Summary
B lymphocytes play a central role in host immune defense. B cell receptor (BCR) signaling regulates survival, proliferation and differentiation of B lymphocytes. Signaling through the BCR signalosome is a multi-component cascade that is tightly regulated and is important in the coordination of B cell differentiation and function. At different stages of development, B cells that have BCRs recognizing self are eliminated to prevent autoimmunity. microRNAs (miRNAs) are small single-stranded non-coding RNAs that contribute to post-transcriptional regulation of gene expression and have been shown to orchestrate cell fate decisions through the regulation of lineage specific transcriptional profiles. Studies have identified miRNAs to be crucial for B cell development in the bone marrow and their subsequent population of the peripheral immune system. In this review, we focus on the role of miRNAs in the regulation of BCR signaling as it pertains to B lymphocyte development and function. In particular, we discuss the most recent studies describing the role of miRNAs in the regulation of both early B cell development and peripheral B cell responses and examine the ways by which miRNAs regulate signals downstream of B cell antigen receptor to prevent aberrant activation and autoimmunity.
Keywords: miRNA, BCR, PI3K, AKT, MAPK, NF-κB
Section 1: Introduction
microRNAs
microRNAs (miRNAs) are evolutionarily conserved, small non-coding RNAs (ncRNAs) that are crucial in the regulation of a myriad of cellular functions via RNA interference (RNAi). In hematopoietic cell lines, multiple studies describe the impact of miRNAs on lineage determination and maturation.1,2 Likewise, because miRNAs regulate many important biological pathways in lymphocytes, their deregulation has been implicated in malignant disease and autoimmunity.3 Transcribed in the nucleus by RNA polymerase II, miRNAs undergo processing by the microprocessor (Drosha and DGCR8) to cleave the double-stranded hairpin primary miRNA into shorter stem-loop precursor miRNAs (pre-miRNAs). In the cytoplasm, Dicer, an RNaseIII enzyme that cleaves double-stranded RNA, further cuts the pre-miRNAs into 20–23 nucleotide duplexes. Mature miRNAs are incorporated into the RNA induced silencing complex and guide the translational repression or degradation of mRNA target genes via miRNA-dependent recognition of 3’untranslated regions (3’UTRs).1,2,4,5
B cell development
B lymphocyte development and maturation is a tightly orchestrated process during which progenitor cells generate unique antigen receptors following rearrangement of immunoglobulin heavy chain (IgH) and light chain (IgL) loci.6 After the induction of critical transcription factors (E2A, EBF, and Pax5), the bone marrow-resident common lymphoid progenitors (CLP) commit to the B cell lineage and enter the pro-B cell stage.7 Here, the enzymatic activity of recombination activating genes 1 and 2 (RAG1 and RAG2) induce V(D)J recombination at the IgH locus.8,9 Successful rearrangement of the IgH locus allows for the progression of the B cells to the large pre-B cell stage where the IgH pairs with a surrogate light chain and is presented on the cell surface as a pre-B cell receptor (pre-BCR).8 Proper signaling through the pre-BCR ensures that the recombinatorial events result in a functional IgH which then leads to the downregulation of RAG1/2 and the initiation of rapid proliferation. The pre-B cell exits the cell cycle as a small pre-B cell and a second wave of RAG1/2 expression promotes VJ recombination at the IgL loci.8,10,11 Two molecules of the IgL associate with two molecules of the IgH to form the B cell receptor (BCR) on the cell surface of immature B lymphocytes. It is the combination of the heavy and light chains that confers BCR specificity for a cognate antigen. This process also has the potential to generate BCRs with self-reactive specificities. Immature B cells with newly minted BCRs undergo negative selection to eliminate self-reactive lymphocytes. If the membrane-bound BCR reacts strongly to a self-antigen, the immature B cell either internalizes the BCR, decreasing signaling and increasing RAG1/2 mediated light chain editing, or undergoes clonal deletion.11,12 Basal tonic signaling through the BCR of the immature B cell restricts RAG1/2 expression and is critical for the maintenance of immature B lymphocytes as they exit the bone marrow to mature in the periphery.13 Throughout this carefully coordinated developmental process in the bone marrow, signaling through the pre-BCR and, in later stages, mature BCR governs the survival and proliferation of B lymphocytes and the B cell antigen receptor continues to play a central role in peripheral B cells.
Peripheral immature B cells, often referred to as transitional B cells, undergo further negative selection wherein cells that receive a strong signal via the BCR in the absence of T cell help or cytokine signals undergo apoptosis or become anergic (enter a state of hypo-responsiveness).14,15 Amplitude of BCR signaling, cytokine stimulation via B cell activating factor (BAFF), and engagement of NOTCH2 and BTK influence which compartment of maturity the B cell will enter: marginal zone (MZ) or follicular (FO) B cells.16–19 FO B cells, which constitute the majority of mature B lymphocytes, reside in follicles within secondary lymphoid organs adjacent to T cell zones and are quickly engaged in response to T cell-dependent antigens. MZ B cells are largely in the white pulp of the spleen and have intrinsic properties that ensure rapid response to blood borne pathogens in a T-independent manner – leading to the generation of plasma cells that secrete natural, polyreactive IgM antibodies (MZ and FO B cell fate is reviewed in Pillai et al. 2009).18 FO B cells form from precursors with stronger BCR signaling and they are dependent on the BCR-dependent induction of BTK.18,20 Conversely, weaker signaling through the BCR and the induction of NOTCH2 favors the development of MZ B cells.20
Upon engagement of its B cell antigen receptor, FO B cells either differentiate into antibody-producing plasma cells or enter specialized microstructures, called germinal centers (GCs) – a fate decision thought to be primarily governed by the strength of BCR signal in response to cognate antigen. Naïve, mature FO B cells that initially respond strongly to their cognate antigen differentiate into plasma cells.21 Weaker BCR signaling promotes GC entry where the Ig loci that encode the antibody undergo somatic hypermutation to optimize the binding affinity of the BCR and where class switch recombination (CSR) that leads to a change in the antibody isotype takes place.21,22 Within GCs, B lymphocytes undergo rapid proliferation and a subsequent step of Darwinian-like selection which is driven by competition for limited antigen.23 Both of these processes require activation-induced cytidine deaminase (AID).21 GC B cells that outcompete other B cell clones through stronger recognition of the cognate antigen receive proliferative and differentiation signals from T follicular helper cells (TFH) cells. This triggers the induction of transcriptional programs which determine whether the B cell will differentiate into a plasma cell or become a memory B cell.24 As a plasma cell, the BCR heavy chain is alternatively spliced and secreted as an antibody.25 Because so many cell fate decisions throughout B cell development are governed by BCR signal strength, it is critical to understand how miRNAs fine-tune this signaling cascade.
Components of the BCR signalosome
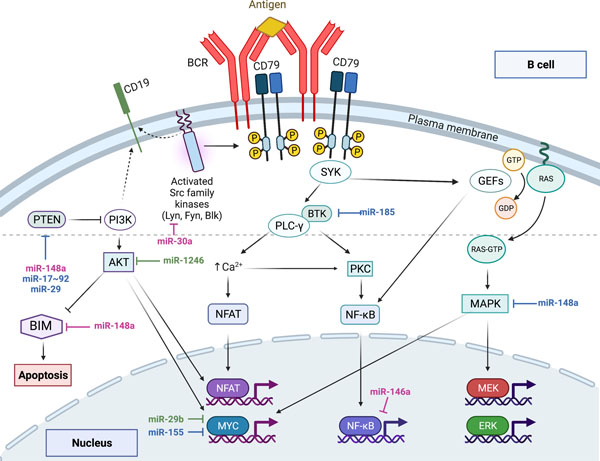
The BCR is a type-I transmembrane immunoglobulin that has two heavy chains and two light chains. Associated with the immunoglobulin is an Igα/Igβ heterodimer that, along with co-receptors, is essential for the intracellular signaling of the BCR.8 After binding of the antigen to the BCR, the cytoplasmic tail of the Igα/Igβ heterodimer and co-receptors are phosphorylated by Lyn kinase.26–28 These events lead to the recruitment of multiple kinases and scaffolding proteins to the inner side of the plasma membrane and form the BCR signalosome.29 Syk is the principle kinase that activates many of the proteins recruited to the inner membrane through the actions of Lyn.30 CD19 co-receptor and BTK promote the aggregation of BCR molecules into microclusters on the plasma membrane, which further amplify B cell activation.31 Signaling through the BCR is critical for the maintenance of mature B cells, and stage specific ablation of surface Ig or Igα/Igβ signaling subunit results in apoptosis of B cells.16,32 Downstream of the BCR signalosome there are three key signaling pathways that activate following BCR engagement: the NF-κB pathway, the RAS-MAPK pathway, and the PI3K-AKT pathway (Figure 1). The cross talk between these three signaling pathways adds to the complexity of BCR signaling. The brief overview of signal transduction through each of these three signaling pathways is critical for appreciating the myriad of nodes where miRNAs exert control over BCR signaling.
Figure 1: Regulation of B cell receptor signaling by microRNAs.
The PI3K-AKT, NF-κB, and RAS-MAPK signaling pathways downstream of the BCR are depicted.
MiRNAs discussed in this review are highlighted in association with relevant targets within the signaling networks. miRNAs in blue were those identified in murine studies, miRNAs in green were identified in human studies, and those in pink have data from both murine and human studies that highlight the role of these miRNA in the regulation of BCR signaling, B cell development, or autoimmunity.
NF-κB:
Activation of the canonical NF-κB pathway is an important survival signal downstream of BCR engagement. Prior to activation, NF-κB is initially bound to IκB in the cytoplasm which prevents its translocation into the nucleus. The kinases BTK and Syk, which are activated upon BCR stimulation, phosphorylate and activate Phospholipase C-γ2 (PLC-γ2) leading to the hydrolyzation of phosphatidylinositol (4,5)-bisphosphate (PIP2) to diaglycerol (DAG) and inositol triphosphate (IP3).29,33 IP3 increases intracellular Ca2+ which, in combination with DAG, activates protein kinase-C (PKC). The activation of PKC and BTK in concert with other recruited factors phosphorylate and activate the inhibitor of IκB kinase (IKK) complex. The activation of IKK then phosphorylates IκB, leading to ubiquitin-mediated degradation of IκB. The unbound NF-κB then translocates to the nucleus to regulate genes responsible for cell proliferation and survival critical to B cell activation.33,34
ERK-MAPK:
The extracellular signal-regulated kinase (ERK) mitogen-activated protein kinase (MAPK) is activated downstream of BCR signaling and is important for B cell proliferation and survival during development and differentiation. The ERK-MAPK pathway is primarily regulated by the G protein RAS. The inactive form of RAS has GDP bound, and guanine nucleotide exchange factors (GEFs) swap GDP for GTP to activate RAS and GTPase-activating proteins (GAPs) deactivate RAS.35 RAS-GTP activates Raf-1/B-Raf which activates MEK1/2 and ERK which translocate to the nucleus, resulting in the activation of a number of transcription factors (Fos, Jun, Ets) which are involved in cell proliferation and survival.29,33
PI3K:
The phosphoinositide 3-kinase (PI3K) signaling cascade is essential for orchestration of B cell development and BCR signaling transduction.36–40 PI3K is recruited to the plasma membrane by the BCR co-receptor, CD19, where it is phosphorylated by LYN, leading to the conversion of phosphatidylinositol (4,5)-biphosphate (PIP2) to phosphatidylinositol (3,4,5)-triphosphate (PIP3).33,41,42 AKT and phosphoinositide-dependent protein kinase1 (PDK1) are recruited to the plasma membrane by PIP3 where AKT is phosphorylated by PDK1 and mTOR. The phosphorylation of AKT in turn, leads to a variety of critical pro-survival, growth, proliferation, and anabolic pathways including the regulation of FOXO1/3, c-Myc, NF-kB, and NFAT.29,33,43–46 In addition to the fact that antigen engagement of the BCR leads to robust PI3K activation, low amplitude PI3K “tonic” signal is essential for the survival of B lymphocytes – blunting of this tonic PI3K signal results in apoptosis of B cells.23,47
Tight regulation of these three pathways is necessary for proper development, as inappropriate BCR signaling can result in malignant transformation and the development of auto-reactive B cells.48 Among the most potent regulators of BCR signaling are those modulators that prevent the formation of the signalosome. The factors FcγRIIb and SHIP work synergistically to prevent the recruitment of PLC-γ2, BTK and AKT.49 Lyn, an important kinase in the propagation of BCR signaling, also has a negative regulatory role in B cell activation through its activation of both SHIP and FcγRIIb. In addition to these negative BCR modulators, each of the three pathways discussed above have additional regulatory molecules involved in mitigating signal strength. As discussed below, miRNA-dependent post-transcriptional regulation of the components of these signaling networks is critical to the proper attenuation of BCR signaling.
In this review, we will focus on miRNA-dependent regulation of BCR signaling as the B cell progresses through various stages of B cell development and discuss its implications for the development of autoimmunity. There are a number of miRNAs that have been implicated in B cell lineage commitment and B cell malignant transformation – these studies are beyond the scope of this review and have been previously reviewed.12,50–55
Section 2: miRNAs in early BCR development and signaling
Early BCR Development
Analysis of a murine model where Dicer1 is selectively ablated in B lymphocytes via mb1-cre, highlights the importance of ncRNAs in early stages of B cell development. Analysis of B cell hematopoeisis in these mice reveals a block in the pro- to pre-B cell transition marked by a significant increase in apoptosis of Dicer-deficient pre-B cells and a near-complete loss of peripheral B lymphocytes in secondary lymphoid organs.56 Mb1-cre mediated deletion of Dgcr8 or Drosha in B cells recapitulates these findings indicating that miRNAs are essential for coordination of early B cell development.57,58 In all three mutant mouse models, the proapoptotic protein BIM was significantly upregulated.56–58 Indeed, Bim has conserved miRNA binding sites in its 3’UTR implicating miRNAs in supressing its expression.56,59 Intriguingly, overexpression of the anti-apoptotic Bcl-2 transgene in miRNA-deficient B cells resulted in only partial rescue (~12%) of B cell development, suggesting that miRNAs contribute to regulation of other critical pathways during B cell differentiation.56,58
A number of these early studies which looked at global miRNA ablation in the hematopoeitic lineage, and later work using conditional ablation of individual miRNA clusters, implicate miRNAs in the coordination of pro- to pre-B cell transition. Differential gene expression analysis in immortalized pro-B cells in which Dicer is inducibly ablated reveals upregulation of gene transcripts with 3’UTRs that have a miR-17~92 binding site.56 Furthermore, conditional deletion of miR-17~92 in B cells leads to a similar phenotype as described above, including a block in pro- to pre-B lymphocyte transition with a concominant increase in apoptosis in the setting of dysregulated BIM and PI3K signaling (discussed in more detail below).59,60 The authors attributed this block in the pro- to pre-B cell transition to miR-17, because in vitro expression of miR-17 alone in HSCs from these mice was sufficient to rescue B cell development.60 Interestingly, ablation of miR-17~92 cluster along with the two paralogue clusters, miR-106a~363 and miR-16b~25, recapitulates the pro- to pre-B cell block even in the absence of BIM dysregulation. This observation suggests that BIM is not the target responsible for the block in B cell development in mice lacking these miRNA clusters and that Bim may not be a physiologically relevant target of miR-17~92 in B cell progenitors. Moreover, a recent study utilizing a hematopoietic lineage-specific cre to mutate the miR-17~92 binding sites within the 3’UTR of Bim only showed a modest increase in BIM protein, with minimal impact on B cell development.61 Furthermore, this developmental block in mice where miR-17~92 was ablated in B cells could not be rescued by a Bcl2-transgene (Tg).60 Collectively, these findings suggest that miRNA likely contribute to the regulation of additional targets and molecular pathways apart from Bim during B cell development that remain to be elucidated.
Inappropriate overexpression of miRNAs can also be detrimental to B cell development. miR-150 and miR-34 are both maintained at low levels throughout early B cell development with miR-150 increasing as the B cell matures.62 Overexpression of miR-150 results in a downregulation of transcription factor c-Myb,63,64 which regulates early B cell development.65 Indeed, through a series of loss-of and gain-of-function experiments Xiao et al showed that upregulation of c-Myb was responsible for the observed failure in appropriate B cell differentiation upon miR-150 overexpression.63 Similarly, gene targeting experiments have demonstrated that constitutitve expression of miR-34, a miRNA normally induced by p53, leads to ihibition of Foxp1 and a subsequent block in B cell development.66 These findings highlight the importance of appropriate temporal expression of individual miRNAs and the presented rescue experiments support the fact that it is often a few crtical targets of any individual miRNA that are responsible for regulatory function of these ncRNAs in any given cellular context.
As previously discussed, successful rearrangement of heavy and light chains, at pro- and pre-B cell stages respectively, is essential for assembly of the BCR which supports B cell survival and differentiation. While the basic mechanisms of V(D)J recombination did not appear to be perturbed in miRNA-deficient B cells, repertoire analyses have revealed that Dicer-dependent ncRNAs do influence BCR repertoire diversity and that while Drosha/DGCR8 miRNAs did not appear to have impact on Ig repertoire, loss of miRNAs were shown to impact the expression level of antigen receptor on B cells.56,58 Amplifying the IgH loci of Dicer-deficient B cells revealed a change in DH element usage compared with controls.56 Additionally, sequence analysis of light chain rearrangements have revealed an increase in the frequency of non-template N nucleotides that corresponded with a prolonged expression of the enzyme TdT throughout the maturation of Dicer KO B cells.56 Consistent with this, a study by Ventura et al. (2008) observed less efficient VH-to-DJH rearrangement in miR-17~92 null pro-B cells compared with controls, however the degree of dysregulation was not reported and further analysis is needed to elucidate a mechanism.59 Furthermore, ablation of either Dicer, Dgcr8 or Drosha in B cells has been found to reduce levels of BCR expression and analysis of mature B cells deficient in miRNAs revealed ongoing BCR editing even in peripheral lymphocytes as a consequence of dysregulated PI3K signaling (discussed in more detail below).57,58 Strikingly, while a transgene encoding a fully rearranged IgH and IgL (IgHEL) led to only partial (~5%) rescue of B cell development, a combination of IgHEL-Tg and Bcl2-Tg was sufficient to fully rescue B cell development in the absence of miRNAs.56,58
Taken together, these findings suggest a central role for miRNAs in choreographing early B cell development, and regulation of signaling pathways critical for B cell survival. In particular, while the review below is on the role of miRNAs in the regulation of signal transcduction downstream of the BCR, ncRNAs also appear to contribute to the assembly of a functional repertoire of B lymphocytes.
PI3K-AKT Pathway
PI3K signaling regulates B cell development and survival and is critical for the coordination of rerrangement events in the bone marrow.36 Pre-BCR and BCR induced PI3K signaling during B cell development provides survival signals and contributes to downregulation of RAG1/2 enzymes leading to the termination of further V(D)J rearrangement.8,10,11,36 In addition to the regulation of survival and rearrangmeent, PI3K-dependent signaling is also required for cell cycle progression and proliferation of developing B lymphocytes.10,11
The PI3K-AKT pathway has been studied in various miRNA-deficient mouse models. An analysis miRNA-deficient B cells that were resuced with a Bcl2-Tg from animals in which Dicer, Drosha, or Dgcr8 were selectively deleted revealed that developing B cells have an increase in the protein levels of the PI3K negative regulator, phosphatase and tensin homolog (PTEN).56,58 The 3’ UTR of PTEN contains numerous miRNA binding sites and PTEN dysregulation in these miRNA-deficient B cells leads to impaired PI3K signaling, a reduction in AKT activation and a corresponding increase in FOXO1 and RAG1/2 expression. Compromised PI3K signaling was also found to persist in peripheral lymhocytes devoid of microprocessor enzymes, such that ongoing light chain editing continued in the peripheral B cells from secondary lymphoid organs of these animals.58
As previously described, developing Dicer and microprocessor deficient B cells exhibited an increase in apoptosis that was initially attributed to direct miRNA dependent dysregulation of BIM protein. However, as BIM is negatively regulated by the PI3K pathway and the ablation of miR-17~92:Bim interactions led to only a modest increase in BIM protein without dysregulation of B cell development, its upregulation in the setting of increased PTEN expression offers an alternative explanation.61 B cells in which miR-17~92 was genetically ablated have also been reported to have an increase in BIM protein in the setting of the upregulation of two PI3K negative regulators, PTEN and Phlpp2.60 However, while PTEN has been confirmed as a direct target of the miR-17~92 family, overexpression of these miRNAs fails to reduce the Phlpp2 reporter activity in renilla-luciferase assays.60 Furthermore, ex vivo DRAQ5 analysis of miRNA-null pre-B cells reveals a decrease in proliferation in the setting of increased PTEN expression, suggesting that PI3K-dependent proliferation of B lymphocytes is also impacted upon upregulation of this negative regulator of the pathway. Collectively, these studies highlight the importance of miRNA regulation of the PTEN-PI3K pathway throughout B cell development.
Section 3: miRNAs in central tolerance and autoimmunity
It is estimated that 50 to 75% of B lymphocytes produced following V(D)J recombination in the bone marrow are autoreactive, but only 3 to 8% of the population develop evident autoimmune disease.67,68 As briefly discussed above, through negative selection, tightly regulated checkpoints ensure that most autoreactive immature B cells are prevented from joining the mature B cell compartment by receptor editing, apoptosis, or anergy.69 Newly minted BCRs in the bone marrow are tested for autoreactivity at the immature B cell stage during a selection and editing process called central tolerance. If a BCR engages with a self-antigen, leading to the activation of signaling pathways above a certain threshold, the immature B cell will either undergo clonal deletion or halt maturation, internalize the BCR and undergo additional light chain editing.70–72
RAS-MAPK Pathway
The role of MAPK signaling in central tolerance remains to be fully elucidated. Increased MAPK signaling has been described in B cells from both lupus patients and mouse models.73–75 Furthermore, constitutively activated RAS signaling has been found to promote the differentiation of autoreactive immature B cells and lead to inceased autoantibody titers.76,77 However, a recent study found that the direct activation of the MAPK pathway alone was insufficient to break central B cell tolerance and hypothesized that its activation in conjunction with other pathways are required.78
Using a functional retroviral screen to overexpress batches of miRNAs in transferred HSPCs, Gonzalez-Martin et al. (2016) identified 7 miRNAs that were enriched in B cells that evaded central tolerance in the IgMb-macroself mouse model.79 This mouse model ubiquitously expresses a membrane-bound superantigen that binds the IgM heavy chain constant region on the surface of immature B lymphocytes, thus mimicing self-recognition by B cells, leading to clonal deletion.80 Although five of the seven miRNAs significantly broke IgMb-macroself central tolerance in a transfer model upon transduction of HSPCs with each of these these miRNAs, overexpression of miR-148a resulted in the most significant break of central tolerance.79 While this study focused specifically on miR-148a, a miRNA shown to compromise B cell tolerance and one that has been implicated in lupus pathogenesis, future studies are needed to fully investigate the importance of the other candidate miRNAs involved in the regulation of B cell central tolerance in this study (miR-26a, miR-26b, miR-342, miR 423 and miR-182).79
Indeed, miR-148a has been shown to be upregulated in a number of autoimmune diseases including lupus, multiple sclerosis, and asthma.9,81,82 Furthermore, miR-148a was found to be highly expressed in pro- and pre-B cells and downregulated at the immature B cell stage where central tolerance occurs.79 miR-148a was also found to be upregulated in human germinal center B cells and plasma cells, suggesting that expression of this miRNA is induced in response to B cell activation.83 The autoimmune suppressor, Gadd45a, a target of p38/JNK pathway induced by DNA-damage response and environmental stress, was found to be significantly upregulated in miR-148a transduced WEHI-231 cell lines compared to controls via RNAseq and qPCR validation.79 Dual luciferase assays validated the 3’UTR of Gadd45α as a direct target of miR-148a. To determine if the break in central tolerance observed upon overexpression of miR-148a was linked to the regulation of Gadd45a, the authors reconstituted IgMb-macroself mice with bone marrow from Gadd45α −/− mice. These mice exhibited a very significant break in central tolerance with a higher percentage of peripheral B lymphocytes than was previously shown with miR-148a overexpression.79 Gadd45α+/− B lymphocytes also had evidence of compromised central tolerance, albeit with lower frequency compared to B cells transduced with miR-148a, suggesting that precise regulation of Gadd45α is essential for proper selection of B cells. Together, these findings suggest that a miR-148a-Gadd45a axis plays an important role in the regulation of central tolerance.79
PI3K-AKT Pathway
A number of studies have established a role for the PI3K cascade in central tolerance. PI3K gain-of-function mutations have been shown to contribute to a predisposition to a wide-range of autoimmune/inflammatory disease. A third of individuals in a well-characterized cohort harboring germline PI3Kδ mutations were found to develop autoimmune and inflammatory manifestations.84 Mice bearing similar mutations recapitulate many of these phenotypes including increased high-avidity autoreactive B cells persisting in the periphery and increased autoantibody production.85–88 Furthermore, deletion of PI3K negative regulators, including PTEN, has been shown to promote the survival of autoreactive B cells in various transgenic mouse models.60,79,89,90
The same group that has described the role of miR-148a in the regulation of MAPK pathway in B cells has also demonstrated miR-148a-dependent regulation of the PI3K pathway and its implications for central tolerance.79 These investigators overexpressed miR-148a in an immature B cell line (WEHI-231) and reported a significant decrease in transcript and protein levels of PTEN and BIM. Dual luciferase assays using 3’ UTRs from these transcripts suggested that both BIM and PTEN are direct targets of miR-148a. Furthermore, when WEHI-231 cells transduced with miR-148a are stimulated with anti-IgM, Annexin V and caspase 3 staining reveal reduced apoptosis in these cells.79 These in vitro findings were consistent with the break in central tolerance observed in miR-148a transduced B cells in vivo. The authors also report a decrease in Bim and Pten transcript and protein levels following transduction of WEHI-231 cells with a miR-26a-Tg, but further studies are needed to evaluate the role of miR-26a in maintenance of B lymphocyte central tolerance.79 Bone marrow reconstitution experiments using IgMb-macroself mice as hosts revealed that BIM and PTEN play a critical role in maintenance of central tolerance, as B cells deficient for these proteins are able to escape negative selection. Together, these studies suggest that appropriate regulation of Gadd45α, Pten and Bcl2l11 (the transcript that encodes BIM) by miR-148a is instrumental in B cell central tolerance, highlighting how a single miRNA can play a central role in regulation of multiple pathways during a complex biological process. The functional consequences of miR-148a-mediated regulation of the target genes that regulate BCR signaling and B cell survival was further highlighted by the investigators when they overexpressed the miRNA in MRL-lpr mice and observed greatly accelerated lethal autoimmune disease in this mouse model of lupus.79
The miR-17–92 cluster has been associated with the manifestation of multiple autoimmune conditions.3,91,92 Utilizing a transgene to overexpress the miR-17~92 cluster in the IgMb-macroself mouse model (introduced above), Lai et al. (2016) described the importance of this miRNA family in controlling central tolerance through PI3K regulation. The authors observed a complete restoration of peripheral B lymphocytes in animals in which this miRNA cluster is selectively expressed in B cells, suggesting that overexpression of miR-17~92 is sufficient to potentiate escape of negative selection.60 These results were recapitulated in a second mouse model used to study central tolerance, IgHEL;mHEL.60,93,94 The authors then dissected the contribution of individual miRNAs from miR-17~92 family to B cell development. Using a lentivirus vector to transduce HSCs, which were then used to reconstitute IgMb-macroself hosts, miR-19 was identified as the specific miRNA from the miR-17~92 cluster that was responsible for the escape of central tolerance at the immature B cell stage.60 As described above, the members of the miR-17~92 family of miRNAs, including miR-19, were shown to directly target Pten. Furthermore, Pten haploinsufficiency was sufficient to break central tolerance. In an effort to determine whether miR-19 controls central tolerance through PTEN dysregulation, the authors co-expressed miR-19 and Pten via lentiviruses in HSCs that were transferred into irradiated IgMb-macroself hosts. The percentage of B cells that escaped central tolerance through the overexpression of both PTEN and miR-19 was comparable to the percentage of B lymphocytes that escaped central tolerance in the IgMb-macroself mouse model controls.60 These findings cement the importance of miR-19 control of central tolerance through PTEN regulation.
Section 4: miRNAs in peripheral BCR signaling and autoimmunity
Introduction to miRNAs in the periphery
As discussed with respect to B cell development, the BCR, composed of heavy and light chains, forms a signaling complex with the Igα and Igβ heterodimers on the B lymphocyte surface. The Igα/Igβ heterodimer plays a critical role in cytoplasmic signaling through the BCR and is responsible for activation of protein tyrosine kinases.34 Following BCR stimulation and phosphorylation of costimulatory molecule CD19, multiple signaling pathways are engaged leading to B cell activation and internalization of the bound antigen.33,95 The nature of BCR engagement and signaling for developing and peripheral B lymphocytes is largely the same, but the context of peripheral B cell antigen receptor signaling is critical for coordinating terminal differentiation of the lymphocytes, antibody-dependent responses, and affinity maturation. BCR antigen affinity and avidity, dictated by the corresponding binding sites on the IgH/IgL chains, establishes the strength of B cell activation.28,96 As previously mentioned, well defined activation pathways downstream of BCR activation include the PI3K-AKT, RAS-MAPK, and NF-κB pathways.33,95,97 Figure 1 details these pathways, and specific miRNAs associated with their regulation that will be discussed in detail below.
B cell activation plays a critical role in host response to infection, however, BCR recognition of self can lead to autoimmunity.33,98,99 For that reason, B cell activation through the antigen receptor is a tightly regulated process. In this section, we will highlight ways in which regulatory miRNAs regulate PI3K-AKT, and MAPK/NF-kB signaling following BCR engagement in peripheral B lymphocytes.
As described earlier, miRNAs are required for B cell development within the bone marrow compartment.51,56–58 In addition to reduced overall B cell numbers, ablation of Dicer1 conditional allele using a CD19-cre driver impairs FO development, and increases transitional and MZ B cells.100 In another study in which Dicer1 was ablated under CD19-cre, the deletion results in a skewed BCR repertoire and features of autoimmunity, with female mice developing systemic lupus erythematous (SLE), Sjogren’s, or scleroderma-like phenotypes.100 These early studies of global miRNA ablation in the B cell lineages suggest that overall loss of miRNA regulatory potential leads to inappropriate selection and/or activation of peripheral B lymphocytes.
A mouse model that contains a conditional deletion of Dicer1 under Aicda-cre, led to ablation of the conditional Dicer allele at a later stage of B cell development than in the mb1-cre and CD19-cre models that were discussed earlier. B cells from Dicerfl/fl Aicda-cre mice have a lower amount of CSR, a reduction in antibody affinity maturation, and compromised plasma cell differentiation when mice are immunized. These mice also display a reduction in germinal center formation, both in the presence and absence of immunization.101 Analysis of B cells from Dicerfl/fl Aicda-cre animals highlights that ablation of miRNA biogenesis using a cre specific to late stage B cell development and activated peripheral B cells impacts: B cell differentiation, germinal center reaction processes, and antibody production—all processes intimately dependent on appropriately tuned BCR signaling.
Analysis of B cells from SLE patients, a disease where pathology is well-established to be dependent on antibody mediated inflammation,102 also reveals that autoantibody production in this disease is linked to loss of regulatory control by miRNAs. In a study that examined miRNA landscape using TaqMan global miRNA arrays in patient biospecimens stratified for being sera-positive for anti-nuclear, anti-double stranded-(ds)-DNA antibodies or both, the results indicate distinct miRNA profiles and transcriptional signatures withing each “sub-type” of SLE.103 This study highlights the critical regulatory role of miRNAs in maintenance of proper B cell selection and activation and the possible failure in this type of post-transcriptional control to autoimmune pathogenesis. Beyond the global impact of miRNAs on fine tuning B cell activation and autoimmune susceptibility, we will highlight specific studies that have identified miRNAs involved in post-transcriptional regulation of the BCR signalosome.
PI3K-AKT Pathway
The PI3K-AKT pathway is activated downstream of the BCR and maintains B cell transcriptional programming. We will discuss how the miRNAs miR-29, miR-1246, and miR-148a target aspects of the PI3K-AKT pathway, and what implications this has on B cell signaling (Figure 1).
A recent study revealed the miR-29 family of miRNAs as a regulator of PI3K signaling in mature B lymphocytes in the periphery. The conditional ablation of miR-29ab1 cluster coupled with the germline deletion of miR-29b2c (mb1-cre miR-29ab1fl/fl miR29b2c−/−) in mice results in an increase in PTEN protein levels with a corresponding dampening of PI3K signaling in mature B cells as evidenced by a decrease in active AKT. Since the PI3K signaling cascade is so vital for maintenance of mature B cells, it is not surprising that these mice were found to have a significant reduction in splenic B cell numbers with a corresponding increase in apoptosis. Furthermore, the surviving FO B cells were found to have low plasma cell differentiation and an increase in CSR. Renilla-luciferase assays confirmed Pten as a target of miR-29. These phenotypes were rescued with a conditional deletion of one copy of Pten (mb1-cre miR-29ab1fl/fl miR29b2c−/− PTENfl/+).104
Another study that examined the role of miR-29 in B cell function, evaluated the impact of miR-29 loss on induction of collagen induced arthritis.105 The authors showed that global loss of miR-29ab1 locus results in a global loss of splenic and thymic cellularity and has a profound impact on T cell differentiation. This likely contributed to the observed defect in germinal center formation. The authors also observed that deletion of miR-29a in vitro is associated with a reduction in B cell proliferation and antibody production, enabling them to make conclusions about the role of miR-29a in these cell-intrinsic processes.105 In all, despite the findings that miR-29 loss was associated with protection against collagen induced arthritis, the study105 is consistent with our conclusions104 that miR-29 is a positive regulator of PI3K signaling (via repression of Pten).
SLE is an autoimmune disease driven by the dysregulation of multiple immune cell types that impacts multiple organ systems.106 Multiple miRNAs have been shown to be differentially expressed in SLE patients.106–108 A miRNA microarray performed on MACS enriched B cells from blood of SLE patients and healthy controls identified six miRNAs with significantly different levels of expression: miR-4286, miR-451, and miR-424 were upregulated and miR-1246, miR-122, and miR-877 were downregulated in B cells from SLE patients. The downregulation of miR-1246 was validated by RT-PCR, and bioinformatic analysis identified the transcription factor Early B cell factor 1 (EBF1) as a predicted target of miR-1246. Inhibition of miR-1246 in primary human B cells, allowed authors to assess EBF1 protein levels and confirm that this putative miRNA target was elevated relative to the control transfection group. Furthermore, flow cytometric analysis revealed that B cells upregulated activation markers CD40, CD80 and CD86 in the presence of the miR-1246 inhibitor. When primary human B cells were transfected with a miR-1246 mimic, there was a reduction in EBF1 and p-AKT as measured by western blot. In addition, the expression of activation markers CD40, CD80 and CD86 was blunted in cells transfected with the miR mimic.107 As miR-1246 is not found in mice, mechanistic studies on the role of this regulatory RNA are, so far, lacking. The study by Lou et al. (2015) implicates low expression of miR-1246 in the dysregulation of B cells in SLE patients, and based on the results of the miR-1246 transfection experiments, it is likely that this miRNA contributes to the regulation of PI3K-AKT pathway, thereby fine-tunning B cell activation and proliferation. Evaluation of miR-1246 expression and function in other B-cell mediated autoimmune diseases may reveal whether the miR-1246-PI3K axis is a consistent player in the regulation of B cell responses.
Another miRNA that has been found to be elevated in a number of autoimmune diseases, including SLE, multiple sclerosis and rheumatoid arthritis, is miR-148 (previously discussed for its role in central tolerance).103,109,110 In a screen of murine B cell subsets from spleen and bone marrow, and human blood-derived B cells, plasmablasts, and plasma cells, miR-148a was identified to have higher expression in plasmablasts and plasma cells and make up 30% of all miRNAs in these cell types.111 miR-148 is also one of the most abundant and specific miRNAs in GC B cells.112 When murine B cells are stimulated in vitro with LPS or anti-IgM, miR-148a is upregulated in a time dependent manner when measurements are made by northern blot and qPCR. miR-148a was found to target the 3’ UTR of Pten, and the transcription factors Bach2 and Mitf which subsequently induce expression of Blimp1 and Irf4 to promote plasma cell differentiation and survival.111 Utilizing a conditional knockout mouse in which the miR-148a allele is flanked with loxP sites, ablation was achieved by either CD19-cre/+ or tamoxifen-inducible Rosa26CreERT2+/–. The deletion of miR-148a results in a reduction of splenic plasma cells and plasmablasts and serum immunoglobulins (IgG, IgA, IgM). The study team also observed a reduction of CD93+ long-lived plasma cells in the bone marrow of mice in which miR-148 was deleted selectively in B cells (assessed by RNA-Seq and confirmed using flow cytometry). Transcriptomics and metabolomics experiments revealed miR-148a deletion led to reduced oxidative phosphorylation, impaired glucose uptake, and altered chemokine expression. As noted by the authors, miR-148 expression coincides with upregulation of Blimp-1 and helps reinforce plasma cell differentiation via repression of GC transcription factors Bach2 and Mitf, along with reducing expression of proapoptotic proteins BIM and PTEN.111,113,114
NZB/WF1 strain is a murine lupus model in which females are more biased to disease, mimicking the known sex bias in lupus patients.102 To explore sex-driven differences in miRNA expression, a TaqMan miRNA assay system was employed to compare miRNA expression between male and female NZB/WF1 splenocytes. miR-148a is observed to be highly expressed in splenocytes from female NZB/WF1 mice when compared to males, and is estrogen responsive, suggesting that expression of this miRNA in lymphocytes contributes to the observed sex-associated differences in lupus incidence in this murine model.115 Along with the finding that miR-148 also promotes lupus-like disease in the MRL-lpr mice,79 these data suggest that the miR-148-PI3K regulatory axis in critical in the regulation of BCR signaling and terminal B cell differentiation. These observations further indicate that miR-148 regulation of this PI3K-AKT signal cascade can contribute to autoimmunity in murine models.
Observations that miR-29, miR-1246, and miR-148a are found to be inappropriately expressed in autoimmune diseases and/or mouse models of lupus are consistent with the critical role these miRNAs play in the regulation of PTEN/PI3K and maintenance of proper B cell selection and differentiation.
NF-kB and RAS-MAPK Pathways
Upon BCR engagement, NF-κB and RAS-MAPK signaling pathways regulate B cell survival and proliferation. The microRNAs −185, −30, −29, −146a, and −155 have been shown to regulate these pathways downstream of BCR signaling.
BTK, described earlier, is a kinase downstream of BCR activation, that has been identified as a target of miR-185.100 Ablation of Dicer1 under CD19-cre, as previously discussed, was found to alter B cell differentiation and favor transitional and MZ differentiation versus FO B cells. A microarray analysis was performed on FO and MZ B cells from CD19-cre Dicer1fl/+, and FO B cells from Dicer1fl/fl mice to identify miRNAs responsible for governing FO cell fate. Using microarray data, 31 miRNAs were found to be upregulated in FO B cells relative to MZ B cells and were validated by RT-PCR.100 When multiple miRNAs are identified in microarray studies, in silico analysis is employed to predict potential miRNA target transcripts so that candidate miRNAs can be selected for downstream validation.116 Using computational predictive binding approaches, Belver et al. (2010) identified the 3’UTR of Btk as a target of miR-185 for further study, but the role of the other 30 differentially expressed miRNAs identified in the course of this study that potentially regulate BCR signaling with respect to B cell differentiation remain to be explored.100 To validate the role of miR-185 in BCR signaling, MACS sorted splenic B cells were transduced with retroviral vectors to overexpress miR-185. The overexpression of miR-185 results in dampened BCR signaling and a reduction in BTK expression. When splenic B cells are transduced to overexpress Btk, it mimics the phenotype of Dicer1 ablation that skews differentiation towards transitional and MZ B cells supporting the role of miR-185 regulation of BTK levels.100
Primary human B cells were isolated from healthy donor PBMCs and stimulated in culture with LPS, anti-IgM, or IL-4 and the investigators were able to show that EphB2, an ephrin receptor tyrosine kinase, was upregulated during B cell activation and played an important role in B cell activation downstream of the BCR.117 When EphB2 expression was reduced in human B cells using siRNA, stimulated B cells had an appreciable reduction in activation.117 Further work to elucidate the precise mechanisms by which EphB2 regulates BCR signaling is still required. Current data indicate that the EphB2 induces Src-p65 and Notch1 signaling, and that this can impact NF-κB expression.117,118 Using the miRWalk database miR-185, miR-661, miR-515–5p, miR-593, miR-204, and miR-211 were identified to have predictive binging sites to the Ephb2 3’UTR. After comparing miRNA expression between stimulated (LPS-treated) and unstimulated human B cells by RT-PCR, miR-185, miR-204 and miR-515p was found to have significantly reduced expression in activated cells. miR-185 had the most significant reduction and was subjected to downstream analysis, while miR-204 and miR-515p were not validated further.117 Other investigators were able to show that miR-204 regulates EphB2 expression in murine hippocampal neurons, so despite not being subject to further experimental validation in B cells, it may, nonetheless, play a role in regulating a target relevant to BCR signaling cascades.119 When Yu et al. (2014) transfected primary human B cells to overexpress miR-185, there was lower B cell activation indicated by lower proliferation and IgG production; and reduced EphB2 and NF-κB expression, determined by western blot. Further, when a Renilla luciferase vector containing the Ephb2 3’UTR was transfected alongside a miR-185 mimic into 293T cells, miR-185 reduced Luciferase reporter activity indicating that this miRNA can directly target the Ephb2 3’UTR.117 Taken together, these studies identify miR-185 as a negative regulator of two tyrosine kinases critical in modulation of the NF-κB pathway downstream of BCR signaling.
A number of studies in humans have associated miR-185 with autoimmunity.120,121 Global miRNA landscape profiling via TaqMan array in the serum of Myasthenia gravis (MG) patients from different disease subgroups and healthy controls implicated loss of miR-185 in disease pathogenesis.120 Extracellular miRNAs were discovered and confirmed to be stable in various biofluids including serum. Profiling human serum samples permits the identification of miRNAs as potential biomarkers for disease and prognosis.122 Although this study was limited by sample size, the results identified 32 differentially expressed miRNAs in MG patients and these targets were validated in a larger cohort using targeted qPCR. This validation confirmed the downregulation of miR-185 in MG patients compared to healthy controls.120 A second study profiled miR-185 expression in circulating B cells of patients with dilated cardiomyopathy (DCM). B cells were targeted as they contribute to DCM by generating anti-heart autoantibodies (anti-adenine nucleotide translocator, anti-β1 adrenergic receptor, anti-myosin heavy chain, and anti-L-type calcium channel antibodies) that lead to heart fibrosis and injury. Measuring miR-185 transcripts using qPCR revealed that DCM patients have higher miR-185 expression than healthy controls, and that DCM patients could be stratified into miR-185 high and low groups, with miR-185-high group having better prognosis and survival.121 Although an interesting association that is consistent with the role of miR-185 dampening BCR signaling, this study did not profile the contributions of other miRNAs and did not identify any specific mechanisms by which miR-185 impacts B cell signaling or activation in DCM patients.
Another miRNA, miR-30a was identified to regulate expression of the gene Lyn. LYN, a member of the SRC family tyrosine kinases that is engaged following B cell activation, is found to have reduced protein and mRNA expression by western blot and PCR in B cells enriched from the PBMCs of SLE patients.123 Mice deficient for LYN have lower peripheral B cells, and with age, develop an autoimmune phenotype including high autoantibody titers, enlarged spleen and lymph nodes, and evident kidney inflammation.124,125 Following a bioinformatic screen, five members of the miR-30 family (a-e) were identified to potentially target the 3’UTR of Lyn. Transfection of miR-30 family mimics into two human B cell lines, enabled investigators to identify miR-30a as the miRNA capable to reducing the expression of LYN protein by western and Lyn mRNA by RT-PCR. When a luciferase reporter construct containing the Lyn 3’UTR was transfected along with miR-30a mimic and compared to a control miRNA, miR-30a was shown to reduce luciferase activity, indicating it could effectively bind the 3’UTR of Lyn. B cells isolated from SLE patient blood were shown to have reduced LYN expression and elevated miR-30a expression with a negative correlation between miR-30a gene expression and LYN protein expression measured by RT-PCR and western blot respectively.126 Altogether, these studies suggest that miR-30a regulation of LYN, a tyrosine kinase linked to MAPK and NF-κB signal transduction, is an important axis in normalization of BCR signaling and loss of this regulatory function contributes to autoimmunity.
Data from human B cells has shown that Aicda has miR-29b binding site in the 3’UTR that is not conserved in mice. Over expression of miR-29b in B cells from human tonsils results in reduced Aicda transcripts and reduced AID protein expression. This overexpression of miR-29b in human B cells also results in reduced Myc gene expression and provides an association in human cells for the miR-29 family regulating the MAPK pathway.127 The role of miR-29 in immune regulation, and particularly in autoimmune disease progression and malignant transformation of B cell lymphomas is reviewed here.128 Given that a number of studies implicate miR-29 in regulation of AKT, MAPK and MYC in human disease, along with mechanistic studies on the regulation of PI3K signaling by miR-29, a picture of the pleotropic role of this miRNA cluster in lymphocyte regulation emerges.
When miRNA, mRNA, and ChiP-seq were utilized contemporaneously to define miRNA expression in FACS sorted mouse and human tonsil hematopoietic cells, miR-146a was found to be upregulated in human GC B cells.112 Additional microarray and gene expression studies of human patients with autoimmune disease such as rheumatoid arthritis, multiple sclerosis, Sjögren’s syndrome, and psoriasis revealed that PBMCs, synovial fibroblasts and tissue, salivary glands, and skin samples from autoimmune patients had upregulated miR-146a expression relative to healthy controls.129–132 To explore the role of miR-146 overexpression in autoimmune disease pathogenesis, a transgenic mouse that carries a miR-146a allele was generated. Young mice with this allele develop spontaneous autoimmunity reminiscent of human autoimmune lymphoproliferative syndrome (ALPS) in which individuals have abnormal regulation of lymphocyte homeostasis.83 miR-146a transgenic animals had greatly enlarged lymph nodes and spleens, along with elevated serum IgG, and increased frequency of GC B cells. Downregulation of FAS on B cells in response to sustained expression of miR-146a was likely responsible for the observed dysregulation of B cells and analysis of the 3’ UTR and luciferase assays suggest that Fas is a direct target of miR-146. Microarray profiling on sorted B cells was used to assess gene expression of miR-146a targets, and several members of the NF-κB pathway were identified among miR-146a targets (as was AP1 and STAT signaling pathways). Furthermore, B cells from miR-146a transgenic mice proliferate to a lesser extent than those from wild-type animals when stimulated with LPS in culture.83 LPS stimulation initiates a BCR independent activation of B cells via Toll like receptor 4 (TLR4) that nonetheless engages both the NF-κb and Ras-MAPK pathways independent of PI3K signaling. In vitro stimulation of B cells via TLR4 is often used to engage signaling networks relevant to antigen receptor signaling.133 The reduced proliferative capacity of B cells overexpressing miR-146a is consistent with a reduction in NF-κB signaling.83 While interpretation of inflammatory phenotypes from these transgenic mice is hampered by the fact that miR-146a is globally overexpressed, the study implicates miR-146a is regulation of BCR signaling of mature B cells. Moreover, it is clear that miR-146a is upregulated in autoimmune conditions, and it is possible that dysregulation of the NF-κB signaling pathway results in altered proliferation capacity downstream of BCR signaling based on the in vitro LPS stimulation conditions. Dysregulation of the NF-κB pathway may also lead to irregular lymphocyte division and frequencies resulting in ALPS like conditions in the transgenic mouse model. Additional work will be required to further elucidate all the mechanisms and specific targets by which miR-146a regulates B cells and BCR signaling.
miR-155, a miRNA that has been previously implicated in a number of B cell malignancies, has been shown to play a critical role in GC B cells and in regulation of terminal differentiation of B cells into plasmablasts.134–137 Using IgHEL transgenic B cell adaptive transfer system, with cells either wild-type for the Bic/miR-155 locus or lacking the miRNA, the authors were able to show that miR-155 deficient plasmablasts displayed reduced proliferation and increased apoptosis. Transcriptome analysis revealed miR-155 regulated DNA replication, and that miR-155−/− B cells had reduced expression of E2F1, E2F2, and Myc.134 Myc is a downstream target of the MAPK pathway. Profiling of miRNAs from lymphocytes of murine lupus models, including MRL-lpr; B6-lpr; NZB/WF1 identified miR-155 as one of the miRNAs highly upregulated in splenic B cells in these autoimmune mouse models.81,115 Additional studies found that miR-155 is also upregulated in splenic B cells following stimulation with LPS and IL-4. Predicative algorithms identified a binding site for miR-155 in the 3’UTR of Aicda and in vitro class-switch recombination assays and transfections with luciferase reporter constructs containing the Aicda 3’UTR further demonstrate that miR-155 contributes to the regulation of AID expression. Using an AID-GFP transgenic mouse model with impaired miR-155 binding in the 3’UTR of Aicda, the loss of the binding site within Aicda leads to increased CSR in vitro after stimulation and a rapid increase in Aicda-Gfp transcripts and GFP fluorescence when measured by flow cytometry.136 Transgenic Aicda-Gfp mice and Aicda-Gfp mice with a mutated Aicda 3’UTR were immunized with NP-CGG; 21 days post immunization the mutant Aicda-Gfp mice had increased GFP expression in GC B cell populations.6,136 This increased AID-GFP expression persisted in the mutant transgenic mice, but not in the control mice that contained the intact Aicda 3’UTR, indicating that miR-155 directly regulates Aicda in post-GC B cells. The Aicda-Gfp mutant mice also display impaired affinity maturation after NP immunization, demonstrating physiological relevance of this miRNA-target interaction.136 Collectively, these data from several mouse models identify roles for miR-155 in the regulation of Myc, DNA replication, and AID upon B cell activation.
All of the aforementioned microRNAs, miR-185, miR-30, miR-29, miR-146a, and miR-155, have data to support their role in fine tuning BCR signaling via different mechanisms in which they reduce the expression of genes critical to B cell activation and proliferation. Further, these miRNAs work in concert with those that regulate the PI3K-AKT signaling pathway to ensure appropriate BCR downstream activation (Figure 1). Due to these critical roles in regulating components of the BCR signal cascade, any kind of alteration in miRNA expression levels can lead to inappropriate BCR signaling that permits the development of autoimmunity and self-reactive B cells.
Section 5: Future directions and outlook for miRNAs and the BCR
Mouse models have enabled mechanistic studies into the role that miRNAs play in orchestrating B cell development in bone marrow and regulating peripheral B cell differentiation and responses.56–58,101 Much of B cell development, differentiation and function is intimately linked to BCR signaling, and we have discussed multiple miRNAs in this review that target components of the BCR signal cascade in early B cell development, central tolerance, and peripheral B cell differentiation. Despite the results of many studies, gaps remain in our knowledge of the precise targets of miRNA regulation and how these specific targets impact signaling downstream of the antigen receptors, with respect to individual pathways involved.
Gene-targeted mouse models in which miRNAs are either deleted or over-expressed, particularly those models that enable manipulation of miRNAs in a cell-type specific manner, are extremely informative for identification of physiologically relevant miRNA-target interaction. A major limitation of these models is that they fail to take into account the pleiotropic nature of miRNA regulatory pathways, even within a given cell type. While we have highlighted some dominant miRNA-target interactions that can be pivotal in the regulation of BCR signaling, it is just as likely that loss of a given miRNA will contribute to dysregulation of multiple pathways, obfuscating results and interpretations. Overexpression models or utilization of miRNA mimics either in vitro or in vivo have provided additional insights into miRNA regulation of the BCR signalosome. But again, these models have limitations as they often exceed physiological parameters of miRNA expression, and the impact of overexpression is often dependent on the abundance of binding sites, genomic context and accessibility within the 3’ UTRs and availability of targeted transcripts. Large miRNA arrays and next-generation sequencing technologies have proved valuable for identifying miRNAs that are differentially expressed, particularly when comparing control and disease states. However, downstream of this analysis, miRNAs of interest are often selected based solely on predicative binding. Incorporation of crosslinking-immunoprecipitation methods such as PAR-CLIP, Ago HITS-CLIP and Halo-Ago2 pull down, allows for characterization of miRNA target relevance in vivo, enabling identification of the most promising miRNA-target interactions.138–140 Distinguishing the most critical miRNA-target relationships in sorted B cells will provide a strong foundation for elucidating the mechanisms of miRNA gene regulation in B cells and BCR signaling.
Future work, as it pertains to BCR signaling, should consider miRNA targets in the context of the signalosome network and validate transcript and protein expression down and upstream of suspected targets. Utilizing systems in which miRNA binding sites within 3’UTRs have been targeted to prevent specific miRNA binding will provide confident assessment of miRNA-target relationships. With improved gene editing tools such as CRISPR, generation of precisely edited animal models with only minimal alterations solely in the miRNA binding motifs is now more feasible than ever. In parallel, next generation transcriptomic and proteomic platforms enable assessment of global effects of individual miRNAs in cell lines and gene-targeted animal models – allowing investigators to appreciate how altering these regulatory networks impacts cellular programs. Integration of precise genetic editing with approaches like transcriptomics and proteomics will continue to advance our understanding of the role of individual miRNAs in B cell biology.
As we have considered in the context of this review, post-transcriptional regulation of BCR signaling pathways by miRNAs plays an important role in autoimmune disease pathogenesis and progression. While the contribution of individual miRNAs in disease etiology remains to be elucidated, given the stability of these short non-coding RNAs, they have inherent potential in clinical applications.122 Dysregulated miRNA expression could have diagnostic or prognostic value, thereby permitting early intervention using traditional therapies.141
Improving our understanding of miRNA contributions to autoimmune disease risk and pathogenesis will further advance translation of laboratory findings to the clinic as therapeutic approaches that dampen expression of individual miRNAs become available. A number of companies are testing miRNA-focused therapeutics in the context of cancer and inflammatory disease (SantarisPharma, MiRagen, etc.), with several in clinical trials.142 In the cancer field, miRNAs are currently being explored to alter tumor gene expression to make drug-resistant tumor cells sensitive to chemotherapeutics.143,144 Targeted delivery of miRNAs to alter tumor gene expression to render the cancer susceptible to the delivered chemotherapeutic represents an opportunity to exploit miRNAs to enhance clinical outcomes.143 The applications of miRNAs being employed to regulate B cell gene transcription to prevent inappropriate activation in the case of autoimmunity holds promise. Although delivery of miRNAs to exploit their therapeutic potential represents a challenge, numerous methods are being tested in current miRNA-focused therapeutics at both the developmental and clinical trial stages (reviewed by Chakraborty et al. 2021).142 Conversely, the impact of endogenous miRNA regulation of BCR signaling could be altered by inhibiting miRNAs or using approaches to target their degradation by exploiting cellular processes such as target RNA-directed miRNA degradation (TRMD).145 As implementation of these miRNA-targeted therapeutics becomes a reality, fundamental understanding of the many nodes in BCR signaling that are targeted by miRNAs will be critical to fully harness the potential for these approaches.
The studies highlighted in our review reveal a diverse array of mechanisms by which individual miRNAs contribute to the regulation of BCR signaling during early B cell development and in the course of peripheral differentiation. Given the central role of B cell antigen receptor in the survival, proliferation and indeed in the very identity of the B cell lymphocytes, it is not surprising that regulation of BCR signaling by miRNAs plays a critical role in the modulation of B cell responses and in the genesis of autoimmune diseases. Given the clinically relevant tools being developed for deployment of miRNA-based therapeutics, and the increased accessibility of genetic engineering in murine models and human cell lines, advancement of our understanding of precisely how miRNAs regulate BCR signaling can have tremendous implications for future therapeutic approaches.
Acknowledgements
Work in the Koralov lab was supported by NIH (R21AI110830, R21AI137752 and R01HL125816), LEO Foundation Grant (LF-OC-20–000351), the Judith and Stewart Colton Center for Autoimmunity Pilot grant, and a Binational Science Foundation grant. T.C.B was supported by NIAID T32AI007180 training grant and M.J.H. was supported by the NYU MSTP T32GM007308 grant. Thank you to Briana Mullins for her critical reading and review of the manuscript. Figures were generated using BioRender.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics. 2010;11(9):597–610. [DOI] [PubMed] [Google Scholar]
- 2.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature cell biology. 2009;11(3):228–234. [DOI] [PubMed] [Google Scholar]
- 3.Singh RP, Massachi I, Manickavel S, et al. The role of miRNA in inflammation and autoimmunity. Autoimmunity reviews. 2013;12(12):1160–1165. [DOI] [PubMed] [Google Scholar]
- 4.Goodnow CC, Crosbie J, Adelstein S, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334(6184):676–682. [DOI] [PubMed] [Google Scholar]
- 5.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annual review of biophysics. 2013;42:217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crouch EE, Li Z, Takizawa M, et al. Regulation of AID expression in the immune response. Journal of Experimental Medicine. 2007;204(5):1145–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busslinger M Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. [DOI] [PubMed] [Google Scholar]
- 8.Rajewsky K Clonal selection and learning in the antibody system. Nature. 1996;381(6585):751–758. [DOI] [PubMed] [Google Scholar]
- 9.Simpson LJ, Patel S, Bhakta NR, et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nature Immunology. 2014;15(12):1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkler TH, Mårtensson I-L. The role of the Pre-B cell receptor in B cell development, repertoire selection, and tolerance. Frontiers in immunology. 2018;9:2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nature immunology. 2008;9(6):613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemazee D Mechanisms of central tolerance for B cells. Nature Reviews Immunology. 2017;17(5):281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verkoczy L, Duong B, Skog P, et al. Basal B cell receptor-directed phosphatidylinositol 3-kinase signaling turns off RAGs and promotes B cell-positive selection. The Journal of Immunology. 2007;178(10):6332–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carsetti R, Köhler G, Lamers MC. Transitional B cells are the target of negative selection in the B cell compartment. The Journal of experimental medicine. 1995;181(6):2129–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petro JB, Gerstein RM, Lowe J, Carter RS, Shinners N, Khan WN. Transitional type 1 and 2 B lymphocyte subsets are differentially responsive to antigen receptor signaling. Journal of Biological Chemistry. 2002;277(50):48009–48019. [DOI] [PubMed] [Google Scholar]
- 16.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igα/β heterodimer. Cell. 2004;117(6):787–800. [DOI] [PubMed] [Google Scholar]
- 17.Cerutti A, Cols M, Puga I. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nature Reviews Immunology. 2013;13(2):118–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pillai S, Cariappa A. The follicular versus marginal zone B lymphocyte cell fate decision. Nature Reviews Immunology. 2009;9(11):767–777. [DOI] [PubMed] [Google Scholar]
- 19.Thomas MD, Srivastava B, Allman D. Regulation of peripheral B cell maturation. Cellular immunology. 2006;239(2):92–102. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman W, Lakkis FG, Chalasani G. B cells, antibodies, and more. Clinical Journal of the American Society of Nephrology. 2016;11(1):137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paus D, Phan TG, Chan TD, Gardam S, Basten A, Brink R. Antigen recognition strength regulates the choice between extrafollicular plasma cell and germinal center B cell differentiation. Journal of experimental medicine. 2006;203(4):1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunological reviews. 2012;247(1):52–63. [DOI] [PubMed] [Google Scholar]
- 23.Srinivasan L, Sasaki Y, Calado DP, et al. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139(3):573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamel KM, Liarski VM, Clark MR. Germinal center B-cells. Autoimmunity. 2012;45(5):333–347. [DOI] [PubMed] [Google Scholar]
- 25.Yabas M, Elliott H, Hoyne GF. The role of alternative splicing in the control of immune homeostasis and cellular differentiation. International journal of molecular sciences. 2016;17(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. B cell antigen receptor signaling 101. Molecular immunology. 2004;41(6–7):599–613. [DOI] [PubMed] [Google Scholar]
- 27.Kurosaki T, Aiba Y, Kometani K, Moriyama S, Takahashi Y. Unique properties of memory B cells of different isotypes. Immunological reviews. 2010;237(1):104–116. [DOI] [PubMed] [Google Scholar]
- 28.Packard TA, Cambier JC. B lymphocyte antigen receptor signaling: initiation, amplification, and regulation. F1000prime reports. 2013;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woyach JA, Johnson AJ, Byrd JC. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120(6):1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fearon DT, Carroll MC. Regulation of B lymphocyte responses to foreign and self-antigens by the CD19/CD21 complex. Annu Rev Immunol. 2000;18:393–422. [DOI] [PubMed] [Google Scholar]
- 31.Merolle MI, Ahmed M, Nomie K, Wang ML. The B cell receptor signaling pathway in mantle cell lymphoma. Oncotarget. 2018;9(38):25332–25341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam K-P, Kühn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90(6):1073–1083. [DOI] [PubMed] [Google Scholar]
- 33.Tanaka S, Baba Y. B Cell Receptor Signaling. In: Wang J-Y, ed. B Cells in Immunity and Tolerance. Singapore: Springer Singapore; 2020:23–36. [Google Scholar]
- 34.Kurosaki T Regulation of BCR signaling. Molecular immunology. 2011;48(11):1287–1291. [DOI] [PubMed] [Google Scholar]
- 35.Tybulewicz VLJ, Henderson RB. Rho family GTPases and their regulators in lymphocytes. Nat Rev Immunol. 2009;9(9):630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdelrasoul H, Werner M, Setz CS, Okkenhaug K, Jumaa H. PI3K induces B-cell development and regulates B cell identity. Scientific reports. 2018;8(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clayton E, Bardi G, Bell SE, et al. A crucial role for the p110δ subunit of phosphatidylinositol 3-kinase in B cell development and activation. The Journal of experimental medicine. 2002;196(6):753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annual review of biochemistry. 1998;67(1):481–507. [DOI] [PubMed] [Google Scholar]
- 39.Okkenhaug K, Bilancio A, Farjot G, et al. Impaired B and T cell antigen receptor signaling in p110δ PI 3-kinase mutant mice. Science. 2002;297(5583):1031–1034. [DOI] [PubMed] [Google Scholar]
- 40.Ramadani F, Bolland DJ, Garcon F, et al. The PI3K isoforms p110α and p110δ are essential for pre–B cell receptor signaling and B cell development. Science signaling. 2010;3(134):ra60–ra60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aiba Y, Kameyama M, Yamazaki T, Tedder TF, Kurosaki T. Regulation of B-cell development by BCAP and CD19 through their binding to phosphoinositide 3-kinase. Blood, The Journal of the American Society of Hematology. 2008;111(3):1497–1503. [DOI] [PubMed] [Google Scholar]
- 42.Okkenhaug K, Vanhaesebroeck B. PI3K in lymphocyte development, differentiation and activation. Nature Reviews Immunology. 2003;3(4):317–330. [DOI] [PubMed] [Google Scholar]
- 43.Alessi DR, James SR, Downes CP, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Current biology. 1997;7(4):261–269. [DOI] [PubMed] [Google Scholar]
- 44.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nature Reviews Immunology. 2009;9(3):195–205. [DOI] [PubMed] [Google Scholar]
- 45.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129(7):1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stokoe D, Stephens LR, Copeland T, et al. Dual role of phosphatidylinositol-3, 4, 5-trisphosphate in the activation of protein kinase B. Science. 1997;277(5325):567–570. [DOI] [PubMed] [Google Scholar]
- 47.Okkenhaug K Rules of engagement: distinct functions for the four class I PI3K catalytic isoforms in immunity. Annals of the New York Academy of Sciences. 2013;1280:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avalos AM, Meyer-Wentrup F, Ploegh HL. Chapter One - B-Cell Receptor Signaling in Lymphoid Malignancies and Autoimmunity. In: Ploegh HL, ed. Advances in Immunology. Vol 123. Academic Press; 2014:1–49. [DOI] [PubMed] [Google Scholar]
- 49.Woyach JA, Johnson AJ, Byrd JC. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120(6):1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao C, Nemazee D, Gonzalez-Martin A. MicroRNA control of B cell tolerance, autoimmunity and cancer. Paper presented at: Seminars in Cancer Biology2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coffre M, Koralov SB. miRNAs in B Cell Development and Lymphomagenesis. Trends in Molecular Medicine. 2017;23(8):721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wigton EJ, Ansel KM. Noncoding RNAs in B cell responses. RNA Biology. 2021;18(5):633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuertes T, Ramiro AR, de Yebenes VG. miRNA-Based Therapies in B Cell Non-Hodgkin Lymphoma. Trends Immunol. 2020;41(10):932–947. [DOI] [PubMed] [Google Scholar]
- 54.Getaneh Z, Asrie F, Melku M. MicroRNA profiles in B-cell non-Hodgkin lymphoma. EJIFCC. 2019;30(2):195–214. [PMC free article] [PubMed] [Google Scholar]
- 55.Solé C, Larrea E, Di Pinto G, Tellaetxe M, Lawrie CH. miRNAs in B-cell lymphoma: Molecular mechanisms and biomarker potential. Cancer Letters. 2017;405:79–89. [DOI] [PubMed] [Google Scholar]
- 56.Koralov SB, Muljo SA, Galler GR, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–874. [DOI] [PubMed] [Google Scholar]
- 57.Brandl A, Daum P, Brenner S, et al. The microprocessor component, DGCR8, is essential for early B‐cell development in mice. European journal of immunology. 2016;46(12):2710–2718. [DOI] [PubMed] [Google Scholar]
- 58.Coffre M, Benhamou D, Rieß D, et al. miRNAs Are Essential for the Regulation of the PI3K/AKT/FOXO Pathway and Receptor Editing during B Cell Maturation. Cell Reports. 2016;17(9):2271–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ventura A, Young AG, Winslow MM, et al. Targeted Deletion Reveals Essential and Overlapping Functions of the miR-17∼92 Family of miRNA Clusters. Cell. 2008;132(5):875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai M, Gonzalez-Martin A, Cooper AB, et al. Regulation of B-cell development and tolerance by different members of the miR-17∼92 family microRNAs. Nature Communications. 2016;7(1):12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Labi V, Peng S, Klironomos F, et al. Context-specific regulation of cell survival by a miRNA-controlled BIM rheostat. Genes & development. 2019;33(23–24):1673–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monticelli S, Ansel KM, Xiao C, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6(8):R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–159. [DOI] [PubMed] [Google Scholar]
- 64.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proceedings of the National Academy of Sciences. 2007;104(17):7080–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23(3):275–286. [DOI] [PubMed] [Google Scholar]
- 66.Rao DS, O’Connell RM, Chaudhuri AA, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33(1):48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodnow CC, Sprent J, de St Groth BF, Vinuesa CG. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature. 2005;435(7042):590–597. [DOI] [PubMed] [Google Scholar]
- 68.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301(5638):1374–1377. [DOI] [PubMed] [Google Scholar]
- 69.Hampe CS. B cells in autoimmune diseases. Scientifica. 2012;2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fields ML, Erikson J. The regulation of lupus-associated autoantibodies: immunoglobulin transgenic models. Current opinion in immunology. 2003;15(6):709–717. [DOI] [PubMed] [Google Scholar]
- 71.Hartley SB, Cooke MP, Fulcher DA, et al. Elimination of self-reactive B lymphocytes proceeds in two stages: arrested development and cell death. Cell. 1993;72(3):325–335. [DOI] [PubMed] [Google Scholar]
- 72.Nemazee D, Hogquist KA. Antigen receptor selection by editing or downregulation of V (D) J recombination. Current opinion in immunology. 2003;15(2):182–189. [DOI] [PubMed] [Google Scholar]
- 73.Grammer AC, Fischer R, Lee O, Zhang X, Lipsky PE. Flow cytometric assessment of the signaling status of human B lymphocytes from normal and autoimmune individuals. Arthritis Res Ther. 2004;6(1):28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wong CK, Wong PT, Tam LS, Li EK, Chen DP, Lam CW. Activation profile of intracellular mitogen-activated protein kinases in peripheral lymphocytes of patients with systemic lupus erythematosus. J Clin Immunol. 2009;29(6):738–746. [DOI] [PubMed] [Google Scholar]
- 75.Wu T, Qin X, Kurepa Z, et al. Shared signaling networks active in B cells isolated from genetically distinct mouse models of lupus. J Clin Invest. 2007;117(8):2186–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rowland SL, DePersis CL, Torres RM, Pelanda R. Ras activation of Erk restores impaired tonic BCR signaling and rescues immature B cell differentiation. Journal of Experimental Medicine. 2010;207(3):607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Teodorovic LS, Babolin C, Rowland SL, et al. Activation of Ras overcomes B-cell tolerance to promote differentiation of autoreactive B cells and production of autoantibodies. Proceedings of the National Academy of Sciences. 2014;111(27):E2797–E2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Greaves SA, Peterson JN, Torres RM, Pelanda R. Activation of the MEK-ERK Pathway Is Necessary but Not Sufficient for Breaking Central B Cell Tolerance. Front Immunol. 2018;9:707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gonzalez-Martin A, Adams BD, Lai M, et al. The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nature immunology. 2016;17(4):433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duong BH, Ota T, Aoki-Ota M, et al. Negative Selection by IgM Superantigen Defines a B Cell Central Tolerance Compartment and Reveals Mutations Allowing Escape. The Journal of Immunology. 2011;187(11):5596–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dai R, Zhang Y, Khan D, et al. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PloS one. 2010;5(12):e14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lindberg RLP, Hoffmann F, Mehling M, Kuhle J, Kappos L. Altered expression of miR-17–5p in CD4+ lymphocytes of relapsing–remitting multiple sclerosis patients. European Journal of Immunology. 2010;40(3):888–898. [DOI] [PubMed] [Google Scholar]
- 83.Guo Q, Zhang J, Li J, et al. Forced miR-146a expression causes autoimmune lymphoproliferative syndrome in mice via downregulation of Fas in germinal center B cells. Blood. 2013;121(24):4875–4883. [DOI] [PubMed] [Google Scholar]
- 84.Coulter TI, Chandra A, Bacon CM, et al. Clinical spectrum and features of activated phosphoinositide 3-kinase δ syndrome: A large patient cohort study. J Allergy Clin Immunol. 2017;139(2):597–606.e594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Avery DT, Kane A, Nguyen T, et al. Germline-activating mutations in PIK3CD compromise B cell development and function. J Exp Med. 2018;215(8):2073–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Greaves SA, Peterson JN, Strauch P, Torres RM, Pelanda R. Active PI3K abrogates central tolerance in high-avidity autoreactive B cells. The Journal of experimental medicine. 2019;216(5):1135–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Preite S, Cannons JL, Radtke AJ, et al. Hyperactivated PI3Kδ promotes self and commensal reactivity at the expense of optimal humoral immunity. Nat Immunol. 2018;19(9):986–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wray-Dutra MN, Al Qureshah F, Metzler G, Oukka M, James RG, Rawlings DJ. Activated PIK3CD drives innate B cell expansion yet limits B cell-intrinsic immune responses. J Exp Med. 2018;215(10):2485–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Browne CD, Del Nagro CJ, Cato MH, Dengler HS, Rickert RC. Suppression of phosphatidylinositol 3, 4, 5-trisphosphate production is a key determinant of B cell anergy. Immunity. 2009;31(5):749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Getahun A, Beavers NA, Larson SR, Shlomchik MJ, Cambier JC. Continuous inhibitory signaling by both SHP-1 and SHIP-1 pathways is required to maintain unresponsiveness of anergic B cells. Journal of Experimental Medicine. 2016;213(5):751–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuo G, Wu C-Y, Yang H-Y. MiR-17–92 cluster and immunity. Journal of the Formosan Medical Association. 2019;118(1):2–6. [DOI] [PubMed] [Google Scholar]
- 92.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nature immunology. 2008;9(4):405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goodnow CC. Transgenic mice and analysis of B-cell tolerance. Annu Rev Immunol. 1992;10:489–518. [DOI] [PubMed] [Google Scholar]
- 94.Goodnow CC, Crosbie J, Adelstein S, et al. Clonal silencing of self-reactive B lymphocytes in a transgenic mouse model. Cold Spring Harb Symp Quant Biol. 1989;54 Pt 2:907–920. [DOI] [PubMed] [Google Scholar]
- 95.Kwak K, Akkaya M, Pierce SK. B cell signaling in context. Nature immunology. 2019;20(8):963–969. [DOI] [PubMed] [Google Scholar]
- 96.Casola S, Otipoby KL, Alimzhanov M, et al. B cell receptor signal strength determines B cell fate. Nature Immunology. 2004;5(3):317–327. [DOI] [PubMed] [Google Scholar]
- 97.Ripperger TJ, Bhattacharya D. Transcriptional and Metabolic Control of Memory B Cells and Plasma Cells. Annual Review of Immunology. 2021;39:345–368. [DOI] [PubMed] [Google Scholar]
- 98.Meinzinger J, Jäck H-M, Pracht K. miRNA meets plasma cells “How tiny RNAs control antibody responses”. Clinical Immunology. 2018;186:3–8. [DOI] [PubMed] [Google Scholar]
- 99.Rickert RC. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nature Reviews Immunology. 2013;13(8):578–591. [DOI] [PubMed] [Google Scholar]
- 100.Belver L, de Yébenes VG, Ramiro AR. MicroRNAs Prevent the Generation of Autoreactive Antibodies. Immunity. 2010;33(5):713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu S, Guo K, Zeng Q, Huo J, Lam K-P. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood. 2012;119(3):767–776. [DOI] [PubMed] [Google Scholar]
- 102.Jacob N, Stohl W. Autoantibody-dependent and autoantibody-independent roles for B cells in systemic lupus erythematosus: past, present, and future. Autoimmunity. 2010;43(1):84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chauhan SK, Singh VV, Rai R, Rai M, Rai G. Differential microRNA profile and post-transcriptional regulation exist in systemic lupus erythematosus patients with distinct autoantibody specificities. Journal of clinical immunology. 2014;34(4):491–503. [DOI] [PubMed] [Google Scholar]
- 104.Hines MJ, Coffre M, Mudianto T, et al. miR-29 Sustains B Cell Survival and Controls Terminal Differentiation via Regulation of PI3K Signaling. Cell Reports. 2020;33(9):108436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van Nieuwenhuijze A, Dooley J, Humblet-Baron S, et al. Defective germinal center B-cell response and reduced arthritic pathology in microRNA-29a-deficient mice. Cellular and Molecular Life Sciences. 2017;74(11):2095–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu H, Chang C, Lu Q. The Epigenetics of Lupus Erythematosus. In: Chang C, Lu Q, eds. Epigenetics in Allergy and Autoimmunity. Singapore: Springer Singapore; 2020:185–207. [DOI] [PubMed] [Google Scholar]
- 107.Luo S, Liu Y, Liang G, et al. The role of microRNA-1246 in the regulation of B cell activation and the pathogenesis of systemic lupus erythematosus. Clinical Epigenetics. 2015;7(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schell SL, Rahman ZSM. miRNA-Mediated Control of B Cell Responses in Immunity and SLE. Frontiers in immunology. 2021;12:683710–683710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pan W, Zhu S, Yuan M, et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. The journal of immunology. 2010;184(12):6773–6781. [DOI] [PubMed] [Google Scholar]
- 110.Te JL, Dozmorov IM, Guthridge JM, et al. Identification of unique microRNA signature associated with lupus nephritis. PloS one. 2010;5(5):e10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Porstner M, Winkelmann R, Daum P, et al. miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors Mitf and Bach2. European Journal of Immunology. 2015;45(4):1206–1215. [DOI] [PubMed] [Google Scholar]
- 112.Kuchen S, Resch W, Yamane A, et al. Regulation of MicroRNA Expression and Abundance during Lymphopoiesis. Immunity. 2010;32(6):828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pracht K, Meinzinger J, Schulz SR, et al. miR-148a controls metabolic programming and survival of mature CD19-negative plasma cells in mice. European Journal of Immunology. 2021;51(5):1089–1109. [DOI] [PubMed] [Google Scholar]
- 114.Tellier J miR‐148a weaves its thread into the plasma cell fate. European Journal of Immunology. 2021;51(5):1076–1079. [DOI] [PubMed] [Google Scholar]
- 115.Dai R, McReynolds S, LeRoith T, Heid B, Liang Z, Ahmed SA. Sex differences in the expression of lupus-associated miRNAs in splenocytes from lupus-prone NZB/WF1 mice. Biology of Sex Differences. 2013;4(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fan X, Kurgan L. Comprehensive overview and assessment of computational prediction of microRNA targets in animals. Briefings in Bioinformatics. 2014;16(5):780–794. [DOI] [PubMed] [Google Scholar]
- 117.Yu M, Liang W, Wen S, et al. EphB2 contributes to human naive B‐cell activation and is regulated by miR‐185. The FASEB Journal. 2014;28(8):3609–3617. [DOI] [PubMed] [Google Scholar]
- 118.Darling TK, Lamb TJ. Emerging Roles for Eph Receptors and Ephrin Ligands in Immunity. Frontiers in Immunology. 2019;10(1473). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Danka Mohammed CP, Rhee H, Phee BK, et al. miR‐204 downregulates EphB2 in aging mouse hippocampal neurons. Aging Cell. 2016;15(2):380–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nogales-Gadea G, Ramos-Fransi A, Suárez-Calvet X, et al. Analysis of Serum miRNA Profiles of Myasthenia Gravis Patients. PLOS ONE. 2014;9(3):e91927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu M, Liang W, Xie Y, et al. Circulating miR-185 might be a novel biomarker for clinical outcome in patients with dilated cardiomyopathy. Scientific Reports. 2016;6(1):33580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research. 2008;18(10):997–1006. [DOI] [PubMed] [Google Scholar]
- 123.Liossis S-NC, Solomou EE, Dimopoulos M-A, Panayiotidis P, Mavrikakis MM, Sfikakis PP. B-cell kinase lyn deficiency in patients with systemic lupus erythematosus. Journal of Investigative Medicine. 2001;49(2):157–165. [DOI] [PubMed] [Google Scholar]
- 124.Chan VWF, Meng F, Soriano P, DeFranco AL, Lowell CA. Characterization of the B Lymphocyte Populations in Lyn-Deficient Mice and the Role of Lyn in Signal Initiation and Down-Regulation. Immunity. 1997;7(1):69–81. [DOI] [PubMed] [Google Scholar]
- 125.Nishizumi H, Taniuchi I, Yamanashi Y, et al. Impaired proliferation of peripheral B cells and indication of autoimmune disease in lyn-deficient mice. Immunity. 1995;3(5):549–560. [DOI] [PubMed] [Google Scholar]
- 126.Liu Y, Dong J, Mu R, et al. MicroRNA-30a Promotes B Cell Hyperactivity in Patients With Systemic Lupus Erythematosus by Direct Interaction With Lyn. Arthritis & Rheumatism. 2013;65(6):1603–1611. [DOI] [PubMed] [Google Scholar]
- 127.Recaldin T, Hobson PS, Mann EH, et al. miR-29b directly targets activation-induced cytidine deaminase in human B cells and can limit its inappropriate expression in naïve B cells. Molecular Immunology. 2018;101:419–428. [DOI] [PubMed] [Google Scholar]
- 128.Horita M, Farquharson C, Stephen LA. The role of miR‐29 family in disease. Journal of cellular biochemistry. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EKL. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Research & Therapy. 2008;10(4):R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stanczyk J, Pedrioli DML, Brentano F, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis & Rheumatism. 2008;58(4):1001–1009. [DOI] [PubMed] [Google Scholar]
- 131.Alevizos I, Illei GG. MicroRNAs in Sjögren’s syndrome as a prototypic autoimmune disease. Autoimmunity Reviews. 2010;9(9):618–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sonkoly E, Wei T, Janson PCJ, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PloS one. 2007;2(7):e610–e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dye JR, Palvanov A, Guo B, Rothstein TL. B cell receptor cross-talk: exposure to lipopolysaccharide induces an alternate pathway for B cell receptor-induced ERK phosphorylation and NF-κB activation. The Journal of Immunology. 2007;179(1):229–235. [DOI] [PubMed] [Google Scholar]
- 134.Arbore G, Henley T, Biggins L, et al. MicroRNA-155 is essential for the optimal proliferation and survival of plasmablast B cells. Life Sci Alliance. 2019;2(3):e201800244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Teng G, Hakimpour P, Landgraf P, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28(5):621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Thai T-H, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–608. [DOI] [PubMed] [Google Scholar]
- 138.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature. 2009;460(7254):479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li X, Pritykin Y, Concepcion CP, et al. High-Resolution In Vivo Identification of miRNA Targets by Halo-Enhanced Ago2 Pull-Down. Molecular Cell. 2020;79(1):167–179.e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Condrat CE, Thompson DC, Barbu MG, et al. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells. 2020;9(2):276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chakraborty C, Sharma AR, Sharma G, Lee S-S. Therapeutic advances of miRNAs: A preclinical and clinical update. Journal of Advanced Research. 2021;28:127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dai X, Tan C. Combination of microRNA therapeutics with small-molecule anticancer drugs: Mechanism of action and co-delivery nanocarriers. Advanced Drug Delivery Reviews. 2015;81:184–197. [DOI] [PubMed] [Google Scholar]
- 144.Wang J, Meng F, Dai E, et al. Identification of associations between small molecule drugs and miRNAs based on functional similarity. Oncotarget. 2016;7(25):38658–38669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sheu-Gruttadauria J, Pawlica P, Klum SM, et al. Structural Basis for Target-Directed MicroRNA Degradation. Molecular Cell. 2019;75(6):1243–1255.e1247. [DOI] [PMC free article] [PubMed] [Google Scholar]