SUMMARY
Here, we present an approach to identify N-linked glycoproteins and deduce their spatial localization using a combination of matrix-assisted laser desorption ionization (MALDI) N-glycan mass spectrometry imaging (MSI) and spatially resolved glycoproteomics. We subjected glioma biopsies to on-tissue PNGaseF digestion and MALDI-MSI and found that the glycan HexNAc4-Hex5-NeuAc2 was predominantly expressed in necrotic regions of high-grade canine gliomas. To determine the underlying sialo-glycoprotein, various regions in adjacent tissue sections were subjected to microdigestion and manual glycoproteomic analysis. Results identified haptoglobin as the protein associated with HexNAc4-Hex5-NeuAc2, thus directly linking glycan imaging with intact glycopeptide identification. In total, our spatially resolved glycoproteomics technique identified over 400 N-, O-, and S- glycopeptides from over 30 proteins, demonstrating the diverse array of glycosylation present on the tissue slices and the sensitivity of our technique. Ultimately, this proof-of-principle work demonstrates that spatially resolved glycoproteomics greatly complement MALDI-MSI in understanding dysregulated glycosylation.
Graphical Abstract
In brief
Malaker et al. use a multi-omics approach, including MALDI-MSI and glycoproteomics, to understand the spatial distribution of glycans and their associated glycoproteins in canine glioma.
INTRODUCTION
Canine glioma is a spontaneous non-human intracranial neoplasm that has recently gained attention as a viable preclinical model for comparative oncology (Bentley et al., 2016; Hubbard et al., 2018; Koehler et al., 2018). Compared with neurosphere cultures and patient-derived homogeneous xenografts transplanted on immune-deficient mice, canine glioma offers a lesser “translational gap” with human glioma. Both human and canine gliomas are highly heterogeneous and present similar histopathological, immunological, molecular, and clinical characteristics. In addition, the animals are immunocompetent and they exhibit similar responses to chemotherapy, radiotherapy, and stereotactic radiosurgery (Mitchell et al., 2019). Furthermore, being household pets, dogs are exposed to similar environmental pressures as their owners and, like humans, are also susceptible to various spontaneous malignancies (Hubbard et al., 2018). Canine glioma can thus serve as an intermediate model that can be exploited for preclinical evaluation of new treatments, providing putative candidates that not only are expected to have better chances of passing phase II clinical trials, but also will be accessible to companion animals. Human glioma is a high-grade brain tumor with poor prognosis and, currently, the only two treatment options are FDA approved: temozolamide and bevacizumab (Dickinson, 2014). In an effort to address the lack of treatment options, the Comparative Brain Tumor Consortium has been established to study the molecular, genetic, and histologic relationship between animal and human malignancies to eliminate barriers in the integration of animals in all aspects of brain tumor research (Hubbard et al., 2018). This work is thus expected to contribute to this effort.
Protein glycosylation is a ubiquitous post-translational modification found on over 50% of the proteome. Classical types of protein glycosylation are N- and O-linked, which modify Asn and Ser/Thr residues, respectively (Moremen et al., 2012; Rudd et al., 2015). N-linked glycosylation can occur when an Asn residue is found in the consensus sequon Asn_X_Ser/Thr, where X symbolizes any amino acid except Pro. N-linked glycans have a core structure consisting of 2 β-N-acetylglucosamine residues (GlcNAc2) connected to three mannose units (GlcNAc2-Man3), which can be extended into complex, hybrid, or high-mannose structures. These glycan structures are important in protein folding, stability, and cell-to-cell interactions outside of the cell (Pinho and Reis, 2015). Extracellular O-linked glycosylation is often initiated by an α-N-acetylgalactosamine (O-GalNAc) on Ser/Thr residues and is commonly referred to as “mucin-type O-glycosylation” since it is frequently found on densely glycosylated mucin domains. O-GalNAc can be extended in monosaccharide units to create large, branched structures that are important for cell adhesion, cell polarization, and, again, cell-to-cell interactions in the extracellular matrix (ECM) (Kufe, 2009; Pinho and Reis, 2015). Other types of O-glycosylation include O-linked β-N-acetylglucosamine (O-GlcNAc) (Hart and Akimoto, 2009; Khidekel et al., 2004), O-mannose (Dobson et al., 2013; Larsen et al., 2019, 2017; Sheikh et al., 2017; Vester-Christensen et al., 2013), O-fucose (Moloney et al., 2000), and O-xylose (Li et al., 2017), among others (Darula and Medzihradszky, 2018). While we are only beginning to understand the latter forms of glycosylation, O-GlcNAc has been extensively described as an intracellular signaling molecule involved in crosstalk with phosphorylation (Hart et al., 2011; Leney et al., 2017). Finally, less commonly, GlcNAc has been reported to modify Cys, creating an S-linked GlcNAc structure (Maynard et al., 2016).
Aberrant glycosylation is a classic hallmark of malignant transformation; this observation has been well-documented and extensively reviewed (Bull et al., 2014; Kufe, 2009; Pearce and Läubli, 2016; Pinho and Reis, 2015; Varki et al., 2015; Veillon et al., 2018). The mechanisms that produce these abnormal glycosylation patterns are broad because glycosylation is a non-template-driven process and is instead reliant on expression of various glycosyltransferases, substrate availability (Tachibana et al., 1994), chaperone function (Hanes et al., 2017), and the cellular milieu (Pearce and Läubli, 2016). As such, diverse glycan structures result from these dysregulated systems. For instance, altered branching (Dennis et al., 1987), increased fucosylation (Noda et al., 1998), and upregulated sialylation (Büll et al., 2018) of N-glycans has been linked to several cancerous processes (Thaysen-Andersen et al., 2016), including the progression of oligodendroglioma to glioblastoma (Furukawa et al., 2015). In addition, mucin-type O-glycans are often truncated and have been typically associated with increased cell migration and tumor metastasis, due to a loss of cell polarization (Kufe, 2009; Pinho and Reis, 2015). These shortened O-glycans comprise oncofetal antigens, neoantigens, and altered levels of normal antigens (Kudelka et al., 2020). Finally, one of the most widely observed changes in glycan structures is an upregulation of sialic acid (Varki et al., 2015; Varki and Gagneux, 2012), which limits complement activation (Blaum et al., 2015), engages inhibitory sialic acid-binding immunoglobulin type lectins (Siglecs) (Hudak et al., 2014; Macauley et al., 2014), and reduces attachment of tumor cells to the basement membrane (Varki, 2008). These changes ultimately allow tumor cells to evade the immune system and increase invasion, causing metastasis. Most of the changes listed here are tumor specific or tumor associated, making them viable biomarker targets for glycan- or glycoprotein-based antibody or antibody-conjugated enzyme therapy (Gray et al., 2020; Posey et al., 2016; Xiao et al., 2016).
The importance of glycosylation in cancer and other pathologies has prompted the in-depth examination of the glycome and the glycoconjugates that they modify. Mass spectrometry (MS) is the premier technique to probe the proteome, glycome, and glycoproteome (Rudd et al., 2015; Thaysen-Andersen et al., 2016). Generally, glycopeptides are then subjected to different types of fragmentation (i.e., tandem MS) to glean information about the peptide and attached glycan (Rudd et al., 2015). Some of the most common fragmentation techniques include higher-energy collision dissociation (HCD) (Olsen et al., 2007), electron transfer dissociation (ETD) (Syka et al., 2004), and supplemental activation with ETD (EThcD [Reiding et al., 2018], AI-ETD [Riley et al., 2017]). With these techniques, it is possible to sequence a peptide, ascertain information about the glycan, and (in some cases) localize the glycosylation site (Riley et al., 2020; Shajahan et al., 2020).
Another MS modality, matrix-assisted laser desorption ionization (MALDI) mass spectrometry imaging (MSI), allows for the determination of the spatial distribution of glycans after enzymatic release from their protein conjugates (Briggs et al., 2016; Drake et al., 2017). In this technique, slices from fresh-frozen or formalin-fixed, paraffin-embedded (FFPE) tissues are applied to slides, followed by antigen retrieval, the addition of de-N-glycanase (PNGaseF) to remove N-linked glycans, and coating of the tissue with matrix (e.g., 1,5-dihydroxybenzoic acid or alpha-cyano-4-hydroxycinnamic acid). The images are generated by rastering a laser across the tissue slices, and the ionized glycans are generally analyzed in a time-of-flight or ion cyclotron mass analyzer (Briggs et al., 2016; Drake et al., 2017). One downside of this technique is that N-glycans are identified primarily based on MS1 exact masses, so that the confidence of assignment is relatively low compared with those techniques employing tandem MS. Another drawback is that only N-glycans can be imaged because there is no enzyme that can perform universal release of O-glycans. Despite these drawbacks, the N-glycan analysis provides an estimation of the spatial distribution of the N-glycome and its changes in a tissue-, disease-, and/or cell type-specific manner. MALDI-MSI of N-glycans has been used on ovarian (Everest-Dass et al., 2016), pancreatic (Powers et al., 2014), and hepatocellular (Powers et al., 2015) cancer tissue samples, and has demonstrated marked N-glycan changes in benign versus tumor regions. Furthermore, it has also been used for the generation of a classification model for colon carcinoma tissue microarrays (Powers et al., 2014). Some groups have also achieved in situ derivatization of different sialic acid linages (Holst et al., 2016), sequential PNGaseF/trypsin digestion of the same tissue (Angel et al., 2017; Heijs et al., 2016), and visualization of ECM proteins (Angel et al., 2018). Taken together, MALDI-MSI is a powerful technique to image the N-glycome.
Recent developments in microextraction strategies in our laboratory coupled with MALDI-MSI have allowed us to perform spatially resolved proteomics on various biological samples from fresh-frozen and FFPE tissues (Quanico et al., 2013; Wisztorski et al., 2013). This spatially resolved proteomics strategy is advantageous in that protein digestion and extraction are performed only on a restricted region of interest defined by a prior MALDI imaging experiment on a consecutive section. The strategy is essentially a solid-liquid extraction method following microdigestion, which is done by depositing picoliter quantities of enzyme using a microspotter (Quanico et al., 2017). This directed digestion and extraction is, in essence, a means of concentrating the analyte of interest by performing digestion only in regions where this analyte is present, thereby minimizing the amount of abundant proteins extracted. The maximum spatial resolution that this strategy can attain is highly dependent on the size of the microdigested spot (Wisztorski et al., 2017).
In this work, we combine the power of MALDI-MSI N-glycan imaging with our microscale proteomics technology to better understand dysregulated glycosylation in canine glioma samples. We first used standard N-glycan imaging techniques to image N-linked glycans from the surface of various canine brain tumors, finding that sialylated glycan structures were more common in the tumor and necrotic regions. We confirmed this result using Sambucus nigra (SNA) lectin staining, which selectively stains sialylated glycans. Then, we applied our microscale proteomics strategy to extract glycopeptides from tissue, which we hypothesized could be used to assign peptide and protein conjugates to better understand our N-glycan imaging results. Using this technique, we identified over 400 unique glycopeptides from 30 glycoproteins, including complex and oligomannose N-glycosylated peptides. Several of the N-glycans were also found in the MALDI-MSI experiments, thus providing us with evidence of directly linked N-glycan imaging with intact glycopeptide sequencing. Surprisingly, we also identified other glycoconjugates, including peptides modified by mucin-type O-GalNAc, O-linked GlcNAc, S-linked GlcNAc, and O-mannose. We used this information to demonstrate that there is a significant increase in sialylated O-GalNAc structures in tumor/necrotic regions compared with benign regions, whereas there is significantly less S- and O-GlcNAc peptides in the cancerous regions. Taken together, this proof-of-principle experiment has demonstrated that we can perform both MALDI-MSI and glycopeptide analysis on FFPE tumor tissue to better understand the glycoproteomic changes in malignant transformation.
RESULTS
MALDI-MSI shows that sialylated glycans are enriched in necrotic regions
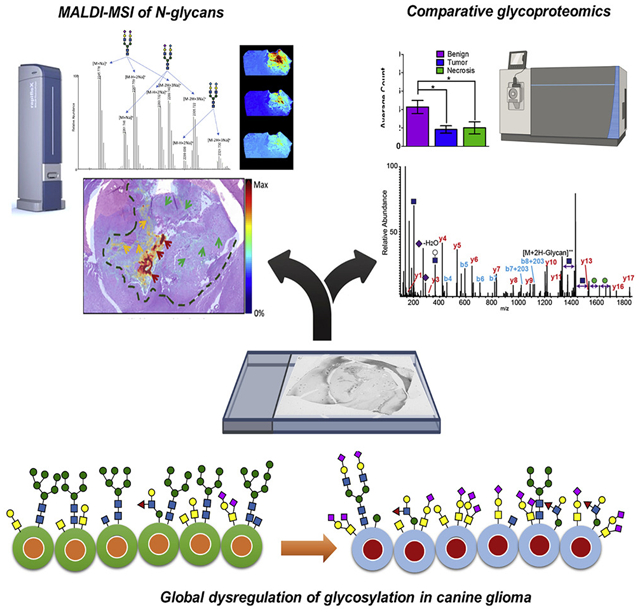
Figure 1A shows the ion distribution of the summed intensities of the sodiated and potassiated sialic acid-containing N-glycan HexNAc4-Hex5-NeuAc2 across different glioma biopsies. Superposition of the summed ion distributions with the H&E-stained section shows that on both anaplastic oligodendroglioma and glioblastoma samples, this glycoform is present in both tumor and necrotic regions but is highly abundant in the latter (Figure 1B). In the anaplastic oligodendroglioma section (Figure 1B, left panel), the distribution extends through the pseudo-glomerular vessels present along the margins of the tumor (indicated by dark blue arrows), suggesting its possible relationship with vigorous and abnormal angiogenesis associated with high-grade glioma. However, regions where both tumor and necrosis are present only show weak distribution (indicated by green arrows). The same observation applies to the glioblastoma section shown in Figure 1B, right panel. In this case, however, the glycoforms are also detected in the pseudo-palisading (indicated by yellow arrows). Magnified views of the H&E-stained images are shown in Data S1. In addition, a plot of the normalized total ion current intensity from spectra taken in various tissue regions, defined based on the H&E-stained images, confirms the upregulation of this glycoform in cancerous regions (Figure 1C).
Figure 1. MALDI-MSI of N-glycans.
(A) Summed ion images of Na+ and K+ adducts of HexNAc4-Hex5-NeuAc2 on canine glioma biopsies. Images were generated using SCiLS Lab.
(B) Superposition of MALDI-MSI glycan images with H&E-stained adjacent sections. Arrows and dashed lines indicate regions annotated by the pathologist. Legend: red, green, yellow, and dark blue arrows represent necrotic regions, tumor regions, pseudo-glomerular vessels, and pseudopallisading necroses, respectively. Dashed lines indicate tumor margins.
(C) Normalized intensity of total ion signals of the combined Na+ and K+ adducts of HexNAc4-Hex5-NeuAc2 species from necrosis, benign, and tumor samples. The boxplot points correspond to relative intensities of m/z 2245-related peaks taken from each spectrum in all images acquired.
(D) PCA loadings of detected glycans in MSI analysis.
(E) MSn spectra of HexNAc4-Hex5-NeuAc2 confirming its structure.
The acquired images were exported in SCiLS and principal-component analysis (PCA) was performed. ROIs where spectra were taken for PCA were defined using histological annotations from the H&E-stained optical images. The loadings plot results (Figure 1D) identified m/z intervals attributed to the aforementioned Na+ and K+ adducts of the biantennary disialylated glycoform HexNAc4-Hex5-NeuAc2 (m/z 2,245, 2,261, 2,267, 2,283, and 2,299) as well as the monosialylated glycoform HexNAc4-Hex5-NeuAc and its K+ adduct (m/z 1,954 and 1,970). The loadings plot also identified m/z enriched in tumor samples, including the fucosylated complex glycoforms HexNAc5-Hex3-Fuc (m/z 1,688) and HexNAc5-Hex3 (m/z 1,542). On the other hand, notable glycoforms enriched in the benign regions include the high-mannose-type HexNac2-Hex5 and its potassiated form (m/z 1,257 and 1,273).
To confirm the glycoform assignments found using GlycoMod searches, we repeated the mass measurements on an LTQ Orbitrap XL instrument equipped with a MALDI source. Collision-based MSn fragmentation was performed where possible (Figure 1E). MS2 of the precursor ion of HexNAc4-Hex5-NeuAc2 ([M + Na]+, m/z 2,245) led to sequential losses of the glycan residues, starting with the first sialic acid producing the Y6α ion with mass similar to HexNAc4-Hex5-NeuAc (m/z 1,954.596, Figure 1E, top panel). This was followed by the loss of the second sialic acid, yielding the Y6α,β ion with a mass similar to HexNAc4-Hex5 (m/z 1,663.580, middle panel). MS4 then led to loss of the terminal GlcNAc (m/z 221.088) yielding the Y6-HexNAc-ol ion. Structures of other glycoforms were also partially elucidated using their MSn spectra and are shown in Data S2, thus confirming their glycan identities as predicted by GlycoMod. The list of all glycolforms detected in the glioma samples is provided in Data S3. In addition, we have compared our results to two studies that performed structural N-glycan information to create a high-confidence glycan list (Briggs et al., 2016; Holst et al., 2017).
SNA lectin staining reveals sialic acid-containing glycans in other tissue regions
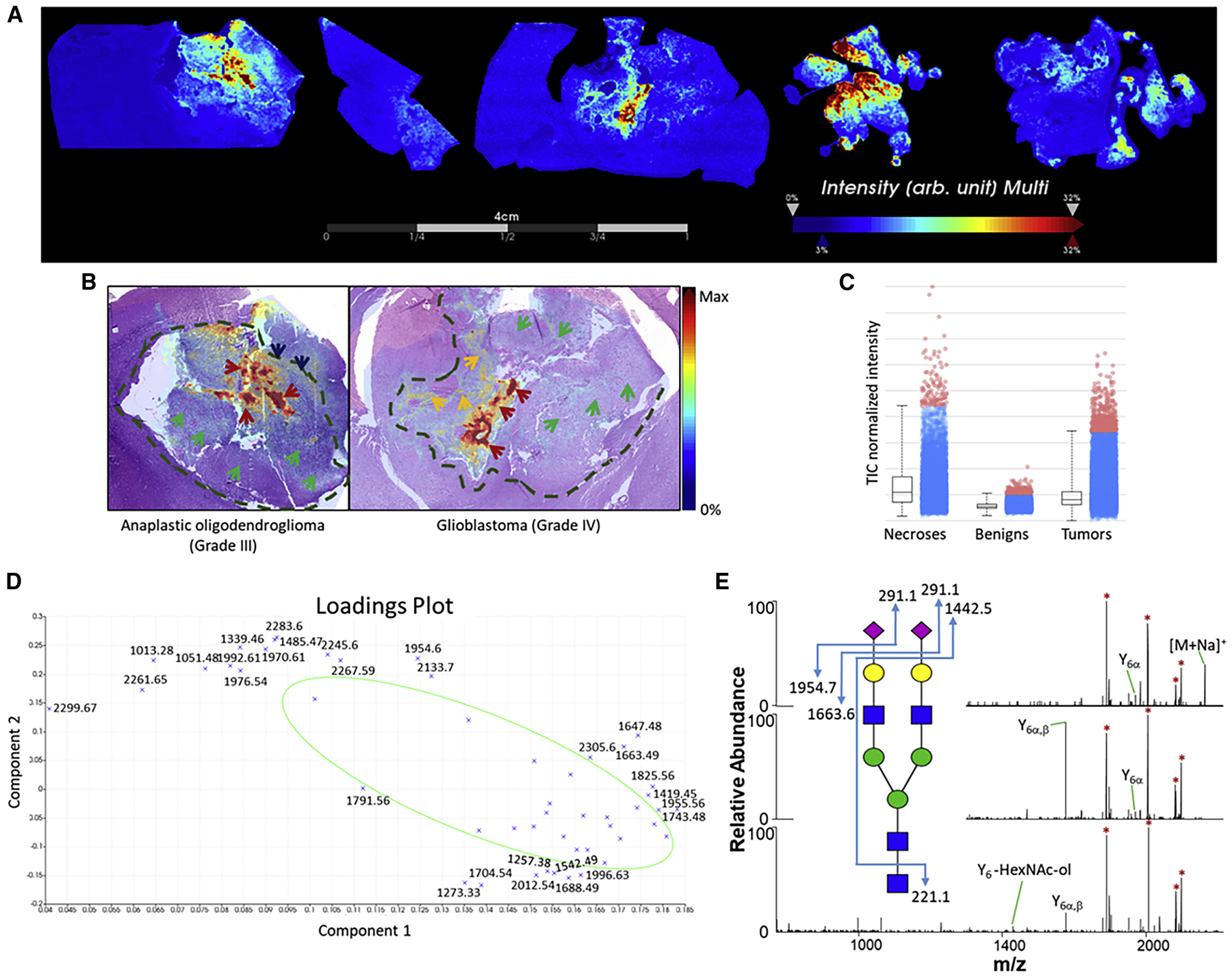
To further verify the distribution of sialylated glycan structures in the necrotic region(s), SNA lectin staining of consecutive slices was performed. Results for all of the biopsies are shown in Data S4. Confocal images taken of the different regions of WHO grade III (Figure 2A) and grade IV (Figure 2B) biopsies demonstrated that sialic acid was present in necrotic zones. Tumor regions and regions marked by the presence of both tumor and necrosis showed positive staining, concomitant with findings from the MALDI imaging experiments. One glioblastoma sample (Figure 2B, bottom panels) showed very intense staining in the tumor region, although the MALDI-MSI only showed moderate distribution of sialic acid-containing glycans in this region. Benign regions, particularly the corpus callosum (Figure 2A, bottom panels) and choroid plexi (Data S4I), likewise showed intense staining, although no sialic acid-containing N-glycans were detected in these zones using MALDI-MSI. SNA stains for 2,6-linked sialic acids and does not discriminate with regard to their origin. Thus, we suspected that the abundant SNA signal was derived from other sources, such as O-linked glycans or glycosphingolipids (Jennemann et al., 1990). To investigate this hypothesis, we turned to the microscale proteomics technique that our laboratory developed.
Figure 2. SNA staining of tissue slices.
Confocal fluorescent images taken at pathologist-annotated regions present in oligodendroglioma (WHO grade III) (A) and glioblastoma (WHO grade IV) (B) samples. H&E stains (left) present for comparison with fluorescent images (right). The sections were incubated in 1% BSA (w/v) in 300 μL of PBS for 30 min at room temperature, then incubated in 10 μg/mL of SNA lectin for 2 h. Slides were then rinsed for 10 min 3× with 1% BSA in PBS, then incubated in approximately 300 μL of DAPI for 20 min. Confocal images were obtained using a fluorescence microscope (Leica Biosystems). Images taken from separate adjacent sections that were incubated without the lectin serve as controls. Scale bars, 100 μm.
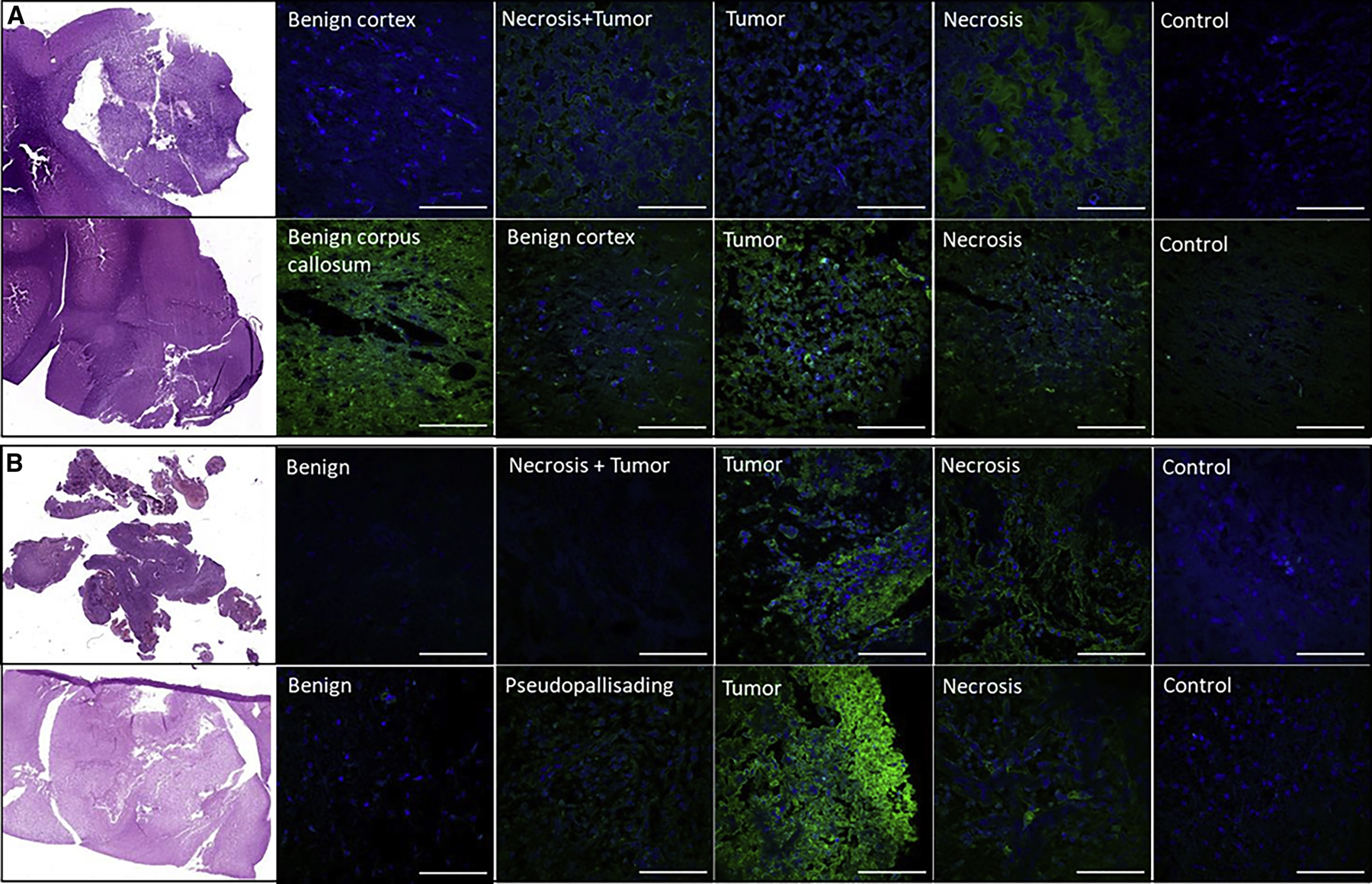
Spatially resolved proteomic analysis reveals hierarchical clustering of benign and cancerous tissue
Regions of interest (ROIs) identified using both N-glycan MALDI imaging and SNA lectin staining were subjected to microdigestion by depositing picoliters of trypsin using a microspotter (Figure 3A). We began with 8 biopsy tissues, which resulted in the microdigestion of 11 benign, 4 margin, 11 tumor, and 7 necrosis samples. The samples were subjected to liquid chromatography-tandem MS (LC-MS/MS) on a Thermo Orbitrap Fusion instrument with HCD, and the raw files were searched using MaxQuant (unmodified peptides) and Byonic (glycopeptides). Hierarchical clustering of the protein groups identified by MaxQuant using the Andromeda search engine and quantified using LFQ intensities showed that samples from the benign regions cluster distinctly from those obtained from the tumor and necrotic zones (Figure 3B), whereas samples taken from the margins do not form a distinct cluster. The overexpressed proteins of the “benign” cluster were associated with pathways related to synaptic processes, such as synaptogenesis, synaptic plasticity, synaptic vesicle endo- and exocytosis, transport, docking and fusion, and long-term synaptic potentiation and depression (Figure 3C). On the other hand, overexpressed proteins in the “tumor and necrosis” cluster were associated with cancer processes, such as cell proliferation, growth, division, metastasis and migration, as well as RNA splicing, apoptosis, and neo-plastic growth (Figure 3D).
Figure 3. Spatially resolved proteomic analysis of tissue slices.
(A) Zoomed optical images of sample microspots from each ROI after microdigestion using the CHIP-1000 printer. Scale bars, 200 μm.
(B) Hierarchical clustering of protein identifications with ANOVA-significant (p < 0.01) differential expression across the different ROIs, using the LFQ intensities calculated by MaxQuant.
(C) Selected overrepresented pathways in the “benign” cluster (pink) (B).
(D) Selected overrepresented pathways in the “necrosis and tumor” cluster (blue) (B).
Spatially resolved glycoproteomic analysis links MALDI glycan imaging with intact N-glycopeptide identification
The microdigested areas were then extracted with different solvent mixtures to maximize peptide extraction and again subjected to LC-MS/MS on a Thermo Orbitrap Fusion instrument. Glycoproteomic analysis identified the glycopeptide MVSHHnLTSGATLINEQWLLTTAK from haptoglobin bearing the HexNAc4-Hex5-NeuAc2 glycan (Figure 4A). Non-glycosylated peptides of haptoglobin were also detected, both in necrotic as well as other regions, but the glycosylated peptide was only observed in necrotic regions. Unfortunately, this was the only sialylated N-glycopeptide that we observed. Other N-glycopeptides detected include those bearing high-mannose motifs from ECM chondroitin sulfate proteoglycans, such as neurocan (AnATLLLGPLR, Figure 4B), members of the immunoglobulin superfamily, such as neurofascin (IgCAMs), limbic system-associated membrane protein (LAMP, IgLONs), contactin 1 (contactin CAMs), Thy-1 cell surface antigen (CD90), and neuroprotective factor prosaposin. Interestingly, while overall numbers of N-glycopeptides were similar between various regions (Figure 5A), the HexNAc2-Hex5 glycan was significantly enriched in the benign regions (66 glycopeptides) when compared with tumor (18 glycopeptides, **p < 0.01) and necrotic regions (25 glycopeptides, *p < 0.05; Figure 5B). This correlated with our MALDI-MSI data, which demonstrated a marked increase in HexNAc2-Hex5 glycans in benign regions. Finally, we observed several glycans modified by fucose, as demonstrated in Figure 4C, with a glycopeptide from the antimicrobial protein myeloperoxidase. No significant change between benign and cancerous fucosylation was observed. Additional annotated spectra can be found in Data S5 and a total list of all N-glycopeptides is provided in Data S6. All raw files, Byonic search results, and hand-annotated Excel files can be found in PRIDE : PXD025537.
Figure 4. Spatially resolved glycoproteomics identifies several types of glycosylation.
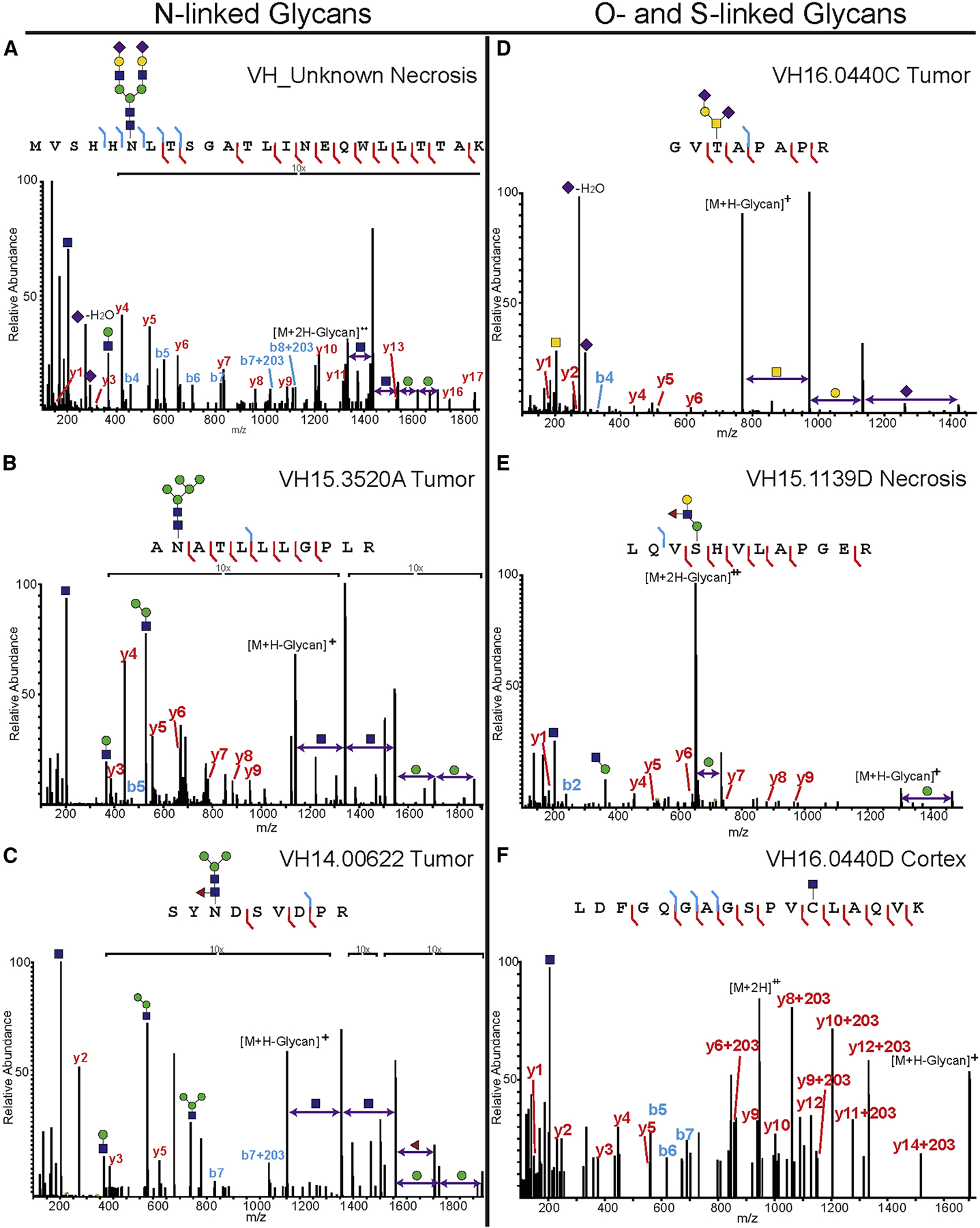
All microdigested samples were subjected to LC-MS/MS analysis on a Thermo Orbitrap Fusion Tribrid, and peptides were fragmented using HCD. Note that, in the case of O-glycosylation, the site of modification is only localized because there was only one possible site of modification. Spectra were annotated manually to confirm glycan composition, peptide sequence, and (if possible) site localize the glycan.
(A) Haptoglobin peptide MVSHHnLTSGATLINEQWLLTTAK bearing the complex, disialylated glycan HexNAc4-Hex5-NeuAc2.
(B) Peptide from neurocan, AnATLLLGPLR, modified with an N-linked high-mannose (HexNAc2-Hex5) structure.
(C) Myeloperoxidase peptide SYnDSVDPR modified with a fucosylated paucimannose N-glycan (HexNAc2-Hex3-Fuc).
(D) Protein tyrosine phosphatase receptor Z1 peptide GVtAPAPR modified with a disialylated, core 1 structure (HexNAc-Hex-NeuAc2).
(E) Another protein tyrosine phosphatase receptor Z1 peptide, LQVsHVLAPEGR, modified with an extended O-mannose glycan (Hex2-HexNAc-Fuc).
(F) Bassoon presynaptic cytomatrix protein peptide LDFGQGAGSPVcLAQVK was modified by an S-linked GlcNAc. For additional spectra and peptide information, please see Data S5 and S6.
Figure 5. Statistical analysis of glycopeptide changes between samples.
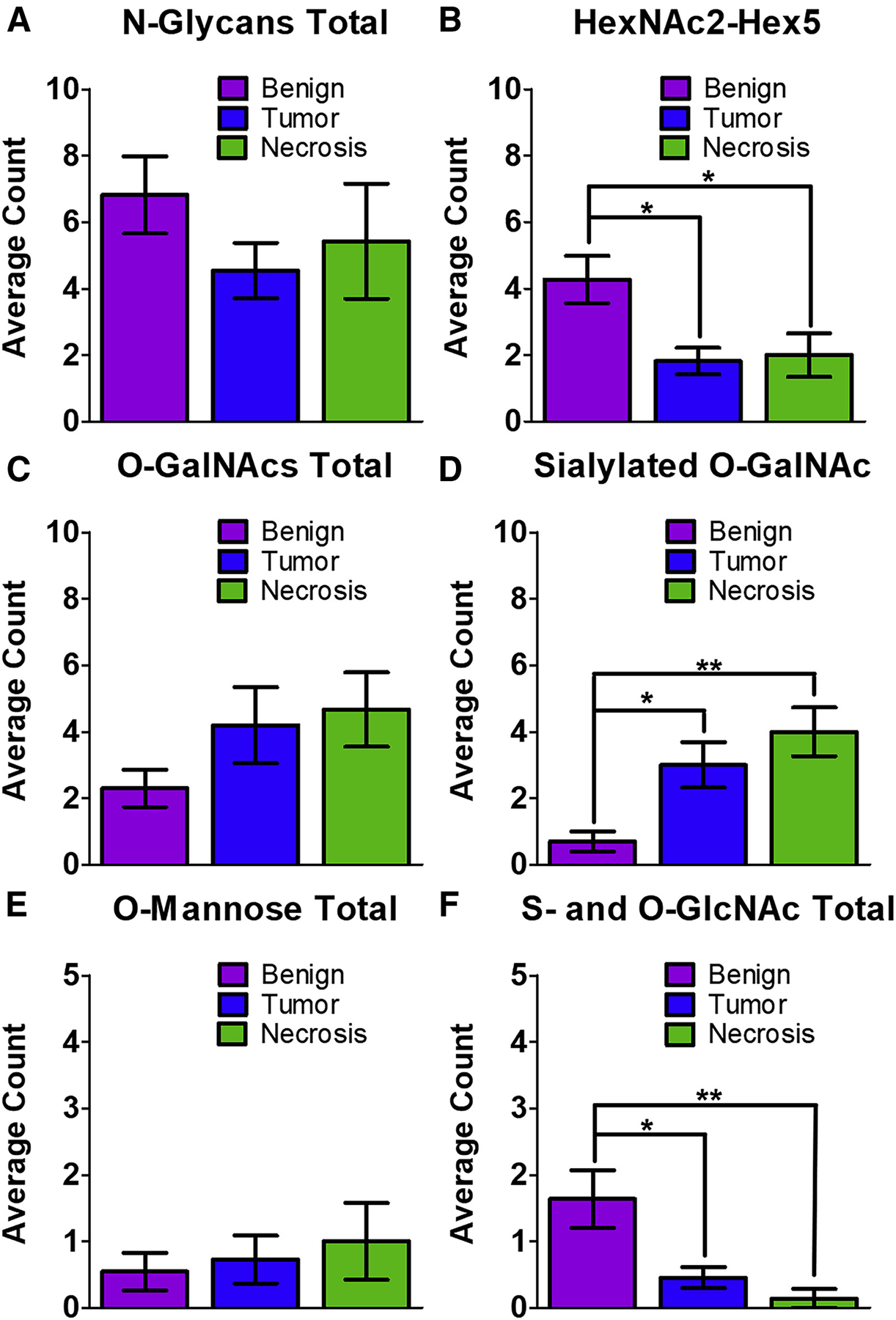
For all samples, a three-way ANOVA test was performed in GraphPad PRISM, mean ± standard error of the mean, *p < 0.05, **p < 0.01.
(A) The total number of N-glycopeptides was not found to be significantly different between the three types of samples.
(B) The number of glycopeptides modified by HexNAc2-Hex5 was found to be significantly higher in the benign samples compared with both tumor and necrotic regions.
(C) The total number of O-GalNAcylated peptides was not was not found to be significantly changed between the three regions.
(D) The number of sialylated O-GalNAc glycopeptides was significantly increased in tumor and necrotic regions when compared with benign regions.
(E) The total number of O-mannosylated peptides was not was not found to be significantly changed between the three regions.
(F) S- and O-linked GlcNAcylated peptides were significantly decreased in both tumor and necrotic regions when compared with benign regions.
Identification of diverse glycopeptides from spatially resolved glycoproteomic experiments
This work was initially focused on finding glycoconjugates modified by the same N-glycans that we observed in MALDI-MSI experiments. However, it was clear after SNA staining and the N-linked glycopeptide analysis that there were other glycosylated species in the samples. Thus, we re-analyzed the data for O-linked mucin-type glycans, which cannot be imaged by MALDI-MSI due to the harsh chemical conditions necessary to release them. We detected a surprisingly large number of O-GalNAcylated peptides in the samples, all of which can be found in Data S6. Most of the glycans that we detected were tumor-associated carbohydrate antigens, such as the T antigen (GalNAc-Gal) and the sialylated T antigen (GalNAc-Gal-NeuAc). One example of this is shown in Figure 4D; the glycopeptide GVtAPAPR from protein phosphatase receptor type Z1 (PTPRZ1, also called phosphacan) was modified by a disialyl-T antigen (GalNac-Hex-NeuAc2) and was found in five of eight tumor biopsies, in three of five necrosis biopsies, and in all margin regions. The same glycopeptide was also detected with the T antigen or sialyl-T antigen in some tissue regions (Data S6). Mono- and disialylated core 1 O-glycans were highly expressed on ECM lecticans, including brevican, versican, and neurocan, and were also observed in fibronectin, fibrinogen, and apolipoprotein E. While we did not detect a statistically significant change in O-GalNAcylated peptides between the benign and tumor/necrotic sections, we did observe a significant increase in sialylated O-GalNAc peptides in the tumor (**p < 0.01) and necrotic (**p < 0.01) regions (Figures 5C and 5D). Finally, while many of the proteins we observed have been noted previously as O-glycoproteins, neurocan, and oligodentocyte myelin glycoproteins had not.
Further analysis of the glycoproteomic data revealed the presence of O-mannosylated glycopeptides, which is perhaps unsurprising because O-mannose accounts for up to 30% of brain O-glycans (Dobson et al., 2013; Larsen et al., 2017; Sheikh et al., 2017). An example is shown in Figure 4E, which displays an annotated glycopeptide spectrum from PTPRZ1. The sequence LQVsHVLAPEGR bears a fucosylated core M1 type O-mannose (Hex2-GlcNAc-Fuc) that contains a Lewis X structure. This glycopeptide was observed in 3 of 11 benign samples, 2 tumor and necrosis samples, and 1 margin sample. PTPRZ1 was previously identified as a substrate for O-mannosyl glycosylation by O-mannose β−1,2-N-acetylglucosaminyltransferase (POMGnT1) (Dwyer et al., 2012). We also observed a peptide from cadherin 13, another known target of O-mannosylation (Vester-Christensen et al., 2013). The three other proteins that we found to be O-mannosylated have heretofore never been described as O-mannosylated. These proteins include albumin, collagen alpha-1 chain, and KH-type splicing regulatory protein (Data S6). We did not detect a statistically significant change in O-mannosylated peptides between benign and tumor/necrotic regions (Figure 5E).
Finally, intracellular O-GlcNAc glycopeptides were also observed, and were detected mainly on synapsin-1 and spectrin beta chain. Synapsin-1 was detected only in the benign regions, while spectrin peptides were observed in the benign, margin, and tumor regions. In addition, a glycopeptide (LDFGQGAGSPVcLAQVK) bearing the rare S-linked glycan attached to a Cys residue, was detected predominantly in benign regions (7 out of 11 samples, Figure 4E). This glycopeptide is part of the bassoon presynaptic cytomatrix protein, a structural protein found in the ribbon synapse, and is involved with presynaptic vesicle release (Rutherford and Pangršič, 2012). The same S-GlcNAcylated glycopeptide, as well as others from bassoon, were described recently by Burlingame and co-workers in the mouse synaptosome (Maynard et al., 2016). Interestingly, and contrary to several other reports, we observed a significant decrease in O- and S-linked GlcNAcylation in the tumor (*p < 0.05) and necrotic (**p < 0.01) regions when compared with benign tissues (Figure 5F). Taken together, these data demonstrate that our spatially resolved glycoproteomics technique can (1) link MALDI-MSI glycan imaging experiments to the associated glycoconjugate (Figures 4A–4C), (2) allow for the detection of a wide array of glycopeptide modifications (Figure 4), and (3) distinguish diverse and significant glycan changes in benign, tumor, and necrotic regions (Figure 5).
DISCUSSION
Malignant glioma is the most common primary tumor in the central nervous system and has an extremely poor prognosis. Canine glioma is a good model system for human brain cancer because of their similar characteristics, including spontaneous origin, high level of invasiveness, and poor clinical outcomes (Hubbard et al., 2018; Mitchell et al., 2019). Since glycosylation is an aberrant feature of all cancers but has been studied infrequently in glioma, this work was initially aimed at developing a MALDI-MSI workflow to visualize N-linked glycans from canine glioma FFPE tissue. We based our protocol on several published studies (Briggs et al., 2016; Drake et al., 2017), which allowed us to easily attain this goal. We noted that a biantennary complex N-linked glycan (HexNAc4-Hex5-NeuAc2) was upregulated in necrotic regions, and a high-mannose N-linked glycan (HexNAc2-Hex5) was enriched in benign regions. We note that we did not include NeuGc in our analyses because it is known to be excluded from rat and mouse brain (Davies and Varki, 2013). That said, it is possible that some of the K+ adducts (+15.9740 with respect to Na+ species) we assigned could be NeuGc species (+15.9949 with respect to Na+ species), and that our methods were not sensitive enough to distinguish between them.
Although mapping of the spatial distribution of N-glycans using MALDI-MSI is useful in many aspects, the information that it can currently provide is limited. By nature of the technique, the N-glycans are removed from the proteins that they modify, thus losing the intact N-glycopeptide/protein information. In an effort to address this limitation, Heijs et al. (2016) developed a successive trypsin protocol after an N-glycan imaging experiment to determine which peptides have been deamidated after PNGaseF treatment. This provided insight into which proteins had been de-N-glycosylated; however, it does not directly link the detected N-glycans with their protein conjugates. This information can only be obtained if the intact glycopeptide itself has been detected and sequenced. In addition, the technique is limited to N-glycans, since these are the only glycans that can be easily and enzymatically liberated from the protein conjugate to be visualized by MALDI-MSI.
Thus, we reasoned that our spatially resolved proteomics method, guided by MALDI-MSI glycan imaging, could potentially allow us to explain the global N-glycosylation changes observed in glioma histological specimens. In these experiments, the importance of spatial localization has to be emphasized. This is exemplified by haptoglobin, where many non-glycosylated peptides were found in all of the tissue sections we studied. However, the glycopeptide MVSHHnLTSGATLINEQWLLTTAK (N107 modified by HexNAc4-Hex5-NeuAc2) from this protein was only detected in the necrotic region, which is also where the glycan HexNAc4-Hex5-NeuAc2 was found to be upregulated in MALDI-MSI experiments. Taken together, we have demonstrated an experiment that directly links glycan imaging with intact glycopeptide identification. Importantly, haptoglobin is a homodimer involved in the scavenging of iron Fe(III) and haptoglobin glycosylation is becoming a major target in cancer research (Zhang et al., 2016). Canine haptoglobin contains three putative N-glycosylation sites and shares conserved sequences with other mammals, including humans. Specifically, N107 of canine haptoglobin shares identical flanking amino acids to N184 of human haptoglobin, which has also been shown to be modified by biantennary complex N-glycans (Fujimura et al., 2008; Zhang et al., 2016). Unfortunately, this was the only sialylated N-glycopeptide that was detected in our glycoproteomic analyses, likely because of the liable nature of sialic acid. Future experiments will include a derivatization step to stabilize and differentiate α-2,3- and α-2,6-linked sialic acids.
Conflicting results of SNA lectin staining with N-glycan MS imaging suggested that other types of glycans could be present on the tissues, since SNA lectin does not discriminate between 2,6-NeuAcs derived from N- and O-glycans. We note that our technique is currently unable to probe for glyco(sphingo)lipids, which could have also contributed to the difference in SNA staining (Jennemann et al., 1990). But with regard to proteins, in addition to performing N-glycopeptide searches, the raw files were exhaustively searched for other potential glycopeptides that were missed by the initial search, and these were manually annotated. Results revealed a diverse set of glycopeptides, including intracellular O-GlcNAc, extracellular O-linked mucin-type glycans, O-mannose, and, surprisingly, even the rare S-GlcNAc. The diversity of the structures detected highlights the importance of using complementary tools to examine canine glioma in addition to N-glycan imaging, and also emphasizes the need for development of other glyco-hydrolases for this approach.
More importantly, the on-tissue spatially resolved glycoproteomics results provided insight into dysregulated glycosylation in glioma. For instance, brevican is the most abundant lectican in the central nervous system and is known to play a role in glioma invasiveness and cell motility (Hu et al., 2008; Lu et al., 2012). In particular, it has been shown that brevican has two glioma-specific isoforms—B/bsia and B/bΔg—that are generated by differential glycosylation and are absent from normal adult brain. While B/bΔg is an underglycosylated proteoform, B/bsia is an oversialylated proteoform expressed by half of the high- and low-grade gliomas that the authors analyzed (Viapiano et al., 2005). Interestingly, though, the authors did not investigate which sites may be modified by the sialylated glycans, leaving it ambiguous as to whether the glycosylation was N- or O-linked. In this study, we report four peptides from brevican modified by O-GalNAc glycans, several of which are modified by sialic acid (Data S6). We did not find any brevican peptides modified by any other type of glycosylation. Thus, our data suggest that the B/bsia proteoform may result from an increase in sialylation of O-GalNAc mucin-type glycosylation.
We also detected several other members of the hyaluronan-binding chondroitin sulfate proteoglycan family (of which brevican is a member), including phosphacan (a splice variant of PTPRZ1), neurocan, and versican. In particular, neurocan and phosphacan bind to neurons and are potent inhibitors of neuronal and glial adhesion. As such, it has been established that the upregulation of both neurocan and phosphacan have negative prognostic implications in glioblastoma (Sim et al., 2009). However, the in-depth impact of their glycosylation patterns remains to be elucidated, especially in the context of glioma. To the best of our knowledge, neurocan was previously unknown to be modified by mucin-type O-glycans. Here, we show that neurocan is modified on at least three different sites by O-GalNAc glycans, and that the majority of the glycans found in tumor and necrotic regions bear sialic acid. As increased sialic acid is linked to increased invasion and metastatic potential, in part due to electrostatic repulsion, it follows that increased sialylation of these proteoglycans could be a mechanism for the characteristically high invasion of glioma (Pearce and Läubli, 2016).
On the other hand, O-GalNAcylation and O-mannosylation of phosphacan has been reported using the SimpleCell technology developed by Clausen and colleagues (Larsen et al., 2019; Steentoft et al., 2013). We corroborate their results in a glioma system, demonstrating that phosphacan is modified in several locations by various O-GalNAc and O-mannose glycans. Defects in O-mannosylation lead to abnormal neuronal migration, and have been associated with a range of muscular dystrophies collectively called a-dystroglycanopathy (Dobson et al., 2013). α-Dystroglycan also forms complexes with ECM glycoproteins particularly laminin, and aberrant O-mannosylation of fully formed a-dystroglycan due to the silencing of like-acetylglucosaminyltransferase, in conjunction with altered integrin expression and regulated ECM degradation, have been reported to contribute to the increased metastatic potential of epithelial cells (de Bernabé et al., 2009).
Finally, S-linked GlcNAcylation is a recently discovered type of glycosylation, and has been reported on a limited number of proteins, including bassoon presynaptic cytomatrix protein. This protein was found in bacterial glycopeptides (Stepper et al., 2011) and mouse and rat synaptosomes (Stepper et al., 2011). Our proteomic data revealed the presence of unmodified bassoon peptides in almost all samples from different regions, but our glycoproteomics analysis demonstrates that the S-GlcNAc glycopeptide was primarily detected in benign regions, and was rarely detected in tumor or necrotic regions. The canine peptide sequence is conserved across mammalia and shares 94% similarity with the human sequence. Bassoon is present in the ribbon synapse and acts as a scaffold that anchors the synaptic ribbon and vesicles to the presynaptic cytomatrix. Recent reports suggest that bassoon has other functions in the cytomatrix and may contribute to synaptic plasticity. The third coiled coil (CC3) of bassoon binds to CtBP1, a transcriptional co-repressor that can be shuttled from the presynaptic compartment to the nucleus during increased neuronal activity. The S-linked glycopeptide LDFGQGAGSPVcLAQVK is located in CC3. Mediation of transcription by glycosylation is a well-known form of epigenetic regulation; thus, it would be interesting to investigate whether S-linked glycans are used for the same function in cancer. If so, they may serve as more selective targets for treatment due to the rarity of S-GlcNAc expression.
Here, we demonstrated the utility of spatially resolved glycoproteomics in complementing MALDI-MSI N-glycan imaging. We first showed that PNGAseF-released N-glycans have differential expression in benign versus cancerous regions using MALDI-MSI on canine glioma FFPE tissues. We then subjected the separate regions to microdigestion followed by LC-MS/MS and exhaustive data analysis to show that: (1) intact N-glycopeptides linked MALDI-MSI experiments to the associated glycoprotein, (2) a wide range of glycopeptide modifications could be identified, and (3) we could distinguish statistically significant glycan changes in benign, tumor, and necrotic regions. We note that this proof-of-principle experiment can be applied to any other FFPE bio-banked samples to identify diagnostic or prognostic indicators of various diseases. We envision that our MALDI N-glycan imaging and spatially resolved glycoproteomics workflow will find use in identifying targets for therapeutic intervention.
STAR★METHODS
RESOURCE AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michel Salzet (michel.salzet@univ-lille.fr).
Materials availability
This study did not generate any new materials.
Data availability
This study did not generate any new unique reagents. The accension number for the MALDI-MSI data files reported in this paper is PRIDE : PXD025826. The accension number for all glycoproteomic raw files and search results is PRIDE : PXD025537.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The study was performed with the approval and in accordance with the guidelines of the Oncovet ethical committee. Biopsies were taken from dog patient samples at the Oncovet Clinic with the approved consent of their owners. The biopsies were immediately subjected to formalin-fixed, paraffin-embedding using standard protocols. An overview of the characteristics of dog patients and collected biopsies can be found in Data S7.
METHOD DETAILS
MALDI-MSI
Sections (8 mm thick) were taken from the FFPE blocks using a microtome (Leica Biosystems, Nanterre, France) and mounted on indium/tin oxide-coated slides (ITO, LaserBioLabs, Sophia-Antipolis, France). The ITO slides were pre-treated by pipetting 1.5 mL of polylysine solution on the conductive surface and incubating at room temperature for 5 min. The treatment was performed twice and dried under a heat gun then dipped in HPLC water. The mounted sections were heated at 60°C on a slide heater for 1 h. While the slides were still hot, the sections were dewaxed in xylene and rehydrated following standard procedures (Angel et al., 2018). The slides were then subjected to antigen retrieval by incubating in 20 mM Tris-HCl (pH = 9) at 95°C for 1h, followed by rinsing in HPLC water. PNGaseF (New England Biosystems) was subjected to dialysis by pipetting 40 μL of PNGaseF onto a PVDF membrane at set on top of 200 mL of water. The buffer exchange was allowed to proceed for 2 h at RT. The PNGaseF was then rehydrated and sprayed onto tissue slices at a rate of 10 μL/min for a total of 15 layers. The slides were placed in a humidified chamber and left overnight at 37°C. After the reaction, DHB matrix was sublimated onto the sections at 140 °C for 10 min, which leads to an average of 0.424 mg matrix deposited per cm2 based on three independent measurements.
MS imaging of tissue slices was performed using a RapiFlex MALDI TOF instrument (Bruker Daltonics, Bremen, Germany). Images were acquired at 70 μm resolution scanning at m/z 700–3,200 with the Smartbeam 3D laser firing at a frequency of 10 kHz. Each spectrum was recorded after accumulating 1,000 laser shots per spot. The laser ablation pattern was set at M5 at a 35 × 35 mm scan range. The ion source voltage was set to 19.984 kV, while the PIE and lens were at 2.608 and 12.366 kV, respectively. The reflectors were set at 20.663, 1.112 and 8.558 kV. Matrix suppression by deflection up to m/z 240 was activated. The reflector detector gain and sampling rate were kept constant on all imaging acquisitions. Additionally, high-resolution MS1 spectra were acquired using a MALDI LTQ orbitrap XL instrument (ThermoFisher Scientific, Bremen, Germany) operated at 30,000 FWHM at m/z 400. Glycans were detected as sodiated or potassiated adducts in positive mode. Spectra were acquired directly on-tissue and were averaged from 10 scans with each scan composed of 1 μscan acquired at 1 μscan/step and 10 laser shots. All MALDI MSI data have been deposited to the ProteomeXchange Consortium via PRIDE: PXD025826.
Following intact MS1 analyses, the Glycomod tool (https://web.expasy.org/glycomod/) was initially used to predict the structure and composition of the observed N-glycans from the full MS spectra recorded at high mass accuracy, with the searches performed at 10 ppm mass tolerance. All monosaccharides were allowed except for NeuGc, which is known to be excluded from rat and mouse brain.(Davies and Varki, 2013) Only the GlycoMod hits that were listed in “GlyConnect” were considered for further analysis. With putative glycan structure information in-hand, the MS images were uploaded on SCiLS Lab version 2016b (Bruker Daltonics). The baseline was calculated using the top hat method. The ion distributions (as sodiated or potassiated adducts) of the glycans were mapped by plotting the signal intensities in all spectra normalized against the total ion count followed by application of weak hotspot removal. These ion distributions were plotted at ±0.250 Da m/z intervals, and the intervals were used for peak alignment and further analysis. Optical images of the H&E-stained adjacent sections were uploaded to the software and co-registered with the corresponding MS images. These H&E images were used to refine the regions of interest (ROIs) in each tissue section where spectra were extracted for Principal Component Analysis (PCA). ROIs corresponding to benign, tumor and necrotic regions, where possible, were taken from each tissue section. The ROIs are shown in Data S8. PCA was performed using the first 5 components, corresponding to approximately 90% of the variance explained, with the first component accounting for more 50% of the variance (Data S8).
MSn
To confirm the glycan structure predicted by GlycoMod, PNGaseF-released samples were subjected to MSn analysis using a MALDI LTQ orbitrap XL instrument (ThermoFisher Scientific, Bremen, Germany) operated at 30,000 FWHM at m/z 400. Glycans were detected as sodiated adducts in positive mode. Spectra were acquired directly on-tissue and were averaged from 10 scans with each scan composed of 1 μscan acquired at 1 μscan/step and 10 laser shots. MSn spectra were acquired directly on tissue using a MALDI LTQ Orbitrap instrument after images had been obtained. The MALDI LTQ Orbitrap XL is equipped with a commercial N2 laser (LTB Lasertechnik, Berlin, Germany) operating at λ = 337 nm with a maximum repetition rate of 60 Hz. The hybrid configuration replaces the heated capillary of the electrospray source with a q00 that sends packets of ions into a linear trap for collision-induced fragmentation (CID), with the fragment ions then being concentrated in a C-trap and transferred to the orbitrap for high-resolution mass analysis. The maximum energy per pulse was set to 12 μJ. Precursor ion isolation was performed using an isolation window between ±1 and ±3 Da and the fragments scanned with a maximum accumulation time of 120 ms. Succeeding MSn of the daughter ions were performed with a maximum accumulation time of 180 ms. External calibration was performed using the ProteoMass MALDI Calibration Kit (Sigma-Aldrich, St. Quentin-Fallavier, France).
H&E and lectin staining
Hematoxylin and eosin (H&E) staining was performed as previously described and slices were scored by a licensed veterinary pathologist (Data S1 for more information). For lectin staining, using a Dako delimiting pen (Agilent Technologies, Santa Clara, CA), dams were created around tissue sections. The sections were then incubated in 1% BSA (w/v) in approximately 300 μL of PBS for 30 min at RT, then incubated in 10 μg/mL of SNA lectin for 2 h. They were then rinsed for 10 min three times with 1% BSA in PBS. The sections were then incubated in approximately 300 μL of DAPI for 20 min and rinsed with PBS for 5 min. Finally, two drops of Vectashield fluorescence mounting medium (Dako, Agilent Technologies) was added and the sections were cover-slipped and sealed with nail polish. Confocal images were obtained using a fluorescence microscope (Leica Biosystems). Adjacent tissue sections incubated in 1% BSA in PBS served as controls. Zeiss LSM700 confocal microscope connected to a Zeiss Axiovert 200 M with an EC Plan-Neofluar 40x/1.30 numerical aperture oil immersion objective (Carl Zeiss AG, Oberkochen, Germany). Processing of the images was performed using Zen software and applied on the entire images as well as on controls. The presented pictures are representative of independent triplicates.
On-tissue microdigestion
Regions of interest (ROIs) discerned using MALDI-MSI, H&E staining, and SNA lectin staining were marked on adjacent tissue sections and optical scans were obtained for reference. Five spots were marked per ROI, and these were digested by microspotting 20 μg/mL of trypsin suspended in 50 mM NH4HCO3 using a chemical inkjet printer (CHIP 1000, Shimadzu, Kyoto, Japan). Each spot was composed of 4 microspots spaced 100 mm center-to-center, with each microspot produced by printing 13 droplets at 200 pL/droplet. By optimizing the waiting time between each pass, this yielded spots with diameters between 450–600 μm. The spots were maintained wet for 2 h, after which, the sections were incubated at 37°C inside an enclosed glass chamber humidified with 1:1 MeOH/H2O for 1h. The sections were then dried under vacuum for 5 min.
The following solutions were used to extract the digested peptides: 0.1% TFA in water, 4:1 ACN/0.1% TFA in water, and 7:3 MeOH/0.1% TFA in water. Each solution (3 μL) was deposited to cover all the digested spots for one region, and the extracts were manually pipetted 10x before recovery. This was repeated once before proceeding with the next solvent system. In cases where the spots were distant from each other, the total volume was divided per spot and extraction was performed separately. The extracts were then frozen in −80°C and dried using a speedvac. The dried extracts were reconstituted in 10 μL 0.1% TFA in water, vortexed for 10 s and sonicated for 5 min. They were then desalted using C18 Ziptips (Pierce, Thermo Fisher Scientific) and taken to dryness in a vacuum concentrator.
Glycoproteomic MS analysis
All of the glycoproteomic samples were analyzed by LC-MS/MS on an Orbitrap Fusion Tribrid (Thermo Fisher Scientific) coupled to a Dionex Ultimate 3000 HPLC. The samples were reconstituted in 7 μL of 0.1% formic acid in water (“buffer A”). Then, a portion of the sample (6.5 μL) was loaded via autosampler isocratically onto a C18 nano pre-column using 0.1% formic acid in water (“Solvent A”). For pre-concentration and desalting, the column was washed with 2% ACN and 0.1% formic acid in water (“loading pump solvent”). Subsequently, the C18 nano pre-column was switched in line with the C18 nano separation column and injected at 0.3 μL/min onto a 75 μm × 250 μm EASY-Spray column (Thermo Fisher Scientific) containing 2 mm C18 beads. The column was held at 40°C using a column heater in the EASY-Spray ionization source (Thermo Fisher Scientific). The samples were eluted at 0.3 μL/min using a 90-min gradient and a 185-min instrument method. Solvent A was comprised of 0.1% formic acid in water, whereas Solvent B was 0.1% formic acid in acetonitrile. The gradient profile was as follows (min:%B) 0:3, 3:3, 93:35, 103:42, 104:98, 109:98, 110:3, 185:3. The instrument method used an MS1 resolution of 60,000 at FWHM 400 m/z, an AGC target of 3e5, and a mass range from 300 to 1,500 m/z. Dynamic exclusion was enabled with a repeat count of 3, repeat duration of 10 s, exclusion duration of 10 s. Only charge states 2–6 were selected for fragmentation. MS2s were generated at top speed for 3 s. HCD was performed on all selected precursor masses with the following parameters: isolation window of 2 m/z, 30% collision energy, Orbitrap detection with a resolution of 30,000, and an AGC target of 1e4 ions. All proteomic raw files have been uploaded to PRIDE: PXD025537.
Glycoproteomic data analysis
Glycoproteomic data analysis was performed as described previously.(Malaker and Ferracane, 2019) Raw files were searched using Byonic by ProteinMetrics against the Uniprot Canis familiaris database (downloaded November 2018). Search parameters included semi-specific cleavage specificity at the C-terminal site of R and K. Mass tolerance was set at 10 ppm for MS1s, 0.1 for MS2s. Methionine oxidation (common 2), asparagine deamidation (common 2), and N-term acetylation (rare 1) were set as variable modifications with a total common max of 3, rare max of 1. Glycosylation was added in three separate searches to minimize search times. In the first search, N-glycans were set as variable modifications (common 2), using the “N-glycan 57 human plasma” database. In the second iteration, O-glycans were set as variable modifications (common 2), using the “O-glycan 6 most common” database. In the final search, an O-mannose database containing (Hex, Hex-HexNAc, Hex-HexNAc2, Hex2-HexNAc-NeuAc, and Hex2-HexNAc-Fuc) was used for a variable modification (common 2). Cysteine carbaminomethylation was set as a fixed modification. Peptide hits were filtered using a 1% FDR. All Byonic glycopeptide search results are uploaded to PRIDE: PXD025537. Note that Byonic searches were used to assist in manual assignment of glycopeptides but were not used to definitively assign glycan structures or the peptide sequence. All peptides were manually validated and/or sequenced using Xcalibur software (Thermo Fisher Scientific). In addition to the Byonic results and raw files, we have uploaded hand-curated excel files for each sample detailing: glycopeptide ID, mass area for relative quantitation, calculated mass ppm, retention time, and protein. These excel files can also be found in PRIDE: PXD025537.
QUANTIFICATION AND STATISTICAL ANALYSIS
The raw files described above were also searched using MaxQuant with the following parameters: fixed cysteine carbaminomethylation, variable deamidation of asparagine, and variable methionine oxidation. The Elsevier’s Pathway Studio version 11.0 (Ariadne Genomics/Elsevier) was used to analyze relationships among differentially expressed proteomics protein candidates using the Ariadne ResNet database.(Bonnet et al., 2009; Yuryev et al., 2009) “Subnetwork Enrichment Analysis” (SNEA) algorithm was selected to extract statistically significant altered biological and functional pathways pertaining to each identified set of protein hits among cluster 1 (overexpressed in tumor and necrotic regions) and cluster 2 (overexpressed in benign). SNEA utilizes Fisher’s statistical test to determine if there are nonrandom associations between two categorical variables organized by specific relationship.(Kobeissy et al., 2016) Integrated Venn diagram analysis was performed using “the InteractiVenn”: a web-based tool for the analysis of complex data sets.(Heberle et al., 2015) See Data S9 for the listed differentially expressed pathways. Each table indicates the Entity designation, Relationship type, and Reference type. For statistical analyses used in Figure 5, one-way ANOVAs were performed for various glycan counts comparing benign, tumor, and necrotic conditions. For instances where p<0.05, Tukey’s Multiple Comparisons posthocs determined significantly different comparisons.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Canine glioma biopsies | Oncovet Clinic | oncovet-clinical-research.com |
| Chemicals, peptides, and recombinant proteins | ||
| Optima LC/MS grade acetonitrile | Fisher | Cat# A955-4 |
| Optima LC/MS grade methanol | Fisher | CAS 67-56-1 |
| Xylene | Fisher | CAS 1330-20-7 |
| Trifluoroacetic acid | Biosolve | CAS 76-05-1 |
| Ammonium bicarbonate | Sigma | CAS 1066-33-7 |
| 2,5 dihydroxybenzoic acid (DHB) | Sigma | CAS 490-79-9 |
| Dako Mounting Medium | Agilent | Cat# CS70330-2 |
| 4′,6-diamidino-2-phenylindole (DAPI) | Sigma | CAS 28718-90-3 |
| poly-D-lysine hydrobromide | Sigma | CAS 27964-99-4 |
| bovine serum albumin (BSA) | Sigma | CAS 9048-46-8 |
| Biotech grade Tris buffer | Interchim | P/N UP158387 |
| Formaldehyde | Sigma | CAS 50-00-0 |
| HCl ampules, sequencing grade | ThermoFisher | Cat# 24308 |
| Formic acid ampules | ThermoFisher | Cat# A11710X1-AMP |
| Fluoresceine-tagged Sambuccus nigra agglutinin (SNA) | Laboratory of Prof. Anne Harduin-Lepers (U Lille); Vector laboratories | Cat # FL-1301-2 |
| Sequence grade modified trypsin | Promega | V5111 |
| Glycerol free PNGaseF | New England Biolabs | P0705S |
| Ethanol, 200 proof | Fisher | CAS 64-17-5 |
| Pierce Ultrapure Water | ThermoFisher | Cat# 51140 |
| Deposited data | ||
| Glycoproteomic.raw files | This Study | PRIDE: PXD025537 |
| Byonic glycoproteomic search results | This Study | PRIDE: PXD025537 |
| Annotated glycoproteomic results | This Study | PRIDE: PXD025537 |
| MALDI-MSI.raw files | This Study | PRIDE: PXD025826 |
| Software and algorithms | ||
| Xcalibur software | ThermoFisher | |
| Byonic | ProteinMetrics | https://proteinmetrics.com/ |
| GlycoMod | ExPasy | web.expasy.org/glycomod/ |
| SCiLS MALDI-MSI software | Bruker | https://scils.de/ |
| Other – Instrumentation | ||
| Rapiflex MALDI-MSI | Bruker | n/a |
| MALDI-LTQ-Orbitrap | Thermo | n/a |
| Orbitrap Fusion Tribrid | Thermo | n/a |
| Dionex Ultimate 3000 HPLC | Thermo | n/a |
| Fluorescence microscope | Leica Biosystems | n/a |
| Centrivap vacuum concentrator | Labconco | n/a |
| Chemical inkjet printer (CHIP 1000) | Shimadzu | n/a |
Highlights.
N-glycan MALDI-MSI revealed a sialylated glycan as upregulated in canine glioma
Spatial glycoproteomics identified haptoglobin as the underlying glycoprotein
Manual assignment of glycopeptides uncovered a diverse array of glycosylation
Observed significant glycopeptide changes between tumor and benign regions
SIGNIFICANCE.
Aberrant glycosylation is a universal feature of cancer; however, we are only beginning to truly understand how cell surfaces change with malignant transformation. MALDI mass spectrometry imaging (MSI) has been gaining attention for investigation of changes in N-glycosylation in various cancers. However, one drawback of this method is that the N-glycans have to be removed from the underlying protein, thus losing that structural information. Here, we overcome this challenge, by first performing MALDI-MSI of N-glycans in canine glioma samples. We then used this information to guide an intact microscale glycoproteomics experiment. In doing so, we were able to directly tie MALDI-MSI N-glycan data to the underlying glycoprotein. Surprisingly, we also found several other types of glycosylation in our experiment, ranging from mucin-type O-GalNAc to intracellular O-GlcNAc. This allowed us to determine statistically significant changes in glycopeptides across cancerous versus non-cancerous regions. Taken together, these experiments represent a unique mechanism to understand spatial changes in altered glycosylation in cancer.
ACKNOWLEDGMENTS
This research was supported by the Institut National de la Sante de la Recherche Medicale (INSERM), l’Institut Universitaire de France, l’I-SITE ULNA, and University de Lille. This work was also supported, in part, by the Howard Hughes Medical Institute and National Institutes of Health (NIH) grant R01 CA200423 awarded to C.R.B. S.A.M. was supported by an NIGMS F32 fellowship and a France-Stanford Center for Interdisciplinary Studies Visiting Scholar fellowship. The authors would also like to thank C.C. Angelakos (Stanford University) for his assistance with statistical analyses.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.chembiol.2021.05.007.
DECLARATION OF INTERESTS
C.R.B. is a co-founder and Scientific Advisory Board member of Lycia Therapeutics, Palleon Pharmaceuticals, Enable Bioscience, Redwood Biosciences (a subsidiary of Catalent), and InterVenn Biosciences, and a member of the Board of Directors of Eli Lilly & Company.
REFERENCES
- Angel PM, Comte-Walters S, Ball LE, Talbot K, Mehta A, Brockbank KGM, and Drake RR (2018). Mapping extracellular matrix proteins in formalin-fixed, paraffin-embedded tissues by MALDI imaging mass spectrometry. J. Proteome Res 17, 635–646. 10.1021/acs.jproteome.7b00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel PM, Mehta A, Norris-Caneda K, and Drake RR (2017). MALDI imaging mass spectrometry of N-glycans and tryptic peptides from the same formalin-fixed, paraffin-embedded tissue section. In Tissue Proteomics, Methods in Molecular Biology, Sarwal MM and Sigdel TK, eds. (Springer; New York: ), pp. 225–241. 10.1007/7651_2017_81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley RT, Ahmed AU, Yanke AB, Cohen-Gadol AA, and Dey M (2016). Dogs are man’s best friend: in sickness and in health. Neuro Oncol. 10.1093/neuonc/now109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaum BS, Hannan JP, Herbert AP, Kavanagh D, Uhrín D, and Stehle T (2015). Structural basis for sialic acid-mediated self-recognition by complement factor H. Nat. Chem. Biol 11, 77–82. 10.1038/nchem-bio.1696. [DOI] [PubMed] [Google Scholar]
- Bonnet A, Lagarrigue S, Liaubet L, Robert-Granié C, SanCristobal M, and Tosser-Klopp G (2009). Pathway results from the chicken data set using GOTM, Pathway Studio and Ingenuity softwares. BMC Proc. 3, S11. 10.1186/1753-6561-3-s4-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MT, Kuliwaba JS, Muratovic D, Everest-Dass AV, Packer NH, Findlay DM, and Hoffmann P (2016). MALDI mass spectrometry imaging of N-glycans on tibial cartilage and subchondral bone proteins in knee osteoarthritis. Proteomics 16, 1736–1741. 10.1002/pmic.201500461. [DOI] [PubMed] [Google Scholar]
- Büll C, Boltje TJ, Balneger N, Weischer SM, Wassink M, van Gemst JJ, Bloemendal VRLJ, Boon L, van der Vlag J, Heise T, et al. (2018). Sialic acid blockade suppresses tumor growth by enhancing T cell-mediated tumor immunity. Cancer Res. 10.1158/0008-5472.CAN-17-3376. [DOI] [PubMed] [Google Scholar]
- Bull C, Stoel MA, den Brok MH, and Adema GJ (2014). Sialic acids sweeten a tumor’s life. Cancer Res. 74, 3199–3204. 10.1158/0008-5472.CAN-14-0728. [DOI] [PubMed] [Google Scholar]
- Darula Z, and Medzihradszky KF (2018). Analysis of mammalian O-glycopeptides—we have made a good start, but there is a long way to go. Mol. Cell Proteomics 17, 2–17. 10.1074/mcp.MR117.000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies LRL, and Varki A (2013). Why is N-glycolylneuraminic acid rare in the vertebrate brain? In SialoGlyco Chemistry and Biology I, Topics in Current Chemistry (Springer; Berlin Heidelberg: ), pp. 31–54. 10.1007/128_2013_419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bernabé DB-V, Inamori K, Yoshida-Moriguchi T, Weydert CJ, Harper HA, Willer T, Henry MD, and Campbell KP (2009). Loss of α-dys-troglycan laminin binding in epithelium-derived cancers is caused by silencing of LARGE. J. Biol. Chem 284, 11279–11284. 10.1074/jbc.C900007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis J, Laferte S, Waghorne C, Breitman M, and Kerbel R (1987). Beta 1–6 branching of Asn-linked oligosaccharides is directly associated with metastasis. Science 236, 582–585. 10.1126/science.2953071. [DOI] [PubMed] [Google Scholar]
- Dickinson PJ (2014). Advances in diagnostic and treatment modalities for intracranial tumors. J. Vet. Intern. Med 28, 1165–1185. 10.1111/jvim.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM, Hempel SJ, Stalnaker SH, Stuart R, and Wells L (2013). O-Mannosylation and human disease. Cell. Mol. Life Sci 70, 2849–2857. 10.1007/s00018-012-1193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake RR, Powers TW, Jones EE, Bruner E, Mehta AS, and Angel PM (2017). MALDI mass spectrometry imaging of N-linked glycans in cancer tissues. In Advances in Cancer Research (Elsevier; ), pp. 85–116. 10.1016/bs.acr.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer CA, Baker E, Hu H, and Matthews RT (2012). RPTPz/phosphacan is abnormally glycosylated in a model of muscle-eye-brain disease lacking functional POMGnT1. Neuroscience 220, 47–61. 10.1016/j.neuroscience.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everest-Dass AV, Briggs MT, Kaur G, Oehler MK, Hoffmann P, and Packer NH (2016). N-Glycan MALDI imaging mass spectrometry on formalin-fixed paraffin-embedded tissue enables the delineation of ovarian cancer tissues. Mol. Cell Proteomics 15, 3003–3016. 10.1074/mcp.M116.059816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura T, Shinohara Y, Tissot B, Pang P-C, Kurogochi M, Saito S, Arai Y, Sadilek M, Murayama K, Dell A, et al. (2008). Glycosylation status of haptoglobin in sera of patients with prostate cancer vs. benign prostate disease or normal subjects. Int. J. Cancer 122, 39–49. 10.1002/ijc.22958. [DOI] [PubMed] [Google Scholar]
- Furukawa J, Tsuda M, Okada K, Kimura T, Piao J, Tanaka S, and Shinohara Y (2015). Comprehensive glycomics of a multistep human brain tumor model reveals specific glycosylation patterns related to malignancy. PLoS One 10, e0128300. 10.1371/journal.pone.0128300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MA, Stanczak MA, Mantuano NR, Xiao H, Pijnenborg JFA, Malaker SA, Miller CL, Weidenbacher PA, Tanzo JT, Ahn G, et al. (2020). Targeted glycan degradation potentiates the anticancer immune response in vivo. Nat. Chem. Biol 10.1038/s41589-020-0622-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes MS, Moremen KW, and Cummings RD (2017). Biochemical characterization of functional domains of the chaperone Cosmc. PLoS One 12, e0180242. 10.1371/journal.pone.0180242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, and Akimoto Y (2009). The O-GlcNAc modification. In Essentials of Glycobiology, Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, and Etzler ME, eds. (Cold Spring Harbor Laboratory Press; ). [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, and Lagerlof O (2011). Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem 80, 825–858. 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H, Meirelles GV, da Silva FR, Telles GP, and Minghim R (2015). InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16, 169. 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijs B, Holst S, Briaire-de Bruijn IH, van Pelt GW, de Ru AH, van Veelen PA, Drake RR, Mehta AS, Mesker WE, Tollenaar RA, et al. (2016). Multimodal mass spectrometry imaging of N-glycans and proteins from the same tissue section. Anal. Chem 88, 7745–7753. 10.1021/acs.analchem.6b01739. [DOI] [PubMed] [Google Scholar]
- Holst S, Belo AI, Giovannetti E, van Die I, and Wuhrer M (2017). Profiling of different pancreatic cancer cells used as models for metastatic behaviour shows large variation in their N-glycosylation. Sci. Rep 7, 16623. 10.1038/s41598-017-16811-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst S, Heijs B, de Haan N, van Zeijl RJM, Briaire-de Bruijn IH, van Pelt GW, Mehta AS, Angel PM, Mesker WE, Tollenaar RA, et al. (2016). Linkage-specific in situ sialic acid derivatization for N-glycan mass spectrometry imaging of formalin-fixed paraffin-embedded tissues. Anal. Chem 88, 5904–5913. 10.1021/acs.analchem.6b00819. [DOI] [PubMed] [Google Scholar]
- Hu B, Kong LL, Matthews RT, and Viapiano MS (2008). The proteoglycan brevican binds to fibronectin after proteolytic cleavage and promotes glioma cell motility. J. Biol. Chem 283, 24848–24859. 10.1074/jbc.M801433200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard ME, Arnold S, Bin Zahid A, McPheeters M, Gerard O’Sullivan M, Tabaran A-F, Hunt MA, and Pluhar GE (2018). Naturally occurring canine glioma as a model for novel therapeutics. Cancer Invest. 36, 415–423. 10.1080/07357907.2018.1514622. [DOI] [PubMed] [Google Scholar]
- Hudak JE, Canham SM, and Bertozzi CR (2014). Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat. Chem. Biol 10, 69–75. 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennemann R, Rodden A, Bauer BL, Mennel HD, and Wiegandt H (1990). Glycosphingolipids of human gliomas. Cancer Res. 50, 7444–7449. [PubMed] [Google Scholar]
- Khidekel N, Ficarro SB, Peters EC, and Hsieh-Wilson LC (2004). Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl. Acad. Sci. U S A 101, 13132–13137. 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobeissy FH, Guingab-Cagmat JD, Zhang Z, Moghieb A, Glushakova OY, Mondello S, Boutté AM, Anagli J, Rubenstein R, Bahmad H, et al. (2016). Neuroproteomics and systems biology approach to identify temporal biomarker changes post experimental traumatic brain injury in rats. Front. Neurol 7. 10.3389/fneur.2016.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler JW, Miller AD, Miller CR, Porter B, Aldape K, Beck J, Brat D, Cornax I, Corps K, Frank C, et al. (2018). A revised diagnostic classification of canine glioma: towards validation of the canine glioma patient as a naturally occurring preclinical model for human glioma. J. Neuropathol. Exp. Neurol 77, 1039–1054. 10.1093/jnen/nly085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudelka MR, Stowell SR, Cummings RD, and Neish AS (2020). Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD. Nat. Rev. Gastroenterol. Hepatol 10.1038/s41575-020-0331-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufe DW (2009). Mucins in cancer: function, prognosis and therapy. Nat. Rev. Cancer 9, 874–885. 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen ISB, Narimatsu Y, Clausen H, Joshi HJ, and Halim A (2019). Multiple distinct O-mannosylation pathways in eukaryotes. Curr. Opin. Struct. Biol 56, 171–178. 10.1016/j.sbi.2019.03.003. [DOI] [PubMed] [Google Scholar]
- Larsen ISB, Narimatsu Y, Joshi HJ, Yang Z, Harrison OJ, Brasch J, Shapiro L, Honig B, Vakhrushev SY, Clausen H, and Halim A (2017). Mammalian O-mannosylation of cadherins and plexins is independent of protein O-mannosyltransferases 1 and 2. J. Biol. Chem 292, 11586–11598. 10.1074/jbc.M117.794487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leney AC, El Atmioui D, Wu W, Ovaa H, and Heck AJR (2017). Elucidating crosstalk mechanisms between phosphorylation and O-GlcNAcylation. Proc. Natl. Acad. Sci. U S A 114, E7255–E7261. 10.1073/pnas.1620529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Fischer M, Satkunarajah M, Zhou D, Withers SG, and Rini JM (2017). Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1). Nat. Commun 8, 185. 10.1038/s41467-017-00255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wu C, Guo L, Liu Y, Mo W, Wang H, Ding J, Wong ET, and Yu M (2012). The role of brevican in glioma: promoting tumor cell motility in vitro and in vivo. BMC Cancer 12, 607. 10.1186/1471-2407-12-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley MS, Crocker PR, and Paulson JC (2014). Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol 14, 653–666. 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaker SA, and Ferracane MJ (2019). Mass spectrometric identification and molecular modeling of glycopeptides presented by MHC class I and II processing pathways. In Immunoproteomics, Methods in Molecular Biology, Fulton KM and Twine SM, eds. (Springer; New York: ), pp. 269–285. 10.1007/978-1-4939-9597-4_17. [DOI] [PubMed] [Google Scholar]
- Maynard JC, Burlingame AL, and Medzihradszky KF (2016). Cysteine S-linked N-acetylglucosamine (S-GlcNAcylation), a new post-translational modification in mammals. Mol. Cell Proteomics 15, 3405–3411. 10.1074/mcp.M116.061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D, Chintala S, Fetcko K, Henriquez M, Tewari BN, Ahmed A, Bentley RT, and Dey M (2019). Common molecular alterations in canine oligodendroglioma and human malignant gliomas and potential novel therapeutic targets. Front. Oncol 9, 780. 10.3389/fonc.2019.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney DJ, Shair LH, Lu FM, Xia J, Locke R, Matta KL, and Haltiwanger RS (2000). Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J. Biol. Chem 275, 9604–9611. 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- Moremen KW, Tiemeyer M, and Nairn AV (2012). Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol 13, 448–462. 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda K, Miyoshi E, Uozumi N, Yanagidani S, Ikeda Y, Gao C, Suzuki K, Yoshihara H, Yoshikawa M, Kawano K, et al. (1998). Gene expression of alpha1-6 fucosyltransferase in human hepatoma tissues: a possible implication for increased fucosylation of alpha-fetoprotein. Hepatology 28, 944–952. 10.1002/hep.510280408. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Macek B, Lange O, Makarov A, Horning S, and Mann M (2007). Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712. 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- Pearce OMT, and Läubli H (2016). Sialic acids in cancer biology and immunity. Glycobiology 26, 111–128. 10.1093/glycob/cwv097. [DOI] [PubMed] [Google Scholar]
- Pinho SS, and Reis CA (2015). Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555. 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- Posey AD, Schwab RD, Boesteanu AC, Steentoft C, Mandel U, Engels B, Stone JD, Madsen TD, Schreiber K, Haines KM, et al. (2016). Engineered CAR T cells targeting the cancer-associated tn-glycoform of the membrane mucin MUC1 control adenocarcinoma. Immunity 44, 1444–1454. 10.1016/j.immuni.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Holst S, Wuhrer M, Mehta A, and Drake R (2015). Two-dimensional N-glycan distribution mapping of hepatocellular carcinoma tissues by MALDI-imaging mass spectrometry. Biomolecules 5, 2554–2572. 10.3390/biom5042554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers TW, Neely BA, Shao Y, Tang H, Troyer DA, Mehta AS, Haab BB, and Drake RR (2014). MALDI imaging mass spectrometry profiling of N-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS One 9, e106255. 10.1371/journal.pone.0106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quanico J, Franck J, Cardon T, Leblanc E, Wisztorski M, Salzet M, and Fournier I (2017). NanoLC-MS coupling of liquid microjunction microextraction for on-tissue proteomic analysis. Biochim. Biophys. Acta 1865, 891–900. 10.1016/j.bbapap.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Quanico J, Franck J, Dauly C, Strupat K, Dupuy J, Day R, Salzet M, Fournier I, and Wisztorski M (2013). Development of liquid microjunction extraction strategy for improving protein identification from tissue sections. J. Proteomics 79, 200–218. 10.1016/j.jprot.2012.11.025. [DOI] [PubMed] [Google Scholar]
- Reiding KR, Bondt A, Franc V, and Heck AJR (2018). The benefits of hybrid fragmentation methods for glycoproteomics. Trac Trends Anal. Chem 108, 260–268. 10.1016/j.trac.2018.09.007. [DOI] [Google Scholar]
- Riley NM, Malaker SA, Driessen M, and Bertozzi CR (2020). Optimal dissociation methods differ for N- and O-glycopeptides. J. Proteome Res 10.1021/acs.jproteome.0c00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley NM, Westphall MS, and Coon JJ (2017). Activated ion-electron transfer dissociation enables comprehensive top-down protein fragmentation. J. Proteome Res 16, 2653–2659. 10.1021/acs.jproteome.7b00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd P, Karlsson NG, Khoo K-H, and Packer NH (2015). Glycomics and glycoproteomics. In Essentials of Glycobiology, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, and Prestegard JH, et al. , eds. (Cold Spring Harbor Laboratory Press; ). [PubMed] [Google Scholar]
- Rutherford MA, and Pangršič T (2012). Molecular anatomy and physiology of exocytosis in sensory hair cells. Cell Calcium 52, 327–337. 10.1016/j.ceca.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Shajahan A, Supekar NT, Gleinich AS, and Azadi P (2020). Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology, cwaa042. 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh MO, Halmo SM, and Wells L (2017). Recent advancements in understanding mammalian O-mannosylation. Glycobiology 27, 806–819. 10.1093/glycob/cwx062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim H, Hu B, and Viapiano MS (2009). Reduced expression of the hyaluronan and proteoglycan link proteins in malignant gliomas. J. Biol. Chem 284, 26547–26556. 10.1074/jbc.M109.013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT-BG, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, et al. (2013). Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488. 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepper J, Shastri S, Loo TS, Preston JC, Novak P, Man P, Moore CH, Havlíček V, Patchett ML, and Norris GE (2011). Cysteine S-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett. 585, 645–650. 10.1016/j.febslet.2011.01.023. [DOI] [PubMed] [Google Scholar]
- Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, and Hunt DF (2004). Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci 101, 9528–9533. 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana H, Taniguchi K, Ushio Y, Teruya K, Osada K, and Murakami H (1994). Changes of monosaccharide availability of human hybridoma lead to alteration of biological properties of human monoclonal antibody. Cytotechnology 16, 151–157. 10.1007/BF00749902. [DOI] [PubMed] [Google Scholar]
- Thaysen-Andersen M, Packer NH, and Schulz BL (2016). Maturing glycoproteomics technologies provide unique structural insights into the N-glycoproteome and its regulation in health and disease. Mol. Cell Proteomics 15, 1773–1790. 10.1074/mcp.O115.057638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A (2008). Sialic acids in human health and disease. Trends Mol. Med 14, 351–360. 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, and Gagneux P (2012). Multifarious roles of sialic acids in immunity: roles of sialic acids in immunity. Ann. NY Acad. Sci 1253, 16–36. 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Kannagi R, Toole B, and Stanley P (2015). Glycosylation changes in cancer. In Essentials of Glycobiology, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, and Prestegard JH, et al. , eds. (Cold Spring Harbor Laboratory Press; ). [PubMed] [Google Scholar]
- Veillon L, Fakih C, Abou-El-Hassan H, Kobeissy F, and Mechref Y (2018). Glycosylation changes in brain cancer. ACS Chem. Neurosci 9, 51–72. 10.1021/acschemneuro.7b00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester-Christensen MB, Halim A, Joshi HJ, Steentoft C, Bennett EP, Levery SB, Vakhrushev SY, and Clausen H (2013). Mining the O-mannose glycoproteome reveals cadherins as major O-mannosylated glycoproteins. Proc. Natl. Acad. Sci. U S A 110, 21018–21023. 10.1073/pnas.1313446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viapiano MS, Bi WL, Piepmeier J, Hockfield S, and Matthews RT (2005). Novel tumor-specific isoforms of BEHAB/brevican identified in human malignant gliomas. Cancer Res. 65, 6726–6733. 10.1158/0008-5472.CAN-05-0585. [DOI] [PubMed] [Google Scholar]
- Wisztorski M, Fatou B, Franck J, Desmons A, Farré I, Leblanc E, Fournier I, and Salzet M (2013). Microproteomics by liquid extraction surface analysis: application to FFPE tissue to study the fimbria region of tubo-ovarian cancer. Prot. Clin. Appl 7, 234–240. 10.1002/prca.201200070. [DOI] [PubMed] [Google Scholar]
- Wisztorski M, Quanico J, Franck J, Fatou B, Salzet M, and Fournier I (2017). Droplet-based liquid extraction for spatially-resolved microproteomics analysis of tissue sections. In Imaging Mass Spectrometry, Methods in Molecular Biology, Cole LM, ed. (Springer; New York: ), pp. 49–63. 10.1007/978-1-4939-7051-3_6. [DOI] [PubMed] [Google Scholar]
- Xiao H, Woods EC, Vukojicic P, and Bertozzi CR (2016). Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc. Natl. Acad. Sci. U S A 113, 10304–10309. 10.1073/pnas.1608069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Kotelnikova E, and Daraselia N (2009). Ariadne’s ChemEffect and Pathway Studio knowledge base. Expert Opin. Drug Discov 4, 1307–1318. 10.1517/17460440903413488. [DOI] [PubMed] [Google Scholar]
- Zhang S, Shang S, Li W, Qin X, and Liu Y (2016). Insights on N-glycosylation of human haptoglobin and its association with cancers. Glycobiology 26, 684–692. 10.1093/glycob/cww016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any new unique reagents. The accension number for the MALDI-MSI data files reported in this paper is PRIDE : PXD025826. The accension number for all glycoproteomic raw files and search results is PRIDE : PXD025537.


