Abstract
Autophagy, an essential cellular process that mediates degradation of proteins and organelles in lysosomes, has been tightly linked to cellular quality control for its role as part of the proteostasis network. The current interest in identifying the cellular and molecular determinants of aging, has highlighted the important contribution of malfunctioning of autophagy with age to the loss of proteostasis that characterizes all old organisms. However, the diversity of cellular functions of the different types of autophagy and the often reciprocal interactions of autophagy with other determinants of aging, is placing autophagy at the center of the aging process. In this work, we summarize evidence for the contribution of autophagy to health- and lifespan and provide examples of the bidirectional interplay between autophagic pathways and several of the so-called hallmarks of aging. This central role of autophagy in aging, and the dependence on autophagy of many geroprotective interventions, has motivated a search for direct modulators of autophagy that could be used to slow aging and extend healthspan. Here, we review some of those ongoing therapeutic efforts and comment on the potential of targeting autophagy in aging.
Keywords: aging, chaperones, lysosomes, organelle turnover, proteostasis, proteolysis
1. Introduction
Autophagy refers to the lysosomal degradation and recycling of all types of intracellular components including, proteins, organelles or even pathogens that reach the cytoplasm (Levine and Klionsky, 2017). The contribution of autophagy to the continuous turnover of the cellular proteome and to the selective removal of malfunctioning or damaged cellular proteins, explains its inclusion as part of the proteostasis network – the subset of chaperones and proteolytic systems that ensure that each protein is at the right time, in the right conformation and in the expected place inside the cells (Sala et al., 2017). This role of autophagy in proteostasis maintenance, along with the early observations of autophagy malfunctioning with age, has linked autophagy to the loss of proteostasis, described as a common feature in all old organisms.
However, functions of autophagy go beyond maintenance of proteostasis (Levine and Klionsky, 2017). This basic cellular process is also essential for maintenance of cellular energetics, cellular response to stress, defense against pathogens, cellular reprograming, maintenance of stemness or fine-tuning of senescence, among others. Consequently, as described in this review, malfunctioning of autophagy with age impinges on many of the cellular and molecular processes known to drive aging (hallmarks of aging) including besides loss of proteostasis, poor response to stress, disrupted cellular energetics, persistent cellular senescence, stem cell exhaustion, epigenetic alterations, defective intercellular communication, accumulation of macromolecular damage and telomere attrition (Kennedy et al., 2014; López-Otín et al., 2013). Growing evidence supports that the hallmarks of aging are interconnected as often, experimental reproduction of one of them is sufficient to elicit some of the others (Lopez-Otin and Kroemer, 2021). Understanding these interconnections is important because, by the same token, interventions that prevent one of these hallmarks, should have a beneficial effect in preventing some of the others.
Here we review the changes with age in the different types of mammalian autophagy, the functional consequences of those changes and the impact that malfunctioning of autophagy has on multiple hallmarks of aging. We propose that the multifunctionality of autophagy makes it an attractive candidate as mediator of the interconnections among the hallmarks of aging and discuss attempts to target autophagy with anti-aging purposes.
2. Autophagic pathways, main molecular players, and regulation
Three major forms of autophagy, known as macroautophagy, chaperone-mediated autophagy (CMA) and microautophagy, have been described to co-exist in all mammalian cells. They differ in their regulation, the nature of cargo that they degrade, and the mechanisms that govern cargo trafficking into the lysosomes.
2.1. Macroautophagy
In macroautophagy, cytoplasmic material is delivered to lysosomes upon its sequestration within double-membraned vesicles named autophagosomes. The central macroautophagy machinery are the autophagy-related proteins (ATGs) that assemble into functional complexes to promote autophagosome formation, trafficking, and fusion with lysosomes (maturation) (Levine and Klionsky, 2017; Mizushima, 2020) (Fig. 1). Depending on the cargo sequestered in autophagosomes, macroautophagy can be “in bulk” (when large portions of the cytoplasm are sequestered withing the forming autophagosome for degradation) or selective (when only specific cellular components are targeted for lysosomal degradation) (Levine and Kroemer, 2019). Selective macroautophagy contributes to turnover of organelles and to protection against external pathogens by targeting aggregates (aggrephagy), ER (ER-phagy), ferritin (ferritinophagy), glycogen (glycophagy), lipid droplets (lipophagy), lysosomes (lysophagy), midbody rings (midbody removal), mitochondria (mitophagy), peroxisomes (pexophagy), ribosomes (ribophagy), nucleic acids (RN/DNautophagy), bacteria and viruses (xenophagy) or zymogen granules (zymophagy) (Lamark and Johansen, 2021). Selective autophagy requires engagement of receptors that recognize specific features in the cargo and link it to the autophagy machinery to facilitate its sequestration (Stolz et al., 2014). Autophagy receptors undergo structural changes (i.e., oligomerization) and post-translational modifications (i.e., phosphorylation, ubiquitination, acetylation) that mediate receptor recruitment to the cargo, docking on the autophagy machinery and their simultaneous degradation with the entrapped cargo (Lamark and Johansen, 2021). Formation of the limiting membrane of the autophagosomes can occur at various locations in the cells including ER, late endosomes, plasma membrane and mitochondria. Membranes of these organelles contribute some of the lipids required for the forming autophagosomes, whereas others are shuttled from the Golgi (Levine and Klionsky, 2017).
Figure. 1. Types of autophagy pathways in mammals.
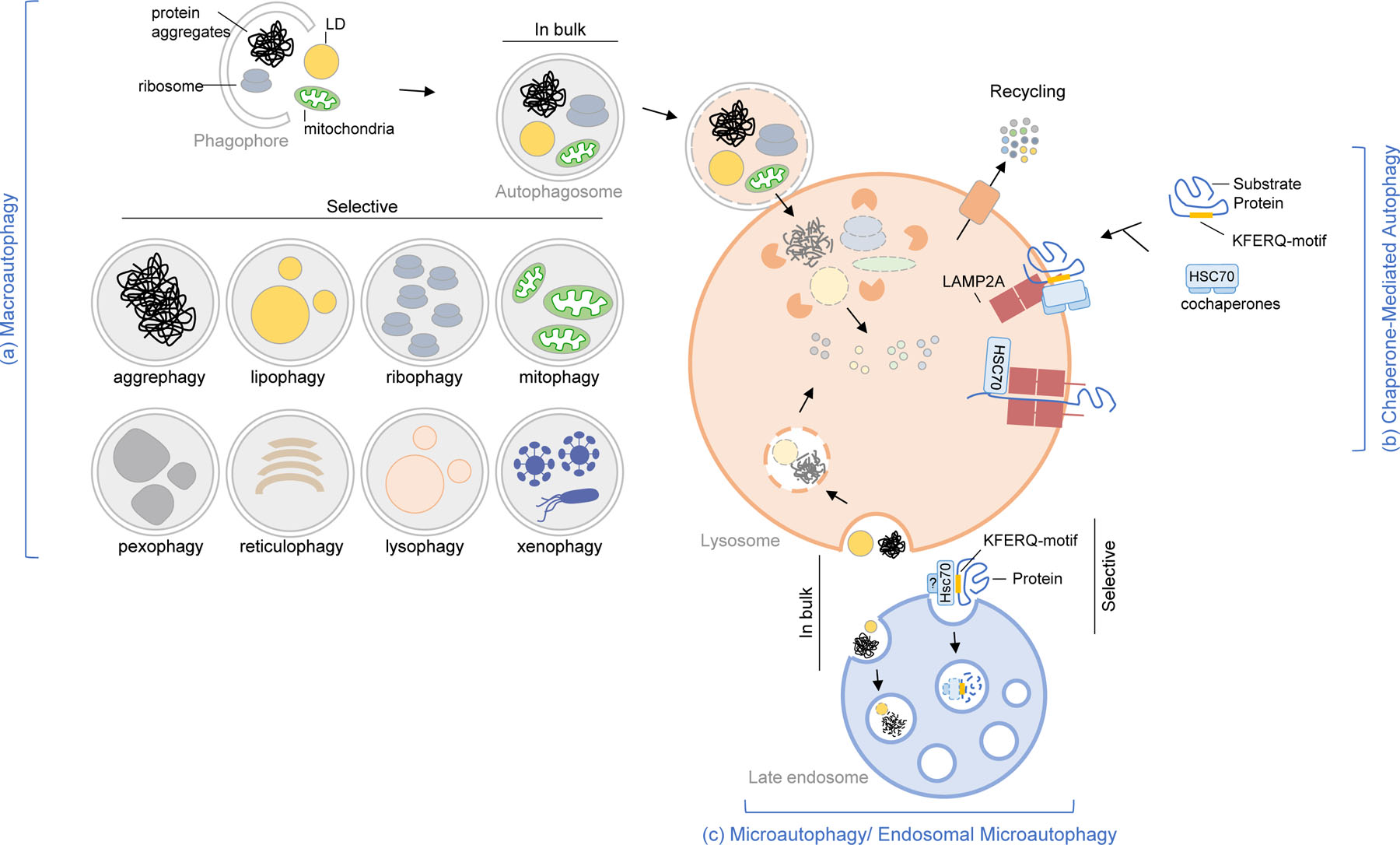
Schematic of the three major types of autophagy in mammals. (a) Macroautophagy is characterized by the sequestration of cargo (proteins, aggregates, organelles, pathogens) inside autophagosomes. In bulk and some forms of selective macroautophagy are depicted. The cargo is degraded in lysosomes upon autophagosome-lysosome fusion. (b) Chaperone-mediated autophagy is characterized by the selective degradation of proteins with KFERQ-motif. The substrates are recognized by HSC70/other chaperones. The protein substrates are degraded in lysosomes upon translocating across the membrane through the receptor multimeric complex (LAMP2A). (c) Microautophagy/Endosomal microautophagy is characterized by the engulfment of the cargo (proteins, aggregates, organelles) by the lysosomal membrane (in microautophagy) or late endosomal membrane (in endosomal microautophagy). The cargo is degraded when the intraluminal membrane is disintegrated giving the lysosomal hydrolases access. Once degraded by the lysosomal hydrolases, the biochemical components are recycled to the cytoplasm. Note: For simplicity, the three pathways are shown to converge on one lysosome; however, the current consensus is that different lysosomal populations cater to different autophagy pathways.
Macroautophagy is induced in response to different stressors such as nutrient or growth factor deprivation, hypoxia, damaged proteins and organelles, genotoxic stress among others (Morishita and Mizushima, 2019). This response is tightly regulated by a variety of signaling pathways that secure appropriate fine-tuning. Two cellular kinases, mechanistic target of rapamycin (mTOR) and AMP-activated protein kinase (AMPK) are central in macroautophagy regulation. mTOR integrates signals from growth factors, stress, energy status, oxygen, and amino acids that trigger its inhibitory effect on macroautophagy (Kim and Guan, 2015). Activation of mTOR (i.e., by nutrients) promotes phosphorylation of ULK1, a kinase required for initiation of the autophagic process, and maintains the ULK1/ATG13/FIP200 complex in an inactive conformation (Mizushima, 2020). This complex is released upon inactivation of mTOR to promote macroautophagy. AMPK instead positively regulates macroautophagy upon its activation under conditions of energy deficiency (elevated AMP/ATP ratio), through a variety of mechanisms including direct phosphorylation of ULK1, RAPTOR and the TSC1-TSC2 complex to inhibit mTOR, of Beclin-1 to activate the Beclin-1–VPS34 complex required for autophagy initiation, and of FoxO3 to activate transcription of autophagy genes such as LC3B, GABARAPL1 and Beclin-1 (Tamargo-Gómez and Mariño, 2018). A gene regulatory network that enhances macroautophagy by increasing lysosomal biogenesis is the transcription factor EB (TFEB), a master regulator that positively controls expression of lysosomal and Atg genes by binding to their promoters (Settembre et al., 2011).
2.2. Chaperone-mediated autophagy
CMA was the first selective form of autophagy described for proteins (Chiang and Dice, 1988). Selectivity of CMA is conferred through a unique mechanism for cargo recognition and internalization into lysosomes. Only proteins bearing a specific targeting motif in their amino acid sequence (biochemically related with the pentapeptide KFERQ) can undergo degradation via CMA (Chiang and Dice, 1988). Almost half of the mammalian proteome contains this motif in their sequence (canonical motif). In addition, in some instances, the targeting motif can be generated through phosphorylation or acetylation, bringing the number of potential CMA substrates to 75% of the proteome (Kirchner et al., 2019). This motif is recognized by a cytosolic chaperone, the heat shock cognate protein of 70KD (HSC70), that brings the substrate to the lysosomal surface. Before internalization for rapid intralysosomal degradation, HSC70 transfers the cargo protein to the lysosome-associated membrane protein type 2A (LAMP2A) that serves as CMA receptor (Cuervo and Dice, 1996). After substrate unfolding, organization of LAMP2A into a multimeric protein complex mediates substrate translocation into the lumen (Bandyopadhyay et al., 2008; Bandyopadhyay et al., 2010) (Fig. 1). A form of HSP90 at the luminal side of the lysosomal membrane stabilizes LAMP2A during this step. Substrate translocation requires a second form of HSC70 located in the lumen of only those lysosomes capable to perform CMA. After internalization of the substrate, the multimeric LAMP2A complex disassembles to start a new cycle of substrate binding and internalization (Bandyopadhyay et al., 2008). A pair of proteins, GFAP and EF1α modulate the stability of multimeric LAMP2A in a GTP-dependent manner (Bandyopadhyay et al., 2010). Although luminal HSC70 is absolutely required for CMA, LAMP2A lysosomal levels and dynamics are a direct determinant of the CMA flux. Both de novo synthesis and regulated degradation of LAMP2A in lipid microdomains at the lysosomal membrane contribute to modulate LAMP2A abundance in lysosomes (Kaushik et al., 2006).
CMA contributes to quality control of oxidized and damaged proteins and to provide amino acids through protein breakdown during conditions of nutrient deprivation (Kaushik and Cuervo, 2018). In addition, fully functional proteins are also targeted for degradation via CMA to terminate their function for regulatory purposes of processes such as lipid and glucose metabolism, cell cycle, transcription and translation, mitochondrial dynamics or stem cell activation, among others (reviewed in (Kaushik and Cuervo, 2018)). All cells display basal CMA activity but can upregulate this autophagic process in response to similar stressors to the ones described for macroautophagy such as starvation (Cuervo et al., 1995), oxidative stress (Kiffin et al., 2004; Valdor et al., 2014), genotoxic damage (Park et al., 2015) and hypoxia (Ferreira et al., 2013; Hubbi et al., 2014).
Identification of effectors and regulators of CMA has been relatively slow because of the inability of using invertebrate model systems since LAMP2A is not present in these species, and consequently they are not able to perform CMA (Kirchner et al., 2019). However, the introduction of mammalian siRNA and CRISPR technology has resulted in considerable progress in the understanding of CMA regulation. Large part of this regulation occurs at the lysosomal membrane through the regulated degradation of LAMP2A in this compartment (Kaushik et al., 2006) and through membrane-associated proteins that regulate the dynamics of assembly and disassembly of the CMA translocation complex (Bandyopadhyay et al., 2010). In addition to the GFAP-EF1α protein pair that controls stability of this complex, the mTORC2 complex at the lysosomal membrane has been shown to act as a constitutive inhibitor of CMA (Arias et al., 2015). mTORC2 inhibitory effect on CMA is counterbalanced, whenever CMA activation is required, through the recruitment of the phosphatase PHLPP1 to lysosomes (Arias et al., 2015). AKT1 is the shared substrate of mTORC2 and PHLPP1 and consequently, the status of AKT1 phosphorylation at the lysosomal membrane is the one ultimately regulating CMA (Arias et al., 2015). CMA is also regulated transcriptionally through different signaling pathways including the negative regulators such as nuclear receptor RARα (Anguiano et al., 2013) glucocorticoid-mediated signaling (Sato et al., 2020) or growth hormone signaling (Endicott et al., 2021) and positive regulators such as NFAT (Valdor et al., 2014), TPD52 (Cai et al., 2021), Class I PI3K (Endicott et al., 2020) among others. Signaling from another organelle, the mitochondria, has been shown to upregulate CMA in a process mediated by the mitochondria peptide humanin (Gong et al., 2018).
2.3. Microautophagy
Although originally described in mammalian liver as the direct engulfment of cytoplasmic material by invaginations of the lysosomal membrane (De Duve and Wattiaux, 1966), the molecular characterization of microautophagy was initiated in yeast. Yeast microautophagy was proven to discriminate cargo, giving rise to terms such as micropexophagy (for peroxisomes) (Sakai et al., 1998), micromitophagy (for mitochondria) (Lemasters, 2014), microlipophagy (for lipid droplets) (Seo et al., 2017) or even portions of the nucleus through piecemeal microautophagy (Roberts et al., 2003). Although most of the molecular components described in yeast did not preserve the same function in mammals, ten years ago, the first form of mammalian microautophagy was described to take place in late endosomes (Sahu et al., 2011) and it is now known as endosomal microautophagy (eMI) (Fig. 1). This process can mediate degradation of proteins “in bulk” or in a selective manner through the recognition of the KFERQ-like motif by cytosolic HSC70. In this case, the chaperone binds to phosphatidylserine and, contrary to CMA, eMI requires neither LAMP2A nor substrate unfolding (Sahu et al., 2011). Cargo is then internalized through membrane deformations and eventually scission in the form of intralumenal vesicles are executed by the ESCRT machinery. Substrate proteins, once incorporated into intraluminal vesicles, can be degraded within late endosomes or upon fusion of late endosomes with lysosomes.
The Drosophila counterpart of eMI (Mukherjee et al., 2016) and other types of mammalian eMI independent of HSC70 (Mejlvang et al., 2018) are activated during starvation, whereas HSC70-dependnet eMI is not upregulated by starvation but responds to proteotoxicity mediated by pathogenic proteins such as tau (Caballero et al., 2018). The regulation of microautophagy is mostly unknown but inhibition of TORC1 upregulates yeast microautophagy (Hatakeyama and De Virgilio, 2019), Drosophila eMI (Mukherjee et al., 2016) and some types of mammalian microautophagy (Sato et al., 2019). More studies are needed to discriminate molecular regulators common to all microautophagy processes from those that are process specific. For example, specific macroautophagy ATG proteins, are necessary for Drosophila eMI (Mukherjee et al., 2016) and yeast piecemeal microautophagy (Krick et al., 2008) but not for mammalian eMI (Sahu et al., 2011).
Yeast microautophagy contributes to turnover of different cellular organelles, quality control of intracellular membranes and of nuclear components (Lemasters, 2014; Roberts et al., 2003; Sakai et al., 1998). eMI in Drosophila regulates neurotransmission by degradation of specific synaptic proteins (Uytterhoeven et al., 2015) and contributes to nutritional regulation through selective degradation of the signaling protein Arouser (Jacomin et al., 2021). Our understanding of the physiological role of mammalian microautophagy is lagging behind due to the lack of specific molecular components selective for eMI that can be genetically modulated without affecting other cellular process in mammals.
2.4. Autophagic crosstalk
All the different types of autophagy described in previous sections coexist in most cells and can compensate for each other, suggesting a coordinated crosstalk among the autophagic processes as well as with other proteolytic systems such as the proteasome. For example, some cells respond to blockage of CMA with upregulation of macroautophagy (Massey et al., 2006) or the proteasome (Schneider et al., 2015), blockage of macroautophagy leads to CMA upregulation (Kaushik et al., 2008) and inhibition of the proteasome to activation of macroautophagy and CMA (Koga et al., 2011). However, this compensatory upregulation may be cell- and tissue-dependent. Thus, although compensatory upregulation of macroautophagy has been reported in mouse models with selective CMA blockage in liver or T cells (Schneider et al., 2015; Valdor et al., 2014), studies in mouse retina, neurons and hematopoietic stem cells show that blockage of CMA in these cells does not evoke a compensatory activation of macroautophagy (Bourdenx et al., 2021; Dong et al., 2021; Rodriguez-Muela et al., 2013). Further studies are needed to understand the peculiarities and molecular mechanisms behind the autophagic crosstalk.
2.5. Autophagy and sexual dimorphism
Despite the extensive progress in our understanding of the molecular basis of the autophagic process, investigations on sex differences on autophagy are lagging behind, and more so in the context of aging. Consequently, except for some interventional studies where sex differences on the modulation of autophagy in aging have been described (see later sections), most of the information on the interplay of autophagy with aging and other hallmarks of aging summarized in this review does not address possible sexual dimorphism on the described effects.
There is however, growing evidence in support of sexual differences in induction, regulation and execution of autophagy (Shang et al., 2021). Sex-biased regulation of autophagy has been attributed, in part, to the effect of steroid hormones and their receptors in this process. Sex hormones androgen and estrogen receptors have been shown to regulate 32 macroautophagy proteins, either at the transcriptional or post-translational level (Türei et al., 2015). Interestingly, the effect of signaling through these receptors on autophagy varies depending on the tissues, likely in relation to the relative abundance of each of them. Further predictors of sexual differences in autophagy is the fact that several genes on the X chromosome participate in autophagy, including genes involved in autophagosome biogenesis such as ATG4 or WDR45/WIPI4, in autophagosome trafficking and fusion such as genes encoding for different Rab proteins or in epigenetic regulation of autophagy, such as HDAC6 (Shang et al., 2021). Interestingly, some of these X-linked genes are integral lysosomal proteins (i.e., ATP6AP2 component of the lysosomal proton pump required for lysosomal acidification) and as such they could be responsible for sexual differences in other autophagic pathways, beyond macroautophagy, that also converge on this organelle. In this respect, the LAMP2 gene, for which one of its spliced variants is a limiting component of CMA, is also X-linked, although sex differences in CMA activity are still poorly characterized.
Increasing evidence also suggest that sex differences in autophagy could be implicated in several human diseases, such as neurodegenerative disorders, cancer, and cardiovascular diseases (Shang et al., 2021). It has been proposed that sex-based differences in autophagy regulation during the lifespan contribute to increased Alzheimer’s disease risk, and greater severity of pathology seen in women (Congdon, 2018). Sexual dimorphism in placental autophagy in response to maternal obesity has been shown to associate with offspring obesity (Muralimanoharan et al., 2016). As described in more detail later in this review, sex differences in intensity and type of autophagy induced have been reported for anti-aging interventions such as caloric restriction (Mitchell et al., 2016).
3. Changes with age in autophagy and relation to life- and health-span
Lysosomal dysfunction in aging has been extensively supported both in invertebrate models and in mammals. A decline in lysosomal proteolytic activity with age was first reported in the nematode Caenorhabditis elegans (Sarkis et al., 1988). This work was followed by studies both in invertebrates and in rodents demonstrating morphological and functional changes in lysosomes, including expansion of the lysosomal compartment, increases in levels of multiple lysosomal proteases (generically known as cathepsins) but reduced enzymatic activity, accumulation of undegraded material inside lysosomes in the form of lipofuscin, etc. (Fig. 2). Early morphological reports in mice liver described accumulation of autophagic vacuoles and defective autophagic response with age to nutritional hormones such as glucagon and insulin (Donati et al., 2001b). In fact, a protective effect of caloric restriction against this gradual functional loss in macroautophagy was demonstrated even before the genetic molecular players of this process were identified (Del Roso et al., 1991).
Figure. 2. Steps in different types of autophagy affected by aging.
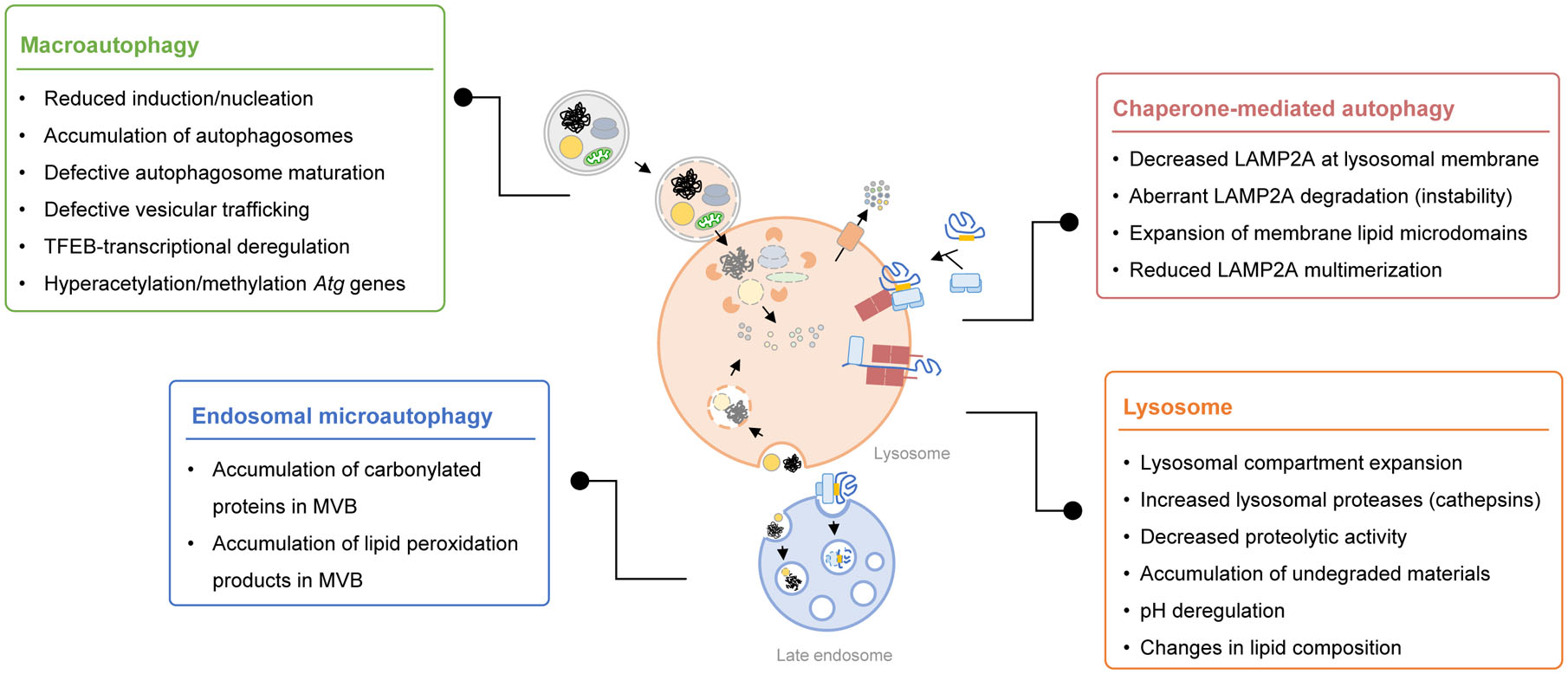
Aging-imposed changes have been described in different steps of each of the autophagic pathways and in lysosomes, the compartment where these pathways converge for degradation of cargo.
Despite the early reports describing changes in autophagy with age it was not until 2003, that a link between autophagy and longevity was experimentally demonstrated in studies in C. elegans (Melendez et al., 2003). This positive relation between lifespan and autophagy activity was later confirmed in D. melanogaster (Simonsen et al., 2008). Studies from invertebrates to mammals, including work in long-lived animals and centenarians supports now the idea that preservation of autophagic capacity is important for longevity, geroprotection and extended healthspan (Madeo et al., 2010; Perez et al., 2009; Raz et al., 2017; Xiao et al., 2018). In this section we review the reported changes in the regulation and activity of the different autophagic pathways with age and the basis for their impact on health- and lifespan.
3.1. Macroautophagy and aging
The molecular dissection of macroautophagy and modulation of autophagy genes strengthened the connections of this process with lifespan. Loss-of-function mutation in ATGs have been shown to decrease lifespan and macroautophagy has proven necessary for the lifespan extension effect of different paradigms such as dietary restriction (Hansen et al., 2008). The use of functional reporters to track autophagic activity has also allowed for a more detailed analysis of the spatiotemporal changes of autophagy with age in experimental models such as C. elegans and have demonstrated that although the decline with age in macroautophagy seems to affect multiple organs, the dependence of life extending interventions on autophagy is organ-specific (Chang et al., 2017). Tissue-specificity may be behind the recently proposed antagonistic pleotropic effect of autophagy, whereby deletion of macroautophagy initiation genes in neurons of post-reproductive worms improves longevity (Wilhelm et al., 2017). Whether this effect translates to mammals has not been investigated yet. Also pending is a better understanding of the contribution of different types of selective macroautophagy to the observed effect on lifespan. Next, we summarize evidence of the relation between macroautophagy and lifespan extension in different experimental models.
3.1.1. C. elegans, macroautophagy and longevity
Studies in C. elegans paved the way to efforts of modulating autophagy as a geroprotective intervention. The tight relation of macroautophagy with the nutritional status and the dependence on autophagy for survival during nutrient deficiency in worms, facilitated connecting changes in nutrient-sensing pathways with age with the autophagic failure in old worms. Upregulation of TOR signaling and insulin/IGF-1 (IIS) pathway has been extensively reported in aging and in fact, inhibition of TOR signaling is enough to promote longevity in an autophagy-dependent manner (Hansen et al., 2008; Kenyon, 2010). Similarly, inhibition of AMPK activity occurs in aging and interventions that activate this signaling mechanism extend C. elegans lifespan also through activation of autophagy (Wu et al., 2021). In addition, of these nutrient sensors, major transcriptional regulators such as the basic helix-loop-helix 30 (HLH-30), C. elegans homologue of the mammalian TFEB, and the stress-inducible transcription factor TP53 also extend lifespan and delay aging in an autophagy-dependent manner in worms (Lapierre et al., 2013; Tavernarakis et al., 2008). In fact, transcriptional upregulation of specific ATGs (atg18, lgg-1, spst-1, bec-1, atg7, and epg-8) with agents such as the calcium blocker Verapamil (Liu et al., 2020) or restoring expression of a single autophagy gene (i.e., atg-18 in atg-18/daf-2 double mutant) (Minnerly et al., 2017) is sufficient for lifespan extension. Studies also in C. elegans have linked the role of autophagy in lipid metabolism with its beneficial effect in lifespan extension (Lapierre et al., 2013; Lapierre et al., 2011).
3.1.2. D. melanogaster, macroautophagy and longevity
Reduced lifespan has been described in D. melanogaster with deficient ATGs expression in brain (Simonsen et al., 2008) and in muscle (Bai et al., 2013). Conversely, overexpression of Atg8a in the same organs extends flies lifespan (Bai et al., 2013; Simonsen et al., 2008). Interestingly, induction of autophagy in the nervous systems of adult flies, improves intestinal barrier function through upregulating autophagy (Ulgherait et al., 2014) suggesting that autophagy induction can result in non-cell autonomous extension of organismal lifespan. As in C. elegans, activation of AMPK or inhibition of TOR and downstream genes such as the transcription factor forkhead box (FOXO) have also been shown to extend lifespan in flies in an autophagy-dependent manner (Bjedov et al., 2010; Demontis and Perrimon, 2010; Lee et al., 2010). Recently, increased levels of Rubicon, a negative regulator of autophagosome-lysosome fusion, have been described with aging across species, and interventions that reduce Rubicon levels extend lifespan in worms and flies along with ameliorating age-associated phenotype in mice (Nakamura et al., 2019).
3.1.3. Mammalian studies on macroautophagy and longevity
Reduced macroautophagy was described more than 20 years ago in livers from aged mice and rats as an accumulation of immature autophagic vacuoles and reduced long-lived protein degradation in lysosomes, both indicative of decreased autophagic flux (Del Roso et al., 2003; Donati et al., 2001a; Terman, 1995) (Fig. 2). Conversely, long-lived mammals such as the naked mole rat (Perez et al., 2009) and centenarian individuals and their progeny (Raz et al., 2017) display preserved autophagic function until very late in life.
The early morphological observations supported age-related problems in autophagosome maturation that have been recently linked to defective intracellular trafficking. In fact, faulty association of molecular motor proteins to autophagosomes and lysosomes prevents their mobilization for the encounter required for fusion (Bejarano et al., 2018). The recent confirmation of the importance of this directional trafficking for autophagosome maturation and the identification of adaptor proteins such as FYCO1 in this intracellular mobilization provides now possible molecular targets to correct the autophagic defect in aging (Nieto-Torres et al., 2021b). However, failure of mammalian macroautophagy occurs at multiple levels. Thus, reduced induction of macroautophagy in response to stressors has been linked to tissue-dependent decreased levels of different autophagy effectors (i.e., Beclin-1, Atg5 and Atg7 in human brain (Lipinski et al., 2010; Shibata et al., 2006); LC3 and Atg7 in mouse hypothalamus (Kaushik et al., 2012)) and to increased levels of negative regulators such as Rubicon in mouse liver and kidney (Nakamura et al., 2019). The determinants of these changes in ATGs levels with age are not fully elucidated, but recent studies support changes with age in TFEB expression, hypusination-mediated activation, and nuclear localization as possible contributors (Madeo et al., 2018; Schneider et al., 2015; Zhang et al., 2019). However, as a note of caution, changes in steady state levels of ATGs do not necessarily correlate with autophagy activity, and in fact in the context of the overall proteostasis loss of aging, intracellular aggregation of some ATGs has been reported (Menzies et al., 2017). Changes in circulating levels of some ATGs, such as reduced Atg5 in the serum of Alzheimer’s disease (AD) patients (Castellazzi et al., 2019), elevated levels of Beclin-1 in the serum (Emanuele et al., 2014) and several ATGs in white blood cells (Xiao et al., 2018) of centenarians have been reported, but the biological relevance of these changes in extracellular ATGs remains unknown.
Mouse models with tissue-specific or inducible systemic deletion of essential Atgs phenocopy aspects of aging such as neurodegeneration and aggregation of intracellular inclusion bodies, lysosomal accumulation of undegraded material in the form of lipofuscin, damaged and dysfunctional mitochondria, accumulation of lipid droplets and overall organ function loss (Hara et al., 2006; Karsli-Uzunbas et al., 2014; Komatsu, 2005; Komatsu et al., 2006; Sato et al., 2018; Singh et al., 2009), as described in more detail in the following sections. Studies directly upregulating autophagy genes in mice as an anti-aging intervention are still limited, but constitutive overexpression of Atg5 (Pyo et al., 2013), and systemic activation of macroautophagy via disruption of the Beclin-1-BCL2 complex (Fernandez et al., 2018) have been shown to extend lifespan and promote healthspan in mice. Whether similar interventions late in life still have beneficial effects needs to be elucidated. However, advances have been made on the use of macroautophagy enhancers in old mice to preserve specific organs’ function (i.e., overexpression of the mitophagy effector Parkin to rescue cardiac aging (Gao et al., 2021) or peptide-mediated activation of Beclin-1-dependent autophagy to improve memory performance (Glatigny et al., 2019)).
3.2. CMA and aging
Decrease in CMA activity during aging has been found in rodents and humans in almost all cell types and tissues (Cuervo and Dice, 2000; Dong et al., 2021; Rodriguez-Muela et al., 2013; Schneider et al., 2015; Valdor et al., 2014; Zhang and Cuervo, 2008), whereas constitutive upregulation of CMA has been recently reported in long-lived mice with reduced growth hormone signaling (Endicott et al., 2021). A decrease in LAMP2A levels is, to date, the main change responsible for the observed decrease in CMA with age. Although in some instances this decrease is due to transcriptional downregulation of the lamp2 gene (Valdor et al., 2014), often, changes in the stability of this receptor at the lysosomal membrane in aging organisms is responsible for reduced CMA. In fact, comparative lipidomic analysis have identified changes with age in the lysosome membrane lipid composition that favor sequestration of LAMP2A in expanded lipid microdomains and promote its degradation (Kiffin et al., 2007; Rodriguez-Navarro et al., 2012) (Fig. 2). This decrease in LAMP2A levels with age seems limiting as genetic restoration of LAMP2A in old mouse liver was efficient in reducing proteotoxicity and preserving hepatic function (Zhang and Cuervo, 2008). Similar intervention in hematopoietic stem cells from old mice has proven efficient in restoring their functionality (Dong et al., 2021).
Both, the capacity of CMA to contribute to quality control and to selectively regulate activity of specific cellular pathways seem to be responsible for the beneficial effects of its restoration in old organisms. Reduced levels of oxidized and aggregated proteins were found in hepatocytes, neurons and in hematopoietic stem cells in mice with genetic or pharmacological enhancement of CMA (Bourdenx et al., 2021; Dong et al., 2021; Zhang and Cuervo, 2008). In addition, part of the beneficial effect was shown due to restoration of other specific CMA-regulated processes such as the linolenic and linoleic acid metabolic pathways, required for switch of hematopoietic cells from the quiescent to the activated state (Dong et al., 2021), or processes such as glycolysis, endocytosis and actin remodeling required for neuronal function (Bourdenx et al., 2021).
The fact that blockage of macroautophagy and CMA often have similar phenotypic consequences (i.e., neuronal degeneration) supports that these pathways are not redundant and that the subset of proteins and cellular components that they degrade is specific for each of them. In fact, comparative proteomic analysis upon blockage of neuronal macroautophagy or CMA has revealed that CMA is responsible from preventing the collapse of the metastable neuronal proteome whereas macroautophagy supports turnover of large protein complexes, structural proteins, membrane receptors and organelles (Bourdenx et al., 2021). Even for organs such as the liver, where in vivo blockage of CMA elicits upregulation of macroautophagy and of the proteasome (Schneider et al., 2014), the presence of other aggravating factors like oxidative stress, lipid challenge or aging, reduces this upregulation of macroautophagy and leads to proteotoxicity (Schneider et al., 2015).
3.3. Microautophagy and aging
The changes and contribution of the different types of microautophagy to the defense against aging remain, for the most part, unknown. Only in the case of mammalian eMI, malfunctioning with age has been proposed in light of the detected accumulation of carbonylated protein and lipid peroxidation products in multivesicular bodies (MVB), which compromises degradation through this type of autophagy (Cannizzo et al., 2012) (Fig. 2). Recent studies support that the blockage of degradation in late endosomes/MVB may favor docking of this compartment with plasma membrane and extracellular release of the undegraded materials, including proteins such as pathogenic forms of tau that fail to be degraded by CMA upon failure of this pathway and are re-routed to eMI instead (Caballero et al., 2021).
4. Impact of autophagy on the hallmarks of aging
The diversity of functions of the different types of autophagy and the growing number of reports supporting failure of most of the autophagic processes with age has provided momentum to revise the basis for the anti-aging effect of autophagy and the contribution of this process to each of the hallmarks of aging (Fig. 3). Next, we review information in support of tight connections of autophagy with several of these hallmarks.
Figure 3. Impact of autophagy on hallmarks of aging.
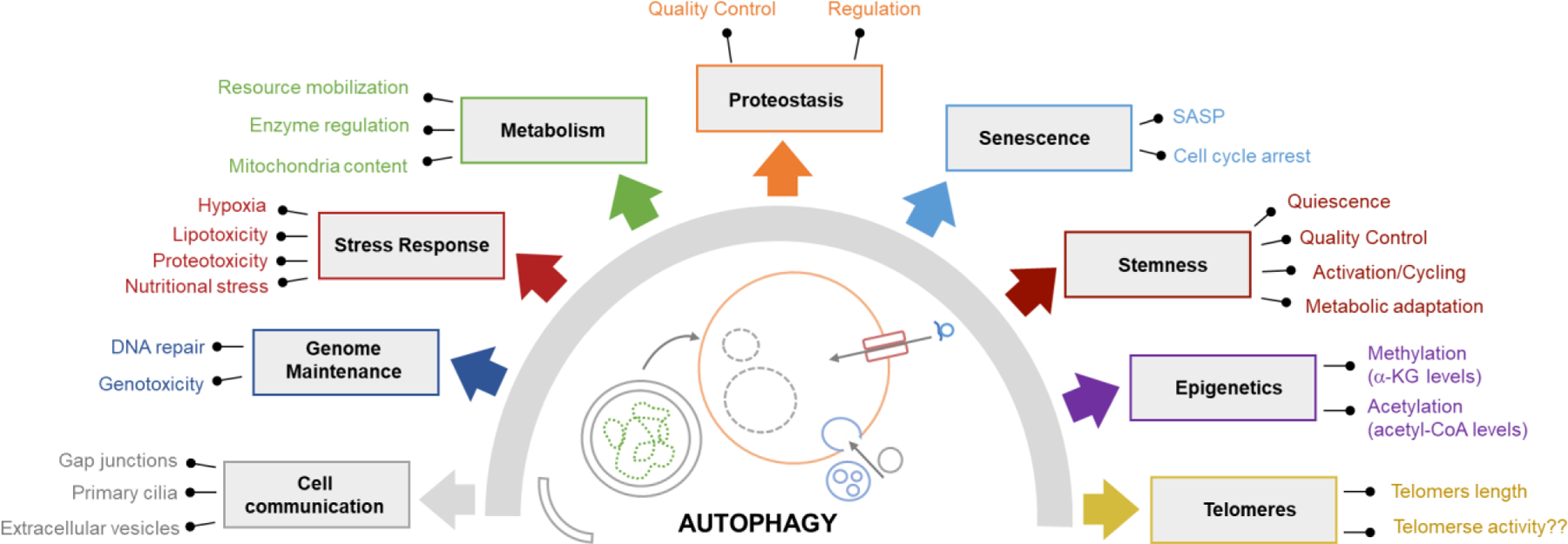
Growing evidence supports bidirectional interactions of the different autophagic pathway with multiple drivers of aging. Here, we illustrate hallmarks of aging affected by changes in autophagic activity and indicate the mechanisms whereby autophagy modulates these processes.
4.1. Autophagy in proteostasis
4.1.1. Cytosolic proteostasis
The contribution of autophagy to maintenance of cellular proteostasis is well-documented. Autophagy is considered an essential component of the proteostasis network that senses and responds to protein misfolding and proteotoxic stress to restore proteome integrity. The proteostasis capacity of many cells and tissues declines with age (Kaushik and Cuervo, 2015; Labbadia and Morimoto, 2015) and loss of proteostasis is also the basis of proteinopathies and protein conformational diseases often aggravated with age (Hartl, 2017).
Chaperones bind unfolded proteins and decide their fate by assisting them with folding or targeting them through degradation by the proteolytic systems, including autophagy (Kim et al., 2013). Chaperone dysfunction in old organisms has been described as a result of undesired posttranslational modifications (Vanhooren et al., 2015), entrapment of cytosolic chaperones inside aggregates reducing their availability (Ben-Zvi et al., 2009) or reduction of ATP levels (required for chaperone function) due to poor cellular energetics with age (Ma and Li, 2015). Despite active contribution of chaperones in selective types of autophagy, changes in chaperone availability or function have not been linked yet to the functional decline of these types of autophagy with age.
The chaperone response most tightly linked to regulation of lifespan is the heat shock response that upon heat stress, rapidly activates the expression of heat shock proteins and other factors of the proteostasis network to maintain the integrity of the proteome. Perturbation of HSF-1 function, major transcription regulator of this response, occurs in aging (Anckar and Sistonen, 2011; Chafekar and Duennwald, 2012) and overexpression of HSF-1 has been shown to extend lifespan in worms in an autophagy-dependent manner (Kumsta et al., 2017).
As described in the previous section, a shared consequence of failure of macroautophagy (Hara et al., 2006; Karsli-Uzunbas et al., 2014; Komatsu, 2005; Komatsu et al., 2006; Sato et al., 2018; Singh et al., 2009) and of CMA (Bourdenx et al., 2021; Dong et al., 2021; Schneider et al., 2014) with age is proteotoxicity due to accumulation of oxidized and misfolded proteins and of aggregates (Fig. 3). Aggregates accumulate in macroautophagy-deficient cells because of failure to directly degrade aggregates through aggrephagy (Lamark and Johansen, 2012). In contrast, CMA is unable to degrade proteins once in aggregation but failure to eliminate the soluble forms of prone-to aggregate proteins, increases their intracellular levels and tilts the balance towards their aggregation (Bourdenx et al., 2021). Loss of proteostasis due to autophagy malfunctioning is particularly detrimental in neurons where experimental models carrying mutations or deletions in macroautophagy-associated genes (Hara et al., 2006; Karsli-Uzunbas et al., 2014; Komatsu et al., 2006; Sato et al., 2018) or in CMA genes (Bourdenx et al., 2021), are sufficient to cause neurodegeneration. Improved proteostasis is one of the common outcomes of the pharmacological and genetic interventions to upregulate macroautophagy or CMA in old organisms described in detail in section 5.
4.1.2. Organelle proteostasis
Autophagic pathways are often upregulated in response to organelle-specific proteotoxicity and contribute to maintenance of proteostasis inside organelles such as ER and mitochondria.
In the case of the ER, accumulation of unfolded nascent proteins triggers the unfolded protein response (UPR) to attenuate protein translation and influx into the ER, enhance ER chaperone expression, modify amino acid metabolism, activate the antioxidant response and autophagy, and ultimately, if unsuccessful, to trigger cellular apoptosis (reviewed in (Hetz et al., 2020)). Unfolded proteins in the ER can be retrotranslocated to the cytosol for degradation by the proteasome (Wu and Rapoport, 2018). Autophagy is emerging as an alternative to the proteasome for clearance of misfolded proteins from the ER through what is known as ER-to-lysosome–associated degradation (ERLAD) (De Leonibus et al., 2019), which includes degradation of ER portions by lysosomes through ER-phagy (Chino and Mizushima, 2020), direct fusion between the lysosomal membrane and ER to degrade ER fragments and luminal cargoes by ERES-microautophagy (Omari et al., 2018), and fusion of single membrane ER-derived vesicles with endolysosomes (Fregno et al., 2018).
Chronic ER stress occurs in aging and is a common contributor of the degeneration observed in different human tissues (Hart et al., 2019; Prell et al., 2019; Tang and Yang, 2015). The ability to activate the UPR decreases with age due to a combination of reduced expression of ER chaperones, elevated basal expression of ER stress and pro-apoptotic mediators such as CHOP, but also to defective macroautophagy (Ghosh et al., 2016). Fully functional UPR is required for lifespan extension interventions and conversely genetic, dietary or pharmacological interventions that improve the capacity to maintain ER proteostasis extend life- and healthspan (Taylor and Hetz, 2020). Although it is anticipated that ER-phagy and the other autophagic mechanisms that contribute to ER proteostasis should also be suitable targets to preserve ER function in aging, the lack of interventions capable to selectively activate these processes have prevented testing this hypothesis.
Mitochondrial dysfunction is central to the pathophysiology of many age-associated diseases and aging (reviewed in (Fakouri et al., 2019)). Here we comment on the mechanisms that maintain proteostasis of the mitochondrial proteome. Since most mitochondrial proteins are nuclear encoded imbalance between the nuclear and mitochondrial genome, excess of protein influx in mitochondria or excessive luminal damage of proteins can all lead to mitochondrial proteotoxicity and induction of the mitochondrial UPR (mtUPR). This mitochondria-to-nucleus communication system activates the expression of mitochondrial chaperones and attenuates mitochondrial protein translation (Zhao, 2002). Macroautophagy contributes indirectly to the regulation of the mtUPR through regulation of SIRT proteins (Xu et al., 2020a), which have been shown to play a role in the mtUPR (Weng et al., 2020).
In addition to the mtUPR, quality control of mitochondrial proteins can be attained through degradation of full or partial portions of mitochondria by mitophagy (reviewed in (Onishi et al., 2021)), sequestration of damaged mitochondria in a parkin-dependent mechanism by early endosomes via the ESCRT machinery (Hammerling et al., 2017) or, through pink1/parkin-dependent budding of vesicles containing the altered proteins from the surface of mitochondria (mitochondria-derived vesicles (MDVs)) and their subsequent fusion with lysosomes for degradation (Roberts et al., 2016). In some instances, MDVs elude intracellular degradation and are secreted as exosomes (Matheoud et al., 2016). CMA has been recently shown to contribute to degradation of mitochondrial proteins in their transit from the cytosol to the mitochondria as a regulatory mechanism of mitochondrial enzymatic load (Schneider et al., 2014) or to regulate mitochondria morphology and dynamics (Nie et al., 2020; Wang et al., 2016a) (Fig. 3).
4.1.3. Non-cell autonomous proteostasis
Besides intracellular proteostasis networks, cells count on non-autonomous mechanisms to coordinate stress responses with neighboring cells and across tissues and organs. These inter-cell stress signaling pathways allow for global regulation of proteostasis at an organismal level and have been shown to impact lifespan (Morimoto, 2020). Both UPR and mtUPR can be transmitted across cell types and tissues to inform of organelle dysfunction and induce the stress response in cells that have not experienced the stress yet.
Autophagy also shows non-cell autonomous effects and regulation. Most examples of non-cell autonomous regulation of macroautophagy and CMA have been described in pathological conditions such as cancer, whereby tumor cells often regulate autophagy in the niche cells to evade the anti-oncogenic immune response (Gewirtz et al., 2018; Valdor et al., 2019). Also, in neurodegenerative conditions such as amyotrophic lateral sclerosis (Madill et al., 2017) and Parkinson’s disease (di Domenico et al., 2019), astrocyte-released factors promote neuronal death by inhibiting macroautophagy in neurons. However, autophagy has also been shown to physiologically contribute to inter-organ communication and influence aging in a non-cell autonomous manner. As mentioned in section 3.1.2. upregulation of AMPK in neurons in Drosophila induces autophagy also in the intestinal epithelium, improving intestinal homeostasis and extending lifespan (Ulgherait et al., 2014). Extracellular release of autophagic material either through autophagosome-mediated unconventional secretion (Kimura et al., 2017), docking of LE/MVB engaged in eMI (Caballero et al., 2021) or through a newly described LC3-dependent extracellular vesicle loading and secretion (Leidal et al., 2020) may all contribute to coordinate inter-organ autophagy.
Sex differences in both the cell-autonomous and non-cell autonomous cellular response to proteotoxicity have been described (Tower et al., 2020), but there is currently no information available on whether differences in autophagy contribute to this sexual dimorphism of the response to stress.
4.2. Autophagy in metabolism and mitochondrial function
Metabolic de-regulation is one of the common characteristics of aging organisms and in fact, many other hallmarks of aging are tightly related to energetic deficiencies (reviewed in (Berry and Kaeberlein, 2021)). The dual interplay between autophagy and metabolism has been known since the early discovery of this process as a mechanism for cell survival under low nutrient conditions that is directly modulated from nutrition-related hormones such as insulin and glucagon (Mortimore et al., 1988). Nowadays, it is well accepted that autophagy contributes to the maintenance and regulation of metabolism through at least three different mechanisms: direct mobilization of energy stores, selective removal of metabolic enzymes for fine-tune regulation of flux through these pathways, and regulation of number and properties of mitochondria in response to the cellular ATP requirements (Singh and Cuervo, 2011)(Fig. 3). We refer interested readers to comprehensive reviews covering the complex interplay between autophagy and metabolism (Lahiri et al., 2019; Morishita and Mizushima, 2019; Singh and Cuervo, 2011). Here, we focus on the role of autophagy in the energetics of aging and expand in more detail on the relation of this process with mitochondria, due to the important role of dysfunction of this organelle with aging.
4.2.1. Metabolism and autophagy
In times of glucose deprivation, liver and muscle glycogen stores are broken down either by cytosolic glycogenolysis via glycogen phosphorylase or through glycophagy (Jiang et al., 2011). Glycophagy generates the rapidly usable non-phosphorylated glucose, unlike glycogenolysis, and requires the action of STBD1 (starch binding domain 1), a glycophagy receptor that binds to glycogen and to the ATG GABARAPL1 (Jiang et al., 2011). Although the need for glycophagy in Drosophila skeletal muscle energetics has been determined (Zirin et al., 2013), most glycophagy studies in mammals have been so far correlative. For example, higher autophagy upregulation with starvation was observed in fast-twitch skeletal muscles, which contain more glycogen stores, as compared to slow-twitch muscles with lower glycogen stores (Mizushima et al., 2004). Only recently, glycophagy has been shown to be intricately linked with lipid droplet biogenesis during brown adipocyte differentiation in mice embryo (Mayeuf-Louchart et al., 2019). In addition to glycogen mobilization, macroautophagy also sustains blood glucose levels by degrading a key suppressor of gluconeogenesis, cryptochrome 1 (Toledo et al., 2018).
Another mechanism by which autophagy controls metabolism is by degrading lipid droplets through lipophagy (Singh et al., 2009), a process whereby lipid droplets are sequestered by autophagosomes and delivered to lysosomes for degradation by lysosomal acid lipases. The generated free fatty acids are utilized for mitochondrial beta-oxidation and energy production. Liver-specific loss of macroautophagy results in marked lipid accumulation in support of contribution of basal macroautophagy to lipid metabolism (Singh et al., 2009). Interestingly, lipophagy in liver and brown adipose tissue requires macroautophagy in hypothalamic neurons (Martinez-Lopez et al., 2016) – pointing to CNS-to-peripheral autophagy axes in regulation of lipid metabolism. Indeed, cold-induced lipophagy in brown adipose tissue is blocked in mice deficient in autophagy (Atg7-null) in proopiomelanocortin (POMC) neurons, which play key roles in energy expenditure. Since autophagy in POMC neurons decreases with age (Kaushik et al., 2012), disruption of this POMC-to-peripheral autophagy axis likely contributes to metabolic aging. Such integrated autophagy axes appear to be important for benefits of dietary interventions, since macroautophagy activation and lipid loss in response to the twice-a-day feeding intervention are blocked in mice lacking Atg7 in POMC neurons (Martinez-Lopez et al., 2017b). Lipophagy also participates in nutrient sensing orexigenic hypothalamic agouti gene-related peptide (AgRP) neurons, which are important for food intake. Indeed, lipophagy activation and lipid breakdown in AgRP neurons during fasting supports AgRP production and feeding (Kaushik et al., 2011).
CMA has also been demonstrated to regulate glucose and lipid metabolism. Numerous glycolytic enzymes are CMA substrates and the role of this autophagy pathway in regulating glucose metabolism has been demonstrated in liver, hematopoietic stem cells and neurons (Bourdenx et al., 2021; Cuervo et al., 1995; Dong et al., 2021; Schneider et al., 2014). CMA-incompetent mice display marked hepatosteatosis and dysfunctional livers, in part, due to its role in degrading key lipogenesis enzymes and lipid carriers (Schneider et al., 2014). Additionally, the removal of lipid droplet coat proteins (PLIN2/3) by CMA is a requirement prior to mobilization of lipid droplets by cytosolic lipase machinery or by lipophagy (Kaushik and Cuervo, 2015). Timely and specific removal of these enzymes by CMA is often achieved though posttranslational modifications. For example, AMPK-dependent phosphorylation of PLIN2 triggers it for CMA degradation (Kaushik and Cuervo, 2016). Furthermore, the recently discovered role of CMA in linolenic and linoleic acid metabolism in hematopoietic stem cells is based on timely removal of acetylated forms of FADS2, the limiting enzyme in those pathways (Dong et al., 2021).
4.2.2. Interplay between mitochondria and autophagy
Mitochondrial function and homeostasis have been tightly linked to longevity as accumulation of non-functional mitochondria with age contributes to the poor energetics and high levels of damaging ROS characteristic of aging tissues (Harman, 1956). Autophagy, and more specifically mitophagy, plays a key role in the maintenance not only of the mitochondrial proteome, as described in 4.1.2, but also in the regulation of mitochondrial dynamics and function through turnover of all mitochondrial components (Onishi et al., 2021).
Mitochondria can be degraded by mitophagy “in bulk” (along with other intracellular components), or they can be selectively targeted for lysosomal degradation through different, often co-existing, mechanisms (Onishi et al., 2021; Palikaras et al., 2018) (Fig. 3). Basal mitophagy is responsible for continuous mitochondrial turnover, but this process can be further induced in response to mitochondria depolarization, oxidative stress, hypoxia, and nitrogen starvation. Mitophagy is key for early embryonic development, stem cell homeostasis, cell differentiation, inflammation, and apoptosis among others and consequently, defects in mitophagy are associated with various pathological conditions such as neurodegeneration, cardiovascular diseases, metabolic disorders and cancer (Onishi et al., 2021).
Since the description of the first mitophagy events in mammals (Rodriguez-Enriquez et al., 2004), different types of mitophagy have been shown to assist cells to accommodate to specific cellular contexts and to respond to different stimuli. Common to all types of mitophagy is the use of mitophagy receptors that link the targeted mitochondria to the autophagic machinery for sequestration inside the forming autophagosome. Mitophagy receptors can be mitochondrial membrane proteins (i.e., Parkin/PINK, BNIP3/Nix) or cytosolic proteins (p62, NBR1, OPTN) and recognition of the target mitochondria can be dependent or not on ubiquitin (Onishi et al., 2021).
The age-dependent decline in mitophagy impacts cellular homeostasis, organismal health and lifespan across species. In C. elegans, reduced mitophagy with age leads to accumulation of damaged mitochondria and cellular function failure (D’Amico et al., 2019; Palikaras et al., 2015; Schiavi et al., 2015). Parkin-null flies and PINK1 mutant flies both show significantly reduced longevity (Pesah et al., 2004; Yang et al., 2006), while overexpression of Parkin in flies promotes lifespan extension (Rana et al., 2013). Generation of a mouse model with the fluorescent reporter mt-Keima to track mitophagy has confirmed that mitophagy also decreases with age in multiple mice tissues (brain, skeletal muscle, adult stem cells, among others) (Sun et al., 2015). These findings have generated a growing interest in interventions enhancing mitophagy (described in section 5.2.1).
4.3. Autophagy and cellular stemness
Adult stem cells (SCs) are unique cell types that exist throughout the organism’s lifespan and have the ability to self-renew and differentiate into specialized cells to replace lost cells and regenerate damaged tissues when needed (Urbán and Cheung, 2021). Under normal physiological conditions, adult SCs remain quiescent but in response to environmental stressors, damage and differentiation signals SCs can rapidly switch to an active state and generate new differentiated cells (Urbán and Cheung, 2021). The balance between stemness and differentiation is essential to maintain SCs homeostasis, and thus imbalance can lead to both, the exhaustion of the stem cell pool and/or to the formation of tumors. Recent multiple lines of evidence support the critical role of autophagy in preserving SCs quiescence, self-renewal, activation and differentiation of embryonic and adult SCs (review in (Boya et al., 2018)) (Fig. 3).
SC exhaustion limits the ability of old organisms to replace lost cells and regenerate properly the tissue after damage. This age-dependent decline of the SCs is, in part, driven by loss of autophagy activity with age. The role of autophagy has been studied in multiple types of SCs but here we will only focus on hematopoietic and muscle stem cells as they are the two SC adult populations where the consequences of changes in autophagy with age have been characterized in more detail.
4.3.1. Autophagic requirements of hematopoietic stem cells (HSC)
Adult HSC reside in quiescence in the bone marrow and can rapidly enter cell cycle for self-renewal or differentiation to ensure the production of all blood cells throughout life. Macroautophagy supports HSC quiescence by preserving their glycolytic function and allowing them to survive in a hypoxic environment (Ho et al., 2017). During this state, mitophagy also has an important role controlling oxidative metabolism and eliminating the active mitochondria, an essential step to inhibit differentiation and maintain stemness (Ho et al., 2017). In absence of macroautophagy, the increased mitochondrial content and subsequent shift in metabolic activity from glycolysis to OXPHOS promotes myeloid differentiation and loss of stemness (Ho et al., 2017). Macroautophagy is also essential in HSC differentiation into progenitor cells, acting as a cell remodeling mechanism, protecting cells from cell death and mediating metabolic reprogramming (Mortensen et al., 2011).
Aged HSC display high levels of mitochondria, accumulation of ROS and DNA damage, and declined proteostasis (Ho et al., 2017; Yahata et al., 2011). The age-dependent decline of macroautophagy activity has an important impact on HSC function and stemness (Warr et al., 2013). Interestingly, aged HSC exhibit heterogeneity in their macroautophagy activity, with about two-thirds of the HSC population showing impaired levels of macroautophagy and the other third showing macroautophagy activity comparable to that in young organisms (Ho et al., 2017). Genetic blockage of macroautophagy in mouse HSCs (conditional Atg12 knock-out) mimic features of premature HSC aging and lead to a phenotype which resembles the myeloid-bias characteristic in old mice. Intriguingly, this phenotype is transferable as transplantation of autophagy-deficient HSCs from aged mice resulted in significantly exacerbated age-related decline and myeloid-biased lineage distribution (Ho et al., 2017).
CMA has also been recently shown key for HSC function; in this case, through maintenance of protein quality control under basal conditions and for the regulation of the switch from glucose to fatty acid metabolism required for HSC activation (Dong et al., 2021). Selective removal of inactive FADS2, the limiting enzyme for linolenic and linoleic metabolism by CMA increases flux through these pathways to accommodate the energetic requirements of HSC activation (Dong et al., 2021). Genetic blockage of CMA in HSC (HSC-selective LAMP2A-KO) leads to pronounced accumulation of oxidized and aggregated proteins as CMA-deficient HSCs are not able to activate the compensatory macroautophagy upregulation observed in other cell types. CMA activity also declines in a fraction of the HSC population of old organisms, the one displaying higher levels of ROS, oxidized and aggregated proteins and reduced activating capability (Dong et al., 2021). Interestingly, old HSCs from both mice and humans display deficiencies in fatty acid metabolism comparable to those of CMA-deficient HSCs. Genetic manipulations that prevent loss of CMA with age in HSCs are effective in preserving their repopulating capability. Furthermore, pharmacological upregulation of CMA in already old HSCs is able to rejuvenate aged HSCs, highlighting CMA as a viable therapeutic targeting in HSC aging (Dong et al., 2021).
4.3.2. Autophagy in muscle stem cells (MSC)
MSC or satellites cells are essential for skeletal muscle formation and to repair and replace damaged muscle tissue. In adult muscle, satellite cells are in a quiescent state until they are activated in response to tissue damage to proliferate and generate new myoblasts. A small number of myoblasts returns to quiescence to maintain the satellite cell pool, while the rest differentiate into myocytes to repair the damaged muscle fibers (Cornelison and Perdiguero, 2017).
Basal macroautophagy is essential to maintain quiescent MSC organelle and protein homeostasis and avoid their transition to senescence (Garcia-Prat et al., 2016). Inhibition of macroautophagy in satellite cells leads to high levels of ROS (mostly due to deficient mitophagy) and triggers the epigenetics-mediated induction of the senescence-promoting gene p16INK4a (Sousa-Victor et al., 2014) that initiates senescence entry (Garcia-Prat et al., 2016). The decline of macroautophagy with age in long-lived quiescent MSC underlies their pronounced decrease in the number, regenerative and self-renewal capacities, which results in severely impaired skeletal muscle regeneration with aging (Garcia-Prat et al., 2016). Genetic or pharmacological reestablishment of macroautophagy in geriatric MSC allows these stem cells to recover their regenerative activity and to escape senescence (Garcia-Prat et al., 2016). Induced macroautophagy has been shown essential for supporting the high bioenergetic demands generated during MSC activation (Tang and Rando, 2014) and for the final stages of myocyte differentiation during fiber fusion (Fortini et al., 2016).
These new described roles of macroautophagy and CMA in stem cell maintenance and function and the encouraging results obtained with experimental drugs that modulate these pathways may set the basis for new therapeutic strategies to pharmacologically restore autophagy to enhance stem cell activity during aging.
Future studies are needed to determine how changes in autophagy contribute to the recently described sexual dimorphism in aging hematopoiesis (So et al., 2020) and in the ability of musculoskeletal stem cells for tissue repair (Knewtson et al., 2021).
4.4. Autophagy in intercellular communication
Loss of intercellular communication, essential for coordinating responses in multicellular organisms, is one of the hallmarks of aging (López-Otín et al., 2013). Intercellular communication occurs through areas of direct cellular contact (i.e., connexins and tight junctions) or through cell factors released to the extracellular space such as extracellular vesicles (EV) that dock on the plasma membrane of the recipient cell to activate intracellular signaling cascades. Autophagy has been shown to modulate both types of intercellular communication (Fig. 3).
4.4.1. Gap junctions
Connexins, essential components of the gap junctions, can undergo ubiquitin-dependent (Bejarano et al., 2012; Lichtenstein et al., 2011) and independent (Catarino et al., 2020) degradation through macroautophagy. This process is triggered under conditions of hypoxia such as ischemia-reperfusion (Chen et al., 2017; Lu et al., 2019; Martins-Marques et al., 2015). Although direct studies on the impact of the malfunctioning of autophagy with aging on gap junction function are still missing, genetic blockage of macroautophagy has been shown to result in accumulation of connexins at the plasma membrane and subsequent cellular leakiness (Bejarano et al., 2012). Conversely, connexins at the plasma membrane negatively modulate autophagosome formation from this compartment by direct sequestration of ATG proteins involved in the early steps of macroautophagy (Bejarano et al., 2014). Arrival of ATG14 to the plasma membrane releases this inhibitory effect and promotes macroautophagy activation (Bejarano et al., 2014). Connexin-dependent autophagy has been shown important for maintenance of mitochondria function in metabolic tissues such as brown adipose tissue (Kim et al., 2017), but the contribution of connexins to the described functional decline of macroautophagy with age has not been investigated yet.
4.4.2. Extracellular vesicles
A similar dual interplay has been described between autophagy and some forms of vesicle-mediated intercellular communication (Xing et al., 2021). The non-cell autonomous regulation of autophagy described in section 4.1.3. has been shown, in part, to be mediated by EV and their contents. Multiple small miRNAs present in exosomes – a type of EV that originate from exocytosis of MVB from LE – display autophagy regulatory properties (Xing et al., 2021). Although most miRNA-mediated regulation of autophagy across cells has been described in the context of cancer, growing number of studies support the relevance of this regulation in the pathobiology of the aging cardiovascular systems (Qi et al., 2020; Tian et al., 2019), ischemic brain injury (Chen et al., 2020), myelin regeneration (Yin et al., 2021), microglial activation (Van den Broek et al., 2020), neurodegeneration (Li et al., 2021a), osteoarthritis (Wang et al., 2021) and kidney disease (Sun et al., 2021). The potential therapeutic value of these extracellular miRNA with autophagy modulatory activity has been recently explored in the context of age-related macular degeneration (Hyttinen et al., 2021). Chemical inhibitors of specific miRNAs that function as autophagy repressors have also proven effective against premature senescence (Zhang et al., 2016). In other instances, rather than autophagy regulatory molecules, autophagic components themselves (i.e., LC3, ATG16L1) are released extracellularly in exosomes (Sirois et al., 2012; Wang et al., 2021) thus, indicating that autophagy components can be rerouted to neighboring cells under stress conditions. Although the functional relevance of ATGs release in exosomes remains unknown, recent studies show that EVs from AD patients show reduced abundance of mitophagy components and link this event to reduced intracellular macroautophagy and progression of disease (Gallart-Palau et al., 2020).
Autophagy also shows active involvement in vesicle-mediated intercellular communication, in part, through the regulation of biogenesis and release of EVs (Xing et al., 2021) and, in part, through direct release of autophagic content to the extracellular media as one of the types of unconventional protein secretion (Padmanabhan and Manjithaya, 2020). Multiple evidence supports a regulatory role for autophagy in MVB/LE fusion with the plasma membranes and exosome secretion (Xing et al., 2021). Although the molecular mechanisms behind this modulation are still poorly understood, the regulated degradation by autophagy of cytoskeleton remodeling proteins such as ROCK2 and RhoC (Hu et al., 2020) has been shown to participate in autophagy-regulated exocytosis. Reduced macroautophagy in the AD brain has been linked to qualitative changes in the content of EVs to include prion proteins and APP but fail to incorporate mitophagy and lysosomal membrane proteins (Gallart-Palau et al., 2020). Although the presence of secretory lysosomes that directly dock at the plasma membrane to release their enzymes to the extracellular media has been known for a long time (reviewed in (Buratta et al., 2020)), more recently, a similar docking and fusion of autophagosomes has been shown to contribute to unconventional secretion. In this case, ATGs engage in formation of double membrane vesicles that are not destined for degradation but rather for docking at the plasma membrane and release of their contents (Nieto-Torres et al., 2021a). Secreted proteins include those lacking secretory signals such as acyl-coA-binding protein, inflammatory cytokines, HMGB1, etc. (Cavalli and Cenci, 2020), but also pathogenic proteins thus contributing to the propagation of proteotoxicity in different age-related disorders (reviewed in (Gonzalez et al., 2020).
Blockage of CMA in neuronal cells has recently been shown to also increase extracellular release of pathogenic forms of tau rerouted to LE/MVB (Caballero et al., 2021). However, whether this is just an aberrant phenomenon occurring only under pathological conditions or it points towards a role of CMA in regulation of secretion requires further investigation.
4.4.3. Primary cilia
Primary cilia are organelles situated on the plasma membrane of most cell types, where they sense and transduce extracellular signals to the intracellular milieu. Signaling receptors at the primary cilium modulate autophagy function in response to extracellular signals, such as hedgehog ligand (Pampliega et al., 2013). Changes in the structure and length of primary cilia have been described in senescence (Breslin et al., 2014) and in the aging brain (Guadiana et al., 2016). How cilia alterations with age could impact autophagy remains unknown, but recent studies in C. elegans have shown defective intraflagellar transport in cilia of aged worms that leads to reduced AMPK signaling and would be predicted to reduce autophagy under these conditions (Zhang et al., 2021).
4.5. Autophagy and senescence
Senescence is a physiological response to stress wherein cells enter a state of growth arrest without undergoing cell death and acquire pro-secretory properties (Gorgoulis et al., 2019). Senescence is transiently activated throughout life (i.e., for wound closure, placenta involution, or in response of damage in organs such a liver and lung) but, with age, the transient nature of senescent cells is lost with the subsequent increase in the number of senescent cells (Gorgoulis et al., 2019). Genetic or pharmacological removal of senescent cells in vivo has shown effective in delaying aging and reducing severity of age-related diseases in mice. Recent studies have highlighted a possible interplay between autophagy and senescence (Fig. 3). Stimuli that activate autophagy can also trigger senescence, and both chronic senescence and poor autophagy associate with accumulating macromolecular damage; however, the interplay between senescence and macroautophagy has proven complex (Young et al., 2021).
Early work reported that activation of autophagy precedes oncogene-induced senescence in fibroblasts (Young et al., 2009) and that macroautophagy blockage delayed but did not prevent senescence (Young et al., 2009). However, later studies showed contradicting effects of blocking macroautophagy on senescence and differences in the status of macroautophagy in senescent cells. Discrepancy may originate from the cell type and senescence stimulus utilized. In general, blockage of macroautophagy in primary untransformed cells (neurons, satellite cells, melanocytes, fibroblasts) increases senescence (Garcia-Prat et al., 2016; Moreno-Blas et al., 2019; Ni et al., 2016; Tai et al., 2017) whereas blockage of macroautophagy in transformed cancer cells prevented oncogene-induced senescence (Leidal et al., 2012; Liu et al., 2014). Conversely, chemical upregulation of macroautophagy reduces senescence in tendon stem cells (Nie et al., 2021). Macroautophagy flux decreases in oxidative stress-induced senescence in fibroblasts (Tai et al., 2017) and in long term cultured neurons (Moreno-Blas et al., 2019), whereas DNA damage-induced senescence causes increased macroautophagy flux (Singh et al., 2012). The type of autophagy engaged may also be important, since anti- and pro-senescence effects have been described for selective and in-bulk macroautophagy, respectively (Kang et al., 2015).
Although macroautophagy is not essential for cells to enter senescence, it is required for resolution of oncogene-induced senescence (Young et al., 2009), but whether this is a universal requirement remains unknown. Often times, autophagy-deficient primary cells also display detrimental effects via persistent SASP factors and associated inflammation (Ni et al., 2016) further exacerbating the aging-associated tissue damage. Recently, enhancing autophagy in old cardiomyocytes improved senescence (Li et al., 2021b). The contribution of other types of autophagy such as CMA or eMI to senescence has not been explored.
4.6. Autophagy and other hallmarks of aging
We briefly summarize in this section evidence of the interplay of autophagy with some additional cellular mechanisms considered to drive aging, but for which direct testing that changes in autophagy with age affect these processes is still limited.
4.6.1. Defective response to stress and genomic stability
Besides protection against proteotoxic, metabolic stress and to organelle stress, reviewed in previous sections, autophagy is also activated in response to environmental stressors such as those imposing genotoxic stress. Upregulation of macroautophagy, CMA and eMI have all been described in response to cellular DNA damage (Juretschke and Beli, 2021; Park et al., 2015; Zhu et al., 2019) (Fig. 3). Although the physiological relevance of eMI upregulation under these conditions remains unknown, in the case of CMA, upregulation of this pathway is required for exiting cellular arrest after DNA repair through selective degradation of Chk1 (Park et al., 2015). In fact, accumulation of DNA damage is observed in CMA-deficient cells and mice (Schneider et al., 2014) whereas preservation of CMA activity in old mice reduces levels of DNA damage (Zhang and Cuervo, 2008). The consequences of macroautophagy activation in response to DNA damage have revealed an important role for this type of autophagy in genome maintenance (Ambrosio and Majello, 2020). Macroautophagy modulates the DNA damage response at different levels, including, through direct degradation of DNA-associated proteins (Chen et al., 2015) and DNA resection proteins (Robert et al., 2011), through degradation of p62 that when in excess can disrupt DNA repair (Wang et al., 2016b) and through control of cell cycle, among others. Upregulation of macroautophagy during DNA damage is mediated through molecules such as ATM (that activates AMPK and the TORC complex inhibitor TSC2 and activates p53), PARP1 (that reduces NAD+ and ATP with the subsequent activation of AMPK) or MAPK8 and NFkB (that induce direct expression of some autophagy genes) (Chen et al., 2015).
4.6.2. Autophagy and telomere attrition
By maintaining chromosomal integrity, telomeres play a key role in the lifespan of a cell. Telomere shortening rate is a good predictor of organismal lifespan and defects in telomere maintenance associate with accelerated aging (López-Otín et al., 2013). Furthermore, decreased telomerase activity is one of the characteristics of cellular and tissue aging (Sahin et al., 2011).
There is intricate interplay between autophagy and telomere maintenance (Fig. 3). Telomere loss activates autophagy in cells and is essential for death of epithelial cells and fibroblasts during replicative crisis, the final checkpoint before oncogenic transformation (Nassour et al., 2019). Telomerase activity is also positively correlated with autophagy (Green et al., 2019). A telomerase subunit, human telomerase reverse transcriptase (hTERT), positively regulates PINK1 function, resulting in increased mitophagy (Shin and Chung, 2020) while downregulation of hTERT impaired autophagy flux (Ali et al., 2016; Ding et al., 2021). TERT also modulates autophagy activity via inhibiting mTORC1 (Ali et al., 2016).
Relatively few studies have analyzed the role of autophagy in telomere maintenance. Agents that activate autophagy preserve telomere length. For example, even when applied late-in-life, spermidine treatment, was able to rescue age-related telomere shortening in mice cardiac tissue among other age-associated phenotypes (Wirth et al., 2021). However, it is not clear if this effect occurs through direct activation of telomerase by autophagy, because recent studies describe reduction in telomerase activity in cancer cells upon Beclin 1 overexpression to upregulate autophagy (Taji et al., 2021). Whether the effect of autophagy on telomerase is different in transformed and non-transformed cells require future investigation. No direct link between CMA or eMI and telomere attrition have been discovered yet.
4.6.3. Autophagy and epigenetics
Epigenetics changes comprised mainly of histone modifications and DNA methylation have been described to be intrinsic to the aging process. Epigenetic modulation of autophagy has been extensively demonstrated (reviewed in (Talebian et al., 2020)). Besides miRNAs (described in section 4.4.2), macroautophagy can be regulated by histone modifications (acetylation and methylation) by enzymes such as CARM1 H3R17 methyltransferase (Shin et al., 2016), G9a H3K9 methyltransferase (Artal-Martinez de Narvajas et al., 2013), EZH2 H3K27 methyltransferase (Wei et al., 2015), SIRT1 H4K16 deacetylase, and KAT8 H4K16 acetyltransferase (Fullgrabe et al., 2013). Among these, SIRT1, whose activity has been tightly connected with aging, shows a complex interplay with autophagy whereby nuclear SIRT1 limits expression of autophagy genes, whereas cytosolic SIRT1 activates ATGs through their deacetylation (Morselli et al., 2010). DNA demethylation (carried out by TET enzymes) can also regulate macroautophagy. For example, CpG demethylation by TET1 regulates Atg expression in glioma cells (Fu et al., 2018) and in vascular endothelial cells (Peng et al., 2016). Furthermore, hypermethylation of autophagy genes in aging cells has also been described (Khalil et al., 2016). So far, epigenetic regulation of CMA and eMI have not been described.
Although it is inferred that regulation of the acetyl-coA pool by autophagy (Berry and Kaeberlein, 2021) will have an impact on acetylation and consequently modulate epigenetic modifications, studies on this reciprocal regulation in the context of aging are still sparse (Fig. 3). Of note, the ability of CMA to regulate levels of α-ketoglutarate through degradation of the enzyme isocitrate dehydrogenase, has been shown essential to sustain the epigenetic traits of pluripotent stem cells (Xu et al., 2020c).
5. Autophagy at the center of interventions for successful aging
5.1. Autophagy as common mechanism of geroprotecting interventions
Many of the longevity pathways converge on the induction of autophagy and in many instances this autophagy induction is required for their beneficial effects in lifespan (Madeo et al., 2010; Madeo et al., 2015). Autophagy upregulation is also a common feature of lifestyle interventions that slow down aging (i.e., dietary restriction, exercise) reviewed extensively in (Escobar et al., 2019) and of drugs and natural products (generically known as geroprotectors) that improve lifespan. Here, we briefly review current knowledge of the functional interplay between autophagy and dietary interventions shown to increase lifespan and then focus in more detail on the effect of geroprotectors on autophagy.
5.1.1. Dietary restriction interventions and autophagy
The tight connection between autophagy and nutrient availability was the trigger for early studies exploring changes in autophagy during the different dietary restriction (DR) paradigms shown to extend lifespan in rodents (Donati et al., 2001a). These first studies on the interaction of age, diet and macroautophagy revealed that either 40% caloric restriction (CR) or every-other-day ad libitum feeding improved macroautophagy activation in response to hormonal activators such as glucagon in old rat livers (Donati et al., 2001a). Genetic confirmation that functional macroautophagy was necessary for lifespan extension by DR in worms (Hansen et al., 2008) awakened interest on the molecular mechanisms behind this effect. Studies in a variety of organisms and tissues have demonstrated upregulated expression of many autophagy-related proteins upon nutrient restriction and tight connection of these changes with mTOR and AMPK signaling, as described in detail in previous sections. Recently, the lifespan extension mediated by the fibroblast growth factor-21, has been linked to the ability of this starvation hormone to activate macroautophagy epigenetically through the histone demethylase Jumonji-D3 (Byun et al., 2020).
Interestingly, the beneficial effect of CR on lifespan extension, although universal, requires fine-tuning of the percentage of calorie intake depending on sex and genetic background, even in the same species (Liao et al., 2010). Along with this observation, although an improvement on autophagy is always noticeable in CR rodent livers, the degree to which macroautophagy is upregulated varies with the degree of CR in a sex- and genetic background-dependent manner (Mitchell et al., 2016). Differences in the type of autophagy upregulated by CR were also noticeable, with lipophagy upregulation being more pronounced, despite similar overall autophagic flux increase, in one of the male groups out of the two mice strains compared (Mitchell et al., 2016). This work was also the first one demonstrating upregulation of CMA by CR in old mice livers, which was attained mostly by recruitment of additional lysosomes (those usually not engaged in CMA) to this pathway (Mitchell et al., 2016).
Autophagy has also been implicated in several of the health benefits of intermittent fasting, a dietary intervention of higher human translational feasibility. Especially attractive on this respect are the anti-aging effects (glucose regulation, increased resistance to stress, reduced oxidative damage and downregulation of inflammation) of the twice-a-day feeding protocols that combine daily cycles of ad libitum eating and fasting (de Cabo and Mattson, 2019). Growing number of studies have demonstrated part of these beneficial effects being dependent on autophagy, including the observed correction of the immune response dysfunction to the SARS-CoV-2 infection (Elsayed et al., 2021; Hannan et al., 2020; Jamshed et al., 2019; Martinez-Lopez et al., 2017a).
Future studies are still needed to determine the specific contribution of autophagy to the already described beneficial effect of dietary restriction in other hallmarks of aging such as nutrient sensing, senescence, stem cell exhaustion or intercellular communication (Erbaba et al., 2021)
5.1.2. Geroprotector compounds and autophagy
Geroprotectors are drugs and natural products already approved for clinical use in other medical conditions that when used at lower doses bring benefits to health and lifespan (Partridge et al., 2020) (Fig. 4). Although many of them are likely to exert direct or indirect effect on autophagy, here we summarize those for which a relation with autophagy has been studied in more detail (Table 1).
Figure 4. Modulation of autophagy for geroprotection.
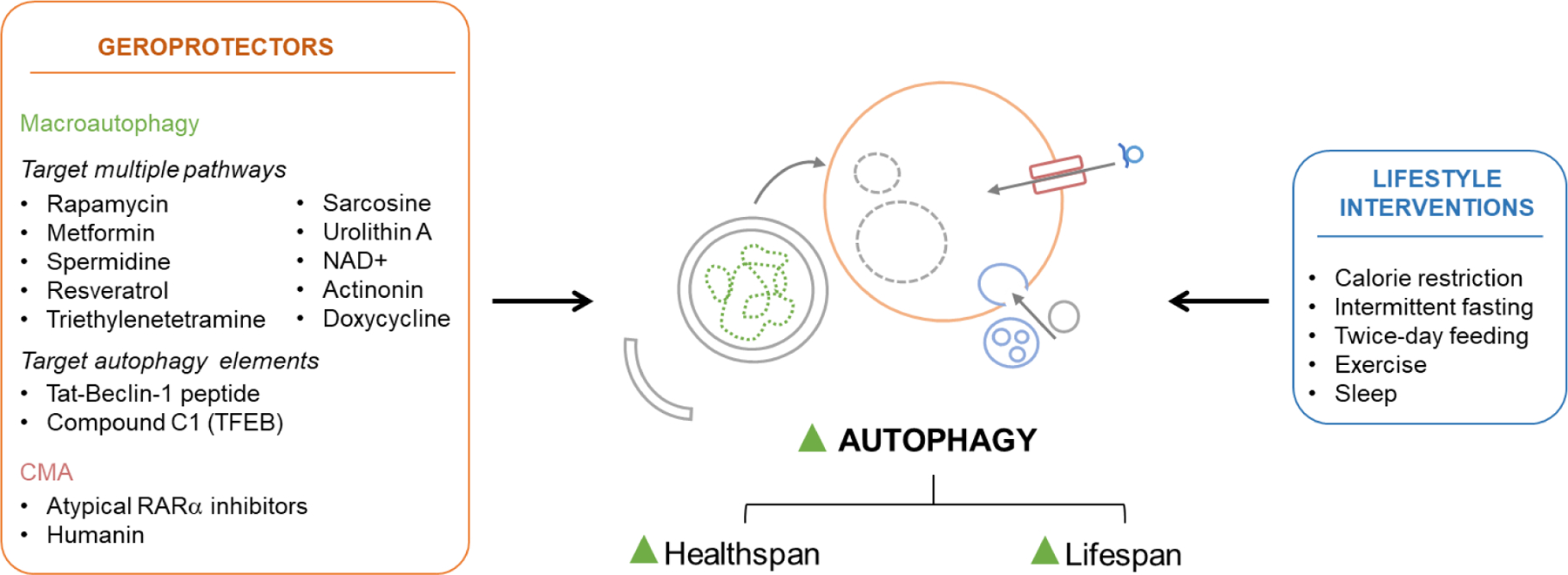
Lifestyle interventions (right) and drugs and natural products under the category of geroprotectors (left) proven to upregulate autophagy and benefit healthspan and lifespan. Geroprotectors with activating effects on macroautophagy are divided in two groups, those that have an effect in multiple cellular processes including autophagy and those designed specifically to target components of autophagy.
Table 1.
Geroprotectors proven to upregulate autophagy and benefit healthspan and lifespan.
| Activators | Mechanism of action | References |
|---|---|---|
| Macroautophagy | ||
| Target multiple cellular pathways (indirect regulation of macroautophagy) | ||
| Rapamycin | inhibition of mTORC1 | (Bjedov et al., 2010; Schinaman et al., 2019) |
| Metformin | activation of AMPK, SIRT1; inhibition of mTORC1; CRM | (Kulkarni et al.,2020; Piskovatska et al., 2019) |
| Spermidine | through multiple mechanisms including inhibition of acetyltransferases, activation of mTORC1, induction of autophagy genes; CRM | (Eisenberg et al., 2016; Eisenberg et al., 2009; Yue et al., 2017) |
| Resveratrol | through different signaling pathways (AMPK, SIRT1, PGC-1α, and inhibition of mTOR); CRM | (Morselli et al., 2010) |
| Triethylenetetramine | protein deacetylation | (Pietrocola et al., 2020) |
| Sarcosine | activation of AMPK | (Walters et al., 2018) |
| Urolithin A | activation of PINK1-dependent and - independent mitophagy | (D’Amico et al., 2021) |
| NAD+ | activation of mitophagy via Nix and ULK1 | (Fang et al., 2019a) |
| Actinonin | activation of mitophagy via mitochondrial ribosomal and RNA decay pathway | (Fang et al., 2019b) |
| Doxycycline | activation of mitophagy | (Xing et al., 2017) |
| Target autophagy elements (direct regulation of macroautophagy) | ||
| Tat-Beclin-1 peptide | Beclin-1 activator | (Shoji-Kawata et al., 2013) |
| Compound C1 | activation of TFEB | (Partridge et al., 2020) |
| CMA | ||
| Atypical RARα inhibitors | inhibition of a subprogram of the RARα signaling | (Bourdenx et al., 2021; Dong et al., 2021) |
| Humanin | Stabilizes substrate/receptor binding during translocation into lysosomes | (Gong et al., 2019) |
Spermidine is a natural polyamine that controls protein acetylation and strongly induces autophagy by acting as a calorie restriction mimetic (Madeo et al., 2019; Madeo et al., 2018). Spermidine also regulates gene expression, oxidative stress response, cell viability, cell growth and proliferation (Pegg, 2016). Physiological levels of spermidine decrease with age in various model organisms and in humans but are higher in centenarians (Eisenberg et al., 2009; Gupta et al., 2013; Pucciarelli et al., 2012). Dietary supplementation of spermidine extends lifespan in yeast, C. elegans, flies and mice in an autophagy-dependent manner (Eisenberg et al., 2016; Eisenberg et al., 2009; Yue et al., 2017). The geroprotective benefits of spermidine feeding ranges from neuroprotection (Gupta et al., 2013; Xu et al., 2020b), cardioprotection (Eisenberg et al., 2016), improved muscle stem cell function (Garcia-Prat et al., 2016) to enhanced immunity (Madeo et al., 2018; Yang et al., 2016). In humans, higher spermidine intake is linked to lower all-cause mortality (Kiechl et al., 2018). Spermidine activates macroautophagy through multiple mechanisms including inhibition of acetyltransferases such as EP300, which suppresses autophagy through acetylation of several ATGs (Mariño et al., 2014) and activation of mTORC1 (Andreux et al., 2019). Spermidine also induces autophagy gene expression by preserving histone acetylation in their promoter region (Madeo et al., 2010) and by favoring hypusination of the translation initiation factor 5A (EIF5A) that promotes TFEB synthesis (Zhang et al., 2019). Besides general macroautophagy, recent evidence shows spermidine induces PINK-1/Parkin mitophagy (Qi et al., 2016). The potent combination of mechanisms by which spermidine activates autophagy supports the potential value of clinical trials increasing intake of spermidine to induce autophagy and delay aging.
The geroprotective effects of the immunosuppressant rapamycin, also the best described pharmacological inducer of macroautophagy, have been well-reported (Arriola Apelo and Lamming, 2016). Rapamycin reduces mTORC1 signaling, which increases with aging in response to cellular damage (Johnson et al., 2013). Despite extensive genetic evidence (section 2.1) supporting that extension of lifespan by modulating mTOR is dependent on autophagy, less studies and only in flies have experimentally demonstrated the contribution of autophagy to the beneficial effects of rapamycin in life-extension (Bjedov et al., 2010; Schinaman et al., 2019). The negative effects related to mTORC2 inhibition upon chronic exposure to rapamycin have motivated the current development of derivatives of rapamycin (rapalogs) with greater specificity (particularly in upregulating autophagy) (Galluzzi et al., 2017). Combinatorial approaches with drugs targeting different hallmarks of aging have included rapamycin with trametinib and lithium, which act synergistically to maximize longevity. This combination has the added advantage of activating autophagy by two non-overlapping mechanisms (Madeo et al., 2010).
Metformin, a drug used for type 2 diabetes, has also shown geroprotective effects in experimental models (Kulkarni et al., 2020; Piskovatska et al., 2019) and has become the first drug approved for testing effects on aging in non-diabetic people in a large clinical trial (TAME: Targeting Aging by Metformin) (Justice et al., 2018). Metformin activates AMPK and Sirtuin 1 and directly inhibits mTORC1 - resulting in robust induction of macroautophagy (Kulkarni et al., 2020; Piskovatska et al., 2019; Piskovatska et al., 2020).
The list of additional products with beneficial effect on autophagy keeps growing and includes supplementation with NAD+ (Fang et al., 2019a), resveratrol (Morselli et al., 2010), triethylenetetramine (Pietrocola et al., 2020) and sarcosine (Walters et al., 2018) (Table 1).
5.2. Autophagy activators as novel geroprotectors
The prominent role that activation of autophagy has as a common mechanism for many longevity promoting interventions justifies current efforts to identify autophagy inducers as a way to accelerate discovery of new geroprotectors (Fig. 4 and Table 1).
5.2.1. Macroautophagy activators
Tat-Beclin-1 peptide is one such available autophagy inducer. This peptide includes a Beclin-1 amino acid sequence recognized by the HIV-1 Nef virulence factor linked to the HIV-1 Tat protein transduction domain to make this peptide cell permeable. Tat-Beclin-1 peptide competes the inhibitory effect of Bcl-2 on Beclin-1, thus enhancing autophagosome nucleation and biogenesis (Shoji-Kawata et al., 2013). This peptide has been shown protective against proteotoxicity (Shoji-Kawata et al., 2013) and infections by virus (i.e., HIV-1, positive-stranded RNA virus) and bacteria such as Listeria monocytogenes that causes listeriosis, an infection of the central nervous system seen primarily in the elderly.
The important role of TFEB as a master transcription regulator of macroautophagy, lysosomal biogenesis and exocytosis, and the studies linking TFEB to lifespan extension in six mechanistically different longevity models in C. elegans (Lapierre et al., 2013) have propelled extensive search for chemical modulators of this transcriptional factor for geroprotective purpose. Growing evidence supports a protective effect of genetic and pharmacological activation of TFEB against neurodegeneration, metabolic diseases and other age-related pathologies (Cortes and La Spada, 2019). mTORC1-dependent phosphorylation of TFEB inhibits its transcription factor activity by preventing its nuclear shuttling whereas dephosphorylation by calcineurin releases this inhibitory effect (Settembre et al., 2011). Starvation activates TFEB by inhibiting mTORC1, inducing Ca2+ release from lysosomes to activate calcineurin and activating AMPK that increases TFEB transcription by enhancing local histone acetylation (Medina et al., 2015; Sakamaki et al., 2018). To avoid undesirable side-effects of long-term mTORC1 inhibition, the search for chemical TFEB activators has been focused on compounds such as a curcumin-derived compound C1 that mediates robust TFEB activation independent of mTORC1 (Song et al., 2016). Although recent studies with curcumin failed to see lifespan extension in mice, possible healthspan benefits have not been ruled out (Partridge et al., 2020). Small molecule TFEB agonist based on Ca2+-dependent mechanisms have shown geroprotective potential by alleviating the metabolic syndrome in mice and extending C. elegans lifespan (Wang et al., 2017). Also promising is the possibility of selectively activating mitophagy through TFEB modulation. Pomegranate polyphenols that extend lifespan in worms and flies (Balasubramani et al., 2014; Zheng et al., 2017) have shown capable to induce TFEB in a novel Ca2+-dependent manner to promote mitophagosome formation and potentiate PINK1/Parkin mitophagy (Tan et al., 2019). Given that TFEB acts as a hub that coordinates lysosomal biology, metabolism and mitochondrial fitness, targeting TFEB regulation may confer broad benefits on healthspan and lifespan.
Growing genetic evidence support that selective targeting of specific types of macroautophagy may be desirable over global upregulation of this process would be desirable, especially when considering long-term treatments. On this respect, interventions to improve mitochondrial quality control by mitophagy can have far-reaching effects for promoting longevity. Urolithin A, a metabolite derived from digestion of pomegranate-enriched ellagitannins by gut microbiota, has proven efficient in inducing mitophagy (Ryu et al., 2016) and in promoting lifespan in worms, improving muscle function in old worms and mice and reducing inflammation and slowing down memory loss in AD mouse models (Gong et al., 2019; Ryu et al., 2016). Oral consumption of urolithin A has proven to be safe in humans and the treated individuals display a molecular imprint of better mitochondrial and cellular health (Andreux et al., 2019). The recently described stimulation of Nrf2-mediated mitochondrial biogenesis by urolithin A (Singh et al., 2019) highlights the potential of using urolithin A to preserve mitochondrial function through coordinated disposal and replacement of this organelle. Actinonin (Fang et al., 2019b) and doxycycline (Xing et al., 2017) are also multi-effect small molecules that are able to induce mitophagy. Pharmacological targeting of mitophagy components has also been recently explored with compounds such as PMI that activates the transcriptional factor Nrf2 to upregulate expression of the mitophagy receptor p62 (East et al., 2014), or inhibitors of deubiquitinase enzymes such as USP30 to enhance ubiquitin labeling of mitochondria by the PINK1/Parkin machinery and promote mitophagy (Nardin et al., 2016).
5.2.2. CMA activators
The promising effects of genetic restoration of CMA in old rodent livers (Zhang and Cuervo, 2008) triggered the search for pharmacological activators of CMA (Table 1). Although CMA has been shown upregulated secondary to exposure to agents that regulate glucose metabolism or change with agents that block translation, those changes on CMA are all secondary and lack selectivity (Finn et al., 2005). Druggable CMA targets include AKT1, mTORC2 and PHLPP1, that act coordinately at the lysosomal membrane in the regulation of CMA activity. However, the numerous additional cellular functions of these targets make them unsuitable as selective modulators of CMA. CMA is also under the negative regulation of the retinoic acid receptor α (RARα), for which pharmacological targeting is also possible. Although chemical inhibition of RARα for CMA activation faces the same issues of selectivity, a screen for atypical RARα -modulating small molecules has identified a subset of retinoid derivatives that only modulate a very restricted part of the RARα-dependent transcriptional program (Anguiano et al., 2013). Optimization of these molecules to acquire drug-like properties for in vivo use has allowed to demonstrate their ability to revert the functional decline of old mouse and human stem cells (Dong et al., 2021) and to reduce neuronal and glial pathology in mouse models of different tauopathies including AD (Bourdenx et al., 2021). Besides the transcriptional upregulation of LAMP2A induced by these novel small molecules, future interventions aiming at stabilizing the preexisting levels of LAMP2A at the lysosomal membrane should also be effective in preventing the age-dependent decline in CMA.
6. Concluding remarks: An exciting future ahead
Framing the study of aging into driving cellular and molecular pathways (Lopez-Otin and Kroemer, 2021) was visionary and has changed the aging research landscape. This pathway-oriented context has allowed systematic validation of the contribution of these processes to aging and testing of their potential value as geroprotective targets. This exercise has highlighted autophagy as one of the central pathways in the protection against the functional loss and increased vulnerability to disease associated with aging. As reviewed in this work, autophagy may exert this influential geroprotective effect by acting on several (if not all) drivers of aging. This realization makes now very attractive the possibility of targeting autophagy to prevent other drivers’ age-dependent effects. The often bidirectional interaction of autophagy with the other hallmarks of aging also offers additional ways of improving autophagy through targeting of those other pathways. Thus, the fluidity and continuous reassessment of the hallmarks of aging should help to further understand interactions among these cellular drivers. Future studies are necessary to determine the possible tissue-dependence and hierarchic organization of this dual interplay of autophagy with the other aging hallmarks, and to identify the molecular players regulating the effect of autophagy on the other pathways and vice versa. Special attention should also be placed on sex differences in autophagy and their implication for those aging features and age-related diseases with sexual dimorphism. In this respect, functional assays for autophagy in multiple tissues should complement the still scarce information available on sex differences mostly relying on steady state measurements of autophagy.
The fact that many of the anti-aging interventions currently under study have been shown to converge on autophagy, has provided momentum to the search for approaches to activate different autophagic pathways in a more selective manner or even through complementary mechanisms. A better understanding of the consequences of long-term activation of autophagy is needed, especially on light of recent reports describing detrimental effects of excessive activation of macroautophagy. This is less of a concern in interventions enhancing the lysosomal or endosomal capability for CMA or eMI, where only substrates primed for degradation by the chaperone will benefit from these interventions. Future development of activators of selective types of macroautophagy may also help overcome this problem. Combining different autophagic strategies may mirror the benefits of compensatory crosstalk between different proteolytic systems seen to some extent in physiological aging. The challenge for combinatorial approaches resides in finding complementary effective dosing, which may also vary among individuals. Applying precision medicine to identify suitable drug-pairing and artificial intelligence-based dosing strategies could expand the feasibility of enhancing autophagy to promote longevity.
Highlights.
Autophagy is central for health- and lifespan
Different autophagic processes malfunction with age
Autophagic failure impinges on multiple hallmarks of aging
Autophagy enhancement is common to most geroprotective interventions
Autophagy modulation shows potential as geroprotective intervention
Acknowledgements
Authors thank the members of their respective groups and colleagues in the field of autophagy and of aging for the valuable feedback and suggestions when preparing this work. We apologize to those esteemed colleagues whose work may have been omitted from this review due to space limitation.
Funding
Work in our laboratories is supported by the National Institutes of Health [AG054108, AG021904, AG017617, AG038072 , AG031782 and NS100717 and DK098408 (to AMC) and P30AG038072 (P&F), P30DK041296 (P&F) and DK124308 (to EA)], Fondo de Investigación Sanitaria-Instituto de Salud Carlos III (Spain)-FEDER [PI20/00728 (to MM-V), Spanish Ministry of Science and Innovation [RTI2018-097948-A-100 and RYC-2016-20480 (to OP)] and the generous support of the JPB Foundation, the Rainwater Charitable Foundation, the Glenn Foundation, the Backus Foundation and Robert and Renee Belfer (to AMC), the Grace Science Foundation (to SK), the Michael J Fox Foundation, the Silverstein Foundation and the BBVA Foundation (to MMV).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
AMC is co-founder and scientific advisor for Life Bioscience and consults for Generian Therapeutics and Cognition. The rest of the authors declare no competing interests.
REFERENCES
- Ali M, Devkota S, Roh JI, Lee J, Lee HW, 2016. Telomerase reverse transcriptase induces basal and amino acid starvation-induced autophagy through mTORC1. Biochem Biophys Res Commun 478, 1198–1204. [DOI] [PubMed] [Google Scholar]
- Ambrosio S, Majello B, 2020. Autophagy Roles in Genome Maintenance. Cancers (Basel) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J, Sistonen L, 2011. Regulation of HSF 1 Function in the Heat Stress Response: Implications in Aging and Disease. Annual Review of Biochemistry 80, 1089–1115. [DOI] [PubMed] [Google Scholar]
- Andreux PA, Blanco-Bose W, Ryu D, Burdet F, Ibberson M, Aebischer P, Auwerx J, Singh A, Rinsch C, 2019. The mitophagy activator urolithin A is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Natural Metabolism 1, 595–603. [DOI] [PubMed] [Google Scholar]
- Anguiano J, Garner TP, Mahalingam M, Das BC, Gavathiotis E, Cuervo AM, 2013. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nature chemical biology 9, 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E, Koga H, Diaz A, Mocholi E, Patel B, Cuervo AM, 2015. Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol Cell 59, 270–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriola Apelo SI, Lamming DW, 2016. Rapamycin: An InhibiTOR of Aging Emerges From the Soil of Easter Island. J Gerontol A Biol Sci Med Sci 71, 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artal-Martinez de Narvajas A, Gomez TS, Zhang JS, Mann AO, Taoda Y, Gorman JA, Herreros-Villanueva M, Gress TM, Ellenrieder V, Bujanda L, Kim DH, Kozikowski AP, Koenig A, Billadeau DD, 2013. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol 33, 3983–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Kang P, Hernandez AM, Tatar M, 2013. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet 9, e1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramani SP, Mohan J, Chatterjee A, Patnaik E, Kukkupuni SK, Nongthomba U, Venkatasubramanian P, 2014. Pomegranate Juice Enhances Healthy Lifespan in Drosophila melanogaster: An Exploratory Study. Frontiers in public health 2, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM, 2008. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol 28, 5747–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay U, Sridhar S, Kaushik S, Kiffin R, Cuervo AM, 2010. Identification of regulators of chaperone-mediated autophagy. Mol Cell 39, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano E, Girao H, Yuste A, Patel B, Marques C, Spray DC, Pereira P, Cuervo AM, 2012. Autophagy modulates dynamics of connexins at the plasma membrane in a ubiquitin-dependent manner. Mol Biol Cell 23, 2156–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano E, Murray JW, Wang X, Pampliega O, Yin D, Patel B, Yuste A, Wolkoff AW, Cuervo AM, 2018. Defective recruitment of motor proteins to autophagic compartments contributes to autophagic failure in aging. Aging Cell 17, e12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano E, Yuste A, Patel B, Stout RF Jr., Spray DC, Cuervo AM, 2014. Connexins modulate autophagosome biogenesis. Nature cell biology 16, 401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI, 2009. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proceedings of the National Academy of Sciences 106, 14914–14919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry BJ, Kaeberlein M, 2021. An energetics perspective on geroscience: mitochondrial protonmotive force and aging. Geroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L, 2010. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab 11, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdenx M, Martin-Segura A, Scrivo A, Rodriguez-Navarro JA, Kaushik S, Tasset I, Diaz A, Storm NJ, Xin Q, Juste YR, Stevenson E, Luengo E, Clement CC, Choi SJ, Krogan NJ, Mosharov EV, Santambrogio L, Grueninger F, Collin L, Swaney DL, Sulzer D, Gavathiotis E, Cuervo AM, 2021. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Codogno P, Rodriguez-Muela N, 2018. Autophagy in stem cells: repair, remodelling and metabolic reprogramming. Development (Cambridge, England) 145, 1–14. [DOI] [PubMed] [Google Scholar]
- Breslin L, Prosser SL, Cuffe S, Morrison CG, 2014. Ciliary abnormalities in senescent human fibroblasts impair proliferative capacity. Cell Cycle 13, 2773–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratta S, Tancini B, Sagini K, Delo F, Chiaradia E, Urbanelli L, Emiliani C, 2020. Lysosomal Exocytosis, Exosome Release and Secretory Autophagy: The Autophagic- and Endo-Lysosomal Systems Go Extracellular. Int J Mol Sci 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun S, Seok S, Kim YC, Zhang Y, Yau P, Iwamori N, Xu HE, Ma J, Kemper B, Kemper JK, 2020. Fasting-induced FGF21 signaling activates hepatic autophagy and lipid degradation via JMJD3 histone demethylase. Nat Commun 11, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero B, Bourdenx M, Luengo E, Diaz A, Sohn PD, Chen X, Wang C, Juste YR, Wegmann S, Patel B, Young ZT, Kuo SY, Rodriguez-Navarro JA, Shao H, Lopez MG, Karch CM, Goate AM, Gestwicki JE, Hyman BT, Gan L, Cuervo AM, 2021. Acetylated tau inhibits chaperone-mediated autophagy and promotes tau pathology propagation in mice. Nat Commun 12, 2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero B, Wang Y, Diaz A, Tasset I, Juste YR, Stiller B, Mandelkow EM, Mandelkow E, Cuervo AM, 2018. Interplay of pathogenic forms of human tau with different autophagic pathways. Aging Cell 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Zou F, Xuan R, Lai XY, 2021. Exosomes from mesenchymal stem cells expressing microribonucleic acid-125b inhibit the progression of diabetic nephropathy via the tumour necrosis factor receptor-associated factor 6/Akt axis. Endocr J. [DOI] [PubMed] [Google Scholar]
- Cannizzo ES, Clement CC, Morozova K, Valdor R, Kaushik S, Almeida LN, Follo C, Sahu R, Cuervo AM, Macian F, Santambrogio L, 2012. Age-related oxidative stress compromises endosomal proteostasis. Cell reports 2, 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellazzi M, Patergnani S, Donadio M, Giorgi C, Bonora M, Bosi C, Brombo G, Pugliatti M, Seripa D, Zuliani G, Pinton P, 2019. Autophagy and mitophagy biomarkers are reduced in sera of patients with Alzheimer’s disease and mild cognitive impairment. Sci Rep 9, 20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarino S, Ribeiro-Rodrigues TM, R SF, Ramalho J, Abert C, Martens S, Girão H, 2020. A Conserved LIR Motif in Connexins Mediates Ubiquitin-Independent Binding to LC3/GABARAP Proteins. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G, Cenci S, 2020. Autophagy and Protein Secretion. J Mol Biol 432, 2525–2545. [DOI] [PubMed] [Google Scholar]
- Chafekar SM, Duennwald ML, 2012. Impaired Heat Shock Response in Cells Expressing Full-Length Polyglutamine-Expanded Huntingtin. PLoS ONE 7, e37929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Kumsta C, Hellman AB, Adams LM, Hansen M, 2017. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang C, Sun L, Wang D-L, Chen L, Huang Z, Yang Q, Gao J, Yang X-B, Chang J-F, Chen P, Lan L, Mao Z, Sun F-L, 2015. RAD6 promotes homologous recombination repair by activating the autophagy-mediated degradation of heterochromatin protein HP1. Molecular and cellular biology 35, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Guo Y, Yang W, Zheng P, Zeng J, Tong W, 2017. Involvement of autophagy in connexin 40 reduction in the late phase of traumatic brain injury in rats. Brain Res Bull 131, 100–106. [DOI] [PubMed] [Google Scholar]
- Chen W, Wang H, Zhu Z, Feng J, Chen L, 2020. Exosome-Shuttled circSHOC2 from IPASs Regulates Neuronal Autophagy and Ameliorates Ischemic Brain Injury via the miR-7670-3p/SIRT1 Axis. Mol Ther Nucleic Acids 22, 657–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HI, Dice JF, 1988. Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J Biol Chem 263, 6797–6803. [PubMed] [Google Scholar]
- Chino H, Mizushima N, 2020. ER-Phagy: Quality Control and Turnover of Endoplasmic Reticulum. Trends in Cell Biology 30, 384–398. [DOI] [PubMed] [Google Scholar]
- Congdon EE, 2018. Sex Differences in Autophagy Contribute to Female Vulnerability in Alzheimer’s Disease. Front Neurosci 12, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelison D, Perdiguero E, 2017. Muscle Stem Cells: A Model System for Adult Stem Cell Biology. Methods in molecular biology 1556, 3–19. [DOI] [PubMed] [Google Scholar]
- Cortes CJ, La Spada AR, 2019. TFEB dysregulation as a driver of autophagy dysfunction in neurodegenerative disease: Molecular mechanisms, cellular processes, and emerging therapeutic opportunities. Neurobiology of disease 122, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF, 1996. A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273, 501–503. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF, 2000. Age-related decline in chaperone-mediated autophagy. J Biol Chem 275, 31505–31513. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Knecht E, Terlecky SR, Dice JF, 1995. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol 269, C1200–1208. [DOI] [PubMed] [Google Scholar]
- D’Amico D, Andreux PA, Valdés P, Singh A, Rinsch C, Auwerx J, 2021. Impact of the Natural Compound Urolithin A on Health, Disease, and Aging. Trends Mol Med 27, 687–699. [DOI] [PubMed] [Google Scholar]
- D’Amico D, Mottis A, Potenza F, Sorrentino V, Li H, Romani M, Lemos V, Schoonjans K, Zamboni N, Knott G, Schneider BL, Auwerx J, 2019. The RNA-Binding Protein PUM2 Impairs Mitochondrial Dynamics and Mitophagy During Aging. Mol Cell 73, 775–787 e710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cabo R, Mattson MP, 2019. Effects of Intermittent Fasting on Health, Aging, and Disease. New England Journal of Medicine 381, 2541–2551. [DOI] [PubMed] [Google Scholar]
- De Duve C, Wattiaux R, 1966. Functions of lysosomes. Annu Rev Physiol 28, 435–492. [DOI] [PubMed] [Google Scholar]
- De Leonibus C, Cinque L, Settembre C, 2019. Emerging lysosomal pathways for quality control at the endoplasmic reticulum. FEBS Letters 593, 2319–2329. [DOI] [PubMed] [Google Scholar]
- Del Roso A, Bombara M, Fierabracci V, Masini M, Masiello P, Pollera M, Bergamini E, 1991. Effect of dietary restriction on the age-related changes in hormone-regulated protein breakdown. Aging (Milano) 3, 407–408. [DOI] [PubMed] [Google Scholar]
- Del Roso A, Vittorini S, Cavallini G, Donati A, Gori Z, Masini M, Pollera M, Bergamini E, 2003. Ageing-related changes in the in vivo function of rat liver macroautophagy and proteolysis. Exp Gerontol 38, 519–527. [DOI] [PubMed] [Google Scholar]
- Demontis F, Perrimon N, 2010. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Domenico A, Carola G, Calatayud C, Pons-Espinal M, Muñoz JP, Richaud-Patin Y, Fernandez-Carasa I, Gut M, Faella A, Parameswaran J, Soriano J, Ferrer I, Tolosa E, Zorzano A, Cuervo AM, Raya A, Consiglio A, 2019. Patient-Specific iPSC-Derived Astrocytes Contribute to Non-Cell-Autonomous Neurodegeneration in Parkinson’s Disease. Stem Cell Reports 12, 213–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Nie Z, She Z, Bai X, Yang Q, Wang F, Wang F, Geng X, 2021. The Regulation of ROS- and BECN1-Mediated Autophagy by Human Telomerase Reverse Transcriptase in Glioblastoma. Oxid Med Cell Longev 2021, 6636510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati A, Cavallini G, Paradiso C, Vittorini S, Pollera M, Gori Z, Bergamini E, 2001a. Age-related changes in the autophagic proteolysis of rat isolated liver cells: effects of antiaging dietary restrictions. J Gerontol A Biol Sci Med Sci 56, B375–383. [DOI] [PubMed] [Google Scholar]
- Donati A, Cavallini G, Paradiso C, Vittorini S, Pollera M, Gori Z, Bergamini E, 2001b. Age-related changes in the regulation of autophagic proteolysis in rat isolated hepatocytes. J Gerontol A Biol Sci Med Sci 56, B288–293. [DOI] [PubMed] [Google Scholar]
- Dong S, Wang Q, Kao YR, Diaz A, Tasset I, Kaushik S, Thiruthuvanathan V, Zintiridou A, Nieves E, Dzieciatkowska M, Reisz JA, Gavathiotis E, D’Alessandro A, Will B, Cuervo AM, 2021. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature 591, 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East DA, Fagiani F, Crosby J, Georgakopoulos ND, Bertrand H, Schaap M, Fowkes A, Wells G, Campanella M, 2014. PMI: a DeltaPsim independent pharmacological regulator of mitophagy. Chem Biol 21, 1585–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Abdellatif M, Schroeder S, Primessnig U, Stekovic S, Pendl T, Harger A, Schipke J, Zimmermann A, Schmidt A, Tong M, Ruckenstuhl C, Dammbrueck C, Gross AS, Herbst V, Magnes C, Trausinger G, Narath S, Meinitzer A, Hu Z, Kirsch A, Eller K, Carmona-Gutierrez D, Büttner S, Pietrocola F, Knittelfelder O, Schrepfer E, Rockenfeller P, Simonini C, Rahn A, Horsch M, Moreth K, Beckers J, Fuchs H, Gailus-Durner V, Neff F, Janik D, Rathkolb B, Rozman J, de Angelis MH, Moustafa T, Haemmerle G, Mayr M, Willeit P, von Frieling-Salewsky M, Pieske B, Scorrano L, Pieber T, Pechlaner R, Willeit J, Sigrist SJ, Linke WA, Mühlfeld C, Sadoshima J, Dengjel J, Kiechl S, Kroemer G, Sedej S, Madeo F, 2016. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med 22, 1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Fröhlich KU, Sinner F, Tavernarakis N, Minois N, Kroemer G, Madeo F, 2009. Induction of autophagy by spermidine promotes longevity. Nature cell biology 11, 1305–1314. [DOI] [PubMed] [Google Scholar]
- Elsayed HRH, El-Nablaway M, Khattab BA, Sherif RN, Elkashef WF, Abdalla AM, El Nashar EM, Abd-Elmonem MM, El-Gamal R, 2021. Independent of Calorie Intake, Short-term Alternate-day Fasting Alleviates NASH, With Modulation of Markers of Lipogenesis, Autophagy, Apoptosis, and Inflammation in Rats. J Histochem Cytochem 69, 575–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele E, Minoretti P, Sanchis-Gomar F, Pareja-Galeano H, Yilmaz Y, Garatachea N, Lucia A, 2014. Can enhanced autophagy be associated with human longevity? Serum levels of the autophagy biomarker beclin-1 are increased in healthy centenarians. Rejuvenation Res 17, 518–524. [DOI] [PubMed] [Google Scholar]
- Endicott SJ, Boynton DN Jr., Beckmann LJ, Miller RA, 2021. Long-lived mice with reduced growth hormone signaling have a constitutive upregulation of hepatic chaperone-mediated autophagy. Autophagy 17, 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott SJ, Ziemba ZJ, Beckmann LJ, Boynton DN, Miller RA, 2020. Inhibition of class I PI3K enhances chaperone-mediated autophagy. J Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbaba B, Arslan-Ergul A, Adams MM, 2021. Effects of caloric restriction on the antagonistic and integrative hallmarks of aging. Ageing Res Rev 66, 101228. [DOI] [PubMed] [Google Scholar]
- Escobar KA, Cole NH, Mermier CM, VanDusseldorp TA, 2019. Autophagy and aging: Maintaining the proteome through exercise and caloric restriction. Aging Cell 18, e12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakouri NB, Hou Y, Demarest TG, Christiansen LS, Okur MN, Mohanty JG, Croteau DL, Bohr VA, 2019. Toward understanding genomic instability, mitochondrial dysfunction and aging. The FEBS Journal 286, 1058–1073. [DOI] [PubMed] [Google Scholar]
- Fang EF, Hou Y, Lautrup S, Jensen MB, Yang B, SenGupta T, Caponio D, Khezri R, Demarest TG, Aman Y, Figueroa D, Morevati M, Lee HJ, Kato H, Kassahun H, Lee JH, Filippelli D, Okur MN, Mangerich A, Croteau DL, Maezawa Y, Lyssiotis CA, Tao J, Yokote K, Rusten TE, Mattson MP, Jasper H, Nilsen H, Bohr VA, 2019a. NAD(+) augmentation restores mitophagy and limits accelerated aging in Werner syndrome. Nat Commun 10, 5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Hou Y, Palikaras K, Adriaanse BA, Kerr JS, Yang B, Lautrup S, Hasan-Olive MM, Caponio D, Dan X, Rocktaschel P, Croteau DL, Akbari M, Greig NH, Fladby T, Nilsen H, Cader MZ, Mattson MP, Tavernarakis N, Bohr VA, 2019b. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat Neurosci 22, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AF, Sebti S, Wei Y, Zou Z, Shi M, McMillan KL, He C, Ting T, Liu Y, Chiang WC, Marciano DK, Schiattarella GG, Bhagat G, Moe OW, Hu MC, Levine B, 2018. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558, 136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JV, Fôfo H, Bejarano E, Bento CF, Ramalho JS, Girão H, Pereira P, 2013. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy 9, 1349–1366. [DOI] [PubMed] [Google Scholar]
- Finn PF, Mesires NT, Vine M, Dice JF, 2005. Effects of small molecules on chaperone-mediated autophagy. Autophagy 1, 141–145. [DOI] [PubMed] [Google Scholar]
- Fortini P, Ferretti C, Iorio E, Cagnin M, Garribba L, Pietraforte D, Falchi M, Pascucci B, Baccarini S, Morani F, Phadngam S, De Luca G, Isidoro C, Dogliotti E, 2016. The fine tuning of metabolism, autophagy and differentiation during in vitro myogenesis. Cell Death and Disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregno I, Fasana E, Bergmann TJ, Raimondi A, Loi M, Soldà T, Galli C, D’Antuono R, Morone D, Danieli A, Paganetti P, Anken E, Molinari M, 2018. ER -to-lysosome-associated degradation of proteasome‐resistant ATZ polymers occurs via receptor‐mediated vesicular transport. The EMBO Journal 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R, Ding Y, Luo J, Huang KM, Tang XJ, Li DS, Guo SW, 2018. Ten-eleven translocation 1 regulates methylation of autophagy-related genes in human glioma. Neuroreport 29, 731–738. [DOI] [PubMed] [Google Scholar]
- Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B, 2013. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 500, 468–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallart-Palau X, Guo X, Serra A, Sze SK, 2020. Alzheimer’s disease progression characterized by alterations in the molecular profiles and biogenesis of brain extracellular vesicles. Alzheimer’s research & therapy 12, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G, 2017. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nature reviews. Drug discovery 16, 487–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Yu W, Lv P, Liang X, Sun S, Zhang Y, 2021. Parkin overexpression alleviates cardiac aging through facilitating K63-polyubiquitination of TBK1 to facilitate mitophagy. Biochim Biophys Acta Mol Basis Dis 1867, 165997. [DOI] [PubMed] [Google Scholar]
- Garcia-Prat L, Martinez-Vicente M, Perdiguero E, Ortet L, Rodriguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, Sandri M, Munoz-Canoves P, 2016. Autophagy maintains stemness by preventing senescence. Nature 529, 37–42. [DOI] [PubMed] [Google Scholar]
- Gewirtz DA, Tyutyunyk-Massey L, Landry JW, 2018. The potentially conflicting cell autonomous and cell non-autonomous functions of autophagy in mediating tumor response to cancer therapy. Biochem Pharmacol 153, 46–50. [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Mau T, O’Brien M, Garg S, Yung R, 2016. Impaired autophagy activity is linked to elevated ER-stress and inflammation in aging adipose tissue. Aging (Albany NY) 8, 2525–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatigny M, Moriceau S, Rivagorda M, Ramos-Brossier M, Nascimbeni AC, Lante F, Shanley MR, Boudarene N, Rousseaud A, Friedman AK, Settembre C, Kuperwasser N, Friedlander G, Buisson A, Morel E, Codogno P, Oury F, 2019. Autophagy Is Required for Memory Formation and Reverses Age-Related Memory Decline. Curr Biol 29, 435–448 e438. [DOI] [PubMed] [Google Scholar]
- Gong Z, Huang J, Xu B, Ou Z, Zhang L, Lin X, Ye X, Kong X, Long D, Sun X, He X, Xu L, Li Q, Xuan A, 2019. Urolithin A attenuates memory impairment and neuroinflammation in APP/PS1 mice. Journal of neuroinflammation 16, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Tasset I, Diaz A, Anguiano J, Tas E, Cui L, Kuliawat R, Liu H, Kuhn B, Cuervo AM, Muzumdar R, 2018. Humanin is an endogenous activator of chaperone-mediated autophagy. J Cell Biol 217, 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez CD, Resnik R, Vaccaro MI, 2020. Secretory Autophagy and Its Relevance in Metabolic and Degenerative Disease. Frontiers in endocrinology 11, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, Campisi J, Collado M, Evangelou K, Ferbeyre G, Gil J, Hara E, Krizhanovsky V, Jurk D, Maier AB, Narita M, Niedernhofer L, Passos JF, Robbins PD, Schmitt CA, Sedivy J, Vougas K, von Zglinicki T, Zhou D, Serrano M, Demaria M, 2019. Cellular Senescence: Defining a Path Forward. Cell 179, 813–827. [DOI] [PubMed] [Google Scholar]
- Green PD, Sharma NK, Santos JH, 2019. Telomerase Impinges on the Cellular Response to Oxidative Stress Through Mitochondrial ROS-Mediated Regulation of Autophagy. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadiana SM, Parker AK, Filho GF, Sequeira A, Semple-Rowland S, Shaw G, Mandel RJ, Foster TC, Kumar A, Sarkisian MR, 2016. Type 3 Adenylyl Cyclase and Somatostatin Receptor 3 Expression Persists in Aged Rat Neocortical and Hippocampal Neuronal Cilia. Front Aging Neurosci 8, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta VK, Scheunemann L, Eisenberg T, Mertel S, Bhukel A, Koemans TS, Kramer JM, Liu KS, Schroeder S, Stunnenberg HG, Sinner F, Magnes C, Pieber TR, Dipt S, Fiala A, Schenck A, Schwaerzel M, Madeo F, Sigrist SJ, 2013. Restoring polyamines protects from age-induced memory impairment in an autophagy-dependent manner. Nat Neurosci 16, 1453–1460. [DOI] [PubMed] [Google Scholar]
- Hammerling BC, Najor RH, Cortez MQ, Shires SE, Leon LJ, Gonzalez ER, Boassa D, Phan S, Thor A, Jimenez RE, Li H, Kitsis RN, Dorn GW II, Sadoshima J, Ellisman MH, Gustafsson AB, 2017. A Rab5 endosomal pathway mediates Parkin-dependent mitochondrial clearance. Nat Commun 8, 14050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan MA, Rahman MA, Rahman MS, Sohag AAM, Dash R, Hossain KS, Farjana M, Uddin MJ, 2020. Intermittent fasting, a possible priming tool for host defense against SARS-CoV-2 infection: Crosstalk among calorie restriction, autophagy and immune response. Immunol Lett 226, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C, 2008. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 4, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N, 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889. [DOI] [PubMed] [Google Scholar]
- Harman D, 1956. Aging: a theory based on free radical and radiation chemistry. J Gerontol 11, 298–300. [DOI] [PubMed] [Google Scholar]
- Hart CR, Ryan ZC, Pfaffenbach KT, Dasari S, Parvizi M, Lalia AZ, Lanza IR, 2019. Attenuated activation of the unfolded protein response following exercise in skeletal muscle of older adults. Aging 11, 7587–7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, 2017. Protein misfolding diseases. Annu. Rev. Biochem 86, 21–26. [DOI] [PubMed] [Google Scholar]
- Hatakeyama R, De Virgilio C, 2019. TORC1 specifically inhibits microautophagy through ESCRT-0. Curr Genet 65, 1243–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Zhang K, Kaufman RJ, 2020. Mechanisms, regulation and functions of the unfolded protein response. Nature Reviews Molecular Cell Biology 21, 421–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, Figueroa ME, Passegué E, 2017. Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu SQ, Zhang QC, Meng QB, Hu AN, Zou JP, Li XL, 2020. Autophagy regulates exosome secretion in rat nucleus pulposus cells via the RhoC/ROCK2 pathway. Exp Cell Res 395, 112239. [DOI] [PubMed] [Google Scholar]
- Hubbi ME, Gilkes DM, Hu H, Kshitiz, Ahmed I, Semenza GL, 2014. Cyclin-dependent kinases regulate lysosomal degradation of hypoxia-inducible factor 1alpha to promote cell-cycle progression. Proc Natl Acad Sci U S A 111, E3325–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttinen JMT, Blasiak J, Felszeghy S, Kaarniranta K, 2021. MicroRNAs in the regulation of autophagy and their possible use in age-related macular degeneration therapy. Ageing Res Rev 67, 101260. [DOI] [PubMed] [Google Scholar]
- Jacomin AC, Gohel R, Hussain Z, Varga A, Maruzs T, Eddison M, Sica M, Jain A, Moffat KG, Johansen T, Jenny A, Juhasz G, Nezis IP, 2021. Degradation of arouser by endosomal microautophagy is essential for adaptation to starvation in Drosophila. Life Sci Alliance 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamshed H, Beyl RA, Della Manna DL, Yang ES, Ravussin E, Peterson CM, 2019. Early Time-Restricted Feeding Improves 24-Hour Glucose Levels and Affects Markers of the Circadian Clock, Aging, and Autophagy in Humans. Nutrients 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Wells CD, Roach PJ, 2011. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun 413, 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Rabinovitch PS, Kaeberlein M, 2013. mTOR is a key modulator of ageing and age-related disease. Nature 493, 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretschke T, Beli P, 2021. Causes and consequences of DNA damage-induced autophagy. Matrix Biol. [DOI] [PubMed] [Google Scholar]
- Justice JN, Niedernhofer L, Robbins PD, Aroda VR, Espeland MA, Kritchevsky SB, Kuchel GA, Barzilai N, 2018. Development of Clinical Trials to Extend Healthy Lifespan. Cardiovascular endocrinology & metabolism 7, 80–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, Lu T, Yankner BA, Campisi J, Elledge SJ, 2015. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 349, aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, White E, 2014. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov 4, 914–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Arias E, Kwon H, Lopez NM, Athonvarangkul D, Sahu S, Schwartz GJ, Pessin JE, Singh R, 2012. Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep 13, 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM, 2015. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nature cell biology 17, 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM, 2016. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy 12, 432–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM, 2018. The coming of age of chaperone-mediated autophagy. Nature Reviews Molecular Cell Biology 19, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Massey AC, Cuervo AM, 2006. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J 25, 3921–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Massey AC, Mizushima N, Cuervo AM, 2008. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell 19, 2179–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, Cuervo AM, Singh R, 2011. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab 14, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, Rando TA, Richardson A, Schadt EE, Wyss-Coray T, Sierra F, 2014. Geroscience: linking aging to chronic disease. Cell 159, 709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ, 2010. The genetics of ageing. Nature 464, 504–512. [DOI] [PubMed] [Google Scholar]
- Khalil H, Tazi M, Caution K, Ahmed A, Kanneganti A, Assani K, Kopp B, Marsh C, Dakhlallah D, Amer AO, 2016. Aging is associated with hypermethylation of autophagy genes in macrophages. Epigenetics 11, 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiechl S, Pechlaner R, Willeit P, Notdurfter M, Paulweber B, Willeit K, Werner P, Ruckenstuhl C, Iglseder B, Weger S, Mairhofer B, Gartner M, Kedenko L, Chmelikova M, Stekovic S, Stuppner H, Oberhollenzer F, Kroemer G, Mayr M, Eisenberg T, Tilg H, Madeo F, Willeit J, 2018. Higher spermidine intake is linked to lower mortality: a prospective population-based study. The American journal of clinical nutrition 108, 371–380. [DOI] [PubMed] [Google Scholar]
- Kiffin R, Christian C, Knecht E, Cuervo AM, 2004. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell 15, 4829–4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey AC, Martinez-Vicente M, Cuervo AM, 2007. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci 120, 782–791. [DOI] [PubMed] [Google Scholar]
- Kim SN, Kwon HJ, Im SW, Son YH, Akindehin S, Jung YS, Lee SJ, Rhyu IJ, Kim IY, Seong JK, Lee J, Yoo HC, Granneman JG, Lee YH, 2017. Connexin 43 is required for the maintenance of mitochondrial integrity in brown adipose tissue. Sci Rep 7, 7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Guan KL, 2015. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest 125, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU, 2013. Molecular chaperone functions in protein folding and proteostasis. Annual review of biochemistry 82, 323–355. [DOI] [PubMed] [Google Scholar]
- Kimura T, Jia J, Claude-Taupin A, Kumar S, Choi SW, Gu Y, Mudd M, Dupont N, Jiang S, Peters R, Farzam F, Jain A, Lidke KA, Adams CM, Johansen T, Deretic V, 2017. Cellular and molecular mechanism for secretory autophagy. Autophagy 13, 1084–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner P, Bourdenx M, Madrigal-Matute J, Tiano S, Diaz A, Bartholdy BA, Will B, Cuervo AM, 2019. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol 17, e3000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knewtson KE, Ohl NR, Robinson JL, 2021. Estrogen Signaling Dictates Musculoskeletal Stem Cell Behavior: Sex Differences in Tissue Repair. Tissue Eng Part B Rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Martinez-Vicente M, Macian F, Verkhusha VV, Cuervo AM, 2011. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat Commun 2, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J.-i.I., Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K, 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T, 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen EL, Millen J, Goldfarb DS, Thumm M, 2008. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell 19, 4492–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni AS, Gubbi S, Barzilai N, 2020. Benefits of Metformin in Attenuating the Hallmarks of Aging. Cell Metab 32, 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumsta C, Chang JT, Schmalz J, Hansen M, 2017. Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nature Communications 8, 14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI, 2015. The biology of proteostasis in aging and disease. Annual Review of Biochemistry 84, 435–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri V, Hawkins WD, Klionsky DJ, 2019. Watch What You (Self-) Eat: Autophagic Mechanisms that Modulate Metabolism. Cell Metab 29, 803–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Johansen T, 2012. Aggrephagy: Selective Disposal of Protein Aggregates by Macroautophagy. International Journal of Cell Biology 2012, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Johansen T, 2021. Mechanisms of Selective Autophagy. Annu Rev Cell Dev Biol. [DOI] [PubMed] [Google Scholar]
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, Dillin A, Hansen M, 2013. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun 4, 2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Gelino S, Meléndez A, Hansen M, 2011. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol 21, 1507–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, Ocorr K, Ellisman MH, Bodmer R, Bier E, Karin M, 2010. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 327, 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal AM, Cyr DP, Hill RJ, Lee PW, McCormick C, 2012. Subversion of autophagy by Kaposi’s sarcoma-associated herpesvirus impairs oncogene-induced senescence. Cell Host Microbe 11, 167–180. [DOI] [PubMed] [Google Scholar]
- Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J, Kai F, Goldsmith J, Liu JY, Huang YH, Monkkonen T, Vlahakis A, Huang EJ, Goodarzi H, Yu L, Wiita AP, Debnath J, 2020. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nature cell biology 22, 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemasters JJ, 2014. Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3). Redox biology 2, 749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ, 2017. Autophagy wins the 2016 Nobel Prize in Physiology or Medicine: Breakthroughs in baker’s yeast fuel advances in biomedical research. Proc Natl Acad Sci U S A 114, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G, 2019. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 176, 11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wang Z, Xing H, Wang Y, Guo Y, 2021a. Exosomes derived from miR-188-3p-modified adipose-derived mesenchymal stem cells protect Parkinson’s disease. Mol Ther Nucleic Acids 23, 1334–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WW, Wang HJ, Tan YZ, Wang YL, Yu SN, Li ZH, 2021b. Reducing lipofuscin accumulation and cardiomyocytic senescence of aging heart by enhancing autophagy. Exp Cell Res 403, 112585. [DOI] [PubMed] [Google Scholar]
- Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF, 2010. Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell 9, 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein A, Minogue PJ, Beyer EC, Berthoud VM, 2011. Autophagy: a pathway that contributes to connexin degradation. J Cell Sci 124, 910–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J, 2010. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A 107, 14164–14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, He Z, Simon HU, 2014. Autophagy suppresses melanoma tumorigenesis by inducing senescence. Autophagy 10, 372–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Lin H, Mao Z, Zhang L, Bao K, Jiang B, Xia C, Li W, Hu Z, Li J, 2020. Verapamil extends lifespan in Caenorhabditis elegans by inhibiting calcineurin activity and promoting autophagy. Aging (Albany NY) 12, 5300–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco M.a., Partridge L, Serrano M, Kroemer G, 2013. The hallmarks of aging. Cell 153, 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Kroemer G, 2021. Hallmarks of health. Cell 184, 1929–1939. [DOI] [PubMed] [Google Scholar]
- Lu Q, Li W, Li Z, Chen Z, Fu W, Jiang Q, Ding S, 2019. Effect of autophagy on cardiomyocyte membrane Cx43 acute remodeling in rats with ischemia-reperfusion. Int J Clin Exp Pathol 12, 2639–2645. [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Li J, 2015. Metabolic shifts during aging and pathology. Comprehensive Physiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Carmona-Gutierrez D, Hofer SJ, Kroemer G, 2019. Caloric Restriction Mimetics against Age-Associated Disease: Targets, Mechanisms, and Therapeutic Potential. Cell Metab 29, 592–610. [DOI] [PubMed] [Google Scholar]
- Madeo F, Eisenberg T, Pietrocola F, Kroemer G, 2018. Spermidine in health and disease. Science 359. [DOI] [PubMed] [Google Scholar]
- Madeo F, Tavernarakis N, Kroemer G, 2010. Can autophagy promote longevity? Nature cell biology 12, 842–846. [DOI] [PubMed] [Google Scholar]
- Madeo F, Zimmermann A, Maiuri MC, Kroemer G, 2015. Essential role for autophagy in life span extension. Journal of Clinical Investigation 125, 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madill M, McDonagh K, Ma J, Vajda A, McLoughlin P, O’Brien T, Hardiman O, Shen S, 2017. Amyotrophic lateral sclerosis patient iPSC-derived astrocytes impair autophagy via non-cell autonomous mechanisms. Mol Brain 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, Zamzami N, Scoazec M, Durand S, Enot DP, Fernández Á F, Martins I, Kepp O, Senovilla L, Bauvy C, Morselli E, Vacchelli E, Bennetzen M, Magnes C, Sinner F, Pieber T, López-Otín C, Maiuri MC, Codogno P, Andersen JS, Hill JA, Madeo F, Kroemer G, 2014. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell 53, 710–725. [DOI] [PubMed] [Google Scholar]
- Martinez-Lopez N, Garcia-Macia M, Sahu S, Athonvarangkul D, Liebling E, Merlo P, Cecconi F, Schwartz GJ, Singh R, 2016. Autophagy in the CNS and Periphery Coordinate Lipophagy and Lipolysis in the Brown Adipose Tissue and Liver. Cell Metab 23, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N, Tarabra E, Toledo M, Garcia-Macia M, Sahu S, Coletto L, Batista-Gonzalez A, Barzilai N, Pessin JE, Schwartz GJ, Kersten S, Singh R, 2017a. System-wide Benefits of Intermeal Fasting by Autophagy. Cell Metab 26, 856–871.e855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez N, Tarabra E, Toledo M, Garcia-Macia M, Sahu S, Coletto L, Batista-Gonzalez A, Barzilai N, Pessin JE, Schwartz GJ, Kersten S, Singh R, 2017b. System-wide Benefits of Intermeal Fasting by Autophagy. Cell Metab 26, 856–871 e855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Marques T, Catarino S, Zuzarte M, Marques C, Matafome P, Pereira P, Girão H, 2015. Ischaemia-induced autophagy leads to degradation of gap junction protein connexin43 in cardiomyocytes. Biochem J 467, 231–245. [DOI] [PubMed] [Google Scholar]
- Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM, 2006. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A 103, 5805–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheoud D, Sugiura A, Bellemare-Pelletier A, Laplante A, Rondeau C, Chemali M, Fazel A, Bergeron JJ, Trudeau LE, Burelle Y, Gagnon E, McBride HM, Desjardins M, 2016. Parkinson’s Disease-Related Proteins PINK1 and Parkin Repress Mitochondrial Antigen Presentation. Cell 166, 314–327. [DOI] [PubMed] [Google Scholar]
- Mayeuf-Louchart A, Lancel S, Sebti Y, Pourcet B, Loyens A, Delhaye S, Duhem C, Beauchamp J, Ferri L, Thorel Q, Boulinguiez A, Zecchin M, Dubois-Chevalier J, Eeckhoute J, Vaughn LT, Roach PJ, Dani C, Pederson BA, Vincent SD, Staels B, Duez H, 2019. Glycogen Dynamics Drives Lipid Droplet Biogenesis during Brown Adipocyte Differentiation. Cell reports 29, 1410–1418 e1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, Di Paola S, Peluso I, Armani A, De Stefani D, Venditti R, Montefusco S, Scotto-Rosato A, Prezioso C, Forrester A, Settembre C, Wang W, Gao Q, Xu H, Sandri M, Rizzuto R, De Matteis MA, Ballabio A, 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nature cell biology 17, 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlvang J, Olsvik H, Svenning S, Bruun JA, Abudu YP, Larsen KB, Brech A, Hansen TE, Brenne H, Hansen T, Stenmark H, Johansen T, 2018. Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J Cell Biol 217, 3640–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B, 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391. [DOI] [PubMed] [Google Scholar]
- Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, Fullgrabe J, Jackson A, Jimenez Sanchez M, Karabiyik C, Licitra F, Lopez Ramirez A, Pavel M, Puri C, Renna M, Ricketts T, Schlotawa L, Vicinanza M, Won H, Zhu Y, Skidmore J, Rubinsztein DC, 2017. Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron 93, 1015–1034. [DOI] [PubMed] [Google Scholar]
- Minnerly J, Zhang J, Parker T, Kaul T, Jia K, 2017. The cell non-autonomous function of ATG-18 is essential for neuroendocrine regulation of Caenorhabditis elegans lifespan. PLoS Genet 13, e1006764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, Fang E, Aon M, González-Reyes JA, Cortassa S, Kaushik S, Gonzalez-Freire M, Patel B, Wahl D, Ali A, Calvo-Rubio M, Burón MI, Guiterrez V, Ward TM, Palacios HH, Cai H, Frederick DW, Hine C, Broeskamp F, Habering L, Dawson J, Beasley TM, Wan J, Ikeno Y, Hubbard G, Becker KG, Zhang Y, Bohr VA, Longo DL, Navas P, Ferrucci L, Sinclair DA, Cohen P, Egan JM, Mitchell JR, Baur JA, Allison DB, Anson RM, Villalba JM, Madeo F, Cuervo AM, Pearson KJ, Ingram DK, Bernier M, de Cabo R, 2016. Effects of Sex, Strain, and Energy Intake on Hallmarks of Aging in Mice. Cell Metab 23, 1093–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, 2020. The ATG conjugation systems in autophagy. Curr Opin Cell Biol 63, 1–10. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y, 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Blas D, Gorostieta-Salas E, Pommer-Alba A, Mucino-Hernandez G, Geronimo-Olvera C, Maciel-Baron LA, Konigsberg M, Massieu L, Castro-Obregon S, 2019. Cortical neurons develop a senescence-like phenotype promoted by dysfunctional autophagy. Aging (Albany NY) 11, 6175–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI, 2020. Cell-nonautonomous regulation of proteostasis in aging and disease. Cold Spring Harbor Perspectives in Biology 12, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita H, Mizushima N, 2019. Diverse Cellular Roles of Autophagy. Annu Rev Cell Dev Biol 35, 453–475. [DOI] [PubMed] [Google Scholar]
- Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, Michaud M, Madeo F, Tavernarakis N, Kroemer G, 2010. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell death & disease 1, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, Stranks AJ, Glanville J, Knight S, Jacobsen S-EW, Kranc KR, Simon AK, 2011. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. The Journal of experimental medicine 208, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore GE, Lardeux BR, Adams CE, 1988. Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. J Biol Chem 263, 2506–2512. [PubMed] [Google Scholar]
- Mukherjee A, Patel B, Koga H, Cuervo AM, Jenny A, 2016. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy 12, 1984–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralimanoharan S, Gao X, Weintraub S, Myatt L, Maloyan A, 2016. Sexual dimorphism in activation of placental autophagy in obese women with evidence for fetal programming from a placenta-specific mouse model. Autophagy 12, 752–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Oba M, Suzuki M, Takahashi A, Yamamuro T, Fujiwara M, Ikenaka K, Minami S, Tabata N, Yamamoto K, Kubo S, Tokumura A, Akamatsu K, Miyazaki Y, Kawabata T, Hamasaki M, Fukui K, Sango K, Watanabe Y, Takabatake Y, Kitajima TS, Okada Y, Mochizuki H, Isaka Y, Antebi A, Yoshimori T, 2019. Suppression of autophagic activity by Rubicon is a signature of aging. Nat Commun 10, 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardin A, Schrepfer E, Ziviani E, 2016. Counteracting PINK/Parkin Deficiency in the Activation of Mitophagy: A Potential Therapeutic Intervention for Parkinson’s Disease. Curr Neuropharmacol 14, 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassour J, Radford R, Correia A, Fuste JM, Schoell B, Jauch A, Shaw RJ, Karlseder J, 2019. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 565, 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni C, Narzt MS, Nagelreiter IM, Zhang CF, Larue L, Rossiter H, Grillari J, Tschachler E, Gruber F, 2016. Autophagy deficient melanocytes display a senescence associated secretory phenotype that includes oxidized lipid mediators. Int J Biochem Cell Biol 81, 375–382. [DOI] [PubMed] [Google Scholar]
- Nie D, Zhang J, Zhou Y, Sun J, Wang W, Wang JH, 2021. Rapamycin Treatment of Tendon Stem/Progenitor Cells Reduces Cellular Senescence by Upregulating Autophagy. Stem Cells Int 2021, 6638249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie T, Tao K, Zhu L, Huang L, Hu S, Yang R, Xu P, Mao Z, Yang Q, 2020. Chaperone-mediated autophagy controls the turnover of E3 ubiquitin ligase MARCHF5 and regulates mitochondrial dynamics. Autophagy, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres JL, Leidal AM, Debnath J, Hansen M, 2021a. Beyond Autophagy: The Expanding Roles of ATG8 Proteins. Trends Biochem Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres JL, Shanahan SL, Chassefeyre R, Chaiamarit T, Zaretski S, Landeras-Bueno S, Verhelle A, Encalada SE, Hansen M, 2021b. LC3B phosphorylation regulates FYCO1 binding and directional transport of autophagosomes. Curr Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omari S, Makareeva E, Roberts-Pilgrim A, Mirigian L, Jarnik M, Ott C, Lippincott-Schwartz J, Leikin S, 2018. Noncanonical autophagy at ER exit sites regulates procollagen turnover. Proceedings of the National Academy of Sciences 115, E10099–E10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Yamano K, Sato M, Matsuda N, Okamoto K, 2021. Molecular mechanisms and physiological functions of mitophagy. EMBO J 40, e104705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S, Manjithaya R, 2020. Facets of Autophagy Based Unconventional Protein Secretion-The Road Less Traveled. Front Mol Biosci 7, 586483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N, 2015. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525–528. [DOI] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N, 2018. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nature cell biology 20, 1013–1022. [DOI] [PubMed] [Google Scholar]
- Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM, 2013. Functional interaction between autophagy and ciliogenesis. Nature 502, 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Suh Y, Cuervo AM, 2015. Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat Commun 6, 6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Fuentealba M, Kennedy BK, 2020. The quest to slow ageing through drug discovery. Nature reviews. Drug discovery 19, 513–532. [DOI] [PubMed] [Google Scholar]
- Pegg AE, 2016. Functions of Polyamines in Mammals. J Biol Chem 291, 14904–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Yang Q, Li AF, Li RQ, Wang Z, Liu LS, Ren Z, Zheng XL, Tang XQ, Li GH, Tang ZH, Jiang ZS, Wei DH, 2016. Tet methylcytosine dioxygenase 2 inhibits atherosclerosis via upregulation of autophagy in ApoE−/− mice. Oncotarget 7, 76423–76436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez VI, Buffenstein R, Masamsetti V, Leonard S, Salmon AB, Mele J, Andziak B, Yang T, Edrey Y, Friguet B, Ward W, Richardson A, Chaudhuri A, 2009. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci U S A 106, 3059–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesah Y, Pham T, Burgess H, Middlebrooks B, Verstreken P, Zhou Y, Harding M, Bellen H, Mardon G, 2004. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development 131, 2183–2194. [DOI] [PubMed] [Google Scholar]
- Pietrocola F, Castoldi F, Madeo F, Kroemer G, 2020. Triethylenetetramine (trientine): a caloric restriction mimetic with a new mode of action. Autophagy 16, 1534–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskovatska V, Stefanyshyn N, Storey KB, Vaiserman AM, Lushchak O, 2019. Metformin as a geroprotector: experimental and clinical evidence. Biogerontology 20, 33–48. [DOI] [PubMed] [Google Scholar]
- Piskovatska V, Storey KB, Vaiserman AM, Lushchak O, 2020. The Use of Metformin to Increase the Human Healthspan. Advances in experimental medicine and biology 1260, 319–332. [DOI] [PubMed] [Google Scholar]
- Prell T, Stubendorff B, Le TT, Gaur N, Tadić V, Rödiger A, Witte OW, Grosskreutz J, 2019. Reaction to Endoplasmic Reticulum Stress via ATF6 in Amyotrophic Lateral Sclerosis Deteriorates With Aging. Frontiers in Aging Neuroscience 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucciarelli S, Moreschini B, Micozzi D, De Fronzo GS, Carpi FM, Polzonetti V, Vincenzetti S, Mignini F, Napolioni V, 2012. Spermidine and spermine are enriched in whole blood of nona/centenarians. Rejuvenation Res 15, 590–595. [DOI] [PubMed] [Google Scholar]
- Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, Jung YK, 2013. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun 4, 2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Qiu Q, Gu X, Tian Y, Zhang Y, 2016. ATM mediates spermidine-induced mitophagy via PINK1 and Parkin regulation in human fibroblasts. Sci Rep 6, 24700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Wu D, Li M, Yan Z, Yang X, Ji N, Wang Y, Zhang J, 2020. The pluripotent role of exosomes in mediating non-coding RNA in ventricular remodeling after myocardial infarction. Life Sci 254, 117761. [DOI] [PubMed] [Google Scholar]
- Rana A, Rera M, Walker DW, 2013. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc Natl Acad Sci U S A 110, 8638–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz Y, Guerrero-Ros I, Maier A, Slagboom PE, Atzmon G, Barzilai N, Macian F, 2017. Activation-Induced Autophagy Is Preserved in CD4+ T-Cells in Familial Longevity. J Gerontol A Biol Sci Med Sci 72, 1201–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, Foiani M, 2011. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 471, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P, Moshitch-Moshkovitz S, Kvam E, O’Toole E, Winey M, Goldfarb DS, 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell 14, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RF, Tang MY, Fon E.a., Durcan TM, 2016. Defending the mitochondria: The pathways of mitophagy and mitochondrial-derived vesicles. The international journal of biochemistry & cell biology 79, 427–436. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, He L, Lemasters JJ, 2004. Role of mitochondrial permeability transition pores in mitochondrial autophagy. Int J Biochem Cell Biol 36, 2463–2472. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Muela N, Koga H, Garcia-Ledo L, de la Villa P, de la Rosa EJ, Cuervo AM, Boya P, 2013. Balance between autophagic pathways preserves retinal homeostasis. Aging Cell 12, 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro JA, Kaushik S, Koga H, Dall’Armi C, Shui G, Wenk MR, Di Paolo G, Cuervo AM, 2012. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci U S A 109, E705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Felix AA, Williams EG, Jha P, Lo Sasso G, Huzard D, Aebischer P, Sandi C, Rinsch C, Auwerx J, 2016. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med 22, 879–888. [DOI] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, Maser RS, Tonon G, Foerster F, Xiong R, Wang YA, Shukla SA, Jaskelioff M, Martin ES, Heffernan TP, Protopopov A, Ivanova E, Mahoney JE, Kost-Alimova M, Perry SR, Bronson R, Liao R, Mulligan R, Shirihai OS, Chin L, DePinho RA, 2011. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L, 2011. Microautophagy of cytosolic proteins by late endosomes. Developmental cell 20, 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S, 1998. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol 141, 625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamaki JI, Long JS, New M, Van Acker T, Tooze SA, Ryan KM, 2018. Emerging roles of transcriptional programs in autophagy regulation. Transcription 9, 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala AJ, Bott LC, Morimoto RI, 2017. Shaping proteostasis at the cellular, tissue, and organismal level. J Cell Biol 216, 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkis GJ, Ashcom JD, Hawdon JM, Jacobson LA, 1988. Decline in protease activities with age in the nematode Caenorhabditis elegans. Mech Ageing Dev 45, 191–201. [DOI] [PubMed] [Google Scholar]
- Sato M, Seki T, Konno A, Hirai H, Kurauchi Y, Hisatsune A, Katsuki H, 2019. Rapamycin activates mammalian microautophagy. J Pharmacol Sci 140, 201–204. [DOI] [PubMed] [Google Scholar]
- Sato M, Ueda E, Konno A, Hirai H, Kurauchi Y, Hisatsune A, Katsuki H, Seki T, 2020. Glucocorticoids negatively regulates chaperone mediated autophagy and microautophagy. Biochem Biophys Res Commun 528, 199–205. [DOI] [PubMed] [Google Scholar]
- Sato S, Uchihara T, Fukuda T, Noda S, Kondo H, Saiki S, Komatsu M, Uchiyama Y, Tanaka K, Hattori N, 2018. Loss of autophagy in dopaminergic neurons causes Lewy pathology and motor dysfunction in aged mice. Scientific Reports 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavi A, Maglioni S, Palikaras K, Shaik A, Strappazzon F, Brinkmann V, Torgovnick A, Castelein N, De Henau S, Braeckman BP, Cecconi F, Tavernarakis N, Ventura N, 2015. Iron-Starvation-Induced Mitophagy Mediates Lifespan Extension upon Mitochondrial Stress in C. elegans. Curr Biol 25, 1810–1822. [DOI] [PubMed] [Google Scholar]
- Schinaman JM, Rana A, Ja WW, Clark RI, Walker DW, 2019. Rapamycin modulates tissue aging and lifespan independently of the gut microbiota in Drosophila. Scientific Reports 9, 7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JL, Suh Y, Cuervo AM, 2014. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell metabolism 20, 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JL, Villarroya J, Diaz-Carretero A, Patel B, Urbanska AM, Thi MM, Villarroya F, Santambrogio L, Cuervo AM, 2015. Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell 14, 249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo AY, Lau PW, Feliciano D, Sengupta P, Gros MAL, Cinquin B, Larabell CA, Lippincott-Schwartz J, 2017. AMPK and vacuole-associated Atg14p orchestrate mu-lipophagy for energy production and long-term survival under glucose starvation. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A, 2011. TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang D, Wang L, Klionsky DJ, Cheng H, Zhou R, 2021. Sex differences in autophagy-mediated diseases: toward precision medicine. Autophagy 17, 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J, 2006. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem 281, 14474–14485. [DOI] [PubMed] [Google Scholar]
- Shin HJ, Kim H, Oh S, Lee JG, Kee M, Ko HJ, Kweon MN, Won KJ, Baek SH, 2016. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature 534, 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin WH, Chung KC, 2020. Human telomerase reverse transcriptase positively regulates mitophagy by inhibiting the processing and cytoplasmic release of mitochondrial PINK1. Cell death & disease 11, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, Huerta C, Virgin HW, Helms JB, Eerland R, Tooze SA, Xavier R, Lenschow DJ, Yamamoto A, King D, Lichtarge O, Grishin NV, Spector SA, Kaloyanova DV, Levine B, 2013. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD, 2008. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4, 176–184. [DOI] [PubMed] [Google Scholar]
- Singh K, Matsuyama S, Drazba JA, Almasan A, 2012. Autophagy-dependent senescence in response to DNA damage and chronic apoptotic stress. Autophagy 8, 236–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Chandrashekharappa S, Bodduluri SR, Baby BV, Hegde B, Kotla NG, Hiwale AA, Saiyed T, Patel P, Vijay-Kumar M, Langille MGI, Douglas GM, Cheng X, Rouchka EC, Waigel SJ, Dryden GW, Alatassi H, Zhang HG, Haribabu B, Vemula PK, Jala VR, 2019. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun 10, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Cuervo AM, 2011. Autophagy in the cellular energetic balance. Cell Metab 13, 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ, 2009. Autophagy regulates lipid metabolism. Nature 458, 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirois I, Groleau J, Pallet N, Brassard N, Hamelin K, Londono I, Pshezhetsky AV, Bendayan M, Hébert MJ, 2012. Caspase activation regulates the extracellular export of autophagic vacuoles. Autophagy 8, 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So EY, Jeong EM, Wu KQ, Dubielecka PM, Reginato AM, Quesenberry PJ, Liang OD, 2020. Sexual dimorphism in aging hematopoiesis: an earlier decline of hematopoietic stem and progenitor cells in male than female mice. Aging (Albany NY) 12, 25939–25955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JX, Sun YR, Peluso I, Zeng Y, Yu X, Lu JH, Xu Z, Wang MZ, Liu LF, Huang YY, Chen LL, Durairajan SS, Zhang HJ, Zhou B, Zhang HQ, Lu A, Ballabio A, Medina DL, Guo Z, Li M, 2016. A novel curcumin analog binds to and activates TFEB in vitro and in vivo independent of MTOR inhibition. Autophagy 12, 1372–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Victor P, Gutarra S, García-Prat L, Rodriguez-Ubreva J, Ortet L, Ruiz-Bonilla V, Jardí M, Ballestar E, González S, Serrano AL, Perdiguero E, Muñoz-Cánoves P, 2014. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316–321. [DOI] [PubMed] [Google Scholar]
- Stolz A, Ernst A, Dikic I, 2014. Cargo recognition and trafficking in selective autophagy. Nature cell biology 16, 495–501. [DOI] [PubMed] [Google Scholar]
- Sun B, Zhai S, Zhang L, Sun G, 2021. The role of extracellular vesicles in podocyte autophagy in kidney disease. J Cell Commun Signal 15, 299–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmstrom KM, Fergusson MM, Yoo YH, Combs CA, Finkel T, 2015. Measuring In Vivo Mitophagy. Mol Cell 60, 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai H, Wang Z, Gong H, Han X, Zhou J, Wang X, Wei X, Ding Y, Huang N, Qin J, Zhang J, Wang S, Gao F, Chrzanowska-Lightowlers ZM, Xiang R, Xiao H, 2017. Autophagy impairment with lysosomal and mitochondrial dysfunction is an important characteristic of oxidative stress-induced senescence. Autophagy 13, 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji F, Abdoli A, Baesi K, Sheikholeslami F, Kouchesfahani HM, 2021. Overexpression of Beclin1 gene leads to reduction of telomerase activity in MDCK cells and enhances apoptosis. J Cancer Res Ther 17, 225–230. [DOI] [PubMed] [Google Scholar]
- Talebian S, Daghagh H, Yousefi B, Ozkul Y, Ilkhani K, Seif F, Alivand MR, 2020. The role of epigenetics and non-coding RNAs in autophagy: A new perspective for thorough understanding. Mech Ageing Dev 190, 111309. [DOI] [PubMed] [Google Scholar]
- Tamargo-Gómez I, Mariño G, 2018. AMPK: Regulation of Metabolic Dynamics in the Context of Autophagy. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Yu CY, Sim ZW, Low ZS, Lee B, See F, Min N, Gautam A, Chu JJH, Ng KW, Wong E, 2019. Pomegranate activates TFEB to promote autophagy-lysosomal fitness and mitophagy. Sci Rep 9, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AH, Rando TA, 2014. Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J 33, 2782–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HZ, Yang LM, 2015. Activation of the unfolded protein response in aged human lenses. Mol Med Rep 12, 389–393. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G, 2008. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy 4, 870–873. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Hetz C, 2020. Mastering organismal aging through the endoplasmic reticulum proteostasis network. Aging Cell 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman A, 1995. The effect of age on formation and elimination of autophagic vacuoles in mouse hepatocytes. Gerontology 41 Suppl 2, 319–326. [DOI] [PubMed] [Google Scholar]
- Tian J, Popal MS, Zhao Y, Liu Y, Chen K, Liu Y, 2019. Interplay between Exosomes and Autophagy in Cardiovascular Diseases: Novel Promising Target for Diagnostic and Therapeutic Application. Aging Dis 10, 1302–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo M, Batista-Gonzalez A, Merheb E, Aoun ML, Tarabra E, Feng D, Sarparanta J, Merlo P, Botre F, Schwartz GJ, Pessin JE, Singh R, 2018. Autophagy Regulates the Liver Clock and Glucose Metabolism by Degrading CRY1. Cell Metab 28, 268–281 e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J, Pomatto LCD, Davies KJA, 2020. Sex differences in the response to oxidative and proteolytic stress. Redox biology 31, 101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Türei D, Földvári-Nagy L, Fazekas D, Módos D, Kubisch J, Kadlecsik T, Demeter A, Lenti K, Csermely P, Vellai T, Korcsmáros T, 2015. Autophagy Regulatory Network - a systems-level bioinformatics resource for studying the mechanism and regulation of autophagy. Autophagy 11, 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulgherait M, Rana A, Rera M, Graniel J, Walker DW, 2014. AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell reports 8, 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbán N, Cheung TH, 2021. Stem cell quiescence: the challenging path to activation. Development (Cambridge, England) 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uytterhoeven V, Lauwers E, Maes I, Miskiewicz K, Melo MN, Swerts J, Kuenen S, Wittocx R, Corthout N, Marrink SJ, Munck S, Verstreken P, 2015. Hsc70-4 Deforms Membranes to Promote Synaptic Protein Turnover by Endosomal Microautophagy. Neuron 88, 735–748. [DOI] [PubMed] [Google Scholar]
- Valdor R, García-Bernal D, Riquelme D, Martinez CM, Moraleda JM, Cuervo AM, Macian F, Martinez S, 2019. Glioblastoma ablates pericytes antitumor immune function through aberrant up-regulation of chaperone-mediated autophagy. Proc Natl Acad Sci U S A 116, 20655–20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdor R, Mocholi E, Botbol Y, Guerrero-Ros I, Chandra D, Koga H, Gravekamp C, Cuervo AM, Macian F, 2014. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nature immunology 15, 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broek B, Pintelon I, Hamad I, Kessels S, Haidar M, Hellings N, Hendriks JJA, Kleinewietfeld M, Brône B, Timmerman V, Timmermans JP, Somers V, Michiels L, Irobi J, 2020. Microglial derived extracellular vesicles activate autophagy and mediate multi-target signaling to maintain cellular homeostasis. Journal of extracellular vesicles 10, e12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhooren V, Navarrete Santos A, Voutetakis K, Petropoulos I, Libert C, Simm A, Gonos ES, Friguet B, 2015. Protein modification and maintenance systems as biomarkers of ageing. Mechanisms of ageing and development 151, 71–84. [DOI] [PubMed] [Google Scholar]
- Walters RO, Arias E, Diaz A, Burgos ES, Guan F, Tiano S, Mao K, Green CL, Qiu Y, Shah H, Wang D, Hudgins AD, Tabrizian T, Tosti V, Shechter D, Fontana L, Kurland IJ, Barzilai N, Cuervo AM, Promislow DEL, Huffman DM, 2018. Sarcosine Is Uniquely Modulated by Aging and Dietary Restriction in Rodents and Humans. Cell reports 25, 663–676 e666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Cai Z, Tao K, Zeng W, Lu F, Yang R, Feng D, Gao G, Yang Q, 2016a. Essential control of mitochondrial morphology and function by chaperone-mediated autophagy through degradation of PARK7. Autophagy 12, 1215–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Niederstrasser H, Douglas PM, Lin R, Jaramillo J, Li Y, Oswald NW, Zhou A, McMillan EA, Mendiratta S, Wang Z, Zhao T, Lin Z, Luo M, Huang G, Brekken RA, Posner BA, MacMillan JB, Gao J, White MA, 2017. Small-molecule TFEB pathway agonists that ameliorate metabolic syndrome in mice and extend C. elegans lifespan. Nat Commun 8, 2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, He SH, Liang X, Zhang XX, Li SS, Li TF, 2021. ATF4-modified serum exosomes derived from osteoarthritic mice inhibit osteoarthritis by inducing autophagy. IUBMB Life 73, 146–158. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang N, Zhang L, Li R, Fu W, Ma K, Li X, Wang L, Wang J, Zhang H, Gu W, Zhu WG, Zhao Y, 2016b. Autophagy Regulates Chromatin Ubiquitination in DNA Damage Response through Elimination of SQSTM1/p62. Mol Cell 63, 34–48. [DOI] [PubMed] [Google Scholar]
- Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, Debnath J, Passegue E, 2013. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494, 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei FZ, Cao Z, Wang X, Wang H, Cai MY, Li T, Hattori N, Wang D, Du Y, Song B, Cao LL, Shen C, Wang L, Wang H, Yang Y, Xie D, Wang F, Ushijima T, Zhao Y, Zhu WG, 2015. Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy 11, 2309–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng H, Ma Y, Chen L, Cai G, Chen Z, Zhang S, Ye Q, 2020. A New Vision of Mitochondrial Unfolded Protein Response to the Sirtuin Family. Current Neuropharmacology 18, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm T, Byrne J, Medina R, Kolundzic E, Geisinger J, Hajduskova M, Tursun B, Richly H, 2017. Neuronal inhibition of the autophagy nucleation complex extends life span in post-reproductive C. elegans. Genes Dev 31, 1561–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth A, Wolf B, Huang CK, Glage S, Hofer SJ, Bankstahl M, Bar C, Thum T, Kahl KG, Sigrist SJ, Madeo F, Bankstahl JP, Ponimaskin E, 2021. Novel aspects of age-protection by spermidine supplementation are associated with preserved telomere length. Geroscience 43, 673–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Liu J, Ma J, Yan Q, Jiang Z, 2021. Neoagarotetraose extends the lifespan of Caenorhabditis elegans through AMPK mediated signaling pathways and activation of autophagy. Journal of Functional Foods 77, 104341. [Google Scholar]
- Wu X, Rapoport TA, 2018. Mechanistic insights into ER-associated protein degradation. Current Opinion in Cell Biology 53, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao FH, Chen XQ, Yu Q, Ye Y, Liu YW, Yan D, Yang LQ, Chen G, Lin R, Yang L, Liao X, Zhang W, Zhang W, Tang NL, Wang XF, Zhou J, Cai WW, He YH, Kong QP, 2018. Transcriptome evidence reveals enhanced autophagy-lysosomal function in centenarians. Genome Res 28, 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Tan J, Miao Y, Lv Y, Zhang Q, 2021. Crosstalk between exosomes and autophagy: A review of molecular mechanisms and therapies. J Cell Mol Med 25, 2297–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Liqi Z, Jian L, Qinghua Y, Qian Y, 2017. Doxycycline Induces Mitophagy and Suppresses Production of Interferon-beta in IPEC-J2 Cells. Front Cell Infect Microbiol 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang L, Fozouni P, Evjen G, Chandra V, Jiang J, Lu C, Nicastri M, Bretz C, Winkler JD, Amaravadi R, Garcia BA, Adams PD, Ott M, Tong W, Johansen T, Dou Z, Berger SL, 2020a. SIRT1 is downregulated by autophagy in senescence and ageing. Nature cell biology 22, 1170–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu TT, Li H, Dai Z, Lau GK, Li BY, Zhu WL, Liu XQ, Liu HF, Cai WW, Huang SQ, Wang Q, Zhang SJ, 2020b. Spermidine and spermine delay brain aging by inducing autophagy in SAMP8 mice. Aging (Albany NY) 12, 6401–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang Y, García-Cañaveras JC, Guo L, Kan M, Yu S, Blair IA, Rabinowitz JD, Yang X, 2020c. Chaperone-mediated autophagy regulates the pluripotency of embryonic stem cells. Science 369, 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahata T, Takanashi T, Muguruma Y, Ibrahim AA, Matsuzawa H, Uno T, Sheng Y, Onizuka M, Ito M, Kato S, Ando K, 2011. Accumulation of oxidative DNA damage restricts the self-renewal capacity of human hematopoietic stem cells. Blood 118, 2941–2950. [DOI] [PubMed] [Google Scholar]
- Yang Q, Zheng C, Cao J, Cao G, Shou P, Lin L, Velletri T, Jiang M, Chen Q, Han Y, Li F, Wang Y, Cao W, Shi Y, 2016. Spermidine alleviates experimental autoimmune encephalomyelitis through inducing inhibitory macrophages. Cell Death Differ 23, 1850–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B, 2006. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A 103, 10793–10798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G, Yu B, Liu C, Lin Y, Xie Z, Hu Y, Lin H, 2021. Exosomes produced by adipose-derived stem cells inhibit schwann cells autophagy and promote the regeneration of the myelin sheath. Int J Biochem Cell Biol 132, 105921. [DOI] [PubMed] [Google Scholar]
- Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM, Narita M, 2009. Autophagy mediates the mitotic senescence transition. Genes Dev 23, 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ARJ, Cassidy LD, Narita M, 2021. Autophagy and senescence, converging roles in pathophysiology as seen through mouse models. Adv Cancer Res 150, 113–145. [DOI] [PubMed] [Google Scholar]
- Yue F, Li W, Zou J, Jiang X, Xu G, Huang H, Liu L, 2017. Spermidine Prolongs Lifespan and Prevents Liver Fibrosis and Hepatocellular Carcinoma by Activating MAP1S-Mediated Autophagy. Cancer research 77, 2938–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cuervo AM, 2008. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med 14, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Alsaleh G, Feltham J, Sun Y, Napolitano G, Riffelmacher T, Charles P, Frau L, Hublitz P, Yu Z, Mohammed S, Ballabio A, Balabanov S, Mellor J, Simon AK, 2019. Polyamines Control eIF5A Hypusination, TFEB Translation, and Autophagy to Reverse B Cell Senescence. Mol Cell 76, 110–125 e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JA, Zhou BR, Xu Y, Chen X, Liu J, Gozali M, Wu D, Yin ZQ, Luo D, 2016. MiR-23a-depressed autophagy is a participant in PUVA- and UVB-induced premature senescence. Oncotarget 7, 37420–37435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Dai Y, Song M, Zhou Y, Zhou J, Yan X, Shen Y, 2021. The decrease of intraflagellar transport impairs sensory perception and metabolism in ageing. Nat Commun 12, 1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, 2002. A mitochondrial specific stress response in mammalian cells. The EMBO Journal 21, 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Heber D, Wang M, Gao C, Heymsfield SB, Martin RJ, Greenway FL, Finley JW, Burton JH, Johnson WD, Enright FM, Keenan MJ, Li Z, 2017. Pomegranate juice and extract extended lifespan and reduced intestinal fat deposition in Caenorhabditis elegans. International journal for vitamin and nutrition research. Internationale Zeitschrift fur Vitamin-und Ernahrungsforschung. Journal international de vitaminologie et de nutrition 87, 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Sun C, Ren J, Wang G, Ma R, Sun L, Yang D, Gao S, Ning K, Wang Z, Chen X, Chen S, Zhu H, Gao Z, Xu J, 2019. Stress-induced precocious aging in PD-patient iPSC-derived NSCs may underlie the pathophysiology of Parkinson’s disease. Cell death & disease 10, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin J, Nieuwenhuis J, Perrimon N, 2013. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol 11, e1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]


