Supplemental Digital Content is available in the text.
Keywords: cardiac myocyte, Hutchinson-Gilford progeria syndrome, smooth muscle, cell
Background:
Hutchinson-Gilford progeria syndrome (HGPS) is a rare disorder characterized by premature aging and death mainly because of myocardial infarction, stroke, or heart failure. The disease is provoked by progerin, a variant of lamin A expressed in most differentiated cells. Patients look healthy at birth, and symptoms typically emerge in the first or second year of life. Assessing the reversibility of progerin-induced damage and the relative contribution of specific cell types is critical to determining the potential benefits of late treatment and to developing new therapies.
Methods:
We used CRISPR-Cas9 technology to generate LmnaHGPSrev/HGPSrev (HGPSrev) mice engineered to ubiquitously express progerin while lacking lamin A and allowing progerin suppression and lamin A restoration in a time- and cell type–specific manner on Cre recombinase activation. We characterized the phenotype of HGPSrev mice and crossed them with Cre transgenic lines to assess the effects of suppressing progerin and restoring lamin A ubiquitously at different disease stages as well as specifically in vascular smooth muscle cells and cardiomyocytes.
Results:
Like patients with HGPS, HGPSrev mice appear healthy at birth and progressively develop HGPS symptoms, including failure to thrive, lipodystrophy, vascular smooth muscle cell loss, vascular fibrosis, electrocardiographic anomalies, and precocious death (median lifespan of 15 months versus 26 months in wild-type controls, P<0.0001). Ubiquitous progerin suppression and lamin A restoration significantly extended lifespan when induced in 6-month-old mildly symptomatic mice and even in severely ill animals aged 13 months, although the benefit was much more pronounced on early intervention (84.5% lifespan extension in mildly symptomatic mice, P<0.0001, and 6.7% in severely ill mice, P<0.01). It is remarkable that major vascular alterations were prevented and lifespan normalized in HGPSrev mice when progerin suppression and lamin A restoration were restricted to vascular smooth muscle cells and cardiomyocytes.
Conclusions:
HGPSrev mice constitute a new experimental model for advancing knowledge of HGPS. Our findings suggest that it is never too late to treat HGPS, although benefit is much more pronounced when progerin is targeted in mice with mild symptoms. Despite the broad expression pattern of progerin and its deleterious effects in many organs, restricting its suppression to vascular smooth muscle cells and cardiomyocytes is sufficient to prevent vascular disease and normalize lifespan.
Clinical Perspective.
What Is New?
We have generated a new Hutchinson-Gilford progeria syndrome–like mouse model that ubiquitously expresses progerin, lacks lamin A and allows progerin suppression and lamin A restoration in a time- and cell type–specific manner on Cre recombinase activation.
Progerin suppression and lamin A restoration extended the lifespan in mice with mild symptoms and even in severely ill animals, although the benefit was much more apparent in mildly symptomatic animals.
Despite the broad expression pattern of progerin and its deleterious effects in many organs, restricting its suppression and lamin A restoration to vascular smooth muscle cells and cardiomyocytes prevented the development of vascular pathology and normalized lifespan in HGPSrev mice.
What Are the Clinical Implications?
Our preclinical studies demonstrate that it is never too late to treat Hutchinson-Gilford progeria syndrome, although the benefit was much more pronounced when progerin suppression and lamin A restoration were achieved at early stages of disease progression.
Strategies to treat Hutchinson-Gilford progeria syndrome through gene therapy or RNA therapy should consider targeting vascular smooth muscle cells and cardiomyocytes.
Hutchinson-Gilford progeria syndrome (HGPS) is an ultrarare genetic disorder (estimated prevalence 1 in 18–20 million people; https://www.progeriaresearch.org/) characterized by accelerated aging and premature death (average lifespan, 14.6 years).1–3 Most patients with HGPS are heterozygous for a de novo synonymous mutation in the LMNA gene (c.1824C>T; p.G608G) that activates the use of a cryptic splice donor site in exon 11.4,5 In normal cells, LMNA expression mostly generates the alternatively spliced isoforms lamin A and lamin C. The HGPS-causing mutation creates an aberrant LMNA mRNA that lacks 150 nucleotides in exon 11; this is translated into progerin, a permanently farnesylated lamin A variant that exerts a dominant-negative effect.1 Growth failure and alopecia manifest in patients with HGPS as the first disease symptoms typically in the first or second year of life. Additional symptoms develop and worsen over time, including dermal and bone abnormalities, joint contractures, and loss of subcutaneous fat. The main medical problem in HGPS is severe cardiovascular disease, including generalized atherosclerosis and vascular calcification and stiffness, which ultimately provoke myocardial infarction, stroke, or heart failure, the causes of death in most patients with HGPS.6,7 The US Food and Drug Administration recently approved the treatment of patients with HGPS with lonafarnib (marketed as Zokinvy by Eiger BioPharmaceuticals), a repurposed farnesyltransferase inhibitor that has extended the lifespan of patients with HGPS by 2.5 years (17% increase).8–10 Nevertheless, there is a great need for better therapies to improve and eventually cure HGPS.
Patients with HGPS are typically diagnosed when symptoms are present or even severe, and treatment has historically been initiated at different disease stages.6 It is therefore important to ascertain how late in life treatment can be initiated in symptomatic individuals while still yielding clinical benefit. Moreover, because progerin is expressed in most differentiated cells, it is vital to identify the cell types that would most benefit from treatment. To address these questions, here we generated the LmnaHGPSrev/HGPSrev (HGPSrev) mouse model, engineered to ubiquitously express progerin and lamin C and lack lamin A, while allowing progerin suppression and lamin A restoration on Cre recombinase activation. We have characterized this model to assess the reversibility of progerin-induced damage by targeting progerin at early and late disease stages. Moreover, we have examined the consequences of suppressing progerin and restoring lamin A specifically in vascular smooth muscle cells (VSMCs) and cardiomyocytes, the major progerin targets.
Methods
Additional methods are provided in the Supplemental Methods.
Data, analytical methods, and study materials will be made available to other researchers for the purposes of reproducing these results or replicating these procedures on reasonable request directed to the authors’ laboratories.
Study Approval and Mouse Models
Mice used in this study were housed in the animal facilities at the Centro Nacional de Investigaciones Cardiovasculares under specific pathogen-free conditions at a constant temperature of 23°C, relative humidity 58%, and a 12-hour dark/light cycle. Mouse health was monitored in a blinded manner at regular intervals throughout the study. Mouse handling and experimental procedures were performed to conform with current European Union guidelines (Directive 2010/63/EU) and Recommendation 2007/526/EC on the protection of animals used for scientific purposes, enforced in Spanish law under Real Decreto 1201/2005; all procedures were approved by the Animal Protection Area of the Comunidad Autónoma de Madrid (PROEX 051/18) and the Centro Nacional de Investigaciones Cardiovasculares Ethics Review Board. To maximize information and minimize the number of animals used, we followed the 3Rs (replace, reduce and refine) principles11 and the ARRIVE guidelines (Animal Research: Reporting of In Vivo Experiments)12 throughout this study.
Studies were carried out in C57BL/6J mice fed a regular rodent chow diet. Equal numbers of females and males were used. Ubc-CreERT2tg/+ mice (B6.Cg Ndor1Tg(UBC-cre/ERT2)1Ejb/1J)13 and LmnaG609G/G609G mice14 were kindly provided by Dr Mariano Barbacid and Dr Carlos López-Otín, respectively. The generation of HGPSrev mice is explained below. HGPSrev mice were crossed with Ubc-CreERT2tg/+ mice to generate LmnaHGPSrev/HGPSrev Ubc-CreERT2tg/+ mice with the Cre transgene in heterozygosis and exhibiting time-conditional Cre activity (referred to as HGPSrev-Ubc-CreERT2 mice). SM22α-Cre mice (B6.Cg-Tg(Tagln-cre)1Her/J15; The Jackson Laboratory; Bar Harbor, ME) were crossed with HGPSrev mice to generate LmnaHGPSrev/HGPSrev-SM22α-Cre mice (HGPSrev-SM22α-Cre mice).
Double-Stranded DNA Donor Template Design for LmnaHGPSrev Strain Generation
For homology directed repair, a 2494-bp double-stranded DNA donor template flanked by EcoRI and NotI recognition sites was synthesized (Figure S1) and inserted into the pcDNA3.1 vector (Genscript; Piscataway, NJ). The double-stranded DNA donor template contains a 938-bp left homology arm (Lmna intron 9, exon 10, and part of intron 10), a 672-bp insert harboring a loxP-flanked cDNA containing exons 11 and 12 from LmnaΔ150 (exon11Δ150 and the coding sequence of exon 12), followed by a bovine growth hormone polyadenylation transcriptional stop signal (BGH-polyA), and an 877-bp right homology arm (part of Lmna intron 10, exon 11, intron 11, and part of exon 12).
Oocyte Microinjection and Implantation Into Pseudopregnant Females
Hormonal superovulation was induced in 10 immature female mice (3–5 weeks old, C57BL/6J genetic background) by intraperitoneal hormone injection. Mice first received an injection of 0.1 mL (5 international units [IU]) of pregnant mare serum gonadotropin, followed 48 hours later by an injection of 0.1 mL (5 IU) of human chorionic gonadotropin. Immediately after the second injection, animals were mated with appropriate stud males. One day after mating, females were checked for vaginal plugs, and those with a positive result were euthanized. Oviducts from euthanized females were extracted and transferred to M2 culture medium (M7167; Sigma-Aldrich; St Louis, MO) containing 350 µg/mL hyaluronidase (H3884; Sigma-Aldrich). Each ampulla was localized and opened to release the cumulus mass, and oocytes were separated after incubation at 37°C for 1 to 2 minutes and then transferred to fresh M2 medium for washing. Zygotes were incubated in Evolve-KSOM culture medium (ZEKS-050; Zenith Biotech; Cork, Ireland) at 37°C in a 5% CO2/5% O2 atmosphere, until they were ready for pronuclear microinjection.16 Zygotes were microinjected with 1 to 2 pL of a microinjection solution containing guide RNAs, double-stranded DNA donor template, and Cas9 endonuclease (Table S1). Zygotes were incubated overnight at 37°C and 5% CO2/5% O2 in Evolve-KSOM medium (ZEKS-050; Zenith Biotech) to reach the 2-cell stage.16 Embryos at the 2-cell stage were transferred to pseudopregnant female mice by passing a sterile glass needle through the infundibulum. Three weeks later, 34 pups were weaned from their gestational mothers.
Identification of Founder HGPSrev Mice
To identify mice carrying the LmnaHGPSrev allele, we extracted genomic DNA from the tails of the 34 mouse pups following a standard protocol with proteinase K (EO0492; Thermo Fisher; Waltham, MA) and performed PCR with specific primers (Table S2, Founders: PCR-1). Additional PCR reactions were run to identify mice carrying a single copy of the mutant allele at the proper location in the Lmna locus (Table S2, Founders: PCR-2 and PCR-3). Genomic DNA from the 4 pups carrying a single copy of the edited allele was amplified by PCR and sequenced at the Sequencing Service of the Centro Nacional de Investigaciones Oncológicas (Madrid, Spain; Figure 1B).
Genotyping of HGPSrev Mice
To genotype HGPSrev, HGPSrev-Ubc-CreERT2, and HGPSrev-SM22α-Cre mice, genomic DNA was extracted from the tail following a standard proteinase K protocol (EO0492; Thermo Fisher), and PCR reactions were performed with specific primers (Table S2, Genotyping).
Isolation, Immortalization, and Transfection of Mouse Embryonic Fibroblasts
Embryos isolated at embryonic day 13.5 were minced and incubated for 20 minutes in 2× trypsin-EDTA (0.5% trypsin, 0.53 mmol/L EDTA•4Na; 15400-054; Invitrogen; Carlsbad, CA). Mouse embryonic fibroblasts (MEFs) from each embryo were plated separately and incubated at 37°C in complete growth medium (DMEM supplemented with 10% heat-inactivated fetal bovine serum, 5% nonessential amino acids, 5% penicillin/streptomycin, and 5% L-glutamine [v/v]). To generate virus to immortalize MEFs, HEK293T cells were seeded and transfected with pCL-Puro-SV40 LT retroviral vector (13970; Addgene; Watertown, MA) and pCL-ECO retroviral packaging plasmid (kindly provided by Dr. Manuel Serrano) using Fugene 6 (E2692; Promega; Madison, WI). Supernatants containing retroviral particles were harvested every 12 hours during 2 days, filtered through 0.45-µm pores, mixed with polybrene (8 µg/mL; 9268-5G; Sigma-Aldrich), and used to infect 2.5 to 5 × 105 MEFs in 100-mm dishes. Infected MEFs were serially passaged to select immortalized cells by adding 2 µg/mL puromycin (P8833; Sigma-Aldrich).
Immortalized MEFs were seeded at ≈80% confluence and were transfected with 1.5 µg pPB CAG ER-Cre-ER IRES Zeocin17 and 4.5 µg pCMV-hyPBase18 mixed with 18 µL TransIT-LT1 Transfection Reagent (MIR2300; GeneFlow; Staffordshire, UK) in DMEM. Zeocin-resistant cells were selected by incubation for at least 10 days in the presence of 400 μg/mL zeocin (R25005; Thermo Fisher).
Longevity Studies
Starting at 4 to 8 weeks of age, mice were weighed and inspected for health and survival twice a week. Health status was examined by a blinded veterinarian. Animals that met humane end point criteria were euthanized and the deaths recorded for the survival curve analysis. The robust regression and outlier removal test was performed in all survival experiments, and identified two outliers: a tamoxifen-injected HGPSrev-Ubc-CreERT2 mouse that died at 34 months and an oil-injected mouse that died at 17 months (in both cases, administration of tamoxifen/oil started at ≈13 months). These mice were excluded from the survival curve and statistical analysis.
Histology and Immunofluorescence
Mouse tissues were fixed in 4% formaldehyde solution (prepared from paraformaldehyde) for 24 h at 4°C, dehydrated through an ascending series of ethanol concentrations, and last embedded in paraffin and cut into serial 4-μm sections.
For immunofluorescence studies, tissue cross-sections were deparaffinized, rehydrated, and washed in PBS. Antigen retrieval was performed by boiling the sections in 10 mmol/L sodium citrate buffer (pH 6) for 20 minutes, and samples were then blocked and permeabilized for 1 hour at room temperature in PBS supplemented with 0.3% Triton X-100, 5% BSA, and 5% normal goat serum (005-000-001; Jackson ImmunoResearch; West Grove, PA). Heart samples were in addition blocked with 100 mmol/L glycine (5001901000; Merck; Kenilworth, NJ). Primary and secondary antibodies were diluted in PBS supplemented with 0.3% Triton X-100, 5% BSA, and 2.5% normal goat serum. Primary antibodies included rat monoclonal anti-CD31 (1:50; DIA-310; Dianova; Hamburg, Germany) and rabbit polyclonal anti-progerin (1:800; generated by the Nourshargh laboratory using peptide immunogens and standard immunization procedures). After overnight incubation at 4°C, samples were washed and then incubated with the appropriate fluorescently labeled secondary antibodies for 2 hours at room temperature (1:400; goat antirat Alexa Fluor 488 A-11006 or 1:400 goat anti-rabbit Alexa Fluor 647 A-21245; Invitrogen) together with anti-α-smooth muscle actin-Cy3 (1:300; C6198; Sigma-Aldrich) and Hoechst nuclear stain (bisBenzimide H 33342 trihydrochloride, B2261; Sigma-Aldrich). Samples were washed and then mounted in Fluoromount-G imaging medium (4958-02; eBioscience; San Diego, CA). Images were acquired with a Leica SP5 DMI 6000B (Leica Microsystems; Wetzlar, Germany) or a LSM 700 Carl Zeiss (Zeiss; Oberkochen, Germany) confocal microscope.
For staining with hematoxylin-eosin and Masson’s trichrome, tissue cross-sections were deparaffinized, rehydrated, and washed in PBS. Stained sections were scanned with a NanoZoomer-RS scanner (Hamamatsu, Japan), and images were exported with NDP.view2 software (Hamamatsu) and quantified with user-customized macros in Fiji software by an operator blinded to genotype. The thickness of the subcutaneous fat layer was qualitatively scored from 1 (thinnest) to 5 (thickest) by 5 independent observers, and mean results are presented for each mouse. For quantitative analysis, regions containing epidermis, hypodermis, and muscle layers were examined by a single blinded manner observer, and the mean thickness was calculated from 10 independent measurements per mouse.
mRNA Isolation and Reverse Transcription for PCR Detection of Lamin A and Progerin
Total RNA was extracted from powdered mouse tissue samples using TriReagent Solution (AM9738; Thermo Fisher) and processed by alcohol precipitation. RNA was quantified in a NanoDrop ND-1000 spectrophotometer, and 1 to 2 µg were transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (4368814; Applied Biosystems; Foster City, CA). Lamin A and progerin mRNAs were detected as previously described.19 cDNA samples (100 ng) were amplified by PCR, and products were separated on a 1.5% agarose gel. Images were acquired with the Molecular Imager Gel Doc XR+ System (Bio-Rad).
Plasma Biochemistry
Blood was extracted from the mandibular sinus of live mice or from the heart (by cardiac puncture) or infrarenal abdominal aorta of euthanized animals. For biochemical analysis, plasma was obtained from blood samples collected in Microvette EDTA tubes (Sarstedt; Newton, NC) by specialized Centro Nacional de Investigaciones Cardiovasculares Animal Facility staff and centrifuged at 180 to 200 g for 15 minutes at 4°C. Biochemical variables were analyzed using a Dimension RxL Max Integrated Chemistry System (Siemens Healthineers; Erlangen, Germany).
ECG
Mice were anesthetized with 1.5% to 2% isoflurane, and 4 ECG electrodes were inserted subcutaneously into the limbs. ECG was recorded in the morning for ≈2 minutes using a MP36R data acquisition workstation (Biopac Systems; Goleta, CA). ECG data were exported with AcqKnowledge software (Biopac Systems) and automatically analyzed using custom R scripts developed to (1) remove noise and baseline fluctuations; (2) detect heartbeats, peaks, and waves; (3) exclude artifacts; and (4) calculate QT intervals and T-wave steepness.
Tamoxifen Administration
Tamoxifen (4-hydroxy-tamoxifen, H6278; Sigma Aldrich) was dissolved in ethanol for cell studies or in corn oil (C8267; Sigma Aldrich) for mouse studies. The corn oil preparation was incubated at 55°C until the tamoxifen was fully dissolved and then passed through a 0.22-µm filter.
Zeocin-resistant wild-type (WT) and HGPSrev MEFs were exposed to 25 nmol/L tamoxifen, and protein lysates were prepared 24, 48, and 72 h after tamoxifen administration. Negative controls were treated with equal volume of vehicle (ethanol) for 72 hours.
Mice were randomized to tamoxifen or oil groups balanced for age and sex. For proof-of-concept studies, ≈3-month-old HGPSrev-Ubc-CreERT2 mice received daily intraperitoneal oil or tamoxifen injections (2 mg/d/mouse) for 10 days; the mice were euthanized 1 week after finishing the treatment, and tissues were extracted for Western blot analysis. For the analysis of HGPSrev-Ubc-CreERT2 mice at different disease stages, tamoxifen administration was commenced at different ages. For longitudinal studies of survival, health status, and body weight evolution, the effect on early disease was assessed by starting oil/tamoxifen administration at ≈6 months of age, coinciding with the beginning of growth failure, whereas late disease was assessed by starting treatment at ≈13 months of age, when mice had developed severe symptoms and were close to maximum survival (Table). For ECG and histopathologic analysis, the effect on intermediate disease (characterized by mild cardiovascular symptoms) was assessed by starting treatment at ≈9 months of age. All mice received daily intraperitoneal injections (1 mg/d/mouse) during 5 days and were euthanized at 14.5 months of age, at which point 4 out of 10 oil-injected animals had died and only 1 out of 12 tamoxifen-injected animals had died.
Table 1.
Age of LmnaHGPSrev-Ubc-CreERT2 Mice at the Time of Initiation of Oil or Tamoxifen Administration
Statistical Analysis
Quantitative data are presented as the mean±SEM unless otherwise stated. Statistical tests were applied after the determination of normal distribution (Shapiro-Wilk normality test) and equality of variances (F test). In experiments with 2 groups and normal distribution, the statistical significance of differences was assessed by unpaired 2-tailed Student’s t test. For nonnormally distributed data in experiments with 2 groups, we used the Mann-Whitney test. In experiments with >2 groups of normally distributed populations, we applied 1-way ANOVA followed by the post hoc Tukey test. For nonnormally distributed data, the nonparametric equivalent Kruskal-Wallis test was performed. When >2 groups were assessed over time without sphericity, we used mixed-effects analysis (2-way ANOVA) with the Geisser-Greenhouse correction and Sidák’s multiple comparisons. Kaplan-Meier survival curves were compared by the log-rank (Mantel-Cox) test. Data on body weight evolution were analyzed by unpaired multiple t tests with the Holm-Sidák correction.
All statistical tests were run in GraphPad Prism 9.0.0. Differences were considered statistically significant when P values were <0.05: *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Results
HGPSrev Mice Develop Progeroid Symptoms and Allow Cre-Dependent Progerin Suppression and Lamin A Restoration
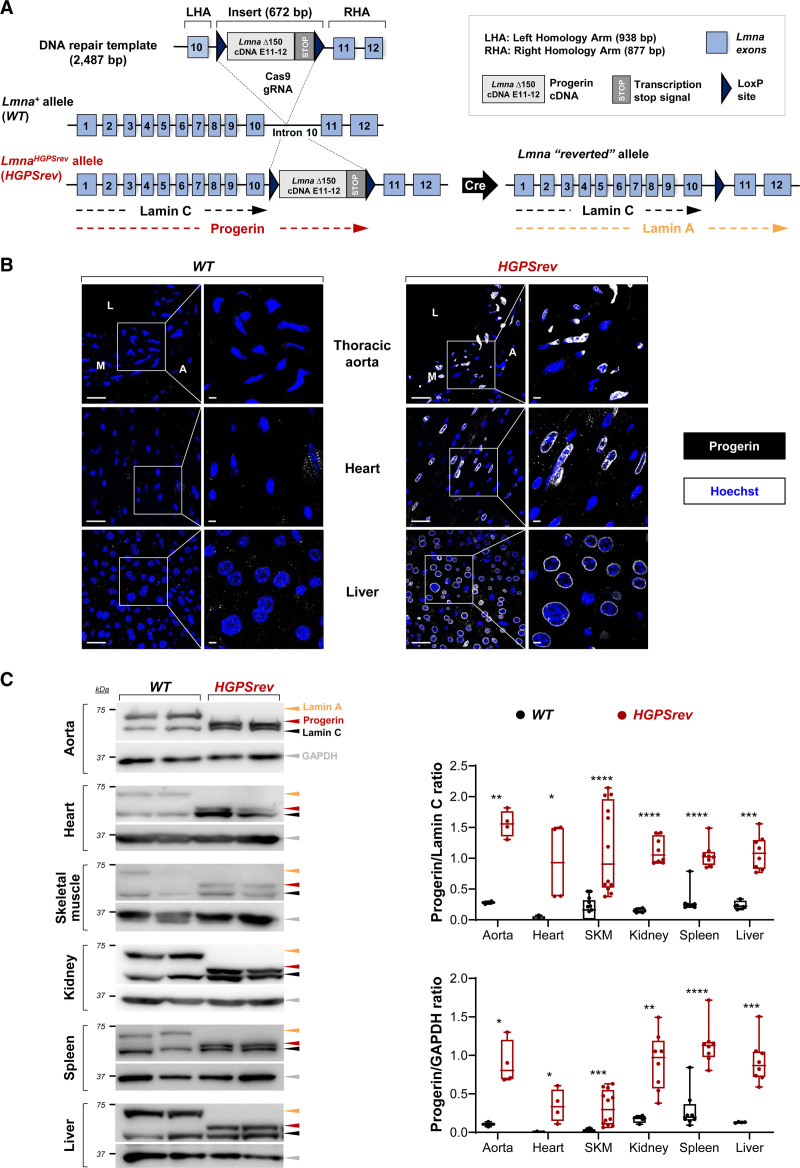
We used the CRISPR-Cas9 strategy to generate the HGPSrev mouse model, engineered to ubiquitously express progerin and lamin C and to lack lamin A, while allowing progerin suppression and lamin A restoration on Cre recombinase activation (Figure 1A, Figure S1A and S1B). Western blot analysis in protein lysates from the tails of founder HGPSrev mice revealed progerin and lamin C expression and undetectable lamin A (Figure S1C). HGPSrev mice obtained after breeding founder mice expressed progerin in all tissues tested, as assessed by reverse transcriptase PCR (Figure S1D: compare WT with HGPSrev), immunofluorescence (Figure 1B, Figure S2: compare WT with HGPSrev), and Western blot (Figure 1C). Analysis of multiple tissues by semiquantitative PCR showed lower progerin mRNA content in HGPSrev mice than in LmnaG609G/G609G mice, a widely used HGPS model14 (Figure S1D: compare HGPSrev with G609G).
Figure 1.
LmnaHGPSrev/HGPSrev (HGPSrev) mice exhibit ubiquitous progerin expression and undetectable lamin A expression. A, CRISPR-Cas9 strategy for generating HGPSrev mice (see details in Methods and Figure S1A). Cre activity generates a Lmna “reverted” allele that causes progerin suppression and lamin A restoration. B, Representative immunofluorescence images showing progerin expression (white) and nuclei (blue) in wild-type (WT) and HGPSrev mice. Scale bar, 25 µm. C, Western blot of lamin A/C, progerin, and GAPDH in 2-month-old WT and HGPSrev mice. Six mice of each genotype were analyzed, and representative images are shown of 2 mice of each genotype. The graphs show the relative amount of progerin normalized using lamin C and GAPDH as controls. (n=3–13 WT mice; n=4–12 HGPSrev mice). Statistical analysis was performed by 2-tailed t test. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. A indicates adventitia; L, lumen; M, media; and SKM, skeletal muscle.
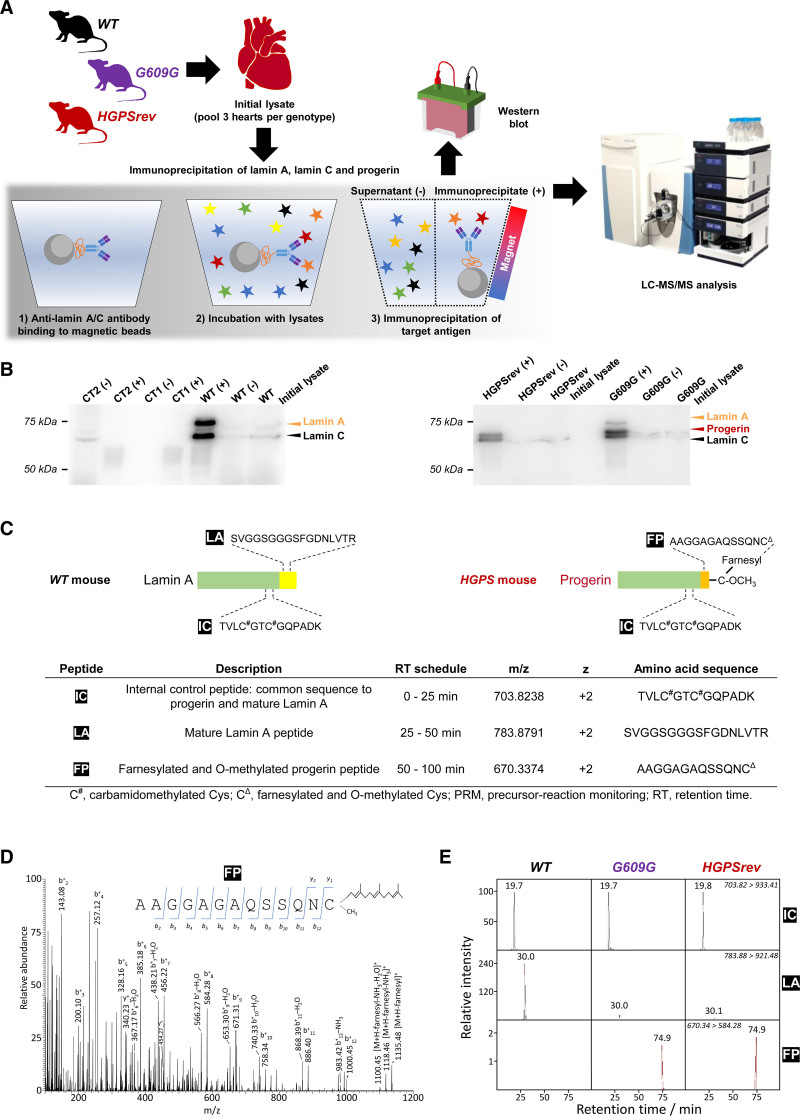
We next examined progerin farnesylation in HGPSrev and LmnaG609G/G609G mice, using WT mice as a negative control. Proteins were immunoprecipitated from heart lysates with an antilamin A/C antibody that recognizes lamin A/C and progerin, and samples were analyzed by Western blot and liquid chromatography coupled to targeted tandem mass spectrometry using a high-resolution precursor-reaction monitoring method (Figure 2A, Supplemental Methods). Compared with the initial lysates, WT heart immunoprecipitates were enriched in lamin A/C (Figure 2B, WT [+] versus WT initial lysate), HGPSrev heart immunoprecipitates were enriched in progerin and lamin C and contained no detectable lamin A (Figure 2B, HGPSrev [+] versus HGPSrev initial lysate), and LmnaG609G/G609G heart immunoprecipitates were enriched in progerin and lamin A/C (Figure 2B, G9609G [+] versus G609G initial lysate). For precursor-reaction monitoring, we monitored a peptide common to lamin A and progerin (internal control) and peptides specific for lamin A and farnesylated progerin (Figure 2C). The tandem mass spectrometry spectra confirmed the presence of the farnesyl moiety in the monitored farnesylated progerin peptide (Figure 2D). MS/MS (tandem mass spectrometry) extracted ion chromatograms obtained from the time-scheduled precursor-reaction monitoring assay showed similar amounts of farnesylated progerin peptide in HGPSrev and LmnaG609G/G609G hearts relative to the total amount of A-type lamin isoforms in these mutant strains, whereas farnesylated progerin peptide was undetectable in WT hearts (Figure 2E). Consistent with this finding, lamin A peptide was strongly depleted in HGPSrev and LmnaG609G/G609G hearts relative to WT hearts (Figure 2E).
Figure 2.
Targeted precursor-reaction monitoring (PRM) analysis to examine progerin farnesylation in mouse heart lysates. A, Workflow for the LC-MS/MS analysis of proteins extracted from mouse hearts and immunoprecipitated with anti-lamin A/C antibodies that recognize lamin A, lamin C, and progerin. For each genotype, each sample was the pool of 3 hearts. WT, wild-type mice; G609G, LmnaG609G/G609G mice; HGPSrev, LmnaHGPSrev/HGPSrev mice. B, Western blots using anti-lamin A/C antibody to check the enrichment of lamin A, lamin C, and progerin in the immunoprecipitated material and supernatant (+, immunoprecipitated; –, supernatant). Controls included samples containing only beads and antibody (CT1) and only beads and protein extract (CT2). A 10-µL aliquot of each sample was loaded onto the gel; see details in Supplemental Material. C, Surrogate peptides used to detect mature lamin A and progerin: IC, internal control peptide (present in both lamin A and progerin); LA, lamin A peptide (specific for lamin A); FP, farnesylated progerin peptide (specific for progerin). D, MS2 fragmentation spectrum from FP obtained in the PRM assay. The insert shows ion ascription to the main fragment-ion series (C-terminal y-series and N-terminal b-series). E, MS/MS (tandem mass spectrometry) extracted ion chromatograms of IC, LA, and FP peptides obtained from the time-scheduled PRM assay for the detection of lamin A and progerin. The ion traces were obtained using fragment ion y+9 from IC, y+8 from LA, and b+8 from FP. LC-MS/MS indicates liquid chromatography coupled to targeted tandem mass spectrometry.
To examine potential off-target (OT) effects in HGPSrev mice, we used the online Off-Spotter tool (https://cm.jefferson.edu/Off-Spotter/). Compared with the 20-mer single guide RNA used for CRISPR-Cas9–dependent editing, this analysis identified 184 mouse genomic sequences containing 3, 4, or 5 mismatches (2, 16, and 166 sequences, respectively; Figure S3A). We selected 3-mismatch sequences (OT-1 and OT-2) and 6 of the 4-mismatch sequences (OT-3, OT-4, OT-5, OT-6, OT-7, and OT-8; Figure S3B), which were amplified by polymerase chain reaction (PCR) using as a template genomic DNA of WT and HGPSrev mice (n=5 per genotype). The PCR products had identical DNA sequences in all mice for all genomic regions examined (Figure S3C), indicating the absence of OT effects.
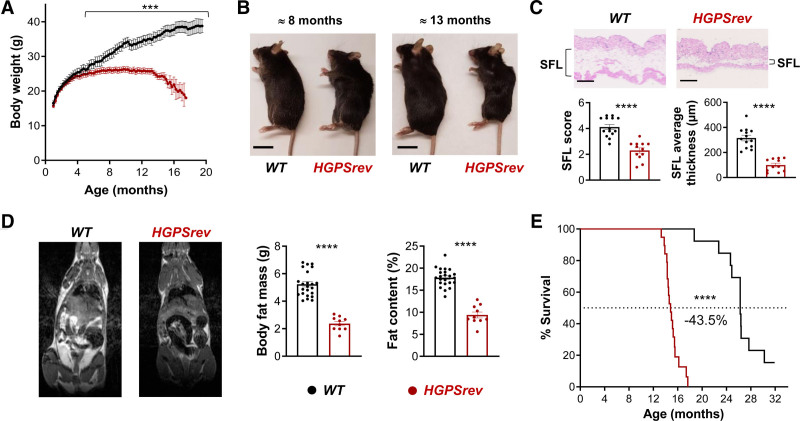
HGPSrev mice looked normal at birth and maintained a normal appearance until aged ≈5 months, when both males and females stopped gaining weight (Figure 3A). From this age, HGPSrev mice also exhibited kyphosis (Figure 3B) and showed a significant fat loss, as assessed from subcutaneous fat histology (Figure 3C) and in vivo magnetic resonance imaging (Figure 3D). At ≈8 months of age, HGPSrev mice had below-normal levels of plasma low-density lipoprotein, but other lipids were normal (Figure S4A). In ≈13-month-old HGPSrev mice, plasma triglycerides, total cholesterol, and high-density lipoprotein levels were reduced (Figure S4B), consistent with observations in other progerin-expressing mouse models.20 Progerin expression was also associated with premature death in HGPSrev mice, which had a median lifespan of 15 months versus 26 months in WT controls (43.5% reduction; Figure 3E).
Figure 3.
Progeroid phenotype in LmnaHGPSrev/HGPSrev (HGPSrev) mice with ubiquitous progerin expression. A, Postnatal body weight curves (n=14 WT; n=22 HGPSrev). Differences were analyzed by unpaired multiple t-tests and the Holm-Sidák correction. B, Representative images of ≈8- and ≈13-month-old mice. Scale bar, 2 cm. C, Representative images of hematoxylin-eosin–stained skin from ≈13-month-old mice and the results of subcutaneous fat layer (SFL) score and thickness quantification (see Methods; n=13 or 14 WT; n=11 or 12 HGPSrev). Statistical analysis was performed by 2-tailed t test. Scale bar, 500 µm. D, Representative images of sagittal whole-body cross-sections obtained by magnetic resonance imaging (fat shown in white) and quantification of body fat mass and percentage fat content in ≈13-month-old mice (n=23 WT; n=10 HGPSrev). Differences were analyzed by 2-tailed t test. E, Kaplan-Meier survival curve (n=13 WT; n=22 HGPSrev). Differences were analyzed by the Mantel-Cox test. ***P<0.001; ****P<0.0001. Data are mean±SEM. Each symbol represents 1 animal.
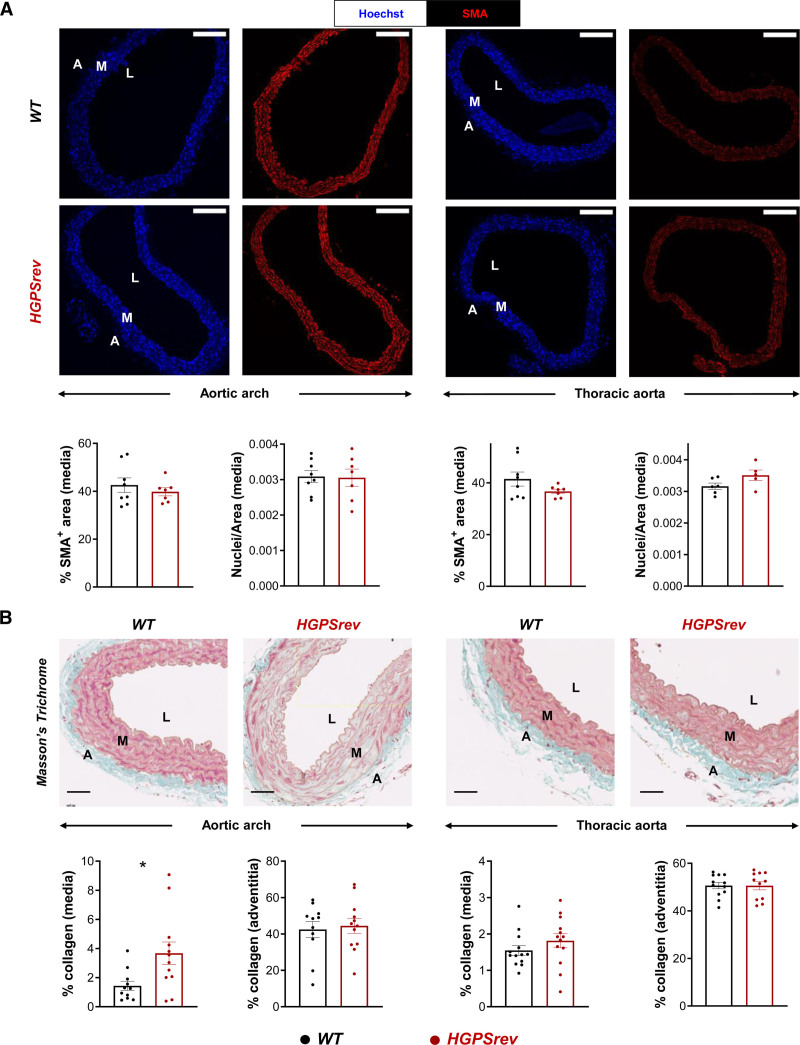
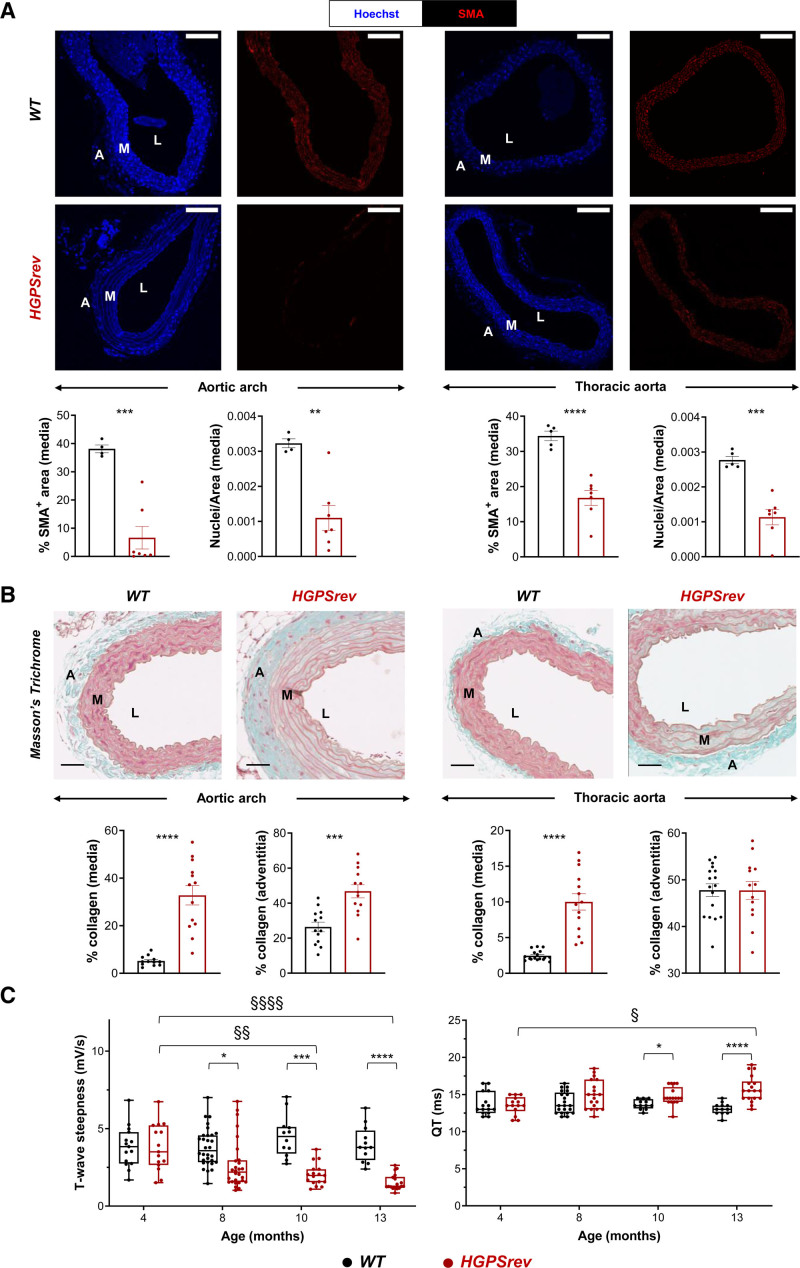
Patients with HGPS and animal models are both characterized by VSMC loss and collagen accumulation in the artery wall, as well as electrocardiographic alterations.7,14,20–30 In ≈8-month-old HGPSrev mice, medial VSMC content appeared normal in both the aortic arch and thoracic aorta (Figure 4A); however, the media of the aortic arch showed increased collagen accumulation compared with age-matched WT mice (Figure 4B). Disease progression manifested as severe VSMC depletion in the aortic arch and thoracic aorta in ≈13-month-old HGPSrev mice (Figure 5A), which also had a significantly elevated collagen content in both the media and adventitia of the aortic arch and in the media of the thoracic aorta (Figure 5B). Likewise, longitudinal ECG assessment of HGPSrev mice revealed an age-dependent reduction in T-wave steepness from 8 months of age and an increase in the QT interval from 10 months (Figure 5C). These findings demonstrate that HGPSrev mice, like patients with HGPS, appear healthy at birth and progressively develop the main features of the human disease, including cardiovascular alterations and premature death.
Figure 4.
Vascular smooth muscle cell (VSMC) content and collagen deposition in the aortas of ≈8-month-old HGPSrev mice. A, Representative immunofluorescence of cross-sections of aortic arch (left) and thoracic aorta (right) stained with anti–smooth muscle α-actin (SMA) antibody (red) and Hoechst 33342 (blue) to visualize vascular smooth muscle cells (VSMCs) and nuclei, respectively. Graphs show quantification of VSMC content in the media as either the percentage of SMA-positive area or nuclear density (n=6–8 WT; n=5–7 HGPSrev). Scale bar, 150 µm. B, Representative images and quantification of Masson’s trichrome staining to visualize medial and adventitial collagen content in cross-sections of aortic arch (left) and thoracic aorta (right; n=11–13 WT; n=11–13 HGPSrev). Scale bar, 50 µm. Data are mean±SEM. Each symbol represents 1 animal. Statistical analysis was performed by 2-tailed t test (*P<0.05). A indicates adventitia; L, lumen; and M, media.
Figure 5.
Cardiovascular abnormalities in ≈13-month-old HGPSrev mice. A, Representative immunofluorescence images of aortic arch (left) and thoracic aorta (right). Specimens were costained with anti–smooth muscle α-actin (SMA) antibody (red) and Hoechst 33342 (blue) to visualize vascular smooth muscle cells (VSMCs) and nuclei, respectively. Graphs show quantification of VSMC content in the media as either the percentage of SMA-positive area or nuclear density (n=4 or 5 WT; n=7 HGPSrev). Scale bar, 150 µm. Data are mean±SEM. Statistical analysis was performed by 2-tailed t test (**P<0.01; ***P<0.001; ****P<0.0001). B, Representative images and quantification of Masson’s trichrome staining to visualize medial and adventitial collagen content in aortic arch (left) and thoracic aorta (right) of ≈13-month-old mice (n=13–17 WT; n=13 or 14 HGPSrev). Scale bar, 50 µm. Data are mean±SEM. Statistical analysis was performed by 2-tailed t test (***P<0.001; ****P<0.0001). C, Longitudinal ECG assessment (n=12–23 WT; n=14–19 HGPSrev). Data are median with interquartile range ± minima and maxima. Differences were analyzed by mixed-effects analysis using the Geisser-Greenhouse correction and Sidák’s multiple comparisons test. Differences over time within each genotype: §P<0.05; §§P<0.01; §§§§P<0.0001. Differences between genotypes at each time point: *P<0.05; ***P<0.001; ****P<0.0001. Each symbol represents 1 animal. A indicates adventitia; M, media; and L, lumen.
Ubiquitous Progerin Suppression and Lamin A Restoration Extends Lifespan When Induced in Mildly Symptomatic and in Severely Ill HGPSrev Mice
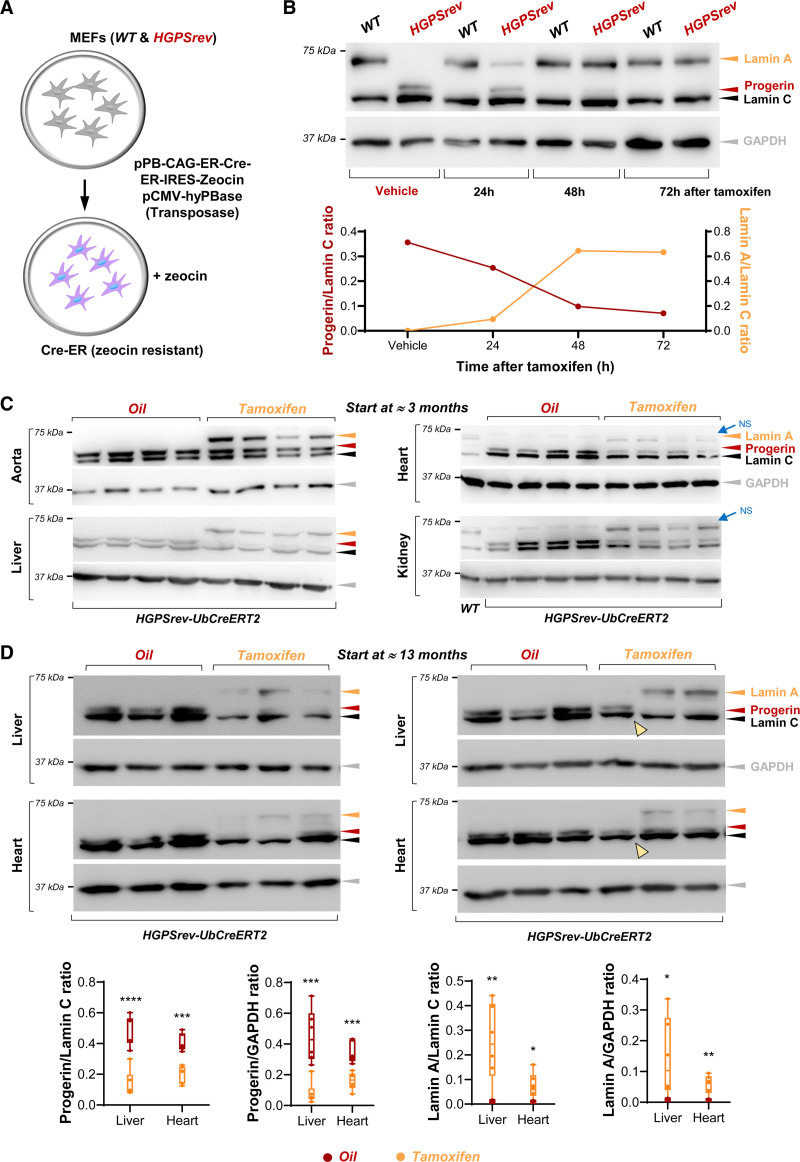
Treatment of patients with HGPS has been initiated at widely differing ages and disease stages,6 but how late treatment can be started and still ameliorate symptoms remains unknown. The HGPSrev model was designed to address this key question by taking advantage of Cre recombinase expression to remove the progerin-expressing cassette and restore lamin A expression (Figure 1A). We first performed in vitro studies in WT and HGPSrev MEFs that were stably transfected with a vector encoding a tamoxifen-inducible Cre recombinase and a zeocin-resistance cassette (Figure 6A). Western blot assays of zeocin-resistant cells showed complete progerin suppression and lamin A restoration in HGPSrev MEFs 72 hours after tamoxifen administration, with no effects in tamoxifen-treated WT MEFs (Figure 6B). To test the system in vivo, we generated HGPSrev-Ubc-CreERT2 mice by crossing HGPSrev mice with transgenic Ubc-CreERT2-tg/+ mice, which ubiquitously express a tamoxifen-inducible Cre recombinase.13 Compared with age-matched oil (vehicle)–injected controls, ≈3-month-old HGPSrev-Ubc-CreERT2 mice injected with tamoxifen and euthanized 1 week later displayed progerin downregulation and lamin A expression in all tissues tested (Figure 6C, quantification in Figure S5). Tamoxifen injection in ≈13-month-old HGPSrev-Ubc-CreERT2 mice also resulted in progerin downregulation and lamin A restoration (Figure 6D).
Figure 6.
In vitro and in vivo tamoxifen-induced Cre-dependent progerin suppression and lamin A restoration. A, Wild-type (WT) and HGPSrev mouse embryonic fibroblasts (MEFs) were cotransfected with plasmids to confer resistance to zeocin and express a tamoxifen-inducible Cre recombinase. B, Zeocin-resistant MEFs were analyzed by Western blot to examine lamin A/C, progerin, and GAPDH expression. Equal volumes of ethanol or tamoxifen (25 nmol/L final concentration) were added to the cells as indicated. The graph shows the relative amount of progerin and lamin A in HGPSrev MEFs (normalized to lamin C content). C and D, Western blot analysis of tissues of LmnaHGPSrev/HGPSrev Ubc-CreERT2-tg/+ mice that received vehicle (oil) or tamoxifen beginning at the age of ≈3 months (C, n=4 each group) and ≈13 months (D, n=6 each group). Mice in C were euthanized 1 week after oil or tamoxifen administration, and mice in D when they met human end point criteria. Yellow arrowheads in D indicate 1 animal in which tamoxifen administration did not suppress progerin or induce lamin A and that died 2 days after the end of tamoxifen administration (see Figure 7B, bottom right). Quantification of the relative amounts of lamin A and progerin in the blots in C is shown in Figure S5). The graphs in D show the relative amount of progerin and lamin A normalized to lamin C and GAPDH content. Statistical analysis to compare genotypes was performed by 2-tailed t test. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. Each symbol represents 1 animal. NS indicates nonspecific band.
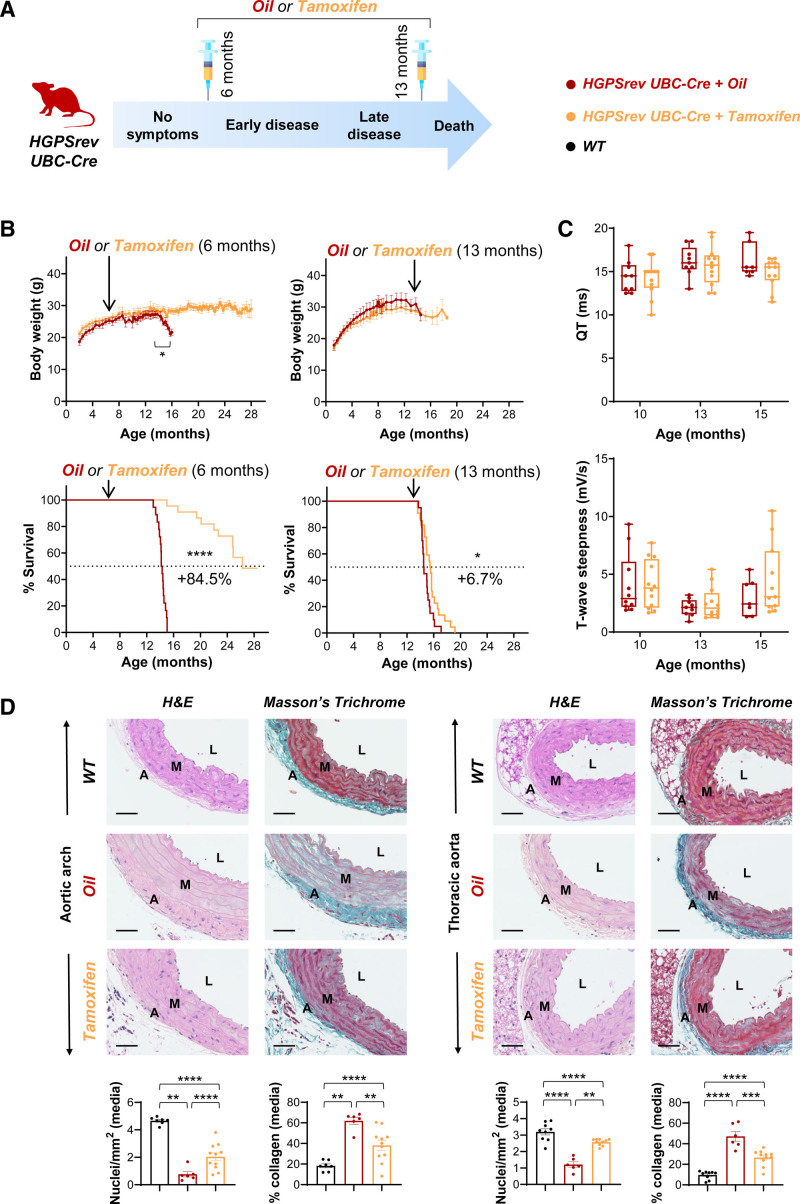
To assess the effect of in vivo systemic progerin suppression and lamin A restoration starting at different stages of HGPS progression, we defined 2 ages for the start of tamoxifen administration, representing early disease (≈6 months: beginning of growth failure, one of the earliest symptoms in patients with HGPS) and late disease (≈13 months: close to maximum survival; Figure 7A, Table). Treating ≈6-month-old HGPSrev-Ubc-CreERT2 mice with tamoxifen prevented the body weight loss observed in oil-injected controls at late disease stages (Figure 7B, top left graph), but body weight remained below WT values in mice >12 months (see WT in Figure 3A). Despite this, progeroid mice injected with tamoxifen at ≈6 months showed an 84.5% increase in median lifespan (P<0.0001 versus oil; experiment ongoing at the time of article submission: all oil-treated controls had died by ≈15 months of age, whereas 13 out of 22 tamoxifen-treated mice were still alive and in good health at ≈27 months of age; Figure 7B, bottom left graph). Beginning tamoxifen administration in ≈13-month-old HGPSrev-Ubc-CreERT2 mice with severe symptoms resulted in progerin downregulation and lamin A expression (Figure 6D), and increased median lifespan by 6.7% (P<0.05 versus oil-treated mice; Figure 7B, bottom right graph). These results demonstrate that administering tamoxifen to HGPSrev-Ubc-CreERT2 mice even at advanced disease stages significantly increases lifespan, although the benefit is much more pronounced when progerin suppression and lamin A restoration are achieved in mildly symptomatic mice.
Figure 7.
Ubiquitous progerin suppression and lamin A restoration extends lifespan in both mildly and severely symptomatic HGPSrev-Ubc-CreERT2 mice. A, Experimental protocol for studies with HGPSrev-Ubc-CreERT2 mice, showing the age at which oil or tamoxifen administration started (details in the Table). B, Oil or tamoxifen were administered at ≈6 months (left: n=18 oil and n=22 tamoxifen) or ≈13 months (right: n=7–20 oil and n=9–23 tamoxifen). The graphs show the results from 2 independent experiments. Differences were analyzed by unpaired multiple t tests and the Holm-Sidák correction in body weight studies and by the Mantel-Cox test in Kaplan-Meier survival curves. C, Oil or tamoxifen was administered at ≈9 months of age, and electrocardiography was performed at the indicated ages (n=7–10 oil; n=11 or 12 tamoxifen). Data are medians with interquartile range ± minima and maxima. Differences were analyzed by mixed-effects analysis with the Geisser-Greenhouse correction and Sidák’s multiple comparisons test. D, Hematoxylin-eosin (H&E) and Masson’s trichrome staining of aortic cross-sections from mice receiving oil/tamoxifen at ≈9 months and euthanized at 14.5 months of age (n=5 or 6 oil; n=11 tamoxifen). A group of age-matched untreated wild-type (WT) mice was included for comparison (n=7–9). Scale bar, 50 µm. Data are mean±SEM. Differences were analyzed by 1-way ANOVA and the post hoc Tukey test. **P<0.01; ***P<0.001; ****P<0.0001. Each symbol represents 1 animal. A indicates adventitia; L, lumen; and M, media.
We next investigated how suppressing progerin and restoring lamin A expression affects the cardiovascular phenotype of HGPSrev-Ubc-CreERT2 mice. Tamoxifen was administered to HGPSrev mice at ≈9 months of age (Table), an intermediate disease stage when the mice already showed a clear reduction in body weight (Figure 3A) and had begun to develop cardiovascular alterations (Figure 4B, Figure 5C). Mice were euthanized at ≈14.5 months of age (at which stage 4 out of 10 oil-injected animals but only 1 out of 12 tamoxifen-injected animals had died). Longitudinal studies showed no statistically significant differences in ECG parameters between both groups of mice at all ages tested (Figure 7C). In contrast, histological analysis revealed increased VSMC number and decreased medial collagen content in the aortic arch and thoracic aorta of tamoxifen-injected HGPSrev-Ubc-CreERT2 mice relative to oil-injected controls, although tamoxifen did not normalize these parameters to the values seen in WT mice (Figure 7D).
Progerin Suppression and Lamin A Restoration in Vascular Smooth Muscle Cells and Cardiomyocytes is Sufficient to Prevent Vascular Damage and to Normalize Lifespan in HGPSrev Mice
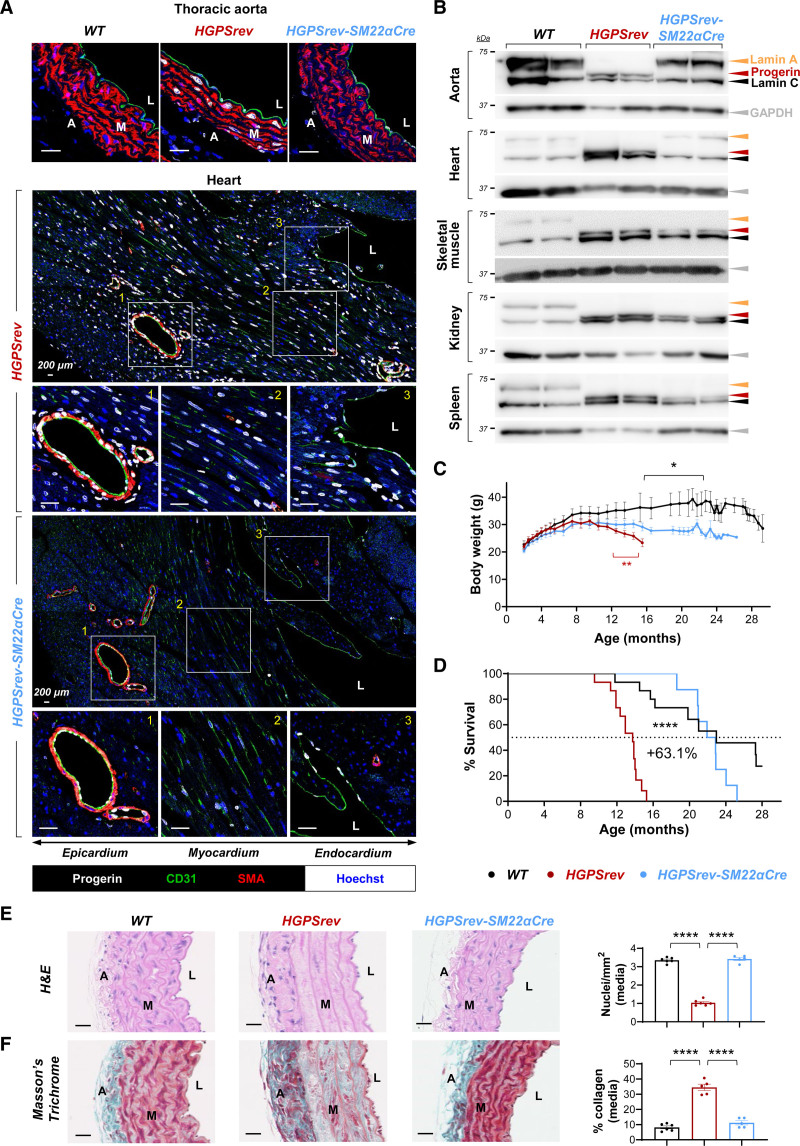
To investigate whether all affected tissues should be targeted to ameliorate disease symptoms, or whether tissue-specific therapies would be effective, we examined the effect of suppressing progerin and restoring lamin A only in VSMCs and cardiomyocytes, major progerin targets in patients with HGPS and animal models.2,6,7,14,20–28 For this analysis, we used the SM22α-Cre transgenic line—which allows Cre-dependent recombination in VSMCs and cardiomyocytes31—to generate HGPSrev-SM22α-Cre mice. Immunofluorescence experiments confirmed that progerin expression was undetectable in arterial VSMCs of HGPSrev-SM22α-Cre mice and was clearly reduced in nonendothelial cardiac cells (69%±3.8% progerin-positive cells in HGPSrev mice versus 17%±2.8% in HGPSrev-SM22α-Cre mice; P<0.0001 by unpaired 2-tailed Student t test), but remained robust in endothelial cells in aorta and heart (Figure 8A) and in noncardiovascular tissues (Figure S2). Progerin elimination and lamin A restoration were also confirmed by Western blot assays in HGPSrev-SM22α-Cre aorta and heart, whereas progerin and lamin C expression and lack of lamin A were observed in skeletal muscle, kidney, and spleen (Figure 8B). Although HGPSrev-SM22α-Cre mice had a higher body weight than HGPSrev controls from 12 months of age, they were significantly thinner than age-matched WT mice from ≈17 months of age (Figure 8C). Despite this, the lifespan (Figure 8D), aortic medial VSMC density (Figure 8E), and collagen content (Figure 8F) of HGPSrev-SM22α-Cre mice were undistinguishable from those of WT mice. These findings complement our recent studies showing that SM22α-Cre–driven progerin expression is sufficient to promote cardiovascular alterations in mice20,29,30,32 and demonstrate that targeting progerin in VSMCs and cardiomyocytes is sufficient to normalize life expectancy despite the reduced body weight caused by progerin expression in other cell types.
Figure 8.
Normal vascular phenotype and lifespan in HGPSrev-SM22α-Cre mice with progerin suppression and lamin A restoration restricted to vascular smooth muscle cells (VSMCs) and cardiomyocytes. A, Representative immunofluorescence images of thoracic aorta and hearts of ≈13-month-old mice. Cross-sections were costained with antibodies against CD31 (green), smooth muscle α-actin (SMA; red) and progerin (white) and with Hoechst 33342 (blue) to visualize endothelial cells, vascular smooth muscle cells (VSMCs), progerin, and nuclei, respectively. B, Western blot of lamin A/C, progerin, and GAPDH in tissues of ≈13-month-old mice. C, Body weight curves (n=9 WT; n=13 HGPSrev; n=11 HGPSrev-SM22α-Cre). Differences were analyzed by unpaired multiple t tests and the Holm-Sídák correction. Red asterisks denote differences between HGPSrev-SM22α-Cre and HGPSrev mice. Black asterisks denote differences between HGPSrev-SM22α-Cre and WT mice. D, Kaplan-Meier survival curve (n=15 WT; n=15 HGPSrev; n=11 HGPSrev-SM22α-Cre). Median lifespan was 13.73 months in HGPSrev mice, 22.4 months in HGPSrev-SM22α-Cre mice, and 22.97 months in WT mice. Differences were analyzed with the Mantel-Cox test. E and F, Representative images of aortic arch stained with hematoxylin-eosin (H&E) and Masson’s trichrome, to quantify VSMCs and fibrosis, respectively, in ≈13-month-old WT mice (n=6), HGPSrev mice (n=5 or 6), and HGPSrev-SM22α-Cre mice (n=5). Differences were analyzed by 1-way ANOVA with the post hoc Tukey test. *P<0.05; ****P<0.0001. Each symbol represents 1 animal. Data are mean±SEM. A indicates adventitia; L, lumen; and M, media. Scale bars, 25 µm (except in the tile scans in A, where they are 200 µm).
Discussion
We report here the generation and characterization of HGSrev mice, a new HGPS model that features ubiquitous progerin and lamin C expression and lack of lamin A, and that allows Cre-dependent progerin suppression and lamin A restoration in a time- and cell type–specific manner. HGPSrev mice progressively develop many symptoms characteristic of the human disease, including growth failure, lipodystrophy, VSMC loss, vascular fibrosis, electrocardiographic anomalies, and premature death. It is interesting that we found increased collagen accumulation in the aortic media of HGPSrev mice at ages when VSMC content still does not differ from that of WT mice. Likewise, previous studies in 8-week-old LmnaG609G/G609G mice that still exhibited normal VSMC content revealed alterations in genes involved in aortic fibrosis32 and higher collagen III and lysyl oxidase expression in carotid arteries,33 suggesting that progerin-induced extracellular matrix alterations precede VSMC death.
Unlike patients with HGPS and LmnaG609G/G609G and LMNA G608G transgenic mice, 2 widely used HGPS models,14,26 HGPSrev mice exhibit undetectable lamin A expression; however, mice that ubiquitously express lamin C and lack lamin A appeared normal,14,34,35 suggesting that lamin A absence is unlikely to provoke progeroid symptoms in HGPSrev mice. Indeed, HGPSrev mice develop disease symptoms more slowly and die later than homozygous LMNA G608G mice36 and LmnaG609G/G609G mice.14 Our proteomic studies in HGPSrev and LmnaG609G/G609G mice revealed similar amounts of farnesylated progerin in both models when normalized to the total amount of A-type lamin isoforms, but HGPSrev mice exhibited lower progerin mRNA level in all tissues tested, which may explain, at least in part, their milder phenotype.
Because HGPS is a progressive disease and children with this condition are diagnosed after the appearance of symptoms,6 it is critical to determine the reversibility of progerin-induced damage and the optimal time window for treatment, chief questions that remain unanswered. In progerin-expressing mice, the progeroid phenotype can be ameliorated and lifespan increased by CRISPR-Cas937,38 and base-editing approaches36 to ubiquitously correct the HGPS-causing mutation or by the delivery of antisense oligonucleotides to block pathogenic splicing of mutant lamin A transcripts14,39,40; however, these treatments were administered to asymptomatic neonates and young animals. We found an 84.5% extension in median lifespan when progerin was suppressed and lamin A restored in mildly symptomatic ≈6-month-old HGPSrev mice. This benefit in survival occurred despite the below-normal body weight of these animals, consistent with previous mouse studies and clinical trials demonstrating lifespan extension without body weight normalization after the administration of various treatments (eg, 3,8,14,32,36–40). Future studies are warranted to test whether early disease stages feature irreversible adipocyte death that causes persistent lipodystrophy. It is important to note that progerin suppression and lamin A restoration also prolonged lifespan in severely affected progeroid HGPSrev mice. Although phenotypic amelioration was much more pronounced when progerin suppression and lamin A restoration were achieved in early disease stages, these results strongly suggest that it is never too late to start treatment for HGPS. Indeed, tipifarnib has been found to prevent the late progression of existing cardiovascular defects when administered to progeroid mice with overt disease symptoms.41 Moreover, lonafarnib improved carotid-femoral pulse wave velocity and other outcome measures in HGPS clinical trial participants who started treatment at advanced disease stages8 and also prolonged survival in a trial population with an average age of enrollment of 8.4 years.3
In a background of normal expression of endogenous wild-type Lmna, the Erikssson laboratory generated transgenic mice with doxycycline-inducible expression of progerin in skin or osteoblasts/odontoblasts, which showed skin or bone/teeth defects on doxycycline administration, respectively.42,43 It is remarkable that the defects in these mutant mice were normalized by turning off progerin expression after the appearance of the phenotype. These seminal studies in mice demonstrated that ectopic progerin expression in skin and bone does not cause irreversible damage to these tissues in the context of normal endogenous lamin A expression. Nonetheless, it is important to note that patients with HGPS express below-normal levels of lamin A and that progerin is expressed in a broad range of tissues. Moreover, a major medical problem in HGPS is severe cardiovascular disease. Thus, uncertainty remained about the ability of progerin suppression and lamin A restoration to halt disease progression and increase lifespan when administered only in cardiovascular cells, the major progerin targets. We found that vascular abnormalities and premature death are both prevented in HGPSrev-SM22α-Cre mice with progerin suppression and lamin A restoration restricted to VSMCs and cardiomyocytes. This benefit occurred despite sustained broad progerin expression in other cell types, which was associated with significantly reduced body weight compared with age-matched WT mice. Although our model does not differentiate between effects in VSMCs and cardiomyocytes, we found that lifespan extension after ubiquitous progerin suppression and lamin A restoration in symptomatic progeroid mice was associated with reductions in both VSMC loss and collagen accumulation in the aortic media but did not ameliorate electrocardiographic defects. Moreover, HGPS is not known to be a cardiomyopathy, and massive VSMC loss and accumulation of extracellular matrix components have been observed in the arterial wall of HGPS mouse models and patients.7,14,20,23,25–30 Taken together, these findings suggest a major role of vascular disease in HGPS. Further studies using the HGPSrev model and Cre transgenic lines specifically targeting VSMCs or cardiomyocytes will conclusively show the relative contribution of these cell types to disease progression and premature death, which may help optimize gene therapy or RNA therapy to treat HGPS.
Article Information
Acknowledgments
The authors thank Simon Bartlett for English editing, and Azim Surani (University of Cambridge, United Kingdom), Steve P. Jackson (University of Cambridge), Manuel Serrano (Institute for Research in Biomedicine, Barcelona, Spain), Mariano Barbacid (Centro Nacional de Investigaciones Oncológicas, Madrid, Spain), Carlos López-Otín (Universidad de Oviedo, Spain) and Enrique Lara-Pezzi (Centro Nacional de Investigaciones Cardiovasculares, Madrid, Spain) for providing the pPB CAG ER-Cre-ER IRES Zeocin, pCMV-hyPBase and pCL-ECO plasmids, the Ubc-CreERT2-tg/+ mice, the LmnaG609G/G609G mice, and the HEK293T cells, respectively. We also thank David Filgueiras (Hospital Clínico San Carlos, Centro Nacional de Investigaciones Cardiovasculares) for advice with electrocardiography, Jacob F. Bentzon (Aarhus University, Denmark; Centro Nacional de Investigaciones Cardiovasculares) for critical reading of the article, Delphine Larrieu and Josep Vicent Forment (University of Cambridge) for advice on MEF experiments, Davide Seruggia and Lluís Montoliu (Centro Nacional de Biotecnología-Consejo Superior de Investigaciones Científicas, Spain) for advice on CRISPR-Cas9 strategy design, and the excellent support of the Centro Nacional de Investigaciones Cardiovasculares Animal Facility (with special thanks to Eva Santos, Marta García, and Belén Ricote), Histology Service, Microscopy Unit (with special thanks to Verónica Labrador), and Advanced Imaging Unit. Author contributions: A.S.-L. and V.A. conceived, designed, and planned the overall study. A.S.-L. generated the HGPSrev mouse model. A.S.-L. and C.E.-E. designed and performed experiments and analyzed and interpreted results. C.G.-G., P.G., M.J.A.-M., and R.R.-B. carried out experiments. V.F. and C.E.-E. designed, analyzed, and interpreted ECG experiments. A.B., L.R., and S.N. generated, characterized, and provided the anti-progerin antibody. E.C. and J.V. designed and performed targeted tandem mass spectrometry and interpreted the results. V.F., M.R.H., L.d.C., B.D., I.B., and A.M. provided advice and scientific discussion. V.A. interpreted the results and supervised the overall study. C.E.-E., I.B., and V.A. wrote the article. All authors discussed the results and critically reviewed article.
Sources of Funding
This study was supported by grants to V.A. from the Spanish Ministerio de Ciencia e Innovación/Agencia Estatal de Investigación/10.13039/501100011033 (grants SAF2016-79490-R, PID2019-108489RB-I00) and the Instituto de Salud Carlos III (grant AC17/00067 as coordinator of TREAT-HGPS, a project in the E-Rare Joint Transnational call, European Union Horizon 2020 Framework Program 2017), with cofunding from Fondo Europeo de Desarrollo Regional (“A Way to Build Europe”). Additional funding came from Ministerio de Ciencia e Innovación (grant SVP-2014-068334) and Asociación Apadrina la Ciencia-Ford España-Ford Motor Company Fund (A.S.L.); Fundación “la Caixa” grants LCF/BQ/DR19/1170012 (C.E.E.) and LCF/BQ/DE14/10320024 (V.F.); Comunidad Autónoma de Madrid grant 2017-T1/BMD-5247 (I.B.); Ministerio de Ciencia e Innovación/Agencia Estatal de Investigación/10.13039/501100011033 grant FJCI-2017-33299 (M.R.H.); and Wellcome Trust grant 098291/Z/12/Z (S.N.). The Centro Nacional de Investigaciones Cardiovasculares is supported by the Ministerio de Ciencia e Innovación, the Instituto de Salud Carlos III, and the Pro–Centro Nacional de Investigaciones Cardiovasculares Foundation, and is a Severo Ochoa Center of Excellence (grant CEX2020-001041-S funded by Ministerio de Ciencia e Innovación/Agencia Estatal de Investigación/10.13039/501100011033). The funders had no role in the design of the study; the collection, analysis, or interpretation of data; the writing of the article; or the decision to publish the results.
Disclosures
None.
Supplemental Material
Supplemental Methods
Tables S1 and S2
Figures S1–S5
References 44–46
Supplementary Material
Nonstandard Abbreviations and Acronyms
- H&E
- hematoxylin-eosin
- HGPS
- Hutchinson-Gilford progeria syndrome
- HGPSrev
- LmnaHGPSrev/HGPSrev mouse
- HRP
- horseradish peroxidase
- IP
- immunoprecipitation
- LC-MS/MS
- liquid chromatography coupled to targeted tandem mass spectrometry
- MRI
- magnetic resonance imaging
- NGS
- normal goat serum
- OT
- off-target
- PRM
- precursor-reaction monitoring
- RT
- room temperature
- SEMS
- spin-echo multislice
- SM22α-Cre
- B6.Cg-Tg(Tagln-cre)1Her/J mouse
- SMA
- smooth muscle α-actin
- TBST-T
- Tris-buffered saline supplemented with 0.2% Tween 20
- Ubc-CreERT2tg/+
- B6.Cg Ndor1Tg(UBC-cre/ERT2)1Ejb/1J mouse
- VSMC
- vascular smooth muscle cell
- WT
- wild-type
A. Sánchez-López and C. Espinós-Estévez contributed equally.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.121.055313.
For Sources of Funding and Disclosures, see page 1792.
Circulation is available at www.ahajournals.org/journal/circ
Contributor Information
Amanda Sánchez-López, Email: amanda.sanchez.asl@gmail.com.
Carla Espinós-Estévez, Email: carla.espinos@cnic.es.
Cristina González-Gómez, Email: cgonzalez@cnic.es.
Pilar Gonzalo, Email: pgonzalo@cnic.es.
María J. Andrés-Manzano, Email: mjandres@cnic.es.
Víctor Fanjul, Email: victorffhh@gmail.com.
Raquel Riquelme-Borja, Email: raquel.inves@gmail.com.
Magda R. Hamczyk, Email: magdarita.hamczyk@cnic.es.
Álvaro Macías, Email: alvaro.macias@cnic.es.
Lara del Campo, Email: laradelcampo@hotmail.com.
Emilio Camafeita, Email: ecamafeita@cnic.es.
Jesús Vázquez, Email: jesus.vazquez@cnic.es.
Anna Barkaway, Email: a.l.barkaway@qmul.ac.uk.
Loïc Rolas, Email: l.rolas@qmul.ac.uk.
Sussan Nourshargh, Email: s.nourshargh@qmul.ac.uk.
Beatriz Dorado, Email: bjdorado@cnic.es.
Ignacio Benedicto, Email: ignacio.benedicto@cnic.es.
References
- 1.Dorado B, Andrés V. A-type lamins and cardiovascular disease in premature aging syndromes. Curr Opin Cell Biol. 2017;46:17–25. doi: 10.1016/j.ceb.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 2.Merideth MA, Gordon LB, Clauss S, Sachdev V, Smith ACM, Perry MB, Brewer CC, Zalewski C, Kim HJ, Solomon B, et al. Phenotype and course of Hutchinson–Gilford progeria syndrome. New Engl J Med. 2008;358:592–604. doi: 10.1056/NEJMoa0706898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon LB, Shappell H, Massaro J, D’Agostino RB, Sr, Brazier J, Campbell SE, Kleinman ME, Kieran MW. Association of lonafarnib treatment vs no treatment with mortality rate in patients with Hutchinson-Gilford progeria syndrome. JAMA. 2018;319:1687–1695. doi: 10.1001/jama.2018.3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A, Le Merrer M, et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125 [DOI] [PubMed] [Google Scholar]
- 5.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ullrich NJ, Gordon LB. Hutchinson-Gilford progeria syndrome. Handb Clin Neurol. 2015;132:249–264. doi: 10.1016/B978-0-444-62702-5.00018-4 [DOI] [PubMed] [Google Scholar]
- 7.Hamczyk MR, del Campo L, Andrés V. Aging in the cardiovascular system: Lessons from Hutchinson-Gilford progeria syndrome. Annu Rev Physiol. 2018;80:27–48. doi: 10.1146/annurev-physiol-021317-121454 [DOI] [PubMed] [Google Scholar]
- 8.Gordon LB, Kleinman ME, Miller DT, Neuberg DS, Giobbie-Hurder A, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland R, Snyder BD, et al. Clinical trial of a farnesyltransferase inhibitor in children with Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2012;109:16666–16671. doi: 10.1073/pnas.1202529109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon LB, Massaro J, D’Agostino RB, Sr, Campbell SE, Brazier J, Brown WT, Kleinman ME, Kieran MW; Progeria Clinical Trials Collaborative. Impact of farnesylation inhibitors on survival in Hutchinson-Gilford progeria syndrome. Circulation. 2014;130:27–34. doi: 10.1161/CIRCULATIONAHA.113.008285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordon LB, Kleinman ME, Massaro J, D’Agostino RB, Sr, Shappell H, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland RH, Nazarian A, et al. Clinical trial of the protein farnesylation inhibitors lonafarnib, pravastatin, and zoledronic acid in children with Hutchinson-Gilford progeria syndrome. Circulation. 2016;134:114–125. doi: 10.1161/CIRCULATIONAHA.116.022188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell WMS, Burch RL. The Principles of Humane Experimental Technique. 1959. Methuen [Google Scholar]
- 12.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osorio FG, Navarro CL, Cadiñanos J, López-Mejía IC, Quirós PM, Bartoli C, Rivera J, Tazi J, Guzmán G, Varela I, et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci Transl Med. 2011;3:106ra107. doi: 10.1126/scitranslmed.3002847 [DOI] [PubMed] [Google Scholar]
- 15.Holtwick R, Gotthardt M, Skryabin B, Steinmetz M, Potthast R, Zetsche B, Hammer RE, Herz J, Kuhn M. Smooth muscle-selective deletion of guanylyl cyclase-A prevents the acute but not chronic effects of ANP on blood pressure. Proc Natl Acad Sci U S A. 2002;99:7142–7147. doi: 10.1073/pnas.102650499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behringer R, Gertsenstein M, Vintersten K, Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual. 2014. 4th ed. Cold Spring Harbor Laboratory Press [Google Scholar]
- 17.Cheetham SW, Gruhn WH, van den Ameele J, Krautz R, Southall TD, Kobayashi T, Surani MA, Brand AH. Targeted DamID reveals differential binding of mammalian pluripotency factors. Development. 2018;145:dev170209. doi: 10.1242/dev.170209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci U S A. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang SH, Qiao X, Farber E, Chang SY, Fong LG, Young SG. Eliminating the synthesis of mature lamin A reduces disease phenotypes in mice carrying a Hutchinson-Gilford progeria syndrome allele. J Biol Chem. 2008;283:7094–7099. doi: 10.1074/jbc.M708138200 [DOI] [PubMed] [Google Scholar]
- 20.Hamczyk MR, Villa-Bellosta R, Gonzalo P, Andrés-Manzano MJ, Nogales P, Bentzon JF, López-Otín C, Andrés V. Vascular smooth muscle-specific progerin expression accelerates atherosclerosis and death in a mouse model of Hutchinson-Gilford progeria syndrome. Circulation. 2018;138:266–282. doi: 10.1161/CIRCULATIONAHA.117.030856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivera-Torres J, Calvo CJ, Llach A, Guzmán-Martínez G, Caballero R, González-Gómez C, Jiménez-Borreguero LJ, Guadix JA, Osorio FG, López-Otín C, et al. Cardiac electrical defects in progeroid mice and Hutchinson-Gilford progeria syndrome patients with nuclear lamina alterations. Proc Natl Acad Sci U S A. 2016;113:E7250–E7259. doi: 10.1073/pnas.1603754113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fanjul V, Jorge I, Camafeita E, Macías Á, González-Gómez C, Barettino A, Dorado B, Andrés-Manzano MJ, Rivera-Torres J, Vázquez J, et al. Identification of common cardiometabolic alterations and deregulated pathways in mouse and pig models of aging. Aging Cell. 2020;19:e13203. doi: 10.1111/acel.13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorado B, Pløen GG, Barettino A, Macías A, Gonzalo P, Andrés-Manzano MJ, González-Gómez C, Galán-Arriola C, Alfonso JM, Lobo M, et al. Generation and characterization of a novel knockin minipig model of Hutchinson-Gilford progeria syndrome. Cell Discov. 2019;5:16. doi: 10.1038/s41421-019-0084-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macías A, Díaz-Larrosa JJ, Blanco Y, Fanjul V, González-Gómez C, Gonzalo P, Andrés-Manzano MJ, da Rocha AM, Ponce-Balbuena D, Allan A, et al. Paclitaxel mitigates structural alterations and cardiac conduction system defects in a mouse model of Hutchinson-Gilford progeria syndrome [published online February 24, 2021]. Cardiovasc Res. doi: 10.1093/cvr/cvab055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stehbens WE, Delahunt B, Shozawa T, Gilbert-Barness E. Smooth muscle cell depletion and collagen types in progeric arteries. Cardiovasc Pathol. 2001;10:133–136. doi: 10.1016/s1054-8807(01)00069-2 [DOI] [PubMed] [Google Scholar]
- 26.Varga R, Eriksson M, Erdos MR, Olive M, Harten I, Kolodgie F, Capell BC, Cheng J, Faddah D, Perkins S, et al. Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2006;103:3250–3255. doi: 10.1073/pnas.0600012103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JM, Nobumori C, Tu Y, Choi C, Yang SH, Jung HJ, Vickers TA, Rigo F, Bennett CF, Young SG, et al. Modulation of LMNA splicing as a strategy to treat prelamin A diseases. J Clin Invest. 2016;126:1592–1602. doi: 10.1172/JCI85908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olive M, Harten I, Mitchell R, Beers JK, Djabali K, Cao K, Erdos MR, Blair C, Funke B, Smoot L, et al. Cardiovascular pathology in Hutchinson-Gilford progeria: correlation with the vascular pathology of aging. Arterioscler Thromb Vasc Biol. 2010;30:2301–2309. doi: 10.1161/ATVBAHA.110.209460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Del Campo L, Sánchez-López A, Salaices M, von Kleeck RA, Expósito E, González-Gómez C, Cussó L, Guzmán-Martínez G, Ruiz-Cabello J, Desco M, et al. Vascular smooth muscle cell-specific progerin expression in a mouse model of Hutchinson-Gilford progeria syndrome promotes arterial stiffness: therapeutic effect of dietary nitrite. Aging Cell. 2019;18:e12936. doi: 10.1111/acel.12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Del Campo L, Sánchez-López A, González-Gómez C, Andrés-Manzano MJ, Dorado B, Andrés V. Vascular smooth muscle cell-specific progerin expression provokes contractile impairment in a mouse model of Hutchinson-Gilford progeria syndrome that is ameliorated by nitrite treatment. Cells. 2020;9:E656. doi: 10.3390/cells9030656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lepore JJ, Cheng L, Min Lu M, Mericko PA, Morrisey EE, Parmacek MS. High-efficiency somatic mutagenesis in smooth muscle cells and cardiac myocytes in SM22alpha-Cre transgenic mice. Genesis. 2005;41:179–184. doi: 10.1002/gene.20112 [DOI] [PubMed] [Google Scholar]
- 32.Hamczyk MR, Villa-Bellosta R, Quesada V, Gonzalo P, Vidak S, Nevado RM, Andrés-Manzano MJ, Misteli T, López-Otín C, Andrés V. Progerin accelerates atherosclerosis by inducing endoplasmic reticulum stress in vascular smooth muscle cells. EMBO Mol Med. 2019;11:e9736. doi: 10.15252/emmm.201809736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Kleeck R, Roberts E, Castagnino P, Bruun K, Brankovic SA, Hawthorne EA, Xu T, Tobias JW, Assoian RK. Arterial stiffness and cardiac dysfunction in Hutchinson-Gilford Progeria Syndrome corrected by inhibition of lysyl oxidase. Life Sci Alliance. 2021;4:e202000997. doi: 10.26508/lsa.202000997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fong LG, Ng JK, Lammerding J, Vickers TA, Meta M, Coté N, Gavino B, Qiao X, Chang SY, Young SR, et al. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J Clin Invest. 2006;116:743–752. doi: 10.1172/JCI27125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez-Mejia IC, de Toledo M, Chavey C, Lapasset L, Cavelier P, Lopez-Herrera C, Chebli K, Fort P, Beranger G, Fajas L, et al. Antagonistic functions of LMNA isoforms in energy expenditure and lifespan. EMBO Rep. 2014;15:529–539. doi: 10.1002/embr.201338126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koblan LW, Erdos MR, Wilson C, Cabral WA, Levy JM, Xiong ZM, Tavarez UL, Davison LM, Gete YG, Mao X, et al. In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature. 2021;589:608–614. doi: 10.1038/s41586-020-03086-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santiago-Fernández O, Osorio FG, Quesada V, Rodríguez F, Basso S, Maeso D, Rolas L, Barkaway A, Nourshargh S, Folgueras AR, et al. Development of a CRISPR/Cas9-based therapy for Hutchinson-Gilford progeria syndrome. Nat Med. 2019;25:423–426. doi: 10.1038/s41591-018-0338-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beyret E, Liao HK, Yamamoto M, Hernandez-Benitez R, Fu Y, Erikson G, Reddy P, Izpisua Belmonte JC. Single-dose CRISPR-Cas9 therapy extends lifespan of mice with Hutchinson-Gilford progeria syndrome. Nat Med. 2019;25:419–422. doi: 10.1038/s41591-019-0343-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erdos MR, Cabral WA, Tavarez UL, Cao K, Gvozdenovic-Jeremic J, Narisu N, Zerfas PM, Crumley S, Boku Y, Hanson G, et al. A targeted antisense therapeutic approach for Hutchinson-Gilford progeria syndrome. Nat Med. 2021;27:536–545. doi: 10.1038/s41591-021-01274-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puttaraju M, Jackson M, Klein S, Shilo A, Bennett CF, Gordon L, Rigo F, Misteli T. Systematic screening identifies therapeutic antisense oligonucleotides for Hutchinson-Gilford progeria syndrome. Nat Med. 2021;27:526–535. doi: 10.1038/s41591-021-01262-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capell BC, Olive M, Erdos MR, Cao K, Faddah DA, Tavarez UL, Conneely KN, Qu X, San H, Ganesh SK, et al. A farnesyltransferase inhibitor prevents both the onset and late progression of cardiovascular disease in a progeria mouse model. Proc Natl Acad Sci U S A. 2008;105:15902–15907. doi: 10.1073/pnas.0807840105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagelius H, Rosengardten Y, Schmidt E, Sonnabend C, Rozell B, Eriksson M. Reversible phenotype in a mouse model of Hutchinson-Gilford progeria syndrome. J Med Genet. 2008;45:794–801. doi: 10.1136/jmg.2008.060772 [DOI] [PubMed] [Google Scholar]
- 43.Strandgren C, Nasser HA, McKenna T, Koskela A, Tuukkanen J, Ohlsson C, Rozell B, Eriksson M. Transgene silencing of the Hutchinson-Gilford progeria syndrome mutation results in a reversible bone phenotype, whereas resveratrol treatment does not show overall beneficial effects. FASEB J. 2015;29:3193–3205. doi: 10.1096/fj.14-269217 [DOI] [PubMed] [Google Scholar]
- 44.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]
- 46.Bonzon-Kulichenko E, Pérez-Hernández D, Núñez E, Martínez-Acedo P, Navarro P, Trevisan-Herraz M, Ramos Mdel C, Sierra S, Martínez-Martínez S, Ruiz-Meana M, et al. A robust method for quantitative high-throughput analysis of proteomes by 18O labeling. Mol Cell Proteomics. 2011;10:M110.003335. doi: 10.1074/mcp.M110.003335 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.