Abstract
An assay that quantifies the amount of human immunodeficiency virus type 1 (HIV-1) DNA in peripheral blood mononuclear cells has been developed. PCR amplification of the HIV-1 DNA is performed in the presence of an internal quantitation standard, and colorimetric detection of the amplified product is performed with microwell plates. The copies of HIV-1 DNA are normalized to total genomic DNA input. The assay has an analytical sensitivity of 10 input copies per amplification reaction and a three-log detection range. In an analysis of sequential samples from patients on combination therapy, HIV-1 DNA was quantifiable for all individuals tested, including those with undetectable plasma HIV-1 RNA. In a separate study, a comparison of HIV-1 DNA levels was made with a group of long-term survivors and progressors. The mean HIV-1 DNA levels were lower in the long-term survivors than in the progressors (P, 0.04). The mean HIV-1 RNA levels were also lower, but the difference was not statistically significant (P, 0.164). A quantitative DNA assay will provide an additional tool to gain insight into the natural history of infection and the continued efficacy of potent antiretroviral therapies.
Quantitation of human immunodeficiency virus type 1 (HIV-1) RNA in plasma is a powerful prognostic tool in clinical practice (15). Further, studies have shown that reductions in plasma viral load correlate with improved clinical outcome in patients receiving antiretroviral therapy (3, 5, 12, 13, 20). In this era of highly active antiretroviral therapy (HAART), reduction of plasma viral load to below the 50-copy/ml detection limit of the AMPLICOR HIV-1 MONITOR test (Roche Molecular Systems, Pleasanton, Calif.) with the ultrasensitive extraction method is often achieved. Consequently, alternative methods are needed to enable further evaluation of the efficacy of therapy in a given individual as well as comparison of different regimens between groups in clinical trials. An assay that quantifies HIV-1 DNA load in the peripheral blood mononuclear cells (PBMCs) of infected individuals is described here. Although other HIV-1 DNA quantitative assays have been previously described, they have required multiple amplifications and/or detection with radiolabeled probes (2, 6, 10). This assay is almost identical to the AMPLICOR HIV-1 MONITOR assay for RNA (Roche Molecular Systems) but differs in (i) the sample preparation method, (ii) the use of plasmid DNA rather than an RNA transcript as the quantitation standard, and (iii) the normalization of DNA load to total cellular input. The assay uses the AMPLICOR HIV-1 MONITOR version 1.5 primers that have a broad subtype detection range (17, 23).
MATERIALS AND METHODS
Clinical specimens.
Nineteen patients from a triple-combination trial (zidovudine, didanosine, and nevirapine) were chosen for HIV-1 DNA quantification. These patients had HIV-1 RNA levels that dropped precipitously after the initiation of therapy, and many were below the detection limit of the ultrasensitive assay.
Five long-term survivors and 10 progressors were selected from San Francisco Men's Health Study participants on the basis of the rate of decline in CD4+ lymphocytes in peripheral blood. Long-term survivors were chosen from deciles 9 and 10, which had the lowest rate of CD4+ lymphocyte decline, as described previously (22). When samples were collected for analysis for HIV-1 DNA and RNA, long-term survivors had been HIV-1 seropositive for more than 10 years.
Sample preparation.
PBMCs are separated from whole blood by Ficoll-Hypaque gradient centrifugation. The purified cells are either processed immediately or stored frozen at −70°C in freezing medium (90% fetal calf serum, 10% dimethyl sulfoxide) until ready for processing. Frozen cells are thawed and separated from freezing medium by centrifugation at 16,000 × g for 3 min. Cells are washed twice with 1 ml of Specimen Wash Buffer (Roche Molecular Systems) and pelleted by centrifugation at 16,000 × g for 3 min. The supernatant is removed, and the final pellet is resuspended in extraction buffer, containing 0.1 mg of proteinase K per ml, 0.05% Nonidet P-40, 0.05% Tween 20, and the quantitation standard (QS). The QS is a linearized pSP64 plasmid DNA in which a 162-bp synthetic DNA fragment that contains the HIV-1 primer binding sites has been inserted. When the QS is amplified, a product of the same length as the HIV-1 target is generated. The probe region for the QS is distinct from that for the HIV-1 target and allows for differentiation by the microwell plate assay in a manner analogous to that used to quantitate HIV-1 RNA copy number in the AMPLICOR HIV-1 MONITOR assay. The amount of extraction reagent varies, but 125 μl is typically used to extract DNA from 106 cells. The cells are then incubated at 60°C for 30 min, after which the proteinase K is inactivated at 100°C for 30 min.
Quantification of total DNA.
The level of HIV-1 DNA is expressed as copies per microgram of cellular DNA. The amount of cellular DNA released by the extraction process is quantified with Hoechst dye (bisbenzimide) as recommended by the manufacturer (Pharmacia). When Hoechst dye, which binds to the minor groove of DNA, is excited by 365-nm light in the presence of DNA, it fluoresces at 458 nm. Fluorescence is measured with a fluorometer that has been calibrated using calf thymus DNA of a known concentration. Two microliters of the extracted sample lysate is diluted in 2 ml of the recommended buffer, and the concentration is determined. The extraction protocol used in our test yields single-stranded DNA. Since the amount of Hoechst dye that binds to single-stranded DNA is half as much as the amount that binds to the double-stranded DNA standard, the DNA concentration determined following extraction is doubled (see Results).
Amplification and detection.
DNA amplification reactions are performed with 50 μl of sample lysate using the AMPLICOR HIV-1 MONITOR master mix (version 1.0) that contains primers SK462 and SK431 (18) or the AMPLICOR HIV-1 MONITOR master mix (version 1.5) that contains primers SK145 and SKCC1B (17). Primers SK145 and SKCC1B have been shown to efficiently amplify the different subtypes of HIV-1 group M (17, 23). Amplifications are performed with rTth DNA polymerase, which has both reverse transcription and DNA polymerase activities (16). To prevent carryover contamination from previous amplifications, dUTP and uracil-N-glycosylase are incorporated into the reactions (14). The cycling parameters used for the version 1.0 master mix are identical to those used for the AMPLICOR HIV-1 MONITOR assay but without the reverse transcription step. The cycling parameters used for the version 1.5 master mix are as follows: 2 min at 50°C; 4 cycles at 95, 52, and 72°C for 10 s each; and 26 cycles at 90, 55, and 72°C for 10 s each.
Following amplification, the PCR products are denatured with NaOH and detected with microwell plates coated with either the HIV-1-specific probe or the QS-specific probe, as for the AMPLICOR HIV-1 MONITOR test (18).
Plasma HIV-1 RNA levels are determined using either the Ultradirect method (19) or the AMPLICOR HIV-1 MONITOR test.
Data reduction.
The DNA copy number is calculated by using the following formula: HIV-1 copies = (total HIV-1 optical density/total QS optical density) × input QS copy number. The HIV-1 copies are then normalized to total genomic DNA and expressed as HIV-1 copies per microgram of extracted DNA.
RESULTS
Effect of sample integrity on Hoechst dye determinations.
Since the lysis procedure involves heating the samples to 100°C for 30 min, we evaluated the capacity of Hoechst dye to bind to denatured and presumably single-stranded DNA. Using calf thymus DNA as a model system, we compared the amount of Hoechst dye that was bound to DNA that was denatured either by heating at 100°C or treatment with 0.1 M NaOH. The amount of dye that bound to the denatured DNA was about 50% the amount that bound to the untreated DNA, regardless of the treatment used. The data showed that the heat treatment was sufficient to completely render the DNA single stranded, as additional treatment with 0.1 M NaOH had no further effect on Hoechst dye-based quantitation. Similar results were obtained with clinical samples treated with either heat alone or heat with NaOH (data not shown). Consequently, a twofold correction factor was used to adjust for differences in dye binding capacity between single- and double-stranded DNAs. The observed twofold difference in this model study was consistent with numerous comparisons of cell counts with Hoechst dye for the clinical specimens (data not shown).
Comparison of Hoechst dye to cell counts.
Using a conversion factor of 1 μg of DNA per 150,000 cells, we compared PBMC counts extrapolated from the Hoechst dye determinations to the actual cell counts determined with a hemocytometer. PBMC aliquots ranging from 107 cells to 78,000 cells were processed in 125 μl of extraction reagent. For each Hoechst dye determination, 2 μl of lysate (representing 1,248 to 160,000 cells) was evaluated. The correlation of the actual cell number with the cell number extrapolated from total DNA determinations with Hoechst dye indicated that the latter method can be used to accurately measure total DNA in cell lysates from 5,000 to 160,000 cells but overestimates the DNA in cell lysates from 1,000 to 5,000 cells. The accuracy of cell counts extrapolated from the Hoechst dye method was compared to that of cell counts measured directly. Of 29 clinical samples evaluated, 24 samples (83%) showed less than a 2-fold difference between the two methods, while the remaining 5 samples (17%) showed between 2- and 2.5-fold differences. The data showed that the cell numbers determined by the Hoechst dye method were comparable to cell counts for clinical specimens.
Extraction efficiency.
To evaluate extraction efficiency as a function of cell density, various amounts of PBMCs from four patients were extracted in triplicate with either 200, 300, or 400 μl of lysis reagent. The number of cells extracted from each patient specimen ranged from 5.7 × 105 to 1.7 × 106 PBMCs. The HIV-1 DNA copies per microgram of total DNA were similar within the patient groups, indicating that efficient lysis was achieved despite differences in cell densities and that a higher cellular input did not interfere with amplification (Table 1).
TABLE 1.
Effect of lysis volume on efficiency of extraction
| Sample | Lysis vol (μl) | No. of HIV-1 DNA copies/PCR | Total DNA concn (μg/PCR) | No. of HIV-1 DNA copies/μg of DNA |
|---|---|---|---|---|
| 79 | 200 | 1,480 | 2.75 | 538 |
| 300 | 1,245 | 1.9 | 655 | |
| 400 | 838 | 1.45 | 578 | |
| 80 | 200 | 472 | 0.95 | 497 |
| 300 | 333 | 0.75 | 444 | |
| 400 | 276 | 0.65 | 425 | |
| 83 | 200 | 663 | 1.65 | 402 |
| 300 | 605 | 1.25 | 484 | |
| 400 | 411 | 0.95 | 433 | |
| 84 | 200 | 1,480 | 2.8 | 529 |
| 300 | 1,014 | 1.9 | 534 | |
| 400 | 536 | 0.95 | 564 |
Sensitivity and linearity range.
To establish the sensitivity and dynamic range of the assay, serial dilutions of a plasmid DNA that contains the HIV-1 sequence were amplified in a background of about 1.3 μg of DNA extracted from a seronegative donor. Linearity was achieved over a range of 10 to 32,000 copies/PCR with an analytical sensitivity of 10 copies of HIV-1 DNA (data not shown).
Assay performance with 8E5 cells.
To determine the ability of the assay to both efficiently extract and quantify HIV-1 DNA in infected cells, we evaluated a dilution panel of 8E5 cells. 8E5 cells, which harbor a single copy of HIV-1 DNA (9), were diluted into negative PBMCs, resulting in expected HIV-1 DNA levels ranging from 400,000 to 10 copies/PCR. Each sample was extracted in 125 μl of extraction reagent and amplified in duplicate. Consistent with the plasmid studies, linearity r2, 0.996; y, 1.0x + 0.07) was achieved at between 10 and 25,000 copies of HIV-1 DNA. Good correlation was observed between the expected numbers based on cell counts and the numbers determined by PCR. In separate experiments, a coded 8E5 cell panel was analyzed, and the results showed that the assay correctly quantified the number of 8E5 cells in each sample and was able to detect as few as 3.2 8E5 cells per 106 cells (R. Coombs, D. Brambilla, J. Bremer, R. Dickover, T. Greenough, S. Kwok, H. Lin, C. Michels, C. Mundy, M. Nowicki, G. Peterson, B. Staes, J. Sullivan, S. Spector, P. Reichelderfer, and B. Yen-Lieberman, Program Abstr. 6th Conf. Retroviruses Opportunistic Infections, abstr. 176, p. 101, 1999).
Assay reproducibility.
To assess the reproducibility of the assay, the percent coefficients of variation (CV) for amplification and detection and for the total DNA concentrations determined with Hoechst dye were calculated for 192 individual clinical samples from 23 different patients. For amplification and detection, the CV were <15% for 105 of the 192 samples, 15 to 30% for 61 samples, and >30% for 26 samples. A CV of <15% was observed for 190 of 192 (99%) of the Hoechst dye determinations, and a CV of >15% was observed for 2 of the samples. To assess the reproducibility of the entire assay, 12 samples from 12 patients were extracted in duplicate, and the percent CV for the final quantitation results was calculated. A CV of <10% was observed for six samples; the CV were 10 to 20% for two samples, 20 to 30% for three samples, and 45% for one sample.
Specificity of the assay for DNA.
The master mix used in this assay contains rTth DNA polymerase which, in the presence of manganese, has both reverse transcription and DNA polymerase activities (16). The extraction procedure used for the assay releases total nucleic acid from the cells. Although the assay does not include a reverse transcription step, intracellular HIV-1 RNA could conceivably be reverse transcribed during the annealing step of the PCR provided the integrity of the RNA was maintained. To determine what contribution intracellular HIV-1 RNA has on DNA quantification, we evaluated (i) the effect of the 100°C heat step (proteinase K inactivation) on the integrity of the RNA and (ii) the extent to which RNA can be reverse transcribed without a 30-min reverse transcription step at 60°C. We found that a purified RNA transcript can be reverse transcribed and amplified after the 100°C treatment. No amplification was observed in the absence of the reverse transcription step. However, when purified RNA was added directly to proteinase K-treated cellular lysates, no amplification was observed regardless of whether the reverse transcription step was incorporated. These results suggest that cellular RNases alone sufficiently degrade intracellular HIV-1 RNA to prevent its amplification in this assay. The elimination of the reverse transcription step further ensures that intracellular HIV-1 RNA will not interfere with HIV-1 DNA quantification. These results were confirmed by amplifications with Taq DNA polymerase, which lacks reverse transcription activity in the presence of magnesium.
Clinical data.
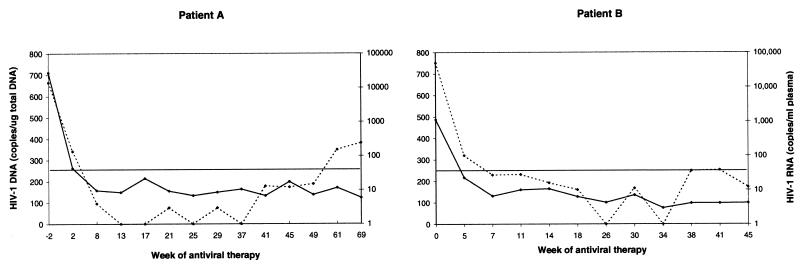
HIV-1 DNA levels in sequential samples from 19 individuals treated with a triple combination of zidovudine, didanosine, and nevirapine were quantified. HIV-1 DNA was detected and quantified for all of the patients tested, including those who had undetectable HIV-1 RNA levels. The data from two representative patients show that the kinetics of DNA decay are slow, even in the presence of nearly maximal suppression of HIV-1 replication in plasma (Fig. 1).
FIG. 1.
Longitudinal evaluation of HIV-1 RNA and DNA levels in the circulation of two patients on HAART with zidovudine, didanosine, and nevirapine. The solid line represents HIV-1 DNA in copies per microgram, and the broken line represents HIV-1 RNA levels in copies per milliliter. The solid horizontal line represents the 50-copy/ml cutoff for HIV-1 RNA levels.
To gain insight into the possible role of HIV-1 DNA levels as prognostic markers in clinical practice, 5 long-term survivors and 10 progressors from the San Francisco Men's Health Study (1, 22) were evaluated. All San Francisco Men's Health Study subjects analyzed were HIV-1 seropositive at cohort entry in 1984. Cryopreserved samples collected at approximately 6 years and 12 years after cohort entry in the progressors and long-term survivors, respectively, were analyzed for HIV-1 DNA in PBMCs. Despite the fact that the long-term survivors had been infected for 6 years longer than the progressors, the CD4+ cell counts were significantly higher (P value determined by unpaired two-tailed t test, <0.00003) in the long-term survivors (582 ± 142) than in the progressors (165 ± 110). The long-term survivors had not been treated with antiretroviral therapy, with the exception of patient F, who had been on zidovudine monotherapy 2 years previously. The mean HIV-1 DNA levels were 175 copies/μg of total DNA for the long-term survivors and 964 copies/μg of total DNA for the progressors (P, 0.04) (Table 2). The median HIV-1 DNA levels were 50 copies/μg of total DNA for the long-term survivors and 638 copies/μg of total DNA for the progressors. The mean HIV-1 RNA levels were 6,748 copies/ml for the survivors and 506,942 copies/ml for the progressors (with the exception of patient O, whose plasma was not available) (P, 0.164). The HIV-1 RNA and DNA levels were highly correlated (P, < 0.001).
TABLE 2.
HIV-1 DNA and RNA levels in the circulation in long-term survivors and progressors
| Group | No. of:
|
||
|---|---|---|---|
| Patient | HIV-1 DNA copies/μg of total DNA | HIV-1 RNA copies/ml | |
| Long-term survivors | A | 50 | 2,174 |
| B | 33 | 7,263 | |
| C | 9 | <200 | |
| D | 708 | 23,667 | |
| E | 76 | 434 | |
| Progressors | F | 1,020 | 270,833 |
| G | 537 | 17,305 | |
| H | 1,111 | 153,930 | |
| I | 548 | 76,688 | |
| J | 1,647 | 1,558,777 | |
| K | 497 | 58,320 | |
| L | 253 | 79,264 | |
| M | 2,746 | 2,008,893 | |
| N | 564 | 338,467 | |
| O | 713 | Not available | |
DISCUSSION
We have developed a prototype assay that quantifies HIV-1 DNA in PBMCs of infected individuals. DNA is extracted from PBMCs with a buffer that contains proteinase K and detergent. A DNA quantitation standard is coamplified with each sample and is used to normalize reaction variability. Quantitation is performed with microwell plates by colorimetric detection, as for the AMPLICOR HIV-1 MONITOR test. The level of HIV-1 DNA in PBMCs is normalized to the amount of genomic DNA determined by Hoechst dye-based quantitation. HIV-1 DNA levels are typically reported as copies/106 cells or as copies per microgram of total DNA; cell counts are determined either with a hemocytometer or with a Coulter counter, and DNA concentrations are determined spectrophotometrically. The Hoechst dye procedure eliminates the need for cell counts, is simple to use, and provides accurate quantitation. The determinations can be performed on crude lysates and eliminate the organic extractions required for accurate spectrophotometric determinations. Additionally, Hoechst dye measures the amount of DNA released, whereas normalization to cell counts assumes complete lysis of the cells.
In this assay, DNA from as many as 3 × 106 cells can be efficiently lysed and amplified. Although about 2.5 μg of DNA (from about 400,000 cells) is typically amplified, the need for a higher input will depend on the level of HIV-1 DNA in the specimen. The results showed that varying the amount of extraction reagent did not affect the final quantitation. The ability to analyze a larger sample size will improve the sensitivity and accuracy of the assay for low-copy-number samples. The current assay has a linear detection range of 10 to 32,000 copies/μg of DNA. In the analyses performed to date, the level of DNA has generally been less than 1,000 copies/μg of DNA. The quantitative HIV-1 DNA assay described here provides a valuable addition to the analytical tools already available to monitor viral replication. Analysis of sequential samples from patients on combination therapy showed that HIV-1 DNA levels in PBMCs remained detectable and quantifiable despite plasma HIV-1 RNA levels that were undetectable for up to 1 year. As reported by other groups (4, 7, 11, 21), an initial decrease in HIV-1 DNA levels was observed immediately upon initiation of therapy, followed by slower decay. The presence of detectable DNA is consistent with the successful isolation of virus from PBMCs of patients with undetectable HIV-1 RNA (8, 24). The data from the long-term survivors suggest that lower HIV-1 DNA levels may be indicative of a better prognosis; however, the sample size in this study was small. Further work is needed with a larger sample size, and longitudinal analysis may be required to determine if there is any correlation between HIV-1 DNA levels and progression.
The availability of this new standardized assay will be important in at least three ways. First, it will facilitate the comparison of the efficacy of different HAART regimens. Second, it will permit evaluation of the true depth of virological suppression in patients with nonquantifiable plasma viral loads, as any increase in HIV-1 DNA levels in this setting will be consistent with ongoing low-level viral replication in tissue stores (or perhaps even in the circulation). Third, as immune-based therapies aimed at purging HIV-1 DNA reservoirs are evaluated, this assay will be most important in quantifying their efficacy. In conclusion, an assay has been developed to accurately quantitate HIV-1 DNA in circulating PBMCs. The insightful application of this assay is expected to improve the use of antiretroviral agents in clinical practice.
ACKNOWLEDGMENTS
We thank Marisa Hong and Kevin Kwok for assistance and John Sninsky for critical review of the manuscript.
We acknowledge the support for the Men's Health Study from the NIH (grants R01 AI40010 and AI-34783). This work was also supported by Roche Molecular Systems, the manufacturer of AMPLICOR HIV-1 MONITOR.
REFERENCES
- 1.Betts M, Krowka J, Kepler T, Davidian M, Christopherson C, Kwok S, Louie L, Eron J, Sheppard H, Frelinger J. Human immunodeficiency virus type-1 (HIV-1) specific cytotoxic T lymphocyte activity is inversely correlated with HIV-1 viral load in HIV-1 infected long-term survivors. AIDS Res Hum Retrovir. 1999;15:1219–1228. doi: 10.1089/088922299310313. [DOI] [PubMed] [Google Scholar]
- 2.Bieniasz P D, Ariyoshi K, Bourelly M A, Bloor S, Foxall R B, Harwood E C, Weber J N. Variable relationship between proviral DNA load and infectious virus titre in the peripheral blood mononuclear cells of HIV-1-infected individuals. AIDS. 1993;7:803–806. doi: 10.1097/00002030-199306000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Bruisten S M, Frissen P H J, Van Swieten P, Harrigan P R, Kinghorn I, Larder B, Weigel H M, De Vries E, Regez R M, Henrichs J H, Koot M, Huisman J G. Prospective longitudinal analysis of viral load and surrogate markers in relation to clinical progression in HIV type 1-infected persons. AIDS Res Hum Retrovir. 1997;13:327–335. doi: 10.1089/aid.1997.13.327. [DOI] [PubMed] [Google Scholar]
- 4.Bruisten S M, Reiss P, Loeliger A E, Van Swieten P, Schuurman R, Boucher C, Weverling G, Huisman J G. Cellular proviral HIV type 1 DNA load persists after long-term RT-inhibitor therapy in HIV type 1 infected persons. AIDS Res Hum Retrovir. 1998;14:1053–1058. doi: 10.1089/aid.1998.14.1053. [DOI] [PubMed] [Google Scholar]
- 5.Brun-Vezinet F, Boucher C, Loveday C, Descamps D, Fauveau V, Izopet J, Jeffries D, Kaye S, Krzyanowski C, Nunn A, Schuurman R, Seigneurin J M, Tamalet C, Tedder R, Weber J, Weverling G J. HIV-1 viral load, phenotype, and resistance in a subset of drug-naive participants from the Delta trial. Lancet. 1997;350:983–990. doi: 10.1016/s0140-6736(97)03380-1. [DOI] [PubMed] [Google Scholar]
- 6.Donovan R M, Dickover R E, Goldstein E, Huth R G, Carlson J R. HIV-1 proviral copy number in blood mononuclear cells from AIDS patients on zidovudine therapy. J Acquir Immune Defic Syndr. 1991;8:766–769. [PubMed] [Google Scholar]
- 7.Finzi D, Blankson J, Siliciano J D, Margolick J B, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn T C, Chaisson R E, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano R F. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 8.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 9.Folks T M, Powell D, Lightfoote M, Koenig S, Fauci A S, Benn S, Rabson A, Daugherty D, Gendelman H E, Hoggan M D, Venkatesan S, Martin M A. Biological and biochemical characterization of cloned LEU-3− cells surviving infection with the acquired immune deficiency syndrome retrovirus. J Exp Med. 1986;164:280–290. doi: 10.1084/jem.164.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta P, Ding M, Cottrill M, Rinaldo C, Kingsley L, Wolinsky S, Mellors J. Quantitation of human immunodeficiency virus type 1 DNA and RNA by a novel internally controlled PCR assay. J Clin Microbiol. 1995;33:1670–1673. doi: 10.1128/jcm.33.6.1670-1673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho D D, Moudgil T, Alam M. Quantitation of human immunodeficiency virus type 1 in the blood of infected persons. N Engl J Med. 1989;321:1621–1625. doi: 10.1056/NEJM198912143212401. [DOI] [PubMed] [Google Scholar]
- 12.Jurriaans S, Van Gemen B, Weverling G J, Van Strijp D, Nara P, Coutinho R, Koot M, Schuitemaker H, Goudsmit J. The natural history of HIV-1 infection: virus load and virus phenotype independent determinants of clinical course? Virology. 1994;204:223–233. doi: 10.1006/viro.1994.1526. [DOI] [PubMed] [Google Scholar]
- 13.Katzenstein D, Hammer S, Hughes M, Gundacker H, Jackson J B, Fiscus S, Rasheed S, Elbeik T, Reichman R, Japour A, Merigan T, Hirsch M. The relation of virological and immunological markers to clinical outcomes after nucleoside therapy in HIV-infected adults with 200 to 500 CD4 cells per cubic millimeter. N Engl J Med. 1996;335:1091–1098. doi: 10.1056/NEJM199610103351502. [DOI] [PubMed] [Google Scholar]
- 14.Longo M C, Berninger M S, Hartley J L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 15.Mellors J W, Munoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C R. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 16.Meyers T W, Gelfand D H. Reverse transcription and DNA amplification by a Thermus thermophilus DNA polymerase. Biochemistry. 1991;30:7661–7666. doi: 10.1021/bi00245a001. [DOI] [PubMed] [Google Scholar]
- 17.Michael N L, Herman S A, Kwok S, Dreyer K, Wang J, Christopherson C, Spadoro J P, Young K K, Polonis V, McCutchan F E, Carr J, Mascola J R, Jagodzinski L L, Robb M L. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J Clin Microbiol. 1999;37:2557–2563. doi: 10.1128/jcm.37.8.2557-2563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulder J, Resnick R, Saget B, Scheibel S, Herman S, Payne H, Harrigan R, Kwok S. A rapid and simple method for extracting human immunodeficiency virus type 1 RNA from plasma: enhanced sensitivity. J Clin Microbiol. 1997;35:1278–1280. doi: 10.1128/jcm.35.5.1278-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Brien W, Hartigan P, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff M, Hamilton J. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 21.Perelson A S, Essunger P, Cao Y, Vesenen M, Hurley A, Saksela K, Markowitz M, Ho D D. Decay characteristics of HIV-1 infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 22.Sheppard H W, Lang W, Ascher M S, Vittinghoff E, Winkelstein W. The characterization of non-progressors: long-term HIV-1 infection with stable CD4+ T-cell levels. AIDS. 1993;7:1159–1166. [PubMed] [Google Scholar]
- 23.Triques K, Coste J, Perret J L, Segarra C, Mpoudi E, Reynes J, Delaporte E, Butcher A, Dreyer K, Herman S, Spadoro J, Peeters M. Efficiencies of four versions of the AMPLICOR HIV-1 MONITOR test for quantification of different subtypes of human immunodeficiency virus type 1. J Clin Microbiol. 1999;37:110–116. doi: 10.1128/jcm.37.1.110-116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1294. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]