Abstract
Purpose:
To develop a three-dimensional Multitasking multi-echo (MT-ME) technique for the comprehensive characterization of liver tissues with 5-minute free-breathing acquisition, whole-liver coverage, a spatial resolution of 1.5 × 1.5 × 6 mm3, and simultaneous quantification of T1, water-specific T1 (T1w), PDFF, and R2*.
Methods:
Six-echo bipolar spoiled GRE readouts following inversion recovery (IR) preparation was performed to generate T1, water/fat and R2* contrast. MR Multitasking was used to reconstruct the MT-ME images with 3 spatial dimensions, one T1 recovery dimension, one multi-echo dimension, and one respiratory dimension. A basis-function-based approach was developed for T1w quantification, followed by the estimation of R2* and T1-corrected PDFF. The intra-session repeatability and the agreement against references of MT-ME measurements were tested on a phantom and 15 clinically healthy subjects. In addition, 4 patients with confirmed liver diseases were recruited and the agreement between MT-ME measurements and references was assessed.
Results:
MT-ME produced high-quality, co-registered T1, T1w, PDFF, and R2* maps with good intra-session repeatability and substantial agreement with references on phantom and human studies. The ICCs of T1, T1w, PDFF, and R2* from the repeat MT-ME measurements on clinically healthy subjects were 0.989, 0.990, 0.999, 0.988, respectively. The ICCs of T1, PDFF, and R2* between the MT-ME and reference measurements were 0.924, 0.987, 0.975 in healthy and 0.980, 0.999, 0.998 in patients. The T1w was independent to PDFF (R=−0.029, P=0.904).
Conclusion:
The proposed MT-ME technique quantifies T1, T1w, PDFF, and R2* simultaneously and is clinically promising for the comprehensive characterization of liver tissue properties.
Keywords: liver T1/PDFF/R2* mapping, MR Multitasking, low-rank tensor, water-specific T1, free-breathing acquisition
1. Introduction
Chronic liver disease is increasing in prevalence and has become a major cause of mortality worldwide in recent years.1,2 Nonalcoholic fatty liver disease (NAFLD) is one of the major types, which is estimated to affect 25% of the world population.2 The progressive form of NAFLD is nonalcoholic steatohepatitis (NASH), which is typically accompanied by liver inflammation and fibrosis2. Liver fibrosis can advance into cirrhosis, an end-stage disease that invariably leads to death3,4. Understanding the complex interplay between liver fat, iron concentration, and fibrosis is of great significance to the management of liver disease. Currently, liver biopsy is the gold standard for the diagnosis and staging of liver disease. However, it is invasive, prone to sampling error, associated with risks of complications, and subject to inter-observer variability5–7. Therefore, an objective, non-invasive tool for comprehensive evaluation of liver tissues is highly desired.
In recent years, quantitative MRI with multiparametric mapping has been a promising approach for the objective, noninvasive assessment of liver diseases7–12. Proton density fat fraction (PDFF) is a widely used MRI marker to quantify tissue fat concentration13–15. R2*, the reciprocal of T2*, is proportional to iron content in the liver10,16. MR elastography (MRE) is a widely-accepted approach to measure liver stiffness as well as fibrosis but requires complicated setups and additional external hardware17. Recently, the role of T1 in the assessment of liver fibrosis and inflammation has gained increased interest. Prior investigations have reported that the T1 relaxation time is significantly lengthened in fibrotic and cirrhotic livers in both animal and human studies with histological confirmation12,18–21. Some works showed that T1 values were significantly correlated with MRE measurements; it also had reduced inter-reader variability and improved reproducibility compared to MRE20,22. These studies indicate that T1 may have the potential to serve as a surrogate biomarker of MRE in the longitudinal monitoring of liver disease with easier setup and improved reliability.
Despite all the encouraging results, a standard clinical protocol including the measurement of PDFF, R2*, and T1 for liver diseases has yet to be established due to demanding technical challenges. First, while PDFF and R2* are measured simultaneously in one 3D acquisition, T1 usually needs to be measured with several repeated 2D acquisitions. Well-validated 3D T1 mapping techniques for liver are lacking. The most frequently used sequences for T1 measurement are 2D modified Look-Locker inversion recovery (MOLLI)23, shortened MOLLI (shMOLLI)24, or saturation-recovery single shot acquisition (SASHA)25, which mandates a number of breath-holds to cover multiple slices. This can cause extra burden on patients, reduced reliability, and misregistration across breath-holds. Second, the breath-hold sequences force compromises between spatial coverage, spatial resolution, and scan time (limited by breath-hold duration). Third, the presence of liver fat confounds the quantification of water T1 and reduces the sensitivity of T1 in the estimation of fibrosis26,27. Previous works have demonstrated that MOLLI/shMOLLI or SASHA produce large and complex T1 errors when applied to mixed water-fat systems28. Some recent publications have developed approaches that can quantify T1, PDFF, and R2* simultaneously within a single scan, and correct T1 confounding from fat by measuring water-specific T1 (T1w)8,9. However, these techniques still require breath-holds and have limited spatial coverage.
To address the above-mentioned limitations, we propose a novel MR technique, Multitasking multi-echo (MT-ME), for the comprehensive characterization of liver tissues. MR Multitasking29 uses a low-rank tensor (LRT) imaging model to exploit the high correlation along and across different image dimensions for a vastly accelerated acquisition, and has been demonstrated for 2D multiparametric mapping in moving organs and 3D multiparametric mapping in the brain30–32. MT-ME is the first Multitasking technique for 3D multiparametric mapping on moving organs. The aim of this study was to describe and validate the MT-ME technique for whole-liver free-breathing acquisition and simultaneous quantification of water-specific T1 (T1w), PDFF, and R2* in a phantom and in vivo.
2. Methods
2.1. Sequence and sampling
A 3D continuous-acquisition sequence was designed based on spoiled gradient echo (spoiled GRE) readouts, consisting of periodic non-selective inversion recovery (IR) preparations and 6-echo bipolar readouts to generate and acquire T1, water/fat, and R2* contrasts, as shown in Figure 1A. Both RF- and gradient-spoiling are used to reduce the residual transverse magnetization at the end of each readout. The TEs were chosen as 1.25, 2.50, 3.75, 5.00, 6.25, 7.50 ms according to Zhong et al14. An axial stack-of-stars sampling pattern with golden-angle rotation in kx-ky plane and Gaussian-density randomized Cartesian reordering in the kz dimension was used to incoherently sample k-space. A perpendicular center k-space line in the head–foot direction (kx= ky = 0) was acquired every 8 readouts (every 74 ms) to form the training data, which were used to determine LRT temporal factors. Positioning the training center k-space lines along the kz direction better captures respiratory motion33. The sampling pattern was demonstrated in Figure 1B.
Figure 1:
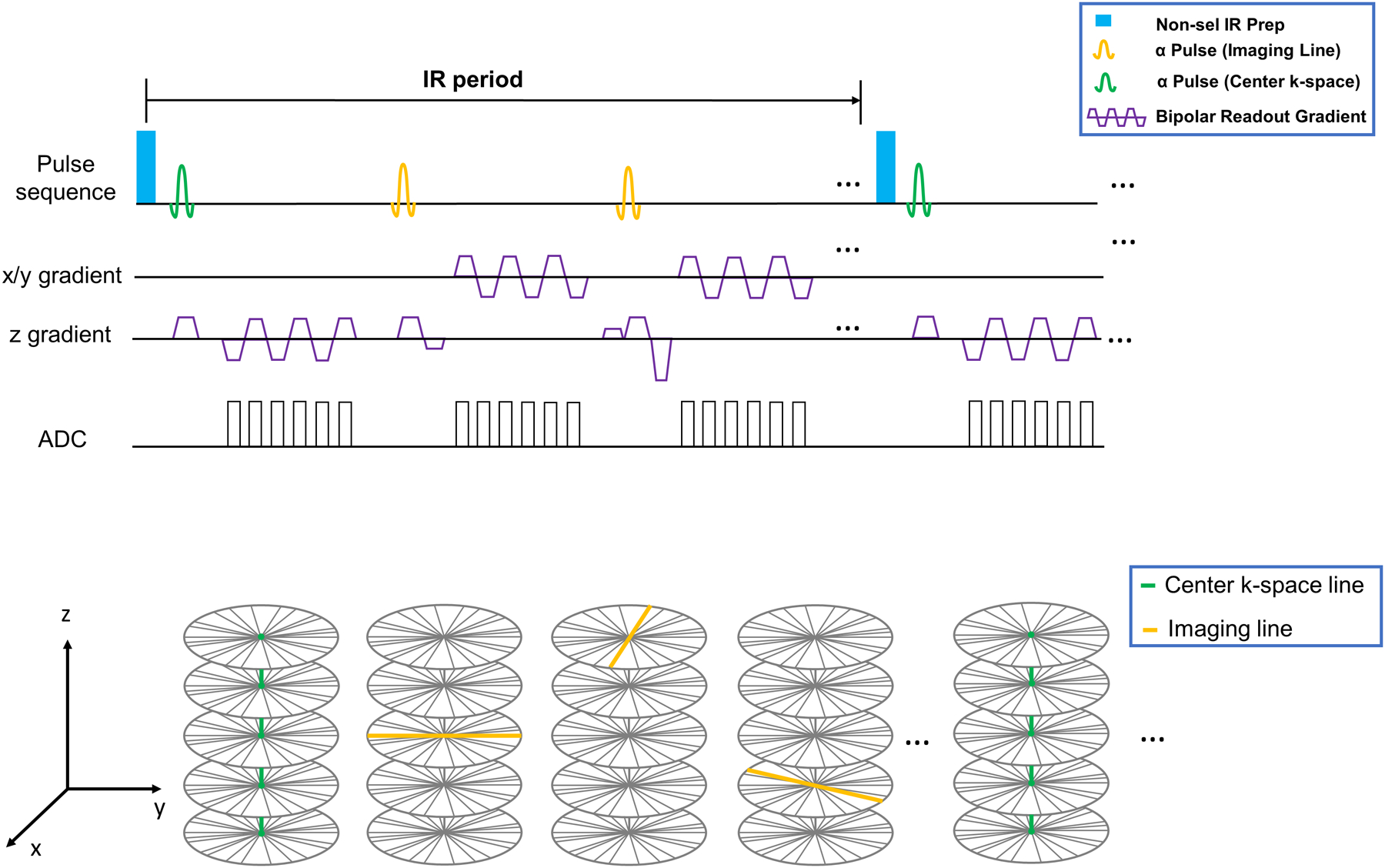
(A) Pulse sequence diagram for the proposed MT-ME technique. Non-selective inversion recovery(IR) preparation pulse is applied periodically followed by 3D SPGR readouts. Six-echo bipolar gradients were used for data collection. The center k-space readout was set on kz (superior-inferior) direction. Pre-phaser and spoiler of readout was not included in the simplified figure. (B): Simplified illustration of k-space sampling strategy. A stack-of-star sampling pattern with golden-angle in-plane and Gaussian-density randomized reordering in kz direction was implemented. The training data formed with center k-space lines was acquired every 8 readouts in kz (superior-inferior) direction for better capture of respiratory motion. Rest of the data forms the imaging data.
2.2. Image reconstruction
The target MT-ME images have three spatial dimensions x, y, z, and three temporal dimensions: IR time TI, multi-echo dimension TE, and respiratory motion bin r. It can be represented as a 4-way tensor by collapsing the three spatial dimensions into voxel index x = [x y z]T. The strong correlation along and across multiple dimensions of induces it to be low-rank, so that it can be decomposed into the product of different basis matrices representing each dimension29,34–36:
| (1) |
where the columns of U, V, W and Q contain the basis functions for spatial, IR, multi-echo, and respiratory dimensions, respectively, is the core tensor, and ×i denotes the ith mode product. The reconstruction of the final images can be framed as the recovery of each basis matrix, which significantly reduces the total degrees of freedom and requires a much shorter scan than without the LRT model.
The first step was to determine V from a pre-generated dictionary describing IR according to the Bloch equation. The dictionary consisted of 101 T1 values logarithmically spaced from 100 ms to 3000 ms, 17 flip angles from 1° to 9° in half-degree increments, and 21 inversion pulse flip angles linearly spaced from 90° to 180°. The IR basis matrix V was directly obtained from the singular value decomposition (SVD) of this dictionary.
The second step was to identify respiratory motion and divide data into 6 respiratory bins using an automatic respiratory binning method described in our previous work29. The prior-generated IR basis matrix V was used to compensate the T1 contrast of the training data. Then a modified k-means clustering algorithm was performed to assign each training data readout to a specific respiratory bin. All the data at different respiratory bins were used for image reconstruction. After the reconstruction, images at all 6 bins are available.
The third step was to recover W, Q, and through the completion of the training data tensor. The acquired (k,t)-space training data dtr were reshaped into a 4-way tensor with one k-space readout dimension, and three temporal modes TI, TE, and r. The training data were not available from every combination of TI, TE, and respiratory state, resulting in an undersampled . A small-scale low-rank tensor completion algorithm was applied to recover the training data tensor using:
| (2) |
where M(·) is the training data sampling pattern, ‖·‖* denotes the nuclear norm, Dtr,(i) denotes the mode-i matricization of the tensor , and R(·) is an optional additional regularization functional, which was chosen as temporal total variation (TV) along the respiratory dimension in this work. Following the recovery of , the core-tensor , multi-echo basis functions W, and respiratory basis functions Q were obtained using the higher-order SVD (HOSVD)37.
The final step was to recover the spatial basis functions U by fitting the temporal basis functions and core tensor into the acquired imaging data d:
| (3) |
where is the product of the core tensor and temporal basis functions, S is the coil sensitivity operator, FΩ is the nonuniform fast Fourier transform (NUFFT) on the undersampling set Ω, and R(·) is an optional regularization functional chosen as a spatial wavelet regularizer. In this work, gpuNUFFT38 was used to compute nonuniform fast Fourier transform.
2.3. Multiparametric quantification
2.3.1. Joint T1
In this work, “Joint T1”, denoted as the T1 with both water and fat components, was quantified based on the first-echo MT-ME images at the end-expiration state using the following equation39:
| (4) |
with amplitude A, inversion effect B, flip angle α, spoiled GRE readout interval TR, and recovery time point n = 1, 2, ⋯ , N (N=192 per IR period) such that τ = nTR. In the quantification, A, B, and T1 were estimated jointly from the signal evolution. The effect of the RF transmit field (B1+) inhomogeneity on the T1 estimation was analyzed and is presented in the Supporting Information Section D.
2.3.2. Water-specific T1 (T1w)
One straightforward way to map T1w would be to perform water/fat separation on the multi-echo images at each inversion time, and then perform the T1 mapping using the water images only. However, for 3D images with 192 inversion times, this approach would be time-consuming. Therefore, we performed a basis-function-based approach8,40. Specifically, a partially-recomposed tensor was generated based on model basis factors:
| (5) |
where the superscript “+” denotes the pseudoinverse. is composed of all model factors except the IR basis V, therefore keeping T1 information embedded and reducing the number of water/fat separations that must be performed: for any given respiratory bin, water/fat separation only needs to be done 5 times (the dimension of V), rather than on each of the 192 inversion times. is a three-way tensor with a spatial dimension, a multi-echo dimension, and a respiratory dimension. The water-fat separation was then performed on the end-expiration images of using a multiple-fat-peak single-R2* model:
| (6) |
where and are the respiratory-motion-resolved water and fat components, respectively, Cl and fl are the weighting and the resonance frequency offsets of the lth fat peak, TE is the echo time, and fB0 represents B0 inhomogeneity. A multi-peak fat model was employed in this work according to Yu et al41. The water-fat separation was performed using an in-house algorithm using variable projection (VARPRO) according to Hernando et al42.
Subsequently, the water images with multiple inversion times were generated as:
| (7) |
T1 quantification was performed using Equation 4, yielding T1w value. The fat-specific T1, denoted as T1f, was also generated similarly on the fat images with multiple inversion times:
| (8) |
To validate the feasibility of the basis-function-based approach, a comparison was performed between the T1w values from the basis-function-based approach and from the straightforward approach where the T1 mapping was conducted on the water images of 192 inversion times, as described in Supporting Information Section A.
2.3.3. PDFF and R2* mapping
Following the estimation of T1w and T1f, PDFF and R2* were fitted together using the multi-echo images of MT-ME at end-expiration at the last TI. The T1 effect of water and fat components43 were also taken into consideration:
| (9) |
where ρw and ρf are the proton density of water and fat, and Mw,last and Mf,last represent the spoiled GRE signal intensity at the last inversion time of water and fat component, which can be calculated with the estimated T1w and T1f according to Equation 4:
The same in-house algorithm as in Section 2.3.2 was used for the estimation of PDFF and R2*. The flow chart in Figure 2 illustrates the entire process of the reconstruction and multiparametric mapping of MT-ME.
Figure 2:
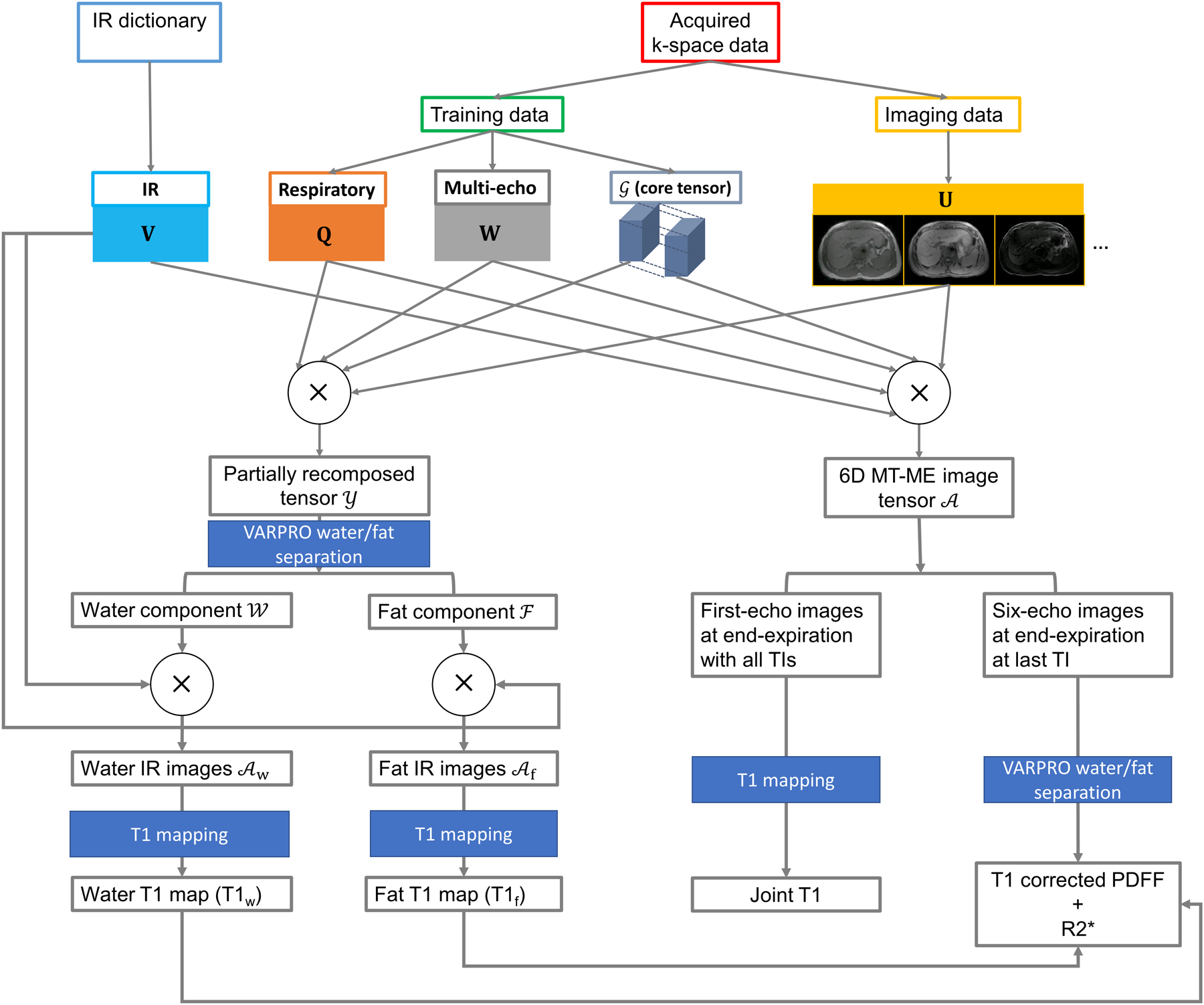
Flow chart of the reconstruction of MT-ME images and the quantification of T1, T1w, PDFF, and R2*.
2.4. Imaging experiments
Experiments were performed on a phantom, clinically healthy subjects (N = 15), and patients with diagnosed liver diseases (N = 4). The in vivo studies were approved by the local institutional review board. All subjects provided written informed consent before scanning. The phantom and volunteer studies were performed on a 3T system (MAGNETOM Vida, Siemens Healthcare, Erlangen, Germany) with an anterior 18-channel flexible array coil in combination with 12–16 elements of the table-mounted spine array. The patient study was performed on another 3T scanner (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany) with the same coil settings. The detailed imaging parameters of the sequences used in the experiments are included in Supporting Information Table S1.
2.4.1. Phantom study
A commercial phantom (Calimetrix, Madison, WI) was used in this work with 15 vials, each of which has a unique combination of T1, PDFF, and R2* values. A prototype single-slice inversion recovery spin echo (IR-SE) without and with water-selective excitation were both acquired at the center slice as references for T1 and T1w, respectively. A product q-DIXON sequence with 3D volumetric interpolated breath-hold examination (VIBE) acquisition and 6-echo bipolar readout was acquired as the reference for PDFF and R2*14. Following the reference scans, the proposed MT-ME sequence was performed twice to assess its feasibility and intra-session repeatability.
In addition, a custom phantom was designed44 to specifically test the ability of MT-ME to map the T1w in the samples with T1 values that match the liver T1 at 3T (700 to 1200 ms9,20,45) in the presence of fat. The details of the phantom study are described in the Supporting Information Section C.
2.4.2. Volunteer study
Fifteen clinically healthy subjects (3 females; age 38±12 years, BMI 24.8±3.4 kg/m2) without a record of liver diseases were recruited as the clinically healthy group. All the scans were performed in transversal orientation. A product 2-echo VIBE-DIXON sequence was performed to generate anatomical water and fat images with following parameters: FOV = 308×380 mm2, matrix = 195×320 mm2, TE = 1.29/2.52 ms, TR = 3.60 ms, slice thickness = 6 mm. The prototype 2D MOLLI sequence was collected under breath-hold at end-expiration for 3 slices at the top, middle and bottom of the liver. The parameters for each slice are: FOV = 306×360 mm2, matrix = 204×240 mm2, TE = 1.17 ms, TR = 2.63 ms, slice thickness = 8 mm. The product q-DIXON VIBE sequence with 6-echo bipolar readout was conducted to provide reference PDFF and R2* values under breath-hold at end-expiration with the parameters: FOV = 359×359 mm2, matrix = 128×128 mm2, TE = 1.25/2.50/3.75/5.00/6.25/7.50 ms, TR = 9.28 ms, slice thickness = 6 mm. Following the breath-hold scans, the proposed free-breathing MT-ME sequence was performed covering the whole liver with the parameters: FOV = 288×288 mm2, matrix = 192×192 mm2, TE = 1.25/2.50/3.75/5.00/6.25/7.50 ms, TR = 9.20 ms, slice thickness = 6 mm, number of slices = 40. The entire protocol was performed twice in the same imaging session half-an-hour apart to assess the intra-session repeatability of MT-ME.
2.4.3. Patient study
Four patients (1 female; age 65±4 years, BMI 31.5±2.3 kg/m2) previously diagnosed with biopsy-confirmed chronic liver diseases were recruited for the study: two with NAFLD and two with NASH. The patients first received a set of clinical standard-of-care MRI scans including 2-echo VIBE-DIXON and 6-echo q-DIXON. Then, MOLLI and MT-ME were performed with the same settings as in the volunteer study. MT-ME was performed one time on each patient due to time limitations.
2.5. Analysis
Image reconstruction and quantitative mapping of T1, T1w, PDFF, and R2* were performed off-line in MATLAB (R2019a, Mathworks, MA, USA) for all MT-ME data. An imaging physician (AK) assessed all the images, including 1) water images, fat images, T1, T1w, PDFF, and R2* maps from MT-ME; 2) water images, fat images, PDFF and R2* maps from q-DIXON; 3) water and fat images from VIBE-DIXON; and 4) T1 maps from MOLLI. For each set of images, the subjective SNR, artifacts, and sharpness were evaluated and a score of overall image quality was given using a 5-point scale: 1-worst (anatomic details were impossible to identify, and there were extremely high noise and artifacts); 2-bad (anatomic details were difficult to identify, and there were significant noise and artifacts); 3-moderate (anatomic structures were visible but not clearly shown, and there were acceptable noise and artifacts); 4-good (the anatomic structures and details were visible but with visible noise and artifacts); 5-excellent (the anatomic structures and details were highly visible and there were no significant noise and artifacts).
The T1, T1w, PDFF, and R2* were measured on phantom and in vivo. In phantom, the ROIs were drawn on a center slice within each vial avoiding the edge. For the in vivo study, the ROIs of MT-ME images were manually drawn on the three slices matching the locations of MOLLI. As much liver tissue as possible was included while avoiding blood vessels, nodules/cysts (if any), and edges. The same ROIs were applied to the maps of T1, T1w, PDFF, and R2*. For reference scans, the ROIs were also drawn on three matched slices avoiding blood vessels, nodules/cysts (if any), and edge. The mean value throughout all pixels was reported for each parameter of each measurement.
The agreement of T1, T1w, PDFF, and R2* measurements from MT-ME and references were evaluated by the sample Pearson correlation coefficient R and intra-class coefficient (ICC) in phantom and in vivo studies. A paired t-test was used to identify the significant difference of parameters measured with the MT-ME against references. A P value less than 0.05 is considered as a significant difference. The repeatability of the MT-ME measurements was assessed by the Bland-Altman plots, ICC and coefficient of variation (CoV) on phantom and volunteer studies.
3. Results
3.1. Phantom study
Vials 1 to 15 are labeled on the gray-scale image in Figure 3A. Vial 14 is a fat-only phantom, whose T1w value is not available from water-excited IR-SE. The quantitative map of T1, T1w, PDFF, and R2* from MT-ME and from references were displayed in Figure 3A. The Bland-Altman plots in Figure 3B shows good repeatability of MT-ME for all the four parameters. The ICCs of T1, T1w, PDFF, and R2* were 0.999, 0.999, >0.999, >0.999, respectively, and the CoVs were 1.4%, 0.5%, 1.1%, 0.9%, respectively. Figure 3C demonstrates the agreement of parameters measured by MT-ME and references. The Pearson Correlation Coefficients (R) were 0.990, 0.996, 0.999, >0.999, respectively, and the ICC were 0.988, 0.994, 0.998, 0.999, respectively, representing good agreements. The paired t-test indicated that the measurement of MT-ME and references showed no significant differences for all the four parameters with P = 0.126, 0.097, 0.319, 0.067. The results of the custom phantom are illustrated in the Supporting Information Figure S3.
Figure 3:
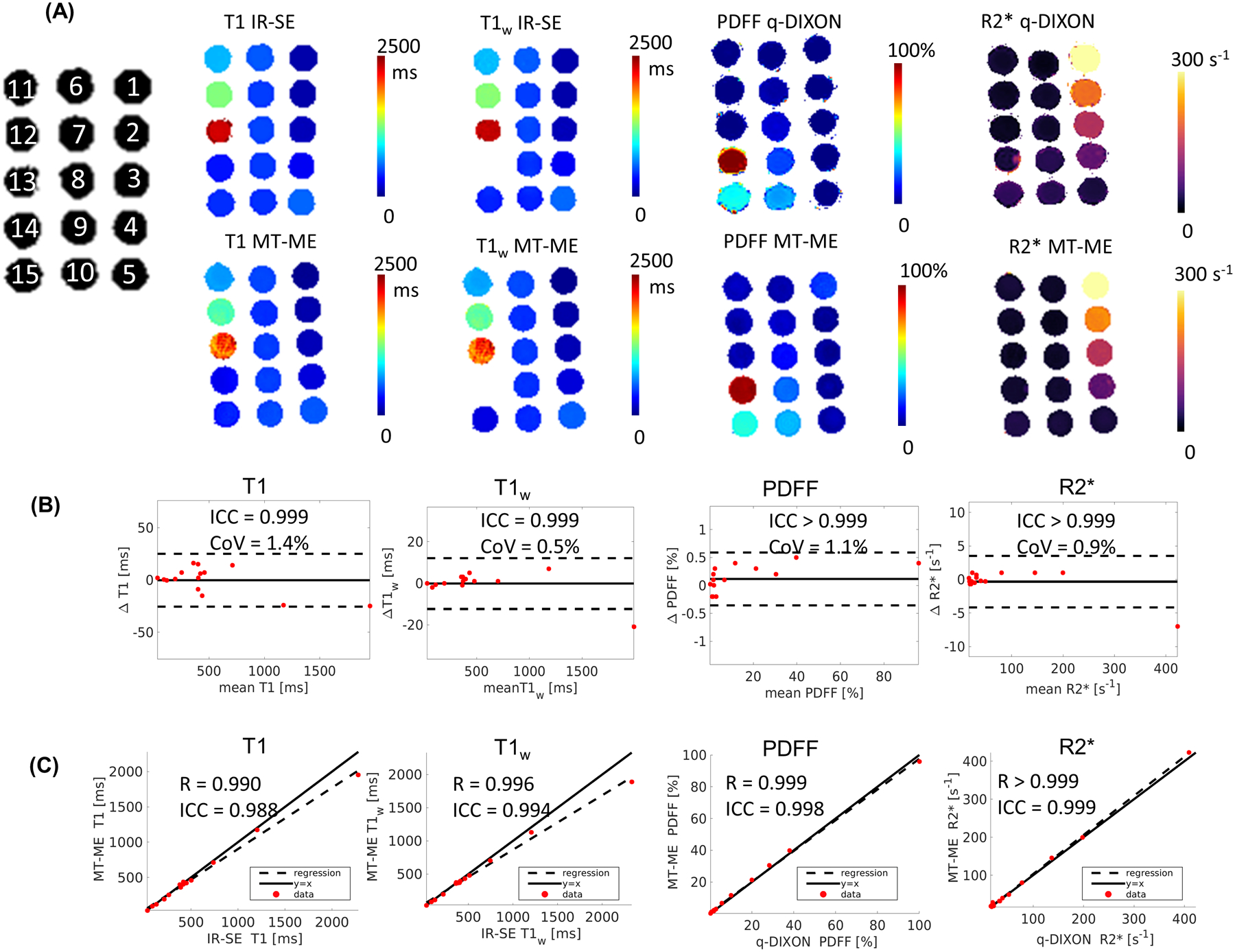
Phantom Measurements. (A) T1, T1w, PDFF, and R2* maps from reference sequences and MT-ME. The Inversion-recovery spin-echo (IR-SE) sequence without and with water excitation were used as reference for joint and water-specific T1. q-DIXON provided the reference for PDFF and R2*. The quantitative maps from MT-ME showed great agreement with references. (B) Bland-Altman plots shows good repeatability of T1, T1w, PDFF, and R2* estimated with MT-ME. (C) Good correlation was demonstrated of T1, T1w, PDFF, and R2* from MT-ME and reference with high Pearson correlation coefficient R and ICC, as labeled on each plot.
3.2. In vivo study
The in vivo protocols were successfully applied to all healthy volunteers and patients, and MT-ME images were reconstructed and processed for all subjects. The reconstructed MT-ME images possess 192 TIs from 9.2 ms to 1766.4 ms with an increment of 9.2 ms, 6 echoes, and 6 respiratory bins, yielding a total of 192×6×6=6912 images, each corresponding to a 3D image volume. Figure 4 is an illustration of the images displayed in either coronal or axial orientation at 3 echoes (echo 1, echo 2, and echo 6), 2 representative TIs (266.8 ms and 1766.4 ms) and 2 respiratory states (end-expiration and end-inspiration). The coronal view with reference lines shows the respiratory motion of the liver, while the transversal view provides clear liver anatomy and contrast. Supporting Information Video S1 further demonstrates the multiple contrasts from MT-ME.
Figure 4:
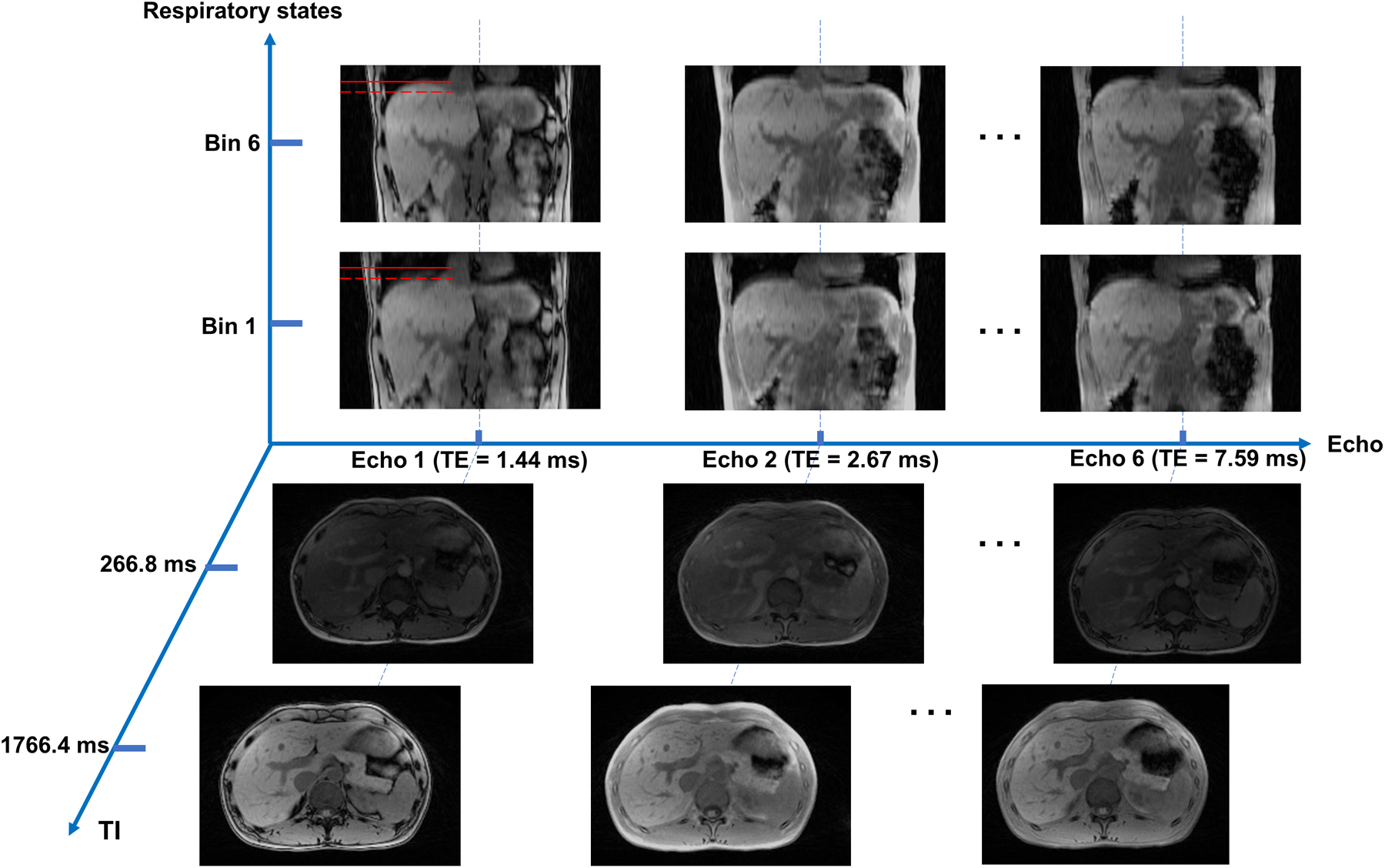
Demonstration of MT-ME images with three spatial dimensions and three temporal dimensions (IR dimension, multi-echo dimension, and respiratory dimension). The images of end-inspiration and end-expiration were displayed in coronal view at the last TI of echo1, echo 2, and echo 6. The red solid reference line on images of echo 1 is the position of liver dome at end-expiration, while the dashed reference line is the position at end-inspiration. Images of different TI times (266.8 ms and 1766.4 ms) were displayed in axial view. The liver and vessels were well delineated.
In clinically healthy subjects, the mean, median, minimum, and maximum values for T1 (ms) are 851.7, 841.0, 718.5, 998.2; for T1w (ms) are 797.6, 814.5, 688.3, 875.4; for PDFF are 7.5%, 4.6%, 1.9%, 19.8%, and for R2* (s−1) are 51.8, 50.5, 37.3, 72.9. In patient group, the mean, median, minimum, and maximum for T1 (ms) are 911.2, 911.8, 792.6, 990.3; for T1w (ms) are 829.8, 869.3, 679.2, 901.5; for PDFF is 10.1%, 9.8%, 4.9%, 15.9%; and for R2* (s−1) is 60.6, 53.3, 44.2, 90.5.
Figure 5 displays the example multiparametric maps from MT-ME and references from a clinically healthy subject. The T1, PDFF, and R2* maps generated by MT-ME are high-quality, co-registered and match well with the available reference maps (an in vivo T1w reference was not available). Figure 6 shows the example multiparametric maps from a 64-year-old patient diagnosed with NASH. Elevated PDFF was detected by q-DIXON and MT-ME.
Figure 5:
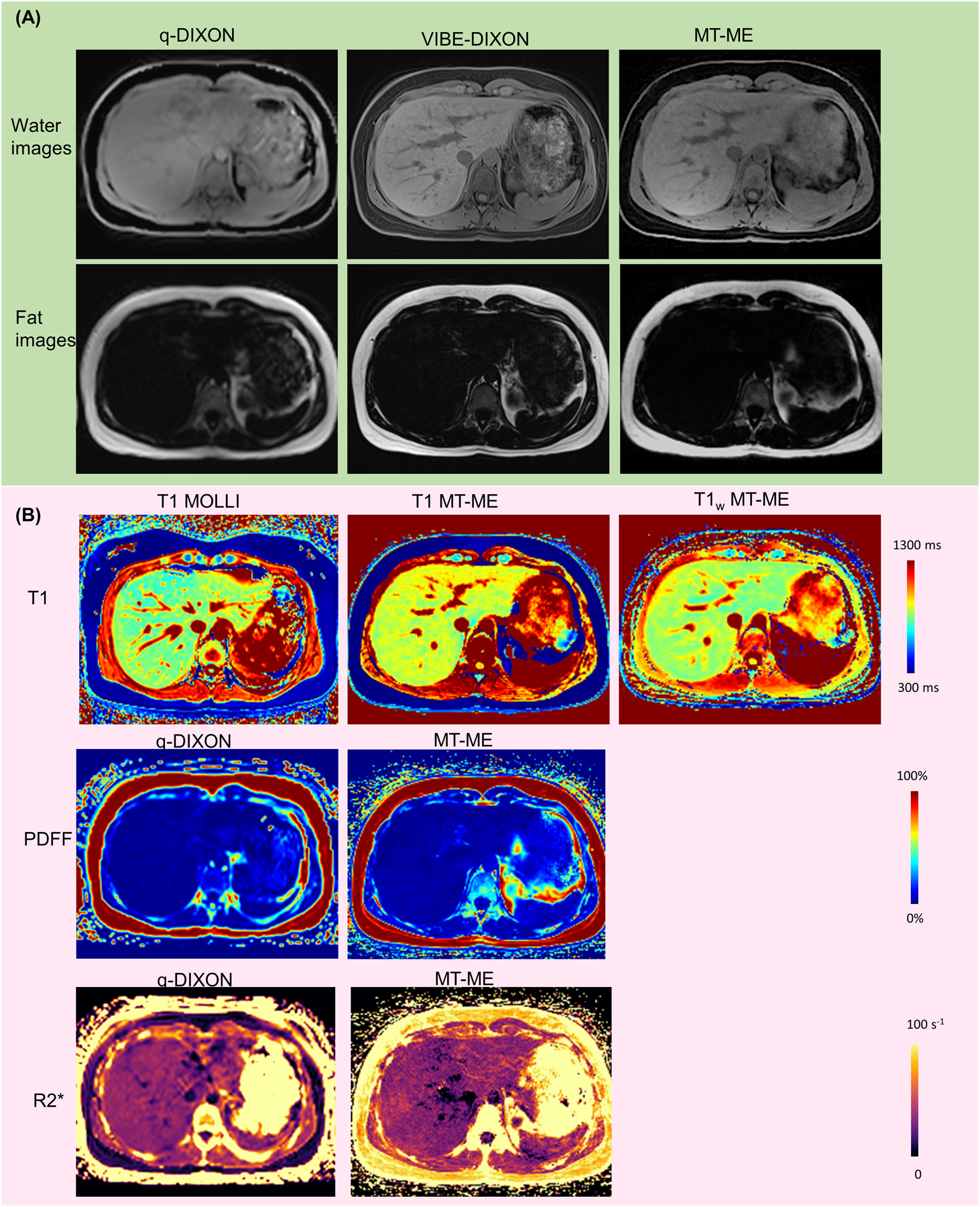
Representative anatomical water/fat images and T1, T1w, PDFF, and R2* maps from a clinically healthy 29-year-old volunteer. (A) Water and fat images from q-DIXON, VIBE-DIXON, and MT-ME. Vessels can be well defined on water images of MT-ME and VIBE-DIXON. (B) Good consistency was showed between MT-ME maps and references. The in vivo T1 reference was 2D MOLLI with a 11-second breath-hold, while the PDFF and R2* reference was q-DIXON with 12-second breath hold. The T1w reference was not available.
Figure 6:
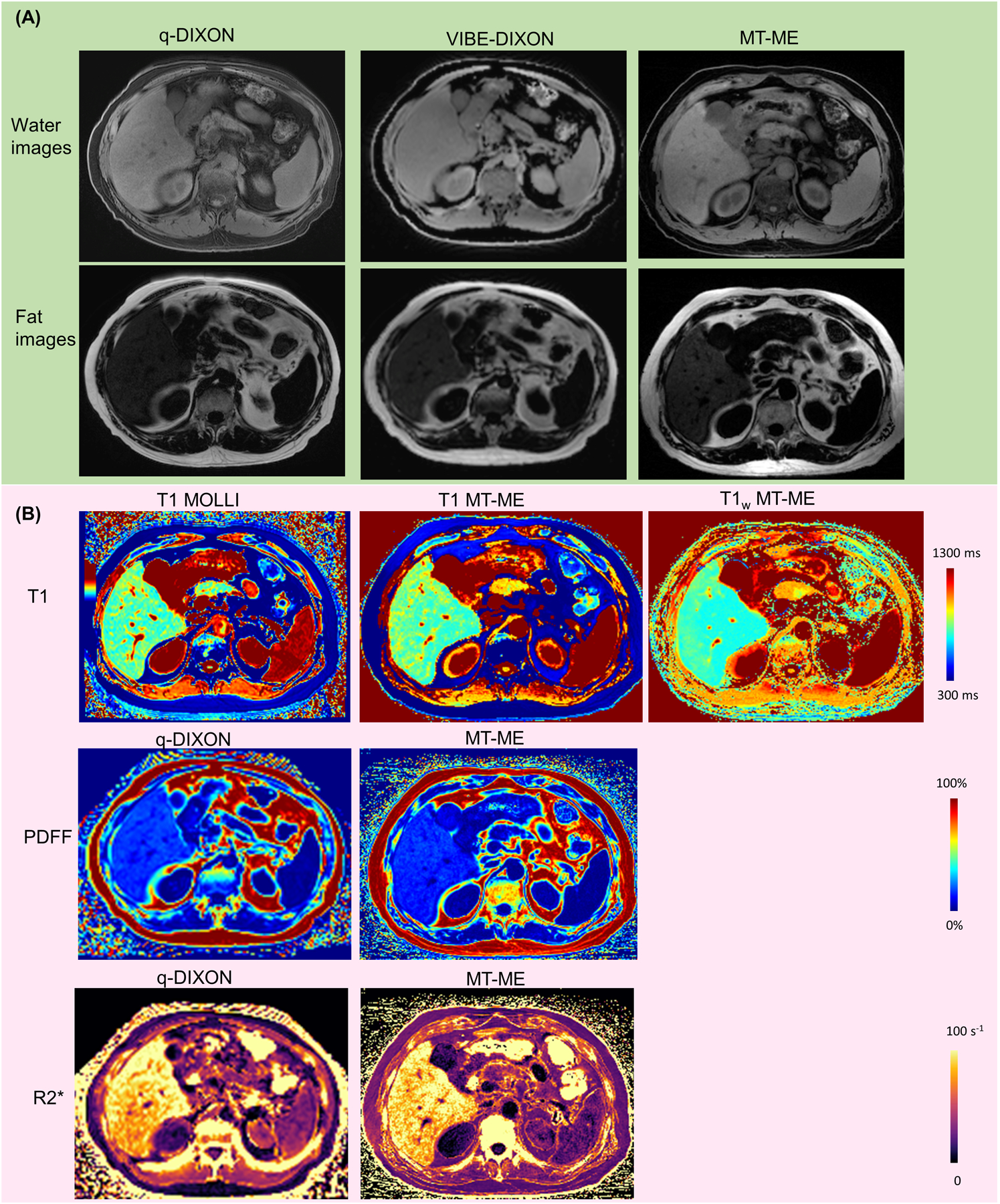
Representative water/fat images and T1, T1w, PDFF, and R2* maps from a 64-year-old patient with NASH. (A) Water and fat images from q-DIXON, VIBE-DIXON, and MT-ME. (B) T1, T1w, PDFF and R2* maps. The mean T1, PDFF, and R2* of reference were 779 ms, 15.5%, 89 s−1, respectively, while the mean T1, T1w, PDFF, and R2* measured from MT-ME were 785 ms, 643 ms, 15.5%, and 85 s−1.
The mean and standard deviation of the scores of water images from MT-ME, VIBE-DIXON, and q-DIXON are 4.7±0.5, 4.8±0.4, and 3.8±0.5, respectively; the scores of fat images in the same order are 4.8±0.4, 4.8±0.3, and 4.1±0.3, respectively. MT-ME produced water and fat images with similar quality compared to VIBE-DIXON, and improved image quality and sharpness compared to q-DIXON. The scores of PDFF maps from MT-ME and q-DIXON are 4.7±0.4, 4.1±0.2, respectively, indicating superior PDFF maps from MT-ME over q-DIXON. The scores of R2* maps from MT-ME and q-DIXON are 3.7±0.5, 3.8±0.3, respectively, indicating that the R2* maps from MT-ME were similar quality to the maps from q-DIXON. The scores of T1, T1w maps from MT-ME and T1 maps from MOLLI are 4.3±0.5, 4.4±0.5 and 4.9±0.3, respectively, which are rated good to excellent. Detailed image quality scores are listed in Supporting Information Table S2 and Table S3.
Supporting Information Figure S1 shows the comparison of the T1w measurements using the proposed basis-function-based approach and using the straightforward approach applying the water/fat separation to the images of each inversion time. For the in vivo data, the ICC of the two measurements is 0.993 with a mean difference at 4.6 ms, indicating that the basis-function-based approach produced similar results compared to the straightforward approach. However, the straightforward approach requires 3D water-fat separation on 192 inversion time images, while the basis-function-based approach requires 3D water-fat separation per each of 5 basis functions, which is 38 times faster.
The example maps of fat-specific T1 (T1f) are displayed in Supporting Information Figure S2. The mean and standard deviation of T1f is 375.8±18.1 in the clinically healthy group, and is 379.5±11.2 in the patient group, which are consistent with literature values45.
The importance of performing T1 correction on the PDFF estimation for the MT-ME technique was demonstrated in Figure 7. Figure 7A and 7B display two representative in vivo cases. Without T1 correction, the water proton density was underestimated due to the longer T1, resulting in overestimated PDFF. After performing T1 correction, the PDFF estimated by MT-ME matched well with the reference from q-DIXON. For all the in vivo datapoints (15 clinically healthy subjects and 4 patients), the T1 corrected PDFF from MT-ME showed increased Pearson correlation coefficient R and ICC with reference values (R = 0.979 and ICC = 0.935 without T1 correction; R = 0.993 and ICC = 0.990 with T1 correction).
Figure 7:
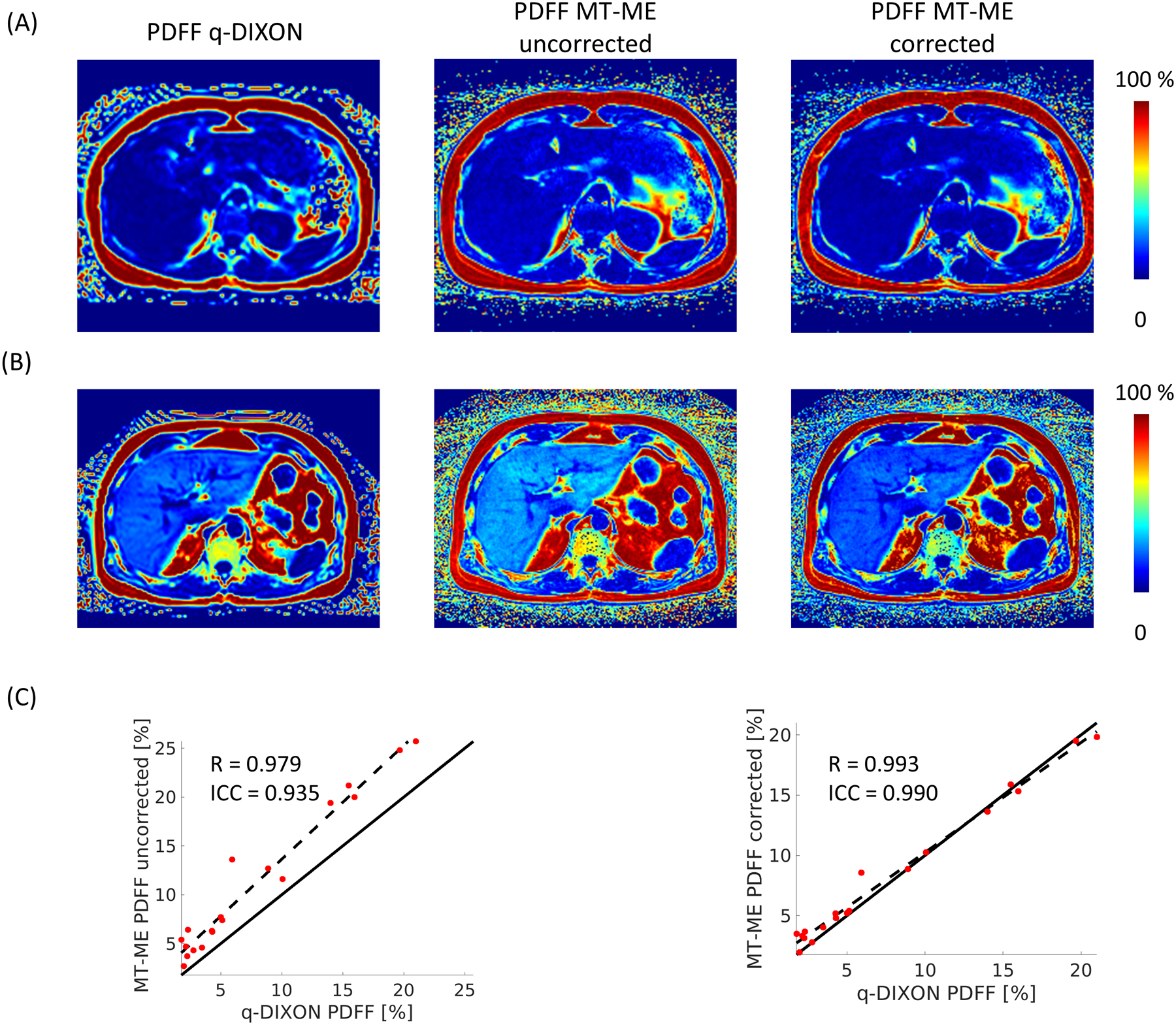
The significance of T1 correction in the estimation of PDFF. Two in vivo examples are display in (A) and (B). Without T1 correction (middle panel of A and B), the PDFF estimated from MT-ME is higher than q-DIXON reference. With correction (right panel of A and B), the PDFF estimated from MT-ME matches well with q-DIXON results. (C) the regression analysis between uncorrected MT-ME PDFF and q-DIXON shows R = 0.979, ICC = 0.935; the regression between corrected MT-ME PDFF and q-DIXON shows improved correlation R = 0.993 and increased ICC = 0.990.
The in vivo intra-session repeatability of T1, T1w, PDFF, and R2* measurements of MT-ME were illustrated by the Bland-Altman plots in Figure 8A. The ICC of T1, T1w, PDFF, and R2* were 0.989, 0.990, 0.999, 0.988, respectively, and the CoV were 0.9%, 0.7%, 1.3%, 1.4%, respectively. The high ICC and low CoV values of all parameters indicated a good intra-session repeatability. Figure 8B shows the regression analysis of MT-ME measurements against in vivo references of the clinically healthy cohort with the Pearson correlation coefficients R = 0.925, 0.992, 0.984 and ICC = 0.924, 0.987, 0.975 for T1, PDFF, and R2*, respectively. The paired t-test showed that there was no significant difference for either PDFF (P = 0.853) or R2* (P = 0.337) measurements. A significant difference was found in T1 measured from MT-ME and MOLLI (P <0.001). The overall T1 from MT-ME was 4.6% higher than the T1 of MOLLI, as listed in Table 1.
Figure 8:
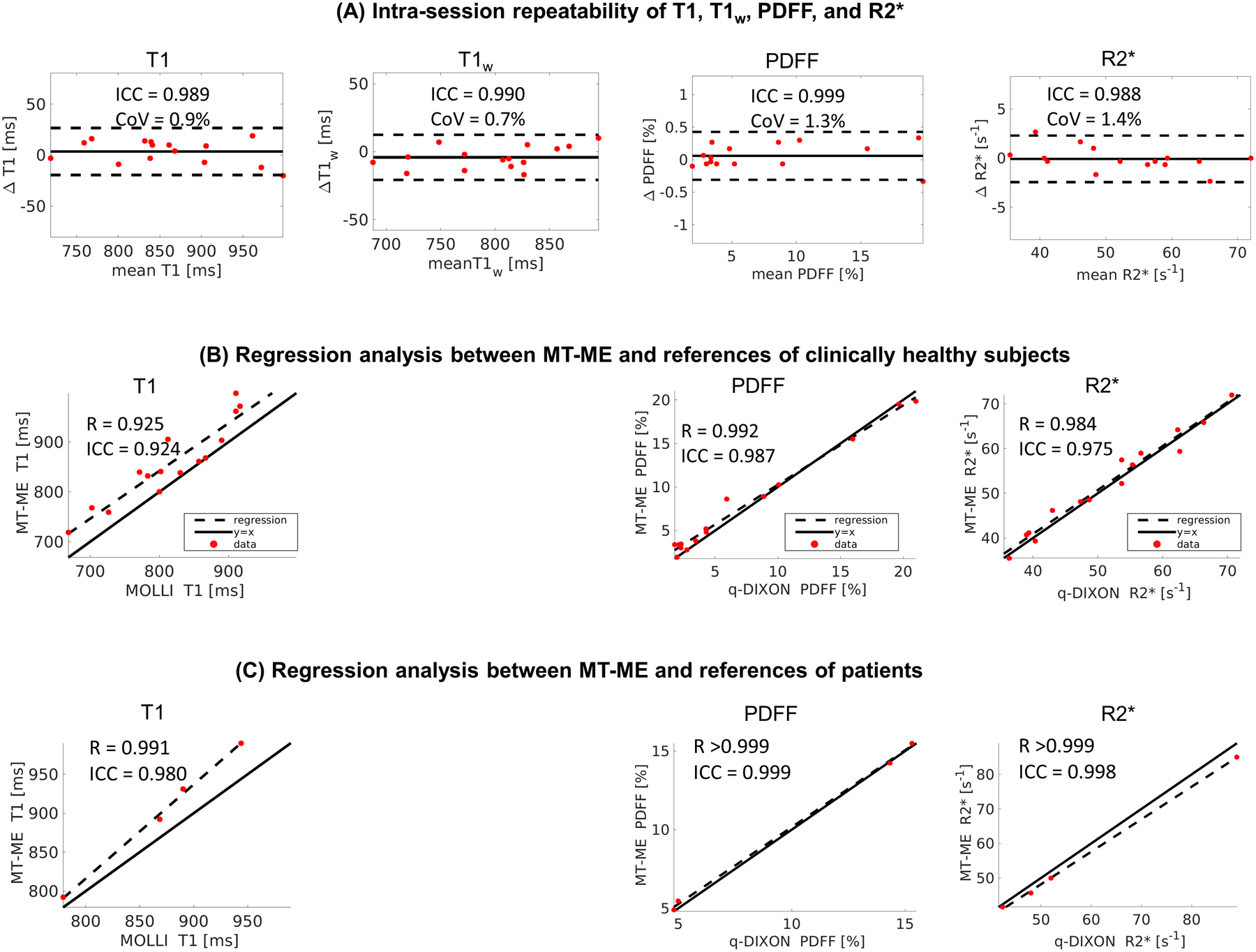
(A) Bland-Altman plots demonstrated a good intra-session repeatability of the multiparametric mapping from MT-ME. The overall intra-session differences for T1 and T1w were less than 3%, while the differences for PDFF and R2* were less than 10%. (B) The regression analysis of T1, PDFF, and R2* measured with MT-ME and references showed good agreement in the clinically healthy subjects. The Pearson correlation coefficient R was 0.990, 0.976, and 0.953, respectively. (C) The regression analysis of T1, PDFF, and R2* measured with MT-ME and references showed good agreement in patients. The Pearson correlation coefficient R was 0.998, >0.999, and >0.999, respectively.
Table 1:
Mean and standard deviation measurements of T1, T1w, PDFF, and R2* from MT-ME and reference, the absolute relative differences between MT-ME reference, and P value of paired t-test are provided for clinically healthy and patient groups.
| T1 [ms] | T1w [ms] | PDFF [%] | R2* [s-1] | ||
|---|---|---|---|---|---|
| Clinically healthy | MT-ME | 851.7±80.2 | 797.6±59.3 | 7.1±6.2 | 52.0±10.7 |
| References | MOLLI | N/A | q-DIXON | q-DIXON | |
| 814.7±78.1 | 7.2±6.6 | 51.6±11.5 | |||
| Absolute relative differences [%] | 4.6% | N/A | 1.4% | 0.8% | |
| P | <0.001* | N/A | 0.853 | 0.337 | |
| Patient | MT-ME | 911.2±83.2 | 829.8±119.5 | 10.0±5.6 | 55.9±20.0 |
| References | MOLLI | N/A | q-DIXON | q-DIXON | |
| 869.3±68.0 | 9.9±5.7 | 57.8±21.2 | |||
| Absolute relative differences [%] | 4.8% | N/A | 1.0% | 3.3% | |
| P | 0.028* | N/A | 0.252 | 0.075 |
indicates statistical significance.
The regression analysis for the multiparametric measurements of patient cohort were shown in Figure 8C, with the Rs = 0.991, > 0.999, >0.999, and ICCs = 0.980, 0.999, 0.998 for T1, PDFF, and R2*, respectively; the P-values from paired t-test were 0.027, 0.252, 0.075, indicating statistical difference in T1 measurement but no differences in PDFF and R2*. Table 1 summarizes the mean and standard deviation measurements of all the parameters from MT-ME and reference for clinically healthy and patient cohorts.
The differences between T1 and T1w for each subject were analyzed using a dot-line diagram, as shown in Figure 9A, including all in vivo datapoints (15 clinically healthy volunteers and 4 patients). The joint T1 were estimated on the first echo images, whose TE = 1.25 ms is close to the out-of-phase of water and fat. Results showed that joint T1 was larger than the T1w by a different portion, which is consistent with previous findings28. The differences between T1 and T1w presented a significantly strong correlation with PDFF with the Pearson correlation coefficient R = 0.945 and P < 0.001, as shown in Figure 9B. On the contrary, the water-specific T1w appeared to be independent to PDFF (R = −0.029, P = 0.904), agreed with the expectation.
Figure 9:
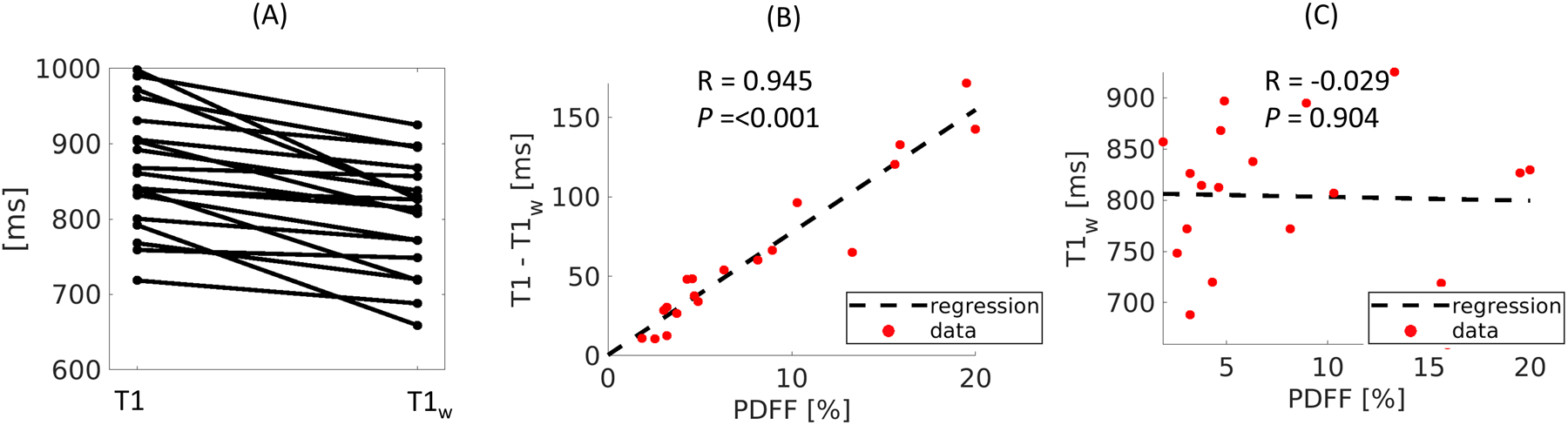
(A) Dot-line diagram of the relation between T1 and T1w estimated on all subjects using MT-ME. The T1w appears lower than joint T1 in all subjects. (B) regression of the differences of T1 and T1w against PDFF. A significantly strong correlation with R = 0.960, P < 0.001 is demonstrated. (C) No correlation was found between T1w against PDFF, with R = 0.016, P = 0.781.
4. Discussion
The potential of multiparametric MRI in the characterization of liver tissues has been demonstrated in a variety of investigations. However, technical challenges, including the requirements for multiple breath-holds, misregistration among separate scans, lack of reliable 3D T1 mapping techniques, and confounder factors among the biomarkers have limited the clinical applications of multiparametric MRI. In this study, a novel technique, MT-ME, was developed, achieving 5-min free-breathing acquisition with whole-liver coverage, a spatial resolution of 1.5 × 1.5 × 6 mm3, and simultaneous, co-registered quantification of T1, T1w, PDFF, and R2*.
A major advantage of the proposed technique is the free-breathing acquisition and motion-resolved reconstruction. In patient populations with breath‐hold difficulties such as serious‐condition, pediatric, sedated, or elderly patients, a free-breathing technique robust to respiratory motion is a desirable and useful clinical option. There are free-breathing techniques available to quantify PDFF and R2* with a stack-of-star sampling pattern46,47 and respiratory averaging. However, the unresolved respiratory motion can lead to reduced image quality and estimation error for PDFF and R2*46,47. In our work, the respiratory motion was captured by training data acquired at the center k-space every 72 ms, and modeled by the Multitasking framework, yielding high-quality images and quantitative maps comparable to breath-hold references.
Another advantage of MT-ME is the 3D coverage with high spatial resolution, which can potentially capture the spatial distribution of fibrosis stage, PDFF and R2* between different segments of liver48–50, which can be used for disease differentiation50. Moreover, the association between the fibrosis and fat concentration and the development of hepatocellular carcinoma are gaining increased attention in recent years51,52. The MT-ME technique with full-liver coverage and high spatial resolution has the potential to capture the nodules and lesions, which can be a promising tool to study this association.
The phantom experiments on a commercial 15-vial fat and R2* phantom showed that the quantification of MT-ME matched well with the gold-standard references. The T1 of vial 13, which is around 2300 ms by IR-SE, was underestimated by MT-ME. This may result from the fact that the sequence parameters were designed for the mapping of liver T1, whose range was typically reported from 700 to 1200 ms on 3T systems9,20,45.
The in vivo experiments demonstrated that T1, PDFF, and R2* from MT-ME were consistent with references for healthy and patient cohorts. q-DIXON is a well-validated product technique on commercial MRI systems from a main MRI vendor for PDFF and R2* quantification with good reproducibility and clinical utility49,53–55. The PDFF and R2* measured by MT-ME were consistent with q-DIXON results with no significant differences. The T1 estimated by MT-ME was 4.6% and 4.8% higher than MOLLI in healthy and patient cohorts with P < 0.001, = 0.028, respectively. However, MOLLI itself can only serve as a suboptimal reference for in vivo T1 measurement due to its tendency to underestimate T156,57. A major factor is the magnetization transfer effect between the free pool and bound pool, which can lead to the underestimation of T1 at 100 to 200 ms58. The high quality of the maps produced by MT-ME and consistency with references demonstrated its capability to achieve respiratory-motion-resolved quantification of T1, PDFF, and R2*, which has a great clinical potential to relief the burdens of breath-hold from patients and improve the mapping quality and reproducibility.
The PDFF estimation in MT-ME used the multi-echo images at the last TI. Given the IR period of 1800 ms, water proton in liver with the T1 around 800–900 ms cannot reach the steady-state at the last TI, while fat proton can almost achieve steady-state. This T1 effect can result in the underestimation of water proton density, leading to the overestimation of PDFF. By introducing T1 correction, the agreement between PDFF of MT-ME and reference was substantially improved with increased Pearson Correlation Coefficient R (from 0.979 to 0.993) and ICC (from 0.935 to 0.990).
Intra-session repeatability is an important indicator of the precision of the quantitative techniques59,60. The phantom studies produced excellent intra-session repeatability for T1, T1w, PDFF, and R2* with high ICC (>=0.999 for all parameters) and low CoV (<1.5% for all parameters). In the volunteer studies, high intra-session repeatability was also demonstrated with ICC > 0.980 and CoV < 1.5% for all parameters with the presence of respiratory motion. As observed in the study, the respiratory pattern can change over time within the same imaging session. The high intra-session repeatability strongly supported that MT-ME is reliable for respiratory-motion-resolved quantification of T1, T1w, PDFF, and R2* and has the potential to serve as a clinical tool for diagnosis, staging, and longitudinal monitoring of liver diseases. Future studies to evaluate the inter-session repeatability and inter-vendor reproducibility of MT-ME will be conducted.
The estimation of liver T1 is affected by the presence of fat. As reported previously, when TE of MOLLI is close to the out-of-phase point of water/fat, the measured T1 tends to be longer than the true T1 of water pool; the bias grows with the increase of PDFF28. This can impair the reliability of T1 in the assessment of fibrosis. It is possible to correct the fat effect on the MOLLI/shMOLLI T1, but at the cost of separate mapping of off-resonance frequency and fat content in combination with numerical simulations26. In this work, the estimation of water-specific T1w was performed efficiently on the reconstructed basis functions. The validation on the phantom study showed that the T1w agreed well with water-excited IR-SE with R = 0.996, ICC = 0.994, and P = 0.097. For the in vivo study, the joint T1 was estimated on the first echo images, the TE of which is 1.25 ms, close to the out-of-phase TE for water and fat. The observation indicated that the joint T1 was longer than T1w for all cases, consistent with the trend reported in Mozes et al28. The differences between joint T1 and T1w showed a strong dependency over PDFF (R = 0.945, P <0.001), while T1w itself was independent to PDFF (R = −0.029, P = 0.904). This is a solid demonstration of the ability of T1w to avoid errors introduced by fat.
A technical limitation of the current MT-ME method is the dependency of T1 and T1w on B1+. It can cause estimation bias and increased intra-subject variation, as analyzed in Supporting Information Section D and Supporting Information Figure S4. To reduce the T1 dependency on B1+, a small flip angle, which is 5°, and a long TR, which is 9.3 ms were chosen in this work61. Supporting information Figure S5 displays the measured intra-subject standard deviation of T1 and T1w for all subjects. The root-mean-squares of the intra-subject standard deviation are 71.6 ms and 59.4 ms for T1 and T1w respectively, larger than the values reported in Thompson et al.9, which corrected B1+ inhomogeneity. Nevertheless, T1w measurements from MT-ME in this work are still sufficient to differentiate normal liver from medium- to high-stage fibrosis or steatosis of liver. According to literature, the differences of T1 is larger than 150 ms between normal liver versus fibrotic liver with fibrosis stage >=3, or between normal liver versus steatosis liver12,62,63, more than two times of the root-mean-squares of the intra-subject standard deviation for both T1 and T1w. In the future, technical improvement to resolve B1+-related variation of T1 and T1w measurements will be investigated. One possible approach is to acquire a separate low-resolution B1+ map. Another possible approach is to incorporate dual-flip angle acquisition to map B1+ and T1 together64–66.
There are several limitations of the study. First, the clinically healthy group comprised subjects without clinical diagnosis of liver diseases. However, given the high prevalence of liver diseases, it is possible that these subjects also possessed liver abnormalities to some extent, which may contribute to the large variation of the T1, PDFF, and R2* values within the group. For the patient study, the sample size is small. The four recruited patients had varied types and stages of diseases (2 patients with NAFLD and 2 with NASH). The overall PDFF values in patient group are relatively low (the median is 9.8% and the maximum is 15.9%), which was unable to show the feasibility of the multiparametric quantification from MT-ME under high-PDFF condition. A possible reason is that the patients had different durations of disease history and a variety of treatments which may have changed the pathological characteristics of liver to a different extent. Furthermore, the histology biomarkers including the fibrosis, fat concentration, and iron concentration were not available for this pilot study. The cross-correlation analysis between quantitative parameters from MT-ME and pathological markers were not accessible. Besides, the intra-session repeatability of MT-ME was not available in the patient group due to the additional time constraints associated with clinical scans.
In the future, studies will be performed on larger patient cohorts with histological validation. The inter-session repeatability and inter-vendor reproducibility of MT-ME will be assessed. Iron is another confounder of T1 measurement in liver. Increased iron concentration can directly reduce T167. Co-registered, simultaneous measurement of T1w and R2* using MT-ME provides the opportunity to develop and incorporate an R2* based iron-compensated algorithm68 into the MT-ME pipeline for the estimation of fat-independent, iron-compensated T1, which may serve as a better indicator of liver fibrosis scores. In addition, advanced shimming techniques to improve B0 field homogeneity can be potentially integrated into the application of MT-ME to further enhance the image quality and precision of the multiparametric quantification69,70.
5. Conclusion
A novel 3D MT-ME technique was developed, enabling simultaneous whole-liver quantification of joint T1, water-specific T1 (T1w), PDFF, and R2* within a 5-minute free-breathing acquisition. The T1, T1w, PDFF, and R2* estimated by MT-ME showed great intra-session repeatability and substantial agreement with reference values in a phantom and in vivo subjects. The proposed MT-ME technique is clinically promising for the comprehensive characterization of liver tissue properties.
Supplementary Material
Supporting information Table S1: List of sequence parameters. IR-SE with or without water excitation share the same imaging parameters. True spatial resolution was listed for all sequences.
Supporting Information Table S2: Image quality scores for MT-ME and reference images. For MT-ME, the water images, fat images, T1 maps, T1w maps, and PDFF maps are scored between good to excellent, and comparable to or better than the references. For R2* maps, the image quality from MT-ME with higher spatial resolution are comparable to the R2* from q-DIXON.
Supporting Information Table S3: The intra-session repeatability of the MT-ME image qualities. The image quality between the two MT-ME scans in the same imaging sessions are very close with ICC>0.9 for all types of images/maps.
Supporting Information Figure S1: the comparison of the T1w quantification the proposed basis-function-based method (T1w,1) and the straightforward method (T1w,2). (A) the comparison on phantom. The mean difference is −2.9 ms, the standard deviation of the differences is 8.6 ms. The ICC is 0.999 with no significant difference between the two measurements. (B) the comparison on in vivo data. The mean difference is −4.6 ms, the standard deviation of the differences is 8.2 ms. The ICC is 0.993 with a P value of 0.030. The overall differences between the T1w measurements from the two methods are small.
Supporting Information Figure S2: (A) An example map of fat-specific T1 (T1f) from a 58-year-old clinically healthy subject. The mean T1f in this case is 362 ms. (B) An example map of fat-specific T1 (T1f) from a 64-year-old NASH patient. The mean T1f in this case is 376 ms.
Supporting Information Figure S3: Measurements on the custom phantom. (A) T1, T1w, PDFF, and R2* maps from reference sequences and MT-ME. (B) Bland-Altman plots shows good repeatability of T1, T1w, PDFF, and R2* estimated with MT-ME. (C) Good correlation was demonstrated of T1, T1w, PDFF, and R2* from MT-ME and reference with high Pearson correlation coefficient R and ICC, as labeled on each plot.
Supporting Information Figure S4: B1+ efficiency maps at multiple slices. The liver is marked by the black boundary.
Supporting Information Figure S5: (A) Mean and standard deviation measurement of liver T1 for all in vivo cases (15 clinically healthy subjects + 4 patients). (B) Mean and standard deviation measurement of liver T1w for all in vivo cases (15 clinically healthy subjects + 4 patients).
Supporting Information Video S1: Demonstration of the MT-ME images (three spatial dimensions, one IR dimension, one multi-echo dimension, and one respiratory dimension).
Acknowledgements
This work was supported by NIH 1R01EB028146 and Doris Duke Charitable Foundation (DDCF) Grant #2020059 (AK). The authors thank Xinheng Zhang for the essential contribution in the construction of the custom phantom.
Reference
- 1.Younossi Z, Anstee QM, Marietti M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastro Hepat. January 2018; 15(1):11–20. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z, Tacke F, Arrese M, et al. Global Perspectives on Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Hepatology. June 2019;69(6):2672–2682. [DOI] [PubMed] [Google Scholar]
- 3.Tsochatzis EA, Bosch J, Burroughs AK. Liver cirrhosis. Lancet. May 17 2014; 383(9930):1749–61. [DOI] [PubMed] [Google Scholar]
- 4.Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. March 8 2008;371(9615):838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rockey DC, Caldwell SH, Goodman ZD, Nelson RC, Smith AD, American Association for the Study of Liver D. Liver biopsy. Hepatology. March 2009;49(3):1017–44. [DOI] [PubMed] [Google Scholar]
- 6.Bedossa P, Dargere D, Paradis V. Sampling variability of liver fibrosis in chronic hepatitis C. Hepatology. December 2003;38(6):1449–57. [DOI] [PubMed] [Google Scholar]
- 7.Bachtiar V, Kelly MD, Wilman HR, et al. Repeatability and reproducibility of multiparametric magnetic resonance imaging of the liver. PLoS One. 2019; 14(4):e0214921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaubert O, Arrieta C, Cruz G, et al. Multi-parametric liver tissue characterization using MR fingerprinting: Simultaneous T-1, T-2, T-2*, and fat fraction mapping. Magn Reson Med. May 13 2020; DOI: 10.1002/mrm.28311 [DOI] [PubMed] [Google Scholar]
- 9.Thompson RB, Chow K, Mager D, Pagano JJ, Grenier J. Simultaneous proton density fat-fraction and R-2* imaging with water-specific T-1 mapping (PROFIT1): application in liver. Magn Reson Med. August 4 2020; DOI: 10.1002/mrm.28434 [DOI] [PubMed] [Google Scholar]
- 10.Hernando D, Levin YS, Sirlin CB, Reeder SB. Quantification of liver iron with MRI: state of the art and remaining challenges. J Magn Reson Imaging. November 2014;40(5):1003–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeder SB, McKenzie CA, Pineda AR, et al. Water-fat separation with IDEAL gradient-echo imaging. J Magn Reson Imaging. March 2007;25(3):644–52. [DOI] [PubMed] [Google Scholar]
- 12.Banerjee R, Pavlides M, Tunnicliffe EM, et al. Multiparametric magnetic resonance for the non-invasive diagnosis of liver disease. J Hepatol. January 2014;60(1):69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reeder SB, Hu HH, Sirlin CB. Proton density fat-fraction: a standardized MR-based biomarker of tissue fat concentration. J Magn Reson Imaging. November 2012;36(5):1011–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong X, Nickel MD, Kannengiesser SA, Dale BM, Kiefer B, Bashir MR. Liver fat quantification using a multi-step adaptive fitting approach with multi-echo GRE imaging. Magn Reson Med. November 2014;72(5):1353–65. [DOI] [PubMed] [Google Scholar]
- 15.Yokoo T, Bydder M, Hamilton G, et al. Nonalcoholic Fatty Liver Disease: Diagnostic and Fat-Grading Accuracy of Low-Flip-Angle Multiecho Gradient-Recalled-Echo MR Imaging at 1.5 T. Radiology. April 2009;251(1):67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood JC, Enriquez C, Ghugre N, et al. MRI R2 and R2* mapping accurately estimates hepatic iron concentration in transfusion-dependent thalassemia and sickle cell disease patients. Blood. August 15 2005;106(4):1460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venkatesh SK, Yin M, Ehman RL. Magnetic resonance elastography of liver: technique, analysis, and clinical applications. J Magn Reson Imaging. March 2013;37(3):544–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luetkens JA, Klein S, Traber F, et al. Quantification of Liver Fibrosis at T1 and T2 Mapping with Extracellular Volume Fraction MRI: Preclinical Results. Radiology. September 2018;288(3):748–754. [DOI] [PubMed] [Google Scholar]
- 19.Hoad CL, Palaniyappan N, Kaye P, et al. A study of T-1 relaxation time as a measure of liver fibrosis and the influence of confounding histological factors. NMR Biomed. June 2015;28(6):706–714. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman DH, Ayoola A, Nickel D, Han F, Chandarana H, Shanbhogue KP. T1 mapping, T2 mapping and MR elastography of the liver for detection and staging of liver fibrosis. Abdom Radiol (NY). March 2020;45(3):692–700. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Sun J, Hu X, et al. Assessment of liver fibrosis by variable flip angle T1 mapping at 3.0T. J Magn Reson Imaging. March 2016;43(3):698–703. [DOI] [PubMed] [Google Scholar]
- 22.Harrison SA, Dennis A, Fiore MM, et al. Utility and variability of three non-invasive liver fibrosis imaging modalities to evaluate efficacy of GR-MD-02 in subjects with NASH and bridging fibrosis during a phase-2 randomized clinical trial. Plos One. September 7 2018;13(9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look‐Locker inversion recovery (MOLLI) for high‐resolution T1 mapping of the heart. Magn Reson Med 2004;52(1):141–146. [DOI] [PubMed] [Google Scholar]
- 24.Piechnik SK, Ferreira VM, Dall’Armellina E, et al. Shortened Modified Look-Locker Inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3 T within a 9 heartbeat breathhold. J Cardiovasc Magn Reson. November 19 2010;12:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chow K, Flewitt JA, Green JD, Pagano JJ, Friedrich MG, Thompson RB. Saturation recovery single-shot acquisition (SASHA) for myocardial T(1) mapping. Magn Reson Med. June 2014;71(6):2082–95. [DOI] [PubMed] [Google Scholar]
- 26.Mozes FE, Tunnicliffe EM, Moolla A, et al. Mapping tissue water T-1 in the liver using the MOLLI T-1 method in the presence of fat, iron and B-0 inhomogeneity. NMR Biomed. February 2019;32(2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larmour S, Chow K, Kellman P, et al. Characterization of T1 bias in skeletal muscle from fat in MOLLI and SASHA pulse sequences: Quantitative fat-fraction imaging with T1 mapping. Magn Reson Med. January 2017;77(1):237–249. [DOI] [PubMed] [Google Scholar]
- 28.Mozes FE, Tunnicliffe EM, Pavlides M, Robson MD. Influence of fat on liver T1 measurements using modified Look-Locker inversion recovery (MOLLI) methods at 3T. J Magn Reson Imaging. July 2016;44(1):105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christodoulou AG, Shaw JL, Nguyen C, et al. Magnetic resonance multitasking for motion-resolved quantitative cardiovascular imaging. Nat Biomed Eng. 2018;2(4):215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw JL, Yang Q, Zhou Z, et al. Free-breathing, non-ECG, continuous myocardial T1 mapping with cardiovascular magnetic resonance multitasking. Magn Reson Med. April 2019;81(4):2450–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma S, Wang N, Fan ZY, et al. Three-dimensional whole-brain simultaneous T1, T2, and T1 rho quantification using MR Multitasking: Method and initial clinical experience in tissue characterization of multiple sclerosis. Magn Reson Med. April 2021; 85(4): 1938–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma S, Nguyen CT, Han F, et al. Three-dimensional simultaneous brain T1, T2, and ADC mapping with MR Multitasking. Magn Reson Med. July 2020; 84(1):72–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang N, Gaddam S, Wang L, et al. Six-dimensional quantitative DCE MR Multitasking of the entire abdomen: Method and application to pancreatic ductal adenocarcinoma. Magn Reson Med. August 2020;84(2):928–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang Z-P. Spatiotemporal imagingwith partially separable functions. IEEE; 2007:988–991. [Google Scholar]
- 35.He J, Liu Q, Christodoulou AG, Ma C, Lam F, Liang ZP. Accelerated High-Dimensional MR Imaging With Sparse Sampling Using Low-Rank Tensors. IEEE Trans Med Imaging. September 2016;35(9):2119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christodoulou AG, Redler G, Clifford B, Liang Z-P, Halpern HJ, Epel B. Fast dynamic electron paramagnetic resonance (EPR) oxygen imaging using low-rank tensors. J Magn Reson. 2016;270:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Lathauwer L, De Moor B, Vandewalle J. A multilinear singular value decomposition. SIAM journal on Matrix Analysis and Applications. 2000;21(4):1253–1278. [Google Scholar]
- 38.Knoll F, Schwarzl A, Diwoky C, et al. gpuNUFFT - An open source GPU library for 3D regridding with direct Matlab interface. Proceedings of the 22nd annual meeting of ISMRM, Milan, Italy 2014;Abstract 4297 [Google Scholar]
- 39.Look DC, Locker DR. Time saving in measurement of NMR and EPR relaxation times. Rev Sci Instrum. 1970;41(2):250–251. [Google Scholar]
- 40.Jaubert O, Cruz G, Bustin A, et al. Water-fat Dixon cardiac magnetic resonance fingerprinting. Magn Reson Med. June 2020;83(6):2107–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu HZ, Shimakawa A, McKenzie CA, et al. Multiecho Water-Fat Separation and Simultaneous R-2* Estimation With Multifrequency Fat Spectrum Modeling. Magn Reson Med. November 2008;60(5):1122–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernando D, Haldar JP, Sutton BP, Ma J, Kellman P, Liang ZP. Joint estimation of water/fat images and field inhomogeneity map. Magn Reson Med. March 2008;59(3):571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu CY, McKenzie CA, Yu H, et al. Fat quantification with IDEAL gradient echo imaging: correction of bias from T(1) and noise. Magn Reson Med. August 2007;58(2):354–64. [DOI] [PubMed] [Google Scholar]
- 44.Hines C, Yu H, Shimakawa A, et al. Validation of fat quantification with T2* correction and accurate spectral modeling in a novel fat-water-iron phantom. Proceedings of the 17th Scientific Annual Meeting of the ISMRM. Honolulu, Hawaii, 2009; Abstract 2707. [Google Scholar]
- 45.Bojorquez J Bricq S, Acquitter C, Brunotte F, Walker PM, Lalande A. What are normal relaxation times of tissues at 3 T?. 2017; 35: 69–80. [DOI] [PubMed] [Google Scholar]
- 46.Wech T, Lemke A, Medway D, et al. Accelerating cine‐MR imaging in mouse hearts using compressed sensing. J Magn Reson Imaging. 2011;34(5):1072–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong X, Armstrong T, Nickel MD, et al. Effect of respiratory motion on free-breathing 3D stack-of-radial liver R 2 * relaxometry and improved quantification accuracy using self-gating. Magn Reson Med. June 2020;83(6):1964–1978. [DOI] [PubMed] [Google Scholar]
- 48.Bonekamp S, Tang A, Mashhood A, et al. Spatial Distribution of MRI-Determined Hepatic Proton Density Fat Fraction in Adults With Nonalcoholic Fatty Liver Disease. J Magn Reson Imaging. June 2014;39(6):1525–1532. doi: 10.1002/jmri.24321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sofue K, Mileto A, Dale BM, Zhong XD, Bashir MR. Interexamination repeatability and spatial heterogeneity of liver iron and fat quantification using MRI-based multistep adaptive fitting algorithm. J Magn Reson Imaging. November 2015;42(5):1281–1290. [DOI] [PubMed] [Google Scholar]
- 50.Shim JH, Yu JS, Chung JJ, et al. Segmental Difference of the Hepatic Fibrosis from Chronic Viral Hepatitis due to Hepatitis B versus C Virus Infection: Comparison Using Dual Contrast Material-Enhanced MRI. Korean J Radiol. Jul-Aug 2011;12(4):431–438. doi: 10.3348/kjr.2011.12.4.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dhamija E, Paul SB, Kedia S. Non-alcoholic fatty liver disease associated with hepatocellular carcinoma: An increasing concern. Indian J Med Res. January 2019;149(1):9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Rourke JM, Sagar VM, Shah T, et al. Carcinogenesis on the background of liver fibrosis: Implications for the management of hepatocellular cancer. World J Gastroentero. October 21 2018;24(39):4436–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bashir MR, Zhong XD, Nickel MD, et al. Quantification of Hepatic Steatosis With a Multistep Adaptive Fitting MRI Approach: Prospective Validation Against MR Spectroscopy. Am J Roentgenol. February 2015;204(2):297–306. [DOI] [PubMed] [Google Scholar]
- 54.Henninger B, Zoller H, Kannengiesser S, Zhong XD, Jaschke W, Kremser C. 3D Multiecho Dixon for the Evaluation of Hepatic Iron and Fat in a Clinical Setting. J Magn Reson Imaging. September 2017;46(3):793–800. [DOI] [PubMed] [Google Scholar]
- 55.Ghasabeh MA, Shaghaghi M, Khoshpouri P, et al. Correlation between incidental fat deposition in the liver and pancreas in asymptomatic individuals (vol 45, pg 203, 2020). Abdom Radiol. October 2020;45(10) [DOI] [PubMed] [Google Scholar]
- 56.Roujol S, Weingartner S, Foppa M, et al. Accuracy, precision, and reproducibility of four T1 mapping sequences: a head-to-head comparison of MOLLI, ShMOLLI, SASHA, and SAPPHIRE. Radiology. September 2014;272(3):683–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tirkes T, Zhao X, Lin C, et al. Evaluation of variable flip angle, MOLLI, SASHA, and IR-SNAPSHOT pulse sequences for T1 relaxometry and extracellular volume imaging of the pancreas and liver. Magn Reson Mater Phys Biol Med. October 2019;32(5):559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robson MD, Piechnik SK, Tunnicliffe EM, et al. T1 measurements in the human myocardium: the effects of magnetization transfer on the SASHA and MOLLI sequences. Magn Reson Med. September 2013;70(3):664–70. doi: 10.1002/mrm.24867 [DOI] [PubMed] [Google Scholar]
- 59.Negrete LM, Middleton MS, Clark L, et al. Inter-examination precision of magnitude-based MRI for estimation of segmental hepatic proton density fat fraction in obese subjects. J Magn Reson Imaging. May 2014;39(5):1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyagi A, Yeganeh O, Levin Y, et al. Intra- and inter-examination repeatability of magnetic resonance spectroscopy, magnitude-based MRI, and complex-based MRI for estimation of hepatic proton density fat fraction in overweight and obese children and adults. Abdom Imaging. October 2015;40(8):3070–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deichmann R, Haase A. Quantification of T1 values by SNAPSHOT-FLASH NMR imaging. J Magn Reson (1969). 1992;96(3):608–612. [Google Scholar]
- 62.Ahn JH, Yu JS, Park KS, et al. Effect of hepatic steatosis on native T1 mapping of 3T magnetic resonance imaging in the assessment of T1 values for patients with non-alcoholic fatty liver disease. Magn Reson Imaging. March 30 2021;80:1–8. [DOI] [PubMed] [Google Scholar]
- 63.Obmann VC, Mertineit N, Marx C, et al. Liver MR relaxometry at 3T-segmental normal T-1 and T-2* values in patients without focal or diffuse liver disease and in patients with increased liver fat and elevated liver stiffness. Sci Rep. May 30 2019;9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Serry FM, Ma S, Li D, et al. Dual Flip-angle IR-FLASH for B1+ Insensitive T1 Mapping: Application to T1 CMR Multitasking. Proceedings of the 28th Scientific Annual Meeting of the ISMRM. Virtual Meeting, 2020. (Abstract 0895) [Google Scholar]
- 65.Zhou R, Weller DS, Yang Y, et al. Dual‐excitation flip‐angle simultaneous cine and T 1 mapping using spiral acquisition with respiratory and cardiac self‐gating. Magn Reson Med. 2021;86(1):82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Serry F, Ma S, Mao X, et al. Dual flip angle (2FA) IR-FLASH with spin history mapping for B1+-corrected T1 mapping: Application to T1 cardiovascular magnetic resonance multitasking. Magn Reson Med. 2021;doi: 10.1002/mrm.28935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wood JC, Otto-Duessel M, Aguilar M, et al. Cardiac iron determines cardiac T2*, T2, and T1 in the gerbil model of iron cardiomyopathy. Circulation. July 26 2005;112(4):535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tunnicliffe EM, Banerjee R, Pavlides M, et al. A model for hepatic fibrosis: the competing effects of cell loss and iron on shortened modified Look-Locker inversion recovery T1 (shMOLLI-T1) in the liver. J Magn Reson Imaging. February 2017;45(2):450–462. [DOI] [PubMed] [Google Scholar]
- 69.Han H, Song AW, Truong TK. Integrated parallel reception, excitation, and shimming (iPRES). Magn Reson Med. July 2013;70(1):241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang H, Stager J, Azab L, et al. Whole Heart High-Order B0 Shimming at 3T Using a UNIfied Coil (UNIC) for RF Receive and Shimming. Proceedings of the 28th Scientific Annual Meeting of the ISMRM Virtual Meeting, 2020. 2020;(Abstract 2183) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information Table S1: List of sequence parameters. IR-SE with or without water excitation share the same imaging parameters. True spatial resolution was listed for all sequences.
Supporting Information Table S2: Image quality scores for MT-ME and reference images. For MT-ME, the water images, fat images, T1 maps, T1w maps, and PDFF maps are scored between good to excellent, and comparable to or better than the references. For R2* maps, the image quality from MT-ME with higher spatial resolution are comparable to the R2* from q-DIXON.
Supporting Information Table S3: The intra-session repeatability of the MT-ME image qualities. The image quality between the two MT-ME scans in the same imaging sessions are very close with ICC>0.9 for all types of images/maps.
Supporting Information Figure S1: the comparison of the T1w quantification the proposed basis-function-based method (T1w,1) and the straightforward method (T1w,2). (A) the comparison on phantom. The mean difference is −2.9 ms, the standard deviation of the differences is 8.6 ms. The ICC is 0.999 with no significant difference between the two measurements. (B) the comparison on in vivo data. The mean difference is −4.6 ms, the standard deviation of the differences is 8.2 ms. The ICC is 0.993 with a P value of 0.030. The overall differences between the T1w measurements from the two methods are small.
Supporting Information Figure S2: (A) An example map of fat-specific T1 (T1f) from a 58-year-old clinically healthy subject. The mean T1f in this case is 362 ms. (B) An example map of fat-specific T1 (T1f) from a 64-year-old NASH patient. The mean T1f in this case is 376 ms.
Supporting Information Figure S3: Measurements on the custom phantom. (A) T1, T1w, PDFF, and R2* maps from reference sequences and MT-ME. (B) Bland-Altman plots shows good repeatability of T1, T1w, PDFF, and R2* estimated with MT-ME. (C) Good correlation was demonstrated of T1, T1w, PDFF, and R2* from MT-ME and reference with high Pearson correlation coefficient R and ICC, as labeled on each plot.
Supporting Information Figure S4: B1+ efficiency maps at multiple slices. The liver is marked by the black boundary.
Supporting Information Figure S5: (A) Mean and standard deviation measurement of liver T1 for all in vivo cases (15 clinically healthy subjects + 4 patients). (B) Mean and standard deviation measurement of liver T1w for all in vivo cases (15 clinically healthy subjects + 4 patients).
Supporting Information Video S1: Demonstration of the MT-ME images (three spatial dimensions, one IR dimension, one multi-echo dimension, and one respiratory dimension).


