin Microbiol Newslett. 1987;19:105–108. [Google Scholar]
- 19.Karmali M A, Steele B T, Petric M, Lim C. Sporadic cases of haemolytic uremic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet. 1983;i:619–620. doi: 10.1016/s0140-6736(83)91795-6. [DOI] [PubMed] [Google Scholar]
- 20.Kehl K S, Havens P, Behnke C E, Acheson D W K. Evaluation of the Premier EHEC assay for detection of Shiga toxin-producing Escherichia coli. J Clin Microbiol. 1997;35:2051–2054. doi: 10.1128/jcm.35.8.2051-2054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konowalchuk J, Speirs J I, Stavric S. Vero response to a cytotoxin of Escherichia coli. Infect Immun. 1977;18:775–779. doi: 10.1128/iai.18.3.775-779.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin D L, MacDonald K L, White K E, Soler J T, Osterholm M T. The epidemiology and clinical aspects of the hemolytic uremic syndrome in Minnesota. N Engl J Med. 1990;323:1161–1167. doi: 10.1056/NEJM199010253231703. [DOI] [PubMed] [Google Scholar]
- 23.Meridian Diagnostics, Inc. Premier EHEC ELISA Product insert. Cincinnati, Ohio: Meridian Diagnostics, Inc.; 1997. [Google Scholar]
- 24.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pai C H, Ahmed N, Lior H, Johnson W M, Sims H V, Woods D E. Epidemiology of sporadic diarrhea due to verotoxin-producing Escherichia coli: a two year prospective study. J Infect Dis. 1988;157:1054–1057. doi: 10.1093/infdis/157.5.1054. [DOI] [PubMed] [Google Scholar]
- 26.Park H C, Vandel N M, Hixon D L. Rapid immunoassay for detection of Escherichia coli O157 directly from stool specimens. J Clin Microbiol. 1996;34:988–990. doi: 10.1128/jcm.34.4.988-990.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paton A W, Paton J C, Goldwater P N, Manning P A. Direct detection of Escherichia coli Shiga-like toxin genes in primary fecal cultures by polymerase chain reaction. J Clin Microbiol. 1993;31:3063–3067. doi: 10.1128/jcm.31.11.3063-3067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierard D, Van Etterijck R, Breynaert J, Moriau L, Lauwers S. Results of screening for verocytotoxin-producing Escherichia coli in faeces in Belgium. Eur J Clin Microbiol Infect Dis. 1990;9:198–201. doi: 10.1007/BF01963837. [DOI] [PubMed] [Google Scholar]
- 29.Riley L W, Remis R S, Helgerson S D, McGee H B, Wells J G, Davis B R, Hebert R J, Olcott E S, Johnson L M, Hargrett N T, Blake P A, Cohen M L. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt H, Beutin L, Karch J. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slutsker L, Ries A A, Greene K D, Wells J G, Hutwagner L, Griffin P M. Escherichia coli O157:H7 diarrhea in the United States: clinical and epidemiologic features. Ann Intern Med. 1997;126:505–513. doi: 10.7326/0003-4819-126-7-199704010-00002. [DOI] [PubMed] [Google Scholar]
- 32.Tarr P I, Neill M A, Clausen C R, Watkins S L, Christie D L, Hickman R O. Escherichia coli O157:H7 and the hemolytic uremic syndrome: importance of early cultures in establishing the etiology. J Infect Dis. 1990;162:553–556. doi: 10.1093/infdis/162.2.553. [DOI] [PubMed] [Google Scholar]
- 33.Volker R. New strategies aimed at E. coli O157:H7. JAMA. 1994;272:503. [PubMed] [Google Scholar]