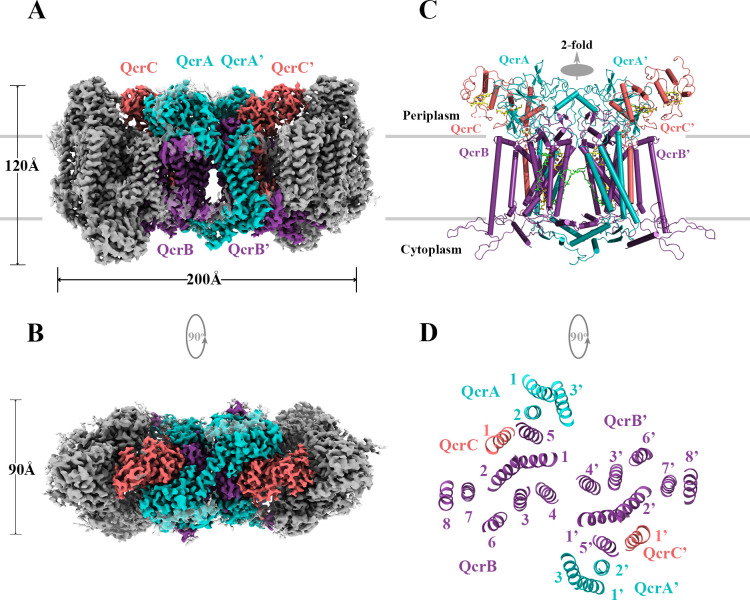
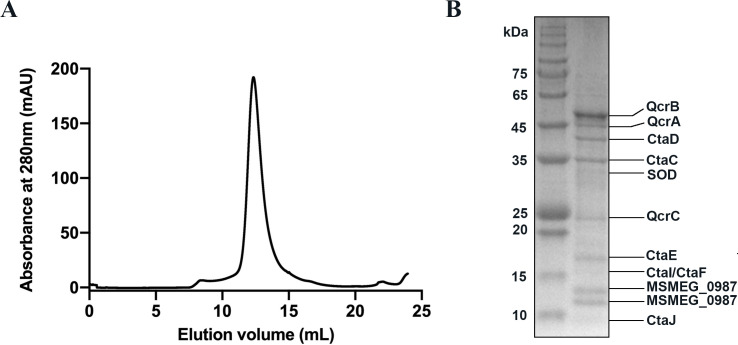
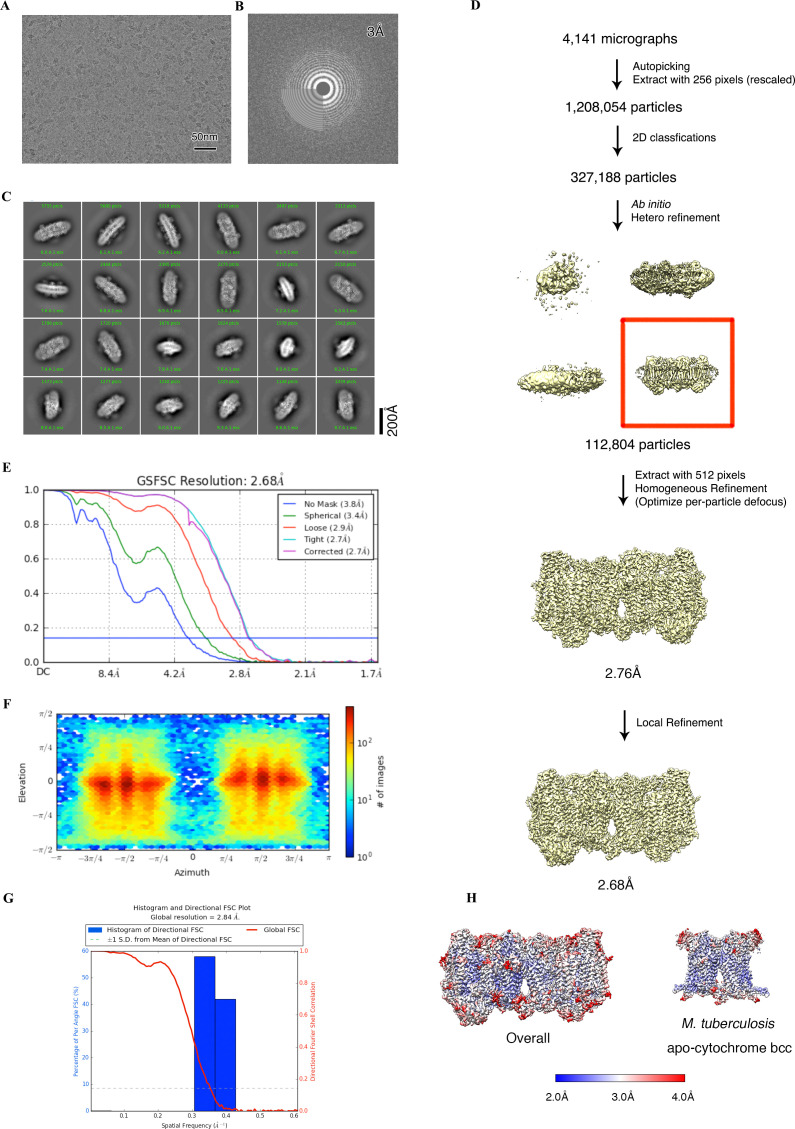
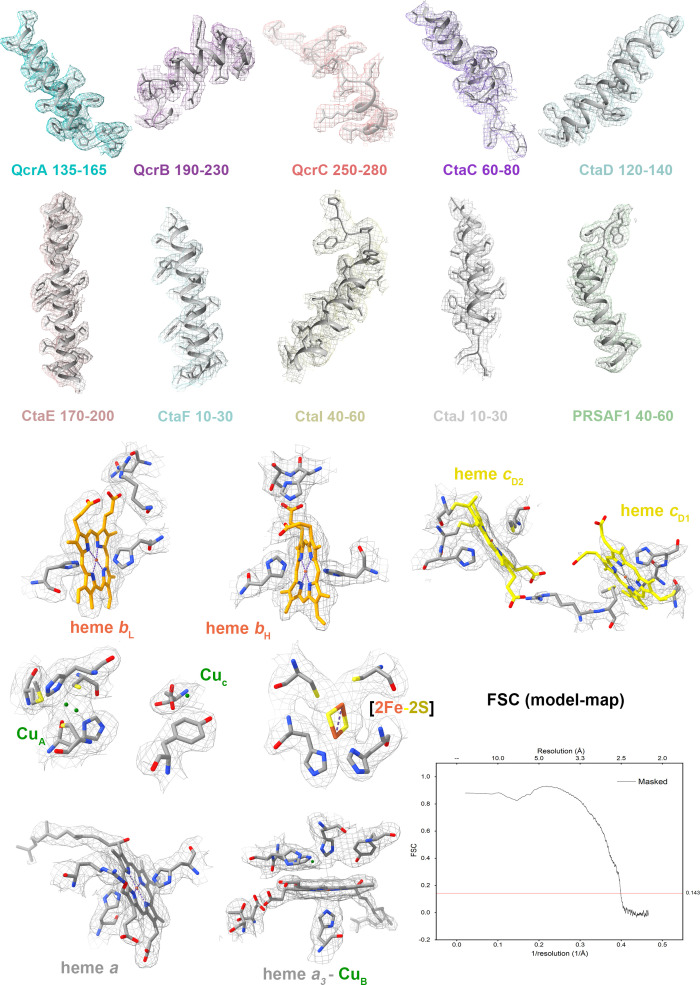
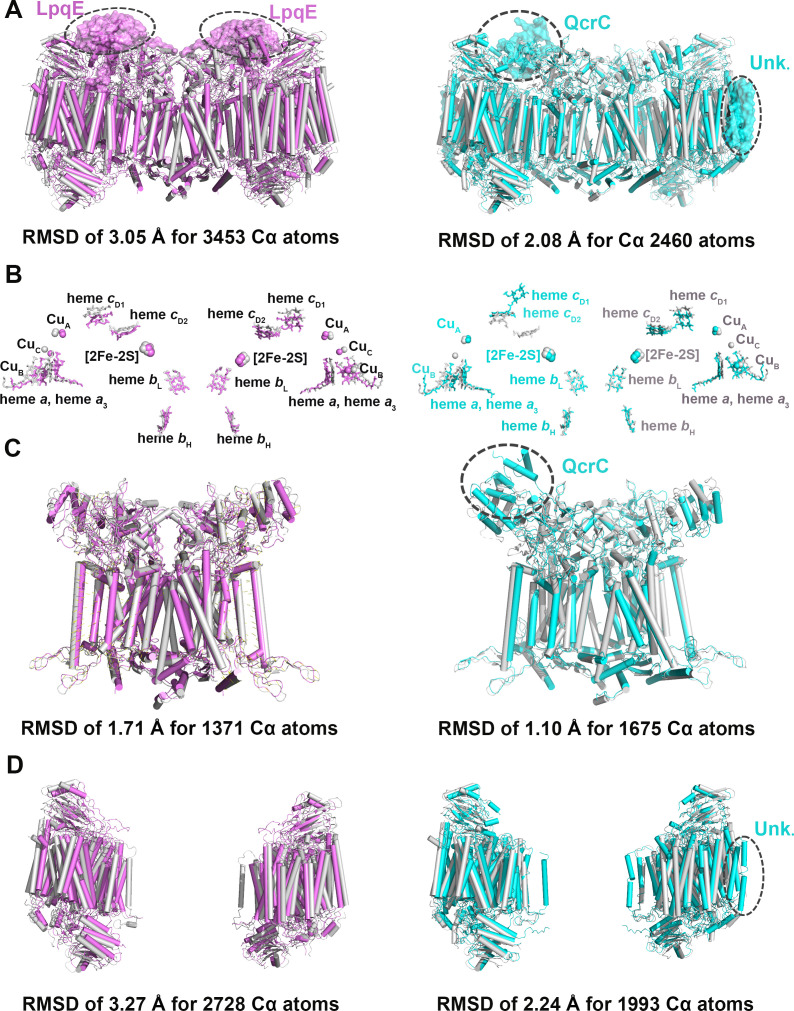
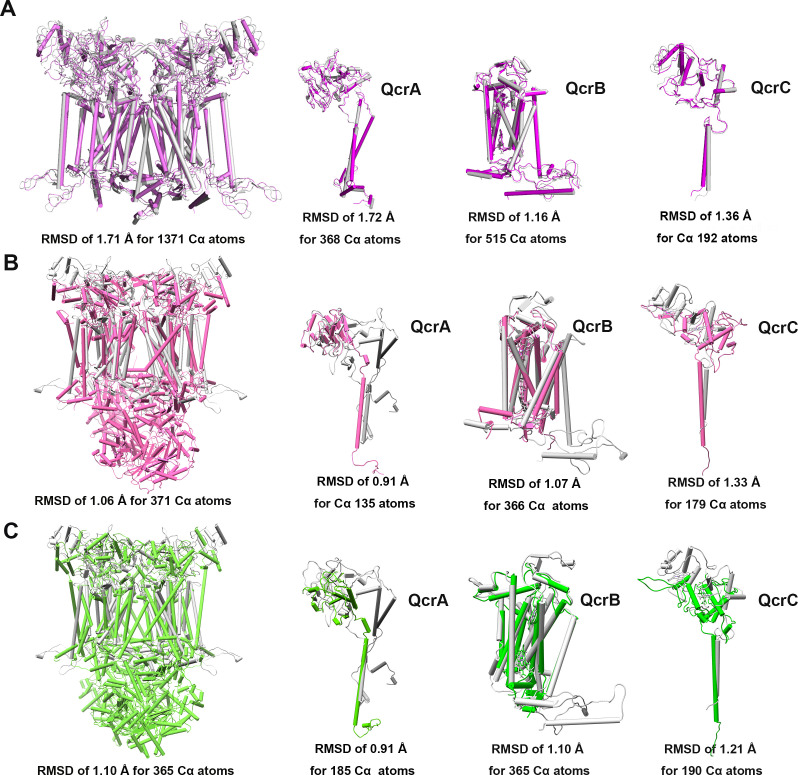
Figure 2. Overall architecture of the hybrid supercomplex.
(A) Front view and (B) top view of the cryo-electron microscopy (cryo-EM) map of hybrid supercomplex at 2.68 Å resolution. QcrA, QcrB, and QcrC of the M. tuberculosis cytochrome bcc are colored teal, purple, and salmon, respectively. Other subunits of the hybrid supercomplex are in gray. (C) Cartoon representation of cytochrome bcc, using the same color scheme as above. The twofold symmetry of the dimer is depicted by the gray axis. The heme groups (bH, bL, cD1, and cD2) and menaquinone/menaquinol (MKP/MKN) are shown as stick models. The [2Fe-2S] clusters are shown as spheres. (D) A cross-sectional view (top) of the helices in the cytochrome bcc dimer.