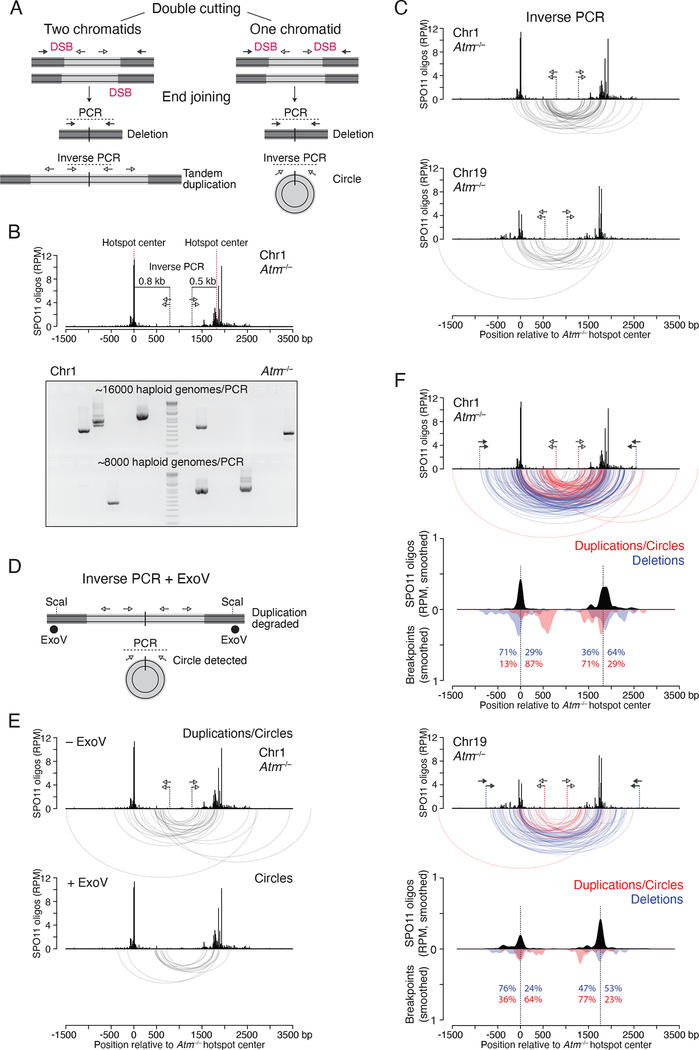
Figure 3. Tandem duplications and circular DNA from double cutting.
(A) Predicted end-joining products from double cutting in one or two chromatids. Lighter grey indicates DNA segments between two DSBs at adjacent hotspots. Inverse PCR detects both tandem duplications and circles.
(B) Inverse PCR assay at the Chr1 hotspot pair. A 1.3-kb PCR product would arise from a tandem duplication or circular DNA from end joining of double cuts at the two hotspot centers. Lower panel, inverse PCR products detected in Atm−/− testis DNA.
(C) Arc diagrams of breakpoints for duplications/circles at the Chr1 (42 events) and Chr19 (22 events) hotspot pairs in Atm−/−.
(D) Strategy to detect only circular DNA products by inverse PCR. ExoV is used to degrade linear DNA containing tandem duplications. ScaI produces entry sites for ExoV. See also Figure S4B.
(E) Arc diagrams of duplications/circles (27 events) and circles (13 events) at the Chr1 hotspot pair, from Atm−/− testis DNA treated without and with ExoV, respectively. Equal numbers of haploid genomes were tested (Figure S4C).
(F) Distinct breakpoint profiles for duplications/circles and deletions at the Chr1 and Chr19 hotspot pairs in Atm−/−. Arc diagrams of duplications/circles (red arcs) and deletions (blue arcs) are overlaid. Below the arc diagrams are smoothed breakpoint and SPO11-oligo profiles (201-bp Hann filter). Duplication/circle profiles are normalized to the number of deletions. Percentages indicate the fraction of breakpoints mapping to the left or right of the hotspot centers. See also Figure S4D.