Abstract
Vancomycin-resistant enterococci (VRE) have emerged as nosocomial pathogens over the last decade, but little is known about their epidemiology. We report on the prevalence of VRE fecal colonization on the basis of a prospective study among patients hospitalized in a hematology intensive care unit and among nonhospitalized subjects living in the local community. A total of 243 rectal swabs from hematology patients and 169 stool samples from the control group were inoculated onto bile-esculin agar plates with and without 6 mg of vancomycin per liter and into an enrichment bile-esculin broth supplemented with 4 mg of vancomycin per liter. A total of 37% of the hospitalized patients and 11.8% of the subjects from the community were found to be VRE carriers. A total of 65 VRE strains were isolated: 12 (18.5%) E. faecium, 46 (70.7%) E. gallinarum, and 7 (10.8%) E. casseliflavus strains. No E. faecalis strains were detected. All the E. faecium strains were of the vanA genotype. Molecular typing by pulsed-field gel electrophoresis revealed a different pattern for each vanA VRE strain that originated from an individual subject. To our knowledge, this is the first study to be carried out in a cattle-rearing region of France. It reports a higher VRE prevalence than that reported in previous European or U.S. studies. A partial explanation is the use of an enrichment broth step which enabled detection of strains which would otherwise have been missed, but the fact that subjects and patients were recruited from a predominantly agricultural area where vancomycin-related antibiotics have recently been used in animal husbandry could also contribute to the high levels of VRE in patients and subjects alike.
Over the last few years, enterococci have grown in importance as nosocomial pathogens (28). Their intrinsic or acquired resistance to many antibiotics, in particular, to glycopeptides, has become a major cause of concern. Vancomycin-resistant enterococci (VRE) were first isolated in 1986 in Europe (24, 37) and in 1987 in the United States (20; A. H. Uttley, C. H. Collins, J. Naidoo, and R. C. George, Letter, Lancet, i:57–58, 1988), and since then, their presence has increasingly been detected throughout the world. According to the Centers for Disease Control and Prevention, in the United States the number of nosocomial enterococcal isolates resistant to vancomycin increased 20-fold between 1989 and 1993 (5). VRE are now the second most common cause of hospital-acquired infections. Since the vanA and vanB vancomycin resistance determinants are transferable, glycopeptide resistance might be passed on to other pathogens such as methicillin-resistant Staphylococcus aureus, thus creating a highly dangerous pathogen difficult to treat with currently available antibiotics.
Before this study, no VRE infection had been reported at our hospital and nothing was known of the epidemiology of VRE inside or outside the hospital wards. We therefore undertook a prospective investigation of VRE intestinal colonization among inpatients and nonhospitalized subjects living in the local community.
MATERIALS AND METHODS
Patients and subjects.
The study was conducted in Limoges, a city with 150,000 inhabitants located in a cattle-rearing area in southwestern France.
For a period of 7 months, from March to September 1997, all patients admitted to the hematology intensive care unit at the Limoges University Teaching Hospital were screened for gastrointestinal carriage of VRE. A rectal swab was taken once weekly until the end of hospitalization. Seventy patients were admitted to the hematology intensive care unit: 30 women and 40 men (age range, 16 to 86 years; mean age, 51.4 years).
During the same period, subjects working in nonclinical hospital units and attending industrial medicine clinics were chosen as the control group: 81 women and 88 men (age range, 15 to 57 years; mean age 36.2 years). A single stool specimen was obtained from each subject.
Culture and identification.
Stool specimens and rectal swabs were inoculated onto bile-esculin agar plates (Oxoid, Dardilly, France) with and without 6 mg of vancomycin (Lilly, Saint-Cloud, France) per liter and into bile-esculin broth (Oxoid) supplemented with 4 mg of vancomycin per liter. The plates and broths were incubated at 37°C for 24 h. All esculin-positive broth cultures were subcultured onto bile-esculin agar plates with and without 6 mg of vancomycin per liter and were incubated at 37°C for 24 h.
Colonies growing on agar with a dark brown halo and morphologically resembling enterococci were primarily identified by Gram staining and by growth in 6.5% NaCl broth (Sanofi Pasteur, Marnes la Coquette, France). Species identification was performed with the API 20 STREP system (bioMérieux, Marcy l'Etoile, France) and was confirmed by PCR analysis. Species identification relies on the amplification of genes coding for ligases specific to each species, ddl (Enterococcus faecium), ddl (Enterococcus faecalis), vanC1, and vanC2, which are specific to E. faecium, E. faecalis, E. gallinarum, and E. casseliflavus, respectively.
Susceptibility testing.
Resistance to vancomycin and teicoplanin was first screened by the E-test method (AB Biodisk, Solna, Sweden). An inoculum with a turbidity equivalent to that of a no. 1 McFarland standard and Mueller-Hinton agar (bioMérieux, La Balme les Grottes, France) were used. The plates were read after incubation at 37°C for 24 h. All isolates for which the MICs of vancomycin were ≥4 mg/liter were further tested by standard agar dilution on Mueller-Hinton agar with a final inoculum of 105 CFU/spot. Vancomycin and teicoplanin were tested in the range of 0.25 to 1,024 mg/liter, and the results were interpreted according to the guidelines set forth by the Comité de l'Antibiogramme de la Société Française de Microbiologie (CA-SFM; for vancomycin and teicoplanin, resistance was an MIC of >16 mg/liter and susceptibility was an MIC of ≤4 mg/liter) (7).
For isolates resistant to vancomycin, susceptibility to amoxicillin, gentamicin, and streptomycin was determined by the disk diffusion technique (with disks from Sanofi Pasteur) with high-content disks for detection of high-level gentamicin and streptomycin resistance. Breakpoints were those laid down by CA-SFM.
E. faecalis V583 of the vanB genotype, E. faecium BM4147 of the vanA genotype, and E. gallinarum BM4174 of the vanC genotype (kindly provided by P. Courvalin, Institut Pasteur, Paris, France) were used as control strains for MIC determinations.
DNA isolation.
Strains were grown overnight at 37°C on Columbia agar supplemented with 5% sheep blood (Becton Dickinson, Meylan, France). The colonies were suspended in 1 ml of sterile water, and the suspension was heated for 15 min at 100°C and then centrifuged at 9,980 × g for 10 min. The supernatant containing DNA was stored at −20°C until further use.
PCR.
Confirmation of species identification and determination of glycopeptide resistance genotypes were performed by PCR. The genes vanA, vanB, vanC1, vanC2, ddl (E. faecium), and ddl (E. faecalis) were amplified with the primers described by Dukta-Malen et al. (10). The PCR amplification mixture consisted of PCR buffer (GIBCO BRL, Cergy-Pontoise, France), 0.2 mM (each) dATP, dCTP, dGTP, and dTTP (Boehringer, Meylan, France), 50 pmol of each primer (Isoprim, Toulouse, France), 2 mM MgCl2, 2 U of Taq DNA polymerase (GIBCO BRL), and 50 ng of enterococcal DNA in a total volume of 50 μl. DNA amplification was carried out with a GeneAmp PCR system 9600 thermal cycler (Perkin-Elmer). The reaction mixtures were heated to 94°C for 3 min, followed by 35 cycles each consisting of 30 s at 94°C, 1 min at 54°C, and 1 min at 72°C. A final elongation step was carried out at 72°C for 10 min. A reagent blank (containing all the components of the reaction mixture except DNA) and positive controls for each van genotype (E. faecalis V583 [vanB], E. faecium BM4147 [vanA], and E. gallinarum BM4174 [vanC1]) were run in every PCR procedure as controls.
The vanD gene, which codes for the VanD resistance phenotype, was detected by the same protocol with the primers described by Perichon et al. (31).
The amplified products were submitted to gel electrophoresis in a 1.5% agarose gel in 0.5× TBE buffer (45 mM Tris base, 45 mM boric acid, 1.25 mM EDTA [pH 8.6]). The gels were stained with ethidium bromide (0.5 mg/liter; Bioprobe systems, Montreuil, France). A 123-bp DNA ladder (GIBCO BRL) was run with each gel, and the VRE genotype was determined by the size of the amplified product.
PFGE.
Strains of the VanA phenotype were tested by pulsed-field gel electrophoresis (PFGE). Overnight cultures grown on brain heart broth (bioMérieux) were centrifuged at 850 × g for 10 min. The pellet was suspended in 5 ml of buffer (10 mM Tris, 10 mM EDTA, 10 mM EGTA, 1 M NaCl [pH 7.6]). An aliquot of this suspension (100 μl) was mixed with 100 μl of 1.8% agarose (Incert agarose; TEBU, Le Perray en Yvelines, France) at 55°C. This mixture was transferred into two 100-μl sample plug molds. The plugs were removed from the molds and were incubated for 18 h at 37°C in 1 ml of lysis solution (6 mM Tris HCl, 1 M NaCl, 100 mM EDTA, 0.5% Brij 58, 0.2% sodium deoxycholate, 0.5% lauroyl sarcosine, 1 mg of lysozyme per ml [pH 7.6]). This lysis solution was replaced by 1 ml of TEP solution (10 mM Tris base, 1 mM EDTA, 10 mg of proteinase K per ml), and the mixture was incubated for 18 h at 50°C. The plugs were then washed for 18 h at 4°C in 5 ml of TE buffer (10 mM Tris base, 1 mM EDTA). A slice of the plug was digested with 20 U of the SmaI restriction enzyme (Boehringer) and was incubated for 6 h at 25°C according to the manufacturer's recommendations. After digestion, the plug was stored in 1 ml of 10 mM Tris base–20 mM EDTA at 4°C.
DNA fragments were separated in a 1% agarose gel (FastLane agarose; TEBU) in 0.5× TBE (Bio-Rad, Ivry sur Seine, France). Electrophoresis was performed with a CHEF DR III apparatus (Bio-Rad). Parameters for electrophoresis were 4.5 V/cm at 14°C for 21 h, with pulse times ramped from 5 to 50 s. The gels were stained with 0.1 mg of ethidium bromide solution (Bioprobe Systems) per liter for 15 min and were then placed onto a UV source. The sizes of the DNA fragments were then determined according to the size of the PFGE I marker (Boehringer).
RESULTS
We collected 243 rectal swab specimens from 70 hematology patients and 169 stool samples from the 169 outpatients in the control group. Sixty-five VRE strains were isolated: 44 from hematology patients and 21 from the control group. Twenty-six hematology patients were VRE carriers (37%), 5 of whom were carriers of two VRE strains of different species, and 20 patients from the control group were also VRE carriers (11.8%), 1 of whom was a carrier of two different species of enterococci.
The genotypes and species of VRE strains isolated from both group of patients are described in Table 1. Among these strains, 12 (18.5%) were identified by PCR as E. faecium, 46 (70.7%) were identified as E. gallinarum, and 7 (10.8%) were identified as E. casseliflavus. No E. faecalis isolates were detected. The results obtained with the API 20 STREP system correlated with the results of PCR for species identification except for 45 of the 46 E. gallinarum strains: 11 were identified as E. faecium and 34 were identified as E. casseliflavus.
TABLE 1.
Species and genotype of VRE strains
| Genotype | Species | No. of strains from:
|
|
|---|---|---|---|
| Hematology patients | Control group | ||
| vanA | E. faecium | 9 | 3 |
| E. faecalis | 0 | 0 | |
| vanB | E. faecium | 0 | 0 |
| E. faecalis | 0 | 0 | |
| vanC1 | E. gallinarum | 32 | 14 |
| vanC2 | E. casseliflavus | 3 | 4 |
| Total | 44 | 21 | |
Twelve strains of the VanA phenotype were isolated: nine from hematology patients and three from the control group. Vancomycin MICs ranged from 256 to 1,024 mg/liter, and teicoplanin MICs ranged from 32 to 256 mg/liter. All these strains were E. faecium and carried the vanA gene, as shown by PCR. Fifty-three VanC phenotype strains were isolated. PCRs confirmed that all E. gallinarum strains had the vanC1 gene and that all E. casseliflavus strains had the vanC2 gene. No vanB or vanD strains were detected.
We compared broth and agar plates for their abilities to detect VRE (Table 2). Forty-nine of the 65 VRE strains (75%) grew only in the bile-esculin broth supplemented with 4 mg of vancomycin per liter. Fifteen (23%) strains were isolated on both broth and agar plates. Only one strain, a vanA E. faecium strain from the control group, grew only on the agar plate.
TABLE 2.
Comparison of performances of broth and agar plate methods for detection of VRE
| Strain (no. of isolates) | No. of strains detected ona:
|
||
|---|---|---|---|
| GBEV6 only | bBEV4 only | Both GBEV6 and bBEV4 | |
| E. faecium vanA (12) | 1 | 8 | 3 |
| E. gallinarum (46) | 0 | 34 | 12 |
| E. casseliflavus (7) | 0 | 7 | 0 |
| Total | 1 | 49 | 15 |
GBEV6, bile-esculin agar plate with 6 mg of vancomycin per liter; bBEV4, bile-esculin broth with 4 mg of vancomycin per liter.
Among the 44 VRE strains isolated from hematology patients, 5 VRE strains (E. faecium vanA) were resistant to amoxicillin; the 39 other strains were susceptible to this antibiotic. Eleven strains showed high-level resistance to streptomycin, including six E. faecium vanA strains. No resistance to high concentrations of gentamicin was detected. All VRE strains isolated from the control group were susceptible to amoxicillin and to high concentrations of gentamicin. Three of them showed high-level resistance to streptomycin.
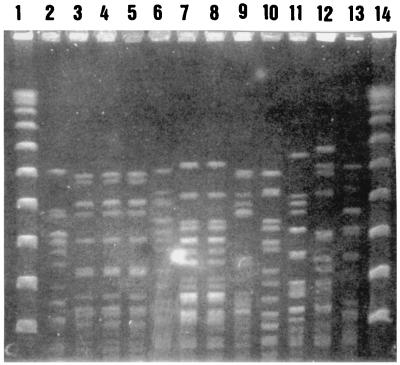
The restriction endonuclease patterns obtained by PFGE with SmaI for the 12 E. faecium strains of the VanA phenotype are presented in Fig. 1. These 12 strains were isolated from nine patients: six in the hematology unit and three in the control group. Strains isolated from the same patient had identical PFGE patterns, but strains isolated from different patients had different patterns.
FIG. 1.
SmaI restriction endonuclease patterns obtained by PFGE for VanA strains. Lanes 1 and 14, PFGE I marker; lane 2, patient 1 (control group); lanes 3, 4, and 5, patient 2 (hematology unit); lane 6, patient 3 (hematology unit); lanes 7 and 8, patient 4 (hematology unit); lane 9, patient 5 (control group); lane 10, patient 6 (control group); lane 11, patient 7 (hematology unit); lane 12, patient 8 (hematology unit); lane 13, patient 9 (hematology unit).
With prior antibiotic exposure being recognized as a risk factor for colonization with VRE, we decided to examine clinical records to determine if antibiotic treatment had been administered in the previous year for each patient hospitalized in the hematology unit; in particular, we looked at oral and/or parenteral vancomycin use and broad-spectrum cephalosporin use. Statistical analysis of these results by the χ2 test showed no significant difference (P < 0.05) between patients who received vancomycin or broad-spectrum cephalosporins and patients who did not (Table 3).
TABLE 3.
Influence of previous administration of vancomycin and/or broad-spectrum cephalosporins on fecal colonization with VRE strains in patients from hematology unit
| Previous antibiotic administration | No. of patients positive/ total no. of patients (%)
|
|
|---|---|---|
| VRE fecal colonization | No VRE fecal colonization | |
| Yes | ||
| Vancomycin | 12/34 (35) | 22/34 (65) |
| C3Ga | 20/53 (38) | 33/53 (62) |
| No | ||
| Vancomycin | 14/36 (39) | 22/36 (61) |
| C3G | 8/17 (47) | 9/17 (53) |
C3G, broad-spectrum cephalosporins.
DISCUSSION
This study documents the prevalence of intestinal colonization among patients from a French teaching hospital's hematology intensive care unit as well as that among nonhospitalized people living in the local community.
VRE were found in 37% of hematology patients. As reported by Gordts et al. (16), these selected patients are at high risk of colonization with VRE because of severe underlying illnesses, neutropenia, intensive therapy with various antibiotics, or length of hospital stay. However, the prevalence of fecal colonization by VRE was higher than that reported in other European studies: 2% in The Netherlands (11), 4.9% in the intensive care units of French general hospitals (3), and 3.5% in Belgium (16). The prevalence found in this study seems closest to the levels observed in the United States: 16% in Texas (8) and 28% in New York (27). However, comparison of the data is very difficult and should be done cautiously since the populations studied differ in size, age, sex ratio, principal diagnosis, possible neutropenia, etc.
VRE were detected in 11.8% of the outpatient control group. VRE colonization rates among community-based subjects vary greatly from one study to another. Low rates were found in The Netherlands (2%) (11), the United Kingdom (2%) (19), and another French study (0.3%) (4). On the other hand, other studies have reported high prevalence rates compatible with our own: 17% in a recent French study (17), 12% in a German study (23), and 28% in a Belgian study (36). In the United States little is known about the presence of VRE in the community at large or the environment, but the limited available data are in contrast to the European data, since VRE appear to be absent or very rare in healthy people outside hospitals and in the environment (8, 15, 26, 34).
Populations are very difficult to compare in all these studies, and in addition, detection methods are not entirely similar. Different media, glycopeptide concentrations, direct plating techniques, and enrichment methods have been used; and this might contribute to an explanation of the various isolation rates recorded by different investigators. No optimal method of screening of stool specimens for VRE has been established, but laboratories commonly use agar supplemented with vancomycin at 6 mg/liter according to the recommendations of the National Committee for Clinical Laboratory Standards (29, 30). In our study we used agar plates, but we also used a bile-esculin broth supplemented with 4 mg of vancomycin per liter, which constitutes an enrichment medium. A total of 75% of the VRE strains grew on this broth and not on the agar plates. If we had used only a vancomycin agar plate, the prevalence rate would have been 10% (7 of 70) instead of 37% for the hematology intensive care unit and 1.8% (3 of 169) instead of 11.8% for the control group: 75% of VRE strains would not have been detected. Several studies have already shown that the use of such enrichment broths increases the rate of recovery of resistant enterococci (13, 37); hence, use of such enrichment media could explain the high rate of recovery of VRE recorded in our study. The selectivities of different media also depend on vancomycin concentrations. Because VanA- and VanB-mediated resistance is inducible with vancomycin, the presence of vancomycin in the medium is essential. A vancomycin concentration that is too high, however, can inhibit growth of low-level vancomycin-resistant strains such as VanB and VanC strains. A high concentration of vancomycin in the medium is probably at the origin of the isolation of only VanA strains in some studies with agar plates with 50 mg (23) or 25 mg (39) of vancomycin per liter. This governed our choice of low vancomycin concentrations (4 mg/liter for the broth) and may explain the high proportion of VanC strains that we isolated.
The majority of the clinical isolates were E. gallinarum (70.7%) (Table 1), while E. faecium accounted for 18.5% and E. casseliflavus accounted for 10.8% of the isolates. No E. faecalis strains were detected. This species distribution was not comparable to those frequently reported in previous studies, in which E. faecium was more frequently detected (11, 16). The same distribution pattern is observed in the United States, which has a predominance of E. faecium isolates (8, 12, 39). E. gallinarum and E. casseliflavus are rarely recovered from clinical specimens. A few European studies report variable isolation rates ranging from 5.9 to 13.6% (3, 11, 16). In the United States, the number of E. gallinarum and E. casseliflavus strains among VRE is very low, from 0.5 to 1% (39). These species are not always taken into account because their resistance to glycopeptides is intrinsic and their pathogenicities are very low. No details on this subject are given in the different studies. However, in our study, even without E. casseliflavus and E. gallinarum species, the prevalence of fecal colonization by VRE would be 8.6% (6 of 70) among hematology patients, which is still higher than the previously reported levels (11, 3, 16).
Molecular typing of the VanA E. faecium strains isolated from different patients showed different patterns, suggesting a genetic unrelatedness of these strains which possibly excludes a common origin of the VRE and which also excludes transmission from patient to patient. Several European studies came to similar conclusions (16, 36, 35). On the other hand, most outbreaks in the United States are caused by the intra- or interhospital spread of clonal strains (14, 18, 32, 39), which suggests that patient-to-patient transmission is the major factor responsible for dissemination of VRE.
The presence of VRE in the stools of nonhospitalized patients suggests that VRE form part of the normal human fecal flora or can be acquired in the community, as confirmed by several other studies (2, 11, 16, 19, 36). A possible source of VRE could be the food chain, since VRE has been reported in the feces of farm animals and in animal product-based foodstuffs (1, 2, 22). The origin of the contamination of meat remains unknown, but it might occur during processing and packaging or through the intestinal flora of slaughtered animals (23). Some European investigators have raised the possibility that the glycopeptide avoparcin, which has been used as a food additive for growth enhancement in animals for nearly 20 years, might have selected VRE strains in animals (2, 11, 21), and in April 1997 the European Community imposed a 2-year ban on the use of avoparcin as an additive in food for animals (6). Our region is a cattle-rearing area where avoparcin had been used until the enforcement of the European ban, and this may partially explain the high VRE prevalence rates recorded in our study. However, some studies carried out in nonagricultural areas, such as in New York City (27), have also shown high VRE prevalence rates, but the epidemiology of VRE in Europe differs from that in the United States.
If VRE colonization came from the ingestion of contaminated meat, the strains would be subjected to a second selection process in the patient's gut by antimicrobial chemotherapy. Administration of vancomycin or antibiotics such as broad-spectrum cephalosporins is frequently reported as a risk factor for VRE infection or colonization (9, 18, 25, 33, 38). However, in our study vancomycin or cephalosporin administration did not appear to influence selection for VRE in fecal flora (Table 3). Schmidt et al. (33) as well as Gordts et al. (16) also failed to find antibiotic administration to be a reliable cause of the presence of VRE strains in fecal flora.
In conclusion, our study reports a high prevalence of VRE colonization of fecal samples from hospitalized patients and healthy nonhospitalized subjects living in the same local community. This prevalence is significantly higher than that reported by other European and U.S. studies. A partial explanation is the use of an enrichment broth step, as it increases the number of VRE by 75%, but the fact that subjects and patients were recruited from a predominantly agricultural area where vancomycin-related antibiotics have been used in animal husbandry may also contribute to the high levels of VRE detected in patients and subjects alike. Further studies are required to clarify the epidemiology of VRE, and they could be usefully complemented by an investigation of the rate of VRE fecal colonization among locally bred animals, one possible source of contamination in the food chain.
ACKNOWLEDGMENT
This work was supported by EP CNRS 118.
REFERENCES
- 1.Aarestrup F M. Occurrence of glycopeptide resistance among E. faecium isolates from conventional and ecological poultry farms. Microb Drug Resist. 1995;1:255–257. doi: 10.1089/mdr.1995.1.255. [DOI] [PubMed] [Google Scholar]
- 2.Bates J, Jordens J Z, Griffiths D T. Farm animal as a putative reservoir for vancomycin-resistant enterococcal infection in man. J Antimicrob Chemother. 1994;34:507–516. doi: 10.1093/jac/34.4.507. [DOI] [PubMed] [Google Scholar]
- 3.Boisivon A, Thibault M, Leclercq R. Colonization by vancomycin-resistant enterococci of the intestinal tract of patients in intensive care units from French general hospitals. Clin Microbiol Infect. 1997;3:175–179. doi: 10.1111/j.1469-0691.1997.tb00594.x. [DOI] [PubMed] [Google Scholar]
- 4.Cavallo J D, Hernandez E, Bouchard P, Debuysere H, Buisson Y. Portage asymptomatique d'entérocoques résistants à la vancomycine en France. Presse Med. 1997;26:807. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin—United States, 1989–1993. Morbid Mortal Weekly Rep. 1993;42:597–599. [PubMed] [Google Scholar]
- 6.Chaslus-Dancla E, Martel J L. Résistance aux antibiotiques chez les animaux d'élevage. Bull Soc Fr Microbiol. 1997;12:152–159. [Google Scholar]
- 7.Comité de l'Antibiogramme de la Société Française de Microbiologie. Communiqué 1998 du Comité de l'Antibiogramme de la Société Française de Microbiologie. Pathol Biol. 1998;46:I–XVI. [Google Scholar]
- 8.Coque T M, Tomayko J F, Ricke S C, Okhyusen P C, Murray B E. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob Agents Chemother. 1996;40:2605–2609. doi: 10.1128/aac.40.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Currie B P, Gnass S, Levi M H. A hospital-based rectal swab culture survey to detect vancomycin-resistant enterococci: utility and application. Int J Infect Dis. 1996;1:87–91. [Google Scholar]
- 10.Dukta-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endtz H P, Van den Braak N, Van Belkum A, Kluytmans J A J W, Koeleman J G M, Spanjaard L, Voss A, Weesink A J L, Vandenbroucke-Grauls C M J E, Buitting A G M, Van Duin A, Verbrugh H A. Fecal carriage of vancomycin-resistant enterococci in hospitalized patients and those living in the community in The Netherlands. J Clin Microbiol. 1997;35:3026–3031. doi: 10.1128/jcm.35.12.3026-3031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontana R, Boaretti M, Grossato A, Tonin E A, Lleo M M, Satta G. Paradoxical response of E. faecalis to the bactericidal activity of penicillin is associated with reduced activity of one autolysine. Antimicrob Agents Chemother. 1990;34:314–320. doi: 10.1128/aac.34.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford M, Perry J D, Gould F K, Orr K E. Neomycin blood agar as a selective medium for vancomycin resistant Enterococcus faecium. J Clin Pathol. 1996;49:437–438. doi: 10.1136/jcp.49.5.437-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frieden T R, Munsiff S S, Low D E, Willey B M, Williams G, Faur Y, Eisner W, Warren S, Kreiswir T H. Emergence of vancomycin-resistant enterococci in New York City. Lancet. 1993;342:76–79. doi: 10.1016/0140-6736(93)91285-t. [DOI] [PubMed] [Google Scholar]
- 15.Glenn Moris J, Shay D K, Hebden J N, McCarter R J, Jr, Perdue B E, Jarvis W, Johnson J A, Dowling T C, Polish L B, Schwalbe R S. Enterococci resistant to multiple antimicrobial agents, including vancomycin. Ann Intern Med. 1995;123:250–259. doi: 10.7326/0003-4819-123-4-199508150-00002. [DOI] [PubMed] [Google Scholar]
- 16.Gordts B, Van Landuyt H, Ieven M, Vandamme P, Goossens H. Vancomycin-resistant enterococci colonizing the intestinal tracts of hospitalized patients. J Clin Microbiol. 1995;33:2842–2846. doi: 10.1128/jcm.33.11.2842-2846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerin F, Perrier-Gros-Claude J D, Foissaud V, Masseron T, Thierry J. Entérocoques résistants à la vancomycine en France. Haute prévalence dans une population ambulatoire de sujets jeunes. Presse Med. 1998;27:1427–1429. [PubMed] [Google Scholar]
- 18.Handwerger S, Raucher B, Altarac D, Monka J, Marchione S, Singh K V, Murray B, Wolff J, Walters B. Nosocomial outbreak due to Enterococcus faecium highly resistant to vancomycin, penicillin, and gentamicin. Clin Infect Dis. 1993;16:750–755. doi: 10.1093/clind/16.6.750. [DOI] [PubMed] [Google Scholar]
- 19.Jordens J Z, Bates J, Griffiths D T. Fecal carriage and nosocomial spread of vancomycin-resistant Enterococcus faecium. J Antimicrob Chemother. 1994;34:515–528. doi: 10.1093/jac/34.4.515. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan R R, Gilligan P H, Facklam R R. Recovery of resistant enterococci during vancomycin prophylaxis. J Clin Microbiol. 1988;26:1216–1218. doi: 10.1128/jcm.26.6.1216-1218.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kjerulf A, Pallesen L, Westh H. Vancomycin-resistant enterococci at a large university hospital in Denmark. APMIS. 1996;104:475–479. doi: 10.1111/j.1699-0463.1996.tb00744.x. [DOI] [PubMed] [Google Scholar]
- 22.Klare I. vanA-mediated high-level glycopeptide resistance in E. faecium from husbandry. FEMS Microbiol Lett. 1995;125:165–172. doi: 10.1111/j.1574-6968.1995.tb07353.x. [DOI] [PubMed] [Google Scholar]
- 23.Klare I, Heier H, Claus H, Bohme G, Marin S, Seltmann G, Hakenbeck R, Antanassova V, Witte W. E. faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animals foodstuffs and fecal samples of humans in the community. Microb Drug Resist. 1995;1:265–272. doi: 10.1089/mdr.1995.1.265. [DOI] [PubMed] [Google Scholar]
- 24.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in E. faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 25.Matushek M, Slaughter S, Rice T, Weinstein R. Epidemiology of colonisation of patients and environment with vancomycin-resistant enterococci. Lancet. 1996;348:1615–1619. doi: 10.1016/S0140-6736(96)02331-8. [DOI] [PubMed] [Google Scholar]
- 26.McDonald L C, Kuehnert M J, Tenover F C, Jarvis W J. Vancomycin-resistant enterococci outside the health-care setting: prevalence, sources, and public health implications. Emerg Infect Dis. 1997;3:311–317. doi: 10.3201/eid0303.970307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montecalvo M A, De Lencastre H, Carraher M, Gedris C, Chung M, Vanhorn K, Wormser G P. Natural history of colonization with vancomycin-resistant Enterococcus faecium. Infect Control Hosp Epidemiol. 1995;16:680–685. doi: 10.1086/647041. [DOI] [PubMed] [Google Scholar]
- 28.Murray B E. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility tests. Approved standard M2-A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 31.Perichon B, Reynolds P, Courvalin P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob Agents Chemother. 1997;41:2016–2018. doi: 10.1128/aac.41.9.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perlada D E, Smulian A G, Cushion M T. Molecular epidemiology and antibiotic susceptibility of enterococci in Cincinnati, Ohio: a prospective citywide survey. J Clin Microbiol. 1997;35:2342–2347. doi: 10.1128/jcm.35.9.2342-2347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt J L, Leclercq R, Scheimberg A, Landauer D. Approche épidémiologique et clinique des entérocoques: résultats d'une enquête. Med Mal Infect. 1994;24(spécial):141–148. [Google Scholar]
- 34.Silverman J, Thal L A, Perri M B, Bostic G, Zervos M J. Epidemiologic evaluation of antimicrobial resistance in community-acquired enterococci. J Clin Microbiol. 1998;36:830–832. doi: 10.1128/jcm.36.3.830-832.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandamme P, Vercauteren E, Lammens C, Pensart N, Ieven M, Pot B, Leclercq R, Goossens H. Survey of enterococcal susceptibility patterns in Belgium. J Clin Microbiol. 1996;34:2572–2576. doi: 10.1128/jcm.34.10.2572-2576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van der Auwera P, Pensart N, Korten V, Murray B E, Leclercq R. Influence of oral glycopeptides on the fecal flora of human volunteers: selection of highly glycopeptide-resistant enterococci. J Infect Dis. 1996;173:1129–1136. doi: 10.1093/infdis/173.5.1129. [DOI] [PubMed] [Google Scholar]
- 37.Van Horn K G, Gedris C A, Rodney K M. Selective isolation of vancomycin-resistant enterococci. J Clin Microbiol. 1996;34:924–927. doi: 10.1128/jcm.34.4.924-927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinstein J W, Roe M, Towns M, Sanders L, Thorpe J J, Corey G R, Sexton D J. Resistant enterococci: a prospective study of prevalence, incidence, and factors associated with colonization in a university hospital. Infect Control Hosp Epidemiol. 1996;17:36–41. doi: 10.1086/647186. [DOI] [PubMed] [Google Scholar]
- 39.Wells C L, Juni B A, Cameron S B, Mason K R, Dunn D L, Ferrieri P, Rhame F S. Stool carriage, clinical isolation, and mortality during an outbreak of vancomycin-resistant enterococci in hospitalized medical and/or surgical patients. Clin Infect Dis. 1995;21:45–50. doi: 10.1093/clinids/21.1.45. [DOI] [PubMed] [Google Scholar]