Abstract
Simple Summary
Uveal melanoma (UM) is the most common primary intraocular malignancy in adults. More than 90% of UMs harbor mutually exclusive activating mutations in G-proteins. The mutations are early events in UM development and considered to be driver mutations in carcinogenesis. Even after treatment of primary uveal melanoma, up to 50% of patients subsequently develop recurrence, predominantly in the liver. GNAQ mutations are not reported to be correlated to survival, while the mutations in GNA11 are reported more frequently in metastatic UM. We investigated the correlation of survival after development of metastasis (Met-to-Death) of metastatic uveal melanoma (MUM) patients with GNA11 and GNAQ mutations. We identified that MUM with mutation patterns of Q209P vs. Q209L in GNA11 and GNAQ might predict survival of MUM patients.
Abstract
Uveal melanoma is the most common primary ocular malignancy in adults, characterized by gene mutations in G protein subunit alpha q (GNAQ) and G protein subunit alpha 11 (GNA11). Although they are considered to be driver mutations, their role in MUM remains elusive. We investigated key somatic mutations of MUM and their impact on patients’ survival after development of systemic metastasis (Met-to-Death). Metastatic lesions from 87 MUM patients were analyzed by next generation sequencing (NGS). GNA11 (41/87) and GNAQ (39/87) mutations were most predominantly seen in MUM. Most GNA11 mutations were Q209L (36/41), whereas GNAQ mutations comprised Q209L (14/39) and Q209P (21/39). Epigenetic pathway mutations BAP1 (42/66), SF3B1 (11/66), FBXW7 (2/87), PBRM1 (1/66), and SETD2 (1/66) were found. No specimen had the EIF1AX mutation. Interestingly, Met-to-Death was longer in patients with GNAQ Q209P compared to GNAQ/GNA11 Q209L mutations, suggesting the difference in mutation type in GNAQ/GNA11 might determine the prognosis of MUM. Structural alterations of the GNAQ/GNA11 protein and their impact on survival of MUM patients should be further investigated.
Keywords: uveal melanoma, metastasis, metastatic uveal melanoma, survival, GNA11, GNAQ, Q209P, Q209L, BAP1, SF3B1
1. Introduction
Uveal melanoma (UM) is the most common primary intraocular malignancy in adults with incidence rate of 5.2 cases per million per year in the United States [1]. Among all melanomas involving the eye, 83% arise from uvea (UM), 5% from conjunctiva, and 10% from other sites [2]. Despite the common embryologic origin of cutaneous and uveal melanocytes, UM has many differing epidemiologic, prognostic, biological features and molecular mechanisms which differ from cutaneous melanoma [3]. For instance, the majority of cutaneous melanoma harbor mutations of B-Raf proto-oncogene, serine/threonine kinase (BRAF), NRAS proto-oncogene, GTPase (NRAS) and neurofibromin 1 (NF1) as well as loss of cyclin dependent kinase inhibitor 2A (CDKN2A) encoding P16NK4a. On the other hand, uveal melanoma rarely harbors these abnormalities. [4,5,6].
Regardless of successful therapy of primary UM, up to 50% of patients subsequently develop systemic recurrence, especially in the liver via the hematogenous route. A bimodal pattern of the mortality displays the first peaks at 2–3 years and the second surge at 8–9 years after enucleation [7]. After development of hepatic metastasis, the median survival of patients is reported to be 12 to 17 months [8].
G protein-coupled receptor (GPCR) pathway mutations in UM has been well documented. Mutations in G protein subunit alpha q (GNAQ) and G protein subunit alpha 11 (GNA11) are the most common in UM and considered to be driver mutations in carcinogenesis [9,10]. Also reported in UM are mutations in cysteinly leukotriene receptor 2 (CYSLTR2) or phospholipase C beta 4 (PLCB4) which are located directly upstream or downstream of the G protein [11,12]. In general, these mutations are mutually exclusive and seen as somatic mutations. Approximately 90% of UM reportedly possesses mutually exclusive GNA11 and GNAQ mutations [9,13]. Mutations are frequently seen at the conserved catalytic glutamine (Q209 in Gαq, exon 5) replaced by either Proline, P, or Leucine, L, which leads to GTPase function deficiency and constitutive activation. Less frequently, mutations were found at position 183 (exon 4) replacing Arginine, R. GNA11 and GNAQ Q209 mutations are considered to send stronger signals to downstream pathways compared to R183 mutations [9,14]. These oncogenic mutations trigger a wide range of cell signaling cascades including the mitogen-activating protein kinases (MAPK), phosphoinositide 3-kinase/serine/threonine protein kinase (PI3K/AKT), and yes-associated protein/transcriptional co-activator with PDZ-binding motif (YAP/TAZ) pathways [15,16]. These mutations arise early in tumor evolution and may promote tumor progression [17]. Although these mutations are the predominant pathways for the development of UM, they are reportedly not associated with overall survival of UM patients [18]. In particular, GNAQ mutations were not correlated with disease-free survival, while the mutations in GNA11 have been reported more frequently in metastatic UM (MUM) [19,20]. It is speculated that mutations in UM with GNA11 mutations may be more aggressive than those with GNAQ mutations.
UM may have secondary somatic mutations affecting BRCA1 associated protein 1 (BAP1), splicing factor 3b subunit 1 (SF3B1), serine and arginine rich splicing factor 2 (SRSF2), or eukaryotic translation initiation factor 1A X-linked (EIF1AX), which tend to occur exclusively from each other [13,21]. These secondary mutations determine the metastatic potential of UM cells. For example, UM with BAP1 alterations tends to develop systemic recurrence earlier compared to those with SF3B1 mutations [22,23]. Tumor cells which harbor EIF1AX mutation tend not to metastasize [23].
These previous studies focused on time from initial diagnosis and treatment of primary UM to development of systemic recurrence or death. These studies did not address prognostic factors to determine time from diagnosis of metastasis to death (Met-to-Death). Here in this study, we aim to identify the correlation of Met-to-Death in MUM patients with commonly seen mutations in MUM. We identified that differences in mutation patterns (Q209P vs. Q209L) in GNAQ and GNA11, rather than GNAQ and GNA11 themselves, might predict the survival of MUM patients.
2. Subject and Methods
2.1. Patients and Clinical Data
Tumor samples were collected and profiled from patients with MUM between the year 2013 and 2018 in this retrospective analysis. This study was approved by the Institutional Review Board (IRB) at Thomas Jefferson University [IRB#18D-183]. Clinical data were obtained from their medical records.
2.2. Tumor Tissue Samples
Tissue specimens from metastatic uveal melanoma patients were retrieved in paraffin-embedded archival core biopsy or surgically removed specimens for mutation analysis. Molecular profiling studies were done at Caris Life Sciences (Headquarters, Irving, TX, USA), except one specimen which was done at Foundation Medicine (Headquarters, Cambridge, MA, USA). In terms of molecular analysis done by Caris Life Sciences, either formalin-fixed paraffin-embedded tissue block or formalin-fixed paraffin-embedded 10 unstained tissue slides that could be enriched to a minimum of 20% tumor by microdissection were sent to Caris Life Sciences. Detailed information on molecular profiling technology is available from the following website (https://www.carislifesciences.com/molecular-profiling-technology (accessed on 28 September 2021)). In brief, tumor DNA was extracted from the tumor specimens obtained by microdissection of the tumor area under the supervision of board-certified pathologists. Tumor samples were analyzed by next-generation sequencing (NGS) of exons using either TruSeq Amplicon 48 Gene Cancer Panel with MiSeq system (Illumina, San Diego, CA, USA) or 592 cancer-relevant genes panel (SureSelect XT, Agilent, Santa Clara, CA, USA) with the NextSeq instrument (Illumina). The overall average depth of coverage was typically >1000×. Variants were called based on a combination of coverage and depth using a sliding scale. The minimum possible variant frequency was approximately 5%. The minimum depth for a called variant was 100×. If coverage fell below 100× in any region of a gene, the entire gene was called “Indeterminate.” Amplification analysis was performed in samples tested by the 592 gene panel. Genes with at least six copies were called amplified. The data for the analysis in this study were obtained from a clinical laboratory system of Thomas Jefferson University that stores all laboratory results ordered by physicians.
2.3. Statistical Analysis
We investigated the correlation between somatic mutations in MUM specimens and survival of patients after development of systemic metastasis. The endpoint of survival analysis was from metastasis to death (Met-to-Death).
Patient characteristics were summarized with frequency counts and percentages for categorical variables and median and interquartile range (IQR) for continuous variables.
For unadjusted comparison of patient characteristics by gene mutation groups, Fisher’s exact test was used for categorical variables, and the Kruskal-Wallis test was used for continuous variables.
The Kaplan-Meier (K-M) survival curves were used to estimate the overall survival (OS) from metastasis and the corresponding median survival time by patient characteristics. Log-rank tests were used to compare the K-M curves.
Univariable Cox proportional hazards models was used to assess the association between survival from metastasis and continuous clinical-pathological factors: age, and time from the primary treatment to the metastases. Multivariable Cox proportional hazards model was used to evaluate the effect of gene mutation type on OS while controlling for other significant predictors of OS. Death due to metastasis was considered an event. All the analyses were performed with SAS 9.4 (SAS Institute Inc., Cary, NC, USA).
3. Results
3.1. Tumor and Patient Characteristics
We submitted 102 samples to commercial companies for analysis; 15 samples were indeterminated for mutation analysis. A total of 87 patients were analyzed for somatic mutations in their metastatic specimens. Of 87 patients, 21 patients were analyzed by NGS with 48 selected cancer genes including GNA11, GNAQ and FBXW7, and 66 patients were analyzed for mutations by NGS with 592 cancer genes including BAP1 and SF3B1 mutations as well as MYC amplification (Supplement Table S1). Median survival time after metastasis was 25 months with a range of 1–137 months. Metastatic specimens were collected within 1 year from diagnosis of metastasis in 72 patients, while 9 specimens were obtained 1–2 years after initial diagnosis of metastasis and 6 specimens were obtained more than 2 years after initial diagnosis of metastasis. No treatment targeting GNAQ/GNA11 mutations was given to any of these patients.
At the end of follow-up, 72 patients died including one unexpected non-cancer event; 2 patients were lost to follow-up; and 13 patients were alive (total 87 patients). There were 46 males and 41 females (Table 1). Median ages at diagnosis of primary and metastatic UM were 58 and 60 years of age, respectively. Most patients were treated with radioactive plaque for their primary UM (74.7%) and 90.8% of patients received a liver-directed therapy in the first and/or second treatment option of metastatic UM [24,25,26,27]. Those diagnosed with the first metastatic lesion in this patient cohort were mostly of the liver, lung and omentum, 84/87 (96.5%), 2/87, (2.3%), 1/87, (1.2%), respectively. Those patients who were diagnosed with only extrahepatic metastases initially had subsequently developed liver metastases (3/3, 100%).
Table 1.
Demographic features of UM patients with metastasis.
| Characteristic | Number |
|---|---|
| Number of Patients | 87 |
| Gender | |
| Male | 46 |
| Female | 41 |
| Age at primary eye diagnosis (years) | |
| Median (Range) | 58 (21–88) |
| Male | 59 (28–88) |
| Female | 55 (21–77) |
| Location of primary uveal melanoma (%) | |
| Choroidal | 67 (77.0) |
| Ciliary | 20 (23.0) |
| Other | 0 |
| Treatment for primary uveal melanoma (%) | |
| Radiopaque | 65 (74.7) |
| Enucleation | 13 (14.9) |
| Others | 7 (8.0) |
| Unknown | 2 (2.2) |
| Age at metastasis diagnosis (years) | |
| Median (Range) | 60 (24–88) |
| Male | 60 (28–88) |
| Female | 60 (24–78) |
| Metastatic site (at the diagnosis) (%) | |
| Liver | 84 (96.5) |
| Lung | 2 (2.3) |
| Omentum | 1 (1.2) |
| M stage (%) | |
| M1a ≤ 3 cm | 69 (79.3) |
| M1b 3.1–8.0 cm | 14 (16.1) |
| M1c ≥ 8.1 cm | 4 (4.6) |
| First and second treatments option at metastatic site (%) | |
| Liver-directed alone | 41 (47.1) |
| Liver-directed + Systemic | 38 (43.7) |
| Systemic alone | 4 (4.6) |
| Recession | 1 (1.1) |
| Unknown | 3 (3.5) |
3.2. Frequency of GNA11 and GNAQ Mutations in Metastatic Uveal Melanoma
Mutations in GNA11 and GNAQ are considered to be driver mutations that lead to constitutive activation of GPCR signaling. We first assessed these known UM driver mutations. The frequent alteration of GNAQ and GNA11 were identified as nearly mutually exclusive in 81 of 87 patients (92.0%) (Table 2). Mutually exclusive GNA11 mutations were found in 41/87 (47.1%) tumors, while GNAQ mutations were found in 39/87 tumors (44.8%). One case (Case 23) harbored two simultaneous mutations of GNA11 Q209L in exon 5 and GNAQ T96S in exon 2, which might indicate the tumor specimen contained a heterozygous population of tumor cells (Supplement Table S2). Six out of 87 tumors were not found to have either GNA11 or GNAQ mutation. As previously reported exon 5 of GNA11 and GNAQ genes, which contains the hotspot mutation leading to the most frequent alteration of the Q209 amino acid, was identified in 74 of 87 patients (85.1%) (Table 2). Frequently, mutations were seen at the position 209 glutamine (Q) to either proline (P) or leucine (L). Of those with GNA11 mutations, the Q209L mutation was found in 37 of these 42 cases (88.1%) while Q209P mutation was found in one case (Case 52) (2.4%). One specimen showed the Q209M mutation (Case 75), one case (Case 74) harbored both the R183C and V344M mutations in GNA11, and two cases had a single mutation at R183C. One specimen had GNA11 Q209L mutation as well as GNAQ T96S mutation (Case 23) (Supplement Table S2).
Table 2.
Frequency of mutations in the two major genes in metastatic uveal melanoma.
| Mutation Status | N |
|---|---|
| GNA11 | 41 (47%) |
| Q209L | 36 |
| Q209P | 1 |
| Q209M | 1 |
| R183C | 2 |
| R183C, V344M | 1 |
| GNAQ | 39 (45%) |
| Q209L | 14 |
| Q209P | 21 |
| R183Q | 2 |
| G48L | 1 |
| R183Q, R338H | 1 |
| Both GNA11 and GNAQ | 1 (1%) |
| GNA11 Q209L, GNAQ T96S | 1 |
| No GNA11 or GNAQ | 6 (7%) |
Of those with GNAQ mutation, the Q209L mutation was found in 14 of these 40 cases (35.0%) while Q209P mutation was found in 21 of 40 cases (52.5%). Two MUM specimens harbored a single mutation at R183Q in exon 4 (2/39, 5.1%). One showed a single mutation at G48L in exon 2 (Case 79). One specimen showed simultaneous mutations of R183Q in exon 4 and R338H in exon 7 (Case 80). One specimen (Case 23) had GNAQ T96S mutation as well as GNA11 Q209L mutation. (Table 2, Supplement Table S2). As expected, the mutations in GNA11 and GNAQ were almost mutually exclusive except for one specimen. These results show that double mutations seem to be rare events in MUM as previously reported in primary UM [9,13].
3.3. Other Somatic Mutations
Among 87 MUM specimens, 66 specimens were analyzed for target-captured deep sequencing of 592 cancer genes. Other mutations found in MUM specimens were mostly related to epigenetic pathways including BAP1 (42/66; 63.6%), SF3B1 (11/66; 16.6%), F-box and WD repeat domain containing 7 (FBXW7) (2/87; 2.3%), protein polybromo-1 (PBRM1) (1/66; 1.5%), and SET domain containing 2, histone lysine methyltransferase (SETD2) (1/66; 1.5%) (Supplement Tables S1 and S2). One case (Case 80) harbored simultaneous alterations of BAP1 and the mutation of SF3B1. In addition, MYC amplifications were assessed and 13 out of 66 specimens (19.7%) were positive for MYC amplifications (Supplement Table S1). No specimen had EIF1AX mutation. NRAS and BRAF mutations, which are commonly seen as driver genes in cutaneous melanoma, were not found either.
Location of BAP1 alterations were variable. SF3B1 mutations including R625C, R625H, R625G, R625L, N626Y, and G742D were detected. SF3B1 mutations were almost always mutually exclusive of the presence of BAP1 alteration in tumor specimens except one specimen.
Two tumors (Cases 34 and 70) had FBXW7 mutations, which has been reported as a tumor suppressor gene [28,29]; one sample carried the mutation in exon 12, c.1856-2A > G; and another sample carried exon 4, c.585-1G > T. One tumor (Case 34) had a mutation (exon 11, c.996-1G > T) of PBRM1, which is the second most common tumor suppressor gene in kidney cancer. The other tumor (Case 73) had a mutation (exon 1, c.68_71 + 1delCTGAG) of SETD2, a histone methyltransferase that mediates trimethylation of lysine 36 on Histone 3 (Supplement Table S2).
3.4. Survival Analysis of Met-to-Death
To examine the possibility for showing a difference in clinical characteristics of tumors with different mutations, we analyzed the Met-to-Death between the mutation of proline (P) and leucine (L) at the position of 209 in GNA11 or GNAQ.
Among 74 patients whose tumors harbored Q209 mutations, 69 patients were analyzed for their survival data. One specimen with mutations in both GNAQ and GNA11 and specimens with GNA11 Q209P (n = 1) and Q209M (n = 1) were excluded from this statistical analysis due to small sample size for statistical justification. Two patients with GNAQ Q209P were also removed from survival analysis due to patients lost in follow-up. Table 3 summarizes the patient characteristics of each group. For unadjusted comparison, there were no statistically significant associations between gene mutation types and patient characteristics (Table 3). Similar proportions of patients with and without BAP1 alterations and SF3B1 mutations were observed across different GNA11/GNAQ mutation types. Other potential confounding factors such as M stage, treatment after metastasis, and time from primary eye treatment to development of metastasis were comparable.
Table 3.
Association between Patients Characteristics and the Type of Gene Mutation.
| Characteristic | ALL (n = 69) |
GNA11/Q209L (n = 36) |
GNAQ/Q209L (n = 14) |
GNAQ/Q209P (n = 19) |
p-Value | |
|---|---|---|---|---|---|---|
| Gender, n (%) | ||||||
| Female | 31 (44.9) | 19 (52.8) | 5 (35.7) | 7 (36.8) | 0.391 | |
| Male | 38 (55.1) | 17 (47.2) | 9 (64.3) | 12 (63.2) | ||
| Primary Site, n (%) | ||||||
| Choroidal | 54 (78.3) | 26 (72.2) | 11 (78.6) | 17 (89.5) | 0.373 | |
| Ciliary | 15 (21.7) | 10 (27.8) | 3 (21.4) | 2 (10.5) | ||
| Age at Primary Dx, median (IQR) | ||||||
| 58.0 (50.0, 66.0) |
59.0 (48.5, 66.5) |
61.5 (57.0, 77.0) |
54.0 (49.0, 61.0) |
0.133 | ||
| Age at Metastasis, median (IQR) | ||||||
| 60.0 (54.0, 69.0) |
60.0 (51.0, 69.0) |
65.0 (60.0, 78.0) |
59.0 (52.0, 63.0) |
0.074 | ||
| Metastasis site, n (%) | ||||||
| Liver | 68 (98.6) | 36 (100.0) | 13 (92.9) | 19 (100.0) | 0.203 | |
| Omentum | 1 (1.4) | 0 (0.0) | 1 (7.1) | 0 (0.0) | ||
| Months from Primary Tx to Met, median (IQR) | ||||||
| 25.0 (11.0, 44.0) |
26.0 (11.0, 37.0) |
27.0 (15.0, 57.0) |
25.0 (14.0, 55.0) |
0.809 | ||
| BAP1—Mutation, n (%) | ||||||
| Yes | 31 (44.9) | 15 (41.7) | 7 (50.0) | 9 (47.4) | 1.000 | |
| No | 19 (27.5) | 10 (27.8) | 4 (28,6) | 5 (26.3) | ||
| N/A | 19 (27.5) | 11 (30.6) | 3 (21.4) | 5 (26.3) | ||
| SF3B1—Mutation, n (%) | Yes | 7 (10.1) | 3 (8.3) | 1 (7.1) | 3 (15.8) | 0.661 |
| No | 43 (62.3) | 22 (61.1) | 10 (71.4) | 11 (57.9) | ||
| N/A | 19 (27.5) | 11 (30.6) | 3 (21.4) | 5 (26.3) | ||
| M stage (tumor size in Liver), n (%) | ||||||
| ≤3 cm | 53 (76.8) | 28 (77.8) | 8 (57.1) | 17 (89.5) | 0.209 | |
| 3.1–8.0 cm | 12 (17.4) | 5 (13.9) | 5 (35.7) | 2 (10.5) | ||
| ≥8.1 cm | 4 (5.8) | 3 (8.3) | 1 (7.1) | 0 (0.0) | ||
| Tx after metastasis (Tx1 + 2), n (%) | ||||||
| Liver direct Tx + Systemic Tx | 31 (45.6) | 17 (47.2) | 5 (38.5) | 9 (47.4) | 0.730 | |
| Liver direct therapy | 33 (48.5) | 17 (47.2) | 7 (53.8) | 9 (47.4) | ||
| Recession | 1 (1.5) | 0 (0.0) | 1 (7.7) | 0 (0.0) | ||
| Systemic Tx | 3 (4.4) | 2 (5.6) | 0 (0.0) | 1 (5.3) | ||
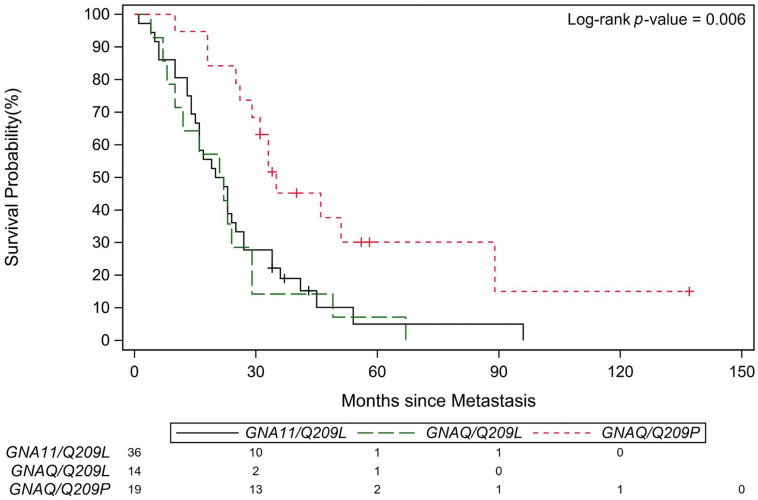
We then analyzed the association of the patient characteristics and the time from Met-to-Death. Kaplan Meier survival (K-M) curve showed patients with GNAQ Q209P mutant tumors had a more favorable outcome than patients with GNA11 Q209L and GNAQ Q 209L mutant tumors after development of metastasis (n = 69, Log-rank test, p = 0.006, Table 4 and Figure 1). The univariable Cox models revealed patients with GNA11 Q209L mutant tumors or GNAQ Q209L mutant tumors showed shorter median survival (Met-to-Death) than patients with GNAQ Q209P mutant tumors. The median survival (Met-to-Death) was 21 months (95% CI: 15–25) for GNA11 Q209L, 21.5 months (95% CI: 8–29) for GNAQ Q209L, and 35 months (95% CI: 26–89) for GNAQ Q209P, respectively (p = 0.006) (Table 4).
Table 4.
Association between Patients Characteristics and Survival from Metastasis.
| Characteristic | N (%) | Median OS (95% CI) | p-Value | |
|---|---|---|---|---|
| Gender, n (%) | ||||
| Female | 31 (44.9) | 31.0 (20.0, 41.0) | 0.122 | |
| Male | 38 (55.1) | 22.5 (15.0, 26.0) | ||
| Primary Dx, n (%) | ||||
| Choroidal | 54 (78.3) | 25.5 (20.0, 33.0) | 0.331 | |
| Ciliary | 15 (21.7) | 21.0 (13.0, 25.0) | ||
| Met Dx site, n (%) | ||||
| Liver | 68 (98.6) | 23.5 (19.0, 29.0) | 0.387 | |
| Omentum | 1 (1.4) | 67.0 * | ||
| Gene Mutation, n (%) | ||||
| GNA11/Q209L | 36 (52.2) | 21.0 (15.0, 25.0) | 0.006 | |
| GNAQ/Q209L | 14 (20.3) | 21.5 (8.0, 29.0) | ||
| GNAQ/Q209P | 19 (27.5) | 35.0 (26.0, 89.0) | ||
| BAP1—Mutation, n (%) | ||||
| Yes | 31 (62.0) | 25.0 (19.0, 31.0) | 0.040 | |
| No | 19 (38.0) | 36.0 (13.0, 89.0) | ||
| 11 | ||||
| SF3B1—Mutation, n (%) | Yes | 7 (14.0) | 89.0 (13.0, 96.0) | 0.011 |
| No | 43 (86.0) | 23.0 (19.0, 29.0) | ||
| M stage (tumor size in Liver), n (%) | ||||
| ≤3 cm | 53 (76.8) | 25.0 (22.0, 33.0) | 0.102 | |
| >3 cm | 16 (23.2) | 12.0 (7.0, 26.0) | ||
| Tx after metastasis # (Tx1 + 2), n (%) | ||||
| Liver direct Tx + Systemic Tx | 31 (44.9) | 33.0 (23.0, 45.0) | 0.104 | |
| Liver direct therapy | 33 (47.8) | 18.0 (14.0, 26.0) | ||
| Recession | 1 (1.4) | 23.0 * | ||
| Systemic Tx | 3 (4.3) | 13.0 * | ||
| Unknown | 1 | |||
| HR (95% CI) | ||||
| Age at Primary Dx | ||||
| 1.04 (1.02, 1.07) | <0.001 | |||
| Age at Metastasis | ||||
| 1.04 (1.01, 1.06) | 0.004 | |||
| Months from Primary Tx to Met (log-transformed) | ||||
| 0.85 (0.69, 1.03) | 0.101 | |||
* The 95% CI is not estimable due to the small number of observations. # Tx1 + 2 = First treatment + Second treatment.
Figure 1.
The Kaplan-Meier Curves for Met-to-Death OS. Kaplan-Meier Curves showing patients carrying tumor mutations with Q209L of GNA11, Q209L of GNAQ and Q209P of GNAQ. The red line represents Q209P of GNAQ (n = 19), the green line represents Q209L of GNAQ (n = 14) and the black line represents Q209L mutation of GNA11 (n = 36).
There was a difference in Met-to-Death OS by BAP1 status. Patients with BAP1 alterations showed shorter median survival than those without BAP1 alterations (Table 4). Although the sample size of patients with known SF3B1 status was small (n = 50), MUM patients whose tumors had SF3B1 mutations (n = 7) had the tendency to live longer [median survival of 89 months (95% CI: 13–96, n = 7)] than those who did not have this mutation [median survival of 23 months (95% CI: 19–29), n = 43)] (p = 0.011) (Table 4). Based on this observation, we reanalyzed the survival between BAP1 alterations and non BAP1 alterations. All patients with SF3B1 mutations were excluded from the non BAP1 alteration group for this analysis. After removing patients with SF3B1 mutations, Met-to-Death OS analysis showed no statistical difference between patients with BAP1 alteration (n = 31) and without BAP1 alteration (n = 12) (p = 0.845) (Supplement Table S3). Additionally, we observed about 4% increase in hazard of death per each additional year of age at eye diagnosis (Hazard Ratio (HR) = 1.04, 95% CI: 1.02–1.07; p < 0.001) and age at metastasis (HR = 1.04, 95% CI: 1.01–1.06; p = 0.004) (Table 4). These were consistent findings as reported by Seeder, et al. [8].
Multivariable Cox models were also performed to evaluate the effect of gene mutation types on Met-to-Death OS with adjusting age at metastasis and log-transformed time from primary treatment to metastasis (Table 5). The analysis revealed that patients with GNA11 Q209L mutant tumors or GNAQ Q209L mutant tumors had shorter Met-to-Death OS as compared to patients with GNAQ Q209P mutant tumors (Hazard Ratio (HR); 3.42, 95% CI: 1.68–6.96, p = 0.001 for GNA11 Q209L vs. GNAQ Q209P, and HR = 3.08, 95% CI: 1.35–7.04, p = 0.008 for GNAQ Q209L vs. GNAQ Q209P). The comparison of Hazard between patients with GNAQ Q209L mutant tumors and patients with GNA11 Q209L mutant tumors did not show a significant difference (HR = 1.11, 95% CI: 0.57–2.15; p = 0.759) (Table 5). Older age at metastasis diagnosis was independently associated with a higher hazard of death (HR = 1.04, 95% CI: 1.01–1.06; p = 0.005). Additionally, longer time from the initial eye treatment to metastasis was associated with lower hazard of death (HR = 0.79, 95% CI: 0.63–0.99; p = 0.041).
Table 5.
Results from the main multivariable Cox Model for OS from Met to Death.
| Factor | HR | (95% CI) | p-Value |
|---|---|---|---|
| Gene Mutation | 0.003 | ||
| GNA11/Q209L vs. GNAQ/Q209P | 3.42 | (1.68, 6.96) | 0.001 |
| GNAQ/Q209L vs. GNAQ/Q209P | 3.08 | (1.35, 7.04) | 0.008 |
| GNA11/Q209L vs. GNAQ/Q209L | 1.11 | (0.57, 2.15) | 0.759 |
| Age at Metastasis (Continuous) | 1.04 | (1.01, 1.06) | 0.005 |
| Time from Primary Tx to Metastasis (log-transformed) | 0.79 | (0.63, 0.99) | 0.041 |
Tumors with BAP1 alterations in UM is a consistent finding for a poor prognosis marker [30]. To confirm the result of the multivariable Cox model in Table 5, we also examined the patients with tumor-analyzing BAP1 status (n = 50). We excluded 19 patients with GNAQ or GNA11 at Q209 mutant tumors who had no data on BAP1 status. Due to sample size limitation, BAP1 wild type specimens with SF3B1 mutations were not excluded from the BAP1 alteration negative population in this analysis. Although patients with BAP1 altered tumors tended to show poorer Met-to-Death survival than patients with BAP1 wild type tumors, the result was not statistically significant (HR = 1.62, 95% CI: 0.75–3.60; p = 0.214) (Supplement Table S4). Meanwhile, controlling for BAP1 status, patients with GNA11 Q209L mutant tumors (n = 29) or GNAQ Q209L mutant tumors (n = 13) still had a poor Met-to-Death survival as compared to patients with GNAQ Q209P mutant tumors (n = 16) (HR = 4.07, 95% CI: 1.62–10.23; p = 0.003 for GNA11 Q209L, and HR = 3.69, 95% CI: 1.31–10.36, p = 0.0013 for GNAQ Q209L) (Supplement Table S4).
4. Discussion
In this study, we investigated the frequency of mutations in MUM specimens and the role of commonly mutated GNA11/GNAQ genes in survival after development of systemic metastasis (Met-to-Death). In 87 MUM patients, we showed that GNA11 and GNAQ mutations were found in 47.1% and 44.8% of patients, respectively. This result was consistent with the analysis performed by other researchers in primary uveal melanoma [9,31], whereas Griewank et al. report on MUM showed more GNA11 mutations than GNAQ mutations in MUM patients [20]. There also has been inconclusive discussion whether prognosis of UM tumors with GNA11 is poorer than that of UM tumors with GNAQ mutations. It is of note that glutamine (Q) at position 209 in GNA11 was commonly replaced with leucine (L) in 97.3% of samples, compared to replacement with proline (P) in 2.7% of these samples [9,20]; therefore, potentially poor prognosis of GNA11 mutated UM tumors might be due to dominant 209L mutation in GNA11. We tested this hypothesis and investigated Met-to-Death survival in patients with MUM after stratifying mutations at position 209 between GNA11 and GNAQ. Interestingly, patients with Q209P in GNAQ mutant tumors significantly correlated with favorable prognosis after development of metastasis. Traditionally, the prediction of prognosis in UM patients has been based on the survival from the date of treatment of primary UM to their death. It has been reported that loss of Chromosome 3 and Chromosome 8q gains in primary UM tumor specimens have been shown to predict survival of UM patients [13,32,33]. In addition, the expression gene profile of class 2 is used for the prediction of prognosis [34]. Currently, other biological prognostic markers are explored with the expression of PRAME or autophagy related proteins in primary uveal melanoma specimens. The level of Beclin-1 expression on primary uveal melanoma correlated with a lower risk of metastasis and higher disease-free survival times [35]. On the other hand, the expression of PRAME identified increasing metastatic risk [36]. None of the above investigations was extended to the analysis on Met-to-Death and the factors to predict survival after development of metastasis remains to be investigated.
GNA11 and GNAQ mutations were considered to occur early and represent initiating events in tumorigenesis [17]. It has been shown that the difference in GNAQ and GNA11 mutations did not affect survival of UM patients after their treatment of primary UM. In this regard, it is rather interesting that our data showed substitutions Q209L vs. Q209P rather than G protein (GNAQ vs. GAN11) impacting the survival of UM patients with metastasis. The substitutions in Q209 might play a role in determining prognosis of MUM patients, especially Met-to-Death.
It remains to be investigated why UM patients with GNAQ Q209P mutant tumors showed favorable outcome after development of metastasis in our study. Since clinical teams does not make any treatment decision based on GNAQ and GNA11 mutation status, there was no difference in their clinical stages and treatment approaches to these patients, including M stage and choice of treatments. There was no difference in frequency of BAP1 or SF3B1 mutations and time from primary eye treatment to development of systemic recurrence among MUM patients with Q209L and Q209P mutations. One of the possibilities is that Q209P mutant tumor may have a signature of higher immunological characteristics than Q209L mutant tumor. Immunogenicity of Q209P mutation might be different from that of Q209L. In this regard, Weeghel et al. reported Q209P or Q209L mutation does not have significant impact on the immunological characteristics of the tumors [37]. It is also possible that the main difference between Q209P mutant and Q209L mutant is that one may have unique structural properties that may impact its ability to bind different interacting partners such as G protein βγ subunits, and Q209P may have a distinct functional feature not shared by Q209L [38]. Different degrees and pathways of downstream signal transduction might result in resistance to treatments and contribute to difference in survival of MUM patients. Investigation on this possibility is underway in our group.
A more widely accepted concept is that UM patients with BAP1 alterations have poor prognosis. Based on our data supported by our basic research experiments using BAP1 altered cell lines, this might not be due to rapid growth of BAP1 altered cells in metastasis. In point of fact, it is reported that BAP1-deficient UM cell lines generally are associated with extremely slow growth characteristics [39]. The distinct slow doubling time that has been seen in established cell lines in vitro has not represented patient outcome. In fact, impact of BAP1 alteration on Met-to-Death is not clearly shown in our survival analysis excluding patients with SF3B1 mutations. It is of note that the detection rate of BAP1 alteration in our subjects is relatively lower than previously published data [5,20]. This is most likely due to differences in detection assay since inactivating BAP1 alterations can be present anywhere along the gene body. The panel sequencings used for our analysis were BAP1 exons assay which did not include intron sequences for BAP1 gene. Furthermore, our NGS assay did not detect large deletions or duplications over ~100 bp. Further studies with more detailed BAP1 gene analyses are needed to investigate the association of BAP1 alteration with Met-to-Death. BAP1 alterations might be an important factor for development of systemic recurrence; however, the role of BAP1 alteration for rapid growth of metastasis or resistant mechanisms for treatments remain to be proven. In this regard, Szalai et al., reported the metastatic pattern might be different between patients with BAP1 altered tumor and SF3B1 mutant tumor. Time to clinically detectable metastases has different peaks among these two mutations. The earlier peaks appear to be associated with BAP1 altered tumors and the later peak is associated with SF3B1 mutated tumors [23]. Although the sample size is small and further investigation with a larger cohort is required, our data also indicate that the presence of SF3B1 mutations might affect the survival of MUM patients (Met-to-Death). These data indicate that BAP1 alterations may be a predictive factor for early systemic recurrence; however, it might not be a prognostic factor for MUM patients who already developed metastasis. Furthermore, future clinical trials might require the stratification of MUM patients based on the status of SF3B1 mutations in addition to type of GNAQ/GNA11 Q209 mutations (P vs. L).
Lastly, our data showed the amplification of MYC was determined in 19.7% of metastatic specimens (13/66). The progression of uveal melanoma has often seen with the amplification of chromosome 8q which is the common region of amplification found to range from 8q24.1 to 8q24.3 [40]. The proto-oncogene MYC is located at 8q24.12-q24.13 and is one candidate for the amplification at this site. Parrella et al., reported 70% of uveal melanoma detected extra copies of the region around the MYC locus by fluorescent in situ hybridization [41]. Later, Ehlers et al. analyzed the region of chromosome 8q with gene expression microarray analysis using Affymetrix Hu133A and B GeneChips. They reported that DDEF1 gene, located at chromosome 8q24, was increased in 8q amplified tumor whereas MYC expression remained unchanged [42]. This indicates that MYC is not always amplified in 8q24 chromosome region in 8q-amplified but other oncogenes residing in chromosome 8q24 might play a role in tumor progression. Since numbers of specimens with MYC amplification were limited, we could not conduct detailed statistical analysis on the role of MYC on Met-to-Death. More studies at 8q24 loci are needed to explore the association of MYC amplification for tumor progression in uveal melanoma.
There are several limitations of this study. First of all, metastatic specimens were obtained more than 2 years from the diagnosis of metastasis in 6 of 87 patients. It is possible that mutational patterns of metastatic tumors had changed with previous treatments. Since there is no treatment given directly targeting GNAQ/GNA11 mutations, we believe it is less likely that the frequency and pattern of GNAQ/GNA11 mutations changed from the diagnosis of metastasis to the time of tumor specimen procurement in this study. Obviously, we do not exclude the possibility of differences in tumor characteristics between primary uveal melanomas and their metastasis.
Another potential confounding factor is the change in analysis methods during the study period. Of 87 patients, 21 patients were analyzed using TruSeq Amplicom 48 Gene Cancer Panel with the MiSeq system for GNAQ and GNA11 mutations, while the rest were analyzed using 592 genes panel with the NexrSeq instrument for NGS assay. Since individual assays were validated for commercial use, we believe the results of GNAQ/GNA11 mutation analysis in individual assays are reliable and consistent. We did not include PLCB4 and CYSLTR2 mutational status for our analysis since those data were not available for analysis in our dataset. Six specimens that had no GNAQ and GNA11 mutations might be identified to have either PLCB4 or CYSLTR2.
In terms of BAP1 alteration analysis, it is possible that our assays underestimate the frequency of dysfunctional BAP1 in tumors since we did not check the intron sequences for BAP1 gene. Since most tissue specimens were exhausted for NGS assay, we were not able to check the expression of BAP1 protein in tissue specimens. The sample size of this study is too small to reliably investigate the role of BAP1 alternation and SF3B1 mutations in survival of metastatic uveal melanoma patients and this remains to be investigated in future studies.
5. Conclusions
This clinical study indicates that MUM tumors with different mutations of Q209 in GNAQ and GNA11 might have different characteristics in terms of survival and response to treatments after development of systemic metastasis. Among patients with MUM, Q209P mutation in their tumor specimens would have a more favorable prognosis than those with Q209L mutation after development of metastases. This might indicate a different signal transduction pattern between these two mutations and well-designed molecular studies should be considered to identify the difference between Q209P compared to GNAQ/GNA11 Q209L mutant UM cells.
Acknowledgments
Authors thank Pamela Walter (Office for Professional Writing Publishing and Communication) at Thomas Jefferson University for carefully reading and editing this manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13225749/s1, Table S1: Frequency of Mutations in Metastatic Uveal Melanoma Tissues, Table S2: Percentage of the Estimated Tumor DNA in Specimens and Frequency of Genetic Variants, Table S3: Association between BAP1 Mutation Status and Survival from Metastasis after Removing Patients with SF3B1 Mutations (n = 43), Table S4: Results from the Multivariable Cox Model for OS from Met to Death in 50 Patients with Known BAP1 Alteration status.
Author Contributions
Conceptualization, M.T. and T.S.; formal analysis, A.S. and I.C.; data collection, M.T., L.H., M.D. and J.S.; writing original draft preparation, M.T. and A.S.; writing—review and editing, M.T., A.S., I.C., L.H., M.D., J.S., M.O., P.B.W., J.L.B., A.E.A. and T.S.; Supervision, T.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Mark Weinzierl Research Fund, the Eye Melanoma Research Fund and the Jill and Kevin Plancher Research Fund at Thomas Jefferson University, and the Ralph and Marian Falk Medical Research Trust Bank of America, N.A., Trustee (U07705). A.E. Aplin was supported by grants from the NIH: R01CA257505 and R01CA253977.
Institutional Review Board Statement
This study was approved by the Institutional Review Board at Thomas Jefferson University (IRB#18D-183).
Informed Consent Statement
This study was approved by the IRB at Thomas Jefferson University (#18D-183). Patient consent was waived due to this study being a retrospective analysis.
Data Availability Statement
The data presented in this study are available in the supplementary material. The further data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
J.S. is employee of Caris Life Sciences. The other authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aronow M.E., Topham A.K., Singh A.D. Uveal Melanoma: 5-Year Update on Incidence, Treatment, and Survival (SEER 1973-2013) Ocul. Oncol. Pathol. 2018;4:145–151. doi: 10.1159/000480640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaliki S., Shields C.L. Uveal melanoma: Relatively rare but deadly cancer. Eye. 2017;31:241–257. doi: 10.1038/eye.2016.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Kooij M.K., Speetjens F.M., van der Burg S.H., Kapiteijn E. Uveal Versus Cutaneous Melanoma; Same Origin, Very Distinct Tumor Types. Cancers. 2019;11:845. doi: 10.3390/cancers11060845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chacon M., Pfluger Y., Angel M., Waisberg F., Enrico D. Uncommon Subtypes of Malignant Melanomas: A Review Based on Clinical and Molecular Perspectives. Cancers. 2020;12:2362. doi: 10.3390/cancers12092362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlsson J., Nilsson L.M., Mitra S., Alsen S., Shelke G.V., Sah V.R., Forsberg E.M.V., Stierner U., All-Eriksson C., Einarsdottir B., et al. Molecular profiling of driver events in metastatic uveal melanoma. Nat. Commun. 2020;11:1894. doi: 10.1038/s41467-020-15606-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russo D., Di Crescenzo R.M., Broggi G., Merolla F., Martino F., Varricchio S., Ilardi G., Borzillo A., Carandente R., Pignatiello S., et al. Expression of P16INK4a in Uveal Melanoma: New Perspectives. Front. Oncol. 2020;10:562074. doi: 10.3389/fonc.2020.562074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demicheli R., Fornili M., Biganzoli E. Bimodal mortality dynamics for uveal melanoma: A cue for metastasis development traits? BMC Cancer. 2014;14:392. doi: 10.1186/1471-2407-14-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seedor R.S., Eschelman D.J., Gonsalves C.F., Adamo R.D., Orloff M., Amjad A., Sharpe-Mills E., Chervoneva I., Shields C.L., Shields J.A., et al. An Outcome Assessment of a Single Institution’s Longitudinal Experience with Uveal Melanoma Patients with Liver Metastasis. Cancers. 2020;12:117. doi: 10.3390/cancers12010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Raamsdonk C.D., Griewank K.G., Crosby M.B., Garrido M.C., Vemula S., Wiesner T., Obenauf A.C., Wackernagel W., Green G., Bouvier N., et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shoushtari A.N., Carvajal R.D. GNAQ and GNA11 mutations in uveal melanoma. Melanoma Res. 2014;24:525–534. doi: 10.1097/CMR.0000000000000121. [DOI] [PubMed] [Google Scholar]
- 11.Moore A.R., Ceraudo E., Sher J.J., Guan Y., Shoushtari A.N., Chang M.T., Zhang J.Q., Walczak E.G., Kazmi M.A., Taylor B.S., et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat. Genet. 2016;48:675–680. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson P., Aoude L.G., Wadt K., Glasson W.J., Warrier S.K., Hewitt A.W., Kiilgaard J.F., Heegaard S., Isaacs T., Franchina M., et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7:4624–4631. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson A.G., Shih J., Yau C., Gibb E.A., Oba J., Mungall K.L., Hess J.M., Uzunangelov V., Walter V., Danilova L., et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell. 2017;32:204–220.e215. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirley M.D., Tang H., Gallione C.J., Baugher J.D., Frelin L.P., Cohen B., North P.E., Marchuk D.A., Comi A.M., Pevsner J. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N. Engl. J. Med. 2013;368:1971–1979. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J.J., Diefenbach R.J., Joshua A.M., Kefford R.F., Carlino M.S., Rizos H. Oncogenic signaling in uveal melanoma. Pigment. Cell Melanoma Res. 2018;31:661–672. doi: 10.1111/pcmr.12708. [DOI] [PubMed] [Google Scholar]
- 16.Chua V., Lapadula D., Randolph C., Benovic J.L., Wedegaertner P.B., Aplin A.E. Dysregulated GPCR Signaling and Therapeutic Options in Uveal Melanoma. Mol. Cancer Res. 2017;15:501–506. doi: 10.1158/1541-7786.MCR-17-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onken M.D., Worley L.A., Long M.D., Duan S., Council M.L., Bowcock A.M., Harbour J.W. Oncogenic mutations in GNAQ occur early in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2008;49:5230–5234. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koopmans A.E., Vaarwater J., Paridaens D., Naus N.C., Kilic E., de Klein A., Rotterdam Ocular Melanoma Study group Patient survival in uveal melanoma is not affected by oncogenic mutations in GNAQ and GNA11. Br. J. Cancer. 2013;109:493–496. doi: 10.1038/bjc.2013.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer J., Kilic E., Vaarwater J., Bastian B.C., Garbe C., de Klein A. Oncogenic GNAQ mutations are not correlated with disease-free survival in uveal melanoma. Br. J. Cancer. 2009;101:813–815. doi: 10.1038/sj.bjc.6605226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griewank K.G., van de Nes J., Schilling B., Moll I., Sucker A., Kakavand H., Haydu L.E., Asher M., Zimmer L., Hillen U., et al. Genetic and clinico-pathologic analysis of metastatic uveal melanoma. Mod. Pathol. 2014;27:175–183. doi: 10.1038/modpathol.2013.138. [DOI] [PubMed] [Google Scholar]
- 21.Field M.G., Durante M.A., Anbunathan H., Cai L.Z., Decatur C.L., Bowcock A.M., Kurtenbach S., Harbour J.W. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat. Commun. 2018;9:116. doi: 10.1038/s41467-017-02428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh A.D., Zabor E.C., Radivoyevitch T. Estimating Cured Fractions of Uveal Melanoma. JAMA Ophthalmol. 2021;139:174–181. doi: 10.1001/jamaophthalmol.2020.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szalai E., Jiang Y., van Poppelen N.M., Jager M.J., de Klein A., Kilic E., Grossniklaus H.E. Association of Uveal Melanoma Metastatic Rate With Stochastic Mutation Rate and Type of Mutation. JAMA Ophthalmol. 2018;136:1115–1120. doi: 10.1001/jamaophthalmol.2018.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valsecchi M.E., Terai M., Eschelman D.J., Gonsalves C.F., Chervoneva I., Shields J.A., Shields C.L., Yamamoto A., Sullivan K.L., Laudadio M., et al. Double-blinded, randomized phase II study using embolization with or without granulocyte-macrophage colony-stimulating factor in uveal melanoma with hepatic metastases. J. Vasc. Interv. Radiol. 2015;26:523–532.e522. doi: 10.1016/j.jvir.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel K., Sullivan K., Berd D., Mastrangelo M.J., Shields C.L., Shields J.A., Sato T. Chemoembolization of the hepatic artery with BCNU for metastatic uveal melanoma: Results of a phase II study. Melanoma Res. 2005;15:297–304. doi: 10.1097/00008390-200508000-00011. [DOI] [PubMed] [Google Scholar]
- 26.Gonsalves C.F., Eschelman D.J., Adamo R.D., Anne P.R., Orloff M.M., Terai M., Hage A.N., Yi M., Chervoneva I., Sato T. A Prospective Phase II Trial of Radioembolization for Treatment of Uveal Melanoma Hepatic Metastasis. Radiology. 2019;293:223–231. doi: 10.1148/radiol.2019190199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eschelman D.J., Gonsalves C.F., Sato T. Transhepatic therapies for metastatic uveal melanoma. Semin. Intervent. Radiol. 2013;30:39–48. doi: 10.1055/s-0033-1333652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luscan A., Just P.A., Briand A., Burin des Roziers C., Goussard P., Nitschke P., Vidaud M., Avril M.F., Terris B., Pasmant E. Uveal melanoma hepatic metastases mutation spectrum analysis using targeted next-generation sequencing of 400 cancer genes. Br. J. Ophthalmol. 2015;99:437–439. doi: 10.1136/bjophthalmol-2014-305371. [DOI] [PubMed] [Google Scholar]
- 29.McCarthy C., Kalirai H., Lake S.L., Dodson A., Damato B.E., Coupland S.E. Insights into genetic alterations of liver metastases from uveal melanoma. Pigment. Cell Melanoma Res. 2016;29:60–67. doi: 10.1111/pcmr.12433. [DOI] [PubMed] [Google Scholar]
- 30.Yavuzyigitoglu S., Koopmans A.E., Verdijk R.M., Vaarwater J., Eussen B., van Bodegom A., Paridaens D., Kilic E., de Klein A., Rotterdam Ocular Melanoma Study G. Uveal Melanomas with SF3B1 Mutations: A Distinct Subclass Associated with Late-Onset Metastases. Ophthalmology. 2016;123:1118–1128. doi: 10.1016/j.ophtha.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 31.Van Raamsdonk C.D., Bezrookove V., Green G., Bauer J., Gaugler L., O’Brien J.M., Simpson E.M., Barsh G.S., Bastian B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shain A.H., Bagger M.M., Yu R., Chang D., Liu S., Vemula S., Weier J.F., Wadt K., Heegaard S., Bastian B.C., et al. The genetic evolution of metastatic uveal melanoma. Nat. Genet. 2019;51:1123–1130. doi: 10.1038/s41588-019-0440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ewens K.G., Kanetsky P.A., Richards-Yutz J., Purrazzella J., Shields C.L., Ganguly T., Ganguly A. Chromosome 3 status combined with BAP1 and EIF1AX mutation profiles are associated with metastasis in uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2014;55:5160–5167. doi: 10.1167/iovs.14-14550. [DOI] [PubMed] [Google Scholar]
- 34.Harbour J.W., Chen R. The DecisionDx-UM Gene Expression Profile Test Provides Risk Stratification and Individualized Patient Care in Uveal Melanoma. PLoS Curr. 2013;5 doi: 10.1371/currents.eogt.af8ba80fc776c8f1ce8f5dc485d4a618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broggi G., Ieni A., Russo D., Varricchio S., Puzzo L., Russo A., Reibaldi M., Longo A., Tuccari G., Staibano S., et al. The Macro-Autophagy-Related Protein Beclin-1 Immunohistochemical Expression Correlates With Tumor Cell Type and Clinical Behavior of Uveal Melanoma. Front. Oncol. 2020;10:589849. doi: 10.3389/fonc.2020.589849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Field M.G., Decatur C.L., Kurtenbach S., Gezgin G., van der Velden P.A., Jager M.J., Kozak K.N., Harbour J.W. PRAME as an Independent Biomarker for Metastasis in Uveal Melanoma. Clin. Cancer Res. 2016;22:1234–1242. doi: 10.1158/1078-0432.CCR-15-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Weeghel C., Wierenga A.P.A., Versluis M., van Hall T., van der Velden P.A., Kroes W.G.M., Pfeffer U., Luyten G.P.M., Jager M.J. Do GNAQ and GNA11 Differentially Affect Inflammation and HLA Expression in Uveal Melanoma? Cancers. 2019;11:1127. doi: 10.3390/cancers11081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maziarz M., Leyme A., Marivin A., Luebbers A., Patel P.P., Chen Z., Sprang S.R., Garcia-Marcos M. Atypical activation of the G protein Galphaq by the oncogenic mutation Q209P. J. Biol. Chem. 2018;293:19586–19599. doi: 10.1074/jbc.RA118.005291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amirouchene-Angelozzi N., Nemati F., Gentien D., Nicolas A., Dumont A., Carita G., Camonis J., Desjardins L., Cassoux N., Piperno-Neumann S., et al. Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Mol. Oncol. 2014;8:1508–1520. doi: 10.1016/j.molonc.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes S., Damato B.E., Giddings I., Hiscott P.S., Humphreys J., Houlston R.S. Microarray comparative genomic hybridisation analysis of intraocular uveal melanomas identifies distinctive imbalances associated with loss of chromosome 3. Br. J. Cancer. 2005;93:1191–1196. doi: 10.1038/sj.bjc.6602834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parrella P., Caballero O.L., Sidransky D., Merbs S.L. Detection of c-myc amplification in uveal melanoma by fluorescent in situ hybridization. Investig. Ophthalmol. Vis. Sci. 2001;42:1679–1684. [PubMed] [Google Scholar]
- 42.Ehlers J.P., Worley L., Onken M.D., Harbour J.W. DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clin. Cancer Res. 2005;11:3609–3613. doi: 10.1158/1078-0432.CCR-04-1941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available in the supplementary material. The further data that support the findings of this study are available from the corresponding author upon reasonable request.