Abstract
Alcohol use produces wide-ranging, diverse effects on the central nervous system. It influences intracellular signaling mechanisms, leading to changes in gene expression, chromatin remodeling and translation. As a result of these molecular alterations, alcohol affects the activity of neuronal circuits. Together, these mechanisms produce long-lasting cellular adaptations in the brain that in turn can drive the development and maintenance of alcohol use disorder. Here, we provide an update on alcohol research, focusing on multiple levels of alcohol-induced adaptations, from intracellular ones to changes in neural circuits. A better understanding of how alcohol affects these diverse and interlinked mechanisms may lead to the identification of novel therapeutic targets and to the development of much-needed novel, efficacious treatment options.
Keywords: alcohol, central nervous system, intracellular signaling, epigenetics, neural circuits
Alcohol-induced neuronal adaptations
Alcohol use disorder (AUD) affects about 10–15% of the global population, causing significant medical, social, and economic burdensi. While most drinkers consume alcohol for years without escalating to excessive use, a subset of people develop harmful drinking patterns [1]. Unfortunately, efficacious treatment options are limited [2], due in part to the complex and multi-faceted ways by which intake of alcohol affects the nervous system. Both acute and chronic alcohol exposure produce molecular and cellular neuroadaptations influencing the activity of discrete brain regions and cell types [3–5]. Consequently, these neuronal adaptations that underlie AUD are influenced by diverse interactions between alcohol and intracellular signaling, the epigenome, neurotransmitters and modulators as well as the activity of neuronal circuits, which in turn drive behaviors such as heavy alcohol use, anxiety, craving, and relapse.
Here, we review recent literature focusing on alcohol-induced neuronal adaptations. We discuss seven distinct, but tightly interlinked levels of alcohol’s effects on the brain, starting from genetic factors that confer susceptibility to AUD (Level 1) through alcohol-induced changes in epigenetic mechanisms (Level 2), transcriptional activity (Level 3), alternative splicing (Level 4), translation (Level 5) and post-translational modifications (Level 6) to circuit-level activity (Level 7). We discuss molecular mechanisms that contribute to the development of this disorder, and describe evidence outlining potential new avenues for medication development for the treatment of AUD. Finally, we consider recent work examining how alcohol-induced plasticity manifests on the level of neural circuit activity and release of neuromodulators to influence decisions of when and how much to drink.
Level 1: Genetic factors in AUD
Common and rare variants in specific genes play a role in both the resilience and susceptibility mechanisms that protect against or promote AUD, respectively [6–9]. Among the most validated functional variants associated with AUD are alcohol dehydrogenase 1B (ADH1B) and aldehyde dehydrogenase 2 (ALDH2), both of which participate in alcohol metabolism in the liver [10]. Interestingly, single nucleotide polymorphisms (SNPs) in both ALDH1B and ALDH2 are protective against the development of AUD [10]. Of note, ALDH2 in cerebellar astrocytes promotes alcohol metabolism, GABA production and intoxication [11]. A recent genome-wide meta-analysis study in 435,563 individuals of European descent has identified SNPs linked to AUD and problem drinking [7]. Among the identified variants are a number of ADH enzymes as well as in genes such as PDE4B, SYNGAP and BDNF [7].
A non-synonymous SNP has also been identified within the brain-derived neurotropic factor (BDNF) gene (G196A, rs6265) that results in the substitution of Valine66 to Methionine66. Carriers of the SNP display early onset of relapse [12], and transgenic knock-in mice carrying the mutation consume excessive amounts of alcohol despite negative consequences [13], a phenotype which is rescued by viral overexpression of wild-type BDNF in the medial prefrontal cortex (mPFC) or via the activation of the BDNF receptor, TrkB [13]. Another example is a SNP in the EE hand-domain containing 2 (EFHD2) gene [14]. This SNP is associated with lifelong drinking as well as a negative association with anxiety phenotypes in healthy adolescence [14], and mice lacking EFhd2 consume more alcohol [14]. These data suggest that both BDNF and EFHD2 are “resilience” genes and that malfunction of either BDNF or EFhd2 contributes to the development of AUD.
Furthermore, an SNP in the gene encoding β-Klotho, a component of the hormone receptors for fibroblast growth factors FGF19 and FGF21, was highly correlated with alcohol consumption in a large genome wide association (GWAS) study of individuals without AUD [15]. Brain-specific knockout of β-Klotho increased alcohol consumption and preference in mice by a yet unknown mechanism [15]. Interestingly, high expression of β-Klotho has previously been observed in mesolimbic regions in mice, such as the nucleus accumbens (NAc) and ventral tegmental area (VTA), which are central to reward seeking behavior [16]. In addition, a polymorphism in Ras suppressor 1 (RSU1), involved in the regulation of neuronal actin dynamics through RAC1, has been shown to influence reward anticipation and alcohol consumption in humans [17]. In contrast, an SNP in TRAF family member-associated NF-κB activator (TANK) is associated with reduced drinking in humans [15]. A study in Tank knockout mice showed that these animals exhibit reduced alcohol drinking and preference, and further studies revealed that Tank is involved in the regulation of alcohol-dependent insular cortical activation of nuclear factor kB (NF-kB) in mice [18].
Level 2: Alcohol-induced alterations in epigenetic regulation
An emerging body of evidence suggests that epigenetic regulation represents an important level of alcohol-induced molecular adaptations in the brain (Figure 1). This umbrella term includes covalent modifications of DNA and histones (e.g., methylation, acetylation) resulting in the condensation or relaxation of chromatin (complex of DNA and associated proteins) leading to repression or enhancement of gene expression, respectively. Importantly, these mechanisms change the landscape of genome-accessibility and the proteome, which in turn contribute to alterations in neuronal cytoarchitecture and activity, and have a critical role in the development of substance use disorders including AUD. For example, alcohol self-administration in macaques was shown to decrease DNA methylation at certain genomic loci in a dose-dependent manner [19]. Interestingly, several of the affected genes were related to synaptic function, including genes encoding proteins that control neurotransmitter release or receptor trafficking [19], providing further evidence that epigenetic regulation plays a role in alcohol-induced synaptic plasticity.
Figure 1. Molecular pathways in alcohol use disorder.

Alcohol binds to a number of transmembrane receptors including glutamate, GABA and dopamine receptors, as well as receptors of different neuropeptides and neurotrophic factors. These in turn affect the activity of several second messenger cascades and intracellular signaling pathways. These pathways mediate long-lasting cellular adaptations affecting, among others, translation and synaptic plasticity, which contribute to neuronal adaptations underlying AUD. In the nucleus of neurons, alcohol has complex effects on the epigenetic regulation of gene expression. These complex and highly interlinked pathways activate specific gene expression programs, which underlie neuronal maladaptations and contribute to the development of alcohol use disorder.
Abbreviations: ACSS2: acetyl-CoA synthetase 2, ADNP: activity dependent neuroprotector homeobox, ALK: anaplastic lymphoma kinase, BDNF: brain-derived neurotrophic factor, cAMP: cyclic adenosine monophosphate, CBP: CREB(cAMP response element-binding protein)-binding protein, CRMP2: collapsing response mediator protein family 2, SRC: sarcoma, D1R: dopamine receptor D1, DNTM1: DNA methyltransferase 1, ERK1/2: extracellular signal-regulated kinase 1/2, FGF2: fibroblast growth factor 2, FGFR1: FGF receptor 1, FMRP: fragile X mental retardation protein, GABAAR: gamma aminobutyric acid receptor A, GDNF: glial cell line-derived neurotrophic factor, GFRa1: GDNF family receptor alpha 1, GluN2B: glutamate N-methyl-D-aspartate receptor subunit 2B, GSK3b: glycogen synthase kinase 3b, IKKb: inhibitor of nuclear factor kappa-B kinase subunit beta, KDM6B: lysine demethylase 6B, LMO4: LIM domain only 4, mTORC1/2: mammalian target of rapamycin complex 1/2, PDE: phosphodiesterase, NFkB: nuclear factor kappa-B, PKA: protein kinase A, PKCe: protein kinase C epsilon, PI3K: phosphoinositide-3-kinase, Prosapip1: Prosap2(prolin-rich synapse-associated protein 2) interacting protein 1, Ret: Rearranged during transfection, Su(H): suppressor of hairless, TNF: tumor necrosis factor, TrkB: tropomyosin receptor kinase B. Figure was created with BioRender.com.
Acute and chronic exposure to alcohol can have opposite effects on epigenetic regulation. For instance, while acute alcohol exposure increased histone acetylation and decreased histone methylation in the central amygdala (CeA), chronic intermittent exposure had opposite effects [20,21]. These findings suggest that the epigenetic landscape undergoes adaptations that might play an important role in the development of AUD.
Epigenetic pathways are tightly interlinked, resulting in increased complexity of alcohol-induced epigenetic dysregulation. For example, chronic exposure to alcohol led to long-lasting reduction of H3K27ac and parallel induction of H3K27me3 at the immediate early gene Arc in the CeA of rats [22]. These acetylation/methylation changes resulted in decreased expression of the non-coding Arc eRNA (enhancer RNA; short non-coding RNAs transcribed from enhancers) and affected Arc transcription [22]. These findings emphasize that alcohol does not affect specific epigenetic mechanisms in a vacuum, and the potential interaction of these regulatory pathways is critical to consider.
Alcohol-induced epigenetic alterations are often mediated by altered expression or activity of epigenetic enzymes, which thus represent a promising new avenue for targeted therapeutic interventions. For example, increased enrichment of DNA methylation in the mPFC was linked to enhanced DNA methyltransferase (Dnmt) activity [23]. Inhibition of Dnmt rescued the methylation and transcriptional changes and prevented the escalation of alcohol intake [23]. Other examples include Cbp and p300 [20], as well as lysine demethylase Lsd1 [21]. Decreased binding of Cbp and lysine demethylase Kdm6b was also shown at specific target genes upon adolescent intermittent alcohol exposure, resulting in anxiety-like behaviors in adult rats [22].
Recently, a previously unanticipated mechanism was identified linking alcohol metabolism to alcohol-induced epigenetic impairments by way of direct incorporation of alcohol-derived acetate into brain histone acetylation [24]. This was driven by the nuclear translocation of metabolic enzyme acetyl-CoA synthetase 2 (Acss2), inhibition of which prevented alcohol-induced changes of histone acetylation and gene expression, and blocked conditioned place preference to alcohol [24]. This and related epigenetic-metabolic pathways [25] represent a radically novel mechanism of alcohol-induced transcriptional changes.
Level 3: Alcohol’s effects on transcriptional activity
Transcription factors often form large multimeric protein complexes that bind to target gene promoters or enhancers to regulate the expression of mRNA. Chronic alcohol exposure in rodents upregulates gene expression in neurons, astrocytes, and microglia [26–28], which raises the possibility that transcription factors serve as one of the master regulators of the neuroadaptations induced by alcohol. The mechanisms that drive alcohol-dependent transcriptional alterations are still being unraveled (Figure 1). For example, the transcriptional activity of NF-κB is controlled through the stimulation of the inhibitor κβ kinase (IKKβ). Using pharmacologic and genetic approaches, Ikkβ was shown to contribute to excessive alcohol intake in mice [29], and its action is localized to neurons at least in the NAc and CeA [29]. Another example is the transcriptional regulator, LIM Domain Only 4 (Lmo4), which was shown to drive vast changes in gene expression in the basolateral amygdala (BLA) of mice in response to repeated exposure to alcohol and to the regulation of alcohol intake [30]. In addition to contributing to the mechanisms that drive excessive drinking (GO signaling), transcription factors are likely to contribute to the gating of alcohol intake (STOP signaling). For example, the activity-dependent neuroprotective protein (Adnp) is a transcription factor that protects against excessive alcohol intake and relapse in female rodents [31].
Level 4: Alternative splicing
An important mechanism underlying phenotypic variability is alternative splicing, which allows for the expression of various transcripts from a single gene. Intriguingly, environmental insults including repeated exposure to alcohol have been shown to impair this mechanism across many species. In postmortem analyses of the striatum and amygdala of individuals with AUD, thousands of transcripts in these brain regions were differentially spliced [32]. Relatedly, hundreds of splice variants were affected by in utero alcohol exposure in fetal cortical tissue [33]. Alcohol-induced impairments of alternative splicing were observed across several species, facilitating the study of these mechanisms in preclinical models of alcohol use. For example, changes in RNA splicing have been identified in the cortex of rodents and monkeys following chronic alcohol exposure [34]. In mice, acute exposure to a single injection of alcohol induced differential expression of over 10,000 exons. Strikingly, these changes showed a large overlap with those observed in mice treated with anti-depressants, potentially outlining a novel mechanism for the rapid antidepressant effects of alcohol [35]. In the brain of fruit flies, lasting changes in the expression of distinct splice variants were linked to alcohol-related associative learning and knock-down of spliceosome-associated proteins prevented the formation of alcohol memories [36]. Further, alcohol was shown to alter dopamine D2R splicing in the reward circuitry of flies via activation of Notch signaling, leading to its interaction with the transcription factor suppressor of hairless Su(H) [37]. This pathway underlies memory formation for alcohol-associated cues.
Splicing of mRNA molecules can also occur at distant cellular compartments including the synapse, thus having a direct effect on the activity of neuronal circuits. Intriguingly, alcohol markedly perturbs the synaptic spliceosome in the cortex of mice, thereby affecting the local translation of proteins involved in synaptic function [38]. These changes are particularly pronounced following repeated exposure to alcohol and were proposed to regulate sensitization [38].
Level 5: Alcohol and protein translation
The kinase mTOR in complex 1 (mTORC1) plays a crucial role in synaptic plasticity, learning and memory by orchestrating the translation of several dendritic proteins [39]. mTORC1 is activated by alcohol in discrete brain regions resulting in the translation of synaptic proteins such as Collapsin response-mediated protein 2 (CRMP2) [40] and ProSap-interacting protein 1 (Prosapip1) [41], as well as Homer1 and PSD-95, GluA2 and Arc [40,42,43]. Through the translation of these transcripts and others, mTORC1 contributes to mechanisms underlying alcohol seeking and drinking as well as reconsolidation of alcohol reward memories and habit [44–46]. Further, protein translation plays a role in additional alcohol-dependent phenotypes (Figure 1). For example, the activity of mRNA binding protein fragile-X mental retardation protein (Fmrp), which plays an important role in translation [47], is enhanced by alcohol in the hippocampus of mice resulting in alteration in the expression of synaptic proteins [48]. Additionally, Fmrp in the hippocampus plays a role in the acute antidepressant actions of alcohol [49]. Interestingly, rapid antidepressants require coordinated actions of Fmrp and mTORC1 [50], raising the possibility that such coordination may also be relevant in the context of alcohol’s actions.
Level 6: The role of posttranslational modifications
Posttranslational modifications such as phosphorylation are core molecular signaling events. Not surprisingly, protein kinases play a central role in AUD (Figure 1) [3]. For instance, the protein tyrosine kinase (PTK) Fyn, through the phosphorylation of GluN2B in the dorsomedial striatum (DMS) of rodents, contributes to molecular and cellular neuroadaptations that drive goal-directed alcohol consumption [51,52]. Interestingly, Fyn also plays a role in heroin use [53], suggesting a more generalized role of the kinase in addiction. Furthermore, GsDREADD-dependent activation of the serine/threonine kinase protein kinase A (Pka) in the DMS of mice activates Fyn specifically in D1R MSNs to enhance alcohol consumption, suggesting that Pka is upstream of Fyn [54]. Indeed, a large body of evidence supports the role of Pka signaling in the actions of alcohol [3]. Interestingly, phosphodiesterase 4 and 10a (Pde4 and Pde10a), enzymes required for the termination of Pka activity [55], have also been implicated in AUD [56]. Furthermore, a genome-wide association study identified PDE4B as a risk factor in elevated alcohol consumption [6,7]. Both Pka’s and Pde’s intracellular compartmentalization are tightly regulated [55], and it is highly likely that this is reflected by the seemingly opposing actions of alcohol on components of the Pka signaling cascade. Repeated alcohol exposure in mice activates another PTK, Src, which in turn stimulates Nf-κB/Tnfα signaling in microglia, resulting in microglia engulfment of mPFC synapses, as well as synaptic pruning and increased anxiety-like behaviors [57]. Another serine/threonine kinase that participates in neuroadaptations underlying AUD is GSK3β [58]. Specifically, Gsk3β in the mPFC participates in mechanisms underlying motivation to consume alcohol and alcohol withdrawal-induced anxiety [58]. Furthermore, genetic analysis in humans indicated that GSK3β is an alcohol dependence risk factor, suggesting a central role of GSK3β in AUD [58]. Surprisingly however, Gsk3β in the NAc is inhibited by alcohol in rats [40], emphasizing the region-specificity of alcohol’s action. Like Fyn, the kinase mTORC2 is specifically activated by alcohol in the DMS of mice [59]. Alcohol-dependent activation of mTORC2 in the DMS promotes F-actin assembly, the formation for mature spines and alcohol intake [59].
Additionally, receptor tyrosine kinases (RTKs) which are activated by growth factors and cytokines play a role in alcohol consumption [60]. For example, alcohol-dependent activation of the anaplastic lymphoma kinase (Alk) in the hippocampus and PFC activates STAT signaling leading to changes in gene expression, and systemic administration of Alk or Stat3 inhibitors attenuates alcohol intake in mice [61,62]. Surprisingly, a number of growth factors/RTKs such as Bdnf and the glial-derived neurotrophic factor (Gdnf) are endogenous factors that limit alcohol use [60,63]. Interestingly, activation of Midkine/Alk signaling also acts to limit alcohol intake in mice [64,65]. In contrast to Bdnf, Gdnf and Midkine, fibroblast growth factor 2 (Fgf2)/Fgf receptor 1 (Fgfr1) signaling promotes excessive drinking in rodents [66,67].
Level 7: Impact of chronic drinking on neuromodulators and neural circuits
Recent advances in neurotechnologies have opened new avenues of investigation into how alcohol-induced alterations in neural circuit activity influence ongoing behaviors and decision-making (Figure 2) [4,68]. Here we will review these advances, focusing on circuit- and receptor-level studies (for review of brain-wide neuronal networks see [69]). Recently, a genome-wide transcriptional assessment of human striatum found that G protein coupled receptors, the primary targets of many neurotransmitters and neuromodulators, were the top canonical pathway affected in striatum of AUD patients [70]. Reverse translation of these findings into a rodent model demonstrated putative therapeutic potential for a positive allosteric modulator of the muscarinic M4 receptor which, when delivered systemically in rats, reduced a wide range of alcohol self-administration behaviors [70].
Figure 2. Neuronal circuits affected by alcohol.
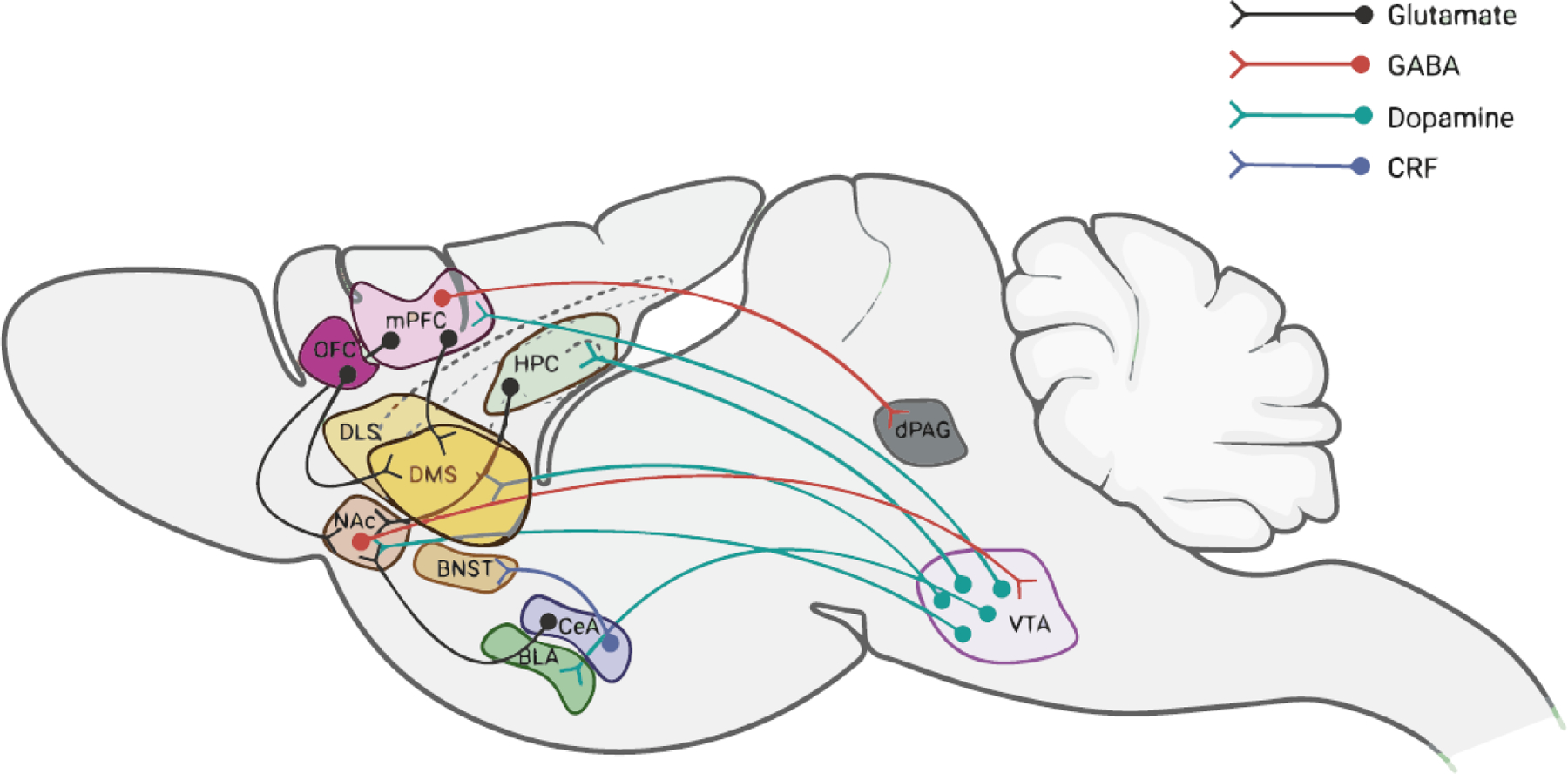
Acute and chronic use of alcohol affects the activity of multiple neuronal circuits, depicted here schematically in the context of a rodent brain. For example, alcohol activates the mesocorticolimbic brain reward circuit, which encompasses dopaminergic projections from the VTA in the midbrain to several forebrain structures including the striatum and cortex. These circuits underlie the rewarding effects of alcohol. Anxiolytic effects of ethanol have been linked to the amygdala. In addition, CRF neurons projecting from the central amygdala to the BNST were shown to contribute to the escalation of alcohol intake. Prefrontal cortical circuits have been implicated in impaired executive control that underlies excessive drinking, as well as weakened cognitive function in AUD. For example, projections from the mPFC to the dorsal striatum have been linked to habitual alcohol drinking and continued use despite negative consequences. Further, neurons projecting from the mPFC to the dPAG play a critical role in compulsive drinking. Strikingly, mice that display inhibitory activity in this circuit during the first alcohol exposure are more likely to develop compulsive drinking behavior.
Abbreviations: BLA: basolateral amygdala; BNST: bed nucleus of the stria terminalis; CeA: central amygdala; CRF: corticotropin releasing factor; DLS: dorsolateral striatum; DMS: dorsomedial striatum; HPC: hippocampus; LA: lateral amygdala; mPFC: medial prefrontal cortex; NAc: nucleus accumbensm; dPAG: dorsal periaqueductal gray; OFC: orbitofrontal cortex; VTA: ventral tegmental area. Figure was created with BioRender.com.
Several recent studies have built on classic literature to further detail the mechanisms by which presynaptic dopamine signaling and postsynaptic activity of medium spiny neurons (MSNs) orchestrate motivated behavior and its dysregulation by chronic alcohol drinking [71,72]. While dopamine signaling in the striatum has long been known to be critical in regulating alcohol drinking behaviors [73], precise monitoring and manipulation of striatal dopamine release with dopamine biosensors and chemogenetics revealed complex, subregion-specific dopamine release patterns underlying alcohol drinking and seeking in rodents [71,72]. In addition, alcohol also engages feeding circuits in the hypothalamus which in turn indirectly modulates dopamine neuron activity [74]. Studies in animal models indicate that following long-term use of alcohol, striatal circuits and receptors undergo a range of adaptations [75,76]. While the specifics vary between males and females and across brain regions, these adaptations are generally thought to be critical determinants in dysregulated drinking behaviors.
Multiple classes of neuropeptide releasing neurons and neuropeptide receptors have been implicated as critical mediators of drinking behaviors, such as neurotensin [77], neuropeptide Y [78], oxytocin [79], opioid peptides [80,81] and corticotrophin-releasing factor (CRF). For instance, in rats and mice, chronic alcohol use alters the activity of the CeA through dysregulation of endocannabinoid, substance P, and corticotrophin releasing factor signaling [82–84]. The bed nucleus of the stria terminalis (BNST) also exhibits plasticity in endocannabinoids and CRF- expressing neurons due to chronic alcohol use, and these alterations modulate drinking, withdrawal-induced negative affect, and stress-induced alcohol seeking in mice [85,86]. Furthermore, the CeA and BNST regions are anatomically connected, and inhibition of CRF neurons projecting from the CeA to the BNST decreases escalation of alcohol intake and somatic withdrawal symptoms in rats [87].
The kappa-opioid receptor (KOR) and its endogenous ligand dynorphin peptide have been an area of great interest. Reduced dynorphin activity or blockade of KORs in several brain regions including the CeA [88,89], BNST [90,91], and the striatum, reduce alcohol consumption in mice and rats. KORs have also been shown to modulate the acute actions of alcohol [92], negative affect during withdrawal [93], and the sensitivity of this receptor is augmented after chronic alcohol use [73]. Fast-acting and selective KOR antagonists have been developed and evaluated in preclinical models using rats, yielding promising results that suggest therapeutic potential for treating AUD [94].
A major theme of recent alcohol research has been to leverage animal models and circuit-analysis approaches to link neural circuit activity with specific aspects of AUD [95]. For example, in mice, chronic alcohol exposure decreased the excitability of OFC outputs to the DMS [96], and alcohol-induced synaptic plasticity in the OFC has been linked to excessive alcohol use in both mice and monkeys models [97,98]. In addition, using a combination of activity dependent genetic tools and chemogenetic manipulations, a small ensemble of mPFC neurons was shown to serve as a memory to cue induced relapse to alcohol use [99]. Interestingly, like the molecular mechanisms that gate the development of AUD [3], STOP mechanisms also occur on the level of circuitries [100]. Specifically, a subset of infralimbic cortical neurons serve to protect against relapse to alcohol use [100].
Projections from mPFC to the striatum have been implicated in mediating specific aspects of drinking behaviors [101–103]. These projections have been targeted to exert bidirectional, long-lasting control of alcohol drinking [103]. Specifically, using optogenetic stimulation protocols to induce plasticity at cortical synapses onto dopamine D1R MSNs in the DMS, recent studies showed that induction of long-term potentiation (LTP) resulted in lasting increases in drinking while induction of long-term depression (LTD) at these synapses produced decreased drinking [101,103]. Furthermore, dysregulation of striatal function can produce pathological drinking behaviors. For instance, manipulations of striatal dopamine D2 receptors (D2Rs), adenosine 2A receptors, or activity of fast-spiking interneurons, among others, alter excessive drinking behaviors [104–106]. Further, disrupted GABAergic transmission in this region is also linked to alcohol-induced cognitive impairments [107]. Together, altered excitability of striatal neurons and upstream cortical regulation of striatal activity influence a diverse range of drinking behaviors, which likely can be attributed to distinct striatal output circuits [108].
Concluding remarks and future perspectives
Advances in neuroscience continue to shed light onto regulatory mechanisms relevant for alcohol use. A striking example is the discovery that certain neurotransmitters, such as serotonin [109] and dopamine [110], can covalently bind to histones and act as epigenetic marks to regulate gene expression. Histone dopaminylation was further shown to influence addiction-like behaviors in the context of cocaine exposure in mice [110]. This novel mechanism could have far reaching implications for other drugs of abuse, including alcohol, which are known to increase dopamine levels in the mesolimbic system [72]. Another example of a recent discovery facilitated by novel approaches is that aldehyde dehydrogenase 2 (ALDH2) in cerebellar astrocytes promotes alcohol metabolism, GABA production and ethanol-induced intoxication in mice [11]. Importantly, the neurobiological basis of AUD appears in many cases to manifest in a sex-specific manner. Understanding convergence and divergence between mechanisms in males and females will continue to be critical moving forward [111,112].
Signaling events that produce transcriptional and translational changes are, in essence, the molecular transducers of the long-lasting cellular adaptations that drive AUD, and targeting some of these signaling molecules could provide an avenue for the development of new therapeutics [12]. An area of focus for drug development efforts has been kinases: over the past 20 years or so, this class of signaling molecules has been one of the leading targets for drug development by pharmaceutical and biotechnology companies [113]. Examples of kinase inhibitors that show promise in preclinical rodent AUD models are the ALK inhibitors NVP-TAE684 and alectinib [61], Fyn kinase inhibitor AZD0530 [52], and mTORC1 inhibitor, Rapamycin [12]. Although Rapamycin is already used in the clinic for indications such as prevention of organ rejection after kidney transplantation [114], its potential use for the treatment of AUD is hampered by adverse effects due to the inhibition of mTORC1 in the periphery [115]. To overcome this limitation, a dual drug strategy which enables the selective inhibition of mTORC1 in the brain while preserving its activity in the periphery was recently developed [116,117]. This strategy, which at least in mice eliminates the side effects that result from prolonged mTORC1 in the periphery, has shown promising results in preclinical alcohol drinking models [117]. Another promising target is epsilon protein kinase C (εPKC), a kinase long known to be linked to AUD [118,119]. New, selective small molecule inhibitors to this kinase have been recently developed, showing promising results in preclinical mouse models of AUD [120]. Other potential inhibitors are MY10, which targets receptor protein tyrosine phosphatase beta/zeta (PTPRZ1) [121], Lacosimide, which prevents CRMP2-dependent microtubule assembly [40], and PDE inhibitors such as NCT02025998, NCT01860807 and ibudilast [56]. Finally, a potential treatment approach that emerged recently in the context of AUD is ketogenic diet, characterized by high fat and low carbohydrate intake. In a study in rats, ketogenic diet has been shown to reduce withdrawal-induced symptoms and craving, as well as alcohol self-administration [122].
Another area requiring further research relates to individual differences in resilience and susceptibility to AUD. Future studies are needed to better understand the mechanisms underlying these individual differences. Studies in animal models provide initial hints to possible contributors to these differences. Studies in outbred rats have shown that about 15% of the animals prefer alcohol to a sweetened solution and display AUD-like phenotypes such as consumption of alcohol despite negative consequences [123], and much of this variance can be explained by differences in excitability of PKCδ expressing interneurons in the CeA [124]. Furthermore, rats undergoing intermittent access to 20% alcohol in 2 bottle choice paradigm exhibit distinct profiles of intake ranging from low alcohol consumers to rats that exhibit slow or rapid escalation of excessive drinking [125]. In a recently-developed behavioral procedure for quantifying the development of compulsive drinking in mice, the activity of a small population of neurons that connect the mPFC to the dorsal periaqueductal gray area were shown to confer vulnerability to compulsive alcohol drinking, and activity in this circuit differentiated subjects prone to compulsive alcohol drinking prior to the expression of the behavior [126].
Together, the studies reviewed earlier illustrate the complexity of AUD, which results from the interaction of the various levels of molecular neuroadaptations in different brain regions and neural circuit changes throughout the brain [127]. The specific molecular pathways and circuits that could serve as the most promising therapeutic targets remain to be delineated (see Outstanding Questions). Finally, the development of cutting-edge tools for neurotransmitter sensing, circuitry mapping and manipulation on a more precise spatial and temporal scale will enable further advances in our understanding of how neural activity and communication are altered by chronic alcohol use to produce excessive drinking behaviors.
Outstanding questions.
Recent studies have outlined several levels of alcohol’s effects on the brain. The precise mechanistic links between these levels, however, remain in many cases unknown. How do the specific levels of alcohol-induced adaptations interact and influence each other to result in complex behavioral changes?
Which molecular pathways and circuits could serve as the most promising potential therapeutic targets for alcohol use disorder (AUD)? Given that AUD is a multifaceted disorder, combinatorial therapeutic approaches that target multiple levels of alcohol-induced maladaptations might be most effective in treating AUD and related conditions.
Which mechanisms determine individual vulnerability to excessive alcohol consumption? Only a small proportion of individuals consuming alcohol go on to develop AUD. A more complete understanding of the molecular pathways, circuits and behavioral traits that contribute to individual and sex differences in susceptibility could guide targeted approaches in treatment and prevention.
Can alcohol-dependent changes in the molecular landscape be explained, at least in part, by spatial and/or temporal alterations of protein compartmentalization? For instance, lipid rafts consisting of membranal lipids and cholesterol serve as important platforms for signaling pathways. Alcohol is known to alter membranal lipid fluidity—does alcohol therefore affect signaling cascades by altering lipid raft composition?
AUD is a polygenic disorder with relatively minor contributions from individual genes. How is it possible that manipulating single genes in animal models can result in animals that are resilient to AUD? Does this limit the translational validity of findings from available animal models?
AUD has a significant heritable component, yet only a relatively small number of single nucleotide polymorphisms have been identified as associated with AUD. Are there additional, currently unknown variants that confer susceptibility for or resilience against developing problem drinking and AUD? An integrative approach in combination with large sample size may reveal additional insights.
Highlights:
Alcohol affects several levels of brain function, from cellular and molecular pathways to circuit-level activity
The effects of alcohol on the various levels of brain function are closely intertwined, and underlie different aspects of alcohol use disorder
Understanding this diverse and interlinked molecular, cellular and circuit landscape will help guide the development of future therapeutic approaches
Acknowledgments:
The authors were supported by the National Institute of Alcohol Abuse and Alcoholism K99AA028577 (GE), R00DA04510 (CAS), R37AA01684 (DR), R01AA027474 (DR) and R01AA027682 (DR), Alkermes Pharmaceuticals (CAS) and the Brain Research Foundation (CAS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests: The authors declare no competing interests in relation to this work.
References
- 1.Koob GF (2021) Drug Addiction: Hyperkatifeia/Negative Reinforcement as a Framework for Medications Development. Pharmacol Rev 73, 163–201. 10.1124/pharmrev.120.000083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvalho AF et al. (2019) Alcohol use disorders. Lancet 394, 781–792. 10.1016/S0140-6736(19)31775-1 [DOI] [PubMed] [Google Scholar]
- 3.Ron D and Barak S (2016) Molecular mechanisms underlying alcohol-drinking behaviours. Nat Rev Neurosci 17, 576–591. 10.1038/nrn.2016.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abrahao KP et al. (2017) Alcohol and the Brain: Neuronal Molecular Targets, Synapses, and Circuits. Neuron 96, 1223–1238. 10.1016/j.neuron.2017.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandey SC et al. (2017) Epigenetic basis of the dark side of alcohol addiction. Neuropharmacology 122, 74–84. 10.1016/j.neuropharm.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke TK et al. (2017) Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry 22, 1376–1384. 10.1038/mp.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H et al. (2020) Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat Neurosci 23, 809–818. 10.1038/s41593-020-0643-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng Q et al. (2019) Genetic loci for alcohol-related life events and substance-induced affective symptoms: indexing the “dark side” of addiction. Transl Psychiatry 9, 71. 10.1038/s41398-019-0397-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y et al. (2021) Identification of novel risk loci with shared effects on alcoholism, heroin, and methamphetamine dependence. Mol Psychiatry 26, 1152–1161. 10.1038/s41380-019-0497-y [DOI] [PubMed] [Google Scholar]
- 10.Edenberg HJ and McClintick JN (2018) Alcohol Dehydrogenases, Aldehyde Dehydrogenases, and Alcohol Use Disorders: A Critical Review. Alcohol Clin Exp Res 42, 2281–2297. 10.1111/acer.13904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin S et al. (2021) Brain ethanol metabolism by astrocytic ALDH2 drives the behavioural effects of ethanol intoxication. Nat Metab 3, 337–351. 10.1038/s42255-021-00357-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ron D and Berger A (2018) Targeting the intracellular signaling “STOP” and “GO” pathways for the treatment of alcohol use disorders. Psychopharmacology (Berl) 235, 1727–1743. 10.1007/s00213-018-4882-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warnault V et al. (2016) The BDNF Valine 68 to Methionine Polymorphism Increases Compulsive Alcohol Drinking in Mice That Is Reversed by Tropomyosin Receptor Kinase B Activation. Biol Psychiatry 79, 463–473. 10.1016/j.biopsych.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mielenz D et al. (2018) EFhd2/Swiprosin-1 is a common genetic determinator for sensation-seeking/low anxiety and alcohol addiction. Mol Psychiatry 23, 1303–1319. 10.1038/mp.2017.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumann G et al. (2016) KLB is associated with alcohol drinking, and its gene product beta-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci U S A 113, 14372–14377. 10.1073/pnas.1611243113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talukdar S et al. (2016) FGF21 Regulates Sweet and Alcohol Preference. Cell metabolism 23, 344–349. 10.1016/j.cmet.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ojelade SA et al. (2015) Rsu1 regulates ethanol consumption in Drosophila and humans. Proc Natl Acad Sci U S A 112, E4085–4093. 10.1073/pnas.1417222112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller CP et al. (2019) The Cortical Neuroimmune Regulator TANK Affects Emotional Processing and Enhances Alcohol Drinking: A Translational Study. Cereb Cortex 29, 1736–1751. 10.1093/cercor/bhy341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cervera-Juanes R et al. (2017) Alcohol-dose-dependent DNA methylation and expression in the nucleus accumbens identifies coordinated regulation of synaptic genes. Transl Psychiatry 7, e994. 10.1038/tp.2016.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H et al. (2018) Adolescent alcohol exposure epigenetically regulates CREB signaling in the adult amygdala. Sci Rep 8, 10376. 10.1038/s41598-018-28415-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyzar EJ et al. (2017) Adolescent alcohol exposure alters lysine demethylase 1 (LSD1) expression and histone methylation in the amygdala during adulthood. Addict Biol 22, 1191–1204. 10.1111/adb.12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyzar EJ et al. (2019) MicroRNA-137 Drives Epigenetic Reprogramming in the Adult Amygdala and Behavioral Changes after Adolescent Alcohol Exposure. eNeuro 6. 10.1523/ENEURO.0401-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barbier E et al. (2015) DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J Neurosci 35, 6153–6164. 10.1523/JNEUROSCI.4571-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mews P et al. (2019) Alcohol metabolism contributes to brain histone acetylation. Nature 574, 717–721. 10.1038/s41586-019-1700-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egervari G et al. (2020) Food for thought. Science 370, 660–662. 10.1126/science.abb4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erickson EK et al. (2018) Astrocyte-specific transcriptome responses to chronic ethanol consumption. Pharmacogenomics J 18, 578–589. 10.1038/s41397-017-0012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brenner E et al. (2020) Single cell transcriptome profiling of the human alcohol-dependent brain. Hum Mol Genet 29, 1144–1153. 10.1093/hmg/ddaa038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warden AS et al. (2020) Microglia Control Escalation of Drinking in Alcohol-Dependent Mice: Genomic and Synaptic Drivers. Biol Psychiatry 88, 910–921. 10.1016/j.biopsych.2020.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truitt JM et al. (2016) Inhibition of IKKbeta Reduces Ethanol Consumption in C57BL/6J Mice. eNeuro 3. 10.1523/ENEURO.0256-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maiya R et al. (2020) Differential regulation of alcohol consumption and reward by the transcriptional cofactor LMO4. Mol Psychiatry. 10.1038/s41380-020-0706-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziv Y et al. (2019) Activity-dependent neuroprotective protein (ADNP) is an alcohol-responsive gene and negative regulator of alcohol consumption in female mice. Neuropsychopharmacology 44, 415–424. 10.1038/s41386-018-0132-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Booven D et al. (2021) Alcohol use disorder causes global changes in splicing in the human brain. Transl Psychiatry 11, 2. 10.1038/s41398-020-01163-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawasawa YI et al. (2017) Genome-wide profiling of differentially spliced mRNAs in human fetal cortical tissue exposed to alcohol. Alcohol 62, 1–9. 10.1016/j.alcohol.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogenpohl JW et al. (2019) Cross-Species Co-analysis of Prefrontal Cortex Chronic Ethanol Transcriptome Responses in Mice and Monkeys. Front Mol Neurosci 12, 197. 10.3389/fnmol.2019.00197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolfe SA et al. (2019) Ethanol and a rapid-acting antidepressant produce overlapping changes in exon expression in the synaptic transcriptome. Neuropharmacology 146, 289–299. 10.1016/j.neuropharm.2018.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petruccelli E et al. (2020) Alcohol Causes Lasting Differential Transcription in Drosophila Mushroom Body Neurons. Genetics 215, 103–116. 10.1534/genetics.120.303101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petruccelli E et al. (2018) Alcohol Activates Scabrous-Notch to Influence Associated Memories. Neuron 100, 1209–1223 e1204. 10.1016/j.neuron.2018.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Brien MA et al. (2018) Ethanol-Induced Behavioral Sensitization Alters the Synaptic Transcriptome and Exon Utilization in DBA/2J Mice. Front Genet 9, 402. 10.3389/fgene.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saxton RA and Sabatini DM (2017) mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976. 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F et al. (2017) mTORC1-dependent translation of collapsin response mediator protein-2 drives neuroadaptations underlying excessive alcohol-drinking behaviors. Mol Psychiatry 22, 89–101. 10.1038/mp.2016.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laguesse S et al. (2017) Prosapip1-Dependent Synaptic Adaptations in the Nucleus Accumbens Drive Alcohol Intake, Seeking, and Reward. Neuron 96, 145–159 e148. 10.1016/j.neuron.2017.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neasta J et al. (2010) Role for mammalian target of rapamycin complex 1 signaling in neuroadaptations underlying alcohol-related disorders. Proc Natl Acad Sci U S A 107, 20093–20098. 10.1073/pnas.1005554107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beckley JT et al. (2016) The First Alcohol Drink Triggers mTORC1-Dependent Synaptic Plasticity in Nucleus Accumbens Dopamine D1 Receptor Neurons. J Neurosci 36, 701–713. 10.1523/JNEUROSCI.2254-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben Hamida S et al. (2019) Mammalian target of rapamycin complex 1 and its downstream effector collapsin response mediator protein-2 drive reinstatement of alcohol reward seeking. Addict Biol 24, 908–920. 10.1111/adb.12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morisot N et al. (2019) mTORC1 in the orbitofrontal cortex promotes habitual alcohol seeking. Elife 8. 10.7554/eLife.51333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barak S et al. (2013) Disruption of alcohol-related memories by mTORC1 inhibition prevents relapse. Nat Neurosci 16, 1111–1117. 10.1038/nn.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darnell JC and Klann E (2013) The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci 16, 1530–1536. 10.1038/nn.3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spencer KB et al. (2016) FMRP Mediates Chronic Ethanol-Induced Changes in NMDA, Kv4.2, and KChIP3 Expression in the Hippocampus. Alcohol Clin Exp Res 40, 1251–1261. 10.1111/acer.13060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfe SA et al. (2016) FMRP regulates an ethanol-dependent shift in GABABR function and expression with rapid antidepressant properties. Nat Commun 7, 12867. 10.1038/ncomms12867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heaney CF et al. (2021) Role of FMRP in rapid antidepressant effects and synapse regulation. Mol Psychiatry. 10.1038/s41380-020-00977-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morisot N and Ron D (2017) Alcohol-dependent molecular adaptations of the NMDA receptor system. Genes Brain Behav 16, 139–148. 10.1111/gbb.12363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morisot N et al. (2019) The Fyn kinase inhibitor, AZD0530, suppresses mouse alcohol self-administration and seeking. Addict Biol 24, 1227–1234. 10.1111/adb.12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Egervari G et al. (2020) Chromatin accessibility mapping of the striatum identifies tyrosine kinase FYN as a therapeutic target for heroin use disorder. Nat Commun 11, 4634. 10.1038/s41467-020-18114-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ehinger Y et al. (2021) cAMP-Fyn signaling in the dorsomedial striatum direct pathway drives excessive alcohol use. Neuropsychopharmacology 46, 334–342. 10.1038/s41386-020-0712-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baillie GS et al. (2019) Therapeutic targeting of 3’,5’-cyclic nucleotide phosphodiesterases: inhibition and beyond. Nat Rev Drug Discov 18, 770–796. 10.1038/s41573-019-0033-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Logrip ML (2015) Phosphodiesterase regulation of alcohol drinking in rodents. Alcohol 49, 795–802. 10.1016/j.alcohol.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Socodato R et al. (2020) Daily alcohol intake triggers aberrant synaptic pruning leading to synapse loss and anxiety-like behavior. Sci Signal 13. 10.1126/scisignal.aba5754 [DOI] [PubMed] [Google Scholar]
- 58.van der Vaart A et al. (2018) Glycogen synthase kinase 3 beta regulates ethanol consumption and is a risk factor for alcohol dependence. Neuropsychopharmacology 43, 2521–2531. 10.1038/s41386-018-0202-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Laguesse S et al. (2018) mTORC2 in the dorsomedial striatum of mice contributes to alcohol-dependent F-Actin polymerization, structural modifications, and consumption. Neuropsychopharmacology 43, 1539–1547. 10.1038/s41386-018-0012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamada K and Lasek AW (2020) Receptor Tyrosine Kinases as Therapeutic Targets for Alcohol Use Disorder. Neurotherapeutics 17, 4–16. 10.1007/s13311-019-00795-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dutton JW 3rd et al. (2017) Anaplastic lymphoma kinase regulates binge-like drinking and dopamine receptor sensitivity in the ventral tegmental area. Addict Biol 22, 665–678. 10.1111/adb.12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hamada K et al. (2021) Binge-like ethanol drinking activates anaplastic lymphoma kinase signaling and increases the expression of STAT3 target genes in the mouse hippocampus and prefrontal cortex. Genes Brain Behav, e12729. 10.1111/gbb.12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barak S et al. (2019) GDNF and alcohol use disorder. Addict Biol 24, 335–343. 10.1111/adb.12628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen H et al. (2017) Midkine in the mouse ventral tegmental area limits ethanol intake and Ccl2 gene expression. Genes Brain Behav 16, 699–708. 10.1111/gbb.12384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mangieri RA et al. (2017) Anaplastic Lymphoma Kinase Is a Regulator of Alcohol Consumption and Excitatory Synaptic Plasticity in the Nucleus Accumbens Shell. Front Pharmacol 8, 533. 10.3389/fphar.2017.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Even-Chen O et al. (2017) Fibroblast Growth Factor 2 in the Dorsomedial Striatum Is a Novel Positive Regulator of Alcohol Consumption. J Neurosci 37, 8742–8754. 10.1523/JNEUROSCI.0890-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Even-Chen O and Barak S (2019) Inhibition of FGF Receptor-1 Suppresses Alcohol Consumption: Role of PI3 Kinase Signaling in Dorsomedial Striatum. J Neurosci 39, 7947–7957. 10.1523/JNEUROSCI.0805-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lovinger DM and Alvarez VA (2017) Alcohol and basal ganglia circuitry: Animal models. Neuropharmacology 122, 46–55. 10.1016/j.neuropharm.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zahr NM et al. (2017) Perspectives on fronto-fugal circuitry from human imaging of alcohol use disorders. Neuropharmacology 122, 189–200. 10.1016/j.neuropharm.2017.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker LC et al. (2020) Acetylcholine Muscarinic M4 Receptors as a Therapeutic Target for Alcohol Use Disorder: Converging Evidence From Humans and Rodents. Biol Psychiatry 88, 898–909. 10.1016/j.biopsych.2020.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Y et al. (2020) The Mesolimbic Dopamine Activity Signatures of Relapse to Alcohol-Seeking. J Neurosci 40, 6409–6427. 10.1523/JNEUROSCI.0724-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valyear MD et al. (2020) Dissociable mesolimbic dopamine circuits control responding triggered by alcohol-predictive discrete cues and contexts. Nat Commun 11, 3764. 10.1038/s41467-020-17543-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siciliano CA et al. (2018) Cross-Species Alterations in Synaptic Dopamine Regulation After Chronic Alcohol Exposure. Handb Exp Pharmacol 248, 213–238. 10.1007/164_2018_106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alhadeff AL et al. (2019) Natural and Drug Rewards Engage Distinct Pathways that Converge on Coordinated Hypothalamic and Reward Circuits. Neuron 103, 891–908 e896. 10.1016/j.neuron.2019.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trantham-Davidson H et al. (2017) Binge-Like Alcohol Exposure During Adolescence Disrupts Dopaminergic Neurotransmission in the Adult Prelimbic Cortex. Neuropsychopharmacology 42, 1024–1036. 10.1038/npp.2016.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salinas AG et al. (2021) Long-term alcohol consumption alters dorsal striatal dopamine release and regulation by D2 dopamine receptors in rhesus macaques. Neuropsychopharmacology. 10.1038/s41386-020-00938-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Torruella-Suarez ML et al. (2020) Manipulations of Central Amygdala Neurotensin Neurons Alter the Consumption of Ethanol and Sweet Fluids in Mice. J Neurosci 40, 632–647. 10.1523/JNEUROSCI.1466-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robinson SL et al. (2019) Medial prefrontal cortex neuropeptide Y modulates binge-like ethanol consumption in C57BL/6J mice. Neuropsychopharmacology 44, 1132–1140. 10.1038/s41386-018-0310-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tunstall BJ et al. (2019) Oxytocin blocks enhanced motivation for alcohol in alcohol dependence and blocks alcohol effects on GABAergic transmission in the central amygdala. PLoS Biol 17, e2006421. 10.1371/journal.pbio.2006421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Munoz B et al. (2018) Alcohol exposure disrupts mu opioid receptor-mediated long-term depression at insular cortex inputs to dorsolateral striatum. Nat Commun 9, 1318. 10.1038/s41467-018-03683-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patton MH et al. (2016) Ethanol Disinhibits Dorsolateral Striatal Medium Spiny Neurons Through Activation of A Presynaptic Delta Opioid Receptor. Neuropsychopharmacology 41, 1831–1840. 10.1038/npp.2015.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khom S et al. (2020) Alcohol dependence potentiates substance P/neurokinin-1 receptor signaling in the rat central nucleus of amygdala. Sci Adv 6, eaaz1050. 10.1126/sciadv.aaz1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Serrano A et al. (2018) Deficient endocannabinoid signaling in the central amygdala contributes to alcohol dependence-related anxiety-like behavior and excessive alcohol intake. Neuropsychopharmacology 43, 1840–1850. 10.1038/s41386-018-0055-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Varodayan FP et al. (2017) Chronic alcohol exposure disrupts CB1 regulation of GABAergic transmission in the rat basolateral amygdala. Addict Biol 22, 766–778. 10.1111/adb.12369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Centanni SW et al. (2019) Endocannabinoid control of the insular-bed nucleus of the stria terminalis circuit regulates negative affective behavior associated with alcohol abstinence. Neuropsychopharmacology 44, 526–537. 10.1038/s41386-018-0257-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salimando GJ et al. (2020) BNST GluN2D-Containing NMDA Receptors Influence Anxiety- and Depressive-like Behaviors and ModulateCell-Specific Excitatory/Inhibitory Synaptic Balance. J Neurosci 40, 3949–3968. 10.1523/JNEUROSCI.0270-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.de Guglielmo G et al. (2019) Inactivation of a CRF-dependent amygdalofugal pathway reverses addiction-like behaviors in alcohol-dependent rats. Nat Commun 10, 1238. 10.1038/s41467-019-09183-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anderson RI et al. (2019) Dynorphin-kappa opioid receptor activity in the central amygdala modulates binge-like alcohol drinking in mice. Neuropsychopharmacology 44, 1084–1092. 10.1038/s41386-018-0294-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bloodgood DW et al. (2020) Kappa opioid receptor and dynorphin signaling in the central amygdala regulates alcohol intake. Mol Psychiatry. 10.1038/s41380-020-0690-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haun HL et al. (2020) Kappa opioid receptors in the bed nucleus of the stria terminalis regulate binge-like alcohol consumption in male and female mice. Neuropharmacology 167, 107984. 10.1016/j.neuropharm.2020.107984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Le AD et al. (2018) Role of kappa-Opioid Receptors in the Bed Nucleus of Stria Terminalis in Reinstatement of Alcohol Seeking. Neuropsychopharmacology 43, 838–850. 10.1038/npp.2017.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Karkhanis AN et al. (2016) Switch from excitatory to inhibitory actions of ethanol on dopamine levels after chronic exposure: Role of kappa opioid receptors. Neuropharmacology 110, 190–197. 10.1016/j.neuropharm.2016.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Erikson CM et al. (2018) Maladaptive behavioral regulation in alcohol dependence: Role of kappa-opioid receptors in the bed nucleus of the stria terminalis. Neuropharmacology 140, 162–173. 10.1016/j.neuropharm.2018.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Domi E et al. (2018) Preclinical evaluation of the kappa-opioid receptor antagonist CERC-501 as a candidate therapeutic for alcohol use disorders. Neuropsychopharmacology 43, 1805–1812. 10.1038/s41386-018-0015-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lovinger DM and Gremel CM (2021) A Circuit-Based Information Approach to Substance Abuse Research. Trends Neurosci 44, 122–135. 10.1016/j.tins.2020.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Renteria R et al. (2018) Chronic alcohol exposure disrupts top-down control over basal ganglia action selection to produce habits. Nat Commun 9, 211. 10.1038/s41467-017-02615-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nimitvilai S et al. (2017) Ethanol Dependence Abolishes Monoamine and GIRK (Kir3) Channel Inhibition of Orbitofrontal Cortex Excitability. Neuropsychopharmacology 42, 1800–1812. 10.1038/npp.2017.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nimitvilai S et al. (2017) Orbitofrontal Neuroadaptations and Cross-Species Synaptic Biomarkers in Heavy-Drinking Macaques. J Neurosci 37, 3646–3660. 10.1523/JNEUROSCI.0133-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Visser E et al. (2020) A persistent alcohol cue memory trace drives relapse to alcohol seeking after prolonged abstinence. Sci Adv 6, eaax7060. 10.1126/sciadv.aax7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laque A et al. (2019) Anti-relapse neurons in the infralimbic cortex of rats drive relapse-suppression by drug omission cues. Nat Commun 10, 3934. 10.1038/s41467-019-11799-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roltsch Hellard E et al. (2019) Optogenetic control of alcohol-seeking behavior via the dorsomedial striatal circuit. Neuropharmacology 155, 89–97. 10.1016/j.neuropharm.2019.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Halladay LR et al. (2020) Prefrontal Regulation of Punished Ethanol Self-administration. Biol Psychiatry 87, 967–978. 10.1016/j.biopsych.2019.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ma T et al. (2018) Bidirectional and long-lasting control of alcohol-seeking behavior by corticostriatal LTP and LTD. Nat Neurosci 21, 373–383. 10.1038/s41593-018-0081-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bocarsly ME et al. (2019) A Mechanism Linking Two Known Vulnerability Factors for Alcohol Abuse: Heightened Alcohol Stimulation and Low Striatal Dopamine D2 Receptors. Cell Rep 29, 1147–1163 e1145. 10.1016/j.celrep.2019.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hong SI et al. (2019) Indirect Medium Spiny Neurons in the Dorsomedial Striatum Regulate Ethanol-Containing Conditioned Reward Seeking. J Neurosci 39, 7206–7217. 10.1523/JNEUROSCI.0876-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Patton MS et al. (2021) Compulsive alcohol consumption is regulated by dorsal striatum fast-spiking interneurons. Neuropsychopharmacology 46, 351–359. 10.1038/s41386-020-0766-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cuzon Carlson VC et al. (2020) Gestational alcohol exposure disrupts cognitive function and striatal circuits in adult offspring. Nat Commun 11, 2555. 10.1038/s41467-020-16385-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gibson GD et al. (2018) Distinct Accumbens Shell Output Pathways Promote versus Prevent Relapse to Alcohol Seeking. Neuron 98, 512–520 e516. 10.1016/j.neuron.2018.03.033 [DOI] [PubMed] [Google Scholar]
- 109.Farrelly LA et al. (2019) Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 567, 535–539. 10.1038/s41586-019-1024-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lepack AE et al. (2020) Dopaminylation of histone H3 in ventral tegmental area regulates cocaine seeking. Science 368, 197–201. 10.1126/science.aaw8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Becker JB and Koob GF (2016) Sex Differences in Animal Models: Focus on Addiction. Pharmacol Rev 68, 242–263. 10.1124/pr.115.011163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vandegrift BJ et al. (2020) Estrogen Receptor alpha Regulates Ethanol Excitation of Ventral Tegmental Area Neurons and Binge Drinking in Female Mice. J Neurosci 40, 5196–5207. 10.1523/JNEUROSCI.2364-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roskoski R Jr. (2020) Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol Res 152, 104609. 10.1016/j.phrs.2019.104609 [DOI] [PubMed] [Google Scholar]
- 114.Podbielski J and Schoenberg L (2001) Use of sirolimus in kidney transplantation. Prog Transplant 11, 29–32. 10.7182/prtr.11.1.5235m177144q114j [DOI] [PubMed] [Google Scholar]
- 115.Li J et al. (2014) Rapamycin: one drug, many effects. Cell Metab 19, 373–379. 10.1016/j.cmet.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang Z et al. (2020) Brain-Restricated mTOR Inhibiton with Binary Pharmacology. BioRxiv, doi: 10.1101/2020.10.12.336677 [DOI] [Google Scholar]
- 117.Ehinger Y et al. (2021) Brain-specific inhibition of mTORC1 eliminates side effects resulting from mTORC1 blockade in the periphery and reduces alcohol intake in mice. Nat Commun 12, 4407. 10.1038/s41467-021-24567-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cozzoli DK et al. (2016) Protein Kinase C Epsilon Activity in the Nucleus Accumbens and Central Nucleus of the Amygdala Mediates Binge Alcohol Consumption. Biol Psychiatry 79, 443–451. 10.1016/j.biopsych.2015.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hodge CW et al. (1999) Supersensitivity to allosteric GABA(A) receptor modulators and alcohol in mice lacking PKCepsilon. Nat Neurosci 2, 997–1002. 10.1038/14795 [DOI] [PubMed] [Google Scholar]
- 120.Blasio A et al. (2018) Novel Small-Molecule Inhibitors of Protein Kinase C Epsilon Reduce Ethanol Consumption in Mice. Biol Psychiatry 84, 193–201. 10.1016/j.biopsych.2017.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Calleja-Conde J et al. (2020) Inhibition of Receptor Protein Tyrosine Phosphatase beta/zeta Reduces Alcohol Intake in Rats. Alcohol Clin Exp Res 44, 1037–1045. 10.1111/acer.14321 [DOI] [PubMed] [Google Scholar]
- 122.Wiers CE et al. (2021) Ketogenic diet reduces alcohol withdrawal symptoms in humans and alcohol intake in rodents. Sci Adv 7. 10.1126/sciadv.abf6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Augier E et al. (2018) A molecular mechanism for choosing alcohol over an alternative reward. Science 360, 1321–1326. 10.1126/science.aao1157 [DOI] [PubMed] [Google Scholar]
- 124.Domi E et al. (2021) A neural substrate of compulsive alcohol use. Sci Adv 7. 10.1126/sciadv.abg9045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ehinger Y et al. (2021) Differential correlation of serum BDNF and microRNA content in rats with rapid or late onset of heavy alcohol use. Addict Biol 26, e12890. 10.1111/adb.12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Siciliano CA et al. (2019) A cortical-brainstem circuit predicts and governs compulsive alcohol drinking. Science 366, 1008–1012. 10.1126/science.aay1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kimbrough A et al. (2020) Brain-wide functional architecture remodeling by alcohol dependence and abstinence. Proc Natl Acad Sci U S A 117, 2149–2159. 10.1073/pnas.1909915117 [DOI] [PMC free article] [PubMed] [Google Scholar]


