To the Editor:
Coagulopathy and intravascular thrombosis are known complications of SARS‐CoV‐2 infection. The pathomechanism of COVID‐19‐associated micro‐ and macrovascular thrombosis likely involves the interplay of the coronavirus with endothelial cells, platelets, and immune cells. 1 Activation of endothelial cells and platelets results in procoagulant imbalance and macrovascular thrombotic events that are associated with disease severity and adverse outcomes. 1 Coagulopathy is marked by altered plasma levels of several coagulation proteins (e.g., D‐dimer, factors V and VIII, von Willebrand factor [vWF], fibrinogen, and ADAMTS13).2, 3, 4 However, previous studies investigated only small numbers of coagulation proteins together, and they were limited in their ability to adjust for confounders.
We leveraged a prospectively collected, previously published 5 acute COVID‐19 biorepository to study the association of plasma levels of 31 coagulation proteins with the occurrence of venous thromboembolic events (VTE). Plasma protein levels of enrolled patients were assayed upon hospital arrival using two independent proteomic platforms (Olink Explore 1536 and Somalogic SomaScan). Given that P‐selectin was independently associated with VTE, we explored its temporal dynamics and potential utility as an early biomarker of thromboembolic complications.
We enrolled 384 patients that presented with respiratory distress to the Emergency Department of an urban, academic hospital in Boston, MA during the first peak of COVID‐19 surge, as described previously. 5 Of the 384 enrolled, 306 patients were confirmed positive for SARS‐CoV‐2 by reverse transcriptase polymerase chain reaction. Specimen collection and banking was performed as previously described. 5 Samples were obtained on the day of admission (day 0) from all patients, and subsequently on days 3 and 7 from COVID‐19‐positive patients still hospitalized (Supplementary Information S1). Clinical data and outcomes were obtained from electronic health records. Patients who were intubated and/or died within 28 days were classified as severe. VTE was defined as pulmonary embolism (PE) or deep venous thrombosis (DVT) diagnosed within 28 days by computed tomography with pulmonary‐phase angiography (CTPE) or venous duplex ultrasonography (US), respectively. Plasma biomarkers were measured using the Olink Explore 1536 Proximity Extension Assay and the Somalogic SomaScan Platform® aptamer assay, as described previously. 5 Thirty‐one coagulation biomarkers were selected a priori from the two protein libraries based on literature and hypothesized relevance (Table S1). All statistical analysis and plotting were done in RStudio (version 4.0.3) using standard packages (Supplementary Information S1).
The clinical characteristics of all enrolled patients are shown in Table S2. Among the 306 COVID‐positive patients enrolled, 13 (4.2%, 95% CI 2.5–7.1) developed PE or DVT during the study period, confirmed by CTPE or US (Table S2). Interestingly, incidence of VTE was similar in the COVID‐19‐negative cohort. Among COVID‐19 patients, those with VTE were older (median age: 69 [interquartile range, IQR 62–78] vs. 58 [IQR 45–75], p = .2) and had significantly higher rates of severe illness (i.e., intubated and/or died in 28 days) than non‐VTE patients (85% vs. 33%, RR 2.53, 95% CI 1.9–3.3). Administration and dosing of anticoagulant medications both prehospital and in‐hospital were not significantly different between groups. Day 0 and day 3 D‐dimer levels were higher in patients who had VTE (Δ +1738 ng/ml at day 0 and Δ +890 ng/ml at day 3 between VTE and non‐VTE patients).
Of the 31 selected coagulation markers, COVID‐19‐positive patients who developed VTE had higher day 0 plasma levels of factors IX and XI, P‐selectin, plasmin, activated protein C, and vWF and lower levels of ADAMTS13, antithrombin III, factor VII, protein C, and prolylcarboxypeptidase, in at least one assay (Table S3). Notably, P‐selectin and vWF levels were significantly higher in both assays, and ADAMTS13 and factor VII were significantly lower.
In multivariable linear regression modeling of each coagulation protein separately—and controlling for possible confounding factors: age, gender, ethnicity, comorbidities, and illness severity—we found that VTE was independently associated with higher day 0 levels of factor XI and P‐selectin in the Somalogic panel and factor IX and P‐selectin in the Olink panel. Factor IX was not significant in the Somalogic panel, while factor XI was not present in the Olink panel. Furthermore, we found independent association of VTE with lower levels of protein C (in the Somalogic panel) and alpha‐2‐antiplasmin (only present in the Somalogic panel) (Figure 1A). P‐selectin was also independently associated with VTE when adding anticoagulation status as a covariate to the model (Figure S1). We analyzed the subset of COVID‐19‐positive patients who had high clinical suspicion of PE or DVT, defined as undergoing CTPE or US during their hospitalization (n = 82 total, n = 13 confirmed VTE), and found an independent association of elevated P‐selectin with VTE (in both assays), factor IX (Olink only), and factor XI (Somalogic only) on day 0 (Figure 1B).
FIGURE 1.
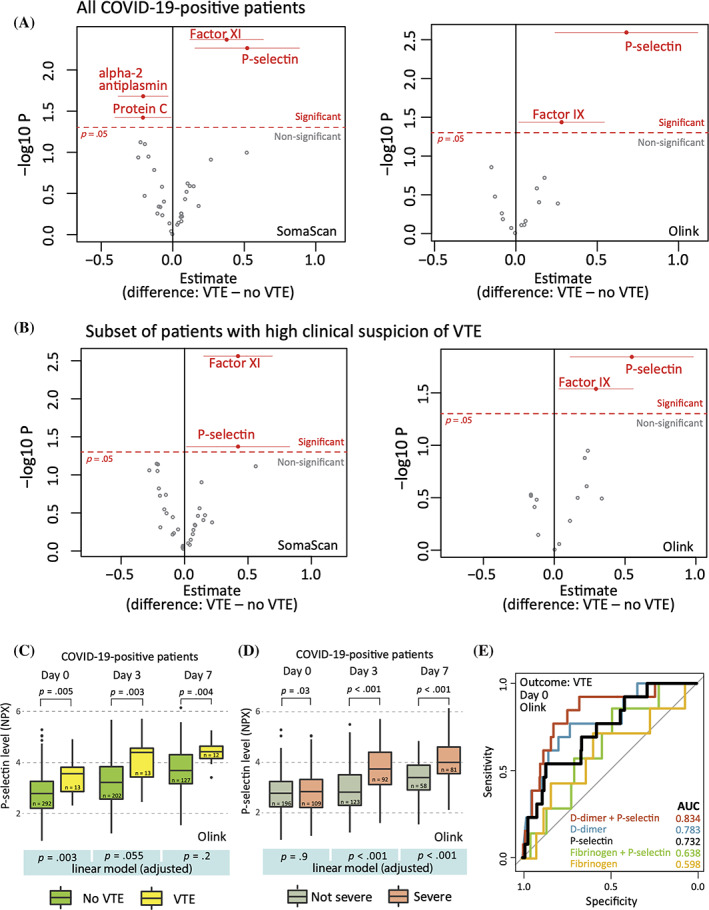
Plasma coagulation protein (incl. P‐selectin) levels and venous thromboembolic events (VTE). (A) Multivariable linear regression models were performed on day 0 data of COVID‐19‐positive patients to identify VTE‐associated proteins, adjusting for age, sex, ethnicity, comorbidities (i.e., heart disease, lung disease, hypertension, diabetes, liver disease), and severity of COVID‐19. Red color marks proteins with significant difference (p < .05). Horizontal lines for significant proteins represent confidence intervals (95%). (B) In a subset of confirmed COVID‐19‐positive patients who underwent computed tomography with pulmonary‐phase angiography or ultrasonography because of clinical suspicion of pulmonary embolism (PE) or deep venous thrombosis (DVT) (n = 82) an association of P‐selectin level with VTE risk was confirmed. (C) Temporal dynamics of plasma P‐selectin levels in COVID‐positive VTE versus non‐VTE patients (Olink assay). (D) Temporal dynamics of P‐selectin in severe versus not severe patients (Olink assay). (E) Predictive performance of P‐selectin, D‐dimer, and fibrinogen for VTE. Receiver operator characteristic (ROC) curves were plotted for logistic regression models that take VTE as outcome and day 0 Olink protein levels of P‐selectin (black), D‐dimer (blue), fibrinogen (yellow), and their combinations (red and green) as predictors. In (C) and (D), univariate tests were nonparametric Kruskal‐Wallis tests. Multivariable linear models (bottom panel, highlighted in blue) were adjusted as in Panel A
Given the independent significance of P‐selectin in both assays, we further analyzed its temporal dynamics. Using the Olink data, we observed a steady increase in the plasma concentration of P‐selectin in VTE patients over time. Median plasma level of P‐selectin in VTE‐positive patients was significantly higher than in VTE‐negative patients at each timepoint (Figure 1C). In adjusted linear regression models, including adjustment for severity, P‐selectin on day 0 was independently associated with VTE, which progressively diminished on days 3 and 7 (Figure 1C bottom panel). This was accompanied by a strong association of illness severity with VTE on days 3 and 7 that was not present on day 0 (Figure 1D). Thus, illness severity may increasingly confound the relationship between P‐selectin and VTE over time, yet P‐selectin had predictive value for VTE early in disease course independent of severity.
To evaluate the predictive performance of P‐selectin in COVID‐19‐positive patients in the context of clinical biomarkers associated with VTE and illness severity, we created logistic regression models using P‐selectin, D‐dimer, and fibrinogen as independent variables and analyzed their receiver operator characteristic (ROC) curves. We found that although the area under the ROC curve (AUC) of the P‐selectin model was lower than that of D‐dimer, the combined P‐selectin + D‐dimer model performed better than D‐dimer alone (AUC 0.834 vs. 0.783, Figure 1E). Both the fibrinogen and fibrinogen + P‐selectin models had lower AUCs than the P‐selectin model alone.
Our study demonstrates a consistent association of serum P‐selectin levels with the development of VTE in a large cohort of patients admitted with COVID‐19, across two independent proteomic platforms. P‐selectin was associated with VTE upon presentation to the hospital after adjusting for age, comorbidities, and illness severity, implicating endothelial activation in the development of VTE. Although not superior to D‐dimer in its ability to discriminate VTE events when analyzed separately, P‐selectin increased the discriminatory ability when used in combination with D‐dimer, compared to D‐dimer alone. Consistent with prior literature, our data show an association of plasma P‐selectin levels with disease severity, although only on hospital days 3 and 7, and not on day 0. This suggests that P‐selectin may be a late marker of illness severity and is consistent with delayed endothelial activity or injury in severe COVID‐19.
P‐selectin is an integral membrane protein of activated platelets and endothelial cells that mediates leukocyte rolling (adhesion to neutrophils and monocytes). Aside from participating in the inflammatory response, P‐selectin contributes to immunothrombosis by facilitating formation of endothelium‐leukocyte and platelet‐leukocyte aggregates and NET‐osis. 6 P‐selectin is shed into the circulation; increased plasma levels are considered a marker of endothelial cell and platelet activation and damage. 2 Increased platelet surface expression of P‐selectin in COVID‐19 has been shown previously. Importantly, a pathogenetic role of P‐selectin in traditional acute respiratory distress syndrome (ARDS) and coronavirus‐induced ARDS has been suggested.
Compared with prior reports, the strengths of our study were that we (1) obtained serial blood samples on prespecified days during hospitalization, (2) enrolled all patients that presented to the hospital during specified times with respiratory distress prior to knowledge of COVID‐19 status, (3) had the power to perform robust, covariate‐adjusted analyses, and (4) enrolled a large sample size with high density of severe illness. Limitations include single‐center sampling, an imperfect outcome measure that likely underrepresents cases of VTE, limitations of protein assay method employed, and lack of causal inference between P‐selectin levels and VTE. 5
The ACTIV‐4, ATTACC, and REMAP‐CAP multiplatform trials showed benefit of early therapeutic‐dose anticoagulation in moderately ill patients. 1 Our finding of elevated P‐selectin for those at‐risk for VTE on hospital admission, adjusted for illness severity, suggests the presence of early endothelial activation for some and thus plausible benefit of anticoagulation prior to development of severe illness. P‐selectin may have a role in the early risk stratification of VTE in COVID‐19 and may be useful in stratifying those for whom anticoagulation is indicated. Moreover, our findings support the hypothesis that P‐selectin blockade may be effective by preventing leucocyte‐platelet adherence. Currently, there is one Phase 2 clinical trial investigating crizanlizumab (a monoclonal antibody against P‐selectin) in COVID‐19 (NCT04435184). Further study of independent cohorts to validate our findings is warranted.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHOR CONTRIBUTIONS
Bánk G. Fenyves: Conceptualization; methodology; formal analysis; writing—original draft; writing—review and editing; investigation. Arnav Mehta: Conceptualization; methodology; writing—review and editing; investigation. Nir Hacohen: Conceptualization; resources; methodology; writing—review and editing; investigation. Michael R. Filbin: Conceptualization; resources; methodology; writing—review and editing; investigation. Marcia B. Goldberg: Conceptualization; resources; methodology; writing—review and editing; investigation. Justin Margolin: investigation. Kyle R. Kays: investigation. Caroline Beakes: investigation.
PATIENT CONSENT
A waiver of informed consent was approved in compliance with the Code of Federal Regulations (45CFR 46, 2018 Common Rule).
Supporting information
Figure S1. Plasma coagulation protein (incl. P‐selectin) levels and VTE—additional analysis. Multivariable linear regression models were performed similarly as in Figure 1A but with two additional confounders: Prehospital and in‐hospital anticoagulation status. Red color marks proteins with significant difference (p < .05). Horizontal lines for significant proteins represent confidence intervals (95%).
Table S1. Supplementary table of analyzed proteins.
Table S2. Demographics, clinical, and laboratory parameters by COVID‐19 status and thromboembolism.
Table S3. Coagulation protein levels of COVID‐19 patients measured by two proteome assays in day 0 samples.
Appendix S1: Supporting Information
ACKNOWLEDGMENTS
Direct funding for this project was provided in part by a grant from the National Institute of Health (N.H., U19 AI082630), an American Lung Association COVID‐19 Action Initiative grant (M.B.G.), and a grant from the Executive Committee on Research at MGH (M.B.G.). We thank Arthur, Sandra and Sarah Irving for a gift that enabled this study and funded the David P. Ryan, MD Endowed Chair in Cancer Research (N.H.). B.G.F was supported by the Rosztoczy Foundation Scholarship. This work was also supported by the Harvard Catalyst/Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences, National Institutes of Health, UL1 TR 002541‐01). We are very grateful for the generous contributions of Olink Proteomics Inc. and Somalogic Inc. for all in this work, without which our findings would not have been possible. In addition, the authors declare the following interests: AM—NIH/NCI T32 2T32CA071345‐21A1 (unrelated to this work). A.M. is a consultant for Third Rock Ventures, Abata Therapeutics, Asher Biotherapeutics and Rheos Medicines, and holds equity in Abata Therapeutics and Asher Biotherapeutics (all unrelated to this work). N.H. holds equity in BioNTech and is a consultant for Related Sciences. MRF—Research grants received from Day Zero Diagnostics, Rapid Pathogen Screening Inc., Nihon Kohden Corporation (all unrelated to this work).
Funding information American Lung Association; David P. Ryan, MD Endowed Chair in Cancer Research; Massachusetts General Hospital; National Institutes of Health, Grant/Award Numbers: UL1 TR 002541‐01, U19 AI082630; Rosztoczy Foundation
Contributor Information
Bánk G. Fenyves, Email: bfenyves@mgh.harvard.edu.
Michael R. Filbin, Email: mfilbin@mgh.harvard.edu.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in the supplementary material of this article. Raw data and scripts used for statistical analysis are available in https://github.com/bank-fenyves/COVIDprot.
REFERENCES
- 1. Leentjens J, van Haaps TF, Wessels PF, Schutgens REG, Middeldorp S. COVID‐19‐associated coagulopathy and antithrombotic agents—lessons after 1 year. Lancet Haematol. 2021;8(7):e524‐e533. doi: 10.1016/s2352-3026(21)00105-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Goshua G, Pine AB, Meizlish ML, et al. Endotheliopathy in COVID‐19‐associated coagulopathy: evidence from a single‐centre, cross‐sectional study. Lancet Haematol. 2020;7(8):e575‐e582. doi: 10.1016/S2352-3026(20)30216-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martín‐Rojas RM, Pérez‐Rus G, Delgado‐Pinos VE, et al. COVID‐19 coagulopathy: an in‐depth analysis of the coagulation system. Eur J Haematol. 2020;105(6):741‐750. doi: 10.1111/ejh.13501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stefely JA, Christensen BB, Gogakos T, et al. Marked factor V activity elevation in severe COVID‐19 is associated with venous thromboembolism. Am J Hematol. 2020;95(12):1522‐1530. doi: 10.1002/ajh.25979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Filbin MR, Mehta A, Schneider AM, et al. Longitudinal proteomic analysis of severe COVID‐19 reveals survival‐associated signatures, tissue‐specific cell death, and cell‐cell interactions. Cell Rep Med. 2021;2(5):100287. doi: 10.1016/j.xcrm.2021.100287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bongiovanni D, Klug M, Lazareva O, et al. SARS‐CoV‐2 infection is associated with a pro‐thrombotic platelet phenotype. Cell Death Dis. 2021;12(1):50. doi: 10.1038/s41419-020-03333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Plasma coagulation protein (incl. P‐selectin) levels and VTE—additional analysis. Multivariable linear regression models were performed similarly as in Figure 1A but with two additional confounders: Prehospital and in‐hospital anticoagulation status. Red color marks proteins with significant difference (p < .05). Horizontal lines for significant proteins represent confidence intervals (95%).
Table S1. Supplementary table of analyzed proteins.
Table S2. Demographics, clinical, and laboratory parameters by COVID‐19 status and thromboembolism.
Table S3. Coagulation protein levels of COVID‐19 patients measured by two proteome assays in day 0 samples.
Appendix S1: Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article. Raw data and scripts used for statistical analysis are available in https://github.com/bank-fenyves/COVIDprot.


