Abstract
We compared the results of human papillomavirus (HPV) detection and typing from 781 cervical samples assayed by three methods: L1 consensus PCR followed by cycle sequencing, L1 consensus PCR with biotinylated primers followed by hybridization to a line blot, and Hybrid Capture assay. Both PCR assays used L1 consensus PCR with primers MY09 and MY11. We evaluated the amplification efficiencies of both PCR assays and also compared the specific HPV types detected by each method. The samples positive by the Hybrid Capture assay were compared to the specific types detected by the PCR-based assays. The concordance between the two PCR assays in producing an HPV amplicon visible by gel electrophoresis or in detecting any HPV type was moderate: kappa values were 0.61 (95% confidence interval [CI] = 0.56 to 0.67) and 0.51 (95% CI = 0.46 to 0.58), respectively. The McNemar test for correlated proportions indicated that biotinylated PCR was less likely to produce a band (P = 0.001) and to detect an HPV type (P = 0.001) than the other PCR assay. In comparing the Hybrid Capture assay results with the HPV types detected by the PCR-based assays, we found that positivity by the Hybrid Capture assay for a number of samples may be due to cross-hybridization with HPV types not included in the Hybrid Capture assay probe cocktails.
Because there are more than 100 types of human papillomaviruses (HPVs), with at least 30 found primarily in the anogenital tract, studying the natural history and epidemiology of HPV is complex. Testing for HPV relies on the detection of viral DNA. Since all HPV types are closely related, assays can be designed to target conserved regions of the genome or regions whose sequences can best be used to discriminate different HPV types. The HPV assay format will determine how many HPV types will be detected as well as the efficiency of type-specific detection and discrimination.
The mucosal HPV types are commonly grouped into “high-risk” and “low-risk” categories on the basis of known epidemiologic associations or, for less common types, by homology to well-studied types. High-risk types are those similar to the types frequently found in anogenital malignancies; low-risk types are those similar to the types found in condylomata. For some applications, this level of HPV distinction may be enough to develop a viral profile in populations of isolates or an isolate from a particular patient. This approach is used in the U.S. Food and Drug Administration (FDA)-approved Hybrid Capture assay format (Digene Corporation, Silver Spring, Md.).
An alternative approach is to identify and specifically type the HPV in each sample. While individual type-specific assays could be used, it is more convenient to use one of several consensus PCR methods (3). Consensus assays use primers directed at relatively conserved regions of the HPV genome. The consensus amplicon is then typed by some method, such as dot blot hybridization with type-specific probes, restriction fragment length polymorphism analysis, or sequencing.
While the performance characteristics of each method of HPV detection is a function of analytic sensitivity and specificity, it is also influenced by the kinds of HPV infections encountered in populations, i.e., the frequencies of novel types and multiple infections. The resultant HPV profile for the study population may shift slightly depending on the assay. Our group currently uses L1 consensus PCR with the MY09-MY11 primer set (4), followed by cycle sequencing of the product. A new assay, also based on the L1 consensus PCR, is in commercial development by Roche Molecular Systems laboratories (2). The assay uses biotinylated primers and a single-tube reaction for both HPV and β-globin amplification. The biotinylated products are then hybridized to a line blot that includes 27 type-specific DNA targets as well as high- and low-copy-number β-globin controls.
The purpose of this study was to examine the performances of three validated HPV assays (the two L1 consensus PCR assays and the FDA-approved Hybrid Capture assay) with cervical samples from almost 800 women in populations with a high probability of having novel HPV types. The potential for detection of multiple and novel types will be different for each of the three assays. While sequencing could potentially identify any HPV isolate that is amplified, its ability to analyze multiple types may be limited. The line blot hybridization assay should easily detect multiple types, but amplicons from novel HPV types would either generate no signal or be misclassified due to cross-hybridization with a closely related type. The Hybrid Capture assay is the only FDA-approved assay, but the design does not distinguish samples with single or multiple types, and it is unclear how much cross-hybridization with novel types could be anticipated. A direct comparison of the results of all three assays is the only way to evaluate overall assay performance. This type of analysis is required to guide the selection of an assay appropriate for a particular application as well as to inform the community about the limits of interpretation of any one assay.
MATERIALS AND METHODS
Study material.
Specimens were collected from three African populations in studies approved by a human subjects review board. Cervical samples were collected with a Dacron swab from women attending either a sexually transmitted disease (STD) or human immunodeficiency virus (HIV) clinic in Johannesburg, South Africa. The samples were placed in 1 ml of Specimen Transport Medium (Digene Diagnostics) and were frozen for shipment to the Centers for Disease Control and Prevention (CDC) in Atlanta, Ga. The thawed samples were vigorously shaken before testing, and the swab was squeezed along the wall of the tube prior to removal. Cervicovaginal lavage samples were collected from women attending the Maternal and Child Health Center in Abidjan, Ivory Coast, and from commercial sex workers invited to a clinic for STD assessment, HIV counseling, and testing. The lavage samples were obtained by using 10 ml of normal saline to irrigate the cervix and vagina. Lavage cellular material was pelleted by low-speed centrifugation, resuspended at a 1:10 dilution in phosphate-buffered saline (volume:volume), frozen, and shipped to CDC. For both types of samples, 100-μl aliquots were digested with proteinase K, and DNA was extracted with phenol-chloroform and was concentrated by ethanol precipitation. The DNA precipitate for each sample was resuspended in 50 μl of water.
In-house HPV PCR assay and cycle sequencing.
HPV DNA was amplified by using L1 consensus primers MY11 and MY09 (4). DNA quantity and integrity were monitored through amplification of part of the β-globin gene in replicate tubes. Five microliters of purified DNA was used in each PCR mixture. Amplification without DNA template was used to monitor contamination in both HPV and β-globin reactions. A 10-μl aliquot of the PCR mixture was visualized by ethidium bromide staining after agarose gel electrophoresis. Samples that revealed an approximately 450-bp band in the HPV reaction were purified, and cycle sequencing in both directions was performed to determine the type of HPV (8). The cycle sequencing procedure was slightly modified so that the BigDye Terminator Cycle Sequencing Kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.) could be used according to the manufacturer's recommendations, and the reaction mixture was analyzed on an ABI Prism 377 DNA Sequencer (Perkin-Elmer Applied Biosystems). We interpreted a sequence to be present if the peaks on the electropherogram were sharp, were well defined, and had sufficient signal height. The University of Wisconsin (Madison) Genetics Computer Group package was used to search all viral sequences in GenBank by using FASTA. If no sequence similarity was detected among the viral sequences, all nucleotide sequences in GenBank were searched. A sequence was considered a match if it had more than 70% nucleotide similarity to an HPV sequence in GenBank. Samples with multiple HPV types were cautiously analyzed so that only sequences with at least 80% similarity to the corresponding HPV sequences in GenBank were considered to be present. For example, the nucleotide sequences of the consensus L1 regions of HPV types 16 and 18 are 72% similar. If a sequence search of a sample yielded 95% similarity to HPV type 16 (HPV-16) and 85% similarity to HPV-18, the sample was considered positive for both types.
Biotinylated HPV PCR assay and line blot assay.
The reagents necessary for the L1 consensus PCR assay and for genotyping of HPV PCR products by the line blot assay were generously provided by Janet Kornegay and Raymond Apple of Roche Molecular Systems, Inc., Alameda, Calif. The amplification with biotinylated HPV consensus, HMB01, and β-globin primers, the line blot assay, and detection were performed as described previously (3). Five microliters of purified DNA was used in the PCR mixture. A 10-μl aliquot of each PCR mixture was evaluated by agarose gel electrophoresis and ethidium bromide staining. The presence or absence of the 450-bp HPV amplicon was noted. A PCR mixture without DNA template was included with each set of reactions to monitor contamination.
Hybrid Capture system.
The commercially available Hybrid Capture Tube (HCT) assay was used to test for high- and low-risk HPV types in all specimens by using probe A (HPV types 6, 11, 42, 43, and 44) and probe B (HPV types 16, 18, 31, 33, 35, 45, 51, 52, and 56). The manufacturer's package insert describes preparation and testing of the cervical swab specimens. The lavage specimens were prepared by a procedure provided by Digene. Briefly, 100 μl of lavage cell concentrate was added to a hybridization tube with 50 μl of denaturation reagent, and the mixture was vortexed. The tubes were incubated at 65°C for 45 min and were then cooled to room temperature. For the remaining steps the instructions in the manufacturer's package insert were followed.
Data analysis.
The PCR assays were performed and the results were interpreted in separate laboratories by different laboratory personnel, and the results were entered into separate Paradox (Borland International, Scotts Valley, Calif.) databases. The results for each sample were tabulated to compare the HPV amplicons visualized by the in-house HPV and biotinylated HPV PCRs as well as the type of HPV detected by the cycle sequencing and line blot assays. For determination of the HPV type(s) that might be detected by the HCT assay, the results were considered concordant if any of the HPV types detected by the line blot assay were represented in either the HCT assay probe A or the HCT assay probe B cocktail. The two-sided McNemar test was used to compare the sensitivity rates obtained either with the two PCR methods or with the biotinylated PCR assay and the HCT assay. The kappa statistic was calculated to determine the level of agreement between pairs of assay methods. In general, kappa values of <0, 0 to 0.2, 0.21 to 0.4, 0.41 to 0.6, 0.61 to 0.8, 0.81 to 0.99, 1.0 indicate poor, slight, fair, moderate, substantial, almost perfect, and perfect agreement, respectively. Statistical analyses were performed with PC-SAS, version 6.10/6.11 (SAS Institute, Cary, N.C.), software.
RESULTS
The performances of the two HPV PCR assays (an in-house PCR with cycle sequencing and a biotinylated PCR with the line blot assay) were compared in four ways. The efficiency of the amplification was compared by evaluating the ability of each assay to produce an HPV amplicon visible by gel electrophoresis and ethidium bromide staining (Table 1). The abilities of hybridization of the biotinylated product and cycle sequencing of the in-house PCR product to detect any HPV type were also compared (Table 2). The type-specific performance of each assay was evaluated by comparing the numbers of samples positive for each HPV type (Table 3). Finally, the overall performance of each assay was evaluated by comparing the HPV type(s) detected in each sample (Table 4).
TABLE 1.
Comparison of amplification efficiency of each PCR assay by visualization of 450-bp amplicon on ethidium bromide-stained agarose gel
| In-house HPV PCR result | No. (%) of samples with the following biotinylated HPV PCR result:
|
|
|---|---|---|
| Positive | Negative | |
| Positive | 344 (44) | 102 (13) |
| Negative | 49 (6) | 286 (36) |
TABLE 2.
Concordance of hybridization of biotinylated product to the line blot versus cycle sequencing of the in-house PCR product for detection of any HPV type
| HPV sequencing result | No. (%) of samples with the following line blot assay result:
|
|
|---|---|---|
| Positive | Negative | |
| Positive | 301 (39) | 137 (18) |
| Negative | 54 (7) | 289 (37) |
TABLE 3.
Detection of HPV types by line blot assay, cycle sequencing, or both
| HPV type | No. of samples positive by:
|
|||
|---|---|---|---|---|
| Cycle sequencing only | Line blot assay only | Both assays | Total | |
| 6 | 0 | 24 | 0 | 24 |
| 11 | 0 | 18 | 0 | 18 |
| 16 | 9 | 36 | 27 | 72 |
| 18 | 12 | 47 | 14 | 73 |
| 26 | 1 | 4 | 0 | 5 |
| 30 | 17 | NTa | NT | 17 |
| 31 | 14 | 15 | 22 | 51 |
| 33 | 17 | 6 | 24 | 47 |
| 35 | 13 | 8 | 2 | 23 |
| 39 | 6 | 15 | 2 | 23 |
| 40 | 0 | 1 | 0 | 1 |
| 42 | 0 | 1 | 0 | 1 |
| 45 | 6 | 14 | 8 | 28 |
| 51 | 4 | 32 | 1 | 37 |
| 52 | 8 | 18 | 16 | 42 |
| 53 | 10 | 43 | 14 | 67 |
| 54 | 1 | 30 | 10 | 41 |
| 55 | 13 | 5 | 1 | 19 |
| 56 | 9 | 15 | 4 | 28 |
| 57 | 0 | 0 | 0 | 0 |
| 58 | 27 | 19 | 33 | 79 |
| 59 | 3 | 8 | 3 | 13 |
| 61 | 28 | NT | NT | 28 |
| 62 | 18 | NT | NT | 18 |
| 64 | 1 | NT | NT | 1 |
| 66 | 18 | 14 | 15 | 47 |
| 67 | 2 | NT | NT | 2 |
| 68 | 5 | 11 | 4 | 20 |
| 69 | 3 | NT | NT | 3 |
| 70 | 14 | NT | NT | 14 |
| 72 | 11 | NT | NT | 11 |
| 73 | 1 | NT | NT | 1 |
| 74 | 8 | NT | NT | 8 |
| CP8304 | 19 | NT | NT | 19 |
| IS039 | 3 | NT | NT | 3 |
| MM4 | 4 | 6 | 5 | 15 |
| MM7 | 2 | 27 | 7 | 36 |
| MM8 | 2 | 31 | 9 | 42 |
| MM9 | 0 | 4 | 2 | 6 |
| SEQ277 | 78 | NT | NT | 78 |
NT, not tested; type not included on the line blot.
TABLE 4.
Detection of HPV types in all samples by cycle sequencing and line blot assay
| HPV type | No. (%) of samples detected by:
|
|
|---|---|---|
| Cycle sequencing | Line blot assay | |
| 16 | 22 (3) | 24 (3) |
| 18 | 10 (1) | 22 (3) |
| 26 | 1 (<1) | 0 |
| 31 | 20 (3) | 15 (2) |
| 33 | 10 (1) | 4 (<1) |
| 35 | 4 (<1) | 0 |
| 39 | 2 (<1) | 6 (<1) |
| 45 | 2 (<1) | 6 (<1) |
| 51 | 0 | 11 (1) |
| 52 | 9 (1) | 10 (1) |
| 55 | 5 (<1) | 3 (<1) |
| 56 | 1 (<1) | 2 (<1) |
| 58 | 27 (3) | 16 (2) |
| 59 | 4 (<1) | 0 |
| 61 | 14 (2) | |
| 62 | 9 (1) | |
| 64 | 1 (<1) | |
| 67 | 1 (<1) | |
| 68 | 2 (<1) | 2 (<1) |
| 69 | 1 (<1) | |
| 70 | 7 (1) | |
| 72 | 3 (<1) | |
| 73 | 0 | |
| CP8304 | 8 (1) | |
| IS039 | 2 (<1) | |
| MM4 | 3 (<1) | 4 (<1) |
| MM7 | 4 (<1) | 9 (1) |
| MM9 | 1 (<1) | 4 (<1) |
| SEQ277 | 62 (8) | |
| 6 | 0 | 8 (1) |
| 11 | 0 | 4 (<1) |
| 30 | 0 | |
| 40 | 0 | 0 |
| 42 | 0 | 0 |
| 53 | 9 (1) | 23 (3) |
| 54 | 7 (1) | 13 (2) |
| 57 | 0 | 0 |
| 66 | 13 (2) | 7 (1) |
| 74 | 1 (<1) | |
| MM8 | 8 (1) | 10 (1) |
| Multiple | 136 (17) | 152 (19) |
| Unknown | 25 (2) | 61 (8) |
| Negative | 347 (44) | 365 (47) |
As shown in Table 1, the concordance between both PCR assays in producing an HPV amplicon visible by gel electrophoresis was substantial (kappa value, 0.61; 95% confidence interval [CI] = 0.56 to 0.67). As shown in Table 2, the concordance of the detection of HPV types between cycle sequencing and the line blot assay was moderate (kappa value, 0.51; 95% CI = 0.46 to 0.58). The McNemar test for correlated proportions indicated that the in-house HPV PCR assay was more likely than the biotinylated PCR assay to produce a visible band (P = 0.001) and to detect any HPV type (P = 0.001).
The HPV type-specific performance of each assay is shown in Table 3. Samples positive for multiple HPV types are included in the totals for each type detected. Cycle sequencing detected 13 HPV types not included in the line blot assay, the most frequent of which was SEQ277 (Genbank accession no. I47614), which was identified in 78 samples. Of the 27 HPV types included in the line blot, the line blot assay generally detected more positive samples than sequencing did. Particularly striking is the absence of HPV types 6 and 11 in the cycle sequencing results. However, sequencing detected HPV types 33, 35, 55, and 58 more frequently than the line blot assay did.
Overall, 492 (63%) of the 781 samples were positive for HPV by one or both assays. An HPV type was detected by cycle sequencing in 446 (57%) samples and by the line blot assay in 393 (50%) samples. Table 4 summarizes the HPV type(s) detected in each sample by both assays. Slightly fewer samples were HPV negative by the sequencing assay (347 samples) than by the line blot assay (365 samples). The unknown category for sequencing includes samples with sequences that fail to match any GenBank HPV sequence (i.e., potentially novel HPV types). Only eight amplicon-positive samples failed to yield a sequence (data not shown), and very few amplicon-negative samples were subjected to cycle sequencing. For the line blot assay, unknown samples are those for which an amplicon was detected on the gel but for which the amplicon did not hybridize to the line blot. Samples with multiple HPV types were grouped as multiple. The line blot assay detected slightly more samples with multiple types (152 samples) than the sequencing assay (136 samples) did. For most of the 27 HPV types included in the line blot assay, similar numbers of samples infected with single HPV types were found by both assays. The exceptions were HPV types 18, 51, 6, 11, and 53, which were found more frequently by the line blot assay, and HPV types 31, 58, and 59, which were found more frequently by the cycle sequencing assay.
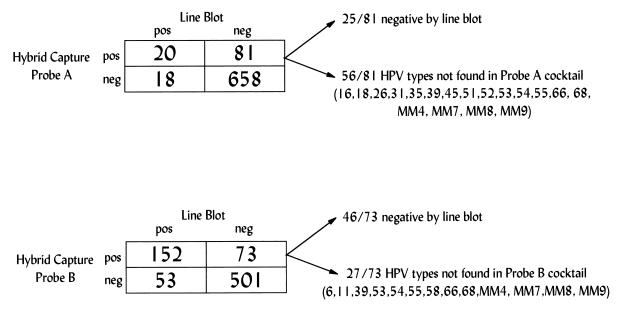
Results of the biotinylated PCR with the line blot assay were also compared to the results of the HCT assay with the low-risk HPV probe A cocktail and the HCT assay with the high-risk HPV probe B cocktail (Fig. 1). The line blot assay results for HPV types not found in each HCT assay probe cocktail were included with results for HPV-negative samples. The concordance between the two assays for the low-risk probe was fair (kappa value, 0.21; 95% CI = 0.11 to 0.31), whereas the concordance for the high-risk probe was moderate (kappa value, 0.56; 95% CI = 0.46 to 0.59). The McNemar test for correlated proportions indicated no significant difference between the two assays for the high-risk probe (P = 0.31); however, the HCT assay with the low-risk probe was more likely than the PCR assay to detect a low-risk HPV type (P = 0.001). To further evaluate why the HCT assay detected more low-risk HPV-positive samples than the line blot PCR assay did, the complete HPV profiles of each HCT probe A-positive and line blot assay-negative sample were determined (Fig. 1). Most of these samples (56 of 81; 69%) were line blot assay positive for HPV types not in the low-risk probe cocktail. The remaining 25 (31%) were negative for HPV by the line blot assay. Of these, only three were cycle sequencing positive, all for types not included in the probe A cocktail. Similar findings are shown in Fig. 1 for probe B.
FIG. 1.
Comparison of positive and negative results by the Hybrid Capture assay and line blot assay for samples infected with different HPV types. The Hybrid Capture assay probe A cocktail contains HPV types 6, 11, 42, 43, and 44 and the probe B cocktail contains HPV types 16, 18, 31, 33, 35, 45, 51, 52, and 56. For a sample to have concordant results by both assays, the type detected (or not detected) by the line blot assay had to be the one included in the probe cocktail and vice versa. All other samples were considered to have discrepant results. The HPV types that may account for the Hybrid Capture assay-positive, line blot assay-negative samples with discrepant results are shown.
DISCUSSION
Both the cycle sequencing assay and the line blot assay involve PCRs that use the MY09 and MY11 primers to amplify a consensus region of the HPV L1 gene and globin primers to determine sample adequacy. The line blot assay coamplifies the β-globin and HPV L1 targets by using biotinylated primers, whereas the cycle sequencing assay uses separate amplifications with unmodified primers. The current line blot assay format detects 27 HPV types, whereas cycle sequencing has the ability to detect all known HPV types as well as potentially novel types that can be amplified with the MY09-MY11 primers. We compared the performances of these two PCR assay formats for the detection and typing of HPV in samples from three African populations.
Amplification efficiency as reflected by positive gel results was slightly greater for the in-house PCR assay. This difference could be explained by the effects of biotinylation of the primers and of the single-tube coamplification of HPV and β-globin DNAs. Discrepancies due to coamplification of HPV and β-globin targets have been reported by others using this line blot assay (1). However, when comparing their abilities to detect specific HPV types, the assays had nearly identical likelihoods of being positive. In some instances the line blot assay outperformed the cycle sequencing assay for particular HPV types. For example, HPV types 6, 16, and 18 were amplified more efficiently with the biotinylated primer mix, while HPV types 33 and 35 were amplified more efficiently with the in-house HPV primers. These differences may be due to variations in the synthesis of the primers at the degenerate nucleotide positions. However, we do know that our in-house primers can efficiently amplify HPV types 6, 11, 16, 18, 31, 33, 35, 52, 56, and 58 individually. It is known that PCR with the MY09-MY11 consensus primers in samples containing multiple HPV types may preferentially amplify certain HPV types and produce inaccurate results (7). Many of our samples contained multiple HPV types, and this may be another reason for the discrepant results.
Many of the samples contained multiple HPV types that were detected either by the line blot assay or by the cycle sequencing assay. Interpretation of results for samples with multiple HPV types was more easily accomplished by looking at the line blot assay results than by deciphering the sequences from the samples. Sequence analysis of samples with multiple types was time-consuming and often required sequence editing. Given the time and the often subjective analysis required for cycle sequencing, the line blot assay proved to be easy and efficient for typing of multiple HPV types obtained by PCR with consensus primers. Despite this limitation of cycle sequencing, the line blot assay and the cycle sequencing assay can be used as complementary assays. The line blot assay can be used to detect the known HPV types in a sample, and cycle sequencing can be used to identify novel types or types not found on the line blot. This practice would require that an aliquot of each PCR mixture be visualized by ethidium bromide staining after electrophoresis. Positive PCR products that give a line blot assay result would not need to be characterized further, whereas positive PCR products that are negative by the line blot assay could be sequenced to determine the HPV type.
The HCT assay uses solution hybridization of RNA probe mixtures to detect low- and high-risk HPV types of DNA in the test sample. We determined which HPV types were hybridizing with the RNA probes in the cocktail by comparing HPV type-specific results by the line blot assay with the results obtained by the HCT assay. Both the low-risk and the high-risk probe cocktails gave positive hybridization results for HPV types not represented in either probe cocktail (Fig. 1). With both probe cocktails, there was cross-hybridization with several HPV types that are genetically similar to the high-risk types present in the probe cocktail. Cross-hybridization has also been observed when comparing the second-generation Hybrid Capture II test with the MY09-MY11 PCR assay, but the types responsible for the cross-hybridization were not characterized (6). Although some cross-hybridization between related types might be expected, it was disturbing to have HCT assay-positive results for samples that were negative by two independent PCR assays. At least three explanations can be given for the results for these HCT assay-positive samples. First, the HPV RNA probes in the cocktail may be hybridizing with novel HPV types or with HPV types that are not amplified by the MY09-MY11 primers. Second, the DNA sample used in the PCR assay may not be as representative of the HPV types present in the sample used in the HCT assay. Third, the HPV RNA probes in the cocktail may be specifically hybridizing with cellular nucleic acids. Several RNA viruses, when used as probes, have been shown to hybridize to cellular rRNAs and to each other (5).
In conclusion, the line blot assay is a very efficient method for typing MY09-MY11 PCR products. Sequence analysis of PCR-positive, line blot assay-negative samples can complement the line blot assay and can be used to identify novel HPV types. Complex probe cocktails may result in false-positive results, and such results should be cautiously interpreted. An understanding of the natural history of HPV-associated disease requires sensitive and specific HPV detection and typing assays that can be used for both routine clinical screening and large-scale epidemiologic studies.
ACKNOWLEDGMENTS
We gratefully acknowledge the investigators in Abidjan, Ivory Coast, and in Johannesburg, South Africa, who provided all the clinical samples. We thank Soheyla Sadeghiani and Donna L. Miller for assistance with processing of the samples and participation in the laboratory testing. We also thank Rosane Nisenbaum for assistance with the statistical analysis.
This work was supported in part by Roche Molecular Systems, which supplied the reagents and supplies for the line blot assay.
REFERENCES
- 1.Coutlee F, Gravitt P, Richardson H, Hankins C, Franco E, Lapointe N, Voyer H the Canadian Women's HIV Study Group. Nonisotopic detection and typing of human papillomavirus DNA in genital samples by the line blot assay. J Clin Microbiol. 1999;37:1852–1857. doi: 10.1128/jcm.37.6.1852-1857.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gravitt P C, Peyton C L, Apple R J, Wheeler C M. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J Clin Microbiol. 1998;36:3020–3027. doi: 10.1128/jcm.36.10.3020-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harnish D G, Belland L M, Scheid E E, Rohan T E. Evaluation of human papillomavirus-consensus primers for HPV detection by the polymerase chain reaction. Mol Cell Probes. 1999;13:9–21. doi: 10.1006/mcpr.1998.0203. [DOI] [PubMed] [Google Scholar]
- 4.Manos M, Ting Y, Wright D K, Lewis A J, Broker T R, Wolinsky S M. Use of polymerase chain reaction amplification for the detection of genital human papillomavirus. Cancer Cells. 1989;7:209–214. [Google Scholar]
- 5.McClure M A, Perrault J. RNA virus genomes hybridization to cellular rRNAs and to each other. J Virol. 1986;57:917–921. doi: 10.1128/jvi.57.3.917-921.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peyton C L, Schiffman M, Lorincz A T, Hunt W E, Mielzynska I, Bratti C, Eaton S, Hildesheim A, Morera L A, Rodriguez A C, Herrero R, Sherman M E, Wheeler C M. Comparison of PCR- and Hybrid Capture-based papillomavirus detection systems using multiple cervical specimen collection strategies. J Clin Microbiol. 1998;36:3248–3254. doi: 10.1128/jcm.36.11.3248-3254.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tucker R A, Johnson P R, Reeves W C, Icenogle J P. Using the polymerase chain reaction to genotype human papillomavirus DNAs in samples containing multiple HPVs may produce inaccurate results. J Virol Methods. 1993;43:321–334. doi: 10.1016/0166-0934(93)90150-p. [DOI] [PubMed] [Google Scholar]
- 8.Unger E R, Vernon S D, Thoms W W, Nisenbaum R, Spann C O, Horowitz I R, Icenogle J P, Reeves W C. Human papillomavirus and disease-free survival in FIGO stage Ib cervical cancer. J Infect Dis. 1995;172:1184–1190. doi: 10.1093/infdis/172.5.1184. [DOI] [PubMed] [Google Scholar]