Abstract
Simple Summary
Squamous cell carcinomas affect different head and neck subsites and, although these tumors arise from the same epithelial lining and share risk factors, they differ in terms of clinical behavior and molecular carcinogenesis mechanisms. Differences between HPV-negative and HPV-positive tumors are those most frequently explored, but further data suggest that the molecular heterogeneity observed among head and neck subsites may go beyond HPV infection. In this review, we explore how alterations of DNA methylation and microRNA expression contribute to head and neck squamous cell carcinoma (HNSCC) development and progression. The association of these epigenetic alterations with risk factor exposure, early carcinogenesis steps, transformation risk, and prognosis are described. Finally, we discuss the potential application of the use of epigenetic biomarkers in HNSCC.
Abstract
Head and neck squamous cell carcinomas (HNSCC) are among the ten most frequent types of cancer worldwide and, despite all efforts, are still diagnosed at late stages and show poor overall survival. Furthermore, HNSCC patients often experience relapses and the development of second primary tumors, as a consequence of the field cancerization process. Therefore, a better comprehension of the molecular mechanisms involved in HNSCC development and progression may enable diagnosis anticipation and provide valuable tools for prediction of prognosis and response to therapy. However, the different biological behavior of these tumors depending on the affected anatomical site and risk factor exposure, as well as the high genetic heterogeneity observed in HNSCC are major obstacles in this pursue. In this context, epigenetic alterations have been shown to be common in HNSCC, to discriminate the tumor anatomical subsites, to be responsive to risk factor exposure, and show promising results in biomarker development. Based on this, this review brings together the current knowledge on alterations of DNA methylation and microRNA expression in HNSCC natural history, focusing on how they contribute to each step of the process and on their applicability as biomarkers of exposure, HNSCC development, progression, and response to therapy.
Keywords: head and neck cancer, DNA methylation, microRNA, field cancerization, biomarker, tobacco, alcohol, HPV
1. Introduction
Head and neck squamous cell carcinomas (HNSCC) are among the ten most frequent and lethal cancers among men worldwide, affecting both developed and developing countries [1]. It is usually associated with lifelong exposure to tobacco and alcohol [2,3,4], but more recently, human papillomavirus (HPV) infection has emerged as a third risk factor [4]. HPV drives oropharyngeal squamous cell carcinomas (OPSCC) with better treatment response and overall survival that led to the establishment of specific guidelines in the last TNM Classification of Malignant Tumors [5]. Nonetheless, for most HNSCC cases, the prognosis is usually poor, with a high incidence of recurrences and second primary tumors (SPT) [4,6]. Therefore, identifying the molecular mechanisms of HNSCC development and progression is of utmost relevance.
HNSCC natural history might differ according to different aspects, such as affected subsite, the treatment offered and patients’ habits. In the 1950s, Slaughter and colleagues evaluated the origin and spreading of tobacco- and alcohol-associated oral squamous cell carcinoma (OSCC), the most frequent and most accessible HNSCC [7]. In this groundbreaking work, they showed that (i) horizontal is more common than vertical spread; (ii) the surrounding benign epithelium was usually microscopically abnormal; (iii) when tumors were ≤1 cm, other in situ cancer foci or isolated islands of invasive carcinoma were found; and (iv) 11% of the cases presented synchronous macroscopic SPT in the mucosa of the upper alimentary and respiratory tracts. These observations led to the proposal of a field of cancerization, in which carcinogens precondition the epithelial lining of these tracts to transformation.
Indeed, tobacco and alcohol-associated HNSCC arise after a lifelong exposure to these risk factors. Although their full onset may take tens of years, the entire process is not completely silent. Precursor lesions can be detected and might be associated with progression risk. For decades, white (leukoplakias) and red (erythroplakias) patches in head and neck linings have been studied, and up to now, it is still difficult to estimate how often they will progress to malignant tumors and the associated factors. Initially, these lesions were evaluated exclusively according to microscopic morphological aspects. In a study from 1987, for example, the authors showed that among 42 patients with white or red patches (or both) at the vocal cords, all presented morphological alterations, ranging from keratosis to microinvasive carcinoma [8]. Among those with keratosis, keratosis with atypia, or atypia, 43%, 42%, and 80%, progressed to carcinoma in situ or microinvasive carcinoma, respectively. The time between the diagnosis of leukoplakias and/or erythroplakias and progression to malignancy also varied between 1 and 30 months. Other relevant observations from this study were the high rate of recurrence of morphological alterations (54.5%), as well as a slightly higher risk of recurrence among patients who did not stop smoking (33.3% versus 26.6% among patients who quit smoking). In a recent meta-analysis, the prevalence of oral potentially malignant disorders was estimated to be 4.47% (95% CI = 2.43–7.08), and varied between populations, with North America showing the lowest prevalence (0.11%) and Asia the highest (10.54%) [9]. The rates of malignant transformation also vary between populations and studies (ranging from 3.2% to 50%), and the strongly associated factors include lesion size, texture, color, tumor subsite, and presence of dysplasia [10]. Nevertheless, besides morphological appearance and clinical aspects, molecular biomarkers might also be used to diagnose and estimate the risk of progression, as addressed in the following sections.
The frequency of HPV-associated HNSCC (HPV-HNSCC) varies worldwide, probably because of the also variable incidence of tobacco and alcohol-associated HNSCC [11]. However, it is undeniable that HPV-HNSCC incidence rates increased in the last years, and it preferably affects younger patients, making the disease a public health issue [12,13]. The most common affected subsites include lingual and palatine tonsils. In contrast to tobacco and alcohol-associated HNSCC, HPV-HNSCC is not associated with dysplasia, lacks keratinization, and shows basaloid morphology [14,15,16]. The discontinuous basement membrane of the reticulated epithelium in tonsils enables HPV assessment to basal keratinocytes in the absence of microabrasions [17], explaining the subsite preference. When these cells replicate, HPV is passed to daughter cells, and through its transcriptome program, it disturbs the differentiation process that would occur in normal conditions [17]. E6 and E7 viral oncoproteins are crucial to high-risk HPV-induced transformation by binding to p53 and Rb, respectively, facilitating their proteasomal degradation [18,19]. When HPV DNA is integrated into the host cell genome, higher levels of viral transcripts are observed [20]. Rb degradation promoted by E7 leads to E2F release, which activates p16INK4a transcription [21]. Although p16 immunostaining is largely applied as a highly sensitive HPV-HNSCC surrogate marker, it is moderately specific [22]. Therefore, novel biomarkers could help discriminate between the two tumor entities.
HNSCC is highly heterogeneous regarding molecular alterations. Although differences according to tumor site and associated risk factors exist, these only partially explain this heterogeneity. In this context, epigenetic alterations have gained attention due to their frequency and site-specific pattern [23,24,25,26], being promising biomarkers. In the following sections, we bring together the current knowledge on alterations of DNA methylation and microRNA expression in HNSCC natural history, focusing on how they contribute to each step of the process and on their applicability as biomarkers of exposure, HNSCC development, progression, and response to therapy.
2. Epigenetic Mechanisms
Epigenetics refers to molecular alterations that do not alter the nucleotide sequence but affect gene expression. These molecular mechanisms are generally stable, reversible, and are inherited during cell division [27]. DNA methylation, noncoding RNAs, and chromatin remodeling are considered epigenetic mechanisms of gene expression control, and their dysregulation is recurrently reported in cancer [28,29]. Since epigenetic marks are overly sensitive to environmental stimuli, intrinsically associated with cell phenotype, tissue-specific, and can be detected in surrogate samples, the development of epigenetic biomarkers in oncology is promising. Much progress has been made in evaluating alterations of DNA methylation and noncoding RNAs in HNSCC. Due to analytical advantages compared with chromatin remodeling, these epigenetic marks are the focus of our discussion here.
DNA methylation is probably the most extensively studied epigenetic mechanism in cancer (Figure 1). It involves the transfer of a methyl group from S-adenosyl methionine (SAM) to the carbon 5 of cytosines followed by guanines (CpG dinucleotides), a reaction catalyzed by DNA-methyltransferases (DNMTs) [30,31]. CpG islands refer to genomic regions with an observed frequency of CpG nucleotides higher than expected, and are mainly located in promoter regions [32]. Although DNA methylation may affect gene expression, both inside and outside CpG islands [33], most studies focus on the analysis of these genomic regions. The impact of this epigenetic mark on gene expression involves the alteration of transcription factor affinity to their consensus sequence in target genes, as well as the recruitment of other proteins, such as methyl-binding domain proteins and chromatin remodelers [32]. When DNA methylation affects promoter regions, the consequence is usually gene silencing, and it is not uncommon to observe their association with heterochromatin [34]. It is also an important mechanism to maintain genomic stability through the shutdown of transposable elements. More recently, due to the widespread use of genome-wide technologies to study DNA methylation, gene bodies, enhancers, and insulators have begun to be addressed and may unveil further interconnections between DNA methylation and genome usage [35].
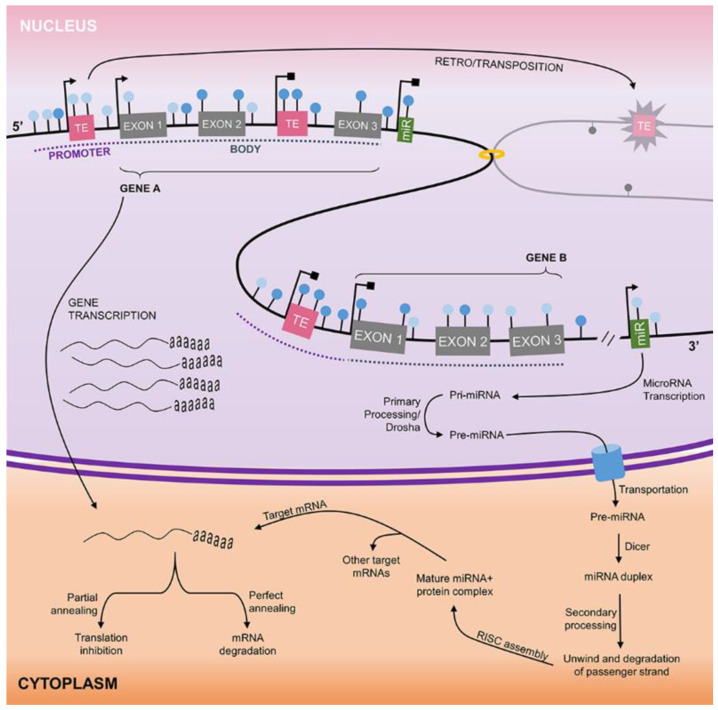
Figure 1.
Overview of gene regulation by DNA methylation and miRNA activity. Genomic DNA, found in the nucleus of eukaryotic cells, contains millions of CpG sites (lollipops) that can be either methylated (dark blue) or demethylated (light blue). These CpG sites are located in gene promoters and bodies, miRNAs, and intergenic regions, as well as in the vicinity of transposable elements (TE). When a promoter region is demethylated (Gene A), gene transcription may take place, leading to mRNA synthesis. However, when the promoter is hypermethylated (Gene B), transcription is inhibited. TEs transcription goes through the same regulatory mechanism. The transcription of some of these elements leads to the production of the transposition machinery, which enables their retrotransposition to a new genomic region, in a “copy–paste” fashion. DNA methylation similarly regulates the expression of miRNAs. Once transcribed, the primary miRNA (Pri-miRNA) is processed still in the nucleus by the RNAse Drosha, forming the precursor miRNA (Pre-miRNA). The Pre-miRNA is exported to the cytoplasm, where it is further processed by the RNAse Dicer, forming the miRNA duplex. After the miRNA duplex unwinds and the degradation of the passenger strand takes place, the lead strand (mature miRNA) assembles to the RNA-induced silencing complex (RISC) and, by nucleotide complementarity, identifies their target mRNAs. A single miRNA may have several mRNA targets and will lead to their degradation (perfect annealing) or translation inhibition (partial annealing).
Among noncoding RNAs, the effects of microRNAs (miRNAs) on gene expression control are probably the most well understood (Figure 1). These molecules are about 22 nucleotides in length and recognize their target mRNAs by complementarity, usually binding to the 3′ untranslated region (UTR) [36,37,38]. MiRNA–mRNA pairing might be complete or incomplete, and the consequences are mRNA degradation and translation inhibition, respectively [37,38]. A single miRNA might have dozens of targets [38], making the impact of their dysregulation on cell phenotype quite dramatic and difficult to predict. Nonetheless, due to their tissue-specific expression pattern, frequently altered expression in cancer, and high stability in biological fluids, miRNAs are promising biomarkers in oncology.
3. HNSCC Etiology and Epigenetics
All etiological factors associated with HNSCC development have been shown to affect epigenetic mechanisms in different tissues. For example, both smoking and alcohol consumption were associated with global hypomethylation in HNSCC [39], while HPV-positive tumors show higher global methylation levels relative to HPV-negative tumors [40]. It is intriguing that while the expression of microRNAs can be regulated by DNA methylation [41], DNA methylation machinery can also be the target of microRNAs [42,43], creating a negative feedback. Additionally, microRNA expression can be influenced by genetic factors such as single nucleotide polymorphisms and mutations [44,45]. However, given the stochasticity of HNSCC carcinogenesis, the order of events is still far from being understood.
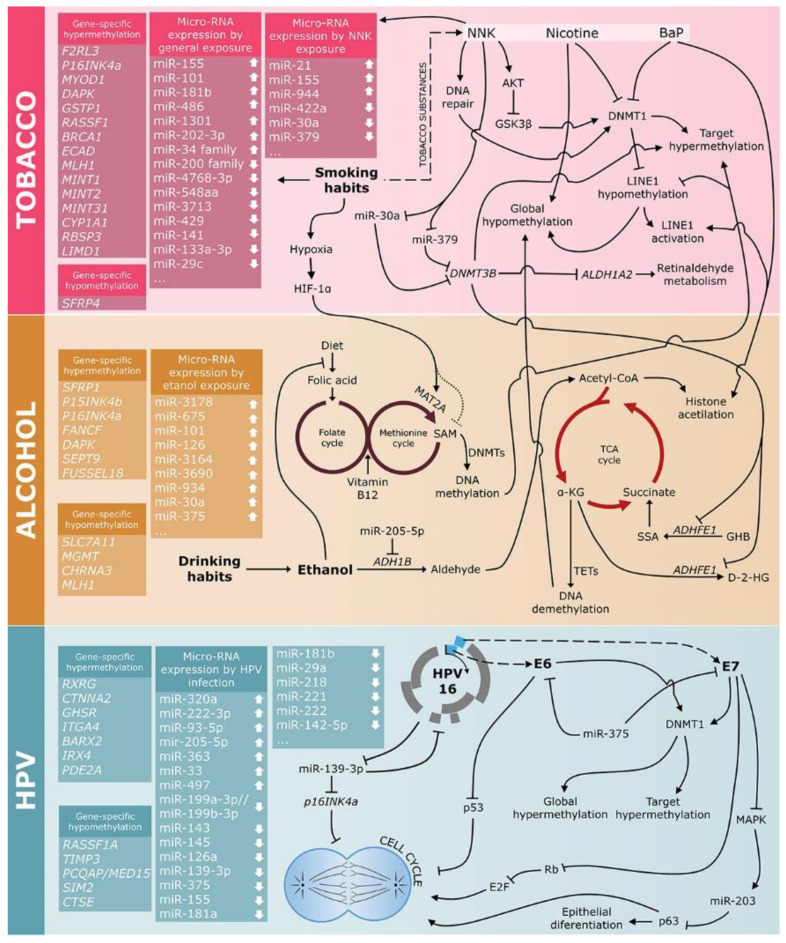
Furthermore, specific microRNA expression and DNA methylation signatures for tobacco [46], alcohol [47,48], and HPV infection [49,50] were established and represent not only a source of exposure biomarkers, but may also add information on carcinogenic mechanisms (Figure 2).
Figure 2.
Risk factors associated with HNSCC development affect DNA methylation and miRNA expression, showing different mechanisms by which tobacco components, alcohol, and HPV infection may lead to aberrant DNA methylation profiles and aberrant miRNA expression. NNK, 4-(methylnitro-samino)-1-(3-pyridyl)-1-butanone; BaP, benzo(a)pyrene; TCA Cycle, tricarboxylic acid cycle.
3.1. Tobacco
Tobacco components have been shown to induce epigenetic alterations (Figure 2 and Table 1). Although the molecular mechanisms are not completely elucidated, some clues have been gathered. In HeLa cells, benzo(a)pyrene (BaP) was shown to induce the deposition of activating histone marks in the promoter of long interspersed element 1 (LINE1) [51]. This was followed by the inhibition of DNMT1 binding to these genomic regions, and by the proteasome-mediated degradation of this enzyme. As a result, BaP induced LINE1 hypomethylation [51]. By binding to nicotinic cholinergic receptors, nicotine was shown to decrease Dnmt1 mRNA and protein levels and induce demethylation of specific loci [52]. Other tobacco components may induce DNA hypermethylation. The tobacco-specific nitrosamine 4-(methylnitro-samino)-1-(3-pyridyl)-1-butanone (NNK) can induce AKT activation that phosphorylates and inhibits GSK3β [53]. Once inactivated, GSK3β can no longer phosphorylate DNMT1, which is not marked for proteasomal degradation, and it is therefore stabilized. Another mechanism by which tobacco components may induce hypermethylation is via DNA damage; Dnmt1 was shown to be recruited to DNA repair sites to restore epigenetic marks [54]. In vitro experiments in HeLa and mouse embryonic stem cells corroborated these findings, showing that following double-strand breaks, half of the strands repaired by homologous recombination become hypermethylated, which is mediated by DNMT1 [55]. Cigarette smoke condensate can also induce Sp1 expression and DNA binding [56]. Since Sp1 is a transcription factor that recognizes CG-rich genomic regions [57], its binding may prevent DNA methylation in these regions [58,59]. Finally, tissue hypoxia induced by cigarette smoke carbon monoxide can induce global DNA hypomethylation. Hypoxia-inducible factor 1α (HIF-1α) induces the expression of methionine adenosyltransferase 2A (MAT2A, involved in SAM synthesis), and as a consequence, SAM levels are decreased, impairing DNA methylation reactions [59,60].
Table 1.
Epigenetic biomarkers in head and neck squamous cell carcinoma.
| Gene/Genomic Element | HNSCC Site | Sample Type | Genomic Region | Molecular Alteration | Ref. |
|---|---|---|---|---|---|
| Risk factor exposure: TOBACCO | |||||
| p16INK4a and MYOD1 | HNSCC | Tissue | Promoter | Hypermethylation | [61] |
| SFRP4 | HNSCC | Tissue | Promoter | Hypermethylation | [62] |
| CYP1A1 | HNSCC | Tissue | Promoter | Hypermethylation | [63] |
| RBSP3, LIMD1 and CDC25A | HNSCC | Tissue | Promoter | Hypermethylation | [64] |
| miR-200 family | OSCC | Tissue | N.A. | Downregulation | [65] |
| miR-155 | OSCC | Tissue | N.A. | Upregulation | [66] |
| miR-29c | LSCC | Tissue | N.A. | Downregulation | [67] |
| miR-202-3p | LSCC | Tissue | N.A. | Upregulation | [68] |
| miR- 4768-3p, miR-548aa and miR-3713 | LSCC | Tissue | N.A. | Downregulation | [68] |
| miR-34 family | HNSCC | Tissue | N.A. | Upregulation | [69] |
| mir-133a-3p | OPSCC | Tissue | N.A. | Downregulation | [70] |
| Risk factor exposure: ALCOHOL | |||||
| p15INK4b | HNSCC | Oral rinse | Promoter | Hypermethylation | [71] |
| FUSSEL18 and SEPT9 | HNSCC | Tissue | Promoter | Hypermethylation | [72] |
| DAPK | HNSCC | Tissue | Promoter | Hypermethylation | [73] |
| SFRP1 | HNSCC | Tissue | Promoter | Hypermethylation | [62] |
| p16INK4a, RASSF1A and FANCF | HNSCC | Tissue | Promoter | Hypermethylation | [61,74] |
| MGMT and CHRNA3 | HNSCC | Tissue | Promoter | Hypomethylation | [61] |
| MLH1 | LSCC | Tissue | Promoter | Hypomethylation | [75] |
| miR-30a, miR-934, miR-3178, miR-675, miR-101, miR-126, miR-3164 and miR-3690 | HNSCC | Tissue | N.A. | Upregulation | [48] |
| miR-375 | HNSCC | Tissue | N.A. | Upregulation | [76] |
| Risk factor exposure: HPV | |||||
| p16INK4a | OSCC, OPSCC and HNSCC | Tissue and Saliva | Promoter | Hypermethylation | [77,78] |
| Hypomethylation | [79,80] | ||||
| RASSF1A, TIMP3 and PCQAP/MED15 | HNSCC | Saliva | Promoter | Hypomethylation | [79] |
| RXRG, CTNNA2, GHSR and ITGA4 | OPSCC | Tissue | Promoter | Hypermethylation | [80] |
| Global methylation/Repetitive Element | HNSCC and OPSCC | Tissue | LINE1 | Hypermethylation | [20] |
| miR-99a-3p and miR-4746-5p | HNSCC | Tissue | N.A. | Upregulation | [81] |
| miR-411-5p | HNSCC | Tissue | N.A. | Downregulation | [81] |
| miR-320a, miR-222-3p, and miR-93-5p | OPSCC | Tissue | N.A. | Upregulation | [82] |
| miR-199a-3p//miR-199b-3p, miR-143, miR-145, and miR-126a | OPSCC | Tissue | N.A. | Downregulation | [82] |
| Cancerization field biomarkers—alterations in premalignant lesions | |||||
| SOCS-3 | HNSCC and dysplasias | Tissue | Promoter | Hypermethylation | [83] |
| p16INK4a and MGMT | OSCC and individuals at risk | Oral rinse and Blood | Promoter | Hypermethylation | [84,85] |
| RARB | OSCC precursor lesions | Tissue | Promoter | Hypermethylation | [86] |
| ZNF582 and PAX1 | OSCC cavity | Tissue | Promoter | Hypermethylation | [87] |
| ZAP70 and GP1BB | Oral squamous intraepithelial lesions | Oral brushing | Exon 3/Exon 1 | Hypermethylation/Hypomethylation | [88] |
| miR-204, miR-31, miR-31*, miR-133a, miR-7, miR-206, and miR-1293 | OSCC precursor lesions | Tissue | N.A. | Upregulation | [89] |
| Diagnostic biomarkers | |||||
| ABO | OSCC | Tissue | Promoter | Hypermethylation | [90] |
| APC | OSCC | Tissue | Promoter | Hypermethylation | [91] |
| C/EBPα | HNSCC | Tissue | Promoter | Hypermethylation | [92] |
| CDH1 | OSCC | Tissue | Promoter | Hypermethylation | [91,93,94,95,96] |
| HNSCC | Tissue | Promoter | Hypermethylation | [97] | |
| LSCC | Tissue | Promoter | Hypermethylation | [98] | |
| Hypopharynx | Tissue | Promoter | Hypermethylation | [98] | |
| CDKN2A | OSCC | Tissue | Promoter | Hypermethylation | [91,93,94,95,96,99,100,101,102] |
| HNSCC | Tissue | Promoter | Hypermethylation | [97,103] | |
| HNSCC | Saliva | Promoter | Hypermethylation | [102,104] | |
| LSCC | Tissue | Promoter | Hypermethylation | [75,98,105] | |
| Hypopharynx | Tissue | Promoter | Hypermethylation | [98] | |
| CHD5 | LSCC | Tissue | Promoter | Hypermethylation | [106] |
| CYGB | OSCC | Tissue | Promoter | Hypermethylation | [100] |
| CYCA1 | OSCC | Tissue | Promoter | Hypermethylation | [93,96] |
| DAPK | OSCC | Tissue | Promoter | Hypermethylation | [73,91,94,99,101,107] |
| OSCC | Blood | Promoter | Hypermethylation | [107] | |
| HNSCC | Tissue | Promoter | Hypermethylation | [73,97,102] | |
| HNSCC | Saliva | Promoter | Hypermethylation | [102,104] | |
| LSCC | Tissue | Promoter | Hypermethylation | [75,98,108] | |
| Hypopharynx | Tissue | Promoter | Hypermethylation | [98] | |
| DKK3 | OSCC | Tissue | Promoter | Hypermethylation | [109] |
| Global methylation/Repetitive Element | OSCC | Blood | LRE1/LINE1 | Hypomethylation | [110] |
| OSCC | Oral rinse | Alu | Hypomethylation | [111] | |
| HOXA9 | OSCC | Tissue and Saliva | Promoter | Hypermethylation | [112] |
| MGMT | OSCC | Tissue | Promoter | Hypermethylation | [91,94,95,96,99,101] |
| HNSCC | Tissue | Promoter | Hypermethylation | [103,113] | |
| HNSCC | Saliva | Promoter | Hypermethylation | [102] | |
| LSCC | Tissue | Promoter | Hypermethylation | [75,98] | |
| Hypopharynx | Tissue | Promoter | Hypermethylation | [98] | |
| MLH1 | LSCC | Tissue | Promoter | Hypermethylation | [75,95] |
| OSCC | Tissue | Promoter | Hypermethylation | ||
| MINT1 | OSCC | Tissue | Promoter | Hypermethylation | [114] |
| NID2 | OSCC | Tissue and Saliva | Promoter | Hypermethylation | [112] |
| p14 | HNSCC | Tissue | Promoter | Hypermethylation | [103,115] |
| p15 | OSCC | Tissue | Promoter | Hypermethylation | [95,116] |
| PCQAP/MED15 | OPSCC | Saliva | Promoter | Hypermethylation | [117] |
| OSCC | Saliva | Promoter | Hypermethylation | [117] | |
| RARβ | OSCC | Tissue | Promoter | Hypermethylation | [93,96] |
| HNSCC | Tissue | Promoter | Hypermethylation | [103] | |
| RARβ2 | Salivary Gland Carcinomas | Tissue | Promoter | Hypermethylation | [118] |
| RASSF1A | OSCC | Tissue | Promoter | Hypermethylation | [119] |
| Salivary Gland Carcinomas | Tissue | Promoter | Hypermethylation | [118] | |
| HNSCC | Tissue | Promoter | Hypermethylation | [97] | |
| HNSCC | Saliva | Promoter | Hypermethylation | [104] | |
| OPSCC | Saliva | Promoter | Hypermethylation | [117] | |
| RUNX3 | OSCC | Tissue | Promoter | Hypermethylation | [94] |
| SFRP1 | OSCC | Tissue | Promoter | Hypomethylation | [109] |
| SFRP2 | OSCC | Tissue | Promoter | Hypermethylation | [109] |
| SFRP4 | OSCC | Tissue | Promoter | Hypermethylation | [109] |
| SFRP5 | OSCC | Tissue | Promoter | Hypermethylation | [109] |
| SOCS-3 | HNSCC | Tissue | Promoter | Hypermethylation | [83] |
| SSTR2 | LSCC | Tissue | Promoter | Hypermethylation | [120] |
| WIF1 | OSCC | Tissue | Promoter | Hypermethylation | [109,119] |
| WRN | OSCC | Tissue | Promoter | Hypermethylation | [91] |
| ZNF14 | HNSCC | Tissue and Saliva | Promoter | Hypermethylation | [121] |
| ZNF160 | HNSCC | Tissue and Saliva | Promoter | Hypermethylation | [121] |
| ZNF420 | HNSCC | Tissue and Saliva | Promoter | Hypermethylation | [121] |
| miR-375 | LSCC | Tissue | N.A. | Downregulation | [122] |
| miR-21 | LSCC | Tissue | N.A. | Upregulation | [122] |
| miR-204 and mir-144 | OSCC | Tissue and plasma | N.A. | Upregulation | [123] |
| miR-193b-5p and miR-370-3p | OSCC | Tissue and plasma | N.A. | Downregulation | [123] |
| miR-657 | LSCC | Tissue | N.A. | Upregulation | [124] |
| miR-1287 | LSCC | Tissue | N.A. | Downregulation | [124] |
| Relapse biomarkers | |||||
| EDNRB and HOXA9 | HNSCC surgical margins | Tissue | Promoter | Hypermethylation | [125] |
| DCC, CCNA1 and p16INK4A | HNSCC surgical margins | Tissue | Promoter | Hypermethylation | [126] |
| PAX5 | HNSCC surgical margins | Tissue | Promoter | Hypermethylation | [127] |
| CDKN2A | LSCC | Tissue | Nonpromoter | Hypomethylation | [128] |
| miR-375 | OSCC precursor lesions | Tissue | N.A. | Downregulation | [129] |
| Biomarkers of poor prognosis | |||||
| KL | HNSCC | Tissue | Promoter | Hypermethylation | [130] |
| HIN1, RASSF1A and RASSF2 | OSCC | Tissue | Promoter | Hypermethylation | [131] |
| DAPK | OSCC | Tissue | Promoter | Hypermethylation | [119] |
| MGMT | OSCC | Tissue | Promoter | Hypermethylation | [95,119,132,133] |
| MINT31 | OSCC | Tissue | Promoter | Hypermethylation | [114] |
| p16INK4A and p14ARF | OSCC | Tissue | Promoter | Hypermethylation | [134] |
| PAX5 and PAX1 | HNSCC | Tissue | Promoter | Hypermethylation | [135] |
| TIMP3 | HNSCC | Saliva | Promoter | Hypermethylation | [136] |
| SEPT9 and SHOX2 | HNSCC | Plasma | Promoter | Hypermethylation | [137] |
| EPHX3 | OSCC | Pre-operative oral brushing | Exon 1 | Hypomethylation | [138] |
| ITGA4 and MIR193A | OSCC | Pre-operative oral brushing | Exon 2/Promoter | Hypermethylation | [138] |
| miR-21 | LSCC | Tissue | N.A. | Upregulation | [122] |
| miR-375 | LSCC | Tissue | N.A. | Downregulation | [122] |
| miR-186-5p and miR-374b-5p | HNSCC | Plasma—before treatment | N.A. | Upregulation | [139] |
| miR-142-3p | HNSCC | Plasma—after treatment | N.A. | Upregulation | [139] |
HNSCC, head and neck squamous cell carcinoma; LSCC, laryngeal squamous cell carcinoma; N.A., not applicable; OPSCC, oropharyngeal squamous cell carcinoma; OSCC, oral squamous cell carcinoma; Ref., references.
Epigenome-wide association studies (EWAS) on different tissue types were carried out to identify tobacco epigenetic signatures. Using blood samples and the HumanMethylation27 assay, Breitling and colleagues identified one CpG locus annotated to F2RL3 that was hypomethylated in heavy smokers relative to never smokers. Former smokers showed methylation patterns similar to never smokers [140]. A following study applying the HumanMethylation450 beadchip, both in lymphoblasts and alveolar macrophages, identified one differentially methylated probe in AHRR gene body when comparing smokers and nonsmokers [141]. Similar findings for AHRR methylation and additional differentially methylated CpGs in CYP1A1 and GFI1 were observed in newborns following maternal smoking [142]. Additionally, in blood-derived DNA analyzed by HumanMethylation 450 assay, a meta-analysis with 15,907 individuals from 16 cohorts identified 18,760 differentially methylated CpGs between current and never smokers, including CpGs mapped to previously identified genes, such as AHRR, RARA, F2RL3, and LRRN3 [143]. A total of 2568 CpGs were also differentially methylated between never and former smokers. The consistent association between the methylation profile of genes involved in xenobiotic metabolism and smoking adds a biological plausibility to the results obtained.
An EWAS carried out in buccal cells from 790 women showed that smoking also leaves a DNA methylation signature in epithelial cells, which can be even more prominent throughout the genome than that determined in blood cells [144]. In this study, the top-ranked CpG sites showed a hypomethylation profile in smokers and affected genes such as AHHR, F2RL3, and CYP1A1, previously identified in blood-based EWAS/smoking studies. While hypomethylated genes were not enriched for specific cell pathways, hypermethylated CpGs were enriched for genes bivalently marked in human embryonic stem cells, and for binding sites of transcription factors associated with stem cell differentiation. These smoking-responsive CpG sites were also differentially methylated in tobacco-associated cancers, suggesting a contribution of these alterations to tumor development.
In OPSCC patients, the blood of 106 never and 303 ever smokers were compared and revealed the differential methylation of 52 CpG sites [145]. Of these, 49 sites had lower methylation levels in smokers, including a CpG mapped to AHRR. A similar analysis was performed to predict survival, and 18 CpG sites annotated to three genes (GFI1, SPEG, and PPT2) were differentially methylated according to both survival and smoking history. Additionally, in OPSCC patients, a DNA methylation score for smoking based on five EWAS was applied on 364 blood samples and was able to explain 51.1% of phenotypic variance [146]. This score showed an accuracy of 0.70 to predict mortality, slightly improving the accuracy of smoking self-report (0.67).
In gene-specific studies, p16INK4a hypermethylation was already reported in 9.7% of oral mucosa and/or tongue of healthy smokers [147]. Similarly, higher methylation levels of p16INK4a and MYOD1 were observed in HNSCC from smokers relative to nonsmokers [61]. SFRP4 hypermethylation seems to be more common in never and former smokers than in current smokers, and it is also independently associated with HPV16 infection [62]. In a study evaluating the methylation profile of 10 genes (p16INK4a, DAPK, GSTP1, RASSF1, BRCA1, ECAD, MLH1, MINT1, MINT2, and MINT31), both the methylation index (determined by the frequency of hypermethylated promoters) and the frequency of CpG island methylator phenotype-high (CIMP-high, at least five hypermethylated genes) cases were higher among smokers relative to nonsmokers [78]. In opposition to the hypomethylation profile observed in the EWAS, CYP1A1 hypermethylation was more frequently observed in HNSCC from smokers relative to healthy tissues from controls [63]. When promoter methylation and gene deletion of RBSP3, LIMD1, and CDC25A were analyzed together in HNSCC, the frequency of alterations was higher in smokers than in nonsmokers [64].
MicroRNAs are also targets of dysregulation by different tobacco components, varying according to route of exposure and head and neck subsite. Immortalized human oral keratinocytes OKF6/TERT exposed to cigarette smoke condensate or chewing tobacco extract showed six and ten differentially expressed microRNAs relative to nontreated cells, respectively, with none in common between treatments [148]. After treatment with smoke condensate, members of the miR-200 family were downregulated. These data are consistent with their downregulation observed in OSCC patients with smoking/chewing tobacco habits [65]. The exposure of oral keratinocytes OK4 to cigarette extract further showed the upregulation of miR-101, miR-181b, miR-486, and miR-1301 [149].
The exposure of HNSCC cells to NNK induced the expression of miR-21 and miR-155, while decreased miR-422a expression. Overexpression of miR-155 was also associated with the habit of chewing tobacco [66]. MiR-21 expression was further associated with MSH2 silencing [150]. MSH2 is a member of the mismatch repair pathway, and its downregulation was associated with poor survival of HNSCC patients [151]. NNK can also induce the expression of miR-944 that targets CISH, a suppressor of the STAT pathway [152,153]. CISH inhibition by miR-944 induces STAT3 activation, modulating the inflammatory response and increasing tumor malignancy [153]. MiR-30a and miR-379 expression levels were downregulated in OSCC cells exposed to NNK. Both miRNAs are downregulated in OSCC relative to adjacent nontumor tissue and target DNMT3B. This can lead to the hypermethylation of ADHFE1 and ALDH1A2 promoters, and their consequent downregulation [154]. Since both genes are involved in alcohol metabolism, these data suggest an interesting potential mechanism for the synergy between alcohol and tobacco in HNSCC.
MiR-29c downregulation was associated with smoking and worse survival in laryngeal squamous cell carcinoma (LSCC) patients [67]. Additionally, in LSCC, smokers showed miR-202-3p upregulation and downregulation of miR- 4768-3p, miR-548aa, and miR-3713 [68]. Among HNSCC patients, miR-34 family was shown to be upregulated in tumors from smokers relative to the adjacent tissues, while no differences were observed for nonsmokers [69]. Smoking or chewing tobacco was associated with miR-429 and miR-141 downregulation in OSCC patients [148]. In OPSCC, a comparison between smokers and nonsmokers HPV-positive patients revealed 38 differentially expressed miRNAs [70]. Mir-133a-3p was among the downregulated miRNAs, and its expression was further shown to be decreased by treatment with cigarette smoke condensate and associated with EGFR and HuR expression in HNSCC cell lines [70].
3.2. HNSCC Etiology and Epigenetics: Alcohol
Epigenetics and metabolism are two substations in the same system, intertwined. This is probably the simplest way to understand how alcohol may induce aberrant DNA methylation (Figure 2, Table 1). Epigenetic writers and erasers are regulated, among other mechanisms, by the availability of their substrates and cofactors. Ten-eleven translocation (TET) enzymes are α-ketoglutarate-dependent oxygenases involved in active DNA demethylation [155]. Due to this cofactor dependency, TET activity is regulated by different factors that affect the tricarboxylic acid cycle (TCA), including hypoxia [156]. For DNMTs activity, the levels of the methyl donor SAM are determinant. SAM is produced by one-carbon metabolism, which relies on the dietary intake of folate and B vitamins. Ethanol is an inhibitor of folate absorption, besides inhibiting different steps of one-carbon metabolism, leading to a reduction of SAM levels. Therefore, alcohol consumption may impair the methylation reactions catalyzed by DNMTs. Indeed, global DNA hypomethylation was observed in the liver of ethanol-fed rats [157]. While the higher intake of folate has already been associated with a decreased risk of HNSCC development [158], the interaction between folate, alcohol, and DNA methylation has been neglected. Nevertheless, several studies have shown an association between DNA methylation patterns and alcohol consumption in different settings, and surprisingly, it is not restricted to DNA hypomethylation.
A recent EWAS, including 13,317 individuals, proposed a 144-CpG signature in whole blood able to discriminate current heavy alcohol drinkers [47]. This model was used to predict all-cause mortality among OPSCC patients, but although it explained 16.5% of phenotypic variance, it showed a similar predictive accuracy to self-reported data [146]. Additionally, in the blood of OPSCC patients, an EWAS identified three new CpG sites and 40 differentially methylated regions associated with alcohol consumption [145]. The gene with the lowest p-value for both analyses was SLC7A11, with the methylation levels of one of its CpG decreasing 0.10% per unit increase in alcohol consumption. This gene plays a role in glutathione synthesis, an antioxidant that can be depleted by excessive ROS production during alcohol metabolism [159].
Other gene-specific studies have suggested an association between alcohol consumption and DNA methylation profiles in HNSCC. P15INK4b methylation was detected in the oral and throat rinses of 48% of HNSCC patients and 68% of healthy individuals who smoked and/or drank alcohol, but in only 8% of healthy nonsmokers and nondrinker individuals [71]. FUSSEL18 and SEPT9 methylation were also associated with alcohol and tobacco consumption history in HNSCC [72]. In a meta-analysis including 18 studies with different sample sources (tumors, liquid biopsies, and buccal scraping), DAPK methylation was significantly associated with alcohol consumption (OR = 1.85, 95% CI = 1.07 ± 3.21) [73]. SFRP1, a member of the secreted frizzled receptor protein family, is hypermethylated in HNSCC and this event is more common in ever drinkers than in never drinkers [62]. Other genes, such as p16INK4a, RASSF1A, and FANCF are also more frequently hypermethylated in HNSCC associated with alcohol consumption [61,74]. On the other hand, MLH1 methylation seems to be more common in individuals who drink less than 100 mL of alcohol/day in comparison with those who drink larger amounts [75].
Recently, alcohol consumption was associated with an aberrant DNA methylation profile in the blood of HNSCC survivors [160]. This dysregulation affected genes encoding histone and mitochondrial ribosomal proteins, as well as genes involved in toll-like receptor signaling, suggesting that alcohol intake can have a systemic effect, even after treatment.
In comparison with data showing an association between alcohol and DNA methylation in HNSCC, data for such an association with microRNA expression are still limited. As observed for DNA methylation, alcohol metabolism is also related to the expression of microRNAs. ADH1B is a key controller of alcohol metabolism, affecting acetaldehyde levels in the body [161], and its expression was shown to be regulated by miR-205-5p in HNSCC [162]. Other genes related to alcohol metabolism, such as ADHFE1 and ALDH1A2, are indirectly regulated by miR-30a and miR-379-mediated DNMT3B downregulation in OSCC [154], as previously mentioned, revealing a crosstalk between the methylation machinery and microRNAs in the xenobiotic response. The dysregulation of genes involved in alcohol metabolism by microRNAs was also associated with aggressive phenotypes. ADHFE1 and ALDH1A2 higher expression, as a consequence of miR-30a and miR-379 induction, were associated with proliferation inhibition of OSCC cell lines [154]. It is important to note that ALDH1A2 does not metabolize acetaldehyde efficiently, but it is important in the metabolism of retinoic acid [163]. In addition, ALDH1A2 is a tumor suppressor candidate in HNSCC [164].
Tumor suppressor genes may also be targets for microRNAs upregulated in HNSCC of alcoholic patients relative to nonalcoholic patients, as shown for miR-30a and miR-934. When miR-30a is inhibited, BNIP3L, PRDM9, and SEPT7 upregulation is observed, while miR-934 inhibition leads to HIPK2, HOXA4, and MLL3 upregulation. MiR-30a and miR-934 inhibition were also associated with increased sensitivity to cisplatin and decreased cell invasion of HNSCC cell lines [48]. Other microRNAs upregulated in HNSCC of alcoholic patients relative to nonalcoholic patients are miR-3178, miR-675, miR-101, miR-126, miR-3164, and miR-3690 [48].
Finally, miR-375, a classically downregulated microRNA in HNSCC [165], was shown to be upregulated in alcoholic patients [76]. MiR-375 targets the AEG-1 [166] and LDHB oncogenes [167] and its lower expression was previously associated with invasion and worse prognosis [168].
3.3. HNSCC Etiology and Epigenetics: HPV
Regarding HPV infection, viral oncoproteins were shown to modulate levels of DNMTs (Figure 2) [169]. E6 knockdown in HPV16 infected cervical cancer cell lines led to decreased DNMT1 expression, which was mediated by higher p53 levels [170]. Similarly, HPV16 E7 oncoprotein was also shown to stimulate Dnmt1 activity, both by promoting E2F activity through Rb downregulation [171], and by direct binding [172], with a consequent decreased E-cadherin expression [173]. Therefore, it is expected that HPV infection affects DNA methylation patterns.
Different studies addressed genome-wide DNA methylation differences between HPV-positive and HPV-negative HNSCC (reviewed in [174]). Hypermethylation was more frequent in HPV-positive HNSCC samples and cell lines relative to HPV-negative counterparts [175,176]. Furthermore, a DNA methylation signature for HPV infection composed of five CpG sites was proposed and showed high specificity to discriminate HPV-positive and HPV-negative HNSCC in different datasets, as well as a capacity to predict survival similar to HPV status, determined by viral mRNA levels [175]. In OPSCC, another DNA methylation signature composed of differentially methylated regions in the promoters of five genes (ALDH1A2, OSR2, GATA4, GRIA4, and IRX4) was shown to predict overall and progression-free survival, independently of HPV status [177].
Promoter methylation of genes recurrently reported altered in HNSCC also seem to differ according to HPV infection. For p16INK4a, results are discordant between studies, including hypermethylation [77,78], hypomethylation [79,80], and no differences found in HPV-positive relative to HPV-negative HNSCC cases [178]. This may be a consequence of both the different sample sources (tumors and saliva) and the different tumor subsites used, including oral cavity, oropharynx, both combined, and even HNSCC generally. Therefore, further studies are needed to resolve these discrepancies. RASSF1A, TIMP3, and PCQAP/MED15 were found to be hypomethylated in the saliva of HPV-positive HNSCC [79], while RXRG, CTNNA2, GHSR, and ITGA4 were reported hypermethylated in HPV-positive oropharyngeal tumors [80]. Global DNA methylation also differs according to HPV status, with LINE1 methylation levels being higher in HPV-positive relative to HPV-negative tumors [179,180,181,182,183]. DNA methylation differences in transposable elements according to HPV status were recently shown to affect gene expression, which was further associated with prognosis [183].
Interestingly, DNA methylation patterns also differ when HPV DNA is integrated or episomal, with integration-positive tumors showing a profile more similar to that of HPV-negative HNSCC [20]. BARX2 and IRX4 were hypermethylated and silenced, while SIM2 and CTSE showed hypomethylation and higher expression in episomal HPV-HNSCC. Finally, differentially methylated genes were enriched for the cAMP response element-binding protein (CREB) pathway, suggesting different carcinogenic biological mechanisms, depending on whether HPV is integrated.
DNA methylation was further associated with miRNA dysregulation in HPV-associated HNSCC [184]. MiR-139-3p was shown to be downregulated in HPV-positive tumors and cell lines relative to their HPV-negative counterparts, and it was shown to target HPV oncoproteins. In vitro experiments with miR-139-3p mimic led to E6 and E7 downregulation, reduced p16, and increased p53 protein levels. Interestingly, miR-139-3p colocalizes with PDE2A, whose promoter is hypermethylated in HPV-positive HNSCC. Corroborating this coregulation, treatment of HPV-positive cell lines with a demethylating agent induced miR-139-3p expression. MiR-375, previously described as a tumor suppressor whose expression is associated with alcohol intake, also seems to be suppressed by DNA methylation in HPV-mediated carcinogenesis [185]. Since HPV E6 and E7 are among miR-375 targets, downregulation of this miRNA contributes to a higher expression of these oncoproteins and a lower expression of their cellular targets, p53 and Rb [186].
Differentiation-associated miRNAs might also take part in HPV-mediated carcinogenesis [187]. As previously mentioned, HPV disturbs the differentiation of infected cells, and this was shown to be mediated, at least in part, by miR-203 downregulation. This miRNA regulates differentiation by targeting p63 and its expression can be downregulated by E7 via the blockage of MAPK pathway [187].
Although the miRNA biogenesis machinery was found to be dysregulated in cervical cancer [188], this is yet to be investigated in HPV-associated HNSCC. Nonetheless, miRNA signatures according to HPV status were reported in HNSCC. By comparing HPV-positive and HPV-negative tumors with adjacent tissue, 282 and 289 differentially expressed miRNAs were identified, respectively [189]. Although these numbers were similar, when specific survival-associated miRNAs were evaluated, the expression of 39 miRNAs was associated with prognosis among HPV-positive patients, 16 among HPV-negative patients, and no miRNA was shared. The miRNA survival signatures also differed in accuracy (0.92 for HPV-positive and 0.72 for HPV-negative) and associated signaling pathways. The poor prognosis miRNA signature for HPV-positive patients was associated with immune-related pathways. In contrast, in HPV-negative cases, it was associated with metabolism and oncogenic pathways, such as WNT and NOTCH signaling.
More recently, the direct comparison of HPV-positive and HPV-negative HNSCC revealed the differential expression of 59 miRNAs, three of which (miR-99a-3p, miR-411-5p, miR-4746-5p) were also associated with overall survival in a multivariate Cox regression analysis [81]. The Gene Set enrichment analysis performed with their potential mRNA targets showed that different pathways were likely to be dysregulated, with genes involved in epithelial–mesenchymal transition being downregulated by miR99a-3p-high/miR-411-5p-low/miR-4746-5p-high. This profile may help to explain the favorable prognosis of these patients.
Oropharyngeal tumors were also compared according to HPV status, revealing the upregulation of miR-320a, miR-222-3p, and miR-93-5p, as well as the downregulation of miR-199a-3p//miR-199b-3p, miR-143, miR-145, and miR-126a in HPV-positive tumors [82]. In tonsillar squamous cell carcinoma, 36 miRNAs were found to be differentially expressed according to HPV status [190].
When comparing HPV-positive HNSCC cell lines with HPV-negative and human foreskin keratinocytes, miR-363, miR-33, and miR-497 were upregulated while miR-155, miR-181a, miR-181b, miR-29a, miR-218, miR-221, miR-222, and miR-142-5p were downregulated [191]. This miRNA dysregulation was associated with the expression of HPV oncoproteins E6 and E7. The analysis of exosomes further revealed that miR-205-5p and miR-1972 are exclusively detected in HPV-positive and HPV-negative HNSCC cell lines, respectively [192].
Regarding minimally invasive biopsies, a miRNA panel composed of miR-9, miR-134, miR-196b, miR-210, and miR-455 evaluated in the saliva of HNSCC patients was shown to discriminate HPV-positive and HPV-negative cases [193].
Not only previously annotated miRNAs show differential expression according to HPV status. By applying a customized in silico pipeline, Rock and colleagues identified 146 novel miRNAs in samples from HNSCC patients, with three (HNnov-miR-2, HNnov-miR-30, and HNnov-miR-125) being significantly downregulated in HPV-positive tumors [194].
Although most studies bring consistent associations between risk factor exposure and aberrant epigenetic profiles in HNSCC, causal effects were not determined. In the attempt to overcome this limitation, in vivo studies should be performed with a focus on head and neck tissues. As an example, the treatment of mice with dibenzo[def,p]chrysene (DBP), a tobacco carcinogen, was able to induce DNA methylation alterations of 30 loci in oral tissues [195]. Fgf3, frequently amplified in HNSCC, was one of the affected genes and showed hypomethylation accompanied by induced expression. Considering that tobacco smoke contains more than 60 different carcinogens [196], alcohol may contribute to carcinogenesis by different mechanisms [197], HPV oncoproteins regulate key epigenetic enzymes [169,170,172,173], and that how all these factors interact is largely unknown, much is yet to be learned considering the effects of risk factor exposure on epigenetic mechanisms in head and neck tissues.
4. Epigenetic Biomarkers in HNSCC
As already mentioned, epigenetic changes coordinate the cellular response to environmental exposures, such as cancer risk factors [198,199]. Thus, even before the histological transformation or acquisition of driver mutations, we can have stochastic or targeted epigenetic changes, the latter sensitizing the cell to transformation, building the pre-cancer epigenetic signatures of the cancerization field (Figure 3) [200,201]. Several of these changes would not only favor the initial gain of a pre-cancer cell phenotype, but would also be maintained or altered after malignant transformation, tumor progression, and metastasis. Thus, epigenetic signatures are not just a consequence of risk factor exposure, or linked to the tissue of origin, but a unique product of these two factors combined [202]. In addition, unlike genetic changes, these signatures are plastic, and can vary from healthy tissue through the cancerization field and tumor progression in a qualitative or quantitative manner depending on the mechanism.
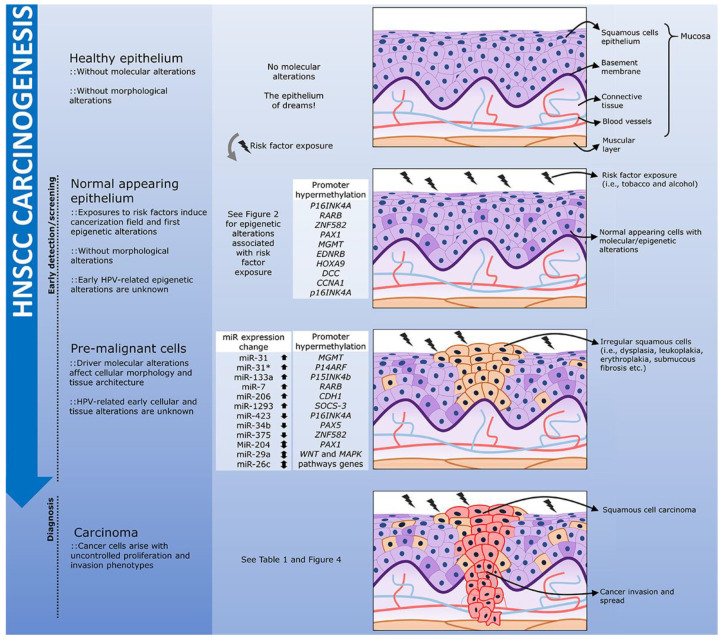
Figure 3.
The natural history of HNSCC development through an epigenetic perspective. The exposure of the head and neck healthy epithelium to risk factors, such as tobacco and alcohol, may lead to the accumulation of DNA methylation alterations and miRNA aberrant expression even before the first morphological changes are observed. This characterizes a molecular cancerization field that may favor the development of pre-malignant lesions. In head and neck, the presence of leukoplakias and erythroplakias, frequently associated with dysplasia, increases the risk of tumor development, and these lesions are characterized by specific epigenetic alterations. The epigenetic aberrations observed in both the cancerization field and in the pre-malignant lesions, have the potential to be used as early markers of HNSCC carcinogenesis in screening programs. Finally, tumor onset and progression may be followed by further aberrant DNA methylation and miRNA expression. HPV-positive HNSCC epigenetic natural history is largely unknown.
Given this scenario, it is impossible not to notice the relevance and potential of epigenetic changes as biomarkers in oncology [203,204]. In the next section, we focus on epigenetic changes as potential biomarkers in clinical use without emphasizing their contribution to gene and biological regulation. Interestingly, due to the aforementioned plastic characteristic of epigenetic marks, it is possible to identify common epigenetic changes, perhaps with different levels, aimed at more than one biomarker objective (e.g., diagnosis and prognosis). Here, we seek to bring a snapshot of epigenetic changes as biomarkers, technical limitations, and advantages, together with future application prospects.
4.1. Biomarkers of the Cancerization Field: From Identification to Surgical Margin and Recurrence
The molecular landscape of HNSCC carcinogenesis has been unraveled, and several candidates for biomarkers to identify pre-cancer lesions in the cancerization field are emerging (Table 1). Except for HPV-positive oropharyngeal tumors, the main genetic alterations in HNSCC are TP53 mutations, present in up to 84% of cases [23]. TP53 mutations have become the focus of many studies (perhaps the majority) aimed at genetic characterization of the cancerization field [205,206,207]. Although less frequently, these changes can already be found in the adjacent nontumoral dysplastic [208,209] and nondysplastic tissues of HNSCC patients [209], becoming a frequent event in key moments of the dysplasia–carcinoma transition [210].
However, TP53 mutations can be detected throughout the entire gene without clear hotspots [211], requiring techniques that are resourceful and time-consuming, such as next-generation sequencing. In any case, we would still have a high number of false negatives due to wild-type TP53 tumors and the HPV-positive group, requiring an even larger gene panel to cover the high genetic heterogeneity of this group [23]. By contrast, studies on esophageal squamous cell carcinoma (ESCC), which shares the same histology, epithelium of origin and concomitant exposure to the same risk factors as HNSCC, suggest not only that epigenetic alterations such as DNA methylation are more frequent, but could even precede the genetic changes in precursor lesions [212,213].
In HNSCC precursor lesions, p16INK4a is one of the first genes inactivated either by deletion or DNA methylation [23,214]. P16INK4a promoter hypermethylation was already extensively evaluated, but results vary widely, probably due to the divergence of the target location within the promoter region, and the methodologies applied, mostly methylation-specific PCR (MSP). Despite cost-effectiveness, it is a technique with a predominant qualitative profile, and may suffer a strong bias due to primer design, for example [215]. Interestingly, the methylation profile of p16INK4a promoter region does not seem to be associated with HPV status assessed by p16 immunohistochemistry, whereas gene body hypermethylation does [216]. However, p16INK4A gene body hypermethylation is likely to occur after its overexpression [217], being more suitable as an infection marker. In addition, deletion is the most frequent p16INK4a inactivation mechanism in HPV-negative HNSCC (57%), and practically nonexistent in HPV-positive tumors [23], in which p16 immunostaining is used as a surrogate biomarker of HPV infection [22].
Other genes with aberrant methylation in precursor lesions include MGMT, p14ARF, p15INK4b, RARB, and SOCS-3 [83,84,86,218]; p16INK4a and MGMT hypermethylation can already be detected in oral rinse [84] and blood [85] of individuals with precursor lesions or a history of OSCC, reducing the invasiveness of the procedure. Higher methylation levels of p16INK4a, p14ARF, and p15INK4b were observed in LSCC precursor lesions [218]. RARB is hypomethylated in 27% of normal adjacent HNSCC tissues and in 53% of oral precursor lesions [86]. For more detailed reviews on the evaluation of these genes as biomarkers of the cancerization field, please refer to [219,220].
ZNF282 and PAX1 hypermethylation were originally proposed as biomarkers of cervical carcinoma precursor lesions [221,222,223]. In the oral cavity, ZNF582 and PAX1 methylation increase as the epithelium goes through transformation, being able to differentiate the visually normal mucosa or with minimal histological changes (hyperplasia or mild dysplasia) from those more advanced pre-cancer changes (moderate or severe dysplasia) and the tumor itself, with sensitivities of 85% and 72% and specificities of 68% and 86%, respectively [87]. Genes involved in the WNT and the MAPK signaling pathways show a similar profile, with aberrant methylation being observed as the precursor lesions progress [224], and may be the focus of future studies.
Although, to the best of our knowledge, whole-genome bisulfite sequencing has not yet been applied to identify DNA methylation alterations throughout HNSCC natural history, next-generation sequencing (NGS) was used to assess the methylation of specific target genes in oral brushing from healthy donors, low-grade squamous intraepithelial lesions (LG-SIL), high-grade SIL (HG-SIL), and OSCC patients [88]. Among the seven targets selected, ZAP70 was hypermethylated in 100% of OSCC and HG-SIL cases, while GP1BB hypomethylation was detected in 90.9% of these individuals. Healthy donors did not show alterations of these genes, while LG-SIL showed an intermediate profile. Although the results were encouraging, this was a preliminary study with a limited number of samples.
In addition to DNA methylation, the expression of microRNAs can be used to identify the cancerization field. MiR-204, miR-31, miR-31*, miR-133a, miR-7, miR-206, and miR-1293 showed overexpression in one or more types of OSCC pre-cancer lesions relative to healthy mucosa [89]. Interestingly, miR-204 is downregulated in oral submucous fibrosis and upregulated in leukoplakia and lichen planus [89]. Similarly, representing this specific pre-cancer lesion expression signature, miR-423 and miR-34b are downregulated in lichen planus, while miR-26c and miR-29a are also downregulated in lichen planus, but overexpressed in leukoplakias [225].
Besides their potential to be applied in the early diagnosis, cancerization field biomarkers can also be used to establish surgical margins, reducing the chance of recurrences. Although histopathological evaluation is widespread, 10–30% of HNSCC patients with free surgical margins will recur [226,227,228], which is one of the main causes of mortality among HNSCC patients [228,229]. Therefore, a molecular screening of cancerization field alterations may predict this phenomenon and, therefore, improve patients’ prognosis [229]. Approximately 25% of HNSCC surgical margins contain genetic alterations [230], and TP53 mutations were already associated with recurrence [231]. Epigenetic alterations, however, also show great potential to predict relapse.
P16INK4A and/or MGMT promoters were found hypermethylated in 32% of HNSCC surgical margins, despite all being considered negative in the histopathological analysis [232]. When evaluated together at the tumor margin, the methylation levels of EDNRB and HOXA9 are predictors of locoregional recurrence-free survival, with their hypermethylation increasing the chance of recurrence approximately three-fold [125].
The analysis of a DNA methylation panel composed of three genes (DCC, CCNA1, and p16INK4A) in surgical margins was able to predict all recurrences in five out of 47 HNSCC patients [126]. In OSCC patients, ZNF582 and PAX1 methylation levels are reduced after treatment, and interestingly they rise again at the recurrence site a few months before the diagnosis [87]. In HNSCC surgical margins, the promoter methylation of another Homeobox gene, PAX5, increases the risk of locoregional recurrence approximately four-fold [127]. While most methylation studies focus on the promoter region of CDKN2A (which encodes both p14ARF and p16INK4A), its gene body hypermethylation and increased p14ARF expression in LSCC were associated with a lower incidence of locoregional recurrence [128]. More recently, a genome-wide DNA methylation screening identified 392 differentially methylated regions associated with recurrence when OSCC free surgical margins were analyzed. Accuracy of up to 98% was achieved to discriminate relapsed and nonrelapsed OSCC patients when a 14-CpG panel signature was applied [233].
MiRNA expression also shows recurrence predictive value. Oral precursor lesions of patients who relapsed showed miR-375 downregulation relative to those who did not relapse, presenting an accuracy of 95.7% and sensitivity of 90% to discriminate the two groups. Despite outstanding discrimination values, it is important to highlight that only six patients were included in the nonrelapsed group [129].
Although little explored in general, the oral cavity has the most well-defined and explored epigenetic cancerization field among HNSCC. This might be a consequence of the relatively easy access to the tumor site and the visual identification of precursor lesions, such as leukoplakias. In addition to the need to gather more data on other head and neck sites and to consistently validate the findings, studies on epigenetic alterations of the cancerization field should adopt more sensitive methodologies, and explore alterations in the mucosa of individuals with low and high exposure to risk factors, different types of precursor lesions, and cancer. Further, large-scale platforms may add new information and help to identify new biomarkers.
4.2. Diagnostic Biomarkers
In comparison to the few common genetic alterations found in HNSCC generally, DNA methylation shows more frequent and subsite-specific changes, an important asset for molecular diagnosis [24,212,213,234]. P16INK4A is hypermethylated in 27–44% of HNSCC [93,97,98,235]; however, it has already been reported that almost half of the samples could not be analyzed due to loss of heterozygosity [98,236]. CDH1 is hypermethylated in 35–43% [93,95,235], DAPK in 43% [73], and MGMT in one-third of HNSCC cases [98,103,113,235,237], and hypermethylation of the latter is independent of HPV status. This hypermethylation profile is not exclusive to the tumor and can be observed in the cancerization field, as already mentioned. For example, adjacent tissues show DAPK hypermethylation in 20% of samples [73]. GNAT1 and SEMA3B are hypermethylated in 75% and 77% of OSCC and 56% and 38% in adjacent tissues, respectively. These and other studies emphasize the importance of applying a sensitive methodology to define and differentiate this hypermethylation status [118,238]. Aberrant methylation in other genes, such as FAM135B and ZNF610 hypermethylation and HOXA9 and DCC hypomethylation, show accuracies of 79%, 93%, 93%, and 93% in distinguishing adjacent nontumor tissues and HNSCC, respectively [239]. Table 1 brings additional candidate diagnostic biomarkers based on aberrant DNA methylation in HNSCC.
MicroRNA dysregulation was also evaluated as a diagnostic biomarker. In LSCC, miR-375 is downregulated, while miR-21 is overexpressed, and their expression ratio has a 99% accuracy to discriminate tumor and adjacent nontumor tissue [122]. In OSCC, miR-204 and mir-144 are overexpressed, while miR-193b-5p and miR-370-3p are downregulated, and have an individual accuracy of 91%, 85%, 57%, and 61%, respectively, and a combined accuracy of 92% to discriminate the tissues [123]. Combining the expression of miR-657 and miR-1287, an accuracy of 97% and sensitivity of 86% were achieved in early stage LSCC [124].
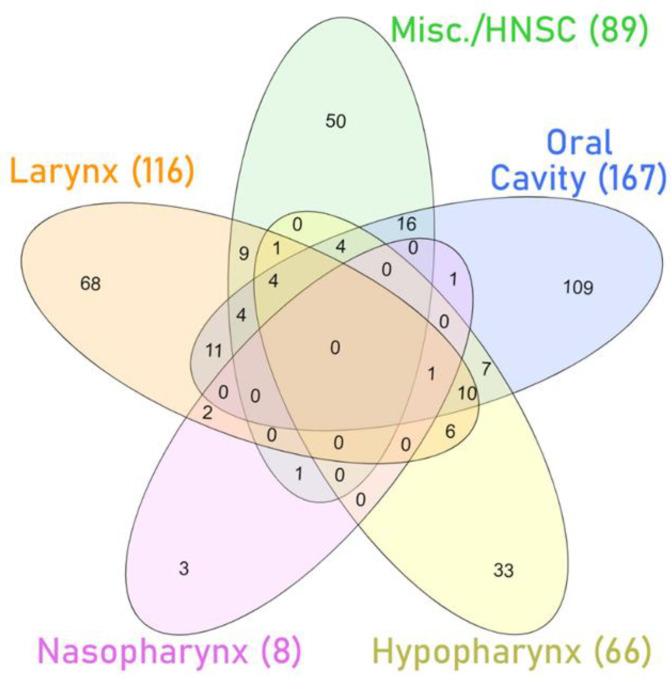
Despite the promising discrimination values mentioned above, few studies share the same findings on miRNA dysregulation in HNSCC (see Table S1 and Figure 4) [240]. This underscores the high intertumoral heterogeneity and the need for establishing subsite-specific biomarkers.
Figure 4.
Venn diagram with the number of microRNAs differentially expressed (upregulated and downregulated) between tumors and nontumor tissues in several studies with specific subsets of HNSCC (larynx [241,242,243,244], nasopharynx [245], hypopharynx [246,247], and oral cavity [248,249,250,251]) or aggregating several sites in the analysis (Misc./HNSCC) [252,253,254]. The list of differentially expressed microRNAs can be found in Table S1.
DNA methylation and microRNAs expression can be sensitively analyzed in fluids, decreasing the diagnosis’s invasiveness and even allowing screening programs in groups at risk. Perhaps the greatest group at risk for HNSCC development is composed of individuals with a previous HNSCC diagnosis, since 9–16% develops a SPT, and 49% in another region of the head and neck [255,256,257]. Noninvasive diagnosis can facilitate the follow-up and SPT early detection, increasing the chances of survival.
With the exception of the oral cavity, access to other sites of the head and neck might be challenging, and less invasive diagnostic techniques are required. The assessment of epigenetic biomarkers in body fluids, such as saliva and blood, has emerged as an alternative and may even impact the biomarker performance [258]. HNSCC patients already show a global hypomethylation in whole blood, estimated by the evaluation of LINE1 transposable elements. However, in addition to being a generic tumor biomarker [259,260,261,262,263,264], global methylation levels showed overlap between HNSCC patients and controls, jeopardizing their discrimination capacity [110]. THF methylation profile was able to differentiate with 93% accuracy (86% sensitivity) individuals without cancer from those with OSCC or OPSCC using oral rinse [265]. The methylation profile of MED15 promoter region in the saliva of HNSCC patients showed an accuracy of 63–70% [266]. In the plasma of LSCC patients and individuals without cancer, 26 differentially expressed microRNAs were identified, 17 overexpressed, and 9 downregulated in patients [243].
In addition to tumor diagnosis, the DNA methylation profile can also help to identify the risk factors associated with tumor development. By evaluating the saliva of HPV-negative HNSCC patients, RASSF1A, p16INK4A, TIMP3, and PCQAP/MED15 genes were found hypermethylated relative to healthy individuals and showed a sensitivity of 71% to discriminate the groups [79]. The same genes are hypomethylated in the saliva of HPV-positive HNSCC patients relative to healthy individuals and distinguished the groups with a sensitivity of 80% [79]. A panel with the methylation profile of p16INK4A, RASSF1A, TIMP3, and PCQAP/MED15 genes in saliva was able to differentiate OSCC and OPSCC patients from individuals without cancer with 91% and 92% sensitivity and 99% and 92% specificity, respectively [117].
A common feature of HNSCC is the marked disparity in the overall survival of patients diagnosed in early stages compared to the more advanced ones. Screenings widely applied in oncology, such as those for breast [267] and cervical [267] cancer early diagnosis have sensitivities of approximately 96%. Studies aimed at tumor screening in the oral cavity showed that despite a good number of pre-cancer lesions were identified (~10%), the number of carcinomas found was low [268], with a sensitivity of 71–76% [269,270]. However, even with the low frequency of tumors, reduced staging and improved overall survival were observed in patients due to screening, showing that it can be worthwhile [269].
4.3. Biomarkers of Treatment Response and Prognosis
Despite advances in oncology, the average five-year survival of HNSCC patients remains the same, 13–57%, varying according to the affected primary site [271,272]. The therapeutic approach depends on the location, resectability, and strategies focused on organ preservation, including surgery, chemotherapy, and radiotherapy [273]. The gold-standard treatment for the disease is surgery, which may be combined with radiotherapy and, in locally advanced cases, chemotherapy based on taxanes and platinum [273].
In HNSCC, molecular changes and etiology can drive treatment choice. Patients with HPV-positive OPSCC respond better to therapy, and treatment deintensification was already proposed [274]. By contrast, smokers and drinkers generally show a poor response to treatment [275,276,277]. In this context, epigenetic changes have been proposed as surrogate markers of therapeutic response and epigenetic drugs were shown to resensitize cell lines to conventional therapy.
DNA methylation alterations have been recently shown to predict locoregional recurrence after surgery, for example. By evaluating the methylation levels of 13 genes by bisulfite NGS in pre-operative OSCC samples collected by oral bushing, Gissi and colleagues showed that five CpG sites in three genes (EPHX3, ITGA4, and MiR193A) were able to predict adverse outcomes with high accuracy (AUC = 0.85) [138].
HNSCC cell lines resistant to irradiation showed global hypermethylation and differential methylation and expression of up to 84 genes relative to sensitive strains [278]. Among these, CCND2, a cell cycle controller, was hypermethylated in the promoter region [278]. DAPK promoter hypermethylation was detected in cell lines resistant to EGFR inhibitors, one of the few target therapies approved by the FDA to treat HNSCC patients [279,280]. Interestingly, DAPK knockout itself is able to induce resistance against these drugs [281]. In taxol-resistant nasopharyngeal carcinoma cell lines, global hypermethylation and 48 differentially methylated genes (30 hypermethylated and 18 hypomethylated) were observed, and treatment with demethylating agents could resensitize cells to chemotherapy [282]. Treatment with demethylating agents also sensitizes HNSCC cell lines to cisplatin [283].
Dozens of miRNAs were also found differentially expressed in HNSCC cell lines resistant to therapy [284,285]. Overexpression of let-7c, let-7d, let-7e, let-7g, and miR-20b [286], and downregulation of miR-200b, miR-15b [287], and miR-181a [288] were associated with platinum resistance.
More recently, FDA also approved immunotherapy based on PD1 and PD-L1 immune checkpoint blockade (pembrolizumab and nivolumab) for treatment of platinum-refractory metastatic HNSCC [289]. PD-L1 and PD-L2 promoter methylation levels are correlated with their expression and associated with HPV infection in HNSCC. Therefore, their assessment might be a promising predictive biomarker for treatment [290]. Furthermore, PD-1 hypermethylation was already associated with a worse prognosis in HNSCC [291].
Regarding prognosis, worse overall and/or disease-free survival in HNSCC were associated with promoter hypermethylation of KL [130], HIN1 [131], RASSF1A/RASSF2 [131], MGMT [95,119,132,133], DAPK [119], MINT31 [114], p16INK4A, and p14ARF [134]. Interestingly, PAX5 and PAX1 promoter methylation is associated with poorer overall survival in African-American patients compared to nonwhite Latinos [135].
A higher miR-21 expression or lower miR-375 expression was associated with a worse prognosis in LSCC [122]. In OPSCC, a panel with miR-142-3p, miR-31, miR-146a, miR-26b, miR-24, and miR-193b can predict survival regardless of HPV status or other clinical characteristics [292]. MiRNA signatures to predict overall survival (miR-107, miR-151, miR-492), disease-free survival (miR-107, miR-182, miR-20b, miR-151, miR-361), and distant relapse-free survival (miR324-5p, miR-492, miR-151, miR-361, miR-152) among OPSCC patients were described in a different study from the same year [293].
Epigenetic biomarkers in liquid biopsies also show promising results in predicting patient prognosis. TIMP3 promoter hypermethylation detected in HNSCC patients’ saliva seems to be an independent marker of recurrence-free survival [136]. Higher SEPT9 and SHOX2 methylation levels in cell-free circulating DNA in the plasma of HNSCC patients were associated with worse overall survival and increased the chances of death approximately 5.2 and 2.3-fold, respectively [137]. When detected in the plasma of HNSCC patients, the high expression of miR-186-5p and miR-374b-5p before treatment and miR-142-3p after treatment were associated with reduced locoregional control. The expression of miR-142-3p was also associated with reduced disease-free progression [139].
4.4. The Epilogue of Epigenetic Biomarkers in HNSCC
The acquisition of epigenetic alterations due to risk factor exposure and transformation in a plastic but stable fashion, and their independence of genetic changes frequency present good potential for epigenetic biomarker development. These changes can be detected early during tumor development, enabling screening programs and increasing survival rates. Furthermore, the same marker has the potential to be used to predict more than one endpoint, such as diagnosis and recurrence, decreasing development time and cost. However, the establishment of the biomarker must be cautious, reproducible, and apply primarily sensitive methodologies.
In the case of DNA methylation, most studies cited here applied MSP or quantitative-MSP (qMSP) for biomarker identification, either directly or after selecting targets from microarray data. Although these techniques are quite user-friendly and generally available, they are subject to amplification biases and usually interrogate single CpG sites, which may impair the interpretation of results. Additionally, MSP does not provide a quantitative measure of methylation levels. This is especially relevant when we consider that pools of cells or of circulating DNA (in liquid biopsies) are usually analyzed. Since loci methylation will vary according to cell type and exposures, DNA methylation levels should be considered as a continuous and not a qualitative variable. Thus, bisulfite pyrosequencing or bisulfite amplicon–NGS could be more suitable for identifying potential biomarkers. Both techniques provide quantitative measures of DNA methylation levels and interrogate a higher number of CpG sites, but have a higher cost and require equipment that is not available in every research or clinical laboratory. Finally, all these techniques require a prior conversion of unmethylated cytosines into uracil by DNA treatment with sodium bisulfite. Despite its widespread use, this treatment might be an additional challenge for the analysis of poor-quality DNA. When these molecules are extracted from formalin-fixed paraffin-embedded (FFPE) tissues or from liquid biopsies, they are usually highly degraded, and the treatment with sodium bisulfite induces further degradation. However, the alternatives, such as the use of methylation-sensitive restriction enzymes or CpG methylation immunoprecipitation suffer from lower sensitivity. Based on these considerations, the ideal combination of techniques to evaluate DNA methylation alterations as biomarkers will depend on sample type, difference range, resources available, and biomarker sensitivity, among other factors. Nevertheless, after the establishment of methylation and discrimination levels, the construction of low-cost and more qualitative tests (such as qMSP) could be valid to decrease time and cost [294].
Although generally less explored than DNA methylation, miRNA-based biomarker development in HNSCC seems more advanced in terms of methodology. While many candidates are identified by microarray, a semiquantitative technique, their validation and assessment in other cohorts and sample sources are usually accomplished by reverse transcription followed by quantitative PCR (RT-qPCR). This is a widespread quantitative approach already applied in the clinics, as recently observed in COVID-19 diagnosis. The major challenge in the development of these biomarkers, however, is probably the definition of cutoffs for distinguishing groups, although this can be overcome by reproducibility studies, necessary for the development of any biomarker.
It is important to note that some genetic factors can affect the performance of the epigenetic biomarker. For example, MTHFR polymorphisms can affect p16INK4A and MGMT methylation levels in the oral cavity [295,296]. Even unexpected factors, such as pregnancy with intrauterine growth restriction, can cause aberrant methylation, such as RASSF1A hypermethylation [297], also observed in HNSCC [298]. In this context, the development of panels can be more sensitive and specific, avoiding interference from different factors and the generation of false negatives. The panel can be applied in minimally invasive approaches such as body fluids, where epigenetic changes are highly stable, or even in tumor biopsies, when available [299,300].
Until now, most of the proposed biomarkers are based on promoter hypermethylation of tumor suppressor genes, the mechanism generally involved in their silencing. Thus, the methylation status of these genes may also be indicative of the use of demethylating agents in the treatment, aiming for their reactivation. This bias towards promoters highlights the need for further studies characterizing the methylation profile in other genomic regions, such as intergenic regions, enhancers, and gene bodies.
Although less explored than DNA methylation and miRNAs, histone modifications have also been reported in HNSCC and have been associated with patients’ outcomes and cancer phenotypes. This is another potential field for the development of biomarkers, not addressed in this review (for further information, please refer to [285,301,302,303,304]), and should be further explored. In OSCC, H3K9ac reduction was associated with the activation of epithelial–mesenchyme transition, cell proliferation, and poor prognosis [305]; also in OSCC, the activation of Wnt/β-catenin signaling and the consequent induction of cell proliferation and invasion were associated with increased H3K27ac through the activation of PLAC2, a long-noncoding RNA [306]. Finally, high H3K27me3 and low H3K4ac levels were associated with poor prognosis [307], while reduced H3K4me3 and increased H3K4me2 levels were observed in OSCC relative to normal tissues [308], suggesting that histone methylation could also be an OSCC biomarker.
As mentioned previously, most studies considered all HNSCC sites as a single entity or focused on the oral cavity, probably due to the easier access and higher tumor occurrence. This might be an additional challenge because of differences in carcinogenesis mechanisms behind each specific subsite. Thus, the epilogue of HNSCC biomarkers is still blurry, and epigenetic changes can be an eye drop.
5. Conclusions
As highlighted in this review, epigenetic alterations are a common feature of HNSCC, and affect all steps in their natural history. In this context, intratumor heterogeneity, a major cause of cancer treatment failure and relapse, cannot be addressed only genetically. Epigenetics confers different phenotypes to subclones and, due to its reversibility, emerges as a potential therapeutic target. Therefore, to improve clinical management, it is of utmost importance that all professionals involved in patient care come to know this somewhat unexplored field.
Thus far, most studies have focused on the methylation of promoters of tumor suppressor genes. Although this approach has already identified putative carcinogenic pathways and biomarkers, much is yet to be learned from epigenetic alterations in HNSCC. Not only the methylation of other genes and other genomic regions, but also other players, including noncoding RNAs and chromatin remodelers, should be explored. For example, global hypomethylation, associated with poor prognosis in other cancer types [189,309,310,311], seems to differ according to HPV status and could help explain the prognosis differences observed. Furthermore, mutations in the histone methyltransferases NSD1 and NSD2 define a group of good prognosis among LSCC patients, but not in other HNSCC subtypes [312]. These data highlight the importance of painting a more detailed canvas of epigenetic landscape in HNSCC. The integration of different epigenetic mechanisms and the comprehension of both inter- and intratumoral heterogeneity may help achieve more personalized HNSCC patient care.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13225630/s1, Table S1: MicroRNAs with altered expression in HNSCC.
Author Contributions
Conceptualization, D.C., T.d.A.S., F.D., L.F.R.P. and S.C.S.-L.; writing—original draft preparation, D.C., S.C.S.-L.; writing—review and editing, T.d.A.S., F.D., L.F.R.P., S.C.S.-L.; visualization, D.C.; supervision, S.C.S.-L.; funding acquisition, S.C.S.-L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. The APC was funded by Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ). S.C.S-L. is a scholar of FAPERJ (#E-26/202.918/2019).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Miranda-Galvis M., Loveless R., Kowalski L.P., Teng Y. Impacts of Environmental Factors on Head and Neck Cancer Pathogenesis and Progression. Cells. 2021;10:389. doi: 10.3390/cells10020389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaenisch R., Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 4.Leemans C.R., Braakhuis B.J., Brakenhoff R.H. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 5.Brierley J.D., Gospodarowicz M.K., Wittekind C. TNM Classification of Malignant Tumours. John Wiley & Sons; Hoboken, NJ, USA: 2017. [Google Scholar]
- 6.Braakhuis B.J., Tabor M.P., Leemans C.R., van der Waal I., Snow G.B., Brakenhoff R.H. Second primary tumors and field cancerization in oral and oropharyngeal cancer: Molecular techniques provide new insights and definitions. Head Neck. 2002;24:198–206. doi: 10.1002/hed.10042. [DOI] [PubMed] [Google Scholar]
- 7.Slaughter D.P., Southwick H.W., Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::AID-CNCR2820060515>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 8.Gillis T.M., Incze J., Strong M.S., Vaughan C.W., Simpson G.T. Natural history and management of keratosis, atypia, carcinoma-in situ, and microinvasive cancer of the larynx. Am. J. Surg. 1983;146:512–516. doi: 10.1016/0002-9610(83)90243-X. [DOI] [PubMed] [Google Scholar]
- 9.Mello F.W., Miguel A.F.P., Dutra K.L., Porporatti A.L., Warnakulasuriya S., Guerra E.N.S., Rivero E.R.C. Prevalence of oral potentially malignant disorders: A systematic review and meta-analysis. J. Oral. Pathol. Med. 2018;47:633–640. doi: 10.1111/jop.12726. [DOI] [PubMed] [Google Scholar]
- 10.Speight P.M., Khurram S.A., Kujan O. Oral potentially malignant disorders: Risk of progression to malignancy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018;125:612–627. doi: 10.1016/j.oooo.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 11.Marur S., D’Souza G., Westra W.H., Forastiere A.A. HPV-associated head and neck cancer: A virus-related cancer epidemic. Lancet Oncol. 2010;11:781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryerson A.B., Peters E.S., Coughlin S.S., Chen V.W., Gillison M.L., Reichman M.E., Wu X., Chaturvedi A.K., Kawaoka K. Burden of potentially human papillomavirus-associated cancers of the oropharynx and oral cavity in the US, 1998–2003. Cancer. 2008;113:2901–2909. doi: 10.1002/cncr.23745. [DOI] [PubMed] [Google Scholar]
- 13.Mahal B.A., Catalano P.J., Haddad R.I., Hanna G.J., Kass J.I., Schoenfeld J.D., Tishler R.B., Margalit D.N. Incidence and Demographic Burden of HPV-Associated Oropharyngeal Head and Neck Cancers in the United States. Cancer Epidemiol. Biomark. Prev. 2019;28:1660–1667. doi: 10.1158/1055-9965.EPI-19-0038. [DOI] [PubMed] [Google Scholar]
- 14.Klussmann J.P., Weissenborn S.J., Wieland U., Dries V., Kolligs J., Jungehuelsing M., Eckel H.E., Dienes H.P., Pfister H.J., Fuchs P.G. Prevalence, distribution, and viral load of human papillomavirus 16 DNA in tonsillar carcinomas. Cancer. 2001;92:2875–2884. doi: 10.1002/1097-0142(20011201)92:11<2875::AID-CNCR10130>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 15.Begum S., Westra W.H. Basaloid squamous cell carcinoma of the head and neck is a mixed variant that can be further resolved by HPV status. Am. J. Surg. Pathol. 2008;32:1044–1050. doi: 10.1097/PAS.0b013e31816380ec. [DOI] [PubMed] [Google Scholar]
- 16.Buexm L.A., Soares-Lima S.C., Brennan P., Fernandes P.V., de Souza Almeida Lopes M., Nascimento de Carvalho F., Santos I.C., Dias L.F., de Queiroz Chaves Lourenço S., Ribeiro Pinto L.F. Hpv impact on oropharyngeal cancer patients treated at the largest cancer center from Brazil. Cancer Lett. 2020;477:70–75. doi: 10.1016/j.canlet.2020.02.023. [DOI] [PubMed] [Google Scholar]
- 17.Faraji F., Zaidi M., Fakhry C., Gaykalova D.A. Molecular mechanisms of human papillomavirus-related carcinogenesis in head and neck cancer. Microbes Infect. 2017;19:464–475. doi: 10.1016/j.micinf.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheffner M., Werness B.A., Huibregtse J.M., Levine A.J., Howley P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 19.Boyer S.N., Wazer D.E., Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 20.Parfenov M., Pedamallu C.S., Gehlenborg N., Freeman S.S., Danilova L., Bristow C.A., Lee S., Hadjipanayis A.G., Ivanova E.V., Wilkerson M.D., et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc. Natl. Acad. Sci. USA. 2014;111:15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khleif S.N., DeGregori J., Yee C.L., Otterson G.A., Kaye F.J., Nevins J.R., Howley P.M. Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc. Natl. Acad. Sci. USA. 1996;93:4350–4354. doi: 10.1073/pnas.93.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prigge E.S., Arbyn M., von Knebel Doeberitz M., Reuschenbach M. Diagnostic accuracy of p16. Int. J. Cancer. 2017;140:1186–1198. doi: 10.1002/ijc.30516. [DOI] [PubMed] [Google Scholar]
- 23.Network C.G.A. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soares-Lima S.C., Mehanna H., Camuzi D., de Souza-Santos P.T., Simão T.A., Nicolau-Neto P., Almeida Lopes M.S., Cuenin C., Talukdar F.R., Batis N., et al. Upper Aerodigestive Tract Squamous Cell Carcinomas Show Distinct Overall DNA Methylation Profiles and Different Molecular Mechanisms behind WNT Signaling Disruption. Cancers. 2021;13:3014. doi: 10.3390/cancers13123014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lokk K., Modhukur V., Rajashekar B., Märtens K., Mägi R., Kolde R., Koltšina M., Nilsson T.K., Vilo J., Salumets A., et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014;15:r54. doi: 10.1186/gb-2014-15-4-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancarella D., Plass C. Epigenetic signatures in cancer: Proper controls, current challenges and the potential for clinical translation. Genome Med. 2021;13:23. doi: 10.1186/s13073-021-00837-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger S.L., Kouzarides T., Shiekhattar R., Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfister S.X., Ashworth A. Marked for death: Targeting epigenetic changes in cancer. Nat. Rev. Drug Discov. 2017;16:241–263. doi: 10.1038/nrd.2016.256. [DOI] [PubMed] [Google Scholar]
- 29.Kelly A.D., Issa J.J. The promise of epigenetic therapy: Reprogramming the cancer epigenome. Curr. Opin. Genet. Dev. 2017;42:68–77. doi: 10.1016/j.gde.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Hermann A., Goyal R., Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 31.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 32.Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neri F., Rapelli S., Krepelova A., Incarnato D., Parlato C., Basile G., Maldotti M., Anselmi F., Oliviero S. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543:72–77. doi: 10.1038/nature21373. [DOI] [PubMed] [Google Scholar]
- 34.Yoo C.B., Jones P.A. Epigenetic therapy of cancer: Past, present and future. Nat. Rev. Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 35.Taberlay P.C., Statham A.L., Kelly T.K., Clark S.J., Jones P.A. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res. 2014;24:1421–1432. doi: 10.1101/gr.163485.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 37.Pu M., Chen J., Tao Z., Miao L., Qi X., Wang Y., Ren J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol. Life Sci. 2019;76:441–451. doi: 10.1007/s00018-018-2940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartel D.P. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith I.M., Mydlarz W.K., Mithani S.K., Califano J.A. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int. J. Cancer. 2007;121:1724–1728. doi: 10.1002/ijc.22889. [DOI] [PubMed] [Google Scholar]
- 40.Lechner M., Frampton G.M., Fenton T., Feber A., Palmer G., Jay A., Pillay N., Forster M., Cronin M.T., Lipson D., et al. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV- tumors. Genome Med. 2013;5:49. doi: 10.1186/gm453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holubekova V., Mendelova A., Jasek K., Mersakova S., Zubor P., Lasabova Z. Epigenetic regulation by DNA methylation and miRNA molecules in cancer. Future Oncol. 2017;13:2217–2222. doi: 10.2217/fon-2017-0363. [DOI] [PubMed] [Google Scholar]
- 42.Duursma A.M., Kedde M., Schrier M., le Sage C., Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E., Liu S., Alder H., Costinean S., Fernandez-Cymering C., et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duan R., Pak C., Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- 45.Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R.M., Okamoto A., Yokota J., Tanaka T., et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 46.Ghantous Y., Schussel J.L., Brait M. Tobacco and alcohol-induced epigenetic changes in oral carcinoma. Curr. Opin. Oncol. 2018;30:152–158. doi: 10.1097/CCO.0000000000000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu C., Marioni R.E., Hedman Å., Pfeiffer L., Tsai P.C., Reynolds L.M., Just A.C., Duan Q., Boer C.G., Tanaka T., et al. A DNA methylation biomarker of alcohol consumption. Mol. Psychiatry. 2018;23:422–433. doi: 10.1038/mp.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saad M.A., Kuo S.Z., Rahimy E., Zou A.E., Korrapati A., Rahimy M., Kim E., Zheng H., Yu M.A., Wang-Rodriguez J., et al. Alcohol-dysregulated miR-30a and miR-934 in head and neck squamous cell carcinoma. Mol. Cancer. 2015;14:181. doi: 10.1186/s12943-015-0452-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boscolo-Rizzo P., Furlan C., Lupato V., Polesel J., Fratta E. Novel insights into epigenetic drivers of oropharyngeal squamous cell carcinoma: Role of HPV and lifestyle factors. Clin. Epigenetics. 2017;9:124. doi: 10.1186/s13148-017-0424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Božinović K., Sabol I., Dediol E., Milutin Gašperov N., Manojlović S., Vojtechova Z., Tachezy R., Grce M. Genome-wide miRNA profiling reinforces the importance of miR-9 in human papillomavirus associated oral and oropharyngeal head and neck cancer. Sci. Rep. 2019;9:2306. doi: 10.1038/s41598-019-38797-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teneng I., Montoya-Durango D.E., Quertermous J.L., Lacy M.E., Ramos K.S. Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation. Epigenetics. 2011;6:355–367. doi: 10.4161/epi.6.3.14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Satta R., Maloku E., Zhubi A., Pibiri F., Hajos M., Costa E., Guidotti A. Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc. Natl. Acad. Sci. USA. 2008;105:16356–16361. doi: 10.1073/pnas.0808699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin R.K., Hsieh Y.S., Lin P., Hsu H.S., Chen C.Y., Tang Y.A., Lee C.F., Wang Y.C. The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. J. Clin. Investig. 2010;120:521–532. doi: 10.1172/JCI40706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mortusewicz O., Schermelleh L., Walter J., Cardoso M.C., Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl. Acad. Sci. USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cuozzo C., Porcellini A., Angrisano T., Morano A., Lee B., Di Pardo A., Messina S., Iuliano R., Fusco A., Santillo M.R., et al. DNA damage, homology-directed repair, and DNA methylation. PLoS Genet. 2007;3:e110. doi: 10.1371/journal.pgen.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Di Y.P., Zhao J., Harper R. Cigarette smoke induces MUC5AC protein expression through the activation of Sp1. J. Biol. Chem. 2012;287:27948–27958. doi: 10.1074/jbc.M111.334375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kadonaga J.T., Carner K.R., Masiarz F.R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 58.Han L., Lin I.G., Hsieh C.L. Protein binding protects sites on stable episomes and in the chromosome from de novo methylation. Mol. Cell Biol. 2001;21:3416–3424. doi: 10.1128/MCB.21.10.3416-3424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee K.W., Pausova Z. Cigarette smoking and DNA methylation. Front. Genet. 2013;4:132. doi: 10.3389/fgene.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Q., Liu L., Zhao Y., Zhang J., Wang D., Chen J., He Y., Wu J., Zhang Z., Liu Z. Hypoxia induces genomic DNA demethylation through the activation of HIF-1α and transcriptional upregulation of MAT2A in hepatoma cells. Mol. Cancer Ther. 2011;10:1113–1123. doi: 10.1158/1535-7163.MCT-10-1010. [DOI] [PubMed] [Google Scholar]
- 61.Mani S., Szymańska K., Cuenin C., Zaridze D., Balassiano K., Lima S.C., Matos E., Daudt A., Koifman S., Filho V.W., et al. DNA methylation changes associated with risk factors in tumors of the upper aerodigestive tract. Epigenetics. 2012;7:270–277. doi: 10.4161/epi.7.3.19306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marsit C.J., McClean M.D., Furniss C.S., Kelsey K.T. Epigenetic inactivation of the SFRP genes is associated with drinking, smoking and HPV in head and neck squamous cell carcinoma. Int. J. Cancer. 2006;119:1761–1766. doi: 10.1002/ijc.22051. [DOI] [PubMed] [Google Scholar]
- 63.Sharma R., Panda N.K., Khullar M. Hypermethylation of carcinogen metabolism genes, CYP1A1, CYP2A13 and GSTM1 genes in head and neck cancer. Oral Dis. 2010;16:668–673. doi: 10.1111/j.1601-0825.2010.01676.x. [DOI] [PubMed] [Google Scholar]
- 64.Sarkar S., Alam N., Mandal S.S., Chatterjee K., Ghosh S., Roychoudhury S., Panda C.K. Differential transmission of the molecular signature of RBSP3, LIMD1 and CDC25A in basal/ parabasal versus spinous of normal epithelium during head and neck tumorigenesis: A mechanistic study. PLoS ONE. 2018;13:e0195937. doi: 10.1371/journal.pone.0195937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arunkumar G., Deva Magendhra Rao A.K., Manikandan M., Prasanna Srinivasa Rao H., Subbiah S., Ilangovan R., Murugan A.K., Munirajan A.K. Dysregulation of miR-200 family microRNAs and epithelial-mesenchymal transition markers in oral squamous cell carcinoma. Oncol. Lett. 2018;15:649–657. doi: 10.3892/ol.2017.7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manikandan M., Deva Magendhra Rao A.K., Rajkumar K.S., Rajaraman R., Munirajan A.K. Altered levels of miR-21, miR-125b-2*, miR-138, miR-155, miR-184, and miR-205 in oral squamous cell carcinoma and association with clinicopathological characteristics. J. Oral Pathol. Med. 2015;44:792–800. doi: 10.1111/jop.12300. [DOI] [PubMed] [Google Scholar]
- 67.Fang R., Huang Y., Xie J., Zhang J., Ji X. Downregulation of miR-29c-3p is associated with a poor prognosis in patients with laryngeal squamous cell carcinoma. Diagn. Pathol. 2019;14:109. doi: 10.1186/s13000-019-0893-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruzgielewicz A., Osuch-Wojcikiewicz E., Niemczyk K., Sieniawska-Buccella O., Siwak M., Walczak A., Nowak A., Majsterek I. Altered Expression of miRNAs Is Related to Larynx Cancer TNM Stage and Patients’ Smoking Status. DNA Cell Biol. 2017;36:581–588. doi: 10.1089/dna.2016.3464. [DOI] [PubMed] [Google Scholar]
- 69.Metheetrairut C., Chotigavanich C., Amornpichetkul K., Keskool P., Ongard S. Expression levels of miR-34-family microRNAs are associated with TP53 mutation status in head and neck squamous cell carcinoma. Eur. Arch. Otorhinolaryngol. 2019;276:521–533. doi: 10.1007/s00405-018-5223-x. [DOI] [PubMed] [Google Scholar]
- 70.House R., Majumder M., Janakiraman H., Ogretmen B., Kato M., Erkul E., Hill E., Atkinson C., Barth J., Day T.A., et al. Smoking-induced control of miR-133a-3p alters the expression of EGFR and HuR in HPV-infected oropharyngeal cancer. PLoS ONE. 2018;13:e0205077. doi: 10.1371/journal.pone.0205077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang H.W., Ling G.S., Wei W.I., Yuen A.P. Smoking and drinking can induce p15 methylation in the upper aerodigestive tract of healthy individuals and patients with head and neck squamous cell carcinoma. Cancer. 2004;101:125–132. doi: 10.1002/cncr.20323. [DOI] [PubMed] [Google Scholar]
- 72.Bennett K.L., Lee W., Lamarre E., Zhang X., Seth R., Scharpf J., Hunt J., Eng C. HPV status-independent association of alcohol and tobacco exposure or prior radiation therapy with promoter methylation of FUSSEL18, EBF3, IRX1, and SEPT9, but not SLC5A8, in head and neck squamous cell carcinomas. Genes Chromosomes Cancer. 2010;49:319–326. doi: 10.1002/gcc.20742. [DOI] [PubMed] [Google Scholar]
- 73.Cai F., Xiao X., Niu X., Zhong Y. Association between promoter methylation of DAPK gene and HNSCC: A meta-analysis. PLoS ONE. 2017;12:e0173194. doi: 10.1371/journal.pone.0173194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marsit C.J., Liu M., Nelson H.H., Posner M., Suzuki M., Kelsey K.T. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: Implications for treatment and survival. Oncogene. 2004;23:1000–1004. doi: 10.1038/sj.onc.1207256. [DOI] [PubMed] [Google Scholar]
- 75.Pierini S., Jordanov S.H., Mitkova A.V., Chalakov I.J., Melnicharov M.B., Kunev K.V., Mitev V.I., Kaneva R.P., Goranova T.E. Promoter hypermethylation of CDKN2A, MGMT, MLH1, and DAPK genes in laryngeal squamous cell carcinoma and their associations with clinical profiles of the patients. Head Neck. 2014;36:1103–1108. doi: 10.1002/hed.23413. [DOI] [PubMed] [Google Scholar]
- 76.Avissar M., McClean M.D., Kelsey K.T., Marsit C.J. MicroRNA expression in head and neck cancer associates with alcohol consumption and survival. Carcinogenesis. 2009;30:2059–2063. doi: 10.1093/carcin/bgp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swangphon P., Pientong C., Burassakarn A., Vatanasapt P., Kleebkaow P., Patarapadungkit N., Treebupachatsakul T., Promthet S., Kongyingyoes B., Ekalaksananan T. Methylation Status of P16Ink4a in Human Papillomavirus-Associated Cancer of Oral Cavity and Oropharynx in Northeastern Thailand. Asian Pac. J. Cancer Prev. 2017;18:699–705. doi: 10.22034/APJCP.2017.18.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choudhury J.H., Ghosh S.K. Promoter Hypermethylation Profiling Identifies Subtypes of Head and Neck Cancer with Distinct Viral, Environmental, Genetic and Survival Characteristics. PLoS ONE. 2015;10:e0129808. doi: 10.1371/journal.pone.0129808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lim Y., Wan Y., Vagenas D., Ovchinnikov D.A., Perry C.F., Davis M.J., Punyadeera C. Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer. 2016;16:749. doi: 10.1186/s12885-016-2785-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakagawa T., Matsusaka K., Misawa K., Ota S., Takane K., Fukuyo M., Rahmutulla B., Shinohara K.I., Kunii N., Sakurai D., et al. Frequent promoter hypermethylation associated with human papillomavirus infection in pharyngeal cancer. Cancer Lett. 2017;407:21–31. doi: 10.1016/j.canlet.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 81.Zhang Q., Chen Y., Hu S.Q., Pu Y.M., Zhang K., Wang Y.X. A HPV16-related prognostic indicator for head and neck squamous cell carcinoma. Ann. Transl. Med. 2020;8:1492. doi: 10.21037/atm-20-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miller D.L., Davis J.W., Taylor K.H., Johnson J., Shi Z., Williams R., Atasoy U., Lewis J.S., Stack M.S. Identification of a human papillomavirus-associated oncogenic miRNA panel in human oropharyngeal squamous cell carcinoma validated by bioinformatics analysis of the Cancer Genome Atlas. Am. J. Pathol. 2015;185:679–692. doi: 10.1016/j.ajpath.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weber A., Hengge U.R., Bardenheuer W., Tischoff I., Sommerer F., Markwarth A., Dietz A., Wittekind C., Tannapfel A. SOCS-3 is frequently methylated in head and neck squamous cell carcinoma and its precursor lesions and causes growth inhibition. Oncogene. 2005;24:6699–6708. doi: 10.1038/sj.onc.1208818. [DOI] [PubMed] [Google Scholar]
- 84.López M., Aguirre J.M., Cuevas N., Anzola M., Videgain J., Aguirregaviria J., Martínez de Pancorbo M. Gene promoter hypermethylation in oral rinses of leukoplakia patients--a diagnostic and/or prognostic tool? Eur. J. Cancer. 2003;39:2306–2309. doi: 10.1016/S0959-8049(03)00550-1. [DOI] [PubMed] [Google Scholar]
- 85.Bhatia V., Goel M.M., Makker A., Tewari S., Yadu A., Shilpi P., Kumar S., Agarwal S.P., Goel S.K. Promoter region hypermethylation and mRNA expression of MGMT and p16 genes in tissue and blood samples of human premalignant oral lesions and oral squamous cell carcinoma. Biomed. Res. Int. 2014;2014:248419. doi: 10.1155/2014/248419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Youssef E.M., Lotan D., Issa J.P., Wakasa K., Fan Y.H., Mao L., Hassan K., Feng L., Lee J.J., Lippman S.M., et al. Hypermethylation of the retinoic acid receptor-beta(2) gene in head and neck carcinogenesis. Clin. Cancer Res. 2004;10:1733–1742. doi: 10.1158/1078-0432.CCR-0989-3. [DOI] [PubMed] [Google Scholar]
- 87.Cheng S.J., Chang C.F., Lee J.J., Chen H.M., Wang H.J., Liou Y.L., Yen C., Chiang C.P. Hypermethylated ZNF582 and PAX1 are effective biomarkers for detection of oral dysplasia and oral cancer. Oral Oncol. 2016;62:34–43. doi: 10.1016/j.oraloncology.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 88.Morandi L., Gissi D., Tarsitano A., Asioli S., Monti V., Del Corso G., Marchetti C., Montebugnoli L., Foschini M.P. DNA methylation analysis by bisulfite next-generation sequencing for early detection of oral squamous cell carcinoma and high-grade squamous intraepithelial lesion from oral brushing. J. Craniomaxillofac. Surg. 2015;43:1494–1500. doi: 10.1016/j.jcms.2015.07.028. [DOI] [PubMed] [Google Scholar]
- 89.Chattopadhyay E., Singh R., Ray A., Roy R., De Sarkar N., Paul R.R., Pal M., Aich R., Roy B. Expression deregulation of mir31 and CXCL12 in two types of oral precancers and cancer: Importance in progression of precancer and cancer. Sci. Rep. 2016;6:32735. doi: 10.1038/srep32735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gao S., Worm J., Guldberg P., Eiberg H., Krogdahl A., Liu C.J., Reibel J., Dabelsteen E. Genetic and epigenetic alterations of the blood group ABO gene in oral squamous cell carcinoma. Int. J. Cancer. 2004;109:230–237. doi: 10.1002/ijc.11592. [DOI] [PubMed] [Google Scholar]
- 91.Supić G., Kozomara R., Branković-Magić M., Jović N., Magić Z. Gene hypermethylation in tumor tissue of advanced oral squamous cell carcinoma patients. Oral Oncol. 2009;45:1051–1057. doi: 10.1016/j.oraloncology.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 92.Bennett K.L., Hackanson B., Smith L.T., Morrison C.D., Lang J.C., Schuller D.E., Weber F., Eng C., Plass C. Tumor suppressor activity of CCAAT/enhancer binding protein alpha is epigenetically down-regulated in head and neck squamous cell carcinoma. Cancer Res. 2007;67:4657–4664. doi: 10.1158/0008-5472.CAN-06-4793. [DOI] [PubMed] [Google Scholar]
- 93.Shaw R.J., Liloglou T., Rogers S.N., Brown J.S., Vaughan E.D., Lowe D., Field J.K., Risk J.M. Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: Quantitative evaluation using pyrosequencing. Br. J. Cancer. 2006;94:561–568. doi: 10.1038/sj.bjc.6602972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Supic G., Jovic N., Kozomara R., Zeljic K., Magic Z. Interaction between the MTHFR C677T polymorphism and alcohol--impact on oral cancer risk and multiple DNA methylation of tumor-related genes. J. Dent. Res. 2011;90:65–70. doi: 10.1177/0022034510385243. [DOI] [PubMed] [Google Scholar]
- 95.Viswanathan M., Tsuchida N., Shanmugam G. Promoter hypermethylation profile of tumor-associated genes p16, p15, hMLH1, MGMT and E-cadherin in oral squamous cell carcinoma. Int. J. Cancer. 2003;105:41–46. doi: 10.1002/ijc.11028. [DOI] [PubMed] [Google Scholar]
- 96.Shaw R.J., Hall G.L., Lowe D., Liloglou T., Field J.K., Sloan P., Risk J.M. The role of pyrosequencing in head and neck cancer epigenetics: Correlation of quantitative methylation data with gene expression. Arch. Otolaryngol. Head Neck Surg. 2008;134:251–256. doi: 10.1001/archoto.2007.50. [DOI] [PubMed] [Google Scholar]
- 97.Hasegawa M., Nelson H.H., Peters E., Ringstrom E., Posner M., Kelsey K.T. Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene. 2002;21:4231–4236. doi: 10.1038/sj.onc.1205528. [DOI] [PubMed] [Google Scholar]
- 98.Dikshit R.P., Gillio-Tos A., Brennan P., De Marco L., Fiano V., Martinez-Peñuela J.M., Boffetta P., Merletti F. Hypermethylation, risk factors, clinical characteristics, and survival in 235 patients with laryngeal and hypopharyngeal cancers. Cancer. 2007;110:1745–1751. doi: 10.1002/cncr.22975. [DOI] [PubMed] [Google Scholar]
- 99.Don K.R., Ramani P., Ramshankar V., Sherlin H.J., Premkumar P., Natesan A. Promoter hypermethylation patterns of P16, DAPK and MGMT in oral squamous cell carcinoma: A systematic review and meta-analysis. Indian J. Dent. Res. 2014;25:797–805. doi: 10.4103/0970-9290.152208. [DOI] [PubMed] [Google Scholar]
- 100.Shaw R.J., Hobkirk A.J., Nikolaidis G., Woolgar J.A., Triantafyllou A., Brown J.S., Liloglou T., Risk J.M. Molecular staging of surgical margins in oral squamous cell carcinoma using promoter methylation of p16(INK4A), cytoglobin, E-cadherin, and TMEFF2. Ann. Surg. Oncol. 2013;20:2796–2802. doi: 10.1245/s10434-012-2713-8. [DOI] [PubMed] [Google Scholar]
- 101.Kulkarni V., Saranath D. Concurrent hypermethylation of multiple regulatory genes in chewing tobacco associated oral squamous cell carcinomas and adjacent normal tissues. Oral Oncol. 2004;40:145–153. doi: 10.1016/S1368-8375(03)00143-X. [DOI] [PubMed] [Google Scholar]
- 102.Rosas S.L., Koch W., da Costa Carvalho M.G., Wu L., Califano J., Westra W., Jen J., Sidransky D. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001;61:939–942. [PubMed] [Google Scholar]
- 103.Maruya S., Issa J.P., Weber R.S., Rosenthal D.I., Haviland J.C., Lotan R., El-Naggar A.K. Differential methylation status of tumor-associated genes in head and neck squamous carcinoma: Incidence and potential implications. Clin. Cancer Res. 2004;10:3825–3830. doi: 10.1158/1078-0432.CCR-03-0370. [DOI] [PubMed] [Google Scholar]
- 104.Ovchinnikov D.A., Cooper M.A., Pandit P., Coman W.B., Cooper-White J.J., Keith P., Wolvetang E.J., Slowey P.D., Punyadeera C. Tumor-suppressor Gene Promoter Hypermethylation in Saliva of Head and Neck Cancer Patients. Transl. Oncol. 2012;5:321–326. doi: 10.1593/tlo.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smigiel R., Sasiadek M., Krecicki T., Ramsey D., Jagielski J., Blin N. Inactivation of the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene in squamous cell carcinoma of the larynx. Mol. Carcinog. 2004;39:147–154. doi: 10.1002/mc.20007. [DOI] [PubMed] [Google Scholar]
- 106.Wang J., Chen H., Fu S., Xu Z.M., Sun K.L., Fu W.N. The involvement of CHD5 hypermethylation in laryngeal squamous cell carcinoma. Oral Oncol. 2011;47:601–608. doi: 10.1016/j.oraloncology.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 107.Liu Y., Zhou Z.T., He Q.B., Jiang W.W. DAPK promoter hypermethylation in tissues and body fluids of oral precancer patients. Med. Oncol. 2012;29:729–733. doi: 10.1007/s12032-011-9953-5. [DOI] [PubMed] [Google Scholar]
- 108.Kong W.J., Zhang S., Guo C., Wang Y., Zhang D. Methylation-associated silencing of death-associated protein kinase gene in laryngeal squamous cell cancer. Laryngoscope. 2005;115:1395–1401. doi: 10.1097/01.MLG.0000166708.23673.3A. [DOI] [PubMed] [Google Scholar]
- 109.Pannone G., Bufo P., Santoro A., Franco R., Aquino G., Longo F., Botti G., Serpico R., Cafarelli B., Abbruzzese A., et al. WNT pathway in oral cancer: Epigenetic inactivation of WNT-inhibitors. Oncol. Rep. 2010;24:1035–1041. doi: 10.3892/or.2010.1035. [DOI] [PubMed] [Google Scholar]
- 110.Hsiung D.T., Marsit C.J., Houseman E.A., Eddy K., Furniss C.S., McClean M.D., Kelsey K.T. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol. Biomark. Prev. 2007;16:108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 111.Puttipanyalears C., Subbalekha K., Mutirangura A., Kitkumthorn N. Alu hypomethylation in smoke-exposed epithelia and oral squamous carcinoma. Asian Pac. J. Cancer Prev. 2013;14:5495–5501. doi: 10.7314/APJCP.2013.14.9.5495. [DOI] [PubMed] [Google Scholar]
- 112.Guerrero-Preston R., Soudry E., Acero J., Orera M., Moreno-López L., Macía-Colón G., Jaffe A., Berdasco M., Ili-Gangas C., Brebi-Mieville P., et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev. Res. (Phila) 2011;4:1061–1072. doi: 10.1158/1940-6207.CAPR-11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jhavar S., Fink D., Rao A., Deb N. MGMT Hypermethylation—Missing Piece of the Puzzle in Primary Head and Neck Squamous Cell Carcinoma? Int. J. Radiat. Oncol. Biol. Phys. 2016;94:939. doi: 10.1016/j.ijrobp.2015.12.265. [DOI] [Google Scholar]
- 114.Ogi K., Toyota M., Ohe-Toyota M., Tanaka N., Noguchi M., Sonoda T., Kohama G., Tokino T. Aberrant methylation of multiple genes and clinicopathological features in oral squamous cell carcinoma. Clin. Cancer Res. 2002;8:3164–3171. [PubMed] [Google Scholar]
- 115.Shaw R. The epigenetics of oral cancer. Int. J. Oral Maxillofac. Surg. 2006;35:101–108. doi: 10.1016/j.ijom.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 116.Yeh K.T., Chang J.G., Lin T.H., Wang Y.F., Tien N., Chang J.Y., Chen J.C., Shih M.C. Epigenetic changes of tumor suppressor genes, P15, P16, VHL and P53 in oral cancer. Oncol. Rep. 2003;10:659–663. [PubMed] [Google Scholar]
- 117.Liyanage C., Wathupola A., Muraleetharan S., Perera K., Punyadeera C., Udagama P. Promoter Hypermethylation of Tumor-Suppressor Genes p16INK4a, RASSF1A, TIMP3, and PCQAP/MED15 in Salivary DNA as a Quadruple Biomarker Panel for Early Detection of Oral and Oropharyngeal Cancers. Biomolecules. 2019;9:148. doi: 10.3390/biom9040148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee E.S., Issa J.P., Roberts D.B., Williams M.D., Weber R.S., Kies M.S., El-Naggar A.K. Quantitative promoter hypermethylation analysis of cancer-related genes in salivary gland carcinomas: Comparison with methylation-specific PCR technique and clinical significance. Clin. Cancer Res. 2008;14:2664–2672. doi: 10.1158/1078-0432.CCR-07-1232. [DOI] [PubMed] [Google Scholar]
- 119.Supic G., Kozomara R., Jovic N., Zeljic K., Magic Z. Prognostic significance of tumor-related genes hypermethylation detected in cancer-free surgical margins of oral squamous cell carcinomas. Oral Oncol. 2011;47:702–708. doi: 10.1016/j.oraloncology.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 120.Shen Z., Chen X., Li Q., Zhou C., Li J., Ye H., Duan S. SSTR2 promoter hypermethylation is associated with the risk and progression of laryngeal squamous cell carcinoma in males. Diagn. Pathol. 2016;11:10. doi: 10.1186/s13000-016-0461-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gaykalova D.A., Vatapalli R., Wei Y., Tsai H.L., Wang H., Zhang C., Hennessey P.T., Guo T., Tan M., Li R., et al. Outlier Analysis Defines Zinc Finger Gene Family DNA Methylation in Tumors and Saliva of Head and Neck Cancer Patients. PLoS ONE. 2015;10:e0142148. doi: 10.1371/journal.pone.0142148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hu A., Huang J.J., Xu W.H., Jin X.J., Li J.P., Tang Y.J., Huang X.F., Cui H.J., Sun G.B. miR-21 and miR-375 microRNAs as candidate diagnostic biomarkers in squamous cell carcinoma of the larynx: Association with patient survival. Am. J. Transl. Res. 2014;6:604–613. [PMC free article] [PubMed] [Google Scholar]
- 123.Pedersen N.J., Jensen D.H., Lelkaitis G., Kiss K., Charabi B.W., Ullum H., Specht L., Schmidt A.Y., Nielsen F.C., von Buchwald C. MicroRNA-based classifiers for diagnosis of oral cavity squamous cell carcinoma in tissue and plasma. Oral Oncol. 2018;83:46–52. doi: 10.1016/j.oraloncology.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 124.Wang Y., Chen M., Tao Z., Hua Q., Chen S., Xiao B. Identification of predictive biomarkers for early diagnosis of larynx carcinoma based on microRNA expression data. Cancer Genet. 2013;206:340–346. doi: 10.1016/j.cancergen.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 125.Hayashi M., Wu G., Roh J.L., Chang X., Li X., Ahn J., Goldsmith M., Khan Z., Bishop J., Zhang Z., et al. Correlation of gene methylation in surgical margin imprints with locoregional recurrence in head and neck squamous cell carcinoma. Cancer. 2015;121:1957–1965. doi: 10.1002/cncr.29303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tan H.K., Saulnier P., Auperin A., Lacroix L., Casiraghi O., Janot F., Fouret P., Temam S. Quantitative methylation analyses of resection margins predict local recurrences and disease-specific deaths in patients with head and neck squamous cell carcinomas. Br. J. Cancer. 2008;99:357–363. doi: 10.1038/sj.bjc.6604478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hayashi M., Guerrero-Preston R., Sidransky D., Koch W.M. Paired box 5 methylation detection by droplet digital PCR for ultra-sensitive deep surgical margins analysis of head and neck squamous cell carcinoma. Cancer Prev. Res. (Phila) 2015;8:1017–1026. doi: 10.1158/1940-6207.CAPR-15-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ben-Dayan M.M., Ow T.J., Belbin T.J., Wetzler J., Smith R.V., Childs G., Diergaarde B., Hayes D.N., Grandis J.R., Prystowsky M.B., et al. Nonpromoter methylation of the CDKN2A gene with active transcription is associated with improved locoregional control in laryngeal squamous cell carcinoma. Cancer Med. 2017;6:397–407. doi: 10.1002/cam4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Harrandah A.M., Fitzpatrick S.G., Smith M.H., Wang D., Cohen D.M., Chan E.K. MicroRNA-375 as a biomarker for malignant transformation in oral lesions. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2016;122:743–752.e1. doi: 10.1016/j.oooo.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 130.Zhu Y., Cao X., Zhang X., Chen Q., Wen L., Wang P. DNA methylation-mediated Klotho silencing is an independent prognostic biomarker of head and neck squamous carcinoma. Cancer Manag. Res. 2019;11:1383–1390. doi: 10.2147/CMAR.S188415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huang K.H., Huang S.F., Chen I.H., Liao C.T., Wang H.M., Hsieh L.L. Methylation of RASSF1A, RASSF2A, and HIN-1 is associated with poor outcome after radiotherapy, but not surgery, in oral squamous cell carcinoma. Clin. Cancer Res. 2009;15:4174–4180. doi: 10.1158/1078-0432.CCR-08-2929. [DOI] [PubMed] [Google Scholar]
- 132.Kato K., Hara A., Kuno T., Mori H., Yamashita T., Toida M., Shibata T. Aberrant promoter hypermethylation of p16 and MGMT genes in oral squamous cell carcinomas and the surrounding normal mucosa. J. Cancer Res. Clin. Oncol. 2006;132:735–743. doi: 10.1007/s00432-006-0122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang S.H., Lee H.S., Mar K., Ji D.D., Huang M.S., Hsia K.T. Loss expression of O6-methylguanine DNA methyltransferase by promoter hypermethylation and its relationship to betel quid chewing in oral squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2010;109:883–889. doi: 10.1016/j.tripleo.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 134.Al-Kaabi A., van Bockel L.W., Pothen A.J., Willems S.M. p16INK4A and p14ARF gene promoter hypermethylation as prognostic biomarker in oral and oropharyngeal squamous cell carcinoma: A review. Dis. Markers. 2014;2014:260549. doi: 10.1155/2014/260549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Guerrero-Preston R., Lawson F., Rodriguez-Torres S., Noordhuis M.G., Pirini F., Manuel L., Valle B.L., Hadar T., Rivera B., Folawiyo O., et al. Variant, Immune Signatures, DNA Methylation, and Social Determinants Linked to Survival Racial Disparities in Head and Neck Cancer Patients. Cancer Prev. Res. (Phila) 2019;12:255–270. doi: 10.1158/1940-6207.CAPR-17-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rettori M.M., de Carvalho A.C., Bomfim Longo A.L., de Oliveira C.Z., Kowalski L.P., Carvalho A.L., Vettore A.L. Prognostic significance of TIMP3 hypermethylation in post-treatment salivary rinse from head and neck squamous cell carcinoma patients. Carcinogenesis. 2013;34:20–27. doi: 10.1093/carcin/bgs311. [DOI] [PubMed] [Google Scholar]
- 137.Schröck A., Leisse A., de Vos L., Gevensleben H., Dröge F., Franzen A., Wachendörfer M., Schröck F., Ellinger J., Teschke M., et al. Free-Circulating Methylated DNA in Blood for Diagnosis, Staging, Prognosis, and Monitoring of Head and Neck Squamous Cell Carcinoma Patients: An Observational Prospective Cohort Study. Clin. Chem. 2017;63:1288–1296. doi: 10.1373/clinchem.2016.270207. [DOI] [PubMed] [Google Scholar]
- 138.Gissi D.B., Fabbri V.P., Gabusi A., Lenzi J., Morandi L., Melotti S., Asioli S., Tarsitano A., Balbi T., Marchetti C., et al. Pre-Operative Evaluation of DNA Methylation Profile in Oral Squamous Cell Carcinoma Can Predict Tumor Aggressive Potential. Int. J. Mol. Sci. 2020;21:6691. doi: 10.3390/ijms21186691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Summerer I., Unger K., Braselmann H., Schuettrumpf L., Maihoefer C., Baumeister P., Kirchner T., Niyazi M., Sage E., Specht H.M., et al. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br. J. Cancer. 2015;113:76–82. doi: 10.1038/bjc.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Breitling L.P., Yang R., Korn B., Burwinkel B., Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am. J. Hum. Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Monick M.M., Beach S.R., Plume J., Sears R., Gerrard M., Brody G.H., Philibert R.A. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012;159B:141–151. doi: 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Joubert B.R., Håberg S.E., Nilsen R.M., Wang X., Vollset S.E., Murphy S.K., Huang Z., Hoyo C., Midttun Ø., Cupul-Uicab L.A., et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ. Health Perspect. 2012;120:1425–1431. doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Joehanes R., Just A.C., Marioni R.E., Pilling L.C., Reynolds L.M., Mandaviya P.R., Guan W., Xu T., Elks C.E., Aslibekyan S., et al. Epigenetic Signatures of Cigarette Smoking. Circ. Cardiovasc. Genet. 2016;9:436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Teschendorff A.E., Yang Z., Wong A., Pipinikas C.P., Jiao Y., Jones A., Anjum S., Hardy R., Salvesen H.B., Thirlwell C., et al. Correlation of Smoking-Associated DNA Methylation Changes in Buccal Cells With DNA Methylation Changes in Epithelial Cancer. JAMA Oncol. 2015;1:476–485. doi: 10.1001/jamaoncol.2015.1053. [DOI] [PubMed] [Google Scholar]
- 145.Langdon R., Richmond R., Elliott H.R., Dudding T., Kazmi N., Penfold C., Ingarfield K., Ho K., Bretherick A., Haley C., et al. Identifying epigenetic biomarkers of established prognostic factors and survival in a clinical cohort of individuals with oropharyngeal cancer. Clin. Epigenetics. 2020;12:95. doi: 10.1186/s13148-020-00870-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Langdon R.J., Beynon R.A., Ingarfield K., Marioni R.E., McCartney D.L., Martin R.M., Ness A.R., Pawlita M., Waterboer T., Relton C., et al. Epigenetic prediction of complex traits and mortality in a cohort of individuals with oropharyngeal cancer. Clin. Epigenetics. 2020;12:58. doi: 10.1186/s13148-020-00850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.von Zeidler S.V., Miracca E.C., Nagai M.A., Birman E.G. Hypermethylation of the p16 gene in normal oral mucosa of smokers. Int. J. Mol. Med. 2004;14:807–811. doi: 10.3892/ijmm.14.5.807. [DOI] [PubMed] [Google Scholar]
- 148.Bhat M.Y., Advani J., Rajagopalan P., Patel K., Nanjappa V., Solanki H.S., Patil A.H., Bhat F.A., Mathur P.P., Nair B., et al. Cigarette smoke and chewing tobacco alter expression of different sets of miRNAs in oral keratinocytes. Sci. Rep. 2018;8:7040. doi: 10.1038/s41598-018-25498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Krishnan A.R., Zheng H., Kwok J.G., Qu Y., Zou A.E., Korrapati A., Li P.X., Califano J.A., Hovell M.F., Wang-Rodriguez J., et al. A comprehensive study of smoking-specific microRNA alterations in head and neck squamous cell carcinoma. Oral Oncol. 2017;72:56–64. doi: 10.1016/j.oraloncology.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Doukas S.G., Vageli D.P., Lazopoulos G., Spandidos D.A., Sasaki C.T., Tsatsakis A. The Effect of NNK, A Tobacco Smoke Carcinogen, on the miRNA and Mismatch DNA Repair Expression Profiles in Lung and Head and Neck Squamous Cancer Cells. Cells. 2020;9:1031. doi: 10.3390/cells9041031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pereira C.S., Oliveira M.V., Barros L.O., Bandeira G.A., Santos S.H., Basile J.R., Guimarães A.L., De Paula A.M. Low expression of MSH2 DNA repair protein is associated with poor prognosis in head and neck squamous cell carcinoma. J. Appl. Oral Sci. 2013;21:416–421. doi: 10.1590/1679-775720130206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tamiya T., Kashiwagi I., Takahashi R., Yasukawa H., Yoshimura A. Suppressors of cytokine signaling (SOCS) proteins and JAK/STAT pathways: Regulation of T-cell inflammation by SOCS1 and SOCS3. Arter. Thromb. Vasc. Biol. 2011;31:980–985. doi: 10.1161/ATVBAHA.110.207464. [DOI] [PubMed] [Google Scholar]
- 153.Peng H.Y., Hsiao J.R., Chou S.T., Hsu Y.M., Wu G.H., Shieh Y.S., Shiah S.G. MiR-944/CISH mediated inflammation via STAT3 is involved in oral cancer malignance by cigarette smoking. Neoplasia. 2020;22:554–565. doi: 10.1016/j.neo.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shiah S.G., Hsiao J.R., Chang H.J., Hsu Y.M., Wu G.H., Peng H.Y., Chou S.T., Kuo C.C., Chang J.Y. MiR-30a and miR-379 modulate retinoic acid pathway by targeting DNA methyltransferase 3B in oral cancer. J. Biomed. Sci. 2020;27:46. doi: 10.1186/s12929-020-00644-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nabel C.S., Manning S.A., Kohli R.M. The curious chemical biology of cytosine: Deamination, methylation, and oxidation as modulators of genomic potential. ACS Chem. Biol. 2012;7:20–30. doi: 10.1021/cb2002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Camuzi D., de Amorim Í., Ribeiro Pinto L.F., Oliveira Trivilin L., Mencalha A.L., Soares Lima S.C. Regulation Is in the Air: The Relationship between Hypoxia and Epigenetics in Cancer. Cells. 2019;8:300. doi: 10.3390/cells8040300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lu S.C., Huang Z.Z., Yang H., Mato J.M., Avila M.A., Tsukamoto H. Changes in methionine adenosyltransferase and S-adenosylmethionine homeostasis in alcoholic rat liver. Am. J. Physiol. Gastrointest Liver Physiol. 2000;279:G178–G185. doi: 10.1152/ajpgi.2000.279.1.G178. [DOI] [PubMed] [Google Scholar]
- 158.Kawakita D., Lee Y.A., Gren L.H., Buys S.S., La Vecchia C., Hashibe M. The impact of folate intake on the risk of head and neck cancer in the prostate, lung, colorectal, and ovarian cancer screening trial (PLCO) cohort. Br. J. Cancer. 2018;118:299–306. doi: 10.1038/bjc.2017.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zakhari S. Alcohol metabolism and epigenetics changes. Alcohol. Res. 2013;35:6–16. [PMC free article] [PubMed] [Google Scholar]
- 160.Moody L., Crowder S.L., Fruge A.D., Locher J.L., Demark-Wahnefried W., Rogers L.Q., Delk-Licata A., Carroll W.R., Spencer S.A., Black M., et al. Epigenetic stratification of head and neck cancer survivors reveals differences in lycopene levels, alcohol consumption, and methylation of immune regulatory genes. Clin. Epigenetics. 2020;12:138. doi: 10.1186/s13148-020-00930-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yan Y., Yang J.Y., Mou Y.H., Wang L.H., Zhang H., Wu C.F. Possible metabolic pathways of ethanol responsible for oxidative DNA damage in human peripheral lymphocytes. Alcohol. Clin. Exp. Res. 2011;35:1–9. doi: 10.1111/j.1530-0277.2010.01316.x. [DOI] [PubMed] [Google Scholar]
- 162.Zhou Z., Liu C., Liu K., Lv M., Li B., Lan Z., Chen W., Kang M. Expression and Possible Molecular Mechanisms of microRNA-205-5p in Patients With Head and Neck Squamous Cell Carcinoma. Technol. Cancer Res. Treat. 2020;19:1533033820980110. doi: 10.1177/1533033820980110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Danquah K.O., Gyamfi D. Molecular Aspects of Alcohol and Nutrition. Elsevier; Amsterdam, The Netherlands: 2016. Alcohol and aldehyde dehydrogenases: Molecular aspects; pp. 25–43. [Google Scholar]
- 164.Seidensaal K., Nollert A., Feige A.H., Muller M., Fleming T., Gunkel N., Zaoui K., Grabe N., Weichert W., Weber K.J., et al. Impaired aldehyde dehydrogenase 1 subfamily member 2A-dependent retinoic acid signaling is related with a mesenchymal-like phenotype and an unfavorable prognosis of head and neck squamous cell carcinoma. Mol. Cancer. 2015;14:204. doi: 10.1186/s12943-015-0476-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kinoshita T., Hanazawa T., Nohata N., Okamoto Y., Seki N. The functional significance of microRNA-375 in human squamous cell carcinoma: Aberrant expression and effects on cancer pathways. J. Hum. Genet. 2012;57:556–563. doi: 10.1038/jhg.2012.75. [DOI] [PubMed] [Google Scholar]
- 166.Nohata N., Hanazawa T., Kikkawa N., Mutallip M., Sakurai D., Fujimura L., Kawakami K., Chiyomaru T., Yoshino H., Enokida H., et al. Tumor suppressive microRNA-375 regulates oncogene AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC) J. Hum. Genet. 2011;56:595–601. doi: 10.1038/jhg.2011.66. [DOI] [PubMed] [Google Scholar]
- 167.Kinoshita T., Nohata N., Yoshino H., Hanazawa T., Kikkawa N., Fujimura L., Chiyomaru T., Kawakami K., Enokida H., Nakagawa M., et al. Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in maxillary sinus squamous cell carcinoma. Int. J. Oncol. 2012;40:185–193. doi: 10.3892/ijo.2011.1196. [DOI] [PubMed] [Google Scholar]
- 168.Harris T., Jimenez L., Kawachi N., Fan J.B., Chen J., Belbin T., Ramnauth A., Loudig O., Keller C.E., Smith R., et al. Low-level expression of miR-375 correlates with poor outcome and metastasis while altering the invasive properties of head and neck squamous cell carcinomas. Am. J. Pathol. 2012;180:917–928. doi: 10.1016/j.ajpath.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Leonard S.M., Wei W., Collins S.I., Pereira M., Diyaf A., Constandinou-Williams C., Young L.S., Roberts S., Woodman C.B. Oncogenic human papillomavirus imposes an instructive pattern of DNA methylation changes which parallel the natural history of cervical HPV infection in young women. Carcinogenesis. 2012;33:1286–1293. doi: 10.1093/carcin/bgs157. [DOI] [PubMed] [Google Scholar]
- 170.Au Yeung C.L., Tsang W.P., Tsang T.Y., Co N.N., Yau P.L., Kwok T.T. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol. Rep. 2010;24:1599–1604. doi: 10.3892/or_00001023. [DOI] [PubMed] [Google Scholar]
- 171.McCabe M.T., Davis J.N., Day M.L. Regulation of DNA methyltransferase 1 by the pRb/E2F1 pathway. Cancer Res. 2005;65:3624–3632. doi: 10.1158/0008-5472.CAN-04-2158. [DOI] [PubMed] [Google Scholar]
- 172.Burgers W.A., Blanchon L., Pradhan S., de Launoit Y., Kouzarides T., Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26:1650–1655. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Laurson J., Khan S., Chung R., Cross K., Raj K. Epigenetic repression of E-cadherin by human papillomavirus 16 E7 protein. Carcinogenesis. 2010;31:918–926. doi: 10.1093/carcin/bgq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ekanayake Weeramange C., Tang K.D., Vasani S., Langton-Lockton J., Kenny L., Punyadeera C. DNA Methylation Changes in Human Papillomavirus-Driven Head and Neck Cancers. Cells. 2020;9:1359. doi: 10.3390/cells9061359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Degli Esposti D., Sklias A., Lima S.C., Beghelli-de la Forest Divonne S., Cahais V., Fernandez-Jimenez N., Cros M.P., Ecsedi S., Cuenin C., Bouaoun L., et al. Unique DNA methylation signature in HPV-positive head and neck squamous cell carcinomas. Genome Med. 2017;9:33. doi: 10.1186/s13073-017-0419-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sartor M.A., Dolinoy D.C., Jones T.R., Colacino J.A., Prince M.E., Carey T.E., Rozek L.S. Genome-wide methylation and expression differences in HPV (+) and HPV (-) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics. 2011;6:777–787. doi: 10.4161/epi.6.6.16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kostareli E., Holzinger D., Bogatyrova O., Hielscher T., Wichmann G., Keck M., Lahrmann B., Grabe N., Flechtenmacher C., Schmidt C.R., et al. HPV-related methylation signature predicts survival in oropharyngeal squamous cell carcinomas. J. Clin. Investig. 2013;123:2488–2501. doi: 10.1172/JCI67010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Allameh A., Moazeni-Roodi A., Harirchi I., Ravanshad M., Motiee-Langroudi M., Garajei A., Hamidavi A., Mesbah-Namin S.A. Promoter DNA Methylation and mRNA Expression Level of p16 Gene in Oral Squamous Cell Carcinoma: Correlation with Clinicopathological Characteristics. Pathol. Oncol. Res. 2019;25:1535–1543. doi: 10.1007/s12253-018-0542-1. [DOI] [PubMed] [Google Scholar]
- 179.Richards K.L., Zhang B., Baggerly K.A., Colella S., Lang J.C., Schuller D.E., Krahe R. Genome-wide hypomethylation in head and neck cancer is more pronounced in HPV-negative tumors and is associated with genomic instability. PLoS ONE. 2009;4:e4941. doi: 10.1371/journal.pone.0004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Poage G.M., Houseman E.A., Christensen B.C., Butler R.A., Avissar-Whiting M., McClean M.D., Waterboer T., Pawlita M., Marsit C.J., Kelsey K.T. Global hypomethylation identifies Loci targeted for hypermethylation in head and neck cancer. Clin. Cancer Res. 2011;17:3579–3589. doi: 10.1158/1078-0432.CCR-11-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Furniss C.S., Marsit C.J., Houseman E.A., Eddy K., Kelsey K.T. Line region hypomethylation is associated with lifestyle and differs by human papillomavirus status in head and neck squamous cell carcinomas. Cancer Epidemiol. Biomark. Prev. 2008;17:966–971. doi: 10.1158/1055-9965.EPI-07-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Furlan C., Polesel J., Barzan L., Franchin G., Sulfaro S., Romeo S., Colizzi F., Rizzo A., Baggio V., Giacomarra V., et al. Prognostic significance of LINE-1 hypomethylation in oropharyngeal squamous cell carcinoma. Clin. Epigenetics. 2017;9:58. doi: 10.1186/s13148-017-0357-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Camuzi D., Buexm L.A., Lourenço S.Q.C., Esposti D.D., Cuenin C., Lopes M.S.A., Manara F., Talukdar F.R., Herceg Z., Ribeiro Pinto L.F., et al. HPV Infection Leaves a DNA Methylation Signature in Oropharyngeal Cancer Affecting Both Coding Genes and Transposable Elements. Cancers. 2021;13:3621. doi: 10.3390/cancers13143621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sannigrahi M.K., Sharma R., Singh V., Panda N.K., Rattan V., Khullar M. Role of Host miRNA Hsa-miR-139-3p in HPV-16-Induced Carcinomas. Clin. Cancer Res. 2017;23:3884–3895. doi: 10.1158/1078-0432.CCR-16-2936. [DOI] [PubMed] [Google Scholar]
- 185.Morel A., Baguet A., Perrard J., Demeret C., Jacquin E., Guenat D., Mougin C., Prétet J.L. 5azadC treatment upregulates miR-375 level and represses HPV16 E6 expression. Oncotarget. 2017;8:46163–46176. doi: 10.18632/oncotarget.17575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Jung H.M., Phillips B.L., Chan E.K. miR-375 activates p21 and suppresses telomerase activity by coordinately regulating HPV E6/E7, E6AP, CIP2A, and 14-3-3ζ. Mol. Cancer. 2014;13:80. doi: 10.1186/1476-4598-13-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Melar-New M., Laimins L.A. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J. Virol. 2010;84:5212–5221. doi: 10.1128/JVI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Snoek B.C., Babion I., Koppers-Lalic D., Pegtel D.M., Steenbergen R.D. Altered microRNA processing proteins in HPV-induced cancers. Curr. Opin. Virol. 2019;39:23–32. doi: 10.1016/j.coviro.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 189.Luo X.J., Zheng M., Cao M.X., Zhang W.L., Huang M.C., Dai L., Tang Y.L., Liang X.H. Distinguishable Prognostic miRNA Signatures of Head and Neck Squamous Cell Cancer With or Without HPV Infection. Front. Oncol. 2020;10:614487. doi: 10.3389/fonc.2020.614487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lajer C.B., Garnæs E., Friis-Hansen L., Norrild B., Therkildsen M.H., Glud M., Rossing M., Lajer H., Svane D., Skotte L., et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: Bridging between HPV-related head and neck cancer and cervical cancer. Br. J. Cancer. 2012;106:1526–1534. doi: 10.1038/bjc.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wald A.I., Hoskins E.E., Wells S.I., Ferris R.L., Khan S.A. Alteration of microRNA profiles in squamous cell carcinoma of the head and neck cell lines by human papillomavirus. Head Neck. 2011;33:504–512. doi: 10.1002/hed.21475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ludwig S., Sharma P., Wise P., Sposto R., Hollingshead D., Lamb J., Lang S., Fabbri M., Whiteside T.L. mRNA and miRNA Profiles of Exosomes from Cultured Tumor Cells Reveal Biomarkers Specific for HPV16-Positive and HPV16-Negative Head and Neck Cancer. Int. J. Mol. Sci. 2020;21:8570. doi: 10.3390/ijms21228570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wan Y., Vagenas D., Salazar C., Kenny L., Perry C., Calvopiña D., Punyadeera C. Salivary miRNA panel to detect HPV-positive and HPV-negative head and neck cancer patients. Oncotarget. 2017;8:99990–100001. doi: 10.18632/oncotarget.21725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rock L.D., Minatel B.C., Marshall E.A., Guisier F., Sage A.P., Barros-Filho M.C., Stewart G.L., Garnis C., Lam W.L. Expanding the Transcriptome of Head and Neck Squamous Cell Carcinoma Through Novel MicroRNA Discovery. Front. Oncol. 2019;9:1305. doi: 10.3389/fonc.2019.01305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sun Y.W., Chen K.M., Imamura Kawasawa Y., Salzberg A.C., Cooper T.K., Caruso C., Aliaga C., Zhu J., Gowda K., Amin S., et al. Hypomethylated Fgf3 is a potential biomarker for early detection of oral cancer in mice treated with the tobacco carcinogen dibenzo[def,p]chrysene. PLoS ONE. 2017;12:e0186873. doi: 10.1371/journal.pone.0186873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hecht S.S. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat. Rev. Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 197.Seitz H.K., Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer. 2007;7:599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- 198.Feil R., Fraga M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 199.Zheng S.C., Widschwendter M., Teschendorff A.E. Epigenetic drift, epigenetic clocks and cancer risk. Epigenomics. 2016;8:705–719. doi: 10.2217/epi-2015-0017. [DOI] [PubMed] [Google Scholar]
- 200.Takeshima H., Ushijima T. Accumulation of genetic and epigenetic alterations in normal cells and cancer risk. NPJ Precis. Oncol. 2019;3:7. doi: 10.1038/s41698-019-0079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hattori N., Ushijima T. Epigenetic impact of infection on carcinogenesis: Mechanisms and applications. Genome Med. 2016;8:10. doi: 10.1186/s13073-016-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hoadley K.A., Yau C., Hinoue T., Wolf D.M., Lazar A.J., Drill E., Shen R., Taylor A.M., Cherniack A.D., Thorsson V., et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 2018;173:291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Asada K., Nakajima T., Shimazu T., Yamamichi N., Maekita T., Yokoi C., Oda I., Ando T., Yoshida T., Nanjo S., et al. Demonstration of the usefulness of epigenetic cancer risk prediction by a multicentre prospective cohort study. Gut. 2015;64:388–396. doi: 10.1136/gutjnl-2014-307094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Deng D., Liu Z., Du Y. Epigenetic alterations as cancer diagnostic, prognostic, and predictive biomarkers. Adv. Genet. 2010;71:125–176. doi: 10.1016/B978-0-12-380864-6.00005-5. [DOI] [PubMed] [Google Scholar]
- 205.Tian D., Feng Z., Hanley N.M., Setzer R.W., Mumford J.L., DeMarini D.M. Multifocal accumulation of p53 protein in esophageal carcinoma: Evidence for field cancerization. Int. J. Cancer. 1998;78:568–575. doi: 10.1002/(SICI)1097-0215(19981123)78:5<568::AID-IJC7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 206.Lavieille J.P., Gazzeri S., Riva C., Reyt E., Brambilla C., Brambilla E. p53 mutations and p53, Waf-1, Bax and Bcl-2 expression in field cancerization of the head and neck. Anticancer Res. 1998;18:4741–4749. [PubMed] [Google Scholar]
- 207.Kanjilal S., Strom S.S., Clayman G.L., Weber R.S., el-Naggar A.K., Kapur V., Cummings K.K., Hill L.A., Spitz M.R., Kripke M.L. p53 mutations in nonmelanoma skin cancer of the head and neck: Molecular evidence for field cancerization. Cancer Res. 1995;55:3604–3609. [PubMed] [Google Scholar]
- 208.el-Naggar A.K., Lai S., Luna M.A., Zhou X.D., Weber R.S., Goepfert H., Batsakis J.G. Sequential p53 mutation analysis of pre-invasive and invasive head and neck squamous carcinoma. Int. J. Cancer. 1995;64:196–201. doi: 10.1002/ijc.2910640309. [DOI] [PubMed] [Google Scholar]
- 209.van Houten V.M., Tabor M.P., van den Brekel M.W., Kummer J.A., Denkers F., Dijkstra J., Leemans R., van der Waal I., Snow G.B., Brakenhoff R.H. Mutated p53 as a molecular marker for the diagnosis of head and neck cancer. J. Pathol. 2002;198:476–486. doi: 10.1002/path.1242. [DOI] [PubMed] [Google Scholar]
- 210.Argiris A., Karamouzis M.V., Raben D., Ferris R.L. Head and neck cancer. Lancet. 2008;371:1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zhou G., Liu Z., Myers J.N. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. J. Cell Biochem. 2016;117:2682–2692. doi: 10.1002/jcb.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lee Y.C., Wang H.P., Wang C.P., Ko J.Y., Lee J.M., Chiu H.M., Lin J.T., Yamashita S., Oka D., Watanabe N., et al. Revisit of field cancerization in squamous cell carcinoma of upper aerodigestive tract: Better risk assessment with epigenetic markers. Cancer Prev. Res. (Phila) 2011;4:1982–1992. doi: 10.1158/1940-6207.CAPR-11-0096. [DOI] [PubMed] [Google Scholar]
- 213.Lima S.C., Hernández-Vargas H., Simão T., Durand G., Kruel C.D., Le Calvez-Kelm F., Ribeiro Pinto L.F., Herceg Z. Identification of a DNA methylome signature of esophageal squamous cell carcinoma and potential epigenetic biomarkers. Epigenetics. 2011;6:1217–1227. doi: 10.4161/epi.6.10.17199. [DOI] [PubMed] [Google Scholar]
- 214.Riese U., Dahse R., Fiedler W., Theuer C., Koscielny S., Ernst G., Beleites E., Claussen U., von Eggeling F. Tumor suppressor gene p16 (CDKN2A) mutation status and promoter inactivation in head and neck cancer. Int. J. Mol. Med. 1999;4:61–65. doi: 10.3892/ijmm.4.1.61. [DOI] [PubMed] [Google Scholar]
- 215.Dahl C., Guldberg P. DNA methylation analysis techniques. Biogerontology. 2003;4:233–250. doi: 10.1023/A:1025103319328. [DOI] [PubMed] [Google Scholar]
- 216.Schlecht N.F., Ben-Dayan M., Anayannis N., Lleras R.A., Thomas C., Wang Y., Smith R.V., Burk R.D., Harris T.M., Childs G., et al. Epigenetic changes in the CDKN2A locus are associated with differential expression of P16INK4A and P14ARF in HPV-positive oropharyngeal squamous cell carcinoma. Cancer Med. 2015;4:342–353. doi: 10.1002/cam4.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ben-Dayan M.M., Li D., Kawachi N., Ow T.J., Belbin T.J., Childs G., Schletch N.F., Prystowsky M.B. HPV-16 E6 and E7 expression drives transcription of p14 (ARF) and p16 (INK4a) prior to Intragenic methylation of CDKN2A. Med. Res. Arch. 2020;8:8. doi: 10.18103/mra.v8i5.2113. [DOI] [Google Scholar]
- 218.Takeshima M., Saitoh M., Kusano K., Nagayasu H., Kurashige Y., Malsantha M., Arakawa T., Takuma T., Chiba I., Kaku T., et al. High frequency of hypermethylation of p14, p15 and p16 in oral pre-cancerous lesions associated with betel-quid chewing in Sri Lanka. J. Oral Pathol. Med. 2008;37:475–479. doi: 10.1111/j.1600-0714.2008.00644.x. [DOI] [PubMed] [Google Scholar]
- 219.Díez-Pérez R., Campo-Trapero J., Cano-Sánchez J., López-Durán M., Gonzalez-Moles M.A., Bascones-Ilundain J., Bascones-Martinez A. Methylation in oral cancer and pre-cancerous lesions (Review) Oncol. Rep. 2011;25:1203–1209. doi: 10.3892/or.2011.1205. [DOI] [PubMed] [Google Scholar]
- 220.Mohan M., Jagannathan N. Oral field cancerization: An update on current concepts. Oncol. Rev. 2014;8:244. doi: 10.4081/oncol.2014.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kong L.Y., Du W., Wang L., Yang Z., Zhang H.S. PAX1 Methylation Hallmarks Promising Accuracy for Cervical Cancer Screening in Asians: Results from a Meta-Analysis. Clin. Lab. 2015;61:1471–1479. doi: 10.7754/Clin.Lab.2015.150232. [DOI] [PubMed] [Google Scholar]
- 222.Chao T.K., Ke F.Y., Liao Y.P., Wang H.C., Yu C.P., Lai H.C. Triage of cervical cytological diagnoses of atypical squamous cells by DNA methylation of paired boxed gene 1 (PAX1) Diagn. Cytopathol. 2013;41:41–46. doi: 10.1002/dc.21758. [DOI] [PubMed] [Google Scholar]
- 223.Lin H., Chen T.C., Chang T.C., Cheng Y.M., Chen C.H., Chu T.Y., Hsu S.T., Liu C.B., Yeh L.S., Wen K.C., et al. Methylated ZNF582 gene as a marker for triage of women with Pap smear reporting low-grade squamous intraepithelial lesions—A Taiwanese Gynecologic Oncology Group (TGOG) study. Gynecol. Oncol. 2014;135:64–68. doi: 10.1016/j.ygyno.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 224.Towle R., Truong D., Hogg K., Robinson W.P., Poh C.F., Garnis C. Global analysis of DNA methylation changes during progression of oral cancer. Oral Oncol. 2013;49:1033–1042. doi: 10.1016/j.oraloncology.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 225.Roy R., Singh R., Chattopadhyay E., Ray A., Sarkar N., Aich R., Paul R.R., Pal M., Roy B. MicroRNA and target gene expression based clustering of oral cancer, precancer and normal tissues. Gene. 2016;593:58–63. doi: 10.1016/j.gene.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 226.Leemans C.R., Tiwari R., Nauta J.J., van der Waal I., Snow G.B. Recurrence at the primary site in head and neck cancer and the significance of neck lymph node metastases as a prognostic factor. Cancer. 1994;73:187–190. doi: 10.1002/1097-0142(19940101)73:1<187::AID-CNCR2820730132>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 227.Huang T.Y., Hsu L.P., Wen Y.H., Huang T.T., Chou Y.F., Lee C.F., Yang M.C., Chang Y.K., Chen P.R. Predictors of locoregional recurrence in early stage oral cavity cancer with free surgical margins. Oral Oncol. 2010;46:49–55. doi: 10.1016/j.oraloncology.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 228.Yanamoto S., Yamada S., Takahashi H., Yoshitomi I., Kawasaki G., Ikeda H., Minamizato T., Shiraishi T., Fujita S., Ikeda T., et al. Clinicopathological risk factors for local recurrence in oral squamous cell carcinoma. Int. J. Oral Maxillofac. Surg. 2012;41:1195–1200. doi: 10.1016/j.ijom.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 229.Thomas Robbins K., Triantafyllou A., Suárez C., López F., Hunt J.L., Strojan P., Williams M.D., Braakhuis B.J.M., de Bree R., Hinni M.L., et al. Surgical margins in head and neck cancer: Intra- and postoperative considerations. Auris Nasus Larynx. 2019;46:10–17. doi: 10.1016/j.anl.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 230.Graveland A.P., Golusinski P.J., Buijze M., Douma R., Sons N., Kuik D.J., Bloemena E., Leemans C.R., Brakenhoff R.H., Braakhuis B.J. Loss of heterozygosity at 9p and p53 immunopositivity in surgical margins predict local relapse in head and neck squamous cell carcinoma. Int. J. Cancer. 2011;128:1852–1859. doi: 10.1002/ijc.25523. [DOI] [PubMed] [Google Scholar]
- 231.van Houten V.M., Leemans C.R., Kummer J.A., Dijkstra J., Kuik D.J., van den Brekel M.W., Snow G.B., Brakenhoff R.H. Molecular diagnosis of surgical margins and local recurrence in head and neck cancer patients: A prospective study. Clin. Cancer Res. 2004;10:3614–3620. doi: 10.1158/1078-0432.CCR-03-0631. [DOI] [PubMed] [Google Scholar]
- 232.Goldenberg D., Harden S., Masayesva B.G., Ha P., Benoit N., Westra W.H., Koch W.M., Sidransky D., Califano J.A. Intraoperative molecular margin analysis in head and neck cancer. Arch. Otolaryngol. Head Neck Surg. 2004;130:39–44. doi: 10.1001/archotol.130.1.39. [DOI] [PubMed] [Google Scholar]
- 233.Sorroche B.P., Talukdar F.R., Lima S.C.S., Melendez M.E., de Carvalho A.C., de Almeida G.C., De Marchi P., Lopes M., Ribeiro Pinto L.F., Carvalho A.L., et al. DNA Methylation Markers from Negative Surgical Margins Can Predict Recurrence of Oral Squamous Cell Carcinoma. Cancers. 2021;13:2915. doi: 10.3390/cancers13122915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Virani S., Light E., Peterson L.A., Sartor M.A., Taylor J.M., McHugh J.B., Wolf G.T., Rozek L.S., Investigators of the University of Michigan Head and Neck Specialized Programs of Research Excellence (SPORE) Program Stability of methylation markers in head and neck squamous cell carcinoma. Head Neck. 2016;38((Suppl. 1)):E1325–E1331. doi: 10.1002/hed.24223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Puri S.K., Si L., Fan C.Y., Hanna E. Aberrant promoter hypermethylation of multiple genes in head and neck squamous cell carcinoma. Am. J. Otolaryngol. 2005;26:12–17. doi: 10.1016/j.amjoto.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 236.López F., Sampedro T., Llorente J.L., Hermsen M., Álvarez-Marcos C. Alterations of p14 ARF, p15 INK4b, and p16 INK4a genes in primary laryngeal squamous cell carcinoma. Pathol. Oncol. Res. 2017;23:63–71. doi: 10.1007/s12253-016-0083-4. [DOI] [PubMed] [Google Scholar]
- 237.Zuo C., Ai L., Ratliff P., Suen J.Y., Hanna E., Brent T.P., Fan C.Y. O6-methylguanine-DNA methyltransferase gene: Epigenetic silencing and prognostic value in head and neck squamous cell carcinoma. Cancer Epidemiol. Biomark. Prev. 2004;13:967–975. [PubMed] [Google Scholar]
- 238.Wang K., Ling T., Wu H., Zhang J. Screening of candidate tumor-suppressor genes in 3p21.3 and investigation of the methylation of gene promoters in oral squamous cell carcinoma. Oncol. Rep. 2013;29:1175–1182. doi: 10.3892/or.2012.2213. [DOI] [PubMed] [Google Scholar]
- 239.Zhou C., Ye M., Ni S., Li Q., Ye D., Li J., Shen Z., Deng H. DNA methylation biomarkers for head and neck squamous cell carcinoma. Epigenetics. 2018;13:398–409. doi: 10.1080/15592294.2018.1465790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Koshizuka K., Hanazawa T., Fukumoto I., Kikkawa N., Okamoto Y., Seki N. The microRNA signatures: Aberrantly expressed microRNAs in head and neck squamous cell carcinoma. J. Hum. Genet. 2017;62:3–13. doi: 10.1038/jhg.2016.105. [DOI] [PubMed] [Google Scholar]
- 241.Zhang Y., Chen Y., Yu J., Liu G., Huang Z. Integrated transcriptome analysis reveals miRNA-mRNA crosstalk in laryngeal squamous cell carcinoma. Genomics. 2014;104:249–256. doi: 10.1016/j.ygeno.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 242.Cao P., Zhou L., Zhang J., Zheng F., Wang H., Ma D., Tian J. Comprehensive expression profiling of microRNAs in laryngeal squamous cell carcinoma. Head Neck. 2013;35:720–728. doi: 10.1002/hed.23011. [DOI] [PubMed] [Google Scholar]
- 243.Ayaz L., Görür A., Yaroğlu H.Y., Ozcan C., Tamer L. Differential expression of microRNAs in plasma of patients with laryngeal squamous cell carcinoma: Potential early-detection markers for laryngeal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2013;139:1499–1506. doi: 10.1007/s00432-013-1469-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sun X., Song Y., Tai X., Liu B., Ji W. MicroRNA expression and its detection in human supraglottic laryngeal squamous cell carcinoma. Biomed. Rep. 2013;1:743–746. doi: 10.3892/br.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wang F., Lu J., Peng X., Wang J., Liu X., Chen X., Jiang Y., Li X., Zhang B. Integrated analysis of microRNA regulatory network in nasopharyngeal carcinoma with deep sequencing. J. Exp. Clin. Cancer Res. 2016;35:17. doi: 10.1186/s13046-016-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kikkawa N., Hanazawa T., Fujimura L., Nohata N., Suzuki H., Chazono H., Sakurai D., Horiguchi S., Okamoto Y., Seki N. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) Br. J. Cancer. 2010;103:877–884. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Fukumoto I., Kinoshita T., Hanazawa T., Kikkawa N., Chiyomaru T., Enokida H., Yamamoto N., Goto Y., Nishikawa R., Nakagawa M., et al. Identification of tumour suppressive microRNA-451a in hypopharyngeal squamous cell carcinoma based on microRNA expression signature. Br. J. Cancer. 2014;111:386–394. doi: 10.1038/bjc.2014.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lajer C.B., Nielsen F.C., Friis-Hansen L., Norrild B., Borup R., Garnæs E., Rossing M., Specht L., Therkildsen M.H., Nauntofte B., et al. Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: A prospective translational study. Br. J. Cancer. 2011;104:830–840. doi: 10.1038/bjc.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Severino P., Brüggemann H., Andreghetto F.M., Camps C., Klingbeil M.e.F., de Pereira W.O., Soares R.M., Moyses R., Wünsch-Filho V., Mathor M.B., et al. MicroRNA expression profile in head and neck cancer: HOX-cluster embedded microRNA-196a and microRNA-10b dysregulation implicated in cell proliferation. BMC Cancer. 2013;13:533. doi: 10.1186/1471-2407-13-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Fukumoto I., Hanazawa T., Kinoshita T., Kikkawa N., Koshizuka K., Goto Y., Nishikawa R., Chiyomaru T., Enokida H., Nakagawa M., et al. MicroRNA expression signature of oral squamous cell carcinoma: Functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br. J. Cancer. 2015;112:891–900. doi: 10.1038/bjc.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Schneider A., Victoria B., Lopez Y.N., Suchorska W., Barczak W., Sobecka A., Golusinski W., Masternak M.M., Golusinski P. Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Sci. Rep. 2018;8:675. doi: 10.1038/s41598-017-18945-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hui A.B., Lenarduzzi M., Krushel T., Waldron L., Pintilie M., Shi W., Perez-Ordonez B., Jurisica I., O’Sullivan B., Waldron J., et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin. Cancer Res. 2010;16:1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 253.Liu C.J., Tsai M.M., Hung P.S., Kao S.Y., Liu T.Y., Wu K.J., Chiou S.H., Lin S.C., Chang K.W. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010;70:1635–1644. doi: 10.1158/0008-5472.CAN-09-2291. [DOI] [PubMed] [Google Scholar]
- 254.Victoria Martinez B., Dhahbi J.M., Nunez Lopez Y.O., Lamperska K., Golusinski P., Luczewski L., Kolenda T., Atamna H., Spindler S.R., Golusinski W., et al. Circulating small non-coding RNA signature in head and neck squamous cell carcinoma. Oncotarget. 2015;6:19246–19263. doi: 10.18632/oncotarget.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.León X., Quer M., Diez S., Orús C., López-Pousa A., Burgués J. Second neoplasm in patients with head and neck cancer. Head Neck. 1999;21:204–210. doi: 10.1002/(SICI)1097-0347(199905)21:3<204::AID-HED4>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 256.Priante A.V., Carvalho A.L., Kowalski L.P. Second primary tumor in patients with upper aerodigestive tract cancer. Braz. J. Otorhinolaryngol. 2010;76:251–256. doi: 10.1590/S1808-86942010000200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Morris L.G., Sikora A.G., Patel S.G., Hayes R.B., Ganly I. Second primary cancers after an index head and neck cancer: Subsite-specific trends in the era of human papillomavirus-associated oropharyngeal cancer. J. Clin. Oncol. 2011;29:739–746. doi: 10.1200/JCO.2010.31.8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ji X., Guan C., Jiang X., Li H. Diagnostic accuracy of DNA methylation for head and neck cancer varies by sample type and number of markers tested. Oncotarget. 2016;7:80019–80032. doi: 10.18632/oncotarget.12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Ogino S., Nosho K., Kirkner G.J., Kawasaki T., Chan A.T., Schernhammer E.S., Giovannucci E.L., Fuchs C.S. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J. Natl. Cancer. Inst. 2008;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Pattamadilok J., Huapai N., Rattanatanyong P., Vasurattana A., Triratanachat S., Tresukosol D., Mutirangura A. LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int. J. Gynecol. Cancer. 2008;18:711–717. doi: 10.1111/j.1525-1438.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- 261.Imperatori A., Sahnane N., Rotolo N., Franzi F., Nardecchia E., Libera L., Romualdi C., Cattoni M., Sessa F., Dominioni L., et al. LINE-1 hypomethylation is associated to specific clinico-pathological features in Stage I non-small cell lung cancer. Lung Cancer. 2017;108:83–89. doi: 10.1016/j.lungcan.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 262.Hong J.H., Jin E.H., Kim S., Song K.S., Sung J.K. LINE-1 hypomethylation is inversely correlated with UHRF1 overexpression in gastric cancer. Oncol. Lett. 2018;15:6666–6670. doi: 10.3892/ol.2018.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Hossain K., Suzuki T., Hasibuzzaman M.M., Islam M.S., Rahman A., Paul S.K., Tanu T., Hossain S., Saud Z.A., Rahman M., et al. Chronic exposure to arsenic, LINE-1 hypomethylation, and blood pressure: A cross-sectional study in Bangladesh. Environ. Health. 2017;16:20. doi: 10.1186/s12940-017-0231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Li S., Yang Q., Hou Y., Jiang T., Zong L., Wang Z., Luo X., Liang W., Zhao H., Ning Y., et al. Hypomethylation of LINE-1 elements in schizophrenia and bipolar disorder. J. Psychiatr. Res. 2018;107:68–72. doi: 10.1016/j.jpsychires.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 265.Puttipanyalears C., Arayataweegool A., Chalertpet K., Rattanachayoto P., Mahattanasakul P., Tangjaturonsasme N., Kerekhanjanarong V., Mutirangura A., Kitkumthorn N. TRH site-specific methylation in oral and oropharyngeal squamous cell carcinoma. BMC Cancer. 2018;18:786. doi: 10.1186/s12885-018-4706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ovchinnikov D.A., Wan Y., Coman W.B., Pandit P., Cooper-White J.J., Herman J.G., Punyadeera C. DNA Methylation at the Novel CpG Sites in the Promoter of MED15/PCQAP Gene as a Biomarker for Head and Neck Cancers. Biomark. Insights. 2014;9:53–60. doi: 10.4137/BMI.S16199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Anderson J., Bourne D., Peterson K., Mackey K. Evidence Brief: Accuracy of Self-Report for Cervical and Breast Cancer Screening. Department of Veterans Affairs (US); Washington, DC, USA: 2019. [PubMed] [Google Scholar]
- 268.Franceschi S., Barzan L., Talamini R. Screening for cancer of the head and neck: If not now, when? Oral Oncol. 1997;33:313–316. doi: 10.1016/S1368-8375(97)00034-1. [DOI] [PubMed] [Google Scholar]
- 269.Sankaranarayanan R., Mathew B., Jacob B.J., Thomas G., Somanathan T., Pisani P., Pandey M., Ramadas K., Najeeb K., Abraham E. Early findings from a community-based, cluster-randomized, controlled oral cancer screening trial in Kerala, India. The Trivandrum Oral Cancer Screening Study Group. Cancer. 2000;88:664–673. doi: 10.1002/(SICI)1097-0142(20000201)88:3<664::AID-CNCR25>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 270.Downer M.C., Evans A.W., Hughes Hallet C.M., Jullien J.A., Speight P.M., Zakrzewska J.M. Evaluation of screening for oral cancer and precancer in a company headquarters. Community Dent. Oral Epidemiol. 1995;23:84–88. doi: 10.1111/j.1600-0528.1995.tb00206.x. [DOI] [PubMed] [Google Scholar]
- 271.Guntinas-Lichius O., Wendt T., Buentzel J., Esser D., Lochner P., Mueller A., Schultze-Mosgau S., Altendorf-Hofmann A. Head and neck cancer in Germany: A site-specific analysis of survival of the Thuringian cancer registration database. J. Cancer Res. Clin. Oncol. 2010;136:55–63. doi: 10.1007/s00432-009-0636-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Pruegsanusak K., Peeravut S., Leelamanit V., Sinkijcharoenchai W., Jongsatitpaiboon J., Phungrassami T., Chuchart K., Thongsuksai P. Survival and prognostic factors of different sites of head and neck cancer: An analysis from Thailand. Asian Pac. J. Cancer Prev. 2012;13:885–890. doi: 10.7314/APJCP.2012.13.3.885. [DOI] [PubMed] [Google Scholar]
- 273.Galbiatti A.L., Padovani-Junior J.A., Maníglia J.V., Rodrigues C.D., Pavarino É., Goloni-Bertollo E.M. Head and neck cancer: Causes, prevention and treatment. Braz. J. Otorhinolaryngol. 2013;79:239–247. doi: 10.5935/1808-8694.20130041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Marur S., Forastiere A.A. Head and Neck Squamous Cell Carcinoma: Update on Epidemiology, Diagnosis, and Treatment. Mayo Clin. Proc. 2016;91:386–396. doi: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 275.Fortin A., Wang C.S., Vigneault E. Influence of smoking and alcohol drinking behaviors on treatment outcomes of patients with squamous cell carcinomas of the head and neck. Int. J. Radiat. Oncol. Biol. Phys. 2009;74:1062–1069. doi: 10.1016/j.ijrobp.2008.09.021. [DOI] [PubMed] [Google Scholar]
- 276.Wu I.-C., Wu C.-C., Lu C.-Y., Hsu W.-H., Wu M.-C., Lee J.-Y., Chou S.-H., Lee J.-M., Chou Y.-P., Wu D.-C. Substance use (alcohol, areca nut and cigarette) is associated with poor prognosis of esophageal squamous cell carcinoma. PLoS ONE. 2013;8:e55834. doi: 10.1371/journal.pone.0055834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.van Imhoff L.C., Kranenburg G.G., Macco S., Nijman N.L., van Overbeeke E.J., Wegner I., Grolman W., Pothen A.J. Prognostic value of continued smoking on survival and recurrence rates in patients with head and neck cancer: A systematic review. Head Neck. 2016;38((Suppl. 1)):E2214–E2220. doi: 10.1002/hed.24082. [DOI] [PubMed] [Google Scholar]
- 278.Chen X., Liu L., Mims J., Punska E.C., Williams K.E., Zhao W., Arcaro K.F., Tsang A.W., Zhou X., Furdui C.M. Analysis of DNA methylation and gene expression in radiation-resistant head and neck tumors. Epigenetics. 2015;10:545–561. doi: 10.1080/15592294.2015.1048953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Bonner J.A., Harari P.M., Giralt J., Azarnia N., Shin D.M., Cohen R.B., Jones C.U., Sur R., Raben D., Jassem J., et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 280.Vermorken J.B., Mesia R., Rivera F., Remenar E., Kawecki A., Rottey S., Erfan J., Zabolotnyy D., Kienzer H.R., Cupissol D., et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N. Engl. J. Med. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 281.Ogawa T., Liggett T.E., Melnikov A.A., Monitto C.L., Kusuke D., Shiga K., Kobayashi T., Horii A., Chatterjee A., Levenson V.V., et al. Methylation of death-associated protein kinase is associated with cetuximab and erlotinib resistance. Cell Cycle. 2012;11:1656–1663. doi: 10.4161/cc.20120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zhang X., Li W., Li H., Ma Y., He G., Tan G. Genomic methylation profiling combined with gene expression microarray reveals the aberrant methylation mechanism involved in nasopharyngeal carcinoma taxol resistance. Anticancer Drugs. 2012;23:856–864. doi: 10.1097/CAD.0b013e3283548d73. [DOI] [PubMed] [Google Scholar]
- 283.Viet C.T., Dang D., Achdjian S., Ye Y., Katz S.G., Schmidt B.L. Decitabine rescues cisplatin resistance in head and neck squamous cell carcinoma. PLoS ONE. 2014;9:e112880. doi: 10.1371/journal.pone.0112880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Yu X., Li Z. MicroRNA expression and its implications for diagnosis and therapy of tongue squamous cell carcinoma. J. Cell Mol. Med. 2016;20:10–16. doi: 10.1111/jcmm.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Castilho R.M., Squarize C.H., Almeida L.O. Epigenetic Modifications and Head and Neck Cancer: Implications for Tumor Progression and Resistance to Therapy. Int. J. Mol. Sci. 2017;18:1506. doi: 10.3390/ijms18071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Yu Z.W., Zhong L.P., Ji T., Zhang P., Chen W.T., Zhang C.P. MicroRNAs contribute to the chemoresistance of cisplatin in tongue squamous cell carcinoma lines. Oral Oncol. 2010;46:317–322. doi: 10.1016/j.oraloncology.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 287.Sun L., Yao Y., Liu B., Lin Z., Lin L., Yang M., Zhang W., Chen W., Pan C., Liu Q., et al. MiR-200b and miR-15b regulate chemotherapy-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting BMI1. Oncogene. 2012;31:432–445. doi: 10.1038/onc.2011.263. [DOI] [PubMed] [Google Scholar]
- 288.Liu M., Wang J., Huang H., Hou J., Zhang B., Wang A. miR-181a-Twist1 pathway in the chemoresistance of tongue squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2013;441:364–370. doi: 10.1016/j.bbrc.2013.10.051. [DOI] [PubMed] [Google Scholar]
- 289.Bauml J.M., Aggarwal C., Cohen R.B. Immunotherapy for head and neck cancer: Where are we now and where are we going? Ann. Transl. Med. 2019;7:S75. doi: 10.21037/atm.2019.03.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Franzen A., Vogt T.J., Müller T., Dietrich J., Schröck A., Golletz C., Brossart P., Bootz F., Landsberg J., Kristiansen G., et al. PD-L1 ( CD274) and PD-L2 ( PDCD1LG2) promoter methylation is associated with HPV infection and transcriptional repression in head and neck squamous cell carcinomas. Oncotarget. 2018;9:641–650. doi: 10.18632/oncotarget.23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Goltz D., Gevensleben H., Dietrich J., Schroeck F., de Vos L., Droege F., Kristiansen G., Schroeck A., Landsberg J., Bootz F., et al. PDCD1 (PD-1) promoter methylation predicts outcome in head and neck squamous cell carcinoma patients. Oncotarget. 2017;8:41011–41020. doi: 10.18632/oncotarget.17354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Gao G., Gay H.A., Chernock R.D., Zhang T.R., Luo J., Thorstad W.L., Lewis J.S., Wang X. A microRNA expression signature for the prognosis of oropharyngeal squamous cell carcinoma. Cancer. 2013;119:72–80. doi: 10.1002/cncr.27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Hui A.B., Lin A., Xu W., Waldron L., Perez-Ordonez B., Weinreb I., Shi W., Bruce J., Huang S.H., O’Sullivan B., et al. Potentially prognostic miRNAs in HPV-associated oropharyngeal carcinoma. Clin. Cancer Res. 2013;19:2154–2162. doi: 10.1158/1078-0432.CCR-12-3572. [DOI] [PubMed] [Google Scholar]
- 294.Consortium B. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat. Biotechnol. 2016;34:726–737. doi: 10.1038/nbt.3605. [DOI] [PubMed] [Google Scholar]
- 295.Ferlazzo N., Currò M., Zinellu A., Caccamo D., Isola G., Ventura V., Carru C., Matarese G., Ientile R. Influence of MTHFR Genetic Background on p16 and MGMT Methylation in Oral Squamous Cell Cancer. Int. J. Mol. Sci. 2017;18:724. doi: 10.3390/ijms18040724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kruszyna Ł., Lianeri M., Rydzanicz M., Gajecka M., Szyfter K., Jagodziński P.P. Polymorphic variants of folate metabolism genes and the risk of laryngeal cancer. Mol. Biol. Rep. 2010;37:241–247. doi: 10.1007/s11033-009-9643-y. [DOI] [PubMed] [Google Scholar]
- 297.Iglesias-Platas I., Martín Trujillo A., Court F., Monk D. Distinct promoter methylation and isoform-specific expression of RASFF1A in placental biopsies from complicated pregnancies. Placenta. 2015;36:397–402. doi: 10.1016/j.placenta.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 298.Meng R.W., Li Y.C., Chen X., Huang Y.X., Shi H., Du D.D., Niu X., Lu C., Lu M.X. Aberrant Methylation of RASSF1A Closely Associated with HNSCC, a Meta-Analysis. Sci. Rep. 2016;6:20756. doi: 10.1038/srep20756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Kang S., Li Q., Chen Q., Zhou Y., Park S., Lee G., Grimes B., Krysan K., Yu M., Wang W., et al. CancerLocator: Non-invasive cancer diagnosis and tissue-of-origin prediction using methylation profiles of cell-free DNA. Genome Biol. 2017;18:53. doi: 10.1186/s13059-017-1191-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Moran S., Martínez-Cardús A., Sayols S., Musulén E., Balañá C., Estival-Gonzalez A., Moutinho C., Heyn H., Diaz-Lagares A., de Moura M.C., et al. Epigenetic profiling to classify cancer of unknown primary: A multicentre, retrospective analysis. Lancet Oncol. 2016;17:1386–1395. doi: 10.1016/S1470-2045(16)30297-2. [DOI] [PubMed] [Google Scholar]
- 301.Romanowska K., Sobecka A., Rawłuszko-Wieczorek A.A., Suchorska W.M., Golusiński W. Head and Neck Squamous Cell Carcinoma: Epigenetic Landscape. Diagnostics. 2020;11:34. doi: 10.3390/diagnostics11010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Gaździcka J., Gołąbek K., Strzelczyk J.K., Ostrowska Z. Epigenetic Modifications in Head and Neck Cancer. Biochem. Genet. 2020;58:213–244. doi: 10.1007/s10528-019-09941-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Le J.M., Squarize C.H., Castilho R.M. Histone modifications: Targeting head and neck cancer stem cells. World J. Stem Cells. 2014;6:511–525. doi: 10.4252/wjsc.v6.i5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Irimie A.I., Ciocan C., Gulei D., Mehterov N., Atanasov A.G., Dudea D., Berindan-Neagoe I. Current Insights into Oral Cancer Epigenetics. Int. J. Mol. Sci. 2018;19:670. doi: 10.3390/ijms19030670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Webber L.P., Wagner V.P., Curra M., Vargas P.A., Meurer L., Carrard V.C., Squarize C.H., Castilho R.M., Martins M.D. Hypoacetylation of acetyl-histone H3 (H3K9ac) as marker of poor prognosis in oral cancer. Histopathology. 2017;71:278–286. doi: 10.1111/his.13218. [DOI] [PubMed] [Google Scholar]
- 306.Chen F., Qi S., Zhang X., Wu J., Yang X., Wang R. lncRNA PLAC2 activated by H3K27 acetylation promotes cell proliferation and invasion via the activation of Wnt/β-catenin pathway in oral squamous cell carcinoma. Int. J. Oncol. 2019;54:1183–1194. doi: 10.3892/ijo.2019.4707. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 307.Chen Y.W., Kao S.Y., Wang H.J., Yang M.H. Histone modification patterns correlate with patient outcome in oral squamous cell carcinoma. Cancer. 2013;119:4259–4267. doi: 10.1002/cncr.28356. [DOI] [PubMed] [Google Scholar]
- 308.Mancuso M., Matassa D.S., Conte M., Colella G., Rana G., Fucci L., Piscopo M. H3K4 histone methylation in oral squamous cell carcinoma. Acta Biochim. Pol. 2009;56:405–410. doi: 10.18388/abp.2009_2473. [DOI] [PubMed] [Google Scholar]
- 309.Chen H.C., Yang C.M., Cheng J.T., Tsai K.W., Fu T.Y., Liou H.H., Tseng H.H., Lee J.H., Li G.C., Wang J.S., et al. Global DNA hypomethylation is associated with the development and poor prognosis of tongue squamous cell carcinoma. J. Oral Pathol. Med. 2016;45:409–417. doi: 10.1111/jop.12381. [DOI] [PubMed] [Google Scholar]
- 310.Zelic R., Fiano V., Grasso C., Zugna D., Pettersson A., Gillio-Tos A., Merletti F., Richiardi L. Global DNA hypomethylation in prostate cancer development and progression: A systematic review. Prostate Cancer Prostatic Dis. 2015;18:1–12. doi: 10.1038/pcan.2014.45. [DOI] [PubMed] [Google Scholar]
- 311.Kitahara H., Okamoto T., Shimamatsu S., Kohno M., Morodomi Y., Tagawa T., Kitao H., Okano S., Oda Y., Maehara Y., et al. Hypomethylation Is Associated With Malignant Traits and Cell Proliferation in Lung Adenocarcinoma. Anticancer Res. 2020;40:5659–5666. doi: 10.21873/anticanres.14579. [DOI] [PubMed] [Google Scholar]
- 312.Peri S., Izumchenko E., Schubert A.D., Slifker M.J., Ruth K., Serebriiskii I.G., Guo T., Burtness B.A., Mehra R., Ross E.A., et al. NSD1- and NSD2-damaging mutations define a subset of laryngeal tumors with favorable prognosis. Nat. Commun. 2017;8:1772. doi: 10.1038/s41467-017-01877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.