Abstract
Genes in Caenorhabditis elegans operons are transcribed as polycistronic pre-mRNAs in which downstream gene products are trans spliced to a specialized spliced leader, SL2. SL2 is donated by a 110-nucleotide RNA, SL2 RNA, present in the cell as an Sm-bound snRNP. SL2 RNA can be conceptually folded into a phylogenetically conserved three-stem-loop secondary structure. Here we report an in vivo mutational analysis of the SL2 RNA. Some sequences can be changed without consequence, while other changes result in a substantial loss of trans splicing. Interestingly, the spliced leader itself can be dramatically altered, such that the first stem-loop cannot form, with only a relatively small loss in trans-splicing efficiency. However, the primary sequence of stem II is crucial for SL2 trans splicing. Similarly, the conserved primary sequence of the third stem-loop plays a key role in trans splicing. While mutations in stem-loop III allow snRNP formation, a single nucleotide substitution in the loop prevents trans splicing. In contrast, the analogous region of SL1 RNA is not highly conserved, and its mutation does not abrogate function. Thus, stem-loop III appears to confer a specific function to SL2 RNA. Finally, an upstream sequence, previously predicted to be a proximal sequence element, is shown to be required for SL2 RNA expression.
The mature 5′ ends of mRNAs in many lower eucaryotes, including trypanosomes (41, 50), nematodes (28), trematodes (43), and Euglena (51), are generated through trans splicing. This process involves the donation of a common spliced leader (SL) exon and its associated modified cap structure to the 5′ ends of mRNAs. The reaction proceeds via two transesterification reactions, analogous to cis splicing of introns, and in fact requires most of the same U snRNP cofactors, including U2, U4, U5, and U6 snRNAs (12, 22, 36, 54). However, because the 5′ exon is initially located on a separately transcribed RNA, the SL RNA, the intermediate of the trans-splicing reaction is a Y-branched molecule instead of the lariat molecule found in cis splicing. The SL RNA exists as an RNP particle (20, 33, 52, 55) that includes the Sm core proteins that are also associated with most of the U snRNAs involved in cis splicing (30). Unlike the other U snRNPs, which are recycled after catalysis, the SL RNP is consumed during the trans-splicing reaction.
In trypanosomes, pre-mRNAs are transcribed polycistronically (26, 40) and the mature mRNAs are generated by trans splicing and 3′ end formation. Approximately 25% of Caenorhabditis elegans genes are also organized in polycistronic transcription units, or operons (57). However, unlike trypanosomes, C. elegans possesses a distinct SL RNA, SL2 RNA (25), that is specifically utilized to process the products of downstream genes in operons (46). Therefore, the discovery of SL2 on an mRNA is taken as an indication that the gene is in an operon (1, 2).
Experiments designed to uncover the mechanism of trans-splicing specificity in C. elegans have shown that SL1 trans splicing is usually specified by an outron, an AU-rich sequence containing a 3′ splice site without a paired 5′ splice site upstream (8). In support of this discovery, a conventional gene becomes trans spliced if an outron is added (6). Conversely, a trans-spliced gene can be converted to a conventional gene by adding a 5′ splice site to the outron (7).
Being encoded by a downstream gene in an operon appears to be the only criterion for pre-mRNAs being trans spliced to SL2 (46). This hypothesis is supported by the discovery that an mRNA normally trans spliced exclusively to SL1 becomes trans spliced to SL2 when placed downstream of another gene (46). SL2 trans splicing has been shown to be associated with 3′ end formation of upstream genes. If the polyadenylation signal, AAUAAA, of an upstream gene is mutated, SL2 trans splicing to the downstream gene product is reduced (29). The molecular mechanism of this functional association has yet to be elucidated.
Despite the fact that SL1 and SL2 are trans spliced to different subsets of mRNAs, the SL1 and SL2 RNAs appear quite similar. Both are small (∼100 and 110 nucleotides [nt], respectively), possess a trimethylguanosine cap, lack a poly(A) tail, and associate with Sm proteins (25, 28, 52, 55). Additionally, both RNAs can be folded into a secondary structure that is conserved throughout trans-splicing RNAs (4). This structure consists of three stem-loops, with the trans-splice site located on the 3′ side of stem I and the Sm binding site located in a single-stranded region between stems II and III. These similarities between the SL1 and SL2 RNAs make the question of substrate specificity even more intriguing.
Recent reports indicate that the SL RNA requirements for trans splicing vary significantly between species. In particular, different groups have reported very different results when they mutate or delete portions of the SL itself (15, 19, 32, 34, 47, 56). In nematodes it is clear that the SL1 sequence can be replaced by unrelated sequences of various lengths without loss of in vitro trans splicing (34), but at least one SL mutation was found to prevent trans splicing in vivo (15). In contrast, mutational analysis of the intron portion of the molecule demonstrated that both stem-loop II and the Sm binding region are crucial for SL RNA function in both trypanosomes and nematodes (11, 23, 49). However, it has been possible to significantly alter the third stem-loop without eliminating trans splicing (32, 49).
Phylogenetic comparative sequence analysis of SL2 RNA genes from the nematodes Dolichorhabditis and C. elegans identified several conserved features that suggest a functional significance (13). These conserved features include the 5′ end of the spliced leader, the trans-splice site, part of stem II, the Sm binding site, and the top of stem-loop III (13). Similarly, previous comparative sequence analysis of regions upstream of each of the U snRNA and SL RNA genes in C. elegans resulted in the discovery of a putative proximal sequence element (PSE), thought to be required for transcription of these genes (53).
Here we tested the idea that the conserved regions are important for trans splicing by in vivo mutational analysis of the C. elegans SL2 RNA. The results of this analysis suggest similarities between the SL2 RNA and other trans-splicing RNAs, but they also demonstrate distinct differences. The entire SL sequence was found to be dispensable for both expression and trans splicing of the SL2 RNA. In contrast, the primary sequence of stem II and the sequence located at the top of stem-loop III were both found to be necessary for polycistronic pre-mRNA processing.
MATERIALS AND METHODS
Worms.
The basic procedures for handling and maintaining worms have been described elsewhere (3). Transgenic worms were generated (39) by injecting young hermaphrodite gonads of strain MT2597 [rol-6 (n1178)] (J. Kramer, personal communication) with both the test plasmid and the selection plasmid, pRF5 [rol-6 su 1006dom)] (7), at 50 to 100 μg/ml in water (16). Plasmid DNAs for injection were prepared as previously described (17).
Mutated SL2 RNA gene construction.
Plasmid pASL2 was generated by digesting pSL2+ (containing an ∼3.8-kb BamHI fragment isolated from λSL2/9 [25] and subcloned into pGEM-3Z+) with BamHI and HindIII and inserting the approximately 2.0-kb fragment containing the SL2α gene into pBluescript (pBS) SK(+). Mutated SL2 RNA genes were generated by PCR using oligonucleotides that incorporated the mutations to be introduced (24). The C17G mutation was generated using pASL2 as template with oligonucleotides 1012 plus T3 primer and 1013 plus T7 primer, respectively. The products were gel purified and fused by performing PCR using a mixture of these products as template and using the T3 and T7 primer pairs. The resulting product was digested with BamHI and HindIII and cloned into pBS SK(+) digested with BamHI and HindIII. The presence of the desired mutation was confirmed by sequencing the SL2 RNA coding region. The U4A, U5A, Stem 3 sub, and ΔPSE mutated RNAs were created as above using the C17G plasmid as template and pairing the T3 and T7 primers with oligonucleotide pairs 1224-1223, 1246-1247, and 1011-1010, respectively (see below for sequences). The SL sub mutation was created as above, using pASL2 as template and oligonucleotides 1325 and 1324 as primers. The Stem 2 sub, G93C,A96C, G93C, and A96C mutated RNAs were created using a single “runaround” PCR (9) with the C17G plasmid as template and SL2 oligonucleotide pairs 1392-1393, 1394-1395, 1394-1397, and 1396-1395 respectively.
Mutated SL1 RNA gene construction.
Plasmid pSL15S [containing a 985-bp BamHI fragment including both the C. elegans SL1 RNA and 5S rRNA genes cloned into pBS II SK(−) (14)] was used as template to generate mSL1 by runaround PCR with oligonucleotide pair 1412-1413. mSL1 then served as template to generate Stem 3 sub mSL1 by runaround PCR with oligonucleotide pair 1414-1415.
Oligonucleotides.
The oligonucleotides used were T3 primer (1015; 5′-AATTAACCCTCACTAAAGGG-3′), T7 primer (1016; 5′-GTAATACGACTCACTATAGGGC-3′, 1012 (5′-CGTACCTTGACTAACTGGGTT-3′), 1013 (5′-AACCCAGTTAGTCAAGGTACG-3′), 1224 (5′-AACTGGGTTATTACCGAAGATA-3′), 1223 (5′-TATCTTCGGTAATAACCCAGTT-3′), 1246 (5′-CGTGTGTACCCTGTCATTTCGAGG-3′), 1247 (5′-CCTCGAAATGACAGGGTACACACG-3′), 1011 (5′-CCGCTCTGCTCCTCTCGGGGGGTAGAGCAA-3′), 1010 (5′-TTGCTCTACCCCCCGAGAGGAGCAGAGCGG-3′), 1325 (5′-CAGCGTACCTAATCATTGACCCAATTTTGGGAAGATACCA-3′), 1324 (5′-TGGTATCTTCCCAAAATTGGGTCAATGATTAGGTACGCTG-3′), 1392 (5′-GAAACTTTCTCAGTAACGACTTTAATTTTTGGAACC-3′), 1393 (5′-CTTTCCAGTGAACTCCAGCGTACCTTGAG-3′), 1394 (5′-GCCCAGCAGAGCGGTTCC-3′), 1395 (5′-TCCTCCGGTAGAGCAAAATAATTG-3′), 1396 (5′-CCCCAGCAGAGCGGTTCC-3′), 1397 (5′-TCATCCGGTAGAGCAAAATAATTG-3′), 1412 (5′-AAGGTAAACATTGAAACTGAC-3′), 1413 (5′-AAACTTGGGTAATTAAACCGA-3′), 1414 (5′-TCATCCAGACAAAAATACTAA-3′), and 1415 (5′-CCCCAGACGTTCCAAAATTTA-3′).
Strains.
Strains are listed in the order in which they appear in the figures. The strain carrying the rol-6 amber mutation (MT2597) is referred to as WT (wild type). All other strains carry mutated SL2 or SL1 RNA genes in extrachromosomal arrays with rol-6 (su1006dom). They are named as follows: BL1300-01, C17G; BL1305-07, U4A,U5A; BL1311-13B, SL sub; BL1317-19, Stem 2 sub; BL1302-04, Stem 3 sub; BL1320-22, G93C,A96C; BL1327-30, G93C; BL1331-33, A96C; BL1308-10, ΔPSE; BL1323-26, mSL1; and BL1334-36, Stem 3 sub mSL1.
RNA isolation.
Large-scale preparations of RNA were made from mixed stages of worms grown on plates as described previously (6).
Primer extensions.
Gel-purified oligonucleotides were end labeled with [γ-32P]ATP and T4 polynucleotide kinase. Labeled oligonucleotides (0.2 ng) were mixed with 20 μg of total RNA in 1× annealing buffer (150 mM KCl, 10 mM Tris [pH 8.3]), heated to 95°C, and annealed at 42°C for 1 h. Reverse transcription was performed for 30 min at 42°C in 1× RT (reverse transcriptase) buffer (12 mM Tris [pH 8.3], 8 mM MgCl2, 4 mM dithiothreitol [DTT], 0.01% NP-40, actinomycin D [50 ng/μl, 1 mM dATP, 1 mM dGTP, 1 mM dTTP, 0.75 mM ddCTP, 5 U of avian myeloblastosis RT [Promega]). The products were ethanol precipitated, resuspended in formamide dye, electrophoresed on 20% polyacrylamide gels containing 7 M urea, and analyzed by autoradiography. Quantitation was performed on a Molecular Dynamics PhosphorImager using ImageQuant software. The primers used were gpd3-5′ end (5′-GCGTTGAACGACTTTCCC-3′), SL2 PE (5′-GGT CAGAACTCCAGCGTACC-3′), SL2 short PE (5′-GAACTCCAGCGTACC-3′), SL1 PE (5′-GGTCAGTTTCAATGTTTACC-3′), and gpd-2 PE (5′ ATTTGGCTTTGGCATGGC-3′)].
Calculating trans-splicing efficiencies.
trans-splicing efficiencies (ratio of trans spliced to expressed) were calculated from the primer extension data shown. The background value at the +5 mSL2 (or +6 mSL1) in the WT lane was subtracted from the experimental lanes to arbitrarily set the mSL2 trans-splicing efficiency in the WT lane at 0%. The trans-splicing efficiency value of the mutated RNA was then normalized to the endogenous RNA level for each strain. Means and standard deviations are shown.
Immunoprecipitations with antibodies against snRNP core proteins.
Protein A-Sepharose (200 μl, packed volume) was incubated at room temperature for 1 h with 100 μl of anti-Sm lupus antiserum (10, 18, 37, 38) in NP-40 lysis buffer (20 mM Tris [pH 8.0], 250 mM NaCl, 1 mM EDTA, 0.5% NP-40) with rotation. The antibody-bead conjugates were washed three times with 500 μl of NP-40 lysis buffer and then blocked with NP-40 lysis buffer–0.1% nonfat dry milk for 1 h at room temperature with rotation. The beads were washed three times with 500 μl of NP-40 lysis buffer and resuspended in 400 μl of NP-40 lysis buffer. BL1304 (Stem 3 sub) extract (50 μl) was incubated rotating at 4°C for 2 h with 150 μl of bead slurry, 1 μl of RNasin (Promega), and 300 μl of NP-40 lysis buffer. The unbound fraction was removed, and the beads were washed three times with NP-40 lysis buffer. The entire unbound, wash, and bound fractions were treated with proteinase K followed by phenol-chloroform extraction and chloroform extraction. The RNAs were ethanol precipitated and resuspended in Tris-EDTA for both ddC primer extension and Northern blot analysis.
Northern analysis.
RNAs were analyzed on 10% denaturing polyacrylamide gels, transferred by electroblotting to Hybond N (Amersham), and hybridized to either a C. elegans U1 snRNA RT-PCR product or a C. elegans 5S rRNA PCR product labeled using a Prime-It II random primer labeling kit (Stratagene). Hybridization was carried out in a mixture of 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1× Denhardt's solution, 50 mM NaPO4, 0.1% sodium dodecyl sulfate (SDS), and 0.2 mg of sheared DNA per ml overnight at 42°C. Wash conditions were 2× SSC–0.2% SDS for 20 min at 24°C followed by 0.1× SSC–0.1% SDS for 30 min at 55°C.
BL1304 crude embryonic extract preparation.
Attempts to integrate the extrachromosomal array of strain BL1304 using γ irradiation (27) were unsuccessful, so the progeny of irradiated worms were screened for high transmittance of the array as assayed by expression of the dominant marker. Rolling worms picked from the best clone were inoculated into 1 liter of liquid S-basal medium and grown at 20°C with Escherichia coli NA22 as a food source. The worms were allowed to form dauers to synchronize the culture. Dauers were purified by treatment with 1% SDS for 30 min at 23°C followed by water washes, sucrose flotation, and water washes. The dauers were then used to inoculate a fresh 1-liter culture of S-basal containing NA22 to release the developmental arrest. Embryos were collected from gravid adults grown in S-basal with NA22 by hypochlorite treatment (0.5 M KOH, 20% bleach) for 10 min at 23°C. Embryos were washed twice with homogenization buffer (10 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 10 mM Tris-HCl [pH 8.0], 50 mM sucrose, 0.05% NP-40, Complete [Boehringer Mannheim] protease inhibitor cocktail tablets [4 tablets/100 ml of homogenization buffer]). The embryos were resuspended in 3 ml of homogenization buffer per ml of packed embryos and homogenized by approximately 20 strokes with a stainless steel Dounce homogenizer. The homogenate was centrifuged for 5 min at high setting on a tabletop clinical centrifuge at 4°C to pellet cellular debris. The supernatant was removed, placed in a glass Dounce homogenizer, and homogenized by approximately 10 strokes with a loose-fitting pestle followed by centrifugation in a Beckman JA-17 rotor at 8,000 rpm (8,817 × g) for 30 min at 4°C. The supernatant was dialyzed for 6 h against 1 liter of dialysis buffer (50 mM KCl, 1 mM DTT, 0.5 mM EDTA, 10% glycerol, 20 mM Tris-HCl [pH 8.0], Complete protease inhibitor cocktail tablets [4 tablets/2 liters of dialysis buffer]). The buffer was changed after 3 h. The dialysate was centrifuged in a Beckman JA-17 rotor at 10,000 rpm (13,776 × g) for 5 min at 4°C and transferred to a fresh tube; 100-μl aliquots were frozen in liquid nitrogen.
RESULTS
Marked SL2 RNA is expressed.
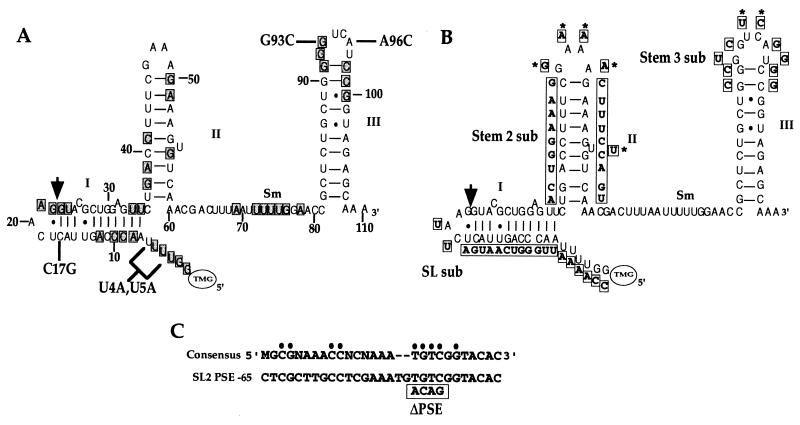
To determine the functional importance of conserved regions of SL2 RNA in C. elegans, a marked copy of the SL2α RNA gene was expressed in vivo from an extrachromosomal tandem array. The marked gene was created by substituting a single nucleotide at position C17 in SL2 to G (Fig. 1A, C17G). The mutation allowed analysis of the fate of the mutated SL2 RNA in a background of wild-type SL2 RNA by primer extension in the presence of ddC. Primer extension products from the mutated SL2 RNA (+5) could easily be resolved from those expected for endogenous SL1 (+2) and SL2 (+9). Furthermore, none of the reported SL2 RNAs contain a G at position 17 (13). Therefore, this mutation should eliminate any background that would otherwise have resulted from SL2 variants (45).
FIG. 1.
Mutations examined in this study. (A) Schematic diagram of SL2 RNA with point mutations. Phylogenetically perfectly conserved positions are shaded (14). The stem-loops are labeled (I, II, and III) as discussed in the text. The arrow marks the site of trans splicing. Point mutations are indicated. The Sm binding site consensus sequence is RAU4–6GR (reviewed in reference 42). (B) Large substitution mutations (boxed) are shown next to the wild-type SL2 RNA structure. Nucleotide positions marked with an asterisk remain unchanged in the mutated SL2 RNA. (C) PSE mutation. The sequences shown are the consensus C. elegans PSE and the proposed PSE from the SL2α gene (53). Invariant positions are indicated by dots (53). The 5′ end of the PSE in this gene is 65 bp upstream of the transcription start site (53). The mutated sequence tested is boxed.
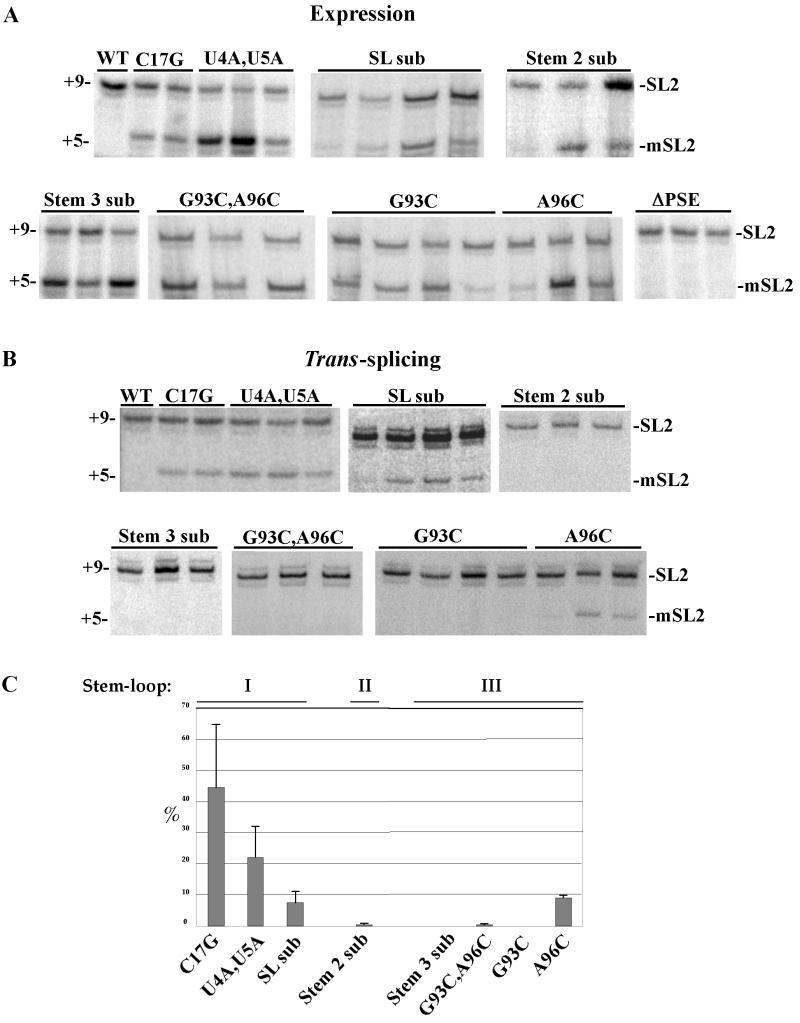
The marked SL2 RNA gene, under the control of its own promoter, was injected into young adult hermaphrodite gonads, along with a rol-6(d) marker gene, and transgenic strains were selected. To determine whether these genes were expressed, we isolated RNA from two strains and performed primer extension using an oligonucleotide (SL2 PE) complementary to bases 22 to 41 of C. elegans SL2α RNA. The transgene was expressed, as evidenced by the primer extension product at position +5 (Fig. 2A, C17G lanes). Primer extension on RNA isolated from an uninjected strain resulted in a product only at the +9 position, as expected from endogenous SL2 RNA (Fig. 2A, WT lane). The quantity of C17G RNA averaged 68% of the amount of endogenous SL2 RNA, indicating that expression of the transgene is quite robust.
FIG. 2.
(A) Expression of mutated SL2 RNAs. Multiple strains carrying each construct were grown, and RNA was extracted. Expression of mutated SL2 RNAs was measured by primer extension in the presence of ddC, using an oligonucleotide that hybridizes to the SL2 RNA just 3′ of the trans-splice site. The band at the +9 position corresponds to endogenous SL2 RNA, while the band at the +5 position corresponds to mutated SL2 RNA. (B) trans splicing of mutated SL2 RNAs to gpd-3 mRNA. trans splicing of wild-type and mutated SL2 RNAs was measured by primer extension in the presence of ddC, using a primer that anneals just 3′ of the trans-splice site of the SL2-accepting gpd-3 mRNA. Multiple strains carrying each construct were tested. The band at the +9 position corresponds to endogenous SL2 RNA trans spliced to gpd-3 mRNA, while the band at the +5 position corresponds to mutated SL2 RNA trans spliced to gpd-3 mRNA. (C) trans-splicing efficiencies of mutated SL2 RNAs, calculated from the data in Fig. 2A and B as described in Materials and Methods. For each strain the trans-splicing efficiency (trans splicing/expression) of the introduced construct was normalized to the calculated efficiency of the endogenous SL2. Means and standard deviations are shown.
Marked SL2 RNA is competent for SL2 trans splicing.
To determine whether the marked SL2 RNA was trans spliced to the product of a downstream gene in an operon, we tested for C17G trans splicing to gpd-3 mRNA, an abundant mRNA that is normally trans spliced almost entirely to SL2 (25). trans splicing was assayed by ddC primer extension using an oligonucleotide (gpd3-5′ end) complementary to bases −1 to +17 relative to the trans-splice site of the gpd-3 mRNA. As can be seen in Fig. 2B, C17G RNA trans spliced efficiently to gpd-3 mRNA. Primer extension on RNA isolated from an uninjected strain resulted in a product only at the +9 position, as expected for endogenous SL2 RNA, while the RNA isolated from the C17G-containing strains produced a strong +5 primer extension product along with the +9 product. trans-splicing efficiency of the C17G RNA was approximately 45% that of the wild type (Fig. 2C). This result confirms the potential utility of the marked SL2 RNA for studying the requirements of the SL2 RNA for trans splicing.
Mutations in the SL still allow trans splicing.
All SL2 RNAs discovered to date begin with the sequence GGUUU (13, 25, 45). To test the functional significance of this conserved 5′ end of SL2, the SL2 sequence was changed to GGUAA in the context of the C17G mutation (Fig. 1A, U4A,U5A). This mutated RNA functioned almost as well as C17G alone. The primer extension product at +5 demonstrates that the mutated SL2 RNA is abundantly expressed and trans spliced to gpd-3 (Fig. 2A and B). The U4A,U5A mutated RNA was trans spliced ∼50% as efficiently as C17G (Fig. 2C).
To determine whether any of the sequences in the SL2 RNA spliced leader were required for function, we changed the entire sequence to its complement, except for the C at position 19, which was changed to U, and the conserved AG found just upstream of the trans-splice site, which was maintained (Fig. 1B, SL sub). Surprisingly, this mutated RNA was expressed abundantly and even retained the ability to be trans spliced to the SL2-accepting substrate (Fig. 2A and B, SL sub lanes). It should be noted that the conserved secondary structure of the SL2 RNA is completely disrupted in the SL sub mutated RNA. Nonetheless, trans splicing occurred at approximately 20% of the level of the C17G construct (Fig. 2C). This result demonstrates that neither the sequence of SL2 nor the presence of a first stem-loop is required for SL2 trans splicing.
The primary sequence of the second stem is important for trans splicing.
The importance of the second stem-loop structure of the SL2 RNA in trans splicing was investigated by changing its primary sequence while maintaining the potential to form a stem (Fig. 1B, Stem 2 sub). This was accomplished by changing each base in the stem to its complement while maintaining the sequence of the GNRA tetraloop and the bulged uridine. Since part of the sequence that hybridized to the SL2 oligonucleotide (SL2 PE) had been changed in this mutated RNA, expression was assayed using a different oligonucleotide (SL2 short PE) complementary to bases 22 to 36 of SL2α RNA, which hybridizes equally well to endogenous and Stem 2 sub RNA. The data in Fig. 2A and B demonstrate that although this mutated RNA is expressed well, it is trans spliced very poorly. One strain consistently exhibited very weak trans splicing (1% of the level of endogenous SL2 RNA), while the other two strains showed no detectable trans splicing (Fig. 2C). This result implies that the primary sequence of the second stem structure is important for SL2 trans splicing.
The top of the third stem-loop is required for SL2 trans splicing.
The importance of the phylogenetically conserved third stem-loop sequence in SL2 trans splicing was tested by replacing the top of the third stem-loop of SL2 RNA with a sequence identical to the top of the third stem-loop of SL1 RNA (Fig. 1B, Stem 3 sub). While this mutated RNA was abundantly expressed, trans splicing to gpd-3 mRNA was not detected (Fig. 2). This result demonstrates a clear requirement for a sequence element located in the top of stem-loop III of SL2 RNA in trans splicing.
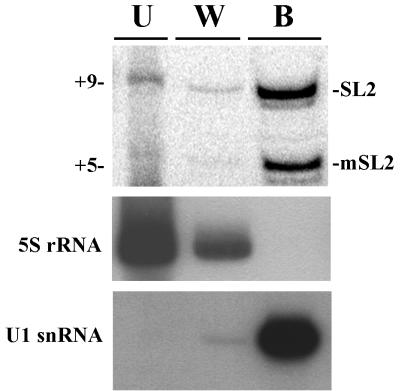
This observation, coupled with the conservation of the primary sequence in this region of the RNA, suggests that an SL2 RNP-specific RNA-RNA or RNA-protein interaction may be disrupted in this mutated RNA. Alternatively, the ability of the mutated SL2 RNA to form a core snRNP might be affected, resulting in a secondary effect on trans splicing. This hypothesis was tested by performing immunoprecipitations from crude embryonic extracts made from a Stem 3 sub mutated RNA strain using a human systemic lupus erythematosus patient serum containing antibodies against the Sm proteins. The SL2 RNAs precipitated by this serum were examined by ddC primer extension. Figure 3 shows that both the endogenous and mutated SL2 snRNPs were efficiently immunoprecipitated by this serum, suggesting that the defect of Stem 3 sub was not simply an inability to form a core snRNP. To control for the specificity of the immunoprecipitation, RNAs precipitated by the serum were subjected to Northern analysis. As expected, U1 snRNA was efficiently precipitated, while 5S rRNA was not (Fig. 3). Similar results were obtained using the monoclonal Sm antibody Y12 (31) (data not shown).
FIG. 3.
Stem 3 sub does not prevent core snRNP formation. RNAs were immunoprecipitated from a Stem 3 sub crude embryonic extract with a human systemic lupus erythematosus patient serum of the Sm type. The RNAs contained in the unbound (U), the first of three washes (W), and bound (B) fractions were analyzed by ddC primer extension to analyze SL2 RNA and by Northern blotting for the U1 and 5S RNA controls. The Northern blot was hybridized to a U1 snRNA probe, stripped, and hybridized to a 5S rRNA probe. 5S RNA, which is not bound to Sm proteins, was not immunoprecipitated, whereas all three snRNAs, U1, SL2, and mSL2, were.
To determine whether the loop nucleotides were required for SL2 trans splicing, a mutation in which two bases in the third loop were changed was tested (Fig. 1A, G93C,A96C). This mutated RNA was highly expressed, but again no trans splicing to gpd-3 was detected (Fig. 2A and B). This result demonstrates that mutation of these two loop nucleotides by themselves is sufficient to prevent the SL2 RNA from participating in trans splicing.
Two additional mutated SL2 RNAs were designed to test the importance of each of these loop nucleotides individually (Fig. 1A, G93C and A96C). Both mutated RNAs, containing a single base change in the context of the C17G RNA, were expressed at a high level (Fig. 2A). Interestingly, no detectable trans splicing was observed for any of the four transgenic strains containing a single base change on the 5′ side of the loop (Fig. 2B, G93C). In contrast, the mutated RNA containing a single base change on the 3′ side of the loop was trans spliced to gpd-3 mRNA in each of the three strains tested (Fig. 2B, A96C). The average trans-splicing efficiencies of G93C and A96C were 0 and 9% of the level of endogenous SL2 RNA, respectively (Fig. 2C). Thus, the G at position 93 is required for SL2 trans splicing, while mutation of the A at position 96 results in a product that is still functional, although at a reduced level, similar to that obtained with the SL sub mutation.
Stem-loop III is necessary but not sufficient for SL2 trans splicing.
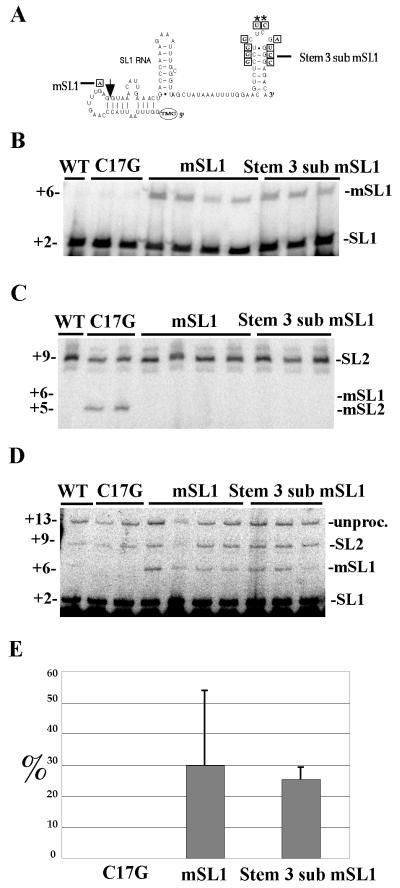
The above experiments demonstrated that the top of the third loop of SL2 RNA is necessary for SL2 trans splicing. To address whether this sequence is also sufficient for SL2 trans splicing, the top of stem-loop III of SL1 RNA was replaced with the sequence from SL2 stem-loop III in a marked SL1 RNA (Fig. 4A, Stem 3 sub mSL1). As a control, strains containing the marked SL1 RNA with the wild-type stem-loop III were also created (Fig. 4A, mSL1). The injected constructs, under the control of the SL1 RNA promoter, contained a single nucleotide substitution in SL1 but were nevertheless expressed. This change, G20 to A, resulted in a shift of the ddC primer extension product from +2 for endogenous SL1 to +6 for the marked SL1. Expression of both constructs was studied by ddC primer extension with the SL1 RNA using an oligonucleotide (SL1 PE) complementary to bases 22 to 41 of C. elegans SL1 RNA. The ddC primer extension product at +6 demonstrates that both mSL1 and Stem 3 sub mSL1 are expressed, although at a lower level than the endogenous SL1 RNA product at +2 (Fig. 4B). trans-splicing specificity was assayed by performing ddC primer extension with the gpd3-5′ end primer. The absence of a detectable product at +6 demonstrated that the sequence of the top of stem-loop III of SL2 RNA is not sufficient to enable SL1 RNA to trans splice onto this SL2-accepting pre-mRNA (Fig. 4C).
FIG. 4.
Analysis of a mutated SL1 RNA containing part of SL2 stem-loop III. (A) Schematic diagram of SL1 RNA with mutations. The predicted secondary structure of the C. elegans SL1 RNA is shown with the mSL1 mutation and the portion of SL1 changed to the equivalent region of SL2 boxed. Nucleotide positions marked with an asterisk remain unchanged in the mutated SL1 RNA. The arrow marks the trans-splice site. (B) Expression of mutated SL1 RNAs. Expression of mutated SL1 RNAs was assayed by primer extension with ddC using a primer that anneals to the SL1 RNA just 3′ of the trans-splice site. The band at the +2 position corresponds to endogenous SL1 RNA, while the band at +6 corresponds to mSL1 RNA. Primer extension from two strains carrying the SL2 mutated C17G RNA is included as a control. (C) trans splicing of Stem 3 sub mSL1 to gpd-3 mRNA. The ability of the mutated SL1 RNAs to trans splice to SL2-accepting mRNAs was determined by primer extension with ddC from the gpd-3 mRNA, as in Fig. 2B. The predicted primer extension terminations at +9, +6, and +5 correspond to endogenous SL2, mSL1, and mSL2, respectively. No trans splicing of the mSL1 or Stem 3 sub mSL1 mutated RNAs to gpd-3 was observed. (D) trans splicing of Stem 3 sub mSL1 to gpd-2 mRNA. The ability of the mutated SL1 RNAs to trans splice to an SL1-accepting mRNA was determined by primer extension with ddC, using a primer that anneals to gpd-2 mRNA just 3′ of the trans-splice site. The bands at the +6 and +2 positions correspond to mSL1 and endogenous SL1 trans splicing, respectively. The bands at the +9 and +13 positions correspond to endogenous SL2 trans-splicing (46) and unprocessed RNA, respectively. trans splicing of mSL2 at +5 can be seen in the C17G lanes upon longer exposure (data not shown). (E) trans-splicing efficiency of mSL1 and Stem 3 sub mSL1 to gpd-2 mRNA. trans-splicing efficiencies (trans splicing/expression) were calculated as described in Materials and Methods. The samples were normalized to the endogenous SL1 trans-splicing levels. Means and standard deviations are shown.
The ability of the mutated SL1 RNAs to trans splice onto a pre-mRNA that normally receives SL1 was assayed by ddC primer extension using an oligonucleotide (gpd-2 PE) complementary to bases −1 to +17 of the abundant and predominantly SL1-accepting gpd-2 mRNA (Fig. 4D). The gpd-2 mRNA is also encoded by a downstream gene in the mai-1/gpd-2/gpd-3 operon and has been shown to be trans spliced to SL2 at some level (46). The results of the experiment depicted in Fig. 4D demonstrate that approximately 10% of endogenous gpd-2 mRNA is trans spliced to SL2 (+9), while 90% is trans spliced to SL1 (+2). The presence of a product at +6 shows that both mSL1 and Stem 3 sub mSL1 form functional snRNPs that participate in SL1 trans splicing. These results demonstrate that the top of the third stem-loop from SL2 RNA is not sufficient to confer SL2 trans-splicing specificity. However, a key difference between SL1 and SL2 trans splicing is revealed by these experiments. While the SL2 RNA is very sensitive to mutations in the third stem-loop, the SL1 RNA is not (Fig. 4E), since the substitution of SL1 sequence for SL2 sequence did not significantly inhibit its ability to function as an SL1 snRNP.
Transcription of the SL2 RNA in vivo requires an intact PSE.
Transcription initiation of snRNA genes requires a PSE (reviewed in reference 42). This element has been found to have very different sequences in different organisms but to be highly conserved between different snRNA genes within the same organism. Based on high conservation, a sequence beginning approximately 65 bp upstream of C. elegans U snRNA genes was predicted to constitute the C. elegans PSE (53). To determine whether the putative PSE of the SL2 α gene was in fact required for SL2 RNA expression, four highly conserved positions of this element were mutated in the marked SL2 RNA gene (Fig. 1C). Three transgenic lines were tested for marked SL2 RNA expression by ddC primer extension with SL2 RNA as template. Expression was 10-fold lower than in the C17G strains (Fig. 2A, ΔPSE lanes). The presence of the transgene was confirmed by PCR (data not shown). This result confirms the hypothesis that this sequence is required for expression of the SL2 RNA gene in vivo and presumably constitutes the C. elegans PSE.
DISCUSSION
The SL RNAs from several trypanosomes and nematodes have been dissected at the molecular level and analyzed for loss of trans-splicing ability. The SL RNA requirements for trans splicing appear to be unique in each of the organisms examined. Here we report the results of the first in vivo mutational analysis of an SL2 RNA, that of C. elegans. This SL RNA is different from the C. elegans SL1 RNA in that it is specifically utilized to process the products of downstream genes in operons. Although trypanosomes also have polycistronic transcription, it appears that they only have a single type of SL RNA. Thus, a mechanism distinguishing trans splicing to upstream gene products or downstream gene products need not exist. In contrast, there must be such a mechanism in C. elegans since the level of SL1 RNA is 7 to 10 times the level of SL2 RNA (S. Kuersten, R. Conrad, and T. Blumenthal, unpublished data), yet many downstream mRNAs are trans spliced almost exclusively to SL2.
Previous in vivo analysis of the C. elegans SL1 RNA took advantage of a genetic mutation (rrs-1) in which all of the SL1 RNA and 5S rRNA genes, which are clustered together in tandem repeats, are deleted (14). Functional requirements of the SL1 RNA were examined by rescue of the rrs-1 mutant with wild-type or mutated SL1 RNA genes (15, 56). A similar genetic tool to examine the SL2 RNA would be extremely difficult to generate because there are multiple, nonidentical SL2 RNA genes scattered throughout the C. elegans genome (13, 25, 45). Therefore, we chose to examine the requirements of SL2 RNA in trans splicing by expressing marked SL2 RNA in transgenic worms. This strategy allowed us to determine the fate of mutated SL2 RNA in the presence of endogenous wild-type SL2 RNA. Thus, in order to be scored as functional, the mutated SL2 RNAs had to be able not only to trans-splice but also to compete successfully with the wild-type SL2 snRNP.
Watson-Crick base pairing across the trans-splice site is not required.
The ability of the marked SL2, C17G, to be trans spliced was intended to serve merely as a marker for transgene SL2. However, our results also suggested that Watson-Crick base pairing between the SL and the trans-splice site might not be required for trans splicing in vivo. SL RNAs from several different organisms were predicted to fold into similar secondary structures composed of three stem-loops (4). In each of these structures the SL was predicted to base pair with the trans-splice site. It had been previously demonstrated that the mechanism of trans splicing did not require the U1 snRNA (22). Therefore, it was hypothesized that the base pairing of the trans-splice site with the SL mimicked the interaction of the 5′ splice site with the U1 snRNA (5). However, the Ascaris SL RNA can function in vitro without such base pairing (34). Indeed, the entire SL exon is dispensable and can be replaced with sequences varying in length from two to 246 nt while maintaining the ability to trans splice (34). The ability of our marked SL2 RNA to function demonstrates that this interaction is not required for SL2 trans splicing in vivo. A close phylogenetic inspection of SL2 RNA genes (13) lends additional support to this conclusion. In those cases where the nucleotide at position +5 is not a C, it is an A, which is not predicted to form a Watson-Crick base pair with the trans-splice site (13).
SL sequence is dispensable for both expression and SL2 trans splicing.
The regions of SL2 that are perfectly conserved in all SL2 RNAs include the very 5′ end of the spliced leader, GGUUU. SL1 also begins with this sequence. In contrast to SL2, the SL1 sequence is almost perfectly conserved throughout all nematodes. The only reported exception is the SL from Meloidogyne incognita, which differs from the canonical 22-nt nematode SL at one position (44). Interestingly, in most SL2 RNAs there is at least a fourth U and in some of the SL2 RNA genes a fifth U following the 5′ GG (13, 25, 45). Additionally, this region of SL2 is predicted to be single stranded by RNA folding programs, while the analogous region of the SL1 RNA is predicted to be base paired in the first stem. The ability of SL2 RNAs with mutations in this conserved region to splice efficiently demonstrates that this region is not required for SL2 trans splicing. The conservation of this region of the RNA may be explained in other ways. For example, this sequence may be conserved due to a selective pressure on trans-splicing efficiency. Alternatively, the conservation could be the result of an unknown role of the leader after splicing, such as translation (35) or mRNA stability.
In contrast to SL1, SL2 has diverged substantially, even within the C. elegans genome. This obvious difference between the two SLs could account for their different trans-splicing specificities. The ability of the SL sub mutated RNA, in which nearly the entire sequence of the SL has been mutated, to be both expressed and trans spliced was therefore surprising. The fact that the RNA is expressed demonstrates that unlike the Ascaris SL RNA or the C. elegans SL1 RNA, the C. elegans SL2 sequence itself is not required for transcription of the SL2 RNA gene (21, 56). While trans-splicing efficiency of the SL sub mutated RNA was only approximately 20% the level of the C17G RNA, it clearly was able to trans splice. Several studies, both in vivo and in vitro, have suggested that the overall length and sequence of the nematode SL1 are not required for SL1 trans splicing (15, 34, 56). However, it was demonstrated that a portion of the leader sequence or the SL1 RNA secondary structure was essential for trans splicing in vivo (15). Again the combination of conserved regions in this sequence and the ability of the SL mutated RNA to trans splice may indicate a role for the SL sequence on the mRNA after splicing. Since we were assaying only for ability to trans splice, if the mutated SL was unable to fulfill a role once transferred to the mRNA, we would not have detected the defect. However, it is clear that the SL2 sequence is not required for the SL2 snRNP to trans splice onto polycistronic pre-mRNAs.
The primary sequence of the second stem is required for SL2 trans splicing.
If there is one requirement for SL RNA trans splicing that seems to be universal, it is the requirement of the stem II structure. However, even this result depends on the RNA being studied. The presence of a Y-structure intermediate demonstrated that the trypanosome Leptomonas collosoma SL RNA was able to participate in the first step of trans splicing with an 8-nt insertion in loop II (19). While the SL RNA from the trypanosome Leptomonas seymouri requires the stem II structure, the primary sequence of the stem can be changed (32). The stem II primary sequence appears to be required for trans splicing in the trypanosome Leishmania tarentolae (49), in the nematode Ascaris lumbricoides in vitro (11, 23), and now in SL2 RNA in C. elegans. Based on the importance of this structure in trans splicing, it would be somewhat surprising if the primary sequence of stem II of SL2 RNA had not been conserved between Dolichorhabditis and C. elegans (13). Indeed, close inspection of the sequences of SL2 RNA in these two nematode genera reveals conservation of the central portion of the second stem, surrounding the bulged nucleotide.
The primary sequence of the third loop is essential for SL2 trans splicing.
A requirement for stem-loop III in a trans-splicing RNA has been demonstrated only once; trans splicing of the L. tarentolae SL RNA is dependent on the formation of stem III, although the primary sequence is unimportant (49). The effect of mutations to L. tarentolae SL RNA stem-loop III could be the result of incorrect 3′ end formation, however (48). The specificity and efficiency of in vitro 3′ end formation of the A. lumbricoides SL RNA also requires stem-loop III sequences (21), although chemical modification interference analysis of the A. lumbricoides SL RNA failed to identify specific positions in stem III that are required for in vitro trans splicing (23). Mutation of Leptomonas collosoma SL RNA by insertion of 8 nt into loop III was tolerated (20). Amazingly, deletion of the entire stem III structure from the trypanosome Leptomonas seymouri SL RNA did not prevent trans splicing (32). Here we present the first evidence that the primary sequence of stem-loop III of a spliced leader RNA is required for trans splicing. Northern analysis failed to detect any aberrantly sized mutated SL2 RNAs (data not shown), consistent with ability of these mutated RNAs to form correct 3′ ends. Replacing the top of SL2 stem-loop III with the sequence from the top of SL1 stem-loop III resulted in a nonfunctional SL2 RNA, even though assembly of a core snRNP by association with Sm proteins was normal. The discovery that even a single base substitution to loop III of SL2 RNA prevented trans splicing suggests an important role for this region. While attempts to determine whether this element is sufficient for conferring SL2 trans-splicing specificity did not show that it is, the ability of SL1 RNA to function with large changes to the same region (Fig. 4) suggests a key difference between SL1 and SL2 RNAs. The stem-loop III region of SL1 RNA was not sensitive to mutation, while stem-loop III of SL2 RNA was very sensitive to mutation.
Transcription of the SL2 RNA requires an intact PSE.
PSEs are located upstream of U snRNA coding regions, specifying the site of transcription initiation, and are essential for U snRNA transcription (reviewed in reference 42). The location of an element upstream of the C. elegans U snRNA genes as well as its conservation suggested that this sequence might be a PSE (53). However, the primary sequence did not resemble the PSE described for other organisms. The results presented here demonstrate that the conserved sequence element, beginning approximately 65 bp upstream of the snRNA genes and previously predicted to be the C. elegans PSE (53), is in fact required for SL2 RNA expression in vivo. It is interesting that the SL1 RNA genes in C. elegans have only a weak match to the PSE consensus (53) and also have a promoter element located within their SLs (56). It is possible that the presence of a promoter element within the spliced leader sequence of SL1 RNA genes alleviates the requirement for a PSE.
Conclusion.
We conclude that the sequence of the entire SL2 exon is dispensable for trans splicing, whereas intron stem-loops II and III are essential. trans-splicing of SL1 RNAs with mutations in stem-loop III suggest a key difference between SL1 and SL2 trans splicing. This region of the RNA may be involved in conferring SL2 trans-splicing specificity. Finally, proper in vivo expression of SL2 RNA requires an intact PSE.
ACKNOWLEDGMENTS
pSL2+, pASL2, and pSL15S were the kind gifts of Xin-Yun Huang and David Hirsh, Atul M. Deshpande, and Kimberly Ferguson and Joel Rothman, respectively. The human autoimmune anti-Sm serum and Y12 monoclonal antibodies were the kind gifts of Scott Kuersten and Iain Mattaj. We are grateful to Atul M. Deshpande, Diego A. R. Zorio, Devin Leake, Izzy Perez, David Caprio, and Margaret MacMorris for helpful comments on the manuscript. We thank Devin Leake for additional assistance with the figures.
This work was supported by grant GM42432 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Blumental T. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 2.Blumenthal T, Stewart K. RNA processing and gene structure. In: Riddle D, Blumenthal T, Meyer B, Priess J, editors. C. elegans II. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 117–145. [PubMed] [Google Scholar]
- 3.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruzik J P, Van Doren K, Hirsh D, Steitz J A. Trans-splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature. 1988;335:559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- 5.Bruzik J P, Steitz J A. Spliced leader RNA sequences can substitute for the essential 5′ end of U1 RNA during splicing in a mammalian in vitro system. Cell. 1990;62:889–899. doi: 10.1016/0092-8674(90)90264-f. [DOI] [PubMed] [Google Scholar]
- 6.Conrad R, Thomas J, Spieth J, Blumenthal T. Insertion of part of an intron into the 5′ untranslated region of Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol Cell Biol. 1991;11:1921–1926. doi: 10.1128/mcb.11.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrad R, Liou R F, Blumenthal T. Conversion of a trans-spliced C. elegans gene into a conventional gene by introduction of a splice donor site. EMBO J. 1993;12:1249–1255. doi: 10.1002/j.1460-2075.1993.tb05766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad R, Lea K, Blumenthal T. SL1 trans-splicing specified by AU-rich synthetic RNA inserted at the 5′ end of Caenorhabditis elegans pre-mRNA. RNA. 1995;1:164–170. [PMC free article] [PubMed] [Google Scholar]
- 9.Coolidge C J, Patton J G. Run-around PCR: a novel way to create duplications using polymerase chain reaction. BioTechniques. 1995;18:763–764. [PubMed] [Google Scholar]
- 10.De Robertis E M, Lienhard S, Parisot R. Intracellular transport of microinjected 5S and small nuclear RNAs. Nature. 1982;295:572–577. doi: 10.1038/295572a0. [DOI] [PubMed] [Google Scholar]
- 11.Denker J A, Maroney P A, Yu Y-T, Kanost R A, Nilsen T W. Multiple requirements for nematode spliced leader RNP function in trans-splicing. RNA. 1996;2:746–755. [PMC free article] [PubMed] [Google Scholar]
- 12.Dungan J M, Watkins K P, Agabian N. Evidence for the presence of a small U5-like RNA in active trans-spliceosomes of Trypanosoma brucei. EMBO J. 1996;15:4016–4029. [PMC free article] [PubMed] [Google Scholar]
- 13.Evans D, Zorio D, MacMorris M, Winter C E, Lea K, Blumenthal T. Operons and SL2 trans-splicing exist in nematodes outside the genus Caenorhabditis. Proc Natl Acad Sci USA. 1997;94:9751–9756. doi: 10.1073/pnas.94.18.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson K C, Heid P J, Rothman J H. The SL1 trans-spliced leader RNA performs an essential embryonic function in Caenorhabditis elegans that can also be supplied by SL2 RNA. Genes Dev. 1996;10:1543–1556. doi: 10.1101/gad.10.12.1543. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson K C, Rothman J H. Alterations in the conserved SL1 trans-spliced leader of Caenorhabditis elegans demonstrate flexibility in length and sequence requirements in vivo. Mol Cell Biol. 1999;19:1892–1900. doi: 10.1128/mcb.19.3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fire A. Integrative transformation of Caenorhabditis elegans. EMBO J. 1986;5:2673–2680. doi: 10.1002/j.1460-2075.1986.tb04550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fire A, Waterston R H. Proper expression of myosin genes in transgenic nematodes. EMBO J. 1989;8:3419–3428. doi: 10.1002/j.1460-2075.1989.tb08506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritz A, Parisot R, Newmeyer D, De Robertis E M. Small nuclear U-ribonucleoproteins in Xenopus laevis development. J Mol Biol. 1984;178:273–285. doi: 10.1016/0022-2836(84)90144-x. [DOI] [PubMed] [Google Scholar]
- 19.Goncharov I, Xu Y-X, Zimmer Y, Sherman K, Michaeli S. Structure-function analysis of the trypanosomatid spliced leader RNA. Nucleic Acids Res. 1998;26:2200–2207. doi: 10.1093/nar/26.9.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goncharov I, Palfi Z, Bindereif A, Michaeli S. Purification of the spliced leader ribonucleoprotein particle from Leptomonas collosoma revealed the existence of an Sm protein in trypanosomes. J Biol Chem. 1999;274:12217–12221. doi: 10.1074/jbc.274.18.12217. [DOI] [PubMed] [Google Scholar]
- 21.Hannon G J, Maroney P A, Ayers D G, Shambaugh J D, Nilsen T W. Transcription of a nematode trans-spliced leader RNA requires internal elements for both initiation and 3′ end-formation. EMBO J. 1990;9:1915–1921. doi: 10.1002/j.1460-2075.1990.tb08318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannon G J, Maroney P A, Nilsen T W. U small nuclear ribonucleoprotein requirements for nematode cis- and trans-splicing in vitro. J Biol Chem. 1991;266:22792–22795. [PubMed] [Google Scholar]
- 23.Hannon G J, Maroney P A, Yu Y T, Hannon G E, Nilsen T W. Interaction of U6 snRNA with a sequence required for function of the nematode SL RNA in trans-splicing. Science. 1992;258:1775–1780. doi: 10.1126/science.1465612. [DOI] [PubMed] [Google Scholar]
- 24.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X-Y, Hirsh D. A second trans-spliced RNA leader sequence in the nematode Caenorhabditis elegans. Proc Natl Acad Sci USA. 1989;86:8640–8644. doi: 10.1073/pnas.86.22.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson P J, Looter J M, Borst P. Inactivation of transcription by UV irradiation of T. brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell. 1987;51:273–281. doi: 10.1016/0092-8674(87)90154-1. [DOI] [PubMed] [Google Scholar]
- 27.Kari C, Seydoux G, White Harrison S, Fire A, Herman R. Gamma ray-induced integration of extrachromosomal arrays. In: Epstein H F, Shakes D C, editors. Caenorhabditis elegans: modern biological analysis of an organism. San Diego, Calif: Academic Press; 1995. pp. 467–468. [Google Scholar]
- 28.Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuersten S, Lea K, MacMorris M, Spieth J, Blumenthal T. Relationship between 3′ end formation and SL2-specific trans-splicing in polycistronic Caenorhabditis elegans pre-mRNA processing. RNA. 1997;3:269–278. [PMC free article] [PubMed] [Google Scholar]
- 30.Lerner M R, Steitz J A. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1979;76:5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lerner E A, Lerner M R, Janeway C A, Jr, Steitz J A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucke S, Xu G-L, Palfi Z, Cross M, Bellofatto V, Bindereif A. Spliced leader RNA of trypanosomes: in vivo mutational analysis reveals extensive and distinct requirements for trans-splicing and cap4 formation. EMBO J. 1996;15:4380–4391. [PMC free article] [PubMed] [Google Scholar]
- 33.Maroney P A, Hannon G J, Denker J A, Nilsen T W. The nematode spliced leader RNA participates in trans-splicing as an Sm snRNP. EMBO J. 1990;9:3667–3673. doi: 10.1002/j.1460-2075.1990.tb07578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maroney P A, Hannon G J, Shambaugh J D, Nilsen T W. Intramolecular base pairing between the nematode spliced leader and its 5′ splice site is not essential for trans-splicing in vitro. EMBO J. 1991;10:3869–3875. doi: 10.1002/j.1460-2075.1991.tb04956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maroney P A, Denker J A, Darzynkiewicz E, Laneve R, Nilsen T W. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA. 1995;1:714–723. [PMC free article] [PubMed] [Google Scholar]
- 36.Maroney P A, Yu Y-T, Jankowska M, Nilsen T W. Direct analysis of nematode cis- and trans-spliceosomes: a functional role for U5 snRNA in spliced leader addition trans-splicing and the identification of novel Sm snRNPs. RNA. 1996;2:735–745. [PMC free article] [PubMed] [Google Scholar]
- 37.Mattaj I W, De Robertis E M. Nuclear segregation of U2 snRNA requires binding of specific snRNP proteins. Cell. 1985;40:111–118. doi: 10.1016/0092-8674(85)90314-9. [DOI] [PubMed] [Google Scholar]
- 38.Matter L, Schopfer K, Wilhelm J A, Nyffenegger T, Parisot R F, De Robertis E M. Molecular characterization of ribonucleoprotein antigens bound by antinuclear antibodies: a diagnostic evaluation. Arthritis Rheum. 1982;25:1278–1283. doi: 10.1002/art.1780251102. [DOI] [PubMed] [Google Scholar]
- 39.Mello C C, Kramer J M, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhich M L, Boothroyd J C. Polycistronic transcripts in trypanosomes and their accumulation during heat shock: evidence for a precursor role in mRNA synthesis. Mol Cell Biol. 1988;8:3837–3846. doi: 10.1128/mcb.8.9.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy W J, Watkins K P, Agabian N. Identification of a novel Y branch structure as an intermediate in trypanosome mRNA processing: evidence for trans-splicing. Cell. 1986;47:517–525. doi: 10.1016/0092-8674(86)90616-1. [DOI] [PubMed] [Google Scholar]
- 42.Parry H D, Scherly D, Mattai I W. 'Snurpogenesis': the transcription and assembly of U snRNP components. Trends Biochem Sci. 1989;14:15–19. [Google Scholar]
- 43.Rajkovic A, Davis R E, Simonsen J N, Rottman R M. A spliced leader is present on a subset of mRNAs from the human parasite Schistosoma mansoni. Proc Natl Acad Sci USA. 1990;87:8879–8883. doi: 10.1073/pnas.87.22.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray C, Abbott A G, Hussey R S. Trans-splicing of a Meloidogyne incognita mRNA encoding a putative esophageal gland protein. Mol Biochem Parasitol. 1994;68:93–101. doi: 10.1016/0166-6851(94)00153-7. [DOI] [PubMed] [Google Scholar]
- 45.Ross L H, Freedman J H, Rubin C S. Structure and expression of novel spliced leader RNA genes in Caenorhabditis elegans. J Biol Chem. 1995;270:22066–22075. doi: 10.1074/jbc.270.37.22066. [DOI] [PubMed] [Google Scholar]
- 46.Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T. Operons in C. elegans: polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- 47.Sturm N R, Fleischmann J, Campbell D A. Efficient trans-splicing of mutated spliced leader exons in Leishmania tarentolae. J Biol Chem. 1998;273:18689–18692. doi: 10.1074/jbc.273.30.18689. [DOI] [PubMed] [Google Scholar]
- 48.Sturm N R, Yu M C, Campbell D A. Transcription termination and 3′-end processing of the spliced leader RNA in kinetoplastids. Mol Cell Biol. 1999;19:1595–1604. doi: 10.1128/mcb.19.2.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sturm N R, Campbell D A. The role of intron structures in trans-splicing and cap 4 formation for the Leishmania spliced leader RNA. J Biol Chem. 1999;274:19361–19367. doi: 10.1074/jbc.274.27.19361. [DOI] [PubMed] [Google Scholar]
- 50.Sutton R E, Boothroyd J C. Evidence for trans-splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tessier L-H, Keller M, Chan R, Fournier R, Weil J H, Imbault P. Short leader sequences may be transferred from small RNAs to pre-mature mRNAs by trans-splicing in Euglena. EMBO J. 1991;10:2621–2625. doi: 10.1002/j.1460-2075.1991.tb07804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas J D, Conrad R C, Blumenthal T. The C. elegans trans-spliced leader RNA is to Sm and has a trimethylguanosine cap. Cell. 1988;54:533–539. doi: 10.1016/0092-8674(88)90075-x. [DOI] [PubMed] [Google Scholar]
- 53.Thomas J, Lea K, Zucker-Aprison E, Blumenthal T. The spliceosomal snRNAs of Caenorhabditis elegans. Nucleic Acids Res. 1990;18:2633–2642. doi: 10.1093/nar/18.9.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tschudi C, Ullu E. Destruction of U2, U4, or U6 small nuclear RNA blocks trans-splicing in trypanosome cells. Cell. 1990;61:459–466. doi: 10.1016/0092-8674(90)90527-l. [DOI] [PubMed] [Google Scholar]
- 55.Van Doren K, Hirsh D. Trans-spliced leader RNA exists as small nuclear ribonucleoprotein particles in Caenorhabditis elegans. Nature. 1988;335:556–559. doi: 10.1038/335556a0. [DOI] [PubMed] [Google Scholar]
- 56.Xie H, Hirsh D. In vivo function of mutated spliced leader RNAs in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:4235–4240. doi: 10.1073/pnas.95.8.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zorio D A R, Cheng N N, Blumenthal T, Spieth J. Operons as a common form of chromosomal organization in C. elegans. Nature. 1994;372:270–272. doi: 10.1038/372270a0. [DOI] [PubMed] [Google Scholar]