Abstract
Spinal ependymomas are the most common spinal cord tumors in adults, but their intratumoral cellular heterogeneity has been less studied, and how spinal microglia are involved in tumor progression is still unknown. Here, our single-cell RNA-sequencing analyses of three spinal ependymoma subtypes dissect the microenvironmental landscape of spinal ependymomas and reveal tumor-associated macrophage (TAM) subsets with distinct functional phenotypes. CCL2+ TAMs are related to the immune response and exhibit a high capacity for apoptosis, while CD44+ TAMs are associated with tumor angiogenesis. By combining these results with those of single-cell ATAC-sequencing data analysis, we reveal that TEAD1 and EGR3 play roles in regulating the functional diversity of TAMs. We further identify diverse characteristics of both malignant cells and TAMs that might underlie the different malignant degrees of each subtype. Finally, assessment of cell-cell interactions reveal that stromal cells act as extracellular factors that mediate TAM diversity. Overall, our results reveal dual functions of TAMs in tumor progression, providing valuable insights for TAM-targeting immunotherapy.
Subject terms: CNS cancer, Cancer in the nervous system
The intratumoural heterogeneity of spinal ependymomas and the role of microglia in tumour progression remain underexplored. Here, the authors perform single-cell RNA- and ATAC-sequencing data analysis of three subtypes and reveal tumour-associated macrophage subsets with distinct functional phenotypes.
Introduction
Ependymal tumors are neuroepithelial malignancies located in both the brain and spinal cord1, and spinal ependymomas are the most common spinal cord tumors in adults, comprising ~60% of all intramedullary neoplasms2. Surgical resection remains the mainstay of treatment for this disease. Due to the invasive growth of tumor cells, it is difficult to completely remove tumors without obvious tumor margins, and aggressive surgical removal is accompanied by potentially severe sensory and motor dysfunction3,4. For patients with subtotal resection who do not receive adjuvant radiotherapy, the recurrence rate of tumors is up to 50–70%4. Therefore, there is an urgent need to identify novel therapeutic targets for spinal ependymomas.
In the past decade, studies focused on genomics, transcriptomic, and methylomics have greatly expanded our understanding of the biology underlying human ependymomas. Extensive genomic and transcriptomic analyses uncovered ependymoma oncogenes and tumor suppressor genes5 and revealed neural stem cells and glial cells as putative cells of origin6,7. The molecular events involving chromosome 22 and mutation of the NF2 gene have long been known as hallmark genetic aberrations of spinal ependymomas8. Genome-wide DNA methylation profiling of ependymomas identified nine molecular subgroups, three in each anatomical compartment of the central nervous system (CNS), including the spinal, posterior fossa, and supratentorial areas, which could help the precise diagnosis and risk stratification of ependymoma patients in the clinic1. Within the spinal ependymoma histopathological subtypes, subependymoma (SE) and ependymoma (EPN) are considered as CNS WHO grade I and II with good prognosis, while anaplastic ependymoma (AEP) are considered as grade III showing aggressive identity9. The previous studies were informed by bulk tumor samples and largely focused on the developmental origins and candidate driver genes of ependymomas.
Single-cell RNA-seq (scRNA-seq) and single-cell ATAC-seq (scATAC-seq) technologies have been successfully applied to characterize tumor heterogeneity at single-cell resolution10 and have revealed the cellular diversity of the tumor microenvironment (TME) in multiple cancer types11–18. Recent studies have begun to reveal the cellular heterogeneity of malignant cells in ependymomas by using scRNA-seq19,20, with samples mainly collected from childhood brain ependymomas. However, the cellular diversity in spinal ependymomas, especially the complexity of stromal and immune cells, remains less studied. Recent advances in cancer treatments targeting tumor-infiltrating immune cells have emerged21. Microglia are the resident macrophages in CNS parenchyma and have emerged as a promising cellular target for tumor immunotherapy considering their roles in neuroinflammation22. We speculate that a deep understanding of tumor-associated macrophages would reveal potential therapeutic targets for ependymomas.
In this work, we perform scRNA-seq, scATAC-seq, and bulk epigenetic ChIP-seq analyses of cells isolated from patients diagnosed with spinal cord ependymomas, determining the diversity of malignant cells and immune cells in these spinal cord ependymomas. Two distinct TAM subsets with different ontogenies and dual functional phenotypes could play crucial roles in tumor growth and invasiveness, which may inform TAM-targeting immunotherapy strategies in human ependymomas and other cancers.
Results
Intratumoral cell types in spinal ependymomas revealed by scRNA-seq and scATAC-seq
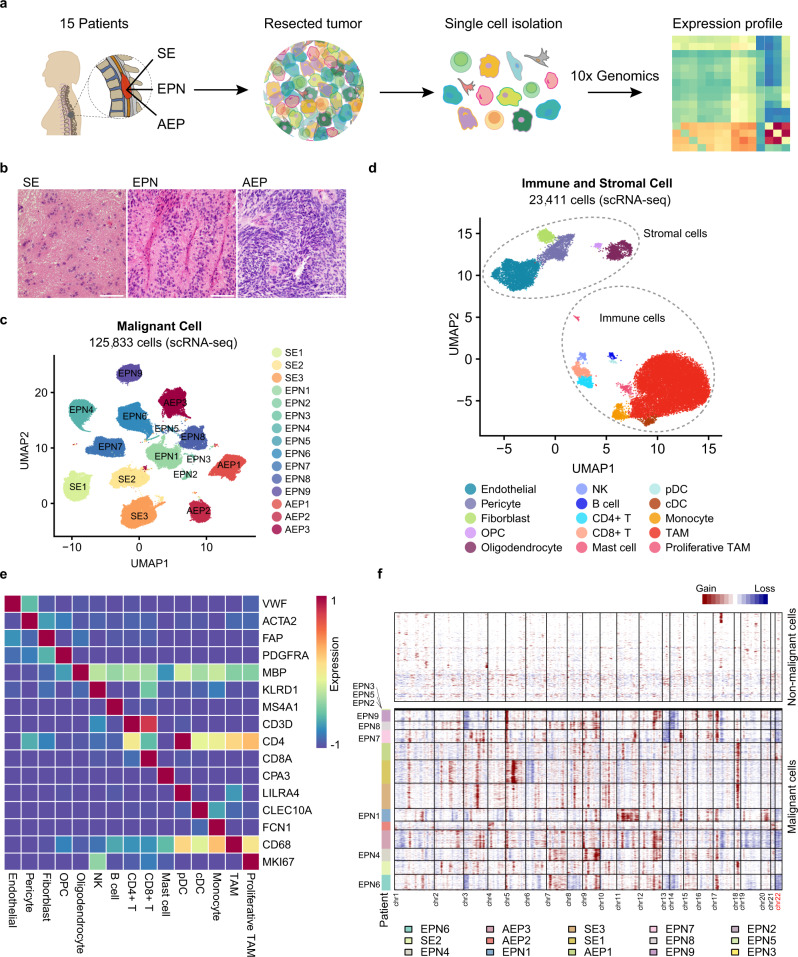
To comprehensively catalog the cellular heterogeneity of spinal ependymomas, we performed droplet-based scRNA-seq (10× Genomics) on cells isolated from tumor tissues of 15 treatment-naive patients diagnosed with different subtypes of spinal ependymoma, including subependymoma (SE, three patients), ependymoma (EPN, 9 patients), and anaplastic ependymoma (AEP, three patients) (Fig. 1a, b and Supplementary Data 1). Notably, SE patients exhibited the lowest degree of malignancy among the three subtypes (Fig. 1b). With strict quality control and filtration (“Methods”), a total of 149,244 cells with a median of 6334 unique molecular identifiers (UMIs) and 2757 genes were retained for downstream analysis (Supplementary Fig. 1a). We first distinguished nonmalignant cells (using canonical gene markers of stromal and immune cells) from malignant cells in the first round of graph-based unsupervised clustering (Supplementary Fig. 1a, b) and found that malignant cells clustered according to tumor origin, while nonmalignant cells clustered by cell type (Fig. 1c and Supplementary Fig. 1c), consistent with previous studies23–25. We then separated nonmalignant cells from malignant cells and identified 14 cell types according to the expression of canonical gene markers, including markers of CD8+ T cells, CD4+ T cells, B cells, mast cells, conventional dendritic cells (cDCs), plasmacytoid dendritic cells (pDCs), monocytes, TAMs, fibroblasts, endothelial cells, and pericytes (Fig. 1d, e, Supplementary Fig. 1d, and Supplementary Data 2). To validate cells classified as malignant, we further inferred large-scale copy-number variations (CNVs) based on scRNA-seq expression profiles with stromal and immune cells as controls23–27 and detected large-scale CNVs in malignant cells from each patient but not in nonmalignant cells, including the reported 22q loss8 in all nine EPN patients (Fig. 1f), which confirmed the rationality of the unsupervised clustering. We then compared the cell-type percentages in each patient and observed that the abundance of malignant cells and the composition of stromal and immune cells varied across patients (Supplementary Fig. 1e, f), suggesting a considerable degree of tumor heterogeneity. These results provide a basic description of the TME cellular composition of human ependymomas.
Fig. 1. Intratumoral cell types in spinal ependymomas revealed by scRNA-seq.
a Scheme of the overall study design. scRNA-seq (10× Genomics) was applied to cells isolated from three subtypes of spinal ependymomas. b Hematoxylin and eosin (HE) staining of tumors from different cancer subtypes. The scale bar represents 30 μm. Images shown are representatives of more than three samples from three independent experiments. c Uniform Manifold Approximation and Projection (UMAP) plot showing the patient distribution of malignant cells, without donor effect correction. d UMAP plot of stromal and immune cell types from all patients (SE1-3, AEP1-3, EPN1-9), donor effect corrected by BBKNN. e Heatmap showing the expression of canonical gene markers of stromal and immune cell types. f Inference of copy-number variations (CNVs) from scRNA-seq. Each row corresponds to a cell. The top panel represents nonmalignant cells, and the bottom panel represents malignant cells, ordered by the patient. See also Supplementary Figs. 1 and 2 and Supplementary Data 1 and 2.
We also generated scATAC-seq data from cells isolated from tumor tissues of two AEP patients to capture the chromatin regulatory landscape that governs transcription dynamics in spinal ependymomas with the highest degree of malignancy. In total, 2854 cells with 635,774 reproducible peaks of chromatin accessibility passed strict quality control and were used for the downstream analysis (Supplementary Fig. 2a). We identified five distinct clusters based on unsupervised clustering and annotated their cell types. Two of those clusters exhibited specifically accessible peaks neighboring OPC markers (OLIG1 and OLIG2) and TAM markers (PTPRC and CD68) (Supplementary Fig. 2b, c). The remaining clusters were regarded as malignant cells that showed high patient specificity and were annotated as astroependymal-like cells, ependymal-like cells, and neuronal-like cells (Supplementary Fig. 2b, c).
Two TAM subsets show distinct functional phenotypes in ependymomas
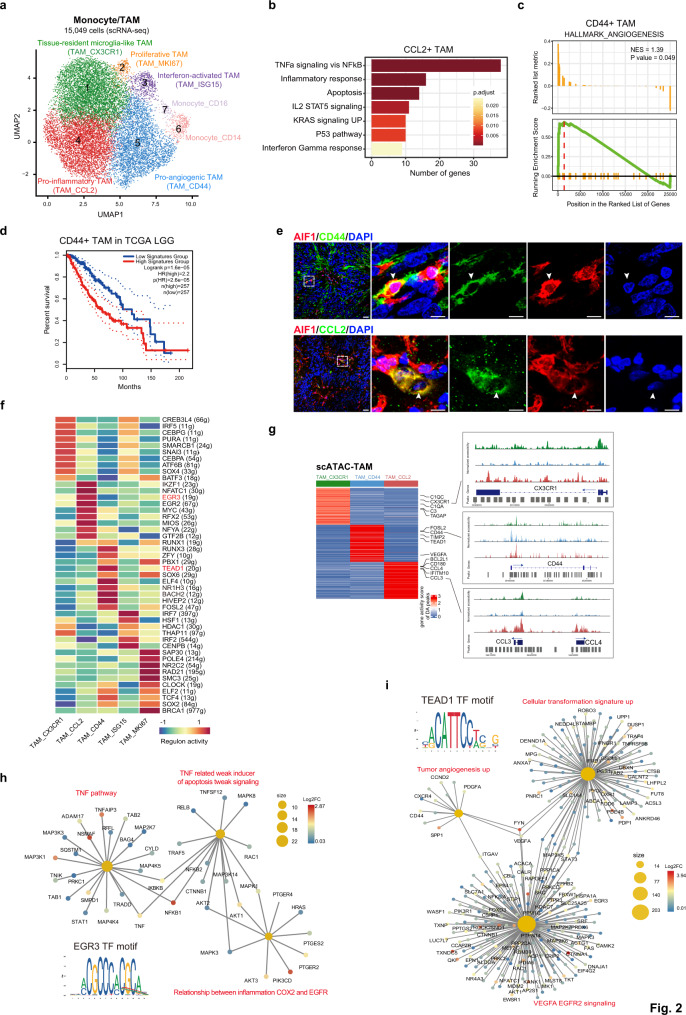
We next focused on the heterogeneity of monocytes and TAMs and performed unsupervised graph-based clustering of their transcriptomes, which resulted in seven subclusters with unique signature genes, including two monocyte subsets (classical CD14+ and nonclassical CD16+ monocytes) and five TAM subsets (Fig. 2a, Supplementary Fig. 3a, and Supplementary Data 3). The tissue-resident microglia-like TAMs in cluster 1 (TAM_CX3CR1) were characterized by high expression of the complement gene C1QC and the microglia homeostatic gene CX3CR1, similar to the reported homeostatic microglia subset in the brain28. The proliferative TAMs in cluster 2 (TAM_MKI67) were characterized by cell cycle gene expression, including expression of MKI67 and TOP2A. The interferon-activated TAMs in cluster 3 (TAM_ISG15), which constituted a small population of TAMs, expressed high levels of interferon response genes, such as ISG15 and IFIT3. The pro-inflammatory TAMs in cluster 4 (TAM_CCL2) showed high expression of the pro-inflammatory mediator IL1B and CNS inflammation-associated chemokines29 (CCL3, CCL4, and CCL2) (Supplementary Fig. 3b), resembling the pre-activated subset reported in normal brain samples from both humans and mice28,30,31. We noticed that the CCL2+ pro-inflammatory TAMs also exhibited high expression of apoptosis-related genes, such as PMAIP1 and NEDD9 (Supplementary Fig. 3b). Enrichment analysis further confirmed that CCL2+ TAMs showed high activity in both inflammatory response and apoptosis pathways (Fig. 2b and Supplementary Data 4). The remaining TAMs, falling into the pro-angiogenic TAM cluster (TAM_CD44, cluster 5), were characterized by high expression of the angiogenesis-associated genes CD4432 and VEGFA33 (Supplementary Fig. 3b). Analysis of the hallmark angiogenesis pathway in CD44+ TAMs using gene set enrichment analysis (GSEA) revealed a significant enrichment of tumor angiogenesis (Fig. 2c), suggesting a pro-tumorigenic role in ependymomas. We then investigated the relationship of CD44+ TAMs with patient prognosis and found that higher expression of signature genes of CD44+ TAMs was associated with a worse clinical outcome in similar cancer types from The Cancer Genome Atlas (TCGA), including brain lower-grade glioma (LGG) and glioblastoma (GBM) (Fig. 2d and Supplementary Fig. 3c). Using immunofluorescence (IF) staining of tumor sections from EPN and AEP patients, we confirmed the presence of CCL2+ TAMs and CD44+ TAMs as subgroups (Fig. 2e and Supplementary Fig. 3e) and observed higher proportions of these two TAM subsets in AEP (Supplementary Fig. 3d). Notably, the functional potentiality of each TAM subcluster was implied by their signature genes and additional validation experiments are required to fully elucidate their biological characteristics in the future.
Fig. 2. Two TAM subsets show distinct functional phenotypes in ependymomas.
a UMAP plot of seven subclusters of monocytes and TAMs. b Bar plot of enriched hallmark pathways for genes upregulated in CCL2+ TAMs. P values were calculated by using enrichr function from R package clusterProfiler with hypergeometric test statistical analyses. Source data are provided as a Source Data file. c Enrichment plot of the hallmark angiogenesis pathway in CD44+ TAMs. P values were calculated by using GESA function from R package clusterProfiler. d Kaplan–Meier plot showing worse clinical outcome for high expression of CD44+ TAMs signature genes in LGG patients from TCGA. +, censored observations. P values were calculated by using both the log-rank test and Cox proportional hazards model. e Representative example of an EPN tumor stained by IF. The upper panel image indicates AIF1+CD44+ TAMs (the scale bar represents 30 μm). The dashed boxes highlight regions shown on the right side and the arrow depicts the CD44+ TAMs in fluorescent images (the scale bar represents 100 μm). The bottom panel image indicates AIF1+CCL2+ TAMs (the scale bar represents 30 μm). The dashed boxes highlight regions shown on the right side and the arrow depicts the CCL2+ TAMs in fluorescent images (the scale bar represents 100 μm). Images shown are representatives of three samples from three independent experiments. f Heatmap showing TF activity for each TAM subsets. The row name showed the regulon gene sets name and gene number is written in the round brackets. The red color marks the regulon of interest. g Heatmap showing cluster-specific ATAC-seq peaks (left). Browser tracks showing ATAC-seq signals for selected marker genes (right). h Network plot of enriched curated gene sets for genes regulated by EGR3 in CCL2+ TAM subset. Nodes for genes were colored by log2FC, and the sizes of nodes for enriched pathways were correlated with the number of genes. i Network plot of enriched curated gene sets for genes regulated by TEAD1 in CD44+ TAM subset. Nodes for genes were colored by log2FC, and the sizes of nodes for enriched pathways were correlated with the number of genes. See also Supplementary Figs. 2–3 and Supplementary Data 3–5.
We then examined the regulatory networks that underlie each TAM subset. Using SCENIC34, we identified specific transcription factor (TF) regulons in each TAM subset (Fig. 2f). We observed that the IKZF1 and EGR3 regulons, which are related to regulating inflammatory genes35,36, were highly activated in CCL2+ TAMs (Fig. 2f). The activated TEAD1 regulon was specifically found in CD44+ TAMs (Fig. 2f), and TEAD1 has been reported to directly promote human glioblastoma cell migration37 and regulate pro-angiogenic activity through YAP1-TEAD1-PGC1α signaling in endothelial cells38. To complement the regulons predicted from the scRNA-seq data, we further analyzed the scATAC-seq data of TAMs. Using the TAM subpopulations derived from the scRNA-seq data as a reference, we identified three TAM subpopulations (TAM_CX3CR1, TAM_CCL2, TAM_CD44) with the scATAC-seq data, and each subpopulation exhibited specific differentially accessible (DA) sites, which could affect marker genes of TAM subpopulations (Fig. 2g, Supplementary Fig. 2d, e, and Supplementary Data 5). We reasoned that due to the limited number of captured cells, other TAM subclusters were missed in our scATAC-seq data. We then applied motif enrichment analysis to cluster-specific accessible peaks to determine potentially key TFs in each TAM subcluster. Strikingly, we observed that the binding motif of TEAD1 was enriched in CD44+ TAMs and that one of its binding sites was located close to the transcription start site (TSS) of VEGFA (Supplementary Fig. 2f). Furthermore, TEAD1 was found to be the upstream regulator of several signaling pathways, including tumor angiogenesis, the cell transformation signature, and VEGFA-EGFR2 signaling (Fig. 2i), indicating its possible role in driving the pro-angiogenic phenotype. For CCL2+ TAMs, we uncovered enrichment of the EGR3 motif, whose target genes were related to the regulation of TNF-related weak inducer of apoptosis (TWEAK) signaling and the TNF pathway (Fig. 2h), suggesting the important role of EGR3 in promoting the antitumor phenotype of CCL2+ TAMs. Overall, our scRNA-seq and scATAC-seq analyses unearthed plausible regulatory mechanisms that shape the functional diversity of TAMs in ependymoma.
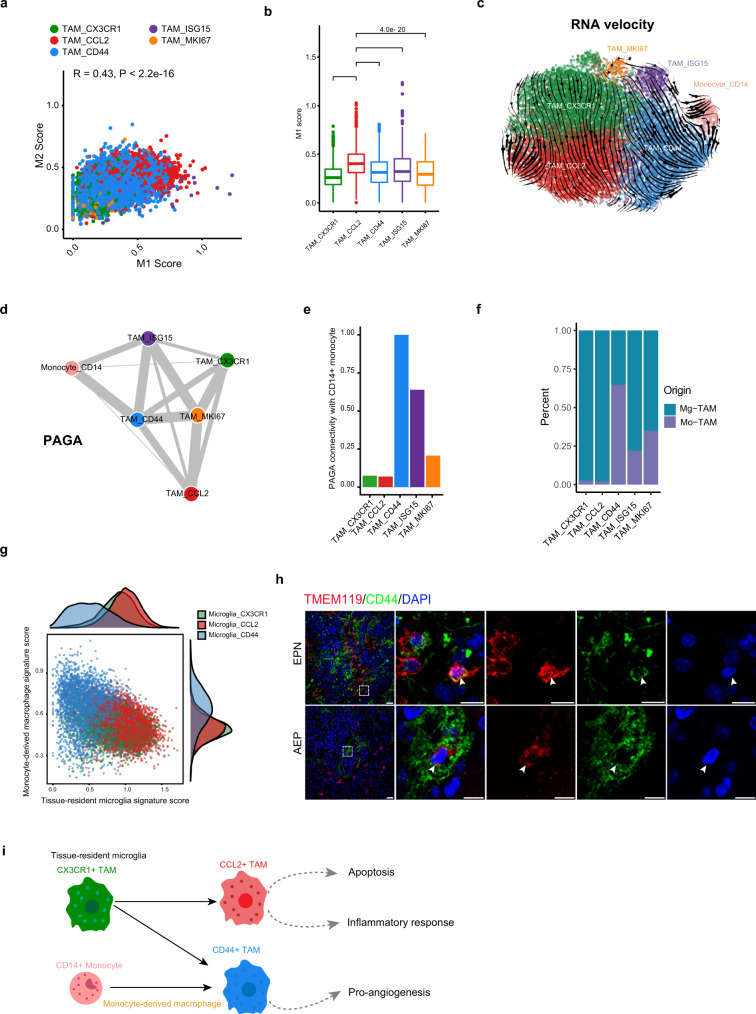
We next examined the expression pattern of the M1 and M2 macrophage signatures in these TAM subsets and observed a coexpression pattern (Fig. 3a). Consistent with previous studies13,17,18,39 and a recent pan-cancer analysis of TAMs40, our results indicated that such a simple macrophage polarization model defined in vitro was also not suitable for TAMs in CNS tumors. We reasoned that the complicated stimuli that coexist within the TME, rather than the one-way stimulus implied by the in vitro model, induce the complex TAM activation status in tumors41–43. Notably, we found that CCL2+ microglia exhibited the highest M1 signature score, suggesting their stronger antitumor capacity (Fig. 3b).
Fig. 3. Developmental trajectory of TAM subsets in spinal ependymomas.
a Scatter plot showing the Pearson correlation between the M1 and M2 signature scores. b Box plot showing the M1 signature score in each TAM subset. P values were calculated by the Wilcoxon test, two-sided comparisons. Multiple hypothesis correction using the Benjamini–Hochberg procedure. n = 15,049 cells. The center line, bounds of box, and whiskers represent mean, 25th to 75th percentile range, and minimum to maximum range in all boxplots. c Steady-state RNA velocity of TAM subsets. d PAGA graph showing the inferred developmental trajectories for TAM subsets. The edge width was correlated with the strength of connectivity between two subclusters. e Bar plot showing PAGA connectivity with CD14+ monocytes. f Bar plot showing the proportion of monocyte origin and tissue-resident microglia origin across each TAM subset (using data reported by Pombo et al. as a reference). Source data are provided as a Source Data file. g Scatter plot showing the scores by average expression of signature genes of tissue-resident microglia versus monocyte-derived macrophages. h Representative examples of tumor section stained by IF. The upper panel image indicates CD44+ TMEM119+ microglia in EPN tumor (the scale bar represents 30 μm). The dashed boxes highlight regions shown on the right side and the arrow depicts the CD44+ microglia in fluorescent images (the scale bar represents 100 μm). The bottom panel image indicates CD44+ TMEM119+ microglia in AEP tumor (the scale bar represents 30 μm). The dashed boxes highlight regions shown on the right side and the arrow depicts the CD44+ microglia in fluorescent images (the scale bar represents 100 μm). Images shown are representatives of three samples from three independent experiments. i Model of the developmental trajectory of monocyte/TAM lineages in spinal ependymomas. See also Supplementary Figs. 3 and 4.
Studies have suggested that TAMs can originate from both tissue-resident macrophages called microglia and tissue-invading monocytes in brain tumors44,45. To explore the ontogenies of TAM subsets in spinal ependymomas, we employed RNA velocity analysis and identified two different origins of CD44+ TAMs (Fig. 3c), including both CD14+ monocytes and CX3CR1+ TAMs. In contrast, our velocity analysis revealed that CCL2+ TAMs originated solely from CX3CR1+ TAMs (Fig. 3c). To complement this prediction, we performed PAGA analysis46. As expected, we observed that CX3CR1+ TAMs and CCL2+ TAMs showed much lower connectivity with CD14+ monocytes (Fig. 3d, e), implying that they were more likely to be TRMs or their derivatives. Moreover, CD44+ TAMs were predicted to have strong connectivity with both CX3CR1+ TAMs and CD14+ monocytes (Fig. 3d). We used SingleR with data from Pombo et al.47 as a reference to predict the origin of TAMs in ependymoma. Consistently, CD44+ TAMs were predicted to have two diverse origins, and CCL2+ TAMs mainly developed from tissue-resident microglia (Fig. 3f). Scoring of cells according to the expression of tissue-resident microglia-specific versus monocyte-derived macrophage-specific genes collected from human brain gliomas48 further supported the diverse ontogenies of the two TAM subsets (Fig. 3g). With immunostaining, we confirmed the presence of CD44+ microglia (also TMEM119+) in EPN and AEP tumor sections, indicating that some CD44+ TAMs were tissue-resident microglia or their derivatives (Fig. 3h). However, fully elucidating the complex trajectories of TAMs in the TME requires additional lineage tracing studies. Collectively, our extensive analyses indicated that CD44+ TAMs might develop from both CD14+ monocytes and CX3CR1+ TRMs, while CCL2+ TAMs might develop from CX3CR1+ TRMs (Fig. 3i).
For a comprehensive description of TAMs in ependymoma, we compared TAMs in our study with microglia in the normal brain49 and TAMs in glioblastoma (GBM)47,50 and IDH-mutant high-grade gliomas (HGG)51. We noticed that microglia from the normal brain showed high expression of CCL2 and CCL3, resembling tissue-resident CCL2+ TAMs in ependymoma (Supplementary Fig. 3f, g). For TAMs from other neurological tumors, we observed that the CCL2+ TAM cluster was present in both GBM and HGG, while the CD44+ TAM cluster was only present in GBM (Supplementary Fig. 4a, b, d). Furthermore, integrated analysis of TAMs from GBM and spinal ependymoma revealed that the CD44+ TAMs from the two tumors were closely related to each other, but they did not overlap in the UMAP plot, indicating that they had their own characteristics in each tumor (Supplementary Fig. 4c, e). Taken together, our analysis results indicated that CD44+ TAMs were not specific to EPNs and could be present in other but not all neurological tumors, which may be related to the diverse tumor microenvironments of different cancer types.
Complex expression heterogeneity of malignant cells in each ependymoma subtype
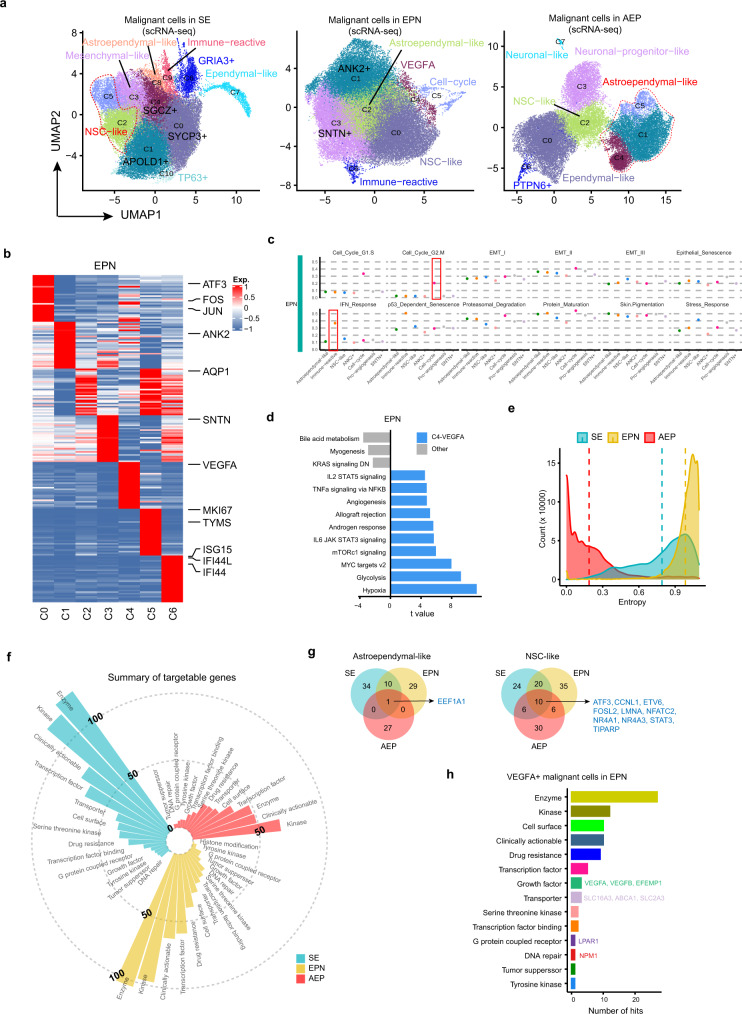
To study potential commonalities and correct for interindividual variations, we applied Harmony alignment52 to malignant cells in each cancer subtype separately. We identified multiple subclusters in each cancer subtype and assigned identities according to their signature genes (Fig. 4a, b, Supplementary Fig. 5a–c, and Supplementary Data 6). Using EPN as an example, our unsupervised clustering gave rise to seven subclusters. The first cluster, C0, was characterized by high expression of ATF3, JUN, and FOS and was termed NSC-like (neural stem cell-like) according to a previous study19. The C2 cluster exhibited high expression of AQP1, a marker of astrocytes, and was named astroependymal-like. The C5 cluster was characterized by specific expression of cell cycle genes, such as MKI67 and TYMS, representing cells undergoing the cell cycle. The C6 cluster, termed immune-reactive, expressed high levels of interferon response genes, including ISG15, suggesting its potential roles in the immune response. The remaining subclusters were named according to their differentially expressed genes. Among the three subtypes, we identified both common (NSC-like and astroependymal-like) and cancer-specific subclusters, such as the immune-reactive subcluster in SE and EPN and the VEGFA+ subcluster in EPN (Fig. 4a). By evaluating the expression patterns of the 12 reported generic tumor cell programs53 across our malignant cells, we observed that the Cell_Cycle-related and IFN_Response programs were highly expressed in specific malignant subsets, but other programs were less different among malignant subsets (Fig. 4c and Supplementary Fig. 5d). Our analysis suggests that the generic tumor cell programs affected some of the malignant cells in our spinal ependymoma dataset.
Fig. 4. Heterogeneity of malignant cells in each ependymoma cancer subtype.
a UMAP plots showing subclusters of malignant cells in each cancer subtype, donor effect corrected by Harmony. b Heatmap showing signature genes for each malignant cell subcluster in EPN. Selected genes were labeled on the right side. c Scatter plots showing average expression of 12 generic tumor cell programs in each malignant subset in EPN. The boxes highlight programs which were expressed in specific subsets. d Bar plot showing different pathways enriched in VEGFA+ C4 and other clusters from EPN scored per cell by gene set variation analysis (GSVA). t values were calculated with limma regression. e Density plot showing the entropy of patient distribution for malignant cells in each cancer subtype. f Circular bar plot showing the number of targetable genes from different categories in each cancer subtype. g Venn plots showing the intersection of targetable genes in astroependymal-like and NSC-like subpopulations across different cancer subtypes. The shared genes between the three cancer subtypes are listed on the right side. h Bar plot showing the number of targetable genes from different categories in VEGFA+ subpopulation of EPN. Source data are provided as a Source Data file. See also Supplementary Fig. 5 and Supplementary Data 6 and 7.
Evaluating hallmark pathways in the VEGFA+ C4 cluster and other clusters from EPN by gene set variation analysis (GSVA) revealed a strong enrichment of angiogenesis and hypoxia in the VEGFA+ C4 cluster (Fig. 4d), suggesting its high capacity to favor tumor progression. We then compared cell fractions in each patient and observed that malignant cells from AEP displayed the highest patient specificity in cell-type composition (Supplementary Fig. 5e), which was further confirmed by our entropy analysis (Fig. 4e). We reasoned that this phenomenon might relate to the different cell types of origin in each AEP patient. Comparing malignant cells from our adult spinal ependymomas with those from pediatric brain ependymomas19, we observed that malignant cells isolated from spinal ependymomas in our study (from SE, EPN, and AEP) were grouped together, while malignant cells collected from pediatric brain ependymomas (from ST and PF) were grouped together, indicating that malignant cells from different anatomic locations exhibited diverse transcriptomic profiles (Supplementary Fig. 5f), highlighting the necessity of studying the transcriptomic characteristics of malignant cells in adult spinal cord ependymomas. These analyses described the heterogeneity of malignant cells and revealed different cellular compositions in each subtype.
To inform future therapeutic approaches targeting malignant cells of spinal ependymomas, we next examined potentially targetable biomarkers specific to the expression signatures of malignant cells in each cancer subtype. By comparing malignant cell population-specific genes with the Drug Gene Interaction database (DGIdb)54, we identified hundreds of pharmacologically targetable genes from different categories in each cancer subtype (Fig. 4f and Supplementary Data 7). Notably, for the common astroependymal-like and NSC-like subpopulations, although we revealed more cancer type-specific druggable vulnerabilities across cancer subtypes, the shared candidates, including the transcription factor gene EEF1A1 and the kinase gene CCNL1, might help unify the pharmacological treatment of spinal ependymomas (Fig. 4g). For the VEGFA+ subpopulation in EPN, our analysis indicated druggable vulnerabilities, including the growth factor genes VEGFA, VEGFB, and EFEMP1, the transporter genes SLC16A3, ABCA1, and SLC2A3, the G protein-coupled receptor gene LPAR1 and the DNA repair gene NPM1 (Fig. 4h). We reasoned that these targetable genes revealed by our scRNA-seq data would provide valuable insights for future therapeutic strategies.
Transcriptional differences reveal cancer subtype-specific characteristics
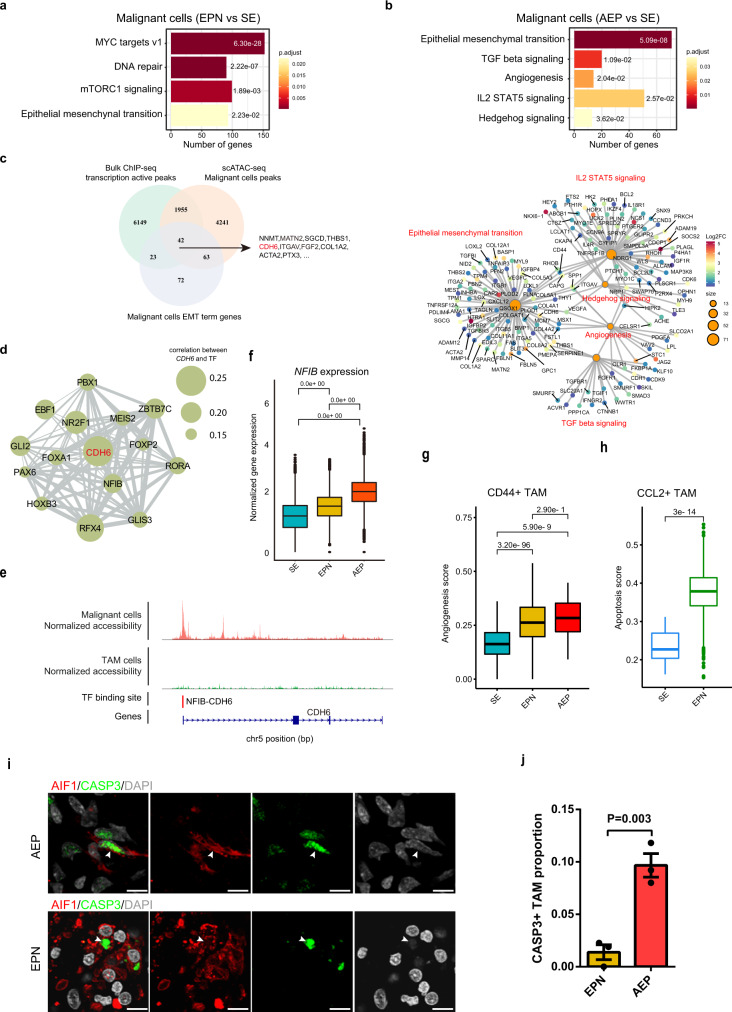
We hypothesized that defining biological programs activated in cells from different cancer subtypes may explain their different degrees of malignancy. We first compared the transcriptomes of malignant cells from different cancer subtypes (Supplementary Data 8), and our enrichment analyses of hallmark pathways revealed specific pathways upregulated in tumor cells from AEP and EPN (Supplementary Data 4). Tumor cells from EPN exhibited an enrichment of the mTORC1 signaling pathway, while tumor cells from AEP showed a significant enrichment of the TGF-β, IL2/STAT5, and Hedgehog signaling pathways (Fig. 5a, b). In addition, tumor cells from AEP showed a significant enrichment of genes from angiogenesis pathways, including the genes VEGFA and SPP1 (Fig. 5b), which play critical roles in controlling the growth of cancer by modulating blood supply in solid tumors55. Notably, one common upregulated pathway in AEP and EPN was the epithelial–mesenchymal transition (EMT) pathway (Fig. 5a, b), an important program that is involved in both cancer development and progression56. Using bulk histone modification ChIP-seq data and scATAC-seq data of malignant cells, we further investigated the epigenetic regulation of the EMT pathway in AEP. Since tumor cells occupied most of the tumor tissues, the bulk ChIP-seq data represented the characteristics of tumor cells. Therefore, we focused the multiomic analysis on only malignant cells and ultimately identified 42 EMT-related genes with active epigenetic signals (Fig. 5c and Supplementary Data 9). Gene CDH6, aberrantly reactivated in cancer and a TGF-β target that can drive the embryonic EMT pathway57, exhibited transcriptional activity around its TSS (Supplementary Fig. 5g) and was upregulated in AEP tumor cells (Fig. 5b). To further investigate the regulatory network associated with CHD6, we predicted its upstream TFs based on scATAC-seq data of tumor cells and revealed that NFIB was one of the upstream regulators (Fig. 5d, e). Moreover, we noticed that the expression of NFIB was upregulated in tumor cells from AEP (Fig. 5f), further implying that NFIB might have crucial effects on the enhanced activity of the EMT pathway in malignant cells.
Fig. 5. Transcriptional differences detected by scRNA-seq analyses reveal cancer subtype-specific characteristics.
a Bar plot of enriched hallmark pathways for genes upregulated in malignant cells from EPN. Adjusted P values were labeled for each pathway. Adjusted P values were calculated by using enrichr function from R package clusterProfiler with hypergeometric test statistical analyses. b Bar plot of enriched hallmark pathways for genes upregulated in malignant cells from AEP (top). Network plot of enriched hallmark pathways for upregulated genes in malignant cells from AEP. Nodes for genes were colored by log2FC, and sizes of nodes for enriched pathways were correlated with the number of genes (bottom). Adjusted P values were labeled for each pathway. Adjusted P values were calculated by using enrichr function from R package clusterProfiler with hypergeometric test statistical analyses. c Venn plot showing the intersection of EMT-related genes with active transcriptional signals. The shared genes are listed on the right side. The red color marks the key genes of interest. d Network plot showing the connection between CDH6 and its upstream TFs. Sizes of circles were related to the correlation value between CDH6 and TFs. e Normalized scATAC-seq profile of CDH6 in AEP across each major subpopulation and NFIB-CDH6 binding site. f Box plot showing the normalized gene expression of NFIB from different cancer subtypes. Adjusted P values were calculated by the Wilcoxon test, two-sided comparisons. n = 122,456 cells. The center line, bounds of box, and whiskers represent mean, 25th to 75th percentile range, and minimum to maximum range in all boxplots. g Box plot showing the angiogenesis signature score of CD44+ TAMs from different cancer subtypes. Adjusted P values were calculated by the Wilcoxon test, two-sided comparisons. n = 3912 cells. The center line, bounds of box, and whiskers represent mean, 25th to 75th percentile range, and minimum to maximum range in all boxplots. h Box plot showing the apoptosis signature score of CCL2+ TAMs from different cancer subtypes. Adjusted P value was calculated by the Wilcoxon test, two-sided comparisons. n = 4647 cells. The center line, bounds of box, and whiskers represent mean, 25th to 75th percentile range, and minimum to maximum range in all boxplots. i Representative example of an EPN and AEP tumor stained by IF. The arrow depicts the CASP3+ TAMs in fluorescent images, and the scale bar represents 30 μm. j Bar plot showing the proportion of CASP3+ TAM in EPN and AEP (n = 3 biologically independent samples). The P value was calculated by t test, Two-way ANOVA analysis. Data are presented as mean values +/− SEM. Source data are provided as a Source Data file. See also Supplementary Fig. 5 and Supplementary Data 4 and 8–10.
We next examined the transcriptomic differences between TAM subsets in different cancer subtypes. Interestingly, we observed that CD44+ TAMs from EPN expressed higher levels of SPP1 (Supplementary Data 10), which has been reported as a marker of angiogenesis-associated TAMs in colon cancer18. We further quantified the score of the angiogenesis pathway in CD44+ TAMs from each cancer subtype and observed significantly increased activity in CD44+ TAMs from EPN and AEP compared with that in CD44+ TAMs from SE (Fig. 5g), suggesting the stronger pro-tumorigenic properties of CD44+ TAMs in EPN and AEP. In addition, for CCL2+ TAMs, higher expression of IL15, which can reduce the immunosuppressive effects of regulatory T cells (Tregs)58, and lower expression of the immunosuppressive gene TREM259 were observed in SE (Supplementary Fig. 5h), suggesting the higher antitumor capacity of CCL2+ TAMs in SE. In addition, we identified lower expression of the pro-apoptotic gene PMAIP160 and higher expression of the anti-apoptotic gene BCL261 in CCL2+ TAMs from SE (Supplementary Fig. 5h), indicating their lower apoptotic activity. Scoring the apoptosis signatures in CCL2+ TAMs in each cancer subtype further supported the notion that CCL2+ TAMs from EPN were more likely to undergo apoptosis (Fig. 5h). We confirmed that the proportion of CASP3+ TAMs was higher in AEP than in EPN by using IF staining (Fig. 5i, j), which complemented the limited number of CCL2+ TAMs in AEP found by our scRNA-seq data. Therefore, our analyses indicate that the CD44+ and CCL2+ TAMs, which played distinctive roles in inflammatory responses and regulating tumor growth, were correlated with the different degrees of malignancy across cancer subtypes, suggesting that the characteristics of TAMs could be used as a marker for different subtypes.
Cell–cell interactions inform stromal cells to regulate TAM subset diversity
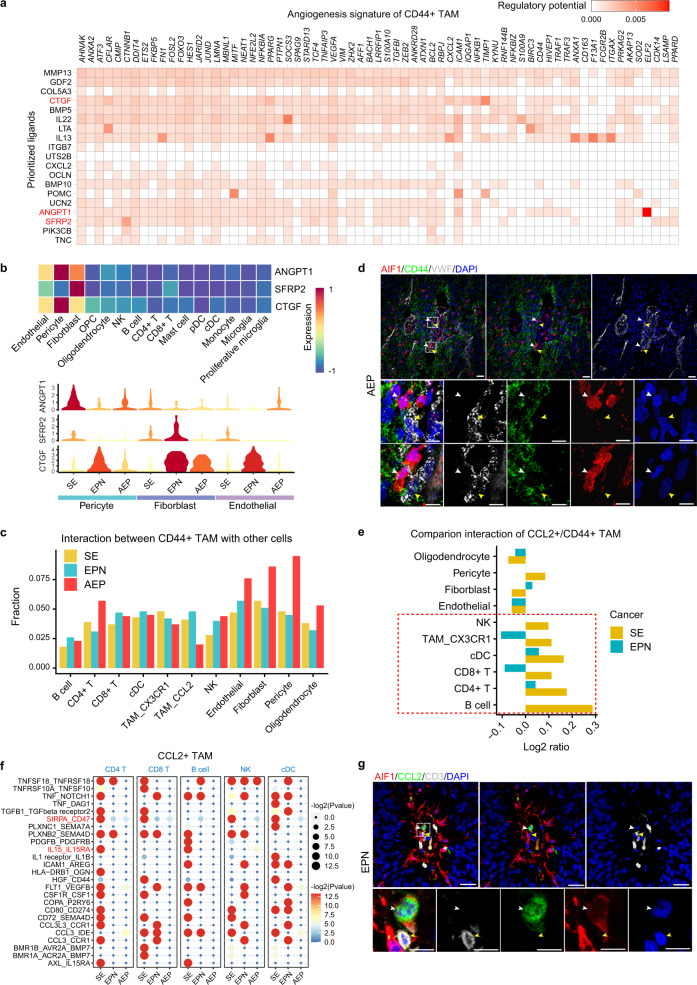
To elucidate the underlying reasons for the functional diversity of TAM subsets, we first performed cell–cell interaction analysis by using CellPhoneDB62 (Supplementary Fig. 6a). Overall, tumor cells in each cancer subtype exhibited more interaction events with fibroblasts, endothelial cells and pericytes than with other cell types (Supplementary Fig. 6b). To explore extracellular signals that drive the special phenotype of CD44+ TAMs, we then performed NicheNet analysis63, which links ligand and target gene expression. Interestingly, NicheNet predicted a panel of ligands that might drive the unique signature genes of CD44+ TAMs (Fig. 6a), including three genes (CTGF, SFRP2, and ANGPT1) associated with angiogenesis64–66. Furthermore, we broadly surveyed the expression of the three ligands and observed that they were highly expressed in fibroblasts, endothelial cells, and pericytes, with diverse patterns across cancer subtypes (Fig. 6b). In addition, we observed more interaction events between CD44+ TAMs and fibroblasts and endothelial cells and pericytes in AEP (Fig. 6c), including the WNT5A-ROR2, JAG1-NOTCH3, and ANGPT2-TEK ligand–receptor (LR) pairs (Supplementary Fig. 6c), which are related to angiogenesis67–69. IF staining of EPN and AEP tumors also showed the physical juxtaposition of CD44+ TAMs and endothelial cells (Fig. 6d and Supplementary Fig. 6d). Our analysis thereby implied that communication with fibroblasts, endothelial cells, and pericytes might induce the angiogenesis phenotype of CD44+ TAMs.
Fig. 6. Cell–cell interaction analyses inform the mechanism of the formation of two TAM subsets.
a Heatmap showing potential ligands driving the signature of CD44+ TAMs. The red color marks the genes of interest. b Heatmap showing the expression of selected ligands in stromal and immune cells (top). Violin plot showing the expression of selected ligands in fibroblasts, endothelial cells and pericytes from each cancer subtype (bottom). c Bar plot showing the fraction distribution of significant interaction events around CD44+ TAMs. The fraction of each interaction pair was calculated by dividing the total number of interaction events related to CD44+ TAMs. d Representative example of an AEP tumor stained by IF with anti-AIF1 (red), CD44 (green), VWF (gray), and DAPI (blue) antibodies. Dashed boxes highlight regions shown in the bottom panel. The white arrow depicts the CD44+ TAMs and the yellow arrow depicts the endothelial cells in fluorescent images. The scale bar in the top panel represents 30 μm, and the scale bar in the bottom panel represents 100 μm. Images shown are representatives of three samples from three independent experiments. e Bar plot showing the ratio of interaction events of CCL2+ TAMs to that of CD44+ TAMs. The fraction of each interaction pair was calculated by dividing the total number of interaction events related to CD44+ and CCL2+ TAMs. Source data are provided as a Source Data file. f Bubble heatmap showing selected significant LR pairs between CCL2+ TAMs and immune cells in each cancer subtype. Each row represents an LR pair, and each column defines a cell–cell interaction pair in a specific cancer subtype. P values were indicated by circle color and size. The red-color marking was the key ligand–receptor pair of interest. P values were calculated by CellPhoneDB. g Representative example of an EPN tumor stained by IF with anti-AIF1 (red), CCL2 (green), CD3 (gray), and DAPI (blue) antibodies. The white arrow depicts the CCL2+ TAMs and the yellow arrow depicts the T cells in fluorescent images. The scale bar in the top panel represents 30 μm and the scale bar in the bottom panel represents 100 μm. Images shown are representatives of three samples from three independent experiments. See also Supplementary Figs. 6 and 7.
We also performed NicheNet analysis to prioritize the ligands that induced the phenotype of CCL2+ TAMs and identified the apoptosis-associated ligand PGF70 (Supplementary Fig. 7a). Furthermore, we noticed that PGF was highly expressed in fibroblasts, endothelial cells and pericytes (Supplementary Fig. 7b), implying that these cells might be involved in inducing the apoptosis of CCL2+ TAMs. We next examined cell–cell interaction events around CCL2+ TAMs. Compared with CD44+ TAMs, CCL2+ TAMs showed more interactions with immune cells (Fig. 6e), consistent with their important roles in the inflammatory response. Close inspection of the LR pairs revealed significant enrichment of the IL15-IL15RA interaction between CCL2+ TAMs and CD4+ T cells in SE (Fig. 6f and Supplementary Fig. 7c), further supporting that the upregulation of IL15 in SE might reduce the immunosuppressive effects of Tregs58. In addition, we identified significant interactions in SE between CCL2+ TAMs and other immune cells, including T cells, through SIRPA-CD47 (Fig. 6f), which might regulate their homeostasis and modulate their immune response71. We confirmed the physical juxtaposition of CCL2+ TAMs and T cells by IF staining (Fig. 6g and Supplementary Fig. 7d). These analyses informed us of a potential mechanism for the higher antitumor capacity of CCL2+ TAMs in SE.
Discussion
Patients diagnosed with spinal ependymomas suffer from multiple symptoms, including neck and back pain, spasticity in the lower extremities, gait ataxia, sensory loss, paresthesias, and paralysis4. Due to the difficulty of surgical removal, new therapeutic strategies for spinal ependymomas are urgently needed clinically. Immunotherapy is believed to be the most promising approach for cancer treatment. Here, our analyses of scRNA-seq and scATAC-seq data characterized the cellular diversity in the TME of spinal ependymomas and highlighted two functionally diverse TAM subsets, providing insights for TAM-targeting immunotherapy.
CCL2+ TAMs, representing the activated TAMs in the TME, were more likely to interact with immune cells and showed a high immune-response capacity, implying their antitumor roles in ependymomas. We noticed that these CCL2+ TAMs exhibited high levels of apoptosis in EPN and AEP. Approaches that inhibit apoptosis of CCL2+ TAMs might help encourage the immune system to attack tumors. In contrast, CD44+ TAMs expressed high levels of angiogenesis-associated genes and tended to interact with endothelial cells, pericytes and fibroblasts to facilitate tumor angiogenesis. Here, we observed a higher pro-angiogenic capacity of CD44+ TAMs in EPN and AEP, providing a plausible explanation for the different degrees of malignancy across cancer subtypes. We reason that strategies that inhibit these CD44+ TAM populations might decelerate the progression of ependymomas. Numerous studies have shown the therapeutic targeting of TAMs can contribute to tumor immunotherapy and the strategies can be roughly divided into three approaches72: eliminating TAMs already present in the TME73,74, inhibition of monocyte recruitment75 and reprogramming of TAMs76. Clodronate-loaded and zoledronate acid-loaded liposomes have been applied to deplete TAMs resulted in decreasing tumor angiogenesis and progression in preclinical solid tumor models successfully, which indicate that these treatments could also be used to deplete CD44+ TAMs in spinal ependymoma.
To investigate the intracellular factors that mediated the functional diversity of TAMs, we further performed scATAC-seq data analysis and revealed that TEAD1 and EGR3 regulated the functional diversity of the CD44+ and CCL2+ TAM subsets, respectively. Previous studies reported that TFs from the TEAD family have a substantial effect on cancer development, progression, and metastasis, suggesting that TEAD proteins could serve as potential therapeutic targets77–79. Here, we discovered the regulatory roles of TEAD1 in CD44+ TAMs, further highlighting its potential application in ependymoma immunotherapy. Moreover, EGR3 motif enrichment analysis showed the strong correlation of EGR3 with the TNF pathway in CCL2+ TAMs; this result indicates that EGR3 plays important role in tumor inflammation, which was consistent with the tumor-suppressive role of EGR3 in certain cancer events80.
For intercellular factors, we uncovered the roles of stromal cells, including fibroblasts, endothelial cells, and pericytes in regulating the functional diversity of TAM subsets in spinal ependymomas. Furthermore, we prioritized multiple ligands that might induce the phenotypes of the CD44+ and CCL2+ TAM subsets. For example, we predicted the regulatory relationship between the ligand PGF and apoptosis-associated genes (NEDD9 and PMAIP1) in CCL2+ TAMs, implying that factors blocking PGF might inhibit the apoptosis of CCL2+ TAMs. These findings invite more investigations to reveal the molecular mechanisms underlying the phenotypic diversity of TAMs, which would further facilitate the development of TAM-targeting immunotherapy.
By comparing malignant cells from our adult spinal ependymoma with those from pediatric brain ependymoma with single-cell transcriptomic analysis, we found that the malignant cells of ependymomas exhibited heterogeneity across anatomic locations, suggesting that knowledge of one type of ependymoma may be unable to be applied to others. By analyzing TAMs isolated from other CNS gliomas, we observed that CD44+ TAMs were also present in GBM, showing a similar expression pattern as those in ependymoma, although they hardly existed in gliomas, indicating that TAM-targeting immunotherapy strategies based on these dual-function TAMs are applicable to multiple types of tumors but not all neurological tumors.
From this study, we were able to summarize potentially targetable biomarkers in malignant cells from each spinal ependymoma subtype, which could inform future therapeutic approaches targeting spinal ependymomas. We also identified diverse activity of the EMT pathway across different spinal ependymoma subtypes, which was associated with their increasing degrees of malignancy. Furthermore, by combining the results of scRNA-seq, scATAC-seq, and bulk ChIP-seq data analyses, we identified the NFIB-CDH6-regulatory program as a potential mediator of the enhanced activity of the EMT pathway.
Overall, our multiomics analyses characterized spinal ependymomas heterogeneity at both the transcriptomic and epigenomic levels. In particular, we highlighted the multifaceted roles of TAMs in tumor progression, indicating that TAM subtype-specific gene expression could be an indicator of the degree of malignancy and that TAM subtype-targeting immunotherapy may have the potential to enhance the immune system to eliminate tumors.
Methods
Patient samples
Fifteen treatment-naïve patients who were pathologically diagnosed with ependymoma subtypes, including seven males and eight females, were enrolled in this study after approval by the Ethics Committee of Beijing Tiantan Hospital, Capital Medical University. All patients provided written informed consent for sample collection and data analyses. We summarized the available clinical characteristics in Supplementary Data 1.
Tumor tissue harvest and dissociation for single-cell suspension
Live-cell isolation from fresh tissue
We collected fresh tumor tissue during surgery and stored in cold phosphate-buffered saline (PBS) to transfer to the laboratory on ice immediately. Tumor tissue was dissociated mechanically with the digestion buffer (2 mg/ml collagenase IV (Gibco), 10 U/ml DNase I (NEB), and 1 mg/ml papain (Sigma) in PBS). After incubation for about 20–30 min at 37 °C on a thermocycler, 10% fetal bovine serum (Gibco) was used to stop the digestion system. Single-cell suspensions dissociated from the tumor were resuspended in 0.04% BSA/PBS and stained with 7-amino-actinomycin D (7-AAD) to sort 7-AAD-negative cells by FACS for single-cell libraries preparation.
Nuclei isolation from frozen tissue
Frozen tumor tissue was homogenized into small cell pellets using a glass tissue grinder (Sigma, Cat #D8938) in ice-cold EZ buffer (Sigma, Cat #NUC-101) and incubated on ice for 5 min. Then nuclei were centrifuged at 500 × g for 5 min at 4 °C. Repeat the wash and centrifuge step again. After that, nuclei were resuspended with PBS, and Debris Removal Solution (MACS) was added to effectively remove cell debris. Isolated nuclei were suspended in Nuclei Suspension Buffer (NSB; consisting of 1× PBS, 0.01% BSA and 0.1% RNase inhibitor (Clontech, Cat #2313A)). A final nuclei suspension was filtered through a 35-μm cell strainer (Corning, Cat #352235) and used for single-cell libraries preparation.
Library preparation and sequencing
Single-cell/nuclei were processed using Chromium Single Cell V2 or V3 Chemistry Library Kits (10× Genomics) and GEM ATAC v1.1 Kits (10× Genomics) according to the manufacturer’s instructions. Thousands of cells/nuclei were partitioned into Gel Beads-in-emulsion (GEMs) on the Chromium Controller, followed by full-length cDNA generated, barcoded, and sequenced on an Illumina NextSeq4000.
Immunohistochemistry
We dropped fresh tumor tissue in 4% paraformaldehyde overnight at 4 °C. After being washed with PBS three times for 10 min, the fixed tissue was placed in 20% and 30% sucrose solution to dehydrate and then was embedded in optimal cutting temperature medium (Thermo Scientific). Embedded tissue was sectioned at 20–30 μm cryosections using Leica CM3050S cryostat. Before IHC staining, tissue slices were subjected to heat-mediated antigen retrieval with Tris/EDTA buffer (PH 9.0). Then, tissue slices were blocked in 5% donkey serum for 2 h. Sections were incubated with primary antibody at the following dilutions: goat anti-IBA1 (1:100, Abcam, ab5076), Rabbit anti-MCP1 (1:100, Abcam, ab73680), mouse anti-CD44 (1:100, Abcam, ab16728), rabbit anti-VWF (1:100, Abcam, ab9378), mouse anti-CD3 (1:50, Abcam, ab699), rabbit anti-cleaved Caspase-3 (1:200, Cell Signaling, #9661), rabbit anti-TMEM119 (1:1000, Abcam, ab185333). After incubation overnight at 4 °C with primary antibody, Alexa Fluor 488, 594, or 647 fluorophore-conjugated secondary antibodies (1:500) (Life Technologies), and DAPI (1:500) were added to slices incubation buffer for 2 h. We used Olympus FV3000 confocal microscope for collecting images.
Chromatin immunoprecipitation-sequencing (ChIP-seq)
CUT& Tag ChIP-seq were performed as described81. Nuclei were washed twice in Wash Buffer (20 mM HEPES pH 7.5; 150 mM Nacl; 0.5 mM Spermidine; 1× Protease inhibitor cocktail). Add 10 μl Concanavalin A-coated magnetic beads (Bangs Laboratories) per sample and incubated at RT for 15 min. Remove supernatant and resuspend in Dig-wash Buffer (20 mM HEPES pH 7.5; 150 mM NaCl; 0.5 mM Spermidine; 1× protease inhibitor cocktail; 0.05% digitonin), supplementing 2 mM EDTA and the primary antibody in 1:50 dilution. Incubate primary antibody overnight at 4 °C and then replaced it with secondary antibody (1:50 diluted in Dig-Wash buffer). Secondary antibody incubation was performed at room temperature (RT) for 30 min. Unbound antibodies in Nuclei was washed using the magnet stand 2–3 times for 5 min. pA-Tn5 adapter complex was diluted in 1:200 using Dig-med Buffer (0.05% digitonin, 20 mM HEPES, pH 7.5, 300 mM NaCl, 0.5 mM Spermidine, 1× protease inhibitor cocktail). Nuclei was resuspended in Dig-med Buffer and incubated with pA-Tn5 at RT for 1 h. After washed, nuclei were resuspended in Tagmentation buffer (10 mM MgCl2 in Dig-med Buffer) and incubated at 37 °C for 1 h. 2.25 µL of 0.5 M EDTA, 2.75 µL of 10% SDS, and 0.5 µL of 20 mg/mL Proteinase K were added to 50 µL of sample to stop tagementation and then inactivate Proteinase K at 70 °C for 20 min. Ampure XP beads were used to extract DNA. DNA was amplified using universal i5 and uniquely barcoded i7 primer and cleaned up by adding 1.1× Ampure XP beads (Beckman Colter). DNA libraries were sequenced on an Illumina NextSeq4000.
scRNA-seq data processing
scRNA-seq experiments were performed on 15 samples (Supplementary Data 1), the raw data of which are accessible under database Genome Sequence Archive (GSA: HRA001112) or database Gene Expression Omnibus (GEO: GSE163686). scRNA-seq data generated from 10× Genomics were processed using the Cell Ranger Single-Cell Software Suite (v3.0.1), which aligned sequencing reads to hg19 human reference genome and quantified gene expression levels of single cells. We merged cells from different tumors using the CellRanger aggr pipeline and the preliminary filtered data were used for downstream analysis. First, we applied further quality control to cells according to three different metrics step by step, including the total UMI count, the number of detected genes, and the proportion of mitochondrial gene count per cell. Specifically, we filtered cells with less than 2000 UMI count or 500 detected genes, as well as cells with more than 20% mitochondrial gene count. To remove potential doublets, we also removed cells with UMI count above 70,000 and the number of detected genes above 10,000. Here, we used a relatively high threshold because malignant cells are thought to express a greater number of genes. Furthermore, we removed potential doublets predicted by Scrublet82. Next, after quality control, we applied the library-size correction method to normalize the raw count matrix by using normalize_total function in SCANPY83. Then the logarithmized normalized count matrix was used for the downstream analysis.
Dimension reduction and unsupervised clustering
We employed the workflow of SCANPY83 to perform dimension reduction and graph-based unsupervised clustering on scRNA-seq data. Briefly, we first selected 2000 highly variable genes (HVGs) for downstream analysis by using scanpy.pp.highly_variable_genes function with parameter “n_top_genes=2000”. Then, effects of the total count per cell, the percentage of mitochondrial gene count and the percentage of count for heat shock protein associated genes (HSP) were regressed out by using scanpy.pp.regress_out function. Principal component analysis (PCA) was performed on selected HVGs by using scanpy.tl.pca function with parameter “svd_solver='arpack', n_comps=100”, and generated a PCA matrix with 100 components. Next, we employed bbknn algorithm with parameter “batch_key='patient', n_pcs=90” to remove the batch effects from different donors and obtain a batch-corrected space. To visualize our scRNA-seq data in a two-dimensional space, Uniform Manifold Approximation and Projection (UMAP) dimension reduction was performed by using scanpy.tl.umap function with default parameter. Last, to cluster single cells by their transcriptional profiles, we used an unsupervised graph-based clustering algorithm implemented in scanpy.tl.leiden function with different resolution parameters adapted to diverse situations. The cluster-specific marker genes were identified by using scanpy.tl.rank_genes_groups function with default parameter. Specifically, we performed two round of dimension reduction and unsupervised clustering for all cells. The first round (“resolution = 2”) was used to distinguish malignant cells from nonmalignant cells and the second round (“resolution = 1”) was applied to nonmalignant and malignant cells respectively. Notably, we used scanpy.pp.neighbors function with default parameter to compute a neighborhood graph instead of bbknn to avoid eliminating the great donor effects in malignant cells. Subclusters within nonmalignant cells were annotated by using canonical markers shown in Fig. 1e. To reveal the cellular diversity in monocytes and TAM, we re-run the dimension reduction and unsupervised clustering with “resolution = 0.6”.
InferCNV analysis
Copy-number variations (CNVs) of cells were estimated by computing a moving average of the relative expression by using InferCNV (inferCNV of the Trinity CTAT Project, https://github.com/broadinstitute/inferCNV)84. Nonmalignant cells including both stomal and immune cells were used as to define a baseline of normal karyotype and their average copy-number value was subtracted from all cells.
Enrichment analysis
We used enrichr and GESA functions from R package clusterProfiler for hypergeometric test and gene set enrichment analysis, respectively. Network plots of enriched pathways were visualized by function cnetplot from R package enrichplot. The R package GSVA from Bioconductor was used for gene set variation analysis. The gene sets (MSigDB) was loaded from R package msigdf and pathways with high difference in activity scores were selected by LIMMA package.
Survival analysis
We used the top 30 genes of CD44+ TAM in Supplementary Data 3 to perform survival analysis by using the GEPIA2 webserver (http://gepia2.cancer-pku.cn/). Two cancer types (LGG and GBM) were selected to test the correlation between the given signature gene list and patients’ survival time with default parameters.
Quantification of gene set score
We used the average expression level of a given gene set to quantify gene set score in every single cell. The M1 and M2 signatures were extracted from the previous publication13. Other gene sets, including angiogenesis and apoptosis were loaded from R package msigdf.
SCENIC analysis
We used SCENIC34 with raw count matrices as input to analyze activated regulons in each TAM subset. Briefly, we inferred the coexpression network by using GRNBoost2 python package and identified regulons by using RcisTarget package. Then, regulon activity for each cell was quantified by AUCell package, and differentially activated regulons in each TAM subset were identified by wilcox.test using other subsets as control. Finally, Benjamini–Hochberg procedure was used to correct the multiple hypothesis.
Developmental trajectory analysis
RNA velocity
We used Velocyto pipeline85 to annotate spliced and unspliced reads and generated the loom file for downstream analysis. Then the python package scVelo86 was used to estimate the steady-state RNA velocities by using default parameters. Finally, the velocities were projected onto UMAP embedding and visualized on the cellular level by using scv.pl.velocity_embedding function. Here we removed CD16+ monocytes because monocyte-derived macrophages were more likely to develop from CD14+ monocytes18.
PAGA
We further used the partition-based graph abstraction method PAGA83 to assess the developmental trajectories for TAMs. The computations were carried out using default parameters. The edge connectivity with CD14+ monocytes between each subpopulation node are further visualized in the bar plot.
Predicting the origins of TAMs by using SingleR
To quantify the fraction of TAMs from different origins (microglia or monocytes), we used SingleR87, a predictor mainly based on transcriptomic similarity with the reference dataset. Here, we used the published data47 with both Mg-TAMs (microglia-derived TAMs) and Mo-TAMs (monocyte-derived TAMs) as a reference.
Integration of TAMs in our study with that from other neurological tumors
We used Harmony52 to integrate TAMs from different studies. Briefly, TAMs were first performed PCA analysis with 100 components by using the top 2000 highly variable genes identified by scanpy.pp.highly_variable_genes function with parameter “n_top_genes=2000”. Then Harmony was applied to the 100 principle components with default parameters to integrate TAMs from different studies and generate a corrected embedding. Next, we used the first 30 Harmony corrected dimensions to compute a neighborhood graph by using scanpy.pp.neighbors function with default parameter. Finally, the UMAP dimension reduction was then performed by using scanpy.tl.umap function with the default parameter.
Data harmonization and unsupervised clustering for malignant cells
We applied Harmony alignment52 to malignant cells from different patients to study their potential commonalities. We adopted two different algorithms to correct different sources of intersample variations. BBKNN uses the mutual nearest neighbors (MNNs) between different batches to correct batch effects, while Harmony deploys a local correction idea that preferentially clusters cells from different batches, thereby better matching the distributions of the shared cell types across donors. For each cancer type, malignant cells were first performed PCA analysis with 100 components as described earlier. Harmony was then applied to the 100 principle components with default parameters to integrate malignant cells from multiple patients and generate a corrected embedding. Next, we used the first 20 Harmony corrected dimensions to compute a neighborhood graph by using scanpy.pp.neighbors function with parameter “n_neighbors=10”. Uniform Manifold Approximation and Projection (UMAP) dimension reduction was then performed by using scanpy.tl.umap function with default parameter. Last, we used the same unsupervised graph-based clustering algorithm to cluster malignant cells and the same function for marker gene identification as described earlier. Notably, we excluded three patients (EPN2, EPN3, and EPN5) in this analysis because of the limited malignant cell numbers.
Evaluation of mixability with entropy metric
We adopted an entropy-based metric to quantify the mixability of malignant cells across different patients after data harmonization13. Specifically, we randomly sampled 25,000 malignant cells and constructed a kNN graph (k = 80) based on the Euclidean distance in UMAP coordinates by using function kNN from R package dbscan. We defined the mixability of cells across patients as Shannon entropy,
| 1 |
where is the fraction of cells from patient d in the 80 nearest neighborhoods of cell i and .
Targetable biomarkers identification
Drug Gene Interaction database (DGIdb)54, a valuable resource that focuses on expert-curated collections of druggable genes, was used to examine potential targetale biomarkers within the specific gene signatures (top 100 genes reported by scanpy.tl.rank_genes_groups function) of each malignant cell subpopulation from each cancer subtype. We excluded DGIdb hits that were categorized as druggable genome, which was predicted on the basis of sequence and structural similarity88.
Differential analysis for malignant cells from different cancer types
Here we first combined the raw count matrix of malignant cells from each cancer type together and then used the sc.pp.normalize_total function from SCANPY to normalize the combined data matrix (library-size corrected), which made the counts comparable among cells. Finally, the logarithmized count matrix was used for differential analysis by the SCANPY function sc.tl.rank_genes_groups using the Wilcoxon test.
Comparison of adult spinal cord ependymoma and pediatric brain ependymoma
To compare malignant cells from our study with that from a published dataset, we first extracted the top 50 signature genes (reported by SCANPY) for each malignant cell subpopulation and then aggregated their averaged expression level in each cell population within each tumor type. Finally, the pairwise correlation of the averaged expression level was calculated by cor function in R with default parameters and visualized in the heatmap.
Cell–cell interaction analysis
We applied CellPhoneDB62 to infer cell–cell interaction between cells in each cancer type. The enriched ligand–receptor interactions between two cell subsets were calculated based on the permutation test. We extracted significant ligand–receptor pairs with P value < 0.05, and summarized the number of interaction events related with tumor cells by the union of significant ligand–receptor pairs. Circle plots depicting the number of interactions between cell types are drawn using R package circlize.
NicheNet analysis
We used NicheNet63 to predict ligands that drive the signature genes of the two TAM subsets. We first calculated DEGs in both CCL2+ and CD44+ TAMs. Then, DEGs with log2FC > 0.2 and adjusted P value < 0.05 in each TAM subset were used as gene sets of interest, respectively. All expressed genes in CCL2+ or CD44+ TAMs were used as the background of genes. Genes were considered as expressed when they have nonzero values in at least 10% of the cells in a cell type. Here, sender cell type was not specified to broadly examine ligand’s activity.
Evaluating expression patterns of the reported generic tumor cell programs across malignant cell subsets
We downloaded the 12 generic tumor cell programs from the pan-cancer single-cell analysis of cancer cell lines53. The average expression of each generic tumor cell program in each malignant cell was calculated and scaled to 0–1. To quantify the aggregated expression level of each program in each malignant subset, we further calculated the average expression scores for each program across each subset and visualized their patterns in scatter plots.
Data processing of single-cell ATAC-seq
scATAC-seq experiments were performed on two samples (Supplementary Data 1), the raw data of which are accessible under database Genome Sequence Archive (GSA: HRA001112) with GSA individual accession number HRI137901 and HRI137902. scATAC-seq raw data for samples HRI137901 and HRI137902 can be obtained under GSA Run accession HRR337425 and HRR337426 respectively. The Cell Ranger software (v3.0.1)89 performed reads filtering, alignment, and transposase cut sites identification. Cell by feature matrix with a window size of 2.5 kb was generated as described previously89 by first reading fragment file into R. Next, the GenomicRanges of the fragments were concatenated for each “start” and “end” followed by identifying all overlaps with the feature by insertions using R function findOverlaps. Then the fraction of Tn5 insertions was computed by dividing the cells-wise sum of the count matrix by the number of insertions for each cell. The processed count matrix was first binarized and then log-normalized for downstream analysis. The binarized count matrix was passed to Signac90 for dimension reduction and clustering. Signac function RunTFIDF and RunSVD were performed with default parameters for dimension reduction analysis. Briefly, the count matrix was the first frequency-inverse document frequency (TF-IDF) transformed by dividing each index by the cellwise sum of the matrix multiplying with the inverse document frequency computed as log(1 + ncol(matrix)/rowSums(matrix)). Singular value decomposition (SVD) was then performed on the TD-IDF matrix for dimension reduction. Clustering was done with the function FindNeighbors and FindClusters. Peak calling for each cluster was performed with fragments from cells in the cluster using MACS291 with parameters ‘–nomodel –shift 75 –extsize 150 –qval 5e-2 -B -f BED –nolambda –keep-dup all –call-summits’ callpeak with MACS2 software (v 2.2.7.1). We retained those peaks only whose q values were less than 0.05 for further analysis and considered these peaks as reproducible peaks. The q value of each peak call is computed adjusting P value to control the false discovery rate with multiple testing correction which is a measure of statistical significance of the peak call. Peaks from each cluster were then merged and a binarized cell-by-peak matrix was constructed by converting nonzero counts to 1. The generated cell-by-peak matrix was then used to create a Signac object. Cells that pass the following quality criterion were kept: (1) the number of fragments in peaks between 1000 and 20,000; (2) the fraction of fragments in peaks greater than 10%; (3) transcriptional start site (TSS) enrichment score greater than 2; (4) ratio reads in genomic blacklist regions smaller than 0.05; (5) nucleosomal signal strength smaller than 4. In total, 4922 cells from two samples were initially profiled, and 2845 cells passed the quality control for further analyses. The filtered cell-by-peak matrix was passed to Signac function RunTFIDF and RunSVD for dimension reduction and function FindNeighbors and FindClusters for cluster identification. FindNeighbors was performed with reduced dimension components 2:30. FindClusters was performed by setting resolution to 0.3. Next, gene activity matrix was generated by quantifying the activity of each gene in the genome by accessing gene-associated chromatin accessibility using function GeneActivity. Clusters were then annotated based on gene activity score profiles of known cell-type markers. TEKT1, GFAP, OLIG2, NEUROD6, and CD68 were used to name major cell types as ependymal-like cells, astrocytependymal-like cells, oligodendrocyte progenitor cell (OPC), neuronal-like cells, and TAMs, respectively.
Calculation of differentially accessible peaks
Differentially accessible peaks were computed with Signac function FindAllMarkers by setting parameters test.use = “LR” and latend.vars = “peak_region_fragments”. Only peaks with adjusted P value smaller than 0.05 were considered as differentially accessible peaks.
Co-accessible regulatory network
To build co-accessible regulatory network, we first identified upstream transcription factors of CDH6 whose peaks were enriched for the malignant cells. Briefly, we computed differentially accessible peaks among the malignant and microglia clusters with Signac function FindAllMarkers, followed by identifying closest genes to each of these peaks with function ClosestFeature. We then performed motif analysis for searching transcription factor motifs closed to the transcription start site (TSS) of CDH6. Next, we computed the intersection of the upstream TFs of CDH6 and the closest genes around those peaks that were more accessible for malignant cells. Then, we computed correlation of the transposed gene activity score matrix (cell in row and gene in the column) containing only CDH6 and those intersected TFs. The correlation matrix was then loaded into software Cytoscape92 for network visualization. Edges with a correlation coefficient smaller than 0.15 were removed. The width of the edges and the size of nodes were scaled with the correlation coefficients.
Integration of scRNA-seq and scATAC-seq data
To integrate scATAC-seq data with snRNA-seq data, we first identified anchors between these two datasets with Signac function FindTransferAnchors based on the expression and gene activity profiles of highly variable genes identified from the scRNA-seq data. We then computed the imputed gene expression for scATAC-seq cells based on the previously computed anchors with function TransferData. To construct co-embedding for the scRNA-seq and scATAC-seq data, we next run Signac function RunPCA and RunUMAP on the imputed gene expression matrix.
Motif enrichment
Motif enrichment was performed with FIMO from the MEME suites93 to identify enriched binding motif in the genes’ regulatory regions. Homer94 function annotatePeaks.pl was then used for motif binding site annotation.
Visualizing differentially accessible peaks
The deeptools95 was applied to visualize pileups of cluster-specific ATAC-seq signals in differentially accessible peaks.
ChIP-seq data processing
ChIP-seq experiments were performed on two samples (Supplementary Data 1), the raw data of which are accessible under database Genome Sequence Archive (GSA: HRA001112) with GSA individual accession number HRI137901 and HRI137902. H3K27ac and H3K4me3 ChIP-seq data for sample HRI137901 can be obtained under GSA Run accession HRR337427 and HRR337429 respectively. H3K27ac and H3K4me3 ChIP-seq data for sample HRI137902 can be obtained under GSA Run accession HRR337428 and HRR337430 respectively. Fastq-files were trimmed for adaptor sequence using cutadapt (3.2)96 and then mapped to hg19 using Bowtie2 (2.4.2)97 with parameters “-3 3 –no-discordant –no-mixed”. SAMtools (1.9) and bedtools (2.30.0) were applied to filter mapped read pairs post alignment. MACS2 (2.2.7.1) was used for peak calling with default parameters98. The visualization of peaks was realized by IGV. For each sample, we identified active promoters by H3K4me3 peaks that overlap H3K27ac within 1000bp to the nearest transcription start site (TSS).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by the National Basic Research Program of China (2019YFA0110101, 2017YFA0103303, 2017YFA0102601), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16020601, XDB32010100), the National Natural Science Foundation of China (NSFC) (81891001, 31771140), BUAA-CCMU Big Data and Precision Medicine Advanced Innovation Center Project (BHME-2019001), Collaborative Research Fund of Chinese Institute for Brain Research, Beijing (2020-NKX-PT-03) and the Open Research Fund of the State Key Laboratory of Cognitive Neuroscience and Learning.
Source data
Author contributions
X.W., Q.W., and W.J. conceived the project, designed the experiments. Y.W., Y.Z., and R.C. collected the clinical samples. Q.Z. and Y.C. performed the single-cell RNA-seq experiment. Q.Z. and Y.L. performed the single-cell ATAC-seq experiment. Z.W. and Q.Z. performed the ChIP-seq experiment. S.C. and Q.Z. analyzed the scRNA-seq data. M.W. and Q.Z. analyzed the scATAC-seq data. Y.L. analyzed the ChIP-seq data. W.L. and B.W. carried out the pathology examination. Y.G. and Q.M. prepared the samples for immunostaining. Q.Z., L.G, Y.C., and M.L. performed immunostaining, imaging, and data analysis. S.C., Q.W., and X.W. wrote the manuscript. All authors edited and proofed the manuscript.
Data availability
The scRNA-seq data generated in this study are available in the Gene Expression Omnibus (GEO) database under accession number GSE163686 and also available in the Genome Sequence Archive (GSA) database under accession number HRA001112. The scATAC-seq and ChIP-seq data generated from two samples in this study are available in the GSA database under accession number HRA001112. In particular, H3K27ac and H3K4me3 ChIP-seq data for sample HRI137901 can be obtained under GSA accession numbers HRR337427 and HRR337429, respectively. H3K27ac and H3K4me3 ChIP-seq data for sample HRI137902 can be obtained under GSA accession numbers HRR337428 and HRR337430, respectively. scATAC-seq data for samples HRI137901 and HRI137902 can be obtained under accession numbers HRR337425 and HRR337426, respectively. The data in GSA are available under restricted access, access can be obtained by contacting Xiaoqun Wang (xiaoqunwang@ibp.ac.cn). The published datasets used in this study can be downloaded from https://www.brainimmuneatlas.org/ or from GEO under accession number GSE16312047; scRNA-seq data for TAMs from glioblastoma, GSE16641851; scRNA-seq data for TAMs from IDH-mutant high-grade gliomas, GSE8446550; scRNA-seq data for TAMs from glioblastoma and GSE6783549; scRNA-seq data for macrophage from normal brain. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks Roman Sankowski and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qianqian Zhang, Sijin Cheng, Yongzhi Wang, Mengdi Wang.
These authors jointly supervised this work: Qian Wu, Wenqing Jia, Xiaoqun Wang.
Contributor Information
Qian Wu, Email: qianwu@bnu.edu.cn.
Wenqing Jia, Email: coffeemd@163.com.
Xiaoqun Wang, Email: xiaoqunwang@ibp.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-27018-9.
References
- 1.Pajtler KW, et al. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27:728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanbali F, et al. Spinal cord ependymoma: radical surgical resection and outcome. Neurosurgery. 2002;51:1162–1172. doi: 10.1097/00006123-200211000-00010. [DOI] [PubMed] [Google Scholar]
- 3.Connolly ID, Ali R, Li Y, Gephart MH. Genetic and molecular distinctions in spinal ependymomas: a review. Clin. Neurol. Neurosurg. 2015;139:210–215. doi: 10.1016/j.clineuro.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Ruda R, Gilbert M, Soffietti R. Ependymomas of the adult: molecular biology and treatment. Curr. Opin. Neurol. 2008;21:754–761. doi: 10.1097/WCO.0b013e328317efe8. [DOI] [PubMed] [Google Scholar]
- 5.Mohankumar KM, et al. An in vivo screen identifies ependymoma oncogenes and tumor-suppressor genes. Nat. Genet. 2015;47:878–887. doi: 10.1038/ng.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson RA, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor MD, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Ebert C, et al. Molecular genetic analysis of ependymal tumors. NF2 mutations and chromosome 22q loss occur preferentially in intramedullary spinal ependymomas. Am. J. Pathol. 1999;155:627–632. doi: 10.1016/S0002-9440(10)65158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louis DN, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 10.Ren X, Kang B, Zhang Z. Understanding tumor ecosystems by single-cell sequencing: promises and limitations. Genome Biol. 2018;19:211. doi: 10.1186/s13059-018-1593-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambrechts D, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 12.Zilionis R, et al. Single-Cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50:1317–1334 e1310. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308 e1236. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young MD, et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018;361:594–599. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng J, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29:725–738. doi: 10.1038/s41422-019-0195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. 2019;176:775–789 e718. doi: 10.1016/j.cell.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845 e820. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020;181:442–459 e429. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 19.Gojo J, et al. Single-cell RNA-seq reveals cellular hierarchies and impaired developmental trajectories in pediatric ependymoma. Cancer Cell. 2020;38:44–59 e49. doi: 10.1016/j.ccell.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillen AE, et al. Single-cell RNA sequencing of childhood ependymoma reveals neoplastic cell subpopulations that impact molecular classification and etiology. Cell Rep. 2020;32:108023. doi: 10.1016/j.celrep.2020.108023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dougan M, Dranoff G, Dougan SK. Cancer immunotherapy: beyond checkpoint blockade. Annu. Rev. Canc Biol. 2019;3:55–75. doi: 10.1146/annurev-cancerbio-030518-055552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao N, Francis NL, Calvelli HR, Moghe PV. Microglia-targeting nanotherapeutics for neurodegenerative diseases. APL Bioeng. 2020;4:030902. doi: 10.1063/5.0013178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puram SV, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624 e1624. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tirosh I, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venteicher, A. S. et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science355, (2017). [DOI] [PMC free article] [PubMed]
- 28.Masuda T, et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution (vol 566, pg 388, 2019) Nature. 2019;568:E4–E4. doi: 10.1038/s41586-019-0924-x. [DOI] [PubMed] [Google Scholar]
- 29.Chui R, Dorovini-Zis K. Regulation of CCL2 and CCL3 expression in human brain endothelial cells by cytokines and lipopolysaccharide. J. Neuroinflam. 2010;7:1. doi: 10.1186/1742-2094-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sankowski R, et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci. 2019;22:2098–2110. doi: 10.1038/s41593-019-0532-y. [DOI] [PubMed] [Google Scholar]
- 31.Geirsdottir L, et al. Cross-species single-cell analysis reveals divergence of the primate microglia program. Cell. 2019;179:1609−+. doi: 10.1016/j.cell.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Ludwig N, et al. CD44(+) tumor cells promote early angiogenesis in head and neck squamous cell carcinoma. Cancer Lett. 2019;467:85–95. doi: 10.1016/j.canlet.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J. Intern Med. 2013;273:114–127. doi: 10.1111/joim.12019. [DOI] [PubMed] [Google Scholar]
- 34.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh KS, et al. Dual roles for Ikaros in regulation of macrophage chromatin state and inflammatory gene expression. J. Immunol. 2018;201:757–771. doi: 10.4049/jimmunol.1800158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baron VT, Pio R, Jia Z, Mercola D. Early growth response 3 regulates genes of inflammation and directly activates IL6 and IL8 expression in prostate cancer. Br. J. Cancer. 2015;112:755–764. doi: 10.1038/bjc.2014.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tome-Garcia J, et al. Analysis of chromatin accessibility uncovers TEAD1 as a regulator of migration in human glioblastoma. Nat. Commun. 2018;9:4020. doi: 10.1038/s41467-018-06258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mammoto A, Muyleart M, Kadlec A, Gutterman D, Mammoto T. YAP1-TEAD1 signaling controls angiogenesis and mitochondrial biogenesis through PGC1α. Microvascular Res. 2018;119:73–83. doi: 10.1016/j.mvr.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chevrier S, et al. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169:736–749 e718. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng S, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184:792–809 e723. doi: 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 41.De Vito A, et al. Overexpression of murine Rnaset2 in a colon syngeneic mouse carcinoma model leads to rebalance of intra-tumor M1/M2 macrophage ratio, activation of T cells, delayed tumor growth, and rejection. Cancers. 2020;12:717. doi: 10.3390/cancers12030717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parisi L, et al. Macrophage polarization in chronic inflammatory diseases: killers or builders? J. Immunol. Res. 2018;2018:e8917804. doi: 10.1155/2018/8917804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Investig. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiss M, Van Gassen S, Movahedi K, Saeys Y, Laoui D. Myeloid cell heterogeneity in cancer: not a single cell alike. Cell Immunol. 2018;330:188–201. doi: 10.1016/j.cellimm.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Croxford AL, et al. The cytokine GM-CSF drives the inflammatory signature of CCR2+ monocytes and licenses autoimmunity. Immunity. 2015;43:502–514. doi: 10.1016/j.immuni.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 46.Wolf FA, et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20:59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pombo Antunes AR, et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat. Neurosci. 2021;24:595–610. doi: 10.1038/s41593-020-00789-y. [DOI] [PubMed] [Google Scholar]
- 48.Muller, S. et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol.18, 1–14 (2017). [DOI] [PMC free article] [PubMed]
- 49.Darmanis S, et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl Acad. Sci. USA. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darmanis S, et al. Single-cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21:1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedrich M, et al. Tryptophan metabolism drives dynamic immunosuppressive myeloid states in IDH-mutant gliomas. Nat. Cancer. 2021;2:723–740. doi: 10.1038/s43018-021-00201-z. [DOI] [PubMed] [Google Scholar]
- 52.Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kinker GS, et al. Pan-cancer single-cell RNA-seq identifies recurring programs of cellular heterogeneity. Nat. Genet. 2020;52:1208–1218. doi: 10.1038/s41588-020-00726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cotto KC, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46:D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishida N, Yano H, Nishida T, Kamura T, Kojiro M. Angiogenesis in cancer. Vasc. Health Risk Manag. 2006;2:213–219. doi: 10.2147/vhrm.2006.2.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Weinberg RA. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front. Med. 2018;12:361–373. doi: 10.1007/s11684-018-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gugnoni M, et al. Cadherin-6 promotes EMT and cancer metastasis by restraining autophagy. Oncogene. 2017;36:667–677. doi: 10.1038/onc.2016.237. [DOI] [PubMed] [Google Scholar]
- 58.Perna SK, et al. Interleukin 15 provides relief to CTLs from regulatory T cell-mediated inhibition: implications for adoptive T cell-based therapies for lymphoma. Clin. Cancer Res. 2013;19:106–117. doi: 10.1158/1078-0432.CCR-12-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katzenelenbogen Y, et al. Coupled scRNA-Seq and intracellular protein activity reveal an immunosuppressive role of TREM2 in cancer. Cell. 2020;182:872–885 e819. doi: 10.1016/j.cell.2020.06.032. [DOI] [PubMed] [Google Scholar]
- 60.Zhao X, Liu X, Su L. Parthenolide induces apoptosis via TNFRSF10B and PMAIP1 pathways in human lung cancer cells. J. Exp. Clin. Cancer Res. 2014;33:3. doi: 10.1186/1756-9966-33-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edlich F. BCL-2 proteins and apoptosis: recent insights and unknowns. Biochem. Biophys. Res. Commun. 2018;500:26–34. doi: 10.1016/j.bbrc.2017.06.190. [DOI] [PubMed] [Google Scholar]
- 62.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 63.Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 64.Liu SC, et al. CTGF increases vascular endothelial growth factor-dependent angiogenesis in human synovial fibroblasts by increasing miR-210 expression. Cell Death Dis. 2014;5:e1485. doi: 10.1038/cddis.2014.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siamakpour-Reihani S, et al. The role of calcineurin/NFAT in SFRP2 induced angiogenesis–a rationale for breast cancer treatment with the calcineurin inhibitor tacrolimus. PLoS ONE. 2011;6:e20412. doi: 10.1371/journal.pone.0020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dai J, et al. Genetic polymorphism in a VEGF-independent angiogenesis gene ANGPT1 and overall survival of colorectal cancer patients after surgical resection. PLoS ONE. 2012;7:e34758. doi: 10.1371/journal.pone.0034758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiu MX, Liu YM, Kuang BH. The oncogenic role of Jagged1/Notch signaling in cancer. Biomed. Pharmacother. 2020;129:110416. doi: 10.1016/j.biopha.2020.110416. [DOI] [PubMed] [Google Scholar]
- 68.Shi YN, et al. Wnt5a and its signaling pathway in angiogenesis. Clin. Chim. Acta. 2017;471:263–269. doi: 10.1016/j.cca.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 69.Yuan HT, Khankin EV, Karumanchi SA, Parikh SM. Angiopoietin 2 is a partial agonist/antagonist of Tie2 signaling in the endothelium. Mol. Cell Biol. 2009;29:2011–2022. doi: 10.1128/MCB.01472-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hou HH, et al. Elastase induces lung epithelial cell autophagy through placental growth factor A new insight of emphysema pathogenesis. Autophagy. 2014;10:1509–1521. doi: 10.4161/auto.29190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nath PR, et al. CD47 expression in natural killer cells regulates homeostasis and modulates immune response to lymphocytic choriomeningitis virus. Front. Immunol. 2018;9:2985. doi: 10.3389/fimmu.2018.02985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiang X, Wang J, Lu D, Xu X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Sig Transduct. Target Ther. 2021;6:1–12. doi: 10.1038/s41392-021-00484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang W, et al. Depletion of tumor-associated macrophages enhances the effect of sorafenib in metastatic liver cancer models by antimetastatic and antiangiogenic effects. Clin. Cancer Res. 2010;16:3420–3430. doi: 10.1158/1078-0432.CCR-09-2904. [DOI] [PubMed] [Google Scholar]
- 74.Lv J, et al. Zoledronic acid inhibits thyroid cancer stemness and metastasis by repressing M2-like tumor-associated macrophages induced Wnt/β-catenin pathway. Life Sci. 2020;256:117925. doi: 10.1016/j.lfs.2020.117925. [DOI] [PubMed] [Google Scholar]
- 75.Dudeck J, et al. Mast-cell-derived TNF amplifies CD8(+) dendritic cell functionality and CD8(+) T cell priming. Cell Rep. 2015;13:399–411. doi: 10.1016/j.celrep.2015.08.078. [DOI] [PubMed] [Google Scholar]
- 76.Xiao Z, et al. Antibody mediated therapy targeting CD47 inhibits tumor progression of hepatocellular carcinoma. Cancer Lett. 2015;360:302–309. doi: 10.1016/j.canlet.2015.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holden JK, Cunningham CN. Targeting the hippo pathway and cancer through the TEAD family of transcription factors. Cancers. 2018;10:81. doi: 10.3390/cancers10030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huh, H. D., Kim, D. H., Jeong, H.-S. & Park, H. W. Regulation of TEAD transcription factors in cancer biology. Cells8, 600 (2019). [DOI] [PMC free article] [PubMed]
- 79.Pobbati AV, Hong W. A combat with the YAP/TAZ-TEAD oncoproteins for cancer therapy. Theranostics. 2020;10:3622–3635. doi: 10.7150/thno.40889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang S, et al. Early growth response 3 inhibits growth of hepatocellular carcinoma cells via upregulation of Fas ligand. Int. J. Oncol. 2017;50:805–814. doi: 10.3892/ijo.2017.3855. [DOI] [PubMed] [Google Scholar]
- 81.Kaya-Okur HS, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291 e289. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tickle, T., Tirosh, I., Georgescu, C., Brown, M. & Haas, B. inferCNV of the trinity CTAT project. https://github.com/broadinstitute/inferCNV (2019).
- 85.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol.38, 1408–1414 (2020). [DOI] [PubMed]
- 87.Aran D, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Finan, C. et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 9, (2017). [DOI] [PMC free article] [PubMed]
- 89.Satpathy AT, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 2019;37:925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stuart T, et al. Single-cell chromatin state analysis with Signac. Nat. Methods. 2021;18:1333–1341. doi: 10.1038/s41592-021-01282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Feng, J., Liu, T. & Zhang, Y. Using MACS to identify peaks from ChIP-Seq data. Curr. Protoc. Bioinforma.Chapter 2, Unit-2.14 (2011). [DOI] [PMC free article] [PubMed]
- 92.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bailey TL, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- 97.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The scRNA-seq data generated in this study are available in the Gene Expression Omnibus (GEO) database under accession number GSE163686 and also available in the Genome Sequence Archive (GSA) database under accession number HRA001112. The scATAC-seq and ChIP-seq data generated from two samples in this study are available in the GSA database under accession number HRA001112. In particular, H3K27ac and H3K4me3 ChIP-seq data for sample HRI137901 can be obtained under GSA accession numbers HRR337427 and HRR337429, respectively. H3K27ac and H3K4me3 ChIP-seq data for sample HRI137902 can be obtained under GSA accession numbers HRR337428 and HRR337430, respectively. scATAC-seq data for samples HRI137901 and HRI137902 can be obtained under accession numbers HRR337425 and HRR337426, respectively. The data in GSA are available under restricted access, access can be obtained by contacting Xiaoqun Wang (xiaoqunwang@ibp.ac.cn). The published datasets used in this study can be downloaded from https://www.brainimmuneatlas.org/ or from GEO under accession number GSE16312047; scRNA-seq data for TAMs from glioblastoma, GSE16641851; scRNA-seq data for TAMs from IDH-mutant high-grade gliomas, GSE8446550; scRNA-seq data for TAMs from glioblastoma and GSE6783549; scRNA-seq data for macrophage from normal brain. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.