Abstract
The Light Cycler technique combines rapid in vitro amplification of DNA in glass capillaries with real-time species determination and quantification of DNA load. We have established a quantitative PCR protocol for two clinically important pathogens, Candida albicans and Aspergillus fumigatus. The sensitivity of the assay was comparable to those of previously described PCR protocols (5 CFU/ml). Specific detection of C. albicans and A. fumigatus could be achieved. The assay showed a high reproducibility of 96 to 99%. The assay was linear in a range between 101 and 104 Aspergillus conidia. As capillaries do not have to be reopened for post-PCR analysis, the risk of carryover contaminations could be minimized. The Light Cycler allowed quantification of the fungal loads in a limited number of clinical specimens from patients with hematological malignancies and histologically proven invasive fungal infections. Five of nine positive samples had fungal loads between 5 and 10 CFU/ml of blood, two of nine positive samples had fungal loads between 10 and 100 CFU/ml of blood, and two of nine samples had fungal loads of more than 100 CFU/ml of blood. All samples were also found to be PCR positive by PCR–enzyme-linked immunosorbent assay analysis.
Invasive fungal infection has become a major cause of morbidity and mortality in immunocompromised patients, for example, neutropenic patients with hematological malignancies and recipients of a allogeneic bone marrow transplants (6). Conventional diagnostic tests like blood culture or serology (10) lack sufficient sensitivity and specificity for the early diagnosis of invasive fungal infections. Thus, diagnostic assays based on in vitro amplification and detection of fungal DNA were developed. For selected groups of patients, PCR-based assays were found to have promising sensitivity and specificity and demonstrated a potential value for the early diagnosis of invasive fungal infections (7, 14). However, some protocols showed poor sensitivity because of the amplification of single-copy genes (5), were limited to the detection of Candida spp. (9), or were time- and labor-intensive (3). Other protocols were not applied to the detection of fungal DNA in clinical samples (2).
Our previously published assay is able to detect a broad range of fungal pathogens in human blood specimens with a high sensitivity and specificity (7). Despite major improvements, the method requires a minimum of 9 h for DNA amplification and oligonucleotide hybridization (11).
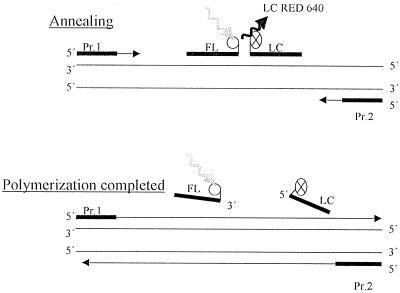
A quantitative PCR assay with the LightCycler (Roche Diagnostics, Mannheim, Germany) amplification and detection system was established. This technology combines rapid thermocycling with glass capillaries with online fluorescence detection of the PCR amplicon. Cycling is achieved by alternating heated air and air of ambient temperature. The detection system is based on fluorescence resonance energy transfer (FRET) with two different specific oligonucleotides. Hybridization probe 1 (FL) is labeled with fluorescein, and hybridization probe 2 (LC) is labeled with the fluorophore Light Cycler Red 640. Both probes can hybridize in a head-to-tail arrangement, bringing the two fluorescent dyes into close proximity. A transfer of energy between the two probes results in emission of red fluorescent light which is measured by photohybrids (Fig. 1). The level of fluorescence is proportional to the amount of DNA generated during the PCR process.
FIG. 1.
Hybridization probe format overview. Pr. 1 and Pr. 2, primers amplifying a conserved region of the 18S rRNA gene. Probe FL is labeled with fluorescein, and probe LC is labeled with Light Cycler Red 640 fluorophore. During annealing, the excitation energy is transferred to the acceptor fluorophore, Light Cycler Red 640 fluorophore. The emitted fluorescence is proportional to the amount of DNA generated during the PCR. After the completion of polymerization, the emission of fluorescence is stopped.
MATERIALS AND METHODS
Fungal cultures.
Fungi (Aspergillus fumigatus DSM 790, Candida albicans DSM 6569) were obtained from the German Collection of Microorganisms (DSM) and were cultured on Sabouraud-glucose-agar for 72 h at 30°C. Serial dilutions of fungal cells were prepared with sterile saline suspensions that were adjusted to a 0.5 McFarland standard (which is equal to 106 CFU/ml).
Collection and handling of samples.
For sensitivity and specificity testing as well as for use as external standards, blood from healthy volunteers was spiked with A. fumigatus conidia (104 to 100/ml, in serial dilutions) or C. albicans cells (104 to 100/ml, in serial dilutions). Samples were extracted and analyzed in parallel.
In addition, we applied this method to a limited number of blood specimens from patients with hematological malignancies. All patients suffered from histologically proven invasive aspergillosis or histologically proven invasive candidiasis (C. albicans) and were included in a weekly screening for the presence of fungal DNA by PCR–enzyme-linked immunosorbent assay (ELISA) (11). All samples were positive either for A. fumigatus or for C. albicans DNA by hybridization in the PCR-ELISA. As controls, 50 samples from patients without clinical evidence of an invasive fungal infection were analyzed.
All DNA specimens had previously been extracted by using the QIAmp Tissue Kit (Qiagen GmbH, Hilden, Germany). Purified DNA was eluted from the QIAmp spin column in a concentrated form in sterile water and was stored at −80°C until retrospective analysis with the Light Cycler system.
Controls.
To monitor for contamination, aliquots of saline or DNA from healthy control persons were prepared concurrently. For each 10 clinical samples analyzed, one extraction control and one PCR-negative control were included.
Specificity of Light Cycler technique.
In order to determine the specificity of the oligonucleotide hybridization based on the FRET technique, DNAs extracted from cultures of A. fumigatus, C. albicans, Candida tropicalis, Candida parapsilosis, Candida krusei, and Candida glabrata as well as DNAs from human fibroblasts from healthy individuals were analyzed.
DNA extraction.
DNA was extracted as described previously (11) by using recombinant lyticase (Sigma, Deissenhofen, Germany) and the QIAmp Tissue Kit (Qiagen, Hilden, Germany).
Light Cycler-based PCR assay.
The Light Cycler PCR and detection system (Roche Diagnostics, Mannheim, Germany) has been used for amplification and online quantification.
PCR was performed in glass capillaries, which ensures rapid equilibration between the air and the reaction components because of the high surface-to-volume ratio of the capillaries.
Primers (5′-ATT GGA GGG CAA GTC TGG TG and 5′-CCG ATC CCT AGT CGG CAT AG; Roth, Karlsruhe, Germany) bind to conserved regions of the fungal 18S rRNA gene as described before (7). For amplicon detection, the Light Cycler DNA Master Hybridization Probes Kit was used as described by the manufacturer. Briefly, two different oligonucleotides hybridize to an internal species-specific sequence of the 18S rRNA gene of A. fumigatus or C. albicans. One probe has been labeled at the 5′ end with the Light Cycler Red 640 fluorophore (5′-TGA GGT TCC CCA GAA GGA AAG GTC CAG C for A. fumigatus; 5′-TGG CGA ACC AGG ACT TTT ACT TTG A for C. albicans) (Tibmolbiol, Berlin, Germany), and the other has been labeled at the 3′ end with fluorescein (5′-GTT CCC CCC ACA GCC AGT GAA GGC for A. fumigatus; 5′-AGC CTT TCC TTC TGG GTA GCC ATT for C. albicans) (Tibmolbiol). During FRET, fluorescein is excited by the light source of the Light Cycler instrument. The excitation energy is transferred to the acceptor fluorophore, Light Cycler Red 640, and the emitted fluorescence is measured after annealing by the photohybrids of the instrument (Fig. 1).
The PCR mixture contained Taq polymerase, 1× Light Cycler hybridization reaction buffer, a deoxynucleoside triphosphate mixture (with dUTP instead of dTTP), 3 mM magnesium chloride, and 12.5 pmol of primers.
Thirty-two samples were run in parallel by performing 45 cycles of repeated denaturation (1 s at 95°C), annealing (15 s at 62°C), and enzymatic chain extension (25 s at 72°C). In order to enhance the specificity of the PCR, TaqStart Antibody (Clontech, Palo Alto, Calif.) was used.
The PCR run was completed within 45 min.
Light Cycler-based quantification of target DNA.
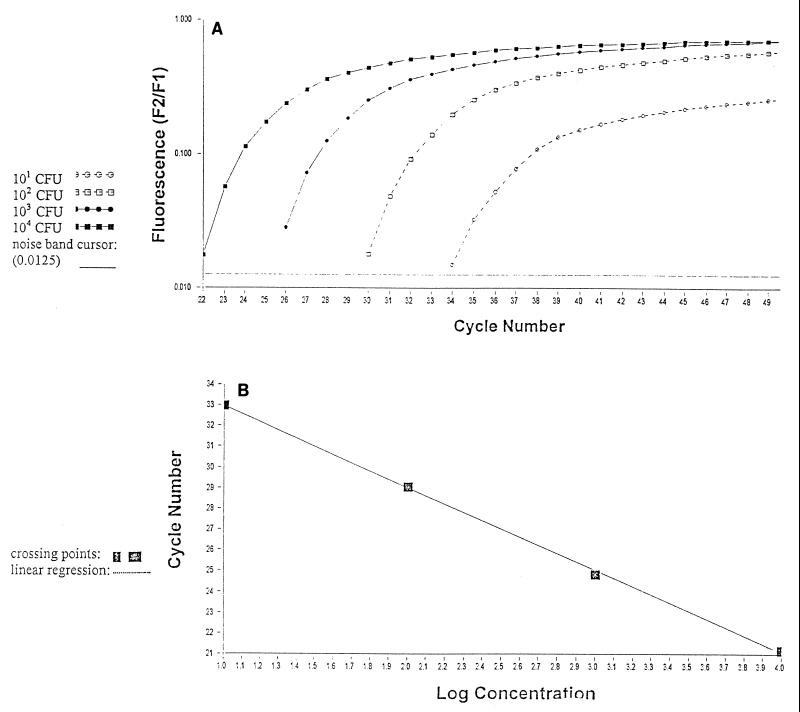
Quantification was performed by online monitoring for identification of the exact time point at which the logarithmic linear phase could be distinguished from the background (crossing point). Serially diluted samples of genomic fungal DNA obtained from A. fumigatus and C. albicans cultures (104 to 100 CFU, corresponding to 100 pg to 10 fg of DNA) were used as external standards in each run (Fig. 2).
FIG. 2.
Quantification of serially diluted A. fumigatus conidia (104 to 101 CFU) by using the Light Cycler-based PCR technique (A) and Light Cycler-based standard curve report for serially diluted A. fumigatus conidia (104 to 101 CFU) (B).
The cycle numbers of the logarithmic linear phase were plotted against the logarithm of the concentration of template DNA. The concentrations of fungal DNA in the clinical samples were calculated by comparing the cycle numbers of the logarithmic linear phase of the samples with the cycle numbers of the external standards.
Gel electrophoresis.
After each Light Cycler run, agarose gel electrophoresis with a TAE (Tris-acetate-EDTA)–2% agarose gel, followed by DNA staining with ethidium bromide, was performed to have an independent validation check of the presence of an amplicon. In order to control the length of the amplicon generated, a 100-bp DNA ladder (Life Technologies, Karlsruhe, Germany) was used (7).
RESULTS
Gel electrophoresis.
Aspergillus and Candida DNAs were successfully amplified with the Light Cycler instrument. All serially diluted samples containing at least 100 CFU/ml showed a single band at 500 bp by gel electrophoresis; the band represented the fungus-specific amplicon (Fig. 3).
FIG. 3.
Agarose gel electrophoresis of serially diluted A. fumigatus conidia (106 to 100 CFU, corresponding to 10 ng to 10 fg of DNA) showing a single, specific band at 500 bp. DNA was extracted as described in the text and was amplified with a conventional thermocycler (lane 2, positive control) and by the Light Cycler™ technique (lanes 3 to 9). Lanes: 1, 100-bp ladder; 2, 106 CFU (conventional thermocycler); 3, 105 CFU (1 ng); 4, 104 CFU (100 pg); 5, 103 CFU (10 pg); 6, 102 CFU (1 pg); 7, 101 CFU (100 fg); 8, 100 CFU (10 fg); 9, negative control (double-distilled H2O).
Sensitivity.
For sensitivity testing, blood from healthy volunteers was spiked with Aspergillus conidia (104 to 100/ml of blood, in serial dilutions) and Candida cells (106 to 100/ml of blood, in serial dilutions). Using the Light Cycler FRET technique, we demonstrated a sensitivity of 5 CFU/ml of blood for Aspergillus conidia and Candida cells (Fig. 4, bar 1). This sensitivity corresponded to that of the amplification of fungal DNA in a conventional thermoblock, followed by conventional hybridization with biotin- or digoxigenin-labeled oligonucleotides (7, 11).
FIG. 4.
Sensitivity and reproducibility of the Light Cycler-based detection of fungal DNA. Bars: 1, 5 CFU (sensitivity control); 2, negative control (double-distilled H2O); 3 to 5, 101 CFU (identical DNA extraction); 6 to 8, 102 CFU (identical DNA extraction); 9 to 11, 103 CFU (identical DNA extraction); 12 to 14, 104 CFU (identical DNA extraction); 15, negative control (double-distilled H2O); 16 to 18, 101 CFU (different dilution series); 19 to 21, 102 CFU (different dilution series); 22 to 24, 103 CFU (different dilution series); 25, negative control (double-distilled H2O); 26 to 34, patient samples.
Specificity of Light Cycler technique.
The oligonucleotide specific for C. albicans hybridized only with DNA extracted from C. albicans cultures. No cross-reaction with A. fumigatus DNA or DNA from other Candida species was observed. The probe specific for A. fumigatus did not hybridize with DNA extracted from Candida species. The negative controls, which consisted of fibroblast DNA from healthy individuals, did not hybridize with either oligonucleotide.
Reproducibility of Light Cycler technique.
Amplification of serially diluted Aspergillus conidia was repeated 10 times with DNA from the same extraction (Fig. 4, bars 3 to 14). The crossing points for runs performed with DNA from identical extractions were calculated and were as follows: for 10 CFU, at 33 cycles (±0.15); for 102 CFU, at 29.6 cycles (±0.13); for 103 CFU, at 25.1 cycles (±0.1); and for 104 CFU, at 21.6 cycles (±0.1).
In order to exclude variations during the extraction procedures, the DNA extraction was repeated five times with five different serial dilutions containing Aspergillus conidia. The sensitivity was also reproducible (10 CFU/ml blood) (Fig. 4, bars 16 to 24). The crossing points were as follows: for 10 CFU, at 32.8 cycles (±0.7); for 102 CFU, at 29.6 cycles (±0.45); and for 103 CFU, at 26.1 cycles (±0.3).
Results from amplification reproducibility studies done on the Light Cycler instrument agree with those for conventional thermal cyclers published previously (7, 11).
Linear range of assay.
The linear range of the assay was from 101 to 104 Aspergillus conidia. Figure 2 plots the results for an Aspergillus standard.
Patient samples.
Seven of seven blood samples from patients with histologically proven invasive aspergillosis and two of two samples from patients with histologically proven invasive candidiasis (C. albicans) were positive by Light Cycler analysis. All nine samples also had positive hybridization results by PCR-ELISA.
The Light Cycler allowed quantification of the fungal loads in these specimens. Five of nine positive samples had fungal loads between 5 and 10 CFU/ml of blood, two of nine positive samples had fungal loads between 10 and 100 CFU/ml of blood, and two of nine samples had with fungal loads of more than 100 CFU/ml of blood (Fig. 4, bars 26 to 34).
Fifty of fifty samples from patients with no clinical evidence of an invasive fungal infection tested negative by both assays.
DISCUSSION
Invasive fungal infections have been reported with an increasing frequency in patients such as bone marrow and solid-organ transplant recipients, patients receiving intense chemotherapy, AIDS patients, patients with cystic fibrosis, neonatal patients, and patients with severe burns (6). The major fungal pathogens that cause invasive disease are still Aspergillus spp. and Candida spp., but other fungal species have also been reported with increasing frequency (10).
As the rate of mortality due to invasive aspergillosis, especially in high-risk patients, is over 90% and clinical signs are often nonspecific, a PCR-based method for the detection of fungi must be rapid to be of clinical benefit (10). However, most published PCR protocols are very time- and labor-intensive and do not allow quantification of the fungal DNA load of a clinical specimen.
Thus, we adapted our previously published PCR assay, based on conventional thermocycling, followed by hybridization by the PCR-ELISA format, to the Light Cycler-FRET detection system with two different specific oligonucleotides (oligonucleotides specific for C. albicans and A. fumigatus). Rapid amplification by alternating heated air and air of ambient temperature and online quantification allow the test to be completed within 45 min. As the DNA extraction procedure including lyticase incubation requires up to 6 h, the whole detection can be completed within 7 h. This may allow high flexibility in routine diagnostic assays.
The Light Cycler optical device is capable of measuring fluorescence in two separate channels simultaneously (Light Cycler Red 640 fluorophore and Light Cycler Red 705 fluorophore), and this allows analysis of different fungal pathogens within a single test tube.
By analyzing genomic Aspergillus DNA extracted from five different serial dilutions (104 to 101 CFU/ml of blood), the Light Cycler-based technique provides a high reproducibility of >95%. The reproducibility was even higher (99%) when the crossing points of dilution series from one identical extraction procedure were analyzed.
The assay is run in closed glass capillaries. Postamplification analysis can be performed without opening the capillaries, minimizing the risk of carryover contaminations.
The value of quantification of the fungal DNA load in clinical samples cannot yet be estimated. However, quantification of the viral load by PCR has been useful for patients with AIDS (13) and Herpes simplex virus (HSV) infection (4). Our study presents a means for the quantification of the fungal load in blood specimens from patients with histologically proven invasive fungal infection. In seven of nine specimens the fungal load was lower than 100 CFU/ml of blood. In comparison with the viral loads in blood specimens (15), these preliminary results show a very small amount of fungal DNA in blood specimens, even for patients suffering from invasive fungal infection. However, these findings have yet to be confirmed in studies involving larger numbers of patients and blood specimens.
Brandt et al. (1) described the automated detection of fungal DNA by using the TaqMan system (Perkin-Elmer, Foster City, Calif.). This system takes advantage of the 5′ exonuclease activity of the Taq polymerase. Upon primer elongation, the specific probe is cleaved, which interrupts the FRET and allows the release of a reporter dye from a quencher dye. The amount of reporter dye released is proportional to the amount of DNA amplified by PCR (8). The assay that they described allowed the detection of 89 of 90 A. fumigatus strains. However, the fungal load in clinical samples was not determined by this technique.
More recently, Ryncarz et al. (12) have shown the development of a quantitative assay for the detection of HSV DNA based on the TaqMan technique. In contrast to the Light Cycler instrument, the TaqMan system is run in a 96-well format, which allows a larger throughput. The fluorescent probe described for HSV allowed the sensitive detection of HSV DNA in culture-positive genital samples. However, the assay was less sensitive than their previously described means of quantification by a competitive PCR method for the detection of HSV DNA either in cerebrospinal fluid or in genital tract specimens.
In conclusion, the Light Cycler technique is standardized, rapid, accurate, and reproducible and combines rapid in vitro amplification with real-time species determination and quantification of the fungal load. The assay allows the use of different specific oligonucleotides in one reaction mixture. Amplification and postamplification analysis are performed in closed glass capillaries, thus minimizing the risk of carryover contamination. Therefore, these assays are valuable tools for the detection of DNA from numerous pathogens in a variety of clinical settings.
ACKNOWLEDGMENTS
We thank M. Klose, Roche Diagnostics, Mannheim, Germany, and O. Landt, Tibmolbiol, Berlin, Germany, for excellent technical support.
REFERENCES
- 1.Brandt M E, Padhye A, Mayer L W, Holloway B P. Utility of random amplified polymorphic DNA PCR and TaqMan automated detection in molecular identification of Aspergillus fumigatus. J Clin Microbiol. 1998;36:2057–2062. doi: 10.1128/jcm.36.7.2057-2062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchman T G, Rossier M, Merz W G, Charache P. Detection of surgical pathogens by in vitro DNA amplification. Part 1. Rapid identification of Candida albicans by in vitro amplification of a fungus-specific gene. Surgery. 1990;108:338–347. [PubMed] [Google Scholar]
- 3.Burgener-Kairuz P, Zuber J P, Jaunin P, Bille J, Rossier M. Rapid detection and identification of Candida albicans and Torulopsis glabrata in clinical specimens by species-specific nested PCR amplification of a cytochrome P-450 lanosterol demethylase (L1A1) gene fragment. J Clin Microbiol. 1994;32:1902–1907. doi: 10.1128/jcm.32.8.1902-1907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cone R W, Hobson A C, Brown Z, Ashley R, Berry S, Ishak L, Winter C, Corey L. Frequent reactivation of genital herpes simplex viruses among pregnant women. JAMA. 1994;272:792–796. [PubMed] [Google Scholar]
- 5.Crampin A C, Matthews R C. Application of the polymerase chain reaction to the diagnosis of candidosis by amplification of an HSP90 gene fragment. J Med Microbiol. 1993;39:233–238. doi: 10.1099/00222615-39-3-233. [DOI] [PubMed] [Google Scholar]
- 6.Denning D W. Invasive aspergillosis. Clin Infect Dis. 1998;26:781–805. doi: 10.1086/513943. [DOI] [PubMed] [Google Scholar]
- 7.Einsele H, Hebart H, Roller G, Löffler J, Rothenhöfer I, Müller C A, Bowden R A, van Burik J-A, Engelhard D, Kanz L, Schumacher U. Detection and identification of fungal pathogens in blood by using molecular probes. J Clin Microbiol. 1997;35:1353–1360. doi: 10.1128/jcm.35.6.1353-1360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holland P, Abramson R, Watson R, Gelfand D. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jordan J A. PCR identification of four medically important Candida species by using a single primer pair. J Clin Microbiol. 1994;32:2962–2967. doi: 10.1128/jcm.32.12.2962-2967.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latgé J P. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Löffler J, Hebart H, Sepe S, Schumacher U, Klingebiel T, Einsele H. Detection of PCR-amplified fungal DNA by using a PCR-ELISA system. Med Mycol. 1998;36:275–279. doi: 10.1080/02681219880000441. [DOI] [PubMed] [Google Scholar]
- 12.Ryncarz A J, Goddard J, Wald A, Huang M-L, Roizman B, Corey L. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J Clin Microbiol. 1999;37:1941–1947. doi: 10.1128/jcm.37.6.1941-1947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tetali S, Bakshi S, Than S, Pahwa S, Abrams E, Romano J, Pahwa S G. Plasma virus load evaluation in relation to disease progression in HIV-infected children. AIDS Res Hum Retrovir. 1998;14:571–577. doi: 10.1089/aid.1998.14.571. [DOI] [PubMed] [Google Scholar]
- 14.Van Burik J-A, Myerson D, Schreckhise R W, Bowden R A. Panfungal PCR assay for detection of fungal infection in human blood specimens. J Clin Microbiol. 1998;36:1169–1175. doi: 10.1128/jcm.36.5.1169-1175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto M, Kimura H, Hironaka T, Hirai K, Hasegawa S, Kuzushima K, Shibata M, Morishima T. Detection and quantification of virus DNA in plasma of patients with Epstein-Barr-virus-associated diseases. J Clin Microbiol. 1995;33:1765–1768. doi: 10.1128/jcm.33.7.1765-1768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]