Abstract
Comprehensive identification of conditionally essential genes requires efficient tools for generating high-density transposon libraries that, ideally, can be analysed using next-generation sequencing methods such as Transposon Directed Insertion-site Sequencing (TraDIS). The Himar1 (mariner) transposon is ideal for generating near-saturating mutant libraries, especially in AT-rich chromosomes, as the requirement for integration is a TA dinucleotide, and this transposon has been used for mutagenesis of a wide variety of bacteria. However, plasmids for mariner delivery do not necessarily work well in all bacteria. In particular, there are limited tools for functional genomic analysis of Pasteurellaceae species of major veterinary importance, such as swine and cattle pathogens, Actinobacillus pleuropneumoniae and Pasteurella multocida, respectively. Here, we developed plasmids, pTsodCPC9 and pTlacPC9 (differing only in the promoter driving expression of the transposase gene), that allow delivery of mariner into both these pathogens, but which should also be applicable to a wider range of bacteria. Using the pTlacPC9 vector, we have generated, for the first time, saturating mariner mutant libraries in both A. pleuropneumoniae and P. multocida that showed a near random distribution of insertions around the respective chromosomes as detected by TraDIS. A preliminary screen of 5000 mutants each identified 8 and 14 genes, respectively, that are required for growth under anaerobic conditions. Future high-throughput screening of the generated libraries will facilitate identification of mutants required for growth under different conditions, including in vivo, highlighting key virulence factors and pathways that can be exploited for development of novel therapeutics and vaccines.
Keywords: Mariner, Transposon, TraDIS, Pasteurellaceae, Actinobacillus pleuropneumoniae, Pasteurella multocida
Background
Actinobacillus pleuropneumoniae, a member of the Pasteurellaceae, is the causative agent of porcine pleuropneumonia, a highly contagious, often fatal, respiratory disease that causes considerable economic losses to the swine industry worldwide (Sassu et al. 2018). Certain virulence factors have been shown to have specific roles in the pathogenesis of A. pleuropneumoniae infection including RTX toxins, capsule, lipopolysaccharide and various outer membrane proteins (Bossé et al. 2002; Chiers et al. 2010). In addition, two signature-tagged mutagenesis (STM) studies identified a large number of genes that contribute to the ability of A. pleuropneumoniae to survive and cause disease in pigs, though neither screen was saturating (Fuller et al. 2000a; Sheehan et al. 2003).
For more than two decades, Tn10 has been the transposon of choice for generating libraries of random mutants of A. pleuropneumoniae for STM and other studies (Tascon et al. 1993; Rioux et al. 1999; Fuller et al. 2000a; Bossé et al. 2001; Sheehan et al. 2003; Grasteau et al. 2011). However, Tn10 has an insertion site preference for GCTNAGC (Bender and Kleckner 1992), and different insertional hotspots were reported in A. pleuropneumoniae STM studies (Fuller et al. 2000a; Sheehan et al. 2003; Bossé et al. 2010), limiting the usefulness of this transposon for creating a fully saturating library. Clearly, a more random transposon mutagenesis system in A. pleuropneumoniae is required to allow genome-wide analysis of fitness using high-throughput sequencing methods such as Transposon Directed Insertion-site Sequencing (TraDIS) that not only precisely map, but also quantitatively measure the relative abundance of each transposon insertion in a pool of mutants (Gawronski et al. 2009; Langridge et al. 2009; van Opijnen et al. 2009).
The Himar1 (mariner) transposon, originally isolated from the horn fly, Haematobia irritans, has been shown to insert randomly into the chromosomes of a wide range of bacteria with a dinucleotide target of “TA” (Picardeau 2010). In particular, mariner has been used for mutagenesis of Haemophilus influenzae (Akerley et al. 1998; Gawronski et al. 2009), another member of the Pasteurellaceae, with a comparatively AT-rich genome, like A. pleuropneumoniae.
Preliminary (unpublished) investigations in our laboratory using the vector pMinihimarRB1 (Bouhenni et al. 2005), a kind gift from Dr. D. Saffarini, revealed that mariner is functional in A. pleuropneumoniae, however disadvantages including high background with kanamycin selection and retention of plasmid in initial transconjugants limited the usefulness of this vector for genome-wide analysis of fitness using TraDIS, a next generation sequencing method for mapping insertion sites (Langridge et al. 2009). We therefore decided to construct a mariner delivery vector incorporating the following desired components: a stringent selection gene carried by the mini-transposon; presence of DNA uptake sequences flanking the selection gene to allow for easy transfer of mutations between different strains via natural transformation; the presence of paired ISceI restriction sites just outside of the mini-transposon element to allow elimination of reads from residual plasmid during TraDIS; sequences to allow mobilization of the vector from a conjugal donor strain; and the C9 hyperactive mutant mariner transposase gene (Lampe et al. 1999), under control of either a constitutive or inducible promoter, encoded adjacent to the mini-transposon element to ensure stability of insertions. As the replication origin of the high copy number T-cloning vector, pGEMT, is the same as that in pBluescript, which we previously showed was not functional in A. pleuropneumoniae (Sheehan et al. 2000), it provided an ideal starting point for rational construction of a suicide mariner delivery system for use in Pasteurellaceae species.
Results
Construction and evaluation of mariner vectors
Two novel mobilizable mariner delivery vectors, pTsodCPC9 and pTlacPC9 (Fig. 1), differing only in the promoter driving expression of the C9 transposase gene, were successfully generated using pGEM-T as vector backbone. Conjugal transfer into A. pleuropneumoniae MIDG2331 from the diaminopimelic acid (DAP)-dependent MFDpir, achieved for both vectors, confirmed that oriT and traJ sequences were sufficient to facilitate mobilization. Comparison of results for A. pleuropneumoniae indicated that although the sodC promoter is constitutively expressed (Bossé et al. 2009), isopropyl-ß-D-galactopyranoside (IPTG) induction of C9 transposase expression from the lac promoter resulted in higher frequencies of transposition (10− 6–10− 8 compared to 10− 7–10− 10). Furthermore, similar frequencies of transposition were found in the genome of Pasteurella multocida MIDG3277 (10− 6–10− 8) using pTlacPC9, whereas no transposant was recovered using pTsodCPC9 in this species. Therefore, the pTlacPC9 vector was used for library construction in both A. pleuropneumoniae and P. multocida.
Fig. 1.
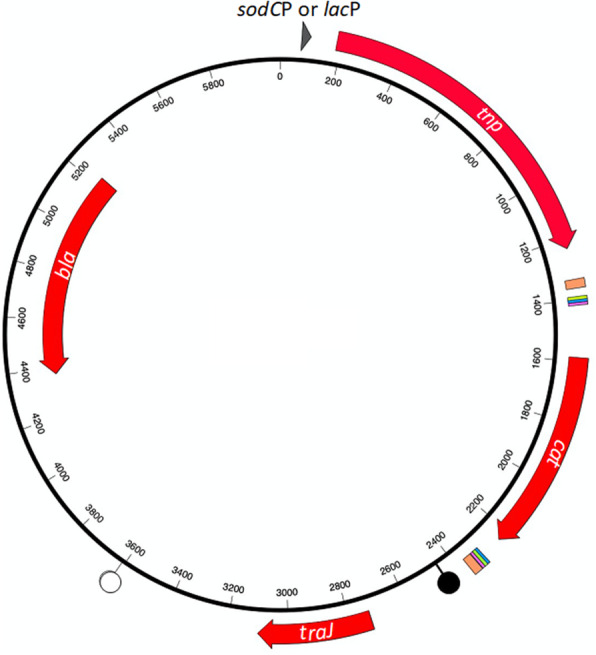
General map of mariner vectors, pTsodCPC9 and pTlacPC9, differing only in the promoter for the tnp gene. Genes tnp (mariner transposase C9 mutant), cat (chloramphenicol resistance), traJ (plasmid transfer gene), and bla (ampicillin resistance) are indicated by the appropriately labelled solid red arrows. The origin of plasmid transfer (oriT) is shown as a filled lollipop ( ), and the origin of plasmid replication (colE1) as a hollow lollipop (
), and the origin of plasmid replication (colE1) as a hollow lollipop ( ). Coloured blocks flanking the cat gene indicate the locations of Himar1 inverted repeats (thick orange), and paired copies of DNA uptake sequences for Neisseria spp. (thin green), H. influenzae (thin pink) and A. pleuropneumoniae (thin blue). Arrowhead upstream of the C9 tnp gene indicates promoter sequences for either A. pleuropneumoniae sodC or E. coli lac genes, depending on the vector (pTsodCPC9 and pTlacPC9, respectively)
). Coloured blocks flanking the cat gene indicate the locations of Himar1 inverted repeats (thick orange), and paired copies of DNA uptake sequences for Neisseria spp. (thin green), H. influenzae (thin pink) and A. pleuropneumoniae (thin blue). Arrowhead upstream of the C9 tnp gene indicates promoter sequences for either A. pleuropneumoniae sodC or E. coli lac genes, depending on the vector (pTsodCPC9 and pTlacPC9, respectively)
Colony PCR revealed that initial transconjugants retained extrachromosomal plasmid, as indicated by amplification of both chloramphenicol (Cm) cassette and oriT/traJ sequences (data not shown). Following subculture on selective agar, only the Cm cassette could be amplified from selected mutants, indicating loss of plasmid and integration of the transposon into the chromosome. Southern blot (not shown) and linker-PCR (Fig. 2) confirmed single insertions in different locations in 12 randomly selected mutants. Insertions were stable in the absence of selection for 20 generations.
Fig. 2.
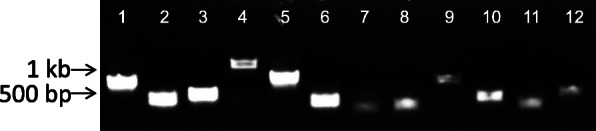
Linker PCR products for 12 randomly selected A. pleuropneumoniae mutants. Sequences flanking mariner insertions were amplified from AluI-digested linker-ligated DNA fragments using primers L-PCR-C and IR-Left_out (for amplification of the left flank). Lanes 1–12 show single amplicons for each of selected mutants, indicating the presence of a single insertion of transposon in each case
TraDIS analysis of A. pleuropneumoniae and P. multocida mariner libraries
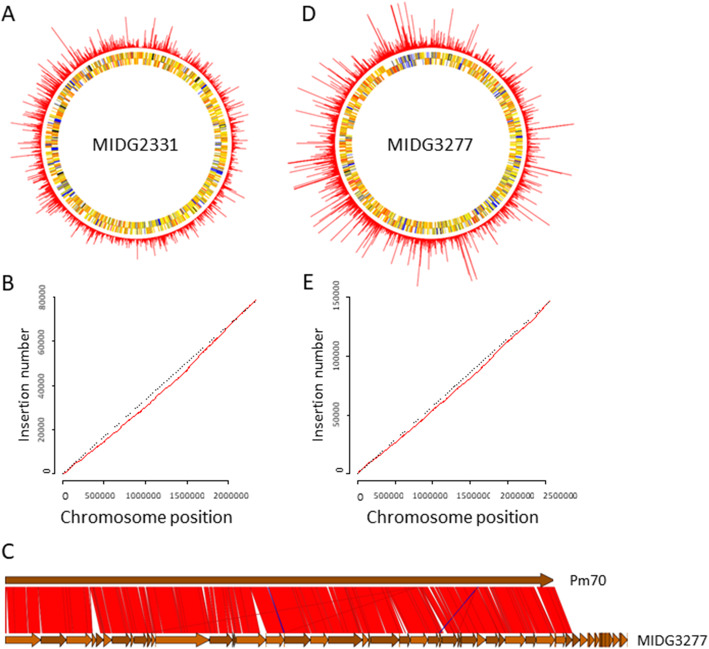
Analysis of linker-PCR products generated from DNA (+/− ISceI digestion) of the pooled A. pleuropneumoniae mariner library containing > 78,000 transconjugants showed a dominant plasmid-specific band only in the sample that was not treated with ISceI (Fig. 3). Both samples showed a strongly stained smear of DNA ranging from 100 to 500 bp in size, indicating a good distribution of insertions in the library. Subsequent TraDIS analysis of ISceI-digested A. pleuropneumoniae library DNA generated 16,565,883 sequence reads from the 5′ end of the mariner transposon. Of these, 15,381,053 (92.8%) were mapped to the complete MIDG2331 reference genome (Bossé et al. 2016), and 99.4% of those (i.e., 15,282,862 reads) corresponded to an insertion at a TA position. These mapped to 78,638 unique insertion sites, representing an insertion approximately every 30 bp on average and occupying 45.8% available TA sites in the genome. The insertion sites were distributed near randomly around the MIDG2331 genome (Fig. 4A), with the exception of a bias towards insertions closer to the origin of replication (Fig. 4B). This is likely due to those regions of the genome being present in multiple copies during transposition and is a commonly observed feature of transposon mutagenesis datasets (Langridge et al. 2009; Chao et al. 2016).
Fig. 3.
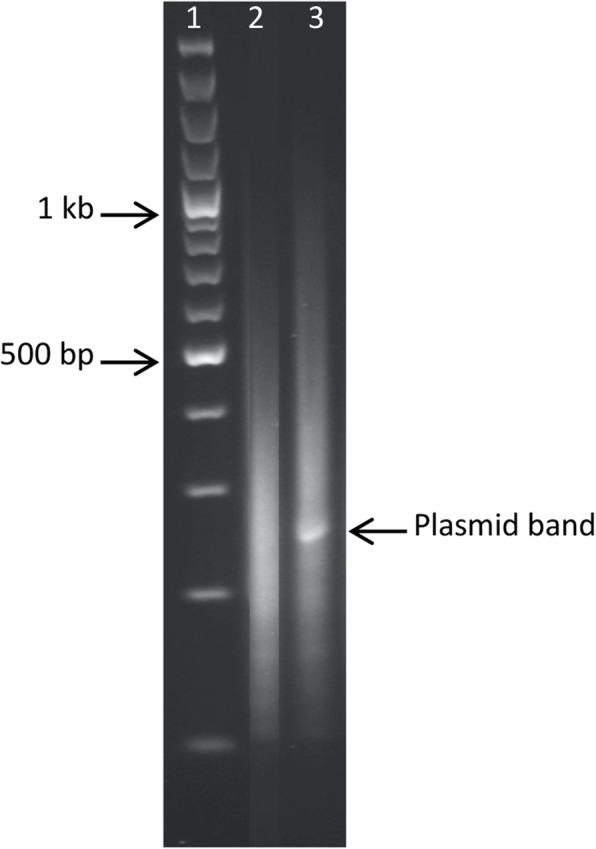
Comparison of linker PCR-products generated from pooled A. pleuropneumoniae library genomic DNA +/− ISceI digestion. Lanes: 1) 100 bp ladder; 2) ISceI-treated sample; 3) untreated sample. Amplification products were generated for the left flanking sequences, as in Fig. 2. The dominant plasmid band in the untreated sample is indicated
Fig. 4.
Distribution of mariner insertions identified in the A. pleuropneumoniae and P. multocida genomes. TraDIS reads for the respective pooled libraries were mapped to A the complete genome of A. pleuropneumoniae MIDG2331 (accession number LN908249); and D the draft genome (pseudochromosome) of P. multocida MIDG3277 (accession number ERZ681052). Each spike plotted around the chromosome represents a single insertion site, with the length of each spike proportional to the number of mapped sequence reads from that insertion site. A total of 78,638 unique insertion sites were identified in the A. pleuropneumoniae MIDG2331 library, and 147,613 in the P. multocida MIDG3277 library. In the MIDG3277 dataset, there were several insertion sites with large numbers of mapped reads. To enable insertions with fewer reads to be seen clearly, read coverage has been capped to a maximum of 50,000 in D (the true maximum coverage at an insertion site was 136,032 reads). The pseudochromosome of P. multocida MIDG3277 was assembled based on ordering of the draft sequence contigs following alignment, using NUCmer 4.0 (43), with the complete genome of P. multocida Pm70, as shown in C. Arrows indicate the position and orientation of the contigs. Red blocks indicate matches in the same orientation, blue blocks indicate matches in the reverse orientation. Plots of the cumulative insertion counts across the MIDG2331 chromosome B and MIDG3277 pseudochromosome E are shown in red, with a dotted line indicating the expected relationship for uniformly distributed insertions. Both libraries deviate from this, with a bias towards insertions close to the origin of replication, but insertions are found across the genome in both libraries
For analysis of the P. multocida mariner library, it was first necessary to generate a draft genome sequence for the isolate used MIDG3277. Sequencing on an Illumina HiSeq 2000 yielded 2,285,355 pairs of 100 bp sequence reads, which were assembled into 160 scaffolds, which were ordered based on a comparison with the P. multocida Pm70 genome (GenBank accession AE004439; Fig. 4C). The assembly had a total size of 2,563,460 bp, with an n50 of 77,141 bp. Contig annotation identified 2,490 coding sequences, together with 35 tRNA genes and 4 rRNA genes. A pseudochromosome sequence of MIDG3277 was obtained by concatenating the ordered scaffolds.
TraDIS analysis of ISceI-digested P. multocida library DNA generated 48,266,652 sequence reads from the 5′ end of the mariner transposon, of which 46,950,914 (97.3%) mapped to the draft genome sequence of MIDG3277. Of these, 99.7% (46,809,272 reads) corresponded to insertions at TA dinucleotides, at 147,613 unique insertion sites. These represent an insertion approximately every 17 bp on average, occupying 85.6% of available TA sites in the genome. The insertion sites were distributed near-randomly across the MIDG3277 pseudochromosome (Fig. 4D), with the exception of a replication cycle bias (Fig. 4E) similar to that observed for the A. pleuropneumoniae MIDG2331 library. The presence of this pattern despite the use of a draft reference genome provides independent confirmation that genomes of P. multocida Pm70 and P. multocida MIDG3277 are largely co-linear.
Preliminary screen for anaerobic mutants
Of approximately 5000 mutants each of A. pleuropneumoniae and P. multocida (from four different matings in both cases) screened for the presence of insertions in genes required for anaerobic growth, 19 and 15 mutants, respectively, were identified that failed to grow. Linker-PCR produced amplicons of different sizes that, when sequenced, mapped to unique TA dinucleotides in eight different genes in the A. pleuropneumoniae MIDG2331 genome (Table 1), and 15 different sites (some intergenic) in the P. multocida MIDG3277 genome (Table 2).
Table 1.
Location of mariner insertions in anaerobic mutants of A. pleuropneumoniae
| Gene | Product of disrupted gene | Gene locationa | Location of inserta |
|---|---|---|---|
| nhaB | Na+/H+ antiporter protein | 363852..365393Cb | 364160 |
| pflB | Formate acetyltransferase | 1178587..1180899C | 1179054 |
| 1179780 | |||
| 1180383 | |||
| atpC | ATP synthase epsilon chain | 1922342..1922761C | 1922665 |
| atpG | ATP synthase gamma chain | 1924188..1925054C | 1924771 |
| 1924970c | |||
| atpA | ATP synthase alpha chain | 1925077..192661C | 1925488 |
| 1925791 | |||
| 1925826c | |||
| 1926363 | |||
| atpB | ATP synthase A chain | 1928081..1928869C | 1928204c |
| 1928575c | |||
| fumC | Fumarate hydratase | 2051573..2052967C | 2052243 |
| MIDG2331_02098 | D-alanyl-D-alanine carboxypeptidase | 2163589..2164257C | 2163863 |
aLocation in the complete genome of MIDG2331 (accession number LN908249)
bStart and end positions of genes are indicated; those on the complementary strand are shown with the suffix C
cInsertions at these locations were mapped in two mutants each
Table 2.
Location of mariner insertions in anaerobic mutants of P. multocida
| Gene | Product of disrupted gene | Gene locationa | Location of inserta |
|---|---|---|---|
| MIDG3277_1258 | Predicted glycosyl transferase | contig21:104818..105990b | contig21:105340 |
| aroG | Deoxy-7-phosphoheptulonate synthase | contig15:68162..69247 | contig15:68149c |
| intergenic | Periplasmic pH-dependent serine endoprotease DegQ | contig 16:46775..48154 | contig16:46700d |
| MIDG3277_1016 | Hypothetical proteine | contig18:76828..77169C | contig18:77129 |
| aroA | 3-phosphoshikimate 1-carboxyvinyltransferase | contig18:79640..80962 | contig18:80029 |
| contig18:80287 | |||
| sucC | Succinate--CoA ligase subunit beta | contig3:86080..87246 | contig3:86908 |
| ccmD | Heme exporter protein CcmD | contig1:16084..16287 | contig1:16136 |
| MIDG3277_1814 | F0F1 ATP synthase subunit I | contig31:36319..36726 | contig31:36262f |
| atpG | F0F1ATP synthase subunit gamma | contig31:40520..41389 | contig31:40497g |
| MIDG3277_0379 | Hypothetical protein | contig6:168..512h | contig6:434 |
| pfeA | Ferric enterobactin receptor | contig27:62820..64826C | contig27:64243 |
| menA | 1,4-dihydroxy-2-naphthoate octaprenyltransferase | contig23:97896..98801C | contig23:98875 |
| menB | 1,4-dihydroxy-2-naphthoyl-CoA synthase | contig23:34911..35768C | contig23:35383 |
| menC | O-succinylbenzoate synthase | contig23:33645..34658C | contig23:34159 |
aLocation in the draft genome of MIDG3277 (accession number ERZ681052)
bStart and end positions of genes are indicated; those on the complementary strand are shown with the suffix C
cInsertion located 13 bases upstream of the start of aroG; insertion likely disrupts transcription of aroG
dInsertion located 75 bases upstream of the start of degQ and 147 bases upstream of the start of MIDG3277_0890 encoding a putative NAD(P)H nitroreductase; insertion likely disrupts transcription of degQ
eThis protein shares 100% identity with PM0836 (Genbank accession AAK02920) which was reported to be in vivo expressed (Hunt et al. 2001)
fInsertion located 57 bases upstream of the start of atpI (MIDG3277_1814); insertion likely disrupts transcription of atpI and remaining genes in the atp operon
gInsertion located 23 bases upstream of atpG; insertion likely disrupts transcription of atpG and remaining genes in the atp operon
hThis predicted CDS is on a small contig and may be part of a larger gene (100% identity with the last 104 AAs of a 818 AA hypothetical protein P1062_0208970; Genbank accession ESQ71762)
Discussion
The mariner transposon does not require host-specific factors for transposition and has a minimal requirement for TA dinucleotides as its target site, allowing for greater distribution in genomes compared to transposons such as Tn5 and Tn10, which have “hot spots” for insertion. Tn10 insertion at hotspots was a severe limitation in our previous A. pleuropneumoniae STM study (Sheehan et al. 2003; Bossé et al. 2010). Given the relatively AT rich genomes of Pasteurellaceae species, mariner should allow for creation of saturating mutant libraries in A. pleuropneumoniae and P. multocida.
Two novel mobilizable mariner vectors, pTsodCPC9 and pTlacPC9, were developed in this study. Both contain mariner mini-transposon elements that carry a Cm resistance gene, known to provide stringent selection of mutants in A. pleuropneumoniae (Bossé et al. 2014), flanked by paired DNA uptake sequences for each of A. pleuropneumoniae, H. influenzae and Neisseria spp. (Redfield et al. 2006; Treangen et al. 2008). Although initially designed for mutagenesis of A. pleuropneumoniae, all features included were designed to facilitate use in other Gram-negative bacteria. As it is often desirable to transfer mutations into wild-type strains to confirm the effect(s) of gene mutation, DNA uptake sequences were designed into the transposon to facilitate transfer of mutations by natural transformation in the species which selectively take up DNA containing these elements (Redfield et al. 2006; Treangen et al. 2008).
Restriction barriers can prevent or decrease efficiency of plasmid delivery by electroporation into various bacteria (Maglennon et al. 2013a; Luan et al. 2013), we therefore added oriT transfer origin and traJ genes from the broad host-range conjugative plasmid RP4 (Ziegelin et al. 1989) to facilitate vector delivery by conjugation. The presence of oriT alone on a plasmid is sufficient for mobilization by conjugal transfer machinery encoded by the tra operon (which includes traJ) present in the chromosome of a suitable donor strain, such as MFDpir used in the current study. However, as TraJ binds to the plasmid oriT to initiate plasmid transfer (Ziegelin et al. 1989) and also acts as a positive regulator for expression of the complete tra operon (Gubbins et al. 2002), a copy of this gene was included in the vectors, to enhance conjugation efficiency. Successful conjugal transfer of both vectors into MIDG2331 confirmed that these sequences were sufficient to allow mobilization from the DAP-dependent Escherichia coli strain MFDpir (Ferrieres et al. 2010). Use of MFDpir facilitates conjugation into wild-type recipient bacteria, instead of antibiotic resistant derivatives, as transconjugant selection is achieved on agar that does not contain DAP which is required for growth of the donor strain (Ferrieres et al. 2010).
The hyperactive C9 mutant transposase gene, previously shown to enhance mariner transposition efficiency (Lampe et al. 1999; Maglennon et al. 2013b) was placed under transcriptional control of either the A. pleuropneumoniae sodC promoter, which we have shown to be active under all conditions investigated so far, both in A. pleuropneumoniae and other Pasteurellaceae species (Bossé et al. 2009), or the E. coli lac promoter which allows induction by IPTG, and which is functional in a variety of bacteria including A. pleuropneumoniae (Sheehan et al. 2003). Although both resulting plasmids, pTsodCPC9 and pTlacPC9, were functional in A. pleuropneumoniae, the former generated a lower frequency of transposants. This may be due to over-production inhibition of expression from the highly active sodC promoter. This phenomenon was described in a recent study by Tellier and Chalmers (2020), where comparison of expression of the Hsmar1 mariner transposase from promoters of different strengths indicated the highest frequency of transposition was achieved with the weakest promoter. We also found that, although the sodC promoter was previously shown to function in P. multocida (Bossé et al. 2009), use of the pTsodCPC9 vector did not yield transposants in this species. In contrast, IPTG induction of C9 transposase gene expression from the lac promoter in the pTlacPC9 vector resulted in similar transposition frequencies in both A. pleuropneumoniae and P. multocida, indicating this vector will likely be a useful genetic tool for other bacteria.
Using the pTlacPC9 vector, saturating libraries were generated for both A. pleuropneumoniae (insertions on average every 30 bp) and P. multocida (insertions on average every 17 bp), with insertions randomly distributed around the respective chromosomes. In both cases, pooled mutant libraries were prepared from initial selective plate cultures of transconjugants (from multiple mating experiments) to limit selective expansion of clones. Under these conditions, extrachromosomal plasmid retained in the library can lead to high numbers of sequencing reads, interfering with TraDIS mapping. By engineering paired ISceI restriction sites (18 bp sequences not usually found in bacterial genomes) flanking the mini-transposon element in our vectors, we were able to effectively eliminate retained plasmid from the libraries prior to sequencing.
Both A. pleuropneumoniae and P. multocida are members of the Pasteurellaceae and are facultative anaerobes that infect the respiratory tracts of animals. Anaerobic growth is known to contribute to virulence of A. pleuropneumoniae (Jacobsen et al. 2004). However, little is known about the importance of genes contributing to anaerobic growth in P. multocida. In this study, we have screened a limited number of mariner mutants of both pathogens to identify genes important for anaerobic growth to further validate the randomness of insertions in these libraries and usefulness of our approach. Of the 19 A. pleuropneumoniae mutants sequenced, 15 unique insertions in eight different genes were identified (Table 1). Of 15 P. multocida mutants sequenced, all were in unique sites in the genome, with some insertions mapped to intergenic sites (Table 2). In both cases, mutants were selected from only four separate mating experiments, so it is possible that the four duplicate insertions mapped for A. pleuropneumoniae mutants were due to clonal expansion following insertion, even though mutants were randomly selected from the primary counter selection plate. As clonal expansion can skew libraries for over representation of more rapidly growing mutants, steps should be taken to limit this prior to TraDIS analysis. For example, multiple independent mating experiments, with shorter incubation periods during conjugal transfer, could be used to generate large libraries whilst minimising expansion of faster growing clones. Additionally, keeping the generated mutants in smaller, less complex pools could help further maximise representation of all fit mutants in a library (Chao et al. 2016).
For both organisms, the atp operon encoding the F0F1 ATP synthase appears to be essential for anaerobic growth. In A. pleuropneumoniae, 13 mutants had disruptions in four genes in atp operon, with nine unique insertion sites. In P. multocida, two of the anaerobic mutants had insertions that mapped to sites upstream of genes in the atp operon (56 bases upstream of atpI, the first gene in the operon, and 24 bases upstream of atpG). These intergenic insertions likely disrupt transcription of all, or part, of the atp operon, and therefore production of functional F0F1 ATP synthase. In the absence of oxidative phosphorylation, the F0F1 enzyme complex extrudes protons at the expense of ATP hydrolysis, to generate the driving force for solute transport and to maintain an acceptable intracellular pH value, and is required for function of certain enzymes involved in anaerobic respiration, such as formate hydrogenlyase (FHL) (Trchounian 2004). Furthermore, the activity of the FHL and F0F1-ATPase systems were found to facilitate the fermentative metabolism of glycerol in E. coli (Gonzalez et al. 2008). Maintenance of membrane potential by the hydrolytic activity of F0F1 ATP synthase has been shown to be vital for anaerobic growth of some bacteria when substrate level phosphorylation is the only source of ATP (Meyrat and von Ballmoos 2019). Although the atp operon has not previously been reported as essential for anaerobic growth of A. pleuropneumoniae or P. multocida, STM studies have identified genes of this operon as important for survival of these bacteria in the host during acute infection (Fuller et al. 2000a; Sheehan et al. 2003; Fuller et al. 2000b), where anaerobic growth is known to be essential.
An insertion in nhaB, encoding a Na+/H+ antiporter, was also found to be important for anaerobic growth of A. pleuropneumoniae in our current screen. As with the atp operon, nhaB has been shown to be required for in vivo growth of A. pleuropneumoniae (Baltes et al. 2007). The importance of interactions between proton and sodium cycles during both aerobic and anaerobic growth has been reported for certain pathogenic bacteria (Häse et al. 2001). Furthermore, Trchounian and Kobayashi (1999) demonstrated that E. coli grown anaerobically was more sensitive to increased extracellular Na+ concentrations than aerobically grown cells, especially at higher pH, and that mutants lacking a functional NhaA/NhaB encoded antiporter showed an even greater growth defect than wild-type strain under these conditions. The relative contributions of proton and sodium cycles to anaerobic growth of A. pleuropneumoniae and P. multocida warrant further investigation.
The importance of menaquinone biosynthesis for anaerobic growth of P. multocida was indicated by single insertions in each of menA, menB, and menC, as well as a single insertion three bases upstream of aroG (likely disrupting transcription), and two in aroA. Menaquinone is involved in anaerobic electron transport, and is derived from chorismate (pathway includes menA, menB and menC), which in turn is derived from shikamate (pathway includes aroA and aroG).
Two of the A. pleuropneumoniae genes identified, pflB and fumC, encode proteins with known roles in anaerobic pathways. There were three separate insertions in pflB encoding pyruvate formate lyase which catalyses generation of formate via decarboxylation of puruvate. As mentioned above, the FHL complex is important in anaerobic respiration where it catalyses the conversion of formate into CO2 and H2 (Sawers 2005). A single insertion was mapped to fumC encoding fumarate hydratase, the enzyme responsible for catalysing conversion of malate to fumarate. Fumarate has previously been shown to be an essential terminal electron acceptor during anaerobic respiration in A. pleuropneumoniae (Jacobsen et al. 2004; Buettner et al. 2008).
Two identified P. multocida genes (ccmD and sucC) also have known anaerobic functions. Genes of the ccm operon are required for maturation of c-type cytochromes, including those involved in electron transfer to terminal reductases of the anaerobic respiratory chain with nitrate, nitrite or TMAO (trimethylamine-N-oxide) as electron acceptors (Schulz et al. 2000). The sucC gene encodes the beta subunit of succinate-CoA ligase, an enzyme in the aerobic citrate (TCA) cycle, where it catalyses hydrolysis of succinyl-CoA to succinate (coupled to the synthesis of either GTP or ATP). This enzyme also mediates the reverse reaction when required for anabolic metabolism, which can be particularly important under anaerobic conditions where the generation of succinyl-CoA via the oxidative pathway from 2-oxoglutarate is repressed (Mat-Jan et al. 1989; Shalel-Levanon et al. 2005).
The remaining mutants in both A. pleuropneumoniae and P. multocida have insertions in (or upstream of) genes not directly linked with anaerobic growth, however further investigation is warranted to determine their possible contributions.
Conclusions
It is clear from this research that we have successfully constructed a mariner mini-transposon delivery vector capable of generating extremely large numbers of random mutants in A. pleuropneumoniae, P. multocida, and likely in other Gram-negative bacteria, which is amenable to genome-wide analysis of fitness using TraDIS. In preliminary experiments, we have established that the pTlacPC9 vector can be used in Aggregatibacter actinomycetemcomitans and Mannheimia haemolytica, though it remains to be determined if the generated libraries are saturating.
The limited number of anaerobic-essential genes (identified individually via phenotypic analysis in the current study) mapped to different insertion sites, including some which had not previously been associated with anaerobic growth of A. pleuropneumoniae and P. multocida. This suggests that TraDIS analysis of the pooled mariner libraries subjected to the same screen will identify many more genes with functions contributing to anaerobic fitness. In future work, we will screen our mariner libraries under different in vitro and in vivo growth conditions, broadening our understanding of conditionally essential genes in the Pasteurellaceae.
Methods
Bacterial strains and culture
For generation of mariner transposon libraries, we chose two different Pasteurellaceae species, A. pleuropneumoniae and P. multocida. The A. pleuropneumoniae clinical serovar 8 isolate, MIDG2331, was previously shown to be genetically tractable and has been fully sequenced (Bossé et al. 2016). The P. multocida isolate used in this study recovered from the respiratory tract of a calf in Scotland in 2008, and was shown to be sequence type 13, and part of clonal complex 13, in a multi-species multilocus sequence typing (MLST) study (Hotchkiss et al. 2011). This isolate, which has been labeled MIDG3277 in our collection, can be found in the pubMLST database (https://pubmlst.org/bigsdb?db=pubmlst_pmultocida_isolates) under the isolate name 22/4. Pasteurellaceae isolates were routinely propagated at 37 °C with 5% CO2 on Brain Heart Infusion (Difco) plates supplemented with 5% horse serum and 0.01% nicotinamide adenine dinucleotide (BHI-S-NAD). E. coli strains used were: Stellar [F−, ara,Δ(lac-proAB) [Φ 80d lacZΔM15], rpsL(str), thi, Δ(mrr-hsdRMS-mcrBC), ΔmcrA, dam, dcm and MFDpir [MG1655 RP4-2-Tc::[ΔMu1::aac(3)IV-ΔaphA-Δnic35-ΔMu2::zeo] ΔdapA::(erm-pir) ΔrecA] (Ferrieres et al. 2010). E. coli strains were maintained in Luria-Bertani (LB). Where appropriate, ampicillin (Amp; 100 μg/ml), chloramphenicol (Cm; 20 and 1 μg/ml for E. coli and A. pleuropneumoniae, respectively), and 0.3 mM DAP (required for growth of the MFDpir strain), were added to media.
Genome sequencing, assembly and annotation
To obtain a draft genome sequence suitable for TraDIS analysis, genomic DNA was extracted from the P. multocida MIDG3277 strain using the FastDNA Spin kit (MP Biomedicals), and sequenced using an Illumina HiSeq 2000 at the Wellcome Sanger Institute. Illumina adapter sequences were trimmed from the reads using Cutadapt V. 1.8.1 (Martin 2011), and the trimmed reads were assembled using SPAdes V. 3.11.0 (Bankevich et al. 2012), using default parameters. The assembled contigs were aligned to the complete genome sequence of P. multocida Pm70 (GenBank accession AE004439) using nucmer V. 4.0.0 (Marçais et al. 2018). Using the alignment, contigs were reordered and reoriented to match the Pm70 reference genome, with one contig manually split where it overlapped the Pm70 origin. The reordered contigs were annotated using Prokka V. 1.11 (Seemann 2014).
Construction of the mariner mini-transposon vectors
All primers are listed in Table 3. CloneAmp HiFi PCR Premix (Takara) was used to amplify sequences for cloning, and the QIAGEN Fast Cycling PCR Kit (Qiagen) was used for verification of clones, using the respective manufacturer’s protocols. When required, blunt PCR products were A-Tailed using 5 U Taq polymerase and 0.2 mM dATP prior to TA cloning into pGEM-T (Promega), according to the manufacturer’s protocol. Also, when required, DpnI digestion was used to remove plasmid DNA template from PCR products prior to cloning. All ligation products and In Fusion cloning products were transformed into E. coli Stellar cells (Clontech) by heat shock, according to the manufacturer’s protocol.
Table 3.
Primers used in this study
| Name | Sequence |
|---|---|
| CmDUSUSS_for | CGCGGATGCCGTCTGAAGTGCGGTACAAGCGGTCGGCAATAGTTACC |
| CmDUSUSS_rev | CGCGAAGTGCGGTATGCCGTCTGAACAAGCGGTTTCAACTAACGGG |
| CmDUSUSS_IRleft | CTGATAAGTCCCCGGTCTGCAGGCGGCCGCACTAGTGATTC |
| CmDUSUSS_IRright | CTGATAAGTCCCCGGTCTCGAAGTGCGGTATGCCGTCTG |
| Himar_IR | TAACAGGTTGGCTGATAAGTCCCCGGTCT |
| ISceI_left | TAGGGATAACAGGGTAATCATGGCCGCGGGATTAACAGGTTG |
| ISceI_right | ATTACCCTGTTATCCCTACGGCCGCACTAGTGATTAACAGG |
| M13_for | GTAAAACGACGGCCAGTG |
| M13_rev | GGAAACAGCTATGACCATG |
| oriTtraJ_left | CCGCCTGCAGGTCGACAAAACAGCAGGGAAGCAGCGCTTTTC |
| oriTtraJ_right | ACTCAAGCTATGCATGCATGGGGACGTGCTTGGCAATC |
| sodCPC9_left | CGAATTGGGCCCGACCGCCAACCGATAAAACCTACATTTTGC |
| sodCPC9_right | CCTCCTTTTCTAGTCGCGGTACCGTCGACTGCAGAATTC |
| lacPC9_left | CGAATTGGGCCCGACGTGAGCGCAACGCAATTAATGTGAGTTAG |
| lacPC9_right | CCTCCTTTTCTAGTCGGCGTAATCATGGTCATAGCTGTTTCC |
| C9sodC_left | AGTCGACGGTACCGCGACTAGAAAAGGAGGATTCCTCATATGG |
| C9lacP_left | GACCATGATTACGCCGACTAGAAAAGGAGGATTCCTCATATGG |
| C9_right | CCGGGAGCATGCGACCCAGTGTGCTGGAATTCGCCCTTAGC |
| L-PCR-C | GATAAGCAGGGATCGGAACC |
| IR-Left_out | CACTTCAGACGGCATCCGCGAATC |
| IR-Right_out | GTTGAAACCGCTTGTTCAGACGGC |
Specific DNA uptake sequences for Neisseria spp. (underlined), H. influenzae (italics), and A. pleuropneumoniae (bold) are indicated in the CmDUSUSS_for and CmDUSUSS_rev sequences
The ISceI restriction site is indicated in bold italic text
The 15 base overhangs for In Fusion cloning are shown in subscript text on the 5′ end of primers oriTtraJ_left to C9_right
A mariner mini-transposon encoding Cm resistance was constructed in stages using pGEM-T as vector backbone. Initially, a Cm cassette flanked by A. pleuropneumoniae uptake signal sequences (USS) was amplified from pUSScat (Bossé et al. 2014) using primers CmDUSUSS_for and CmDUSUSS_rev, which further added DNA uptake sequences for Neisseria spp. and H. influenzae on both sides of the cassette. The resulting 956 bp amplicon was A-tailed and cloned into pGEM-T yielding the plasmid pTCmDUSUSS. The mariner inverted repeat (IR) sequence (TAACAGGTTGGCTGATAAGTCCCCGGTCT) was then added to either side of the CmDUSUSS cassette in two subsequent rounds of PCR amplification and cloning into pGEM-T. In the first round, primers CmDUSUSS_IRleft and CmDUSUSS_IRright added the last 19 bases of the mariner inverted repeat to either side of the cassette. The full IR sequence was then used as a primer (Himar_IR) to amplify the transposon cassette prior to cloning into pGEM-T to yield pTHimarCm. Finally, paired I-SceI restriction sites were added to either side of the transposon by PCR using primers ISceI_left and ISceI_right, and the product was cloned into pGEM-T to yield pTISceHimarCm. The full insert was sequenced using M13_for and M13_rev primers prior to further modifications of the plasmid.
All further cloning steps were performed using the In Fusion HD cloning kit (Clontech), according to the manufacturer’s protocol. A sequence containing oriT and traJ gene was amplified from pBMK1 (Oswald et al. 1999), a generous gift from Gerald Gerlach, using primers oriTtraJ_left and oriTtraJ_right, and cloned into NsiI/SalI cut pTISceHimarCm to yield pTISceHimarCmoriT. The C9 hyperactive Himar1 transposase gene (Lampe et al. 1999), amplified from pCAM45 (May et al. 2004) using primers C9_right and either C9sodC_left or C9lacP_left, was fused by overlap-extension (OE-PCR) PCR to either the A. pleuropneumoniae sodC promoter amplified from pMK-Express (Bossé et al. 2009) using primers sodCPC9_left and sodCPC9_right, or to the lac promoter amplified from pBluescript II KS (Agilent Technologies) using primers lacPC9_left and lacPC9_right. The sodCP-C9 and lacP-C9 OE-PCR products were cloned into ZraI cut pTISceHimarCmoriT to yield the mariner mini-transposon delivery vectors pTsodCPC9 and pTlacPC9, respectively. All inserts were confirmed by sequencing.
Purified pTsodCPC9 and pTlacPC9 plasmids were electroporated into E. coli conjugal donor strain MFDpir (Ferrieres et al. 2010), a generous gift from Jean-Marc Ghigo, with transformants recovered on LB containing 20 μg/ml Cm and 0.3 mM DAP.
Bacterial mating and generation of mariner mutant libraries in A. pleuropneumoniae and P. multocida
Initially, the two different mariner mini-transposon delivery vectors, pTsodCPC9 and pTlacPC9, were evaluated for their ability to produce Cm-resistant mutants in A. pleuropneumoniae serovar 8 strain MIDG2331 following conjugal transfer from DAP-dependent E. coli MFDpir donor strain. For mating experiments, donor and recipient bacteria were grown separately in broth culture to an optical density at 600 nm (OD600) of approximately 1.0 (cultures were adjusted to equivalent OD600). Two hundred microliters of recipient strain were mixed with 0 to 200 μL donor strain (to give ratios of 1:1, 1:2, 1:4 and 1:8, as well as a control of recipient only). The bacteria were pelleted and re-suspended in 200 μL 10 mM MgSO4, and 20 μL aliquots were spotted onto 0.45 μm nitrocellulose filters (Millipore) placed onto BHI-S-NAD agar supplemented with DAP and, when required for induction of the lac promoter, 1 mM IPTG. Plates were incubated overnight at 37 °C with 5% CO2, after which bacteria were recovered in 1 mL sterile phosphate-buffered saline and 100 μL aliquots were plated onto BHI-S-NAD agar supplemented with 2 μg/mL Cm. In preliminary experiments, selected transconjugants were tested, both before and after subculture on selective agar, by colony PCR for the presence of the Cm cassette using primers CmDUSUSS_for and CmDUSUSS_rev, and for presence of plasmid backbone using primers oriTtraJ_left and oriTtraJ_right. Southern blot and linker-PCR using AluI-digested DNA were performed as previously described (Chaudhuri et al. 2009; Maglennon et al. 2013b) to confirm single random insertion of the transposon in selected transconjugants.
For A. pleuropneumoniae mariner library construction, a total of 14 conjugations were performed using the MFDpir + pTlacPC9 donor. For each mating, between 1,400 and 6,200 MIDG2331 transconjugants were collected from the selective agar plates and resuspended into respective 3 mL aliquots of BHI broth. Aliquots of each separate mating pool were stored as individual 2 mL freezer stocks containing a final concentration of 25% glycerol. A combined pooled library of transconjugants was generated by mixing equal 0.5 mL aliquots of each of the individual mutant pools prior to preparation of 2 mL freezer stocks containing a final concentration of 25% glycerol. All conjugations were performed at the same time and transconjugants were collected into pools directly from the selective agar plates, without further subculture, to avoid expansion of clones with increased fitness. Freezer stocks were stored at − 80 °C.
The pTlacPC9 vector was further assessed for the ability to generate Cm-resistant mutants in the related bacterium, P. multocida. In total, nine separate matings were performed using MFDpir + pTlacPC9 donor and P. multocida MIDG3277 recipient, with between 2,388 and 20,640 transconjugants selected per mating. Mutants were stored at − 80 °C as separate pools, and as a combined pool, in 2 mL aliquots in BHI broth containing a final concentration of 25% glycerol, as above.
TraDIS analysis of the A. pleuropneumoniae and P. multocida mariner libraries
Genomic DNA was extracted from the pooled libraries of mutants using the FastDNA Spin kit (MP Biomedicals), according to the manufacturer’s protocol for bacterial cells. To assess the distribution of insertions in the pools prior to TraDIS, linker-PCR was performed as previously described (Maglennon et al. 2013b) with the exception that, for PCR amplification, primer L-PCR-C was paired with either IR_left_out or IR_right_out (see Table 3) in place of L-PCR-L or L-PCR-R, respectively. For comparison, 2.5 μg genomic DNA that were either untreated, or digested with ISceI to remove reads from residual plasmid, were used for linker PCR amplification of the left flank sequences, and the products were separated on 2% Nusieve agarose. Subsequently, 2 μg ISceI-digested DNA was used to prepare Illumina libraries for TraDIS analysis, as previously described (Luan et al. 2013).
TraDIS libraries were sequenced on an Illumina HiSeq 2500 at the Wellcome Sanger Institute. For the A. pleuropneumoniae library, TraDIS reads were mapped to the closed whole genome sequence of MIDG2331 (accession number LN908249). P. multocida TraDIS reads were mapped to the draft genome of MIDG3277 (accession number ERZ681052), constructed as described above. Reads were mapped using BWA mem (Li 2013) V. 0.7.17-r1188, with an increased penalty for 5′ clipping (−L 100,5). To reduce background noise, aligned reads which did not match a TA site at the 5′ end of alignment were excluded from further analysis using a custom Perl script, and the locations of insertions were extracted from the BAM file using SAMtools V. 1.13 (https://academic.oup.com/bioinformatics/article/25/16/2078/204688). Subsequent data analysis was performed using R V. 4.1.1 (https://www.R-project.org/).
Preliminary screen for anaerobic mutants
Approximately 5,000 mutants each for A. pleuropneumoniae and P. multocida (from four separate mutant pools) were screened for insertions in genes required for survival during anaerobic growth. The mutant pools were plated on BHI-S-NAD supplemented with 2 μg/mL Cm at a density of 75 to 150 colonies per plate. Following overnight incubation at 37 °C with 5% CO2, colonies were transferred by replica plating onto two fresh selective plates. One plate was incubated with 5% CO2, and the other was placed in an anaerobic jar, at 37 °C overnight. Mutants that failed to grow anaerobically were re-tested to confirm the growth defect, and the site of transposon insertion was determined for each by direct sequencing of linker-PCR products.
Acknowledgements
The BRaDP1T consortium comprises: Duncan J. Maskell, Alexander W. (Dan) Tucker, Sarah E. Peters, Lucy A. Weinert, Jinhong (Tracy) Wang, Shi-Lu Luan, Roy R. Chaudhuri (University of Cambridge); Andrew N. Rycroft, Gareth A. Maglennon, Jessica Beddow (Royal Veterinary College); Brendan W. Wren, Jon Cuccui, Vanessa S. Terra (London School of Hygiene and Tropical Medicine); and Janine T. Bossé, Yanwen Li and Paul R. Langford (Imperial College London).
Abbreviations
- TraDIS
Transposon Directed Insertion-site Sequencing
- STM
Signature-tagged mutagenesis
- DAP
Diaminopimelic acid
- IPTG
Isopropyl-ß-D-galactopyranoside
- Cm
Chloramphenicol
- FHL
Formate hydrogenlyase
- TMAO
Trimethylamine-N-oxide
- MLST
Multilocus sequence typing
- BHI-S-NAD
Brain Heart Infusion supplemented with 5% horse serum and 0.01% nicotinamide adenine dinucleotide
- LB
Luria-Bertani
- Amp
Ampicillin
- OD600
Optical density at 600 nm
- ENA
European Nucleotide Archive
Authors’ contributions
All authors contributed to the conception and design of the study. JTB, YL, LGL, JW and SEP performed the experiments. JTB and RRC analysed the data. JTB wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by a Longer and Larger (LoLa) grant from the Biotechnology and Biological Sciences Research Council (BBSRC; grant numbers BB/G020744/1, BB/G019177/1, BB/G019274/1 and BB/G018553/1), the UK Department for Environment, Food and Rural Affairs and Zoetis awarded to the Bacterial Respiratory Diseases of Pigs-1 Technology (BRaDP1T) consortium. Funding for LZ was provided by the BBSRC (grant number BB/C508193/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The complete sequences of the two mariner delivery vectors, pTsodCPC9 and pTlacPC9, have been deposited in GenBank under accession numbers MH644834 and MH644835, respectively. The plasmids are available upon request from the corresponding authors. The draft genome sequence of the porcine clinical respiratory isolate of P. multocida, MIDG3277, has been deposited in the European Nucleotide Archive (ENA) under the accession number ERZ681052, with the raw reads available under the accession number ERR200085. The raw TraDIS reads for A. pleuropneumoniae are available in ENA under the accession number ERR271132. The raw TraDIS reads for P. multocida are available in ENA under the accession numbers ERR744003, ERR744016, ERR752316, ERR752329, ERR755725 and ERR755738.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Author Paul R. Langford was not involved in the journal’s review or decisions related to this manuscript. The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Janine T. Bossé and Yanwen Li contributed equally to this work.
Contributor Information
Janine T. Bossé, Email: j.bosse@imperial.ac.uk
Paul R. Langford, Email: p.langford@imperial.ac.uk
on behalf of the BRaDP1T consortium:
Duncan J. Maskell, Alexander W. ( Dan) Tucker, Sarah E. Peters, Lucy A. Weinert, Jinhong ( Tracy) Wang, Shi-Lu Luan, Roy R. Chaudhuri, Andrew N. Rycroft, Gareth A. Maglennon, Jessica Beddow, Brendan W. Wren, Jon Cuccui, Vanessa S. Terra, Janine T. Bossé, Yanwen Li, and Paul R. Langford
References
- Akerley BJ, Rubin EJ, Camilli A, Lampe DJ, Robertson HM, Mekalanos JJ. Systematic identification of essential genes by in vitro mariner mutagenesis. Proceedings of the National Academy of Sciences USA. 1998;95(15):8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes N, Buettner FFR, Gerlach G-F. Selective capture of transcribed sequences (SCOTS) of Actinobacillus pleuropneumoniae in the chronic stage of disease reveals an HlyX-regulated autotransporter protein. Veterinary Microbiology. 2007;123(1–3):110–121. doi: 10.1016/j.vetmic.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J, Kleckner N. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proceedings of the National Academy of Sciences USA. 1992;89(17):7996–8000. doi: 10.1073/pnas.89.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossé JT, Chaudhuri RR, Li Y, Leanse LG, Fernandez Crespo R, Coupland P, Holden MTG, Bazzolli DM, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, Langford PR, on behalf of the BRaDP1T consortium Complete genome sequence of MIDG2331, a genetically tractable serovar 8 clinical isolate of Actinobacillus pleuropneumoniae. Genome Announcements. 2016;4(1):e01667–e01615. doi: 10.1128/genomeA.01667-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossé JT, Durham AL, Rycroft AN, Kroll JS, Langford PR. New plasmid tools for genetic analysis of Actinobacillus pleuropneumoniae and other Pasteurellaceae. Applied and Environmental Microbiology. 2009;75(19):6124–6131. doi: 10.1128/AEM.00809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossé JT, Gilmour HD, MacInnes JI. Novel genes affecting urease acivity in Actinobacillus pleuropneumoniae. Journal of Bacteriology. 2001;183(4):1242–1247. doi: 10.1128/JB.183.4.1242-1247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossé JT, Janson H, Sheehan BJ, Beddek AJ, Rycroft AN, Kroll JS, Langford PR. Actinobacillus pleuropneumoniae: Pathobiology and pathogenesis of infection. Microbes and Infection. 2002;4(2):225–235. doi: 10.1016/s1286-4579(01)01534-9. [DOI] [PubMed] [Google Scholar]
- Bossé JT, Sinha S, Li M-S, O'Dwyer CA, Nash JHE, Rycroft AN, Kroll JS, Langford PR. Regulation of pga operon expression and biofilm formation in Actinobacillus pleuropneumoniae by sigmaE and H-NS. Journal of Bacteriology. 2010;192(9):2414–2423. doi: 10.1128/JB.01513-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossé JT, Soares-Bazzolli DM, Li Y, Wren BW, Tucker AW, Maskell DJ, Rycroft AN, Langford PR, on behalf of the BRaDP1T consortium The generation of successive unmarked mutations and chromosomal insertion of heterologous genes in Actinobacillus pleuropneumoniae using natural transformation. PLoS One. 2014;9(11):e111252. doi: 10.1371/journal.pone.0111252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhenni R, Gehrke A, Saffarini D. Identification of genes involved in cytochrome c biogenesis in Shewanella oneidensis, using a modified mariner transposon. Applied and Environmental Microbiology. 2005;71(8):4935–4937. doi: 10.1128/AEM.71.8.4935-4937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner FFR, Bendallah IM, Bossé JT, Dreckmann K, Nash JHE, Langford PR, Gerlach G-F. Analysis of the Actinobacillus pleuropneumoniae ArcA regulon identifies fumarate reductase as a determinant of virulence. Infection and Immunity. 2008;76(6):2284–2295. doi: 10.1128/IAI.01540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MC, Abel S, Davis BM, Waldor MK. The design and analysis of transposon-insertion sequencing experiments. Nature Reviews. Microbiology. 2016;14(2):119–128. doi: 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH) BMC Genomics. 2009;10(1):291. doi: 10.1186/1471-2164-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiers K, De Waele T, Pasmans F, Ducatelle R, Haesebrouck F. Virulence factors of Actinobacillus pleuropneumoniae involved in colonization, persistence and induction of lesions in its porcine host. Veterinary Research. 2010;41(5):65. doi: 10.1051/vetres/2010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrieres L, Hemery G, Nham T, Guerout AM, Mazel D, Beloin C, Ghigo JM. Silent mischief: Bacteriophage mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. Journal of Bacteriology. 2010;192(24):6418–6427. doi: 10.1128/JB.00621-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller TE, Kennedy MJ, Lowery DE. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microbial Pathogenesis. 2000;29(1):25–38. doi: 10.1006/mpat.2000.0365. [DOI] [PubMed] [Google Scholar]
- Fuller TE, Martin S, Teel JF, Alaniz GR, Kennedy MJ, Lowery DE. Identification of Actinobacillus pleuropneumoniae virulence genes using signature-tagged mutagenesis in a swine infection model. Microbial Pathogenesis. 2000;29(1):39–51. doi: 10.1006/mpat.2000.0364. [DOI] [PubMed] [Google Scholar]
- Gawronski JD, Wong SMS, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proceedings of the National Academy of Sciences USA. 2009;106(38):16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R, Murarka A, Dharmadi Y, Yazdani SS. A new model for the anaerobic fermentation of glycerol in enteric bacteria: Trunk and auxiliary pathways in Escherichia coli. Metabolic Engineering. 2008;10(5):234–245. doi: 10.1016/j.ymben.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Grasteau A, Tremblay YDN, Labrie J, Jacques M. Novel genes associated with biofilm formation of Actinobacillus pleuropneumoniae. Veterinary Microbiology. 2011;153(1–2):134–143. doi: 10.1016/j.vetmic.2011.03.029. [DOI] [PubMed] [Google Scholar]
- Gubbins MJ, Lau I, Will WR, Manchak JM, Raivio TL, Frost LS. The positive regulator, TraJ, of the Escherichia coli F plasmid is unstable in a cpxA* background. Journal of Bacteriology. 2002;184(20):5781–5788. doi: 10.1128/JB.184.20.5781-5788.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häse CC, Fedorova ND, Galperin MY, Dibrov PA. Sodium ion cycle in bacterial pathogens: Evidence from cross-genome comparisons. Microbiology and Molecular Biology Reviews. 2001;65(3):353–370. doi: 10.1128/MMBR.65.3.353-370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss EJ, Hodgson JC, Lainson FA, Zadoks RN. Multilocus sequence typing of a global collection of Pasteurella multocida isolates from cattle and other host species demonstrates niche association. BMC Microbiology. 2011;11(1):115. doi: 10.1186/1471-2180-11-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt ML, Boucher DJ, Boyce JD, Adler B. In vivo-expressed genes of Pasteurella multocida. Infection and Immunity. 2001;69(5):3004–3012. doi: 10.1128/IAI.69.5.3004-3012.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen I, Hennig-Pauka I, Baltes N, Trost M, Gerlach GF. Enzymes involved in anaerobic respiration appear to play a role in Actinobacillus pleuropneumoniae virulence. Infection and Immunity. 2004;73(1):226–234. doi: 10.1128/IAI.73.1.226-234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. Hyperactive transposase mutants of the Himar1 mariner transposon. Proceedings of the National Academy of Sciences USA. 1999;96(20):11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge, G.C., M.-D. Phan, D.J. Turner, T.T. Perkins, L. Parts, J. Haase, I. Charles, D.J. Maskell, S.E. Peters, G. Dougan, J. Wain, J. Parkhill, and A.K. Turner. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Research 19 (12): 2308–2316. 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN] 2013. [Google Scholar]
- Luan, S.-L., R.R. Chaudhuri, S.E. Peters, M. Mayho, L.A. Weinert, S.A. Crowther, J. Wang, P.R. Langford, A. Rycroft, B.W. Wren, A.W. Tucker, D.J. Maskell, and on behalf of the BRaDp1T consortium. 2013. Generation of a Tn5 transposon library in Haemophilus parasuis and analysis by transposon-directed insertion-site sequencing (TraDIS). Veterinary Microbiology 166 (3–4): 558–566. 10.1016/j.vetmic.2013.07.008. [DOI] [PubMed]
- Maglennon GA, Cook BS, Deeney AS, Bossé JT, Peters SE, Langford PR, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, on behalf of the BRaDP1T consortium Transposon mutagenesis in Mycoplasma hyopneumoniae using a novel mariner-based system for generating random mutations. Veterinary Research. 2013;44(1):124. doi: 10.1186/1297-9716-44-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglennon GA, Cook BS, Matthews D, Deeney AS, Bossé JT, Langford PR, Maskell DJ, Tucker AW, Wren BW, Rycroft AN, on behalf of the BRaDP1T consortium Development of a self-replicating plasmid system for Mycoplasma hyopneumoniae. Veterinary Research. 2013;44(1):63. doi: 10.1186/1297-9716-44-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. MUMmer4: A fast and versatile genome alignment system. PLoS Computational Biology. 2018;14(1):e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Mat-Jan F, Williams CR, Clark DP. Anaerobic growth defects resulting from gene fusions affecting succinyl-CoA synthetase in Escherichia coli K12. Molecular & General Genetics. 1989;215(2):276–280. doi: 10.1007/BF00339728. [DOI] [PubMed] [Google Scholar]
- May JP, Walker CA, Maskell DJ, Slater JD. Development of an in vivo Himar1 transposon mutagenesis system for use in Streptococcus equi subsp. equi. FEMS Microbiology Letters. 2004;238(2):401–409. doi: 10.1016/j.femsle.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Meyrat A, von Ballmoos C. Atp synthesis at physiological nucleotide concentrations. Scientific Reports. 2019;9(1):3070. doi: 10.1038/s41598-019-38564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald W, Tonpitak W, Ohrt G, Gerlach G. A single-step transconjugation system for the introduction of unmarked deletions into Actinobacillus pleuropneumoniae serotype 7 using a sucrose sensitivity marker. FEMS Microbiology Letters. 1999;179(1):153–160. doi: 10.1111/j.1574-6968.1999.tb08721.x. [DOI] [PubMed] [Google Scholar]
- Picardeau M. Transposition of fly mariner elements into bacteria as a genetic tool for mutagenesis. Genetica. 2010;138(5):551–558. doi: 10.1007/s10709-009-9408-5. [DOI] [PubMed] [Google Scholar]
- Redfield RJ, Findlay WA, Bossé JT, Kroll JS, Cameron AD, Nash JH. Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evolutionary Biology. 2006;6(1):82. doi: 10.1186/1471-2148-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux S, Galarneau C, Harel J, Frey J, Nicolet J, Kobisch M, Dubreuil JD, Jacques M. Isolation and characterization of mini-Tn10 lipopolysaccharide mutants of Actinobacillus pleuropneumoniae serotype 1. Canadian Journal of Microbiology. 1999;45(12):1017–1026. doi: 10.1139/w99-107. [DOI] [PubMed] [Google Scholar]
- Sassu EL, Bossé JT, Tobias TJ, Gottschalk M, Langford PR, Hennig-Pauka I. Update on Actinobacillus pleuropneumoniae—Knowledge, gaps and challenges. Transboundary and Emerging Diseases. 2018;65(Suppl 1):72–90. doi: 10.1111/tbed.12739. [DOI] [PubMed] [Google Scholar]
- Sawers RG. Formate and its role in hydrogen production in Escherichia coli. Biochemical Society Transactions. 2005;33(pt 1):42–46. doi: 10.1042/BST0330042. [DOI] [PubMed] [Google Scholar]
- Schulz H, Pellicioli EC, Thöny-Meyer L. New insights into the role of CcmC, CcmD and CcmE in the haem delivery pathway during cytochrome c maturation by a complete mutational analysis of the conserved tryptophan-rich motif of CcmC. Molecular Microbiology. 2000;37(6):1379–1388. doi: 10.1046/j.1365-2958.2000.02083.x. [DOI] [PubMed] [Google Scholar]
- Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Shalel-Levanon S, San K-Y, Bennett GN. Effect of oxygen, and ArcA and FNR regulators on the expression of genes related to the electron transfer chain and the TCA cycle in Escherichia coli. Metabolic Engineering. 2005;7(5–6):364–374. doi: 10.1016/j.ymben.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Sheehan BJ, Bossé JT, Beddek AJ, Rycroft AN, Kroll JS, Langford PR. Identification of Actinobacillus pleuropneumoniae genes important for survival during infection in its natural host. Infection and Immunity. 2003;71(7):3960–3970. doi: 10.1128/IAI.71.7.3960-3970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan BJ, Langford PR, Rycroft AN, Kroll JS. [Cu,Zn]-superoxide dismutase mutants of the swine pathogen Actinobacillus pleuropneumoniae are unattenuated in infections of the natural host. Infection and Immunity. 2000;68(8):4778–4781. doi: 10.1128/IAI.68.8.4778-4781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tascon RI, Rodriguez-Ferri EF, Gutiérrez-Martín CB, Rodriguez-Barbosa I, Berche P, Vazquez-Boland JA. Transposon mutagenesis in Actinobacillus pleuropneumoniae with a Tn10 derivative. Journal of Bacteriology. 1993;175(17):5717–5722. doi: 10.1128/jb.175.17.5717-5722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier M, Chalmers R. Compensating for over-production inhibition of the Hsmar1 transposon in Escherichia coli using a series of constitutive promoters. Mobile DNA. 2020;11(1):5. doi: 10.1186/s13100-020-0200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trchounian A. Escherichia coli proton-translocating F0F1-ATP synthase and its association with solute secondary transporters and/or enzymes of anaerobic oxidation-reduction under fermentation. Biochemical and Biophysical Research Communications. 2004;315(4):1051–1057. doi: 10.1016/j.bbrc.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Trchounian A, Kobayashi H. Fermenting Escherichia coli is able to grow in media of high osmolarity, but is sensitive to the presence of sodium ion. Current Microbiology. 1999;39(2):109–114. doi: 10.1007/s002849900429. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Ambur OH, Tonjum T, Rocha EPC. The impact of the neisserial DNA uptake sequences on genome evolution and stability. Genome Biology. 2008;9(3):R60. doi: 10.1186/gb-2008-9-3-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A. Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nature Methods. 2009;6(10):767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelin G, Fürste JP, Lanka E. TraJ protein of plasmid RP4 binds to a 19-base pair invert sequence repetition within the transfer origin. The Journal of Biological Chemistry. 1989;264(20):11989–11994. doi: 10.1016/S0021-9258(18)80164-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete sequences of the two mariner delivery vectors, pTsodCPC9 and pTlacPC9, have been deposited in GenBank under accession numbers MH644834 and MH644835, respectively. The plasmids are available upon request from the corresponding authors. The draft genome sequence of the porcine clinical respiratory isolate of P. multocida, MIDG3277, has been deposited in the European Nucleotide Archive (ENA) under the accession number ERZ681052, with the raw reads available under the accession number ERR200085. The raw TraDIS reads for A. pleuropneumoniae are available in ENA under the accession number ERR271132. The raw TraDIS reads for P. multocida are available in ENA under the accession numbers ERR744003, ERR744016, ERR752316, ERR752329, ERR755725 and ERR755738.