Abstract
Vibrio parahaemolyticus O3:K6 strains responsible for the increase in the number of cases of diarrhea in Calcutta, India, beginning in February 1996 and those isolated from Southeast Asian travelers beginning in 1995 were shown to belong to a unique clone characterized by possession of the tdh gene but not the trh gene and by unique arbitrarily primed PCR (AP-PCR) profiles (J. Okuda, M. Ishibashi, E. Hayakawa, T. Nishino, Y. Takeda, A. K. Mukhopadhyay, S. Garg, S. K. Bhattacharya, G. B. Nair, and M. Nishibuchi, J. Clin. Microbiol. 35:3150–3155, 1997). Evidence supporting a hypothesis that this clone emerged only recently and is spreading to many countries was obtained in this study. Of 227 strains isolated in a hospital in Bangladesh between 1977 and 1998, only 22 strains isolated between 1996 and 1998 belonged to the new O3:K6 clone (defined by the serovar, the tdh and trh typing, and AP-PCR profiles). The O3:K6 strains isolated from clinical sources in Taiwan, Laos, Japan, Thailand, Korea, and the United States between 1997 and 1998 were also shown to belong to the new O3:K6 clone. The clonality of the new O3:K6 strains was also confirmed by analysis of the toxRS sequence, which has been shown to be useful for phylogenetic analysis of the members of the genus Vibrio. The toxRS sequences of the representative strains of the new O3:K6 clone differed from those of the O3:K6 strains isolated before 1995 at least at 7 base positions within a 1,346-bp region. A new PCR method targeted to 2 of the base positions unique to the new O3:K6 clone was developed. This PCR method could clearly differentiate all 172 strains belonging to the new O3:K6 clone from other O3:K6 strains isolated earlier. One hundred sixty-six strains belonging to 28 serovars other than O3:K6 were also examined by the new PCR method. The tdh-positive and trh-lacking strains that belonged to the O4:K68 and O1:K untypeable serovars and were isolated in three countries and from international travelers beginning in 1997 gave positive results. The AP-PCR profiles of these strains were nearly identical to those of the new O3:K6 clone, and their toxRS sequences were 100% identical to that of the new O3:K6 clone. The results suggest that these strains may have diverged from the new O3:K6 clone by alteration of the O:K antigens. In conclusion, this study presents strong evidence for the first pandemicity in the history of V. parahaemolyticus and reports a novel toxRS-targeted PCR method that will be useful in epidemiological investigation of the cases associated with the current pandemic spread.
Some strains of Vibrio parahaemolyticus, a marine bacterium, can cause gastroenteritis in humans through consumption of seafood. It was reported in the late 1960s that almost all clinical strains, but very few environmental strains, manifest Kanagawa phenomenon (KP), β-type hemolysis on Wagatsuma agar (8, 19). KP is caused by high-level production of thermostable direct hemolysin. Thermostable direct hemolysin is encoded by the tdh gene (13, 17), which was detected almost exclusively in clinical strains in an early study (11). The role of thermostable direct hemolysin in enterotoxigenicity was demonstrated by construction and examination of the tdh-deficient mutant of a KP-positive strain (10). Investigation of an outbreak in the Maldives in 1985 revealed that some clinical strains do not possess the tdh gene but carry the tdh-related hemolysin (trh) gene (14). The trh sequence was approximately 70% identical to the tdh sequence. There is much greater strain-to-strain divergence among trh sequences than among tdh sequences. The trh sequences in different strains, however, can be clustered into two groups represented by the trh1 and trh2 genes, which have 84% sequence identity (5). Strains possessing either the tdh gene, the trh gene, or both were shown to be strongly associated with gastroenteritis (5, 20).
Surveillance for V. parahaemolyticus infection was initiated in January 1994 in Calcutta, India. A group of strains belonging to serovar O3:K6 and possessing the tdh gene but not the trh gene appeared suddenly in February 1996 and was shown to be responsible for the high incidence of V. parahaemolyticus infection since then in Calcutta (16). Serovar O3:K6 was not isolated before February 1996 in Calcutta. In addition, the O3:K6 strains isolated in Calcutta were shown to exhibit unique profiles in an arbitrarily primed PCR (AP-PCR) analysis (16). Strains belonging to the same group, i.e., O3:K6 strains possessing the tdh gene but not the trh gene and showing the unique AP-PCR profiles, were also detected among those isolated from travelers arriving in Japan from Southeast Asian countries from 1995 on (16). Thus, the Calcutta O3:K6 strains and the above strains from the travelers were considered to belong to a single clone (16). These results suggested that this unique clone, referred to below as a new O3:K6 clone, might have emerged recently and become prevalent not only in Calcutta, India, but also in other parts of the world.
We examined this hypothesis, and we present evidence in this study for the first pandemicity in the history of V. parahaemolyticus. Clinical strains isolated over 22 years, starting from 1977, in a hospital in Bangladesh were available. The emergence of the new O3:K6 clone in 1996 but not earlier was demonstrated by examination of these strains. Next, we showed by AP-PCR analysis that the clinical strains of serovar O3:K6 isolated in six other countries, including the United States, from 1997 on belong to the same clone. We then developed a novel PCR method to identify the strains belonging to the new O3:K6 clone. We utilized the toxRS operon sequence to develop this PCR method. The toxR and toxS genes in the toxRS operon encode transmembrane proteins involved in the regulation of virulence-associated genes and are well conserved in the genus Vibrio (3, 7, 18; J. H. Rhee, S. E. Lee, S. Y. Kim, S. H. Shin, C. M. Kim, P. Y. Ryu, K. C. Leong, S. H. Choi, and S. S. Chung, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., abstr. B-171, p. 84, 1998; V. Vuddhakul, T. Nakai, C. Matsumoto, T. Oh, T. Nishino, M. Nishibuchi, and J. Okuda, submitted for publication). We used the intraspecies variation of the toxRS sequence to develop a cluster-specific PCR method that allowed for confirmation of the clonality of the new O3:K6 strains. By using this PCR method in our investigation, we found emerging strains that were almost indistinguishable from the new O3:K6 clone, although the strains belonged to different serovars.
MATERIALS AND METHODS
Bacterial strains.
V. parahaemolyticus strains were isolated from clinical specimens of patients with diarrhea in eight countries. In Bangladesh, V. parahaemolyticus strains were isolated at the International Centre for Diarrheal Diseases Research, Bangladesh, located in Dhaka, between 1977 and 1998. Stool specimens were plated directly, after enrichment in bile peptone broth, onto taurocholate-tellurite-gelatin agar (9). The colonies selected were screened for V. parahaemolyticus by a battery of biochemical tests: positive results in the tests for fermentation without gas production of arabinose, glucose, mannitol, and mannose, for lysine and ornithine decarboxylase, and for growth in 8% NaCl; negative results for esculin hydrolysis, for fermentation of salicin, inositol, and sucrose, for arginine dihydrolase, and for growth in 0% NaCl. Presumptively identified strains were lyophilized and stored at room temperature until further characterization. In Taiwan, the strains were isolated during outbreaks in Kaohsiung in 1993 and in Taipei, Hsinchu, Tao-Yuan, Kie-men, and Kee-lung in 1997. The strains were presumptively identified with API 20E strips (bioMérieux, Marcy-l'Étoile, France). Indian strains were isolated in the Infectious Diseases Hospital in Calcutta, and the strains were presumptively identified as described previously (16). Laotian strains were isolated during diarrhea outbreaks in four hospitals in Vientiane in August and September 1997 (24). Thai strains were isolated in Songklanagarind Hospital and Hat-Yai Hospital, Hat-Yai, in September and October 1998 (V. Vuddhakul, A. Chowdhury, N. Patararungrong, P. Pungrasamee, P. Thianmontri, V. Laohaprertthisan, M. Ishibashi, and M. Nishibuchi, unpublished data). Korean strains were isolated in Pusan University Hospital, Pusan, South Korea, in August 1998. The strains were initially identified with API 20E. U.S. strains were isolated during outbreaks in Texas (S. S. Barth, L. S. Del Rosario, T. Baldwin, M. Kingsley, V. Headley, B. Ray, K. Wiles, A. DePaola, D. Cook, C. Kaysner, N. Puhr, N. Daniels, L. Kornstein, and M. Nishibuchi, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. C-57, p. 116, 1999), New York State, and Connecticut in the summer of 1998. Japanese strains were isolated in the following locations and years: Osaka Prefecture, 1997 and 1998; Wakayama Prefecture, 1997, supplied by the Wakayama Prefectural Research Center of Environment and Public Health; Fukuoka Prefecture, 1997 and 1998, supplied by the Fukuoka Institute of Health and Environmental Sciences; and Kyoto Prefecture, 1998, supplied by the Kyoto Prefectural Institute of Hygienic and Environmental Sciences.
The strains isolated from travelers arriving in Japan from abroad were obtained from Osaka Airport Quarantine Station (strains isolated between 1982 and 1994) and Kansai Airport Quarantine Station (strains isolated between 1995 and 1999).
The complete identification of the V. parahaemolyticus strains was carried out by standard biochemical tests and by a PCR method targeted to the V. parahaemolyticus toxR gene as described previously (4).
O:K serovar.
The O:K serovar of each test strain was determined by agglutination tests with specific antisera as described previously (21).
Detection of hemolysin genes.
The presence or absence of the tdh, trh1, and trh2 genes in each test strain was determined by the DNA colony hybridization method using specific DNA probes as described previously (15).
AP-PCR.
AP-PCR was performed as described previously (15, 16) except that two different PCR machines were used in this study. Briefly, 25 ng of purified total DNA, 25 pmol of a primer (primer 1, 2, or 4 included in the RAPD Analysis Primer Set [Pharmacia Biotech, Inc., Uppsala, Sweden]), 2.5 U of polymerase (Ex Taq; Takara, Shiga, Japan), 10× buffer containing 20 mM MgCl2 (Ex Taq buffer; Takara), and 0.125 mM each deoxynucleoside triphosphate were used for each amplification reaction. The thermal cycling was set at one cycle of 95°C for 4 min, followed by 45 cycles of amplification consisting of denaturation at 95°C for 1 min, annealing at 36°C for 1 min, and extension at 72°C for 2 min, and then followed by one cycle of 72°C for 7 min. A Hybaid (Middlesex, England) model HB-TR1L thermal reactor was used when Bangladeshi strains were compared. A Zymoreactor II (model AB-1820; Atto Co., Tokyo, Japan) was used when the strains isolated in many other countries were examined.
toxRS sequence determination.
The toxRS region of the test DNA was amplified by a PCR method, and the nucleotide sequence of the amplicon was determined as described below. The test strain was grown in Luria-Bertani (LB) broth containing 1% NaCl at 37°C with shaking (160 rpm) overnight. One milliliter of the culture was boiled for 10 min and transferred onto ice immediately. The supernatant was then obtained by centrifugation (at 14,000 rpm) on a tabletop centrifuge (Centrifuge 5415C; Eppendorf, Hamburg, Germany) at room temperature. The supernatant was diluted 10-fold in distilled water and used as the template solution for PCR. A pair of PCR primers (toxRS.1 and toxRS.2) was designed so that the amplified sequence covered most coding regions of the toxR and toxS genes of strain AQ3815 (Fig. 1). The sequences of the sense primer (toxRS.1) and the antisense primer (toxRS.2) were 5′-TATCTCCCATGCGCAAACGTA-3′ (positions 73 to 93 in the work of Lin et al. [7] and GenBank accession no. L11929) and 5′-ACAGTACCGTAGAACCGTGAT-3′ (positions 1542 to 1522 in the work of Lin et al. [7] and GenBank accession no. L11929), respectively. The PCR mixture consisted of 2 μl of Thermophilic DNA Polymerase 10× Buffer (magnesium free, containing 100 mM Tris-HCl [pH 9.0], 500 mM KCl, and 1% Triton X-100; Promega Corp., Madison, Wis.), 1.5 mM MgCl2, 0.125 mM each deoxynucleoside triphosphate, 0.5 μM each primer, 2.5 μl of the template solution (supernatant of the boiled culture diluted 1:10), and 0.5 U of Taq DNA polymerase in Storage Buffer A (Promega Corp.) in a 20-μl volume. The amplification conditions were set at one cycle of 96°C for 5 min, followed by 30 cycles of amplification consisting of denaturation at 96°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min, and then followed by one cycle of 72°C for 7 min. The PCR-amplified mixture was resolved by electrophoresis in a 1% agarose gel, and 1,470-bp amplicons were extracted from the gel pieces with QiaexII (Qiagen GmbH, Dusseldorf, Germany) according to the manufacturer's instructions. Approximately 50 ng of the purified amplicons were used as the template for sequence determination. The toxRS.1 primer, the toxRS.2 primer, and 11 other nested primers were used to determine the sequence in both directions with an ABI PRISM Dye Terminator Cycle Sequencing Ready Kit and ABI PRISM 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems, Foster City, Calif.).
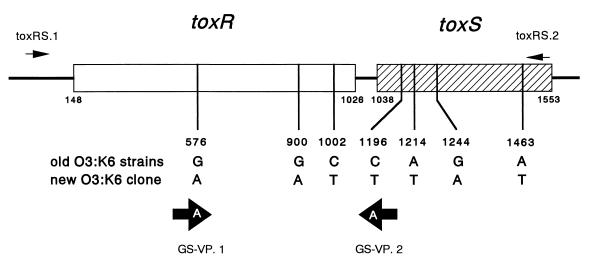
FIG. 1.
Target positions of the PCR primers used to amplify the toxRS sequences and the essential base difference in the toxRS sequence between the old O3:K6 strain group and the new O3:K6 clone. The bases described are those in the coding strand, except that the bases in the GS-VP.1 and GS-VP.2 primers are those incorporated in the actual oligonucleotides. Numerals indicate the base positions that correspond to those in the reported toxRS sequence of strain AQ3815 (Lin et al. [7] and GenBank accession no. L11929).
Group-specific PCR.
A PCR method to specifically detect the toxRS sequence of the new O3:K6 clone was established by a modification of the method of Wu et al. (23). The 16-mer primers designated GS-VP.1 (5′-TAATGAGGTAGAAACA-3′) and GS-VP.2 (5′-ACGTAACGGGCCTACA-3′) were designed to contain a group-specific base at the 3′ end (Fig. 1). The sequences of the primers were identical with (GS-VP.1) or complementary to (GS-VP.2) the deposited toxRS sequence of strain VP81 (the base positions correspond to 561 to 576 and 1211 to 1196 of the toxRS sequences of strain AQ3815 [reference 7 and GenBank accession no. L11929], respectively). The 1:10 diluted supernatant of the boiled LB broth culture prepared as described above was used as the template solution. The PCR mixture consisted of 2 μl of Thermophilic DNA Polymerase 10× Buffer (magnesium free, containing 100 mM Tris-HCl [pH 9.0], 500 mM KCl, and 1% Triton X-100; Promega Corp.), 1.5 mM MgCl2, 0.125 mM each deoxynucleoside triphosphate, 0.2 μM each primer, 2.5 μl of the template solution (supernatant of the boiled culture diluted 1:10), and 0.5 U of Taq DNA polymerase in Storage Buffer A (Promega Corp.) in a 20-μl volume. The amplification conditions were set at one cycle of 96°C for 5 min, followed by 25 cycles of amplification consisting of denaturation at 96°C for 1 min, annealing at 45°C for 2 min, and extension at 72°C for 3 min, and then followed by one cycle of 72°C for 7 min. The PCR-amplified mixture was resolved by electrophoresis in a 1% agarose gel to detect 651-bp amplicons.
Nucleotide sequence accession numbers.
The nucleotide sequence data of the toxRS operon of V. parahaemolyticus strains reported in this paper will appear in the DDJB/EMBL/GenBank databases with the following accession numbers (strain names are given in parentheses): AB029911 (VP81), AB029913 (JKY-VP6), AB029903 (BE-98-2062), AB029912 (VP108), AB029904 (FIHES98V14-1), AB029915 (U-5474), AB029909 (AQ4901), AB029907 (AQ3810), AB029908 (DOH272), AB029905 (AN-5034), AB029906 (AN-16000), AB029910 (Y-27669), and AB029914 (AN-8917).
RESULTS AND DISCUSSION
Emergence of O3:K6 strains in Bangladesh.
Of the strains presumptively identified as V. parahaemolyticus between 1977 and 1998, 227 were finally identified as V. parahaemolyticus. These strains were characterized and are listed in Table 1. The number of strains isolated each year approximates the yearly incidence of V. parahaemolyticus infection, although some strains became nonviable during storage. Like those found in Calcutta (16), many strains belonging to the O3:K6 serovar and carrying the tdh gene but not the trh gene were isolated between 1996 and 1998. The O3:K6 strains possessing only the tdh gene were also detected in 1980 and 1981. Therefore, we examined the possibility that the O3:K6 strains isolated in the 1980-to-1981 and 1996-to-1998 periods in Bangladesh may be genetically related to the new O3:K6 clone isolated in Calcutta. These strains were compared by the AP-PCR method that has been used to identify the new O3:K6 clone (16). Two different primers (designated primer 1 and primer 2) were used, and the results obtained with the two primers were essentially the same (Fig. 2). The AP-PCR profiles of the strains isolated in 1980 and 1981 in Bangladesh were the same but were distinct from that of the new O3:K6 clone (Fig. 2, A1 and A2). On the other hand, the representative strains isolated between 1996 and 1998 showed AP-PCR profiles that were identical with that of the new O3:K6 clone (Fig. 2, B1 and B2). The results indicate that the new O3:K6 clone first appeared in 1996 in Bangladesh.
TABLE 1.
Characteristics of V. parahaemolyticus strains isolated from patients with diarrhea at the International Centre for Diarrheal Diseases Research, Bangladesh, between 1977 and 1998
| Yr of isolation | Presence of genea:
|
No. of O:K serovars | No. of total strains | No. of strains belonging to serovar:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| tdh | trh1 | trh2 | O3:K6 | O4:K68 | O1:KUTb | Otherc | |||
| 1977 | + | − | − | 7 | 11 | 0 | 0 | 0 | 11 |
| − | − | + | 3 | 3 | 0 | 0 | 0 | 3 | |
| 1979 | + | − | − | 2 | 2 | 0 | 0 | 0 | 2 |
| 1980 | + | − | − | 13 | 24 | 6 | 0 | 0 | 18 |
| − | − | + | 5 | 6 | 0 | 0 | 1 | 5 | |
| 1981 | + | − | − | 10 | 19 | 2 | 0 | 0 | 17 |
| − | + | − | 1 | 1 | 0 | 0 | 1 | 0 | |
| − | − | + | 4 | 6 | 0 | 0 | 2 | 4 | |
| 1982 | + | − | − | 11 | 26 | 0 | 0 | 0 | 26 |
| − | − | + | 3 | 3 | 0 | 0 | 1 | 2 | |
| 1983 | + | − | − | 11 | 20 | 0 | 0 | 0 | 20 |
| − | − | + | 3 | 4 | 0 | 0 | 0 | 4 | |
| − | − | − | 2 | 3 | 0 | 0 | 0 | 3 | |
| 1984 | + | − | − | 1 | 1 | 0 | 0 | 0 | 1 |
| 1986 | + | − | − | 2 | 3 | 0 | 0 | 0 | 3 |
| − | − | + | 2 | 2 | 0 | 0 | 0 | 2 | |
| 1987 | + | − | − | 4 | 6 | 0 | 0 | 0 | 6 |
| − | − | − | 1 | 1 | 0 | 0 | 0 | 1 | |
| 1988 | + | − | − | 3 | 3 | 0 | 0 | 0 | 3 |
| 1989 | + | − | − | 1 | 1 | 0 | 0 | 0 | 1 |
| − | − | − | 1 | 1 | 0 | 0 | 0 | 1 | |
| 1990 | + | − | − | 1 | 2 | 0 | 0 | 0 | 2 |
| 1991 | + | − | − | 1 | 1 | 0 | 0 | 0 | 1 |
| − | − | − | 1 | 1 | 0 | 0 | 0 | 1 | |
| 1992 | + | − | − | 1 | 3 | 0 | 0 | 0 | 3 |
| 1993 | + | − | − | 4 | 5 | 0 | 0 | 0 | 5 |
| 1994 | + | − | − | 7 | 9 | 0 | 0 | 0 | 9 |
| − | − | + | 1 | 2 | 0 | 0 | 0 | 2 | |
| 1995 | + | − | − | 5 | 5 | 0 | 0 | 0 | 5 |
| − | − | + | 1 | 1 | 0 | 0 | 1 | 0 | |
| 1996 | + | − | − | 2 | 18 | 14 | 0 | 0 | 4 |
| − | − | − | 1 | 1 | 0 | 0 | 0 | 1 | |
| 1997 | + | − | − | 3 | 8 | 4 | 0 | 0 | 4 |
| − | − | − | 1 | 1 | 0 | 0 | 0 | 1 | |
| 1998 | + | + | − | 4 | 6 | 0 | 0 | 0 | 6 |
| + | − | − | 3 | 17 | 4 | 12 | 1 | 0 | |
| − | − | + | 1 | 1 | 0 | 0 | 1 | 0 | |
+, present; −, absent.
UT, untypeable.
Other, strains belonging to serovars other than O3:K6, O1:KUT, and O4:K68.
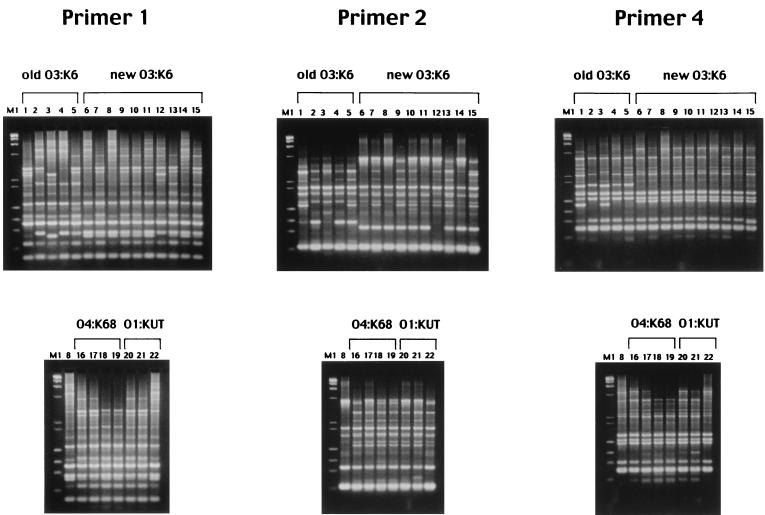
FIG. 2.
Results of AP-PCR assay for O3:K6 strains isolated in Bangladesh. The results obtained with primer 1 are shown in A1 and B1, and those obtained with primer 2 are presented in A2 and B2. (A1 and A2) Lanes: 1 and 13, molecular size markers (phage λ DNA digested with HindIII); 2 and 14, molecular size markers (phage φX174 DNA digested with HaeIII); 3 and 12, strain VP47 (a control strain isolated in Calcutta in 1996 [16]); 4 through 9, Bangladeshi strains isolated in 1980; 10 and 11, Bangladeshi strains isolated in 1981. (B1 and B2) Lanes: 1 and 15, molecular size markers (phage λ DNA digested with HindIII); 2 and 14, molecular size markers (phage φX174 DNA digested with HaeIII); 3 and 13, VP47; 4 through 10, Bangladeshi strains isolated in 1996; 11 and 12, Bangladeshi strains isolated in 1997.
Detection of the new O3:K6 clone in other countries.
After the report of the new O3:K6 clone in the literature (16), O3:K6 strains have been isolated in countries other than India and Bangladesh. We examined our hypothesis that the new O3:K6 clone also caused infection in these countries by isolating O3:K6 strains from patients with diarrhea in six other countries. The number of strains isolated in Taiwan, Laos, Japan, Thailand, Korea, and the United States totaled 119 (Table 2). With the exception of a Taiwanese strain isolated in 1993, these O3:K6 strains were isolated between 1997 and 1999. All of these strains, with the exception of a Japanese strain, had the tdh gene. None had the trh1 or the trh2 gene (Table 2). These strains were compared with representative O3:K6 strains isolated in India, in Japan (from Southeast Asian travelers) (16), and in Bangladesh. Fifty-nine strains were selected from various groups of serovar O3:K6 listed in Table 2 and were examined by AP-PCR with primer 2 first. All O3:K6 strains isolated from 1995 on, including the standard strains of the new O3:K6 clone, showed essentially identical AP-PCR profiles, whereas the strains isolated between 1980 and 1993, collectively referred to below as the old O3:K6, exhibited profiles different from those of the strains isolated from 1995 on (data not shown). Representative strains of various O3:K6 groups listed in Table 2 (experimental strain numbers 1 through 15) were then examined by AP-PCR with two other primers (primers 1 and 4). These results also confirmed that the strains isolated from 1995 on shared nearly identical AP-PCR profiles and thus belong to the new O3:K6 clone. The results obtained with the three primers for the representative stains are presented in Fig. 3. An exceptional tdh-deficient strain, FIHES98V1-32-4, that was isolated in 1998 in Japan showed AP-PCR profiles that diverged slightly from those of other strains when examined with primers 1 and 2 (Fig. 3, lanes 12) (discussed below).
TABLE 2.
Characteristics and GS-PCR results of V. parahaemolyticus strains isolated from clinical sources
| O:K serovar and yr of isolation | No. of strains | Location of isolation or source of isolate | Presence of genea:
|
GS-PCR result | Representative strain(s)b
|
|||
|---|---|---|---|---|---|---|---|---|
| tdh | trh1 | trh2 | Experimental no. | Designation | ||||
| O3:K6 | ||||||||
| 1980–1981 | 8 | Bangladesh | + | − | − | − | 1 | U-5474* |
| 1982–1993 | 8 | Int. travel.c | − | + | − | − | 2 | AQ4901* (THA) |
| 1983 | 1 | Int. travel.c | + | − | − | − | 3 | AQ3810* (SING) |
| 1985–1992 | 4 | Int. travel.c | − | − | + | − | 4 | AQ4037 (MVD) |
| 1993 | 1 | Taiwan | − | − | − | − | 5 | DOH272* |
| 1995–1996 | 5 | Int. travel.d | + | − | − | + | 6 | KX-V225 (THA) |
| 1996–1998 | 22 | Bangladesh | + | − | − | + | 7 | AN-8373 |
| 1996–1998 | 26 | India | + | − | − | + | 8 | VP81* |
| 1997 | 4 | Taiwan | + | − | − | + | 9 | DOH958 15 |
| 1997 | 36 | Laos | + | − | − | + | 10 | 97LVP2 |
| 1997–1998 | 35 | Japan | + | − | − | + | 11 | JKY-VP6* |
| 1998 | 1 | Japan | − | − | − | + | 12 | FIHES98V1-32-4 |
| 1998 | 20 | Thailand | + | − | − | + | 13 | VP47 |
| 1998 | 2 | Korea | + | − | − | + | 14 | VP2 |
| 1998 | 21 | United States | + | − | − | + | 15 | BE-98-2062* |
| O4:K68 | ||||||||
| 1997–1999 | 23 | Int. travel.d | + | − | − | + | 16 | KX-V532 (THA) |
| 1998 | 12 | Bangladesh | + | − | − | + | 17 | AN-5034* |
| 1998 | 7 | India | + | − | − | + | 18 | VP232 |
| 1998 | 5 | Japan | + | − | − | + | 19 | OP-424 |
| O1:KUTe | ||||||||
| 1980–1998 | 6 | Bangladesh | − | − | + | − | ||
| 1981 | 1 | Bangladesh | − | + | − | − | ||
| 1986–1994 | 2 | Int. travel.c | − | − | + | − | ||
| 1986–1995 | 16 | Int. travel.cd | + | + | − | − | ||
| 1989–1994 | 7 | Int. travel.c | + | − | + | − | ||
| 1994 | 1 | India | + | − | + | − | ||
| 1994–1996 | 2 | India | − | − | − | − | ||
| 1994–1998 | 2 | India | − | − | + | − | ||
| 1996–1997 | 3 | India | + | + | − | − | ||
| 1997–1998 | 11 | India | + | − | − | + | 20 | VP185 |
| 1998 | 1 | Bangladesh | + | − | − | + | 21 | AN-16000* |
| 1998 | 2 | United States | + | + | − | − | ||
| 1999 | 1 | Int. travel.d | + | − | − | + | 22 | KX-V737 (THA) |
| Other than O3:K6, O4:K68, and O1:KUT (26 serovars) | ||||||||
| 1978–1999 | 6 | Int. travel.cd | + | − | − | − | ||
| 1981–1998 | 9 | Bangladesh | + | − | − | − | Y-27669*, AN-8917* | |
| 1983–1993 | 5 | Int. travel.c | + | + | − | − | ||
| 1993 | 1 | Japan | + | + | − | − | ||
| 1994–1998 | 28 | India | + | − | − | − | ||
| 1996 | 1 | India | + | − | + | − | ||
| 1996 | 1 | Korea | + | + | − | − | ||
| 1997 | 1 | Taiwan | + | − | − | − | ||
| 1997 | 1 | Taiwan | − | − | − | − | ||
| 1998 | 2 | India | − | − | + | − | ||
| 1998 | 2 | India | − | − | − | − | ||
| 1998 | 2 | Thailand | − | − | − | − | ||
| 1998 | 1 | Thailand | + | + | − | − | ||
| 1998 | 2 | United States | + | + | − | − | ||
| 1998 | 1 | United States | − | − | + | − | ||
| 1998 | 1 | United States | − | − | − | − | ||
+, present; −, absent.
Experimental strain numbers (1 through 22) correspond to lane designations in Fig. 3 and 4. Asterisks indicate strains for which the toxRS sequence was submitted to the DDJB/EMBL/GenBank databases. For strains isolated from international travelers (Int. travel.), the travelers' origins are shown in parentheses, as follows: THA, Thailand; SING, Singapore; MVD, the Maldives.
International traveler(s) arriving at Osaka Airport, Osaka, Japan.
International traveler(s) arriving at Kansai Airport, Osaka, Japan.
UT, untypeable.
FIG. 3.
Results of AP-PCR assay for representative O3:K6, O4:K68, and O1:KUT strains isolated in various geographical locations and from international travelers. Results obtained with different primers are shown in different panels as indicated. For all panels, lane designations correspond to the experimental strain numbers listed in Table 2. M1, molecular size markers (mixture of phage λ DNA digested with HindIII and phage φX174 DNA digested with HaeIII).
Development of a toxRS-targeted PCR method and confirmation of the clonality of the new O3:K6 strains.
The above AP-PCR method has been shown to be useful for the detection of the new O3:K6 clone. However, it is cumbersome, and one has to be careful when using this test. AP-PCR profiles can be influenced by factors such as the quality and concentration of template DNA and primers, the quality of Taq polymerase, and the conditions of the PCR apparatus. Therefore, we decided to develop a new PCR method for detection of the new O3:K6 clone that is easier to perform and less influenced by the above factors.
The nucleotide sequences of conserved genes such as rRNA genes and the gyrB gene encoding the DNA gyrase B subunit are often used for phylogenetic analysis. The toxR gene was first discovered in Vibrio cholerae, and this gene, when assisted by the toxS gene located immediately downstream, has been shown to be involved in the regulation of many virulence-associated genes in this organism (3). The toxR gene was also detected at least in V. parahaemolyticus, Vibrio fischeri, Vibrio vulnificus, and Vibrio hollisae, and the sequences of these genes were analyzed (7, 18; Rhee et al., Abstr. 98th Gen. Meet. Am. Soc. Microbiol., 1998; Vuddhakul et al., submitted). We therefore presumed that the toxR gene is a global regulatory gene conserved in the members of the genus Vibrio. The toxR sequence identity between V. parahaemolyticus and other Vibrio species is considerably lower than the sequence identities for the 16S rRNA gene and the gyrB gene; for example, the percent identities for the toxR gene, the 16S rRNA gene, and the gyrB gene between V. parahaemolyticus and V. hollisae are 59, 95, and 80%, respectively (6, 7; Vuddhakul et al., submitted). We thus developed toxR-targeted PCR methods for identification of V. parahaemolyticus and V. hollisae at the species level (4; Vuddhakul et al., submitted). We presumed in this study that variation in the toxR sequence may also be used for differentiating phylogenetically distinct clusters in V. parahaemolyticus.
We selected representative strains and compared their toxRS operon sequences. A 1,470-bp sequence covering 99.2% of the toxRS coding regions was amplified by a PCR method and compared by digestion with selected restriction endonucleases first. The results suggested the possibility of group-specific base alterations (data not shown). We therefore determined the nucleotide sequences of the amplified toxRS sequence for the following representative strains: five strains belonging to the new O3:K6 clone (VP81, JKY-VP6, BE-98-2062, VP108 [isolated in India in 1996; strain name not shown in Table 2], and FIHES98V14-1 [isolated in Japan in 1998; strain name not shown in Table 2]) and four strains of the old O3:K6 (U-5474, AQ4901, AQ3810, and DOH272). The nucleotide sequence of a 1,364-bp region covering 95.4% of the toxRS coding regions was determined for all the strains. The nucleotide sequences of the five strains of the new O3:K6 clone were 100% identical. There was strain-to-strain variation among the sequences of the old O3:K6 strains. The difference in the sequence between the new O3:K6 clone and the old O3:K6 strains ranged from 11 to 14 bp within the 1,364-bp region, and the sequences of the two groups differed invariably at 7 base positions as illustrated in Fig. 1. Conservation of the 7 bases in the new O3:K6 clone was further confirmed by determining the stretches of the sequences including these 7 bases for 11 more strains (AN-8373, DOH958 15, 97LVP2, and VP47, as well as one Taiwanese strain isolated in 1997, three Japanese strains isolated in 1997 and 1998, one Thai strain isolated in 1998, and two U.S. strains isolated in 1998 for which strain names are not listed in Table 2).
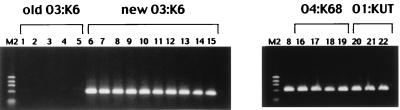
On the basis of the result, we attempted to establish a PCR method to specifically detect the toxRS sequence of the new O3:K6 clone next. We named this method GS-PCR, for group-specific PCR. Restriction endonuclease analyses for a limited number of O3:K6 strains isolated from 1995 on indicated that 2 of the 7 unique bases in the toxRS sequence detected above can be useful for GS-PCR (data not shown). These two bases were incorporated in designing the primer pair GS-VP.1 and GS-VP2 (Fig. 1). The primers were designed and amplification conditions were determined according to the recommendations for an allele-specific PCR method (23). All 194 strains of serovar O3:K6 listed in Table 2 were examined by GS-PCR. All O3:K6 strains isolated from 1995 on, including FIHES98V1-32-4, were positive, whereas all strains of the old O3:K6 group were negative. Examples of the gel electrophoresis of the PCR products are presented in Fig. 4 (left panel). Therefore, we concluded that GS-PCR can be used to distinguish the new O3:K6 clone from the old O3:K6 strains. The results of the toxRS sequence analysis and the GS-PCR test provided additional evidence that the O3:K6 strains isolated from 1995 on in eight countries belong to the same clone.
FIG. 4.
GS-PCR results for representative strains of serovars O3:K6, O4:K68, and O1:KUT, isolated in various geographical locations and from international travelers. M2, molecular size markers (phage φX174 DNA digested with HaeIII). Lane numbers correspond to the experimental strain numbers listed in Table 2.
Divergence of the new O3:K6 clone.
The tdh-positive and trh-lacking clinical strains that were isolated in Calcutta in 1996 belonged to seven serovars, including O3:K6. Of these strains, non-O3:K6 strains were shown to exhibit various AP-PCR profiles that were distinct from that of the new O3:K6 clone in our previous study (16). We did not anticipate that the strains belonging to non-O3:K6 serovars would be phylogenetically close to the new O3:K6 clone. To our surprise, when we examined strains representing serovars other than O3:K6, those belonging to the O4:K68 and O1:K untypeable (KUT) serovars gave positive GS-PCR results. Accordingly, we further examined all available strains of the O4:K68 and O1:KUT serovars by GS-PCR (Table 2). Examples of the gel electrophoresis of the PCR products are presented in Fig. 4 (right panel). The GS-PCR-positive strains of the O4:K68 and O1:KUT serovars shared important characteristics with the new O3:K6 clone. They were positive for the tdh gene and lacked the trh1 and trh2 genes (Table 2). Representative strains of the GS-PCR-positive O4:K68 and O1:KUT groups (Table 2, experimental strain numbers 16 to 19 and 20 to 22, respectively) were examined by AP-PCR. The AP-PCR profiles of these strains were very similar to or indistinguishable from that of the new O3:K6 clone (Fig. 3). The GS-PCR-negative O1:KUT strains gave AP-PCR profiles distinct from those of the GS-PCR-positive O1:KUT strains (data not shown). Furthermore, the toxRS sequences of strains AN-5034 and AN-16000, representing the GS-PCR-positive O4:K68 and O1:KUT groups, respectively (Table 2), were 100% identical with that of the new O3:K6 clone. In contrast, the toxRS sequences of two strains belonging to serovar O8:K22, Y-27669 and AN-8917 (isolated in 1983 and 1998, respectively) (Table 2), differed from that of the new O3:K6 clone by 17 bp in the 1,364-bp region, and the discrepant bases in the O8:K22 strains contained all 7 bases unique to the old O3:K6 strains (Fig. 1). The results indicate that the GS-PCR-positive strains belonging to the O4:K68 and O1:KUT serovars are genetically very close to the new O3:K6 clone. The O4:K68 serovar has never existed in the list of known O:K serovars before. The strains of this serovar were first isolated in 1997 from international travelers and were subsequently detected in India, Bangladesh, and Japan. Although strains of serovar O1:KUT have been detected since 1980, GS-PCR-positive O1:KUT strains first appeared in India in 1997 and were subsequently detected in Bangladesh and from an international traveler. Therefore, the GS-PCR-positive O4:K68 and O1:KUT strains may have diverged from the new O3:K6 clone by alteration of the genes associated with the O:K antigens and followed a spreading pattern similar to that of the new O3:K6 clone. It is interesting that detection of the strains belonging to the O3:K6, O1:KUT, and O4:K68 serovars in Bangladesh was chronologically closely linked (Table 1). It is known that epidemic clones of V. cholerae are phylogenetically very close but can have different O antigens (2). Comparison of the O3:K6, O4:K68, and O1:KUT strains by methods other than the AP-PCR and toxRS analyses would further support our hypothesis on the divergence of O4:K68 and O1:KUT strains.
In this study, we demonstrated that the new O3:K6 clone includes not only the strains isolated in India and those from Southeast Asian travelers but also those isolated from clinical sources in seven other countries including Japan and the United States. This is the first demonstration of pandemicity in the history of V. parahaemolyticus. This new clone may have first appeared in 1995 (16), after which it spread to various parts of the world and variants began to be seen. The strains of the new O3:K6 clone isolated in Calcutta could be subtyped by ribotyping and pulsed-field gel electrophoresis methods (1). One exceptional strain of the new O3:K6 group examined in this study, FIHES98V1-32-4, lacked the tdh gene and exhibited AP-PCR profiles slightly different from the typical profiles of the new O3:K6 clone. A GS-PCR-positive O3:K6 strain lacking the tdh gene was also isolated from seafood implicated in a food poisoning case in Japan that was different from the FIHES98V-1-32-4-associated case (not included in Table 2). The AP-PCR profiles of FIHES98V1-32-4 and the seafood isolate were indistinguishable, and the toxRS sequence of the seafood isolate was 100% identical with those of the new O3:K6 clone (data not shown). The tdh gene can be lost spontaneously in V. parahaemolyticus (11). Although the tdh genes are present in transposon-like structures, the putative transposase gene is mutated (22). In agreement with previous findings with KP-positive strains, the strains of new O3:K6 clone carry two tdh genes (tdh1 and tdh2) that have 97% sequence identity (1, 12; M. Nishibuchi, unpublished data). The two tdh genes do not appear to have been duplicated by a crossover event and are not located in close proximity (they are at least 8 kb apart) in the chromosome (M. Nishibuchi, unpublished data). If FIHES98V1-32-4 originally had the tdh1 and the tdh2 genes, the two tdh genes may have been lost by homologous recombination between the two genes and the intervening sequence may have been lost by a looping-out mechanism. Such presumed genetic rearrangement could be one possible explanation for the slight divergence in the AP-PCR profile.
The GS-PCR test provided unambiguous results, and they were well correlated with the results of AP-PCR analysis. In addition, the GS-PCR is less likely to miss possible variants of the new O3:K6 clone. Therefore, this method will be very useful for the study of the current pandemic spread of V. parahaemolyticus. We found many strains of the new O3:K6 clone in India and Bangladesh in 1996 (Table 2). However, the new O3:K6 clone was isolated from one international traveler originating in Indonesia in 1995 (Table 2) (16). Therefore, it is not clear whether the new O3:K6 clone originated in the India-Bangladesh area. Examination of the distribution of GS-PCR-positive strains in the Asian environment and detailed comparison of the environmental and clinical GS-PCR-positive strains by various genetic fingerprinting methods may provide useful information for the elucidation of the origin of the new O3:K6 clone.
Our previous study (16) and this study indicated that the pandemic clone is responsible for recent V. parahaemolyticus infections in many countries. It has been considered that the majority of V. parahaemolyticus infections are acquired through the consumption of local seafood. A possibility that a considerable portion of V. parahaemolyticus infections are associated with imported seafood, including secondary contaminations in food-processing facilities, may have to be included in future epidemiological investigations.
We do not know why the new clone is responsible for the current pandemic. The level of thermostable direct hemolysin production of the pandemic clone is not very different from that of classical KP-positive strains, and the pandemic clone is sensitive to representative antibiotics (16). The new clone may be more potent than classical KP-positive strains in persisting in the environment or establishing infection. Future study on this aspect is needed to establish control measures.
ACKNOWLEDGMENTS
This research was supported in part by the COE program on “Making Regions: Proto-Areas, Transformations and New Formations in Asia and Africa,” by a Grant-in Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture, Japan, by “Research for the Future” Program of The Japan Society for the Promotion of Science (JSPS-RFTF97L00706), by the US-Japan Cooperative Medical Science Program, Cholera and Related Diarrheal Diseases, Japanese Panel, by the International Centre for Diarrheal Diseases Research, Bangladesh, and by the Japan International Cooperation Agency (JICA/NICED Project 054-1061-E-O).
We thank those who supplied some of the V. parahaemolyticus strains used in this study. We are grateful to Mutsumi Hashimoto and Yohko Takeda for technical assistance.
REFERENCES
- 1.Bag P K, Nandi S, Bhadra R K, Ramamurthy T, Bhattacharya S K, Nishibuchi M, Hamabata T, Yamasaki S, Takeda Y, Nair G B. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J Clin Microbiol. 1999;37:2354–2357. doi: 10.1128/jcm.37.7.2354-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltrán P, Delgado G A, Navarro A, Trujillo F, Selander R K, Cravioto A. Genetic diversity and population structure of Vibrio cholerae. J Clin Microbiol. 1999;37:581–590. doi: 10.1128/jcm.37.3.581-590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiRita V J. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae. Mol Microbiol. 1992;6:451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y B, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1999;37:1173–1177. doi: 10.1128/jcm.37.4.1173-1177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kishishita M, Matsuoka N, Kumagai K, Yamasaki S, Takeda Y, Nishibuchi M. Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl Environ Microbiol. 1992;58:2449–2457. doi: 10.1128/aem.58.8.2449-2457.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kita-Tsukamoto K, Oyaizu H, Nanba K, Shimidu U. Phylogenetic relationships of marine bacteria, mainly members of the family Vibrionaceae, determined on the basis of 16S rRNA sequences. Int J Syst Bacteriol. 1993;43:8–19. doi: 10.1099/00207713-43-1-8. [DOI] [PubMed] [Google Scholar]
- 7.Lin Z, Kumagai K, Baba K, Mekalanos J J, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol. 1993;175:3844–3855. doi: 10.1128/jb.175.12.3844-3855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyamoto Y, Kato T, Obara Y, Akiyama S, Takizawa K, Yamai S. In vitro hemolytic characteristic of Vibrio parahaemolyticus: its close correlation with human pathogenicity. J Bacteriol. 1969;100:1147–1149. doi: 10.1128/jb.100.2.1147-1149.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monsur K A. A highly selective gelatin-taurocholate-tellurite medium for the isolation of Vibrio cholerae. Trans R Soc Trop Med Hyg. 1961;55:440–442. doi: 10.1016/0035-9203(61)90090-6. [DOI] [PubMed] [Google Scholar]
- 10.Nishibuchi M, Fasano A, Russel R G, Kaper J B. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect Immun. 1992;60:3539–3545. doi: 10.1128/iai.60.9.3539-3545.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishibuchi M, Ishibashi M, Takeda Y, Kaper J B. Detection of the thermostable direct hemolysin gene and related DNA sequences in Vibrio parahaemolyticus and other Vibrio species by the DNA colony hybridization test. Infect Immun. 1985;49:481–486. doi: 10.1128/iai.49.3.481-486.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishibuchi M, Kaper J B. Duplication and variation of the thermostable direct haemolysin (tdh) gene in V. parahaemolyticus. Mol Microbiol. 1990;4:87–99. doi: 10.1111/j.1365-2958.1990.tb02017.x. [DOI] [PubMed] [Google Scholar]
- 13.Nishibuchi M, Kumagai K, Kaper J B. Contribution of the tdh1 gene of Kanagawa phenomenon-positive Vibrio parahaemolyticus to production of extracellular thermostable direct hemolysin. Microb Pathog. 1991;11:453–460. doi: 10.1016/0882-4010(91)90042-9. [DOI] [PubMed] [Google Scholar]
- 14.Nishibuchi M, Taniguchi T, Misawa T, Khaeomanee-Iam V, Honda T, Miwatani T. Cloning and nucleotide sequence of the gene (trh) encoding the hemolysin related to the thermostable direct hemolysin of Vibrio parahaemolyticus. Infect Immun. 1989;57:2691–2697. doi: 10.1128/iai.57.9.2691-2697.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okuda J, Ishibashi M, Abbott S L, Janda J M, Nishibuchi M. Analysis of the thermostable direct hemolysin (tdh) gene and the tdh-related hemolysin (trh) genes in urease-positive strains of Vibrio parahaemolyticus isolated on the West Coast of the United States. J Clin Microbiol. 1997;35:1965–1971. doi: 10.1128/jcm.35.8.1965-1971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay A K, Garg S, Bhattacharya S K, Nair G B, Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J Clin Microbiol. 1997;35:3150–3155. doi: 10.1128/jcm.35.12.3150-3155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuda J, Nishibuchi M. Manifestation of the Kanagawa phenomenon, the virulence-associated phenotype, of Vibrio parahaemolyticus depends on a particular single base change in the promoter of the thermostable direct hemolysin. Mol Microbiol. 1998;30:499–511. doi: 10.1046/j.1365-2958.1998.01072.x. [DOI] [PubMed] [Google Scholar]
- 18.Reich K A, Schoolnik G K. The light organ symbiont Vibrio fischeri possesses a homolog of the Vibrio cholerae transmembrane transcriptional activator ToxR. J Bacteriol. 1994;176:3085–3088. doi: 10.1128/jb.176.10.3085-3088.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakazaki R, Tamura K, Kato T, Obara Y, Yamai S, Hobo K. Studies of the enteropathogenic, facultatively halophilic bacteria, Vibrio parahaemolyticus. III. Enteropathogenicity. Jpn J Med Sci Biol. 1968;21:325–331. doi: 10.7883/yoken1952.21.325. [DOI] [PubMed] [Google Scholar]
- 20.Shirai H, Ito H, Hirayama T, Nakabayashi Y, Kumagai K, Takeda Y, Nishibuchi M. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun. 1990;58:3568–3573. doi: 10.1128/iai.58.11.3568-3573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suthienkul O, Ishibashi M, Iida T, Nettip N, Supavej S, Eampokalap B, Makino M, Honda T. Urease production correlates with possession of the trh gene in Vibrio parahaemolyticus strains isolated in Thailand. J Infect Dis. 1995;172:1405–1408. doi: 10.1093/infdis/172.5.1405. [DOI] [PubMed] [Google Scholar]
- 22.Terai A, Baba K, Shirai H, Yoshida O, Takeda Y, Nishibuchi M. Evidence for insertion sequence-mediated spread of the thermostable direct hemolysin gene among Vibrio species. J Bacteriol. 1991;173:5036–5046. doi: 10.1128/jb.173.16.5036-5046.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu D Y, Ugozzoli L, Pal B K, Wallace R B. Allele-specific enzymatic amplification of β-globin genomic DNA for diagnosis of sickle cell anemia. Proc Natl Acad Sci USA. 1989;86:2757–2760. doi: 10.1073/pnas.86.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamashiro T, Insisiengmay S, Honma Y, Higa N, Enami M, Iwanaga M. Bacteriological study on Vibrio parahaemolyticus isolated from the outbreaks of diarrhea in Laos, an inland country. Jpn J Trop Med Hyg. 1998;26:319–322. [Google Scholar]