Abstract
A molecular epidemiological study was performed to investigate the prevalence of GB virus C/hepatitis G virus (GBV-C/HGV) infection among various populations in Surabaya, Indonesia. The prevalence of GBV-C/HGV RNA, determined by reverse transcription-PCR for a portion of the NS3 region of the viral genome, was 2.7% (4 of 150) among randomly collected blood donor sera, which were all negative for both hepatitis B virus surface antigen and antibodies against hepatitis C virus (HCV). On the other hand, the prevalence among anti-HCV-positive blood donors was 17.8% (13 of 73), with the ratio being significantly higher than that observed with the anti-HCV-negative blood donors (P < 0.001). A high prevalence of GBV-C/HGV infection was also observed among patients with chronic liver disease, such as chronic hepatitis (5.7%), liver cirrhosis (11.5%), and hepatocellular carcinoma (7.0%), and patients on maintenance hemodialysis (29.0%). No correlation was observed between GBV-C/HGV viremia and serum alanine aminotransferase levels in the populations tested, suggesting the possibility that GBV-C/HGV does not cause apparent liver injury. Phylogenetic analysis of sequences of a portion of the 5′ untranslated region and the E1 region of the viral genome identified, in addition to a previously reported then novel group of GBV-C/HGV variants (group 4), another novel group of variants (group 5). This result suggests that GBV-C/HGV can be classified into at least five genetic groups. GBV-C/HGV isolates of group 4 and group 5 were each shown to comprise approximately 40% of the total Indonesian isolates.
Although it has been well documented that hepatitis C virus (HCV) is a major etiologic agent of posttransfusion or sporadic non-A, non-B viral hepatitis worldwide (16), further detailed study has suggested the possible presence of a hepatitis virus(es) different from hepatitis A, B, C, D, or E virus (non-A-E hepatitis virus) (1). Recently, two independent research groups reported the identification of new infectious agents of human GB virus C (GBV-C) (33) and hepatitis G virus (HGV) (22). These viruses were molecularly cloned from the sera of patients with suspected viral hepatitis. Sequence analysis of the viral genome has demonstrated that these isolates show 86% identity with each other at the nucleotide level and 95% identity at the amino acid level and that they are different isolates of the same virus (9). In the present paper, this virus is referred to as GBV-C/HGV. The viral genome is single-strand, positive-sense RNA of about 9,400 bases and contains a large open reading frame flanked by untranslated regions at both the 5′ and the 3′ ends (5′UTR and 3′UTR, respectively) (22). The open reading frame encodes a polyprotein precursor of about 2,900 amino acids that consists of structural and nonstructural proteins in the order NH2-(C)-E1-E2/p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH (9, 20, 22). The structure of the genome is typical of that of the Flaviviridae family and, more specifically, is similar to that of HCV. Unlike the HCV genome, however, most, if not all, of the core region of the GBV-C/HGV genome is missing (9, 20, 22).
While some researchers suggested the presence of only a single genotype for GBV-C/HGV (38), other researchers have reported that GBV-C/HGV is classified into three genetic groups (or genotypes), which can be further divided into a number of subgroups (or subtypes), e.g., 1a, 1b, 2a, and 2b, on the basis of the sequence diversity of the full-length genome (18, 29) or partial genomic sequences (4, 21, 25, 26, 34, 40). The distribution of each group or subgroup of GBV-C/HGV has been reported to vary with different geographical areas (4, 19, 25, 26, 34). Similarly, the distribution of each type or subtype of HCV, which apparently shares the same transmission route with GBV-C/HGV, has been known to differ with different geographical areas (8, 24, 35). Through the molecular epidemiological study of HCV infection among various populations in Chiang Mai, Thailand, and Surabaya, Indonesia, we have identified unique HCV subtypes that are not normally found in other areas (5, 8, 14, 15, 35). It is possible, therefore, that a unique sequence variant(s) of GBV-C/HGV is prevailing in those areas. Indeed, we have identified a group of novel sequence variants of GBV-C/HGV in Thailand (19).
Apart from the search for a novel group(s) of sequence variant, it is also important to know the prevalence of GBV-C/HGV infection in order to understand the clinicoepidemiological features of the virus. GBV-C/HGV has been reported to cause persistent infection in various populations, such as patients with liver disease (3, 9–13, 19, 22, 28, 31), individuals at high risk of contracting blood-borne infections (2, 10, 19, 23, 37, 40), and healthy individuals (6, 10, 19, 28, 39). However, on the basis of observations that there are individuals who are infected with GBV-C/HGV but who do not present with liver injury, the possibility has been suggested that GBV-C/HGV is nonpathogenic (2, 3, 28).
In the present study, we have determined the prevalence of GBV-C/HGV infection in various populations in Surabaya, Indonesia, and analyzed the relationship between the virus infection and liver injury. We also report that, in addition to the unique group of Thai variants (tentatively designated group 4), another novel group of GBV-C/HGV (designated as group 5) prevails in Indonesia.
MATERIALS AND METHODS
Serum samples.
Sera were collected from 223 healthy blood donors (BDs) at the Red Cross Blood Transfusion Unit in Surabaya, Indonesia. Of those BD sera, 150 serum samples were collected randomly and the remaining 73 serum samples were obtained from anti-HCV-positive individuals who were chosen intentionally. Sera were also collected from 69 patients on maintenance hemodialysis (HD) and 202 patients with chronic liver disease, including 53 patients with chronic hepatitis (CH), 78 patients with liver cirrhosis (LC), and 71 patients with hepatocellular carcinoma (HCC), at Dr. Soetomo Hospital, Airlangga University, Surabaya, Indonesia. All sera were stored at −80°C until they were used.
The sera were tested for anti-HCV antibodies by a second-generation enzyme-linked immunosorbent assay (Ortho HCV Ab ELISA Test II; Ortho Diagnostics, Inc.) and for hepatitis B surface antigen (HBsAg; subtypes ad and ay) by using an AUSAB enzyme immunoassay (Diagnostics Division, Abbott Laboratories). Serum alanine aminotransferase (ALT) levels were determined by using the ALT (ALAT/GPT) test kit (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer's instruction. Normal ALT levels were ≤29 U/liter for men and ≤22 U/liter for women when testing was done at 30°C.
Reverse transcription and PCR.
RNA extracted from the sera (120 μl of each serum sample) with TRizol LS (Life Technologies, Gaithersburg, Md.) was reverse transcribed into cDNA by using Rous-associated virus type 2 reverse transcriptase (Takara Shuzo, Co., Ltd., Kyoto, Japan) and random hexamer primer (Takara Shuzo). The resultant cDNA was subjected to the first-round PCR over 35 cycles, with each cycle consisting of 1 min at 94°C, 1 min at 45°C, and 2 min at 72°C, followed by the second-round PCR under the same conditions described above. The primers used to amplify a portion of the NS3 region were described previously (19). The primers used to amplify portions of the 5′UTR and the E1 region were selected on the basis of the sequences that had been reported to be conserved among many GBV-C/HGV isolates (9, 18, 20, 22, 25, 29). The sequences and positions of the primers used are shown in Table 1. The PCR products were electrophoresed in an agarose gel containing ethidium bromide and were visualized by UV illumination.
TABLE 1.
Sequences and positions of primers used for amplification of GBV-C/HGV genome
| Primera | Region | Positionb | Polarity | Sequencec (5′ to 3′) | Reference |
|---|---|---|---|---|---|
| GB1 (S) | NS3 | 4269–4288 | Sense | 5′-ATCCCCTTTTATGGGCATGG-3′ | 19 |
| GB2 (R) | NS3 | 4416–4235 | Antisense | 5′-TCTTTGATGATGGAACTGTC-3′ | 19 |
| GB3 (S) | NS3 | 4335–4354 | Sense | 5′-CATTCCAAGGCGGAGTGCGA-3′ | 19 |
| GB4 (R) | NS3 | 4398–4417 | Antisense | 5′-TCTTTACCCCTATAATAGGC-3′ | 19 |
| GB5′-5S | 5′UTR | 102–121 | Sense | 5′-GCCAAAAGGTGGTGGATGGG-3′ | |
| GB5′-4R | 5′UTR | 457–477 | Antisense | 5′-CGGAGCTGGGTGGCCCCATGC-3′ | 25 |
| GB5′-7S | 5′UTR | 134–153 | Sense | 5′-TGGTAGGTCGTAAATCCCGG-3′ | |
| GB5′-6R | 5′UTR | 376–395 | Antisense | 5′-TGGTCCTTGTCAACTCGCCG-3′ | |
| GBE1-1S | E1 | 549–568 | Sense | 5′-ATCATGGCAGTCCTTCTGCT-3′ | |
| GBE1-2R | E1 | 969–988 | Antisense | 5′-TCARTCCATCTCCAAAACTC-3′ | |
| GBE1-3S | E1 | 630–649 | Sense | 5′-GGGCAATATTTSCTCACAAA-3′ | |
| GBE1-4R | E1 | 957–976 | Antisense | 5′-CAAAACTCACTTTCCCACTT-3′ |
R, reverse; S, sense.
The numbering system conforms to that for HGV-1 (accession no. U44402).
R, A and G; S, C and G.
Sequencing and phylogenetic analyses.
The nucleotide sequences of the amplified fragments were determined with the Taq DyeDeoxy Terminator Cycle Sequencing kit (Perkin-Elmer) and ABI 373A DNA sequencer (Applied Biosystems, Inc.), and the amino acid sequences deduced. With the nucleotide sequences thus obtained and those reported previously, phylogenetic trees were constructed by the six-parameter neighbor-joining method, as described previously (8, 14). In brief, nucleotide sequences of the 5′UTR or the E1 region of the GBV-C/HGV genome obtained in this study and those available from international DNA data banks (GenBank, EMBL, DDBJ) were compared by multiple sequence alignment. On the basis of these reliable estimates, a phylogenetic tree was constructed by using a supercomputer at DDBJ, National Institute of Genetics, Mishima, Japan, with the computer program ODEN, version 1.1.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank/EMBL/DDBJ nucleotide sequence databases (accession nos. AB026058 to AB026099 and AB027116 to AB027120).
RESULTS AND DISCUSSION
Prevalence of GBV-C/HGV infection and ALT titers in BDs in Surabaya, Indonesia.
Serum samples randomly obtained from 150 healthy BDs (143 males, 7 females; mean age, 36.3 years; age range, 18 to 58 years) were first tested. To determine the prevalence of GBV-C/HGV viremia, we used a reverse transcription-PCR method to amplify a portion of the NS3 region of the viral genome (19). These BD sera, which were all negative for both HBsAg and anti-HCV antibodies, were classified into two groups; those with normal ALT levels and those with elevated ALT levels, according to the criteria described in Materials and Methods. GBV-C/HGV RNA was detected in 3 (2.3%) of 131 BD with normal ALT levels and 1 (5.3%) of 19 BD with elevated ALT levels (Table 2). There was no significant difference in the GBV-C/HGV prevalence between the two groups, the result being consistent with those in previous reports (6, 10, 17). In total, 4 (2.7%) of 150 serum samples randomly obtained from BDs were positive for GBV-C/HGV RNA. In this connection, the prevalence of GBV-C/HGV viremia has been reported to be 0.8% in Japan (28), 1.0% each in Italy (11) and Thailand (19), 1.7% in the United States (22), 2.0% in the southern part of China (40), and 2.1% in Taiwan (39).
TABLE 2.
Prevalence of GBV-C/HGV, ALT levels, and GBV-C/HGV genotype distribution among BDs in Surabaya, Indonesia
| BD groupa | No. positiveb/no. tested (%) | ALT level (IU/liter)c in BDs with the following GBV-C/HGV status:
|
RNA GBV-C/HGV genotype distributiond | |
|---|---|---|---|---|
| Positive | Negative | |||
| Anti-HCV-negative BDs | ||||
| Normal ALT levels | 3/131 (2.6) | 15 (11–24) | 16 (10–29) | Group 4 (2) |
| Elevated ALT levels | 1/19 (5.3) | 76 | 46 (30–70) | Group 5 (1) |
| Total | 4/150 (2.7)e | 30 (11–76) | 20 (10–70) | Group 4 (2) and group 5 (1) |
| Anti-HCV-positive BDs | ||||
| Normal ALT levels | 2/28 (7.1) | 18 (16–19) | 18 (6–26) | Group 3 (1) |
| Elevated ALT levels | 11/45 (24.4) | 52 (31–100) | 54 (30–190) | Group 2 (1), group 3 (1) and group 4 (3) |
| Total | 13/73 (17.8)e | 47 (16–100) | 38 (6–190) | Group 2 (1), group 3 (2) and group 4 (3) |
All BD sera were negative for HBsAg.
The presence of GBV-C/HGV RNA was determined by successful amplification of the NS3 region of the virus genome.
Values are means and ranges (in parentheses).
Determined on the basis of the 5′UTR sequences. The number of isolates is shown in parentheses.
P < 0.001 (χ2 test with Yates' correction).
It was reported previously that the increase in ALT levels in BDs with a single GBV-C/HGV infection was mild, reaching at most 2 to 3 times the upper limit of normal, in contrast to that in BDs with combined HCV and GBV-C/HGV infections, whose ALT levels reached a peak 14 times the upper limit of normal (6). Three of four serum samples from BDs with a single GBV-C/HGV infection (anti-HCV-negative) had normal ALT levels, and the remaining one had an ALT level of 76 U/ml, 2.6 times the upper limit of normal (Table 2). The mean titer and the range of ALT levels for BDs with a single GBV-C/HGV infection were not different from those observed for BDs who were negative for GBV-C/HGV. This observation supports the idea that infection with GBV-C/HGV alone is unlikely to cause hepatocyte injury.
A high prevalence of GBV-C/HGV RNA among HCV or HBV carriers has been reported (10, 19, 22). In our previous report, the prevalence of anti-HCV antibodies among BDs in Surabaya was estimated to be 2.3% (n = 2,233) (7). As stated above, all the 150 BD serum samples collected randomly were negative for anti-HCV antibodies in the present study, probably due to small sample size. Therefore, we collected an additional 73 BD serum samples that were positive for anti-HCV antibodies (67 males, 6 females; mean age, 46.3 years; age range, 22 to 63 years) and tested them for GBV-C/HGV RNA. The result revealed that 2 (7.1%) of 28 serum samples obtained from anti-HCV-positive BD with normal ALT levels and 11 (24.4%) of 45 serum samples obtained from anti-HCV-positive BDs with elevated ALT levels were positive for GBV-C/HGV RNA. In total, 13 (17.8%) of 73 anti-HCV-positive BD serum samples were positive for GBV-C/HGV RNA. This prevalence ratio was significantly higher than that among the anti-HCV-negative BDs (P < 0.001).
Prevalence of GBV-C/HGV infection among patients with liver disease in Surabaya, Indonesia.
Sera were obtained from 53 CH patients (37 males and 16 females; mean age, 51.7 years; age range, 19 to 70 years), 78 LC patients (46 males and 32 females; mean age, 57.4 years; age range, 31 to 78 years), and 71 HCC patients (55 males and 16 females; mean age, 52.9 years; age range, 13 to 85 years) and were analyzed for HBsAg, anti-HCV antibodies, and GBV-C/HGV RNA. The prevalences of GBV-C/HGV RNA in CH, LC, and HCC patients were 5.7% (3 of 53), 11.5% (9 of 78), and 7.0% (5 of 71), respectively (Table 3). Altogether, the prevalence of GBV-C/HGV RNA in patients with chronic liver disease (CH, LC, and HCC) was 8.4% (17 of 202) and was significantly higher than that for randomly sampled BDs (P < 0.05). There was no significant difference in GBV-C/HGV RNA prevalence between HBsAg-positive and HBsAg-negative patients or between anti-HCV-positive and anti-HCV-negative patients. The latter result appeared to be inconsistent with the result obtained with BDs (Table 2) and previous reports by other investigators that the prevalence of GBV-C/HGV infection was higher in anti-HCV-positive liver disease patients than in anti-HCV-negative patients (19, 22, 28, 41). The reason for this discrepancy remains unknown.
TABLE 3.
Prevalence of GBV-C/HGV, ALT levels, and GBV-C/HGV genotype distribution among CH, LC, and HCC patients in Surabaya, Indonesia
| Patient groupa | No. positiveb/no. tested (%) | ALT levels (IU/liter)c in BD with the following GBV-C/HGV RNA status:
|
GBV-C/HGV genotype distributiond | |
|---|---|---|---|---|
| Positive | Negative | |||
| CH | ||||
| HBsAg−, HCV Ab− | 0/6 (0) | 59 (32–127) | ||
| HBsAg−, HCV Ab+ | 1/38 (2.6) | 190 | 67 (26–272) | NDe |
| HBsAg+, HCV Ab− | 1/5 (20.0) | 204 | 72 (30–140) | Group 5 (1) |
| HBsAg+, HCV Ab+ | 1/4 (25.0) | 32 | 133 (49–289) | ND |
| Total | 3/53 (5.7) | 142 (32–204) | 70 (26–289) | |
| LC | ||||
| HBsAg−, HCV Ab− | 1/14 (7.1) | 18 | 22 (10–60) | Group 5 (1) |
| HBsAg−, HCV Ab+ | 4/48 (8.3) | 20 (17–22) | 32 (9–170) | Group 2 (1) and group 5 (2) |
| HBsAg+, HCV Ab− | 4/15 (26.7) | 24 (12–40) | 36 (11–200) | Group 2 (1) and group 5 (2) |
| HBsAg+, HCV Ab+ | 0/1 (0) | 14 | ||
| Total | 9/78 (11.5) | 21 (12–40) | 31 (9–200) | |
| HCC | ||||
| HBsAg−, HCV Ab− | 1/15 (6.7) | 75 | 47 (10–112) | Group 4 (1) |
| HBsAg−, HCV Ab+ | 2/25 (8.0) | 53 (35–70) | 53 (15–199) | Group 4 (1) |
| HBsAg+, HCV Ab− | 2/27 (7.4) | 45 (24–66) | 57 (9–291) | ND |
| HBsAg+, HCV Ab+ | 0/4 (0) | 60 (21–101) | ||
| Total | 5/71 (7.0) | 54 (24–75) | 54 (9–291) | |
Ab, antibody; −, negative; +, positive.
The presence of GBV-C/HGV RNA was determined by successful amplification of the NS3 region of the virus genome.
Values are means and ranges (in parentheses).
Determined on the basis of the 5′UTR sequences. The number of isolates is shown in parentheses.
ND, not determined due to unsuccessful amplification of the 5′UTR.
A single infection with GBV-C/HGV in the absence of HCV or HBV infection was reported to be uncommon in patients with liver disease (40). Indeed, our results revealed that only 2 (5.7%) of 35 non-B, non-C viral hepatitis patients with liver disease (CH, n = 6; LC, n = 14; and HCC, n = 15) were positive for GBV-C/HGV RNA (Table 3). The hepatopathogenicity of GBV-C/HGV is still controversial. Some researchers reported the possible importance of GBV-C/HGV in the etiology of acute, fulminant, or chronic non-A-E hepatitis (11, 13, 42). On the other hand, it was reported that GBV-C/HGV was unlikely to cause clinically defined hepatitis (2, 12, 28, 31, 41) or to affect the clinical course of patients with hepatitis A, B, or C (3, 12). In our study, mean titers and the ranges of ALT levels did not appear to differ between GBV-C/HGV-positive and -negative patients with chronic liver disease (Table 3). This result supports the idea that GBV-C/HGV infection does not either cause clinically overt hepatitis or affect the severity of hepatitis B or C. At present, however, we cannot exclude the possibility that infection with different strains of GBV-C/HGV would result in different severities of illness. In fact, it was reported that particular mutations in the GBV-C/HGV genome were possibly associated with the occurrence of fulminant hepatitis (13). Another possibility should also be taken into consideration: that GBV-C/HGV infection aggravates concurrent infection with HCV in some but not all patients. This hypothesis might be related to our present observation that the prevalence of GBV-C/HGV RNA tended to be higher in anti-HCV-positive BDs with elevated ALT levels than in those with normal ALT levels, although the difference was not statistically significant, possibly due to the small sample size (Table 2). Further study is needed to elucidate these issues.
Prevalence of GBV-C/HGV infection among HD patients in Surabaya, Indonesia.
In general, HD patients are at high risk of contracting blood-borne infections such as hepatitis C (17, 19, 23, 32). To assess this matter, sera were obtained from 69 HD patients (56 males and 13 females; mean age, 48.7 years, age range, 11 to 72 years) and tested. GBV-C/HGV RNA was detected in 20 (29.0%) of 69 HD patients (Table 4). The prevalence was significantly higher in HD patients than in BDs (P < 0.001) and in CH (P < 0.005), LC (P < 0.05), and HCC (P < 0.005) patients. It was reported that the prevalence of GBV-C/HGV RNA in HD patients was 3.1% in Japan (23), 6.8% in Germany (10), 19% in Italy (32), 22.5% in Australia (17), 25.0% in Thailand (19), and 55% in Yogjakarta, Indonesia (37). Thus, HD patients are at increased risk for GBV-C/HGV infection, but the prevalence rate appears to differ widely from hospital to hospital, probably depending on the precautions taken to prevent intraunit transmission of blood-borne diseases. Again, the mean titers and the ranges of the ALT levels were not different between GBV-C/HGV-positive and -negative HD patients (Table 4).
TABLE 4.
Prevalence of GBV-C/HGV, ALT levels, and GBV-C/HGV genotype distribution among HD patients in Surabaya, Indonesia
| HD groupa | No. positiveb/no. tested (%) | ALT levels (IU/liter)c in BDs with the following GBV-C/HGV RNA status:
|
GBV-C/HGV genotype distributiond | |
|---|---|---|---|---|
| Positive | Negative | |||
| HBsAg−, HCV Ab− | 7/22 (31.8) | 34 (9–68) | 23 (7–68) | Group 5 (4) |
| HBsAg−, HCV Ab+ | 11/41 (26.8) | 30 (7–126) | 42 (8–283) | Group 4 (2) and group 5 (5) |
| HBsAg+, HCV Ab− | 1/3 (33.3) | 131 | 53 (49–56) | Group 5 (1) |
| HBsAg+, HCV Ab+ | 1/3 (33.3) | 24 | 26 (19–32) | NDe |
| Total | 20/69 (29.0) | 36 (7–131) | 36 (7–283) | |
Ab, antibody; −, negative; +, positive.
The presence of GBV-C/HGV RNA was determined by successful amplification of the NS3 region of the virus genome.
Values are means and ranges (in parentheses).
Determined on the basis of 5′UTR sequences. The number of isolates is shown in parentheses.
ND, not determined due to unsuccessful amplification of 5′UTR.
Phylogenetic analysis of 5′UTR and E1 region sequences of the GBV-C/HGV genome.
GBV-C/HGV has tentatively been classified into three groups, which can be further divided into a number of subgroups, on the basis of the sequence diversity of the viral genome (4, 18, 21, 25, 26, 29, 34, 40). Group 1 isolates, including the original GBV-C isolate, are found almost exclusively in West Africa. Group 2 isolates are commonly found in North America and Europe, while group 3 isolates are found in East Asia (25).
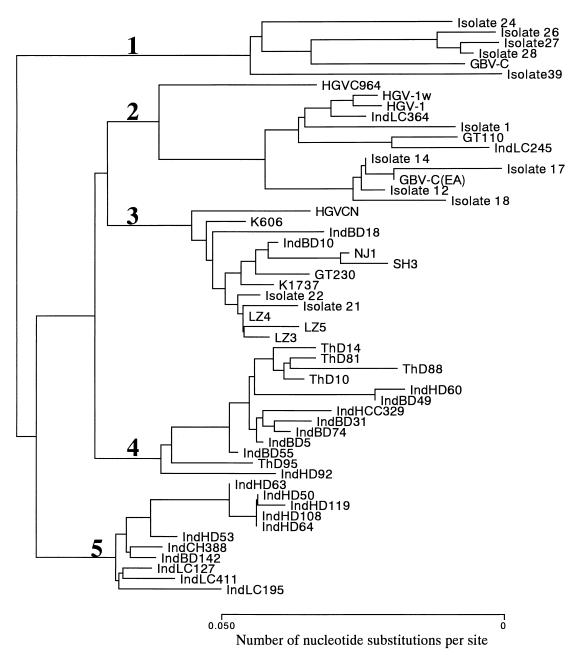
We previously reported on the identification of novel sequence variants of GBV-C/HGV in Thailand that might constitute group 4 on the basis of phylogenetic analysis of 5′UTR sequences of the viral genome (19). In order to determine which group(s) of GBV-C/HGV is prevailing in Surabaya and also in order to see whether a novel group(s) of GBV-C/HGV is found in Surabaya, 5′UTR sequences were amplified from the Indonesian sera and analyzed. Phylogenetic analysis of the 5′UTR sequences classified the Indonesian isolates into four clusters (Fig. 1): (i) IndLC245 and IndLC364 belong to group 2, (ii) IndBD10 and IndBD18 belong to group 3, (iii) IndHD60, IndBD49, IndHCC329, etc., are closely related to a group of Thai variants such as ThD10, and (iv) IndHD53, IndHD63, IndLC195, etc., constitute a novel group of variants. We have tentatively designated the last two groups as group 4 and group 5, respectively. Approximately 40% of the Indonesian GBV-C/HGV isolates tested were shown to belong to group 4 and 40% were shown to belong to novel group 5 (Table 5). Group 2 and group 3 isolates each comprised about 10% of the total isolates. Not a single isolate of group 1 was found in Surabaya.
FIG. 1.
Phylogenetic analysis of 5′UTR sequences of the GBV-C/HGV genome. GBV-C/HGV groups are indicated by numerals. The GenBank/EMBL/DDBJ accession numbers for the sequence data for the isolates in the phylogenetic tree are as follows: Isolate 24, U59541; isolate 26, U59543; isolate 27, U59544; isolate 28, U59545; GBV-C, U36380; isolate 39, U59555; HGVC964, U75356; HGV-1w, D87255; HGV1, U44402; IndLC364, AB026059; isolate 1, U59518; GT110, D90600; IndLC245, AB026058; isolate 14, U59531; isolate 17, U59534; GBV-C(EA), U63715; isolate 12, U59529; isolate 18, U59535; HGVCN, U94695; K606, D87708; IndBD18, AB026061; IndBD10, AB026060; NJ1, U86154; SH3, U86158; GT230, D90601; K1737, D87709; isolate 22, U59539; isolate 21, U59538; LZ4, U86151; LZ5, U86152; LZ3, U86150; ThD14, AB027117; ThD81, AB027118; ThD88, AB027119; ThD10, AB027116; IndHD60, AB026068; IndBD49, AB026064; IndHCC329, AB026067; IndBD31, AB026063; IndBD74, AB026066; IndBD5, AB026062; IndBD55, AB026065; ThD95, AB027120; IndHD92, AB026069; IndHD63, AB026077; IndHD50, AB026075; IndHD119, AB026080; IndHD108, AB026079; IndHD64, AB026078; IndHD53, AB026076; IndCH388, AB026071; IndBD142, AB026070; IndLC127, AB026072; IndLC411, AB026074; IndLC195, AB026073.
TABLE 5.
Prevalence of each genotypic group of GBV-C/HGV among total isolates in Surabaya, Indonesia
| GBV-C/HGV groupa | No. of isolates/no. tested (%) | Patient group(s) (no. of isolates) |
|---|---|---|
| 1 | 0/31 (0) | |
| 2 | 3/31 (9.7) | BD (1) and LC (2) |
| 3 | 2/31 (6.5) | BD (2) |
| 4 (new) | 13/31 (41.9) | BD (5), HCC (2), and HD (6) |
| 5 (new) | 13/31 (41.9) | BD (1), CH (1), LC (5) and HD (6) |
Determined on the basis of the 5′UTR sequences.
In order to quantify the degrees of variation within and between different GBV-C/HGV groups, mean evolutionary distances were calculated. The mean intragroup distances were all shorter than the intergroup distances (Table 6). It thus appears that GBV-C/HGV can be classified into five genetic groups. Those evolutionary distances, however, were shorter than those observed with HCV (30), suggesting that GBV-C/HGV is less variable than HCV. In this connection, it should be noted that only a single genotype was proposed for GBV-C/HGV because the divergence in the nucleotide and amino acid sequences of the entire GBV-C/HGV genome among different isolates fell within a narrow range (38).
TABLE 6.
Mean values for nucleotide evolutionary distances within and between groups of GBV-C/HGV on the basis of 5′UTR sequences
| GBV-C/HGV group | Mean nucleotide evolutionary distance
|
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| 1 | 0.067 | 0.152 | 0.132 | 0.135 | 0.118 |
| 2 | 0.049 | 0.088 | 0.094 | 0.097 | |
| 3 | 0.026 | 0.074 | 0.077 | ||
| 4 | 0.029 | 0.079 | |||
| 5 | 0.023 | ||||
The sequences of the primers used to amplify the 5′UTR in this study were chosen from the sequences that are conserved among many GBV-C/HGV isolates that have been reported (9, 18, 20, 22, 25, 29). However, use of the 5′UTR was unsuccessful with some Indonesian sera that were positive for NS3 amplification. We cannot exclude the possibility, therefore, that another group(s) of GBV-C/HGV variants still remains to be identified.
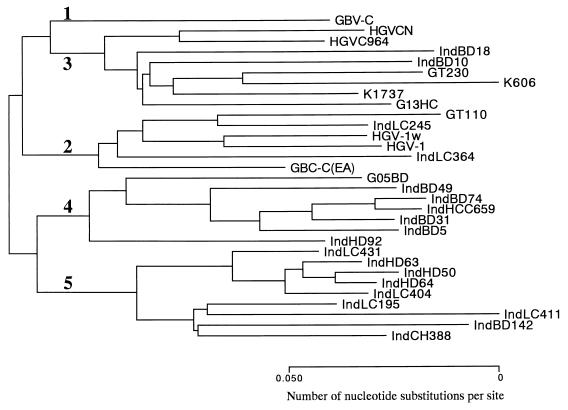
To see whether the same phylogenetic relationship is observed with another region of the viral genome, a portion of the E1 region was amplified and analyzed. Phylogenetic analysis of E1-region sequences also classified the Indonesian isolates into four clusters (groups 2 to 5) (Fig. 2), although somewhat less discriminatingly than analysis with the 5′UTR sequences did. Again, from phylogenetic analysis of E1-region sequences, it is evident that the Indonesian isolates of group 4 and group 5 are distinct from the isolates of groups 1 to 3, with the grouping being consistent with that based on 5′UTR sequences.
FIG. 2.
Phylogenetic analysis of the E1 region sequences of the GBV-C/HGV genome. GBV-C/HGV groups are indicated by numerals. GenBank/EMBL/DDBJ accession numbers for the sequence data for the isolates in the phylogenetic tree are as follows: GBV-C, U36380; HGVCN, U94695; HGVC964, U75356; IndBD18, AB026084; IndBD10, AB026083; GT230, D90601; K606, D87708; K1737, D87709; G13HC, AB000165; GT110, D90600; IndLC245 AB026081; HGV-1w, D87255; HGV1, U44402; IndLC364, AB026082; GBV-C(EA), U63715; G05BD, AB000161; IndBD49, AB026087; IndBD74, AB026088; IndHCC659, AB026089; IndBD31, AB026086; IndBD5, AB026085; IndHD92, AB026090; IndLC431, AB026096; IndHD63, AB026098; IndHD50, AB026097; IndHD64, AB026099; IndLC404, AB026094; IndLC195, AB026093; IndLC411, AB026095; IndBD142, AB026091; IndCH388, AB026092.
In conclusion, we have determined the prevalence of GBV-C/HGV viremia among various populations, such as BDs, patients with CH, LC, and HCC, and HD patients, in Surabaya, Indonesia, and found that GBV-C/HGV infection is prevalent among individuals at high risk of blood-borne diseases, especially HD patients. However, no correlation was observed between GBV-C/HGV viremia and serum ALT levels in the populations tested. This result suggests the possibility that GBV-C/HGV does not cause apparent liver injury. As for the genotype classification, we have identified a novel group of sequence variants of GBV-C/HGV (group 5) in Indonesia, in addition to a previously reported unique group of variants (group 4) isolated in Thailand (19), Myanmar (27), and Japan (36). It would be important to determine whether the novel group of GBV-C/HGV is different from the other groups in terms of pathogenicity, antigenicity, and other virological features.
ACKNOWLEDGMENTS
We are grateful to K. Hachida for assistance in preparing the manuscript.
This work was carried out during a large-scale cooperative study between Southeast Asian countries and Japan conducted by Japan Society for the Promotion of Science. This work was also supported in part by a grant-in-aid from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Alter H J, Bradley D W. Non-A, non-B hepatitis unrelated to the hepatitis C virus (non-ABC) Semin Liver Dis. 1995;15:110–120. doi: 10.1055/s-2007-1007268. [DOI] [PubMed] [Google Scholar]
- 2.Alter H J, Nakatsuji Y, Melpolder J, Wages J, Wisley R, Shih J W-K, Kim J P. The incidence of transfusion-associated hepatitis G virus infection and its relation to liver disease. N Engl J Med. 1997;336:747–754. doi: 10.1056/NEJM199703133361102. [DOI] [PubMed] [Google Scholar]
- 3.Alter M J, Gallagher M, Morris T T, Moyer L A, Meeks E L, Krawczynski K, Kim J P, Margolis H S. Acute non-A-E hepatitis in the United States and the role of hepatitis G virus infection. N Engl J Med. 1997;336:741–746. doi: 10.1056/NEJM199703133361101. [DOI] [PubMed] [Google Scholar]
- 4.An P, Wei L, Wu X, Yuhki N, O'Brien S J, Winkler C. Evolutionary analysis of the 5′ terminal region of hepatitis G virus isolated from different region in China. J Gen Virol. 1997;78:2477–2482. doi: 10.1099/0022-1317-78-10-2477. [DOI] [PubMed] [Google Scholar]
- 5.Apichartpiyakul C, Chittivudikarn C, Miyajima H, Homma M, Hotta H. Analysis of hepatitis C virus isolates among healthy blood donors and drug addicts in Chiang Mai, Thailand. J Clin Microbiol. 1994;32:2276–2279. doi: 10.1128/jcm.32.9.2276-2279.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorkman P, Sundstrom G, Widell A. Hepatitis C virus and GB virus C/hepatitis G virus viremia in Swedish blood donors with different alanine aminotransferase levels. Transfusion. 1998;38:378–384. doi: 10.1046/j.1537-2995.1998.38498257377.x. [DOI] [PubMed] [Google Scholar]
- 7.Darmadi S, Soetjipto, Handajani R, Lusida M I, Soemarto, Sakugawa H, Ishido S, Hotta H. Hepatitis C virus infection-associated markers in sera from blood donors in Surabaya, Indonesia. Microbiol Immunol. 1996;40:401–405. doi: 10.1111/j.1348-0421.1996.tb01086.x. [DOI] [PubMed] [Google Scholar]
- 8.Doi H, Apichartpiyakul C, Ohba K, Mizokami M, Hotta H. Hepatitis C virus (HCV) subtype prevalence in Chiang Mai, Thailand, and identification of novel subtypes of HCV major type 6. J Clin Microbiol. 1996;34:569–574. doi: 10.1128/jcm.34.3.569-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erker J C, Simons J N, Muerhoff A S, Leary T P, Chalmers M L, Desai S M, Mushawar K. Molecular cloning and characterization of a GB virus C isolate from a patient with non-A-E hepatitis. J Gen Virol. 1996;77:2713–2720. doi: 10.1099/0022-1317-77-11-2713. [DOI] [PubMed] [Google Scholar]
- 10.Feucht H H, Zollner B, Polywka S, Knodler B, Schroter M, Nolte H, Laufs R. Prevalence of hepatitis G viremia among healthy subjects, individuals with liver diseases, and persons at risk for parenteral transmission. J Clin Microbiol. 1997;35:767–768. doi: 10.1128/jcm.35.3.767-768.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiordalisi G, Zanella I, Mantero G, Bettinardi A, Stellini R, Paraninfo G, Cadeo G, Primi D. High prevalence of GB virus C infection in a group of Italian patients with hepatitis of unknown etiology. J Infect Dis. 1996;174:181–183. doi: 10.1093/infdis/174.1.181. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi J, Ueno K, Kawakami Y, Kishihara Y, Ariyama I, Furusho N, Sawayama Y, Etoh Y, Kashiwagi S. Clinical course of chronic hepatitis C virus infection is not influenced by concurrent hepatitis G infection. Dig Dis Sci. 1999;44:618–623. doi: 10.1023/a:1026677912187. [DOI] [PubMed] [Google Scholar]
- 13.Heringlake S, Osterkamp S, Trautwein C, Tillman H L, Boker K, Muerhoff S, Mushahwar I K, Hunsmann G, Manns M P. Association between fulminant hepatic failure and a strain of GBV virus C. Lancet. 1996;348:1626–1629. doi: 10.1016/S0140-6736(96)04413-3. [DOI] [PubMed] [Google Scholar]
- 14.Hotta H, Doi H, Hayashi T, Purwanta M, Soemarto W, Mizokami M, Ohba K, Homma M. Analysis of the core and E1 envelope region sequences of a novel variant of hepatitis C virus obtained in Indonesia. Arch Virol. 1994;136:53–62. doi: 10.1007/BF01538816. [DOI] [PubMed] [Google Scholar]
- 15.Hotta H, Handajani R, Lusida M I, Soemarto W, Doi H, Miyajima H, Homma M. Subtype analysis of hepatitis C virus in Indonesia on the basis of NS5b region sequences. J Clin Microbiol. 1994;32:3049–3051. doi: 10.1128/jcm.32.12.3049-3051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1035–1058. [Google Scholar]
- 17.Hyland C A, Mison L, Solomon N, Cockerill J, Wang L, Hunt L A, Selvey L A, Faoagali J, Cooksley W G E, Young I F, Trowbridge R, Borthwick I, Gowans E J. Exposure to GB virus type C or hepatitis G virus in selected Australian adult and children populations. Transfusion. 1998;38:821–827. doi: 10.1046/j.1537-2995.1998.38998409001.x. [DOI] [PubMed] [Google Scholar]
- 18.Katayama K, Kageyama T, Fukushi S, Hoshino F B, Kurihara C, Ishiyama N, Okamura H, Oya A. Full-length GBV-C/HGV genomes from nine Japanese isolates: characterization by comparative analyses. Arch Virol. 1998;143:1063–1075. doi: 10.1007/s007050050356. [DOI] [PubMed] [Google Scholar]
- 19.Katayama Y, Apichartpiyakul C, Handajani R, Ishido S, Hotta H. GB virus C/hepatitis G virus (GBV-C/HGV) infection in Chiang Mai, Thailand, and identification of variants on the basis of 5′-untranslated region sequences. Arch Virol. 1997;142:2433–2445. doi: 10.1007/s007050050253. [DOI] [PubMed] [Google Scholar]
- 20.Leary T P, Muerhoff A S, Simons J N, Pilot-Matias T J, Erker J C, Chalmers M L, Schlauder G G, Dawson G J, Desai S M, Mushahwar I K. Sequence and genomic organization of GBV-C: a novel member of the Flaviviridae associated with human non-A-E hepatitis. J Med Virol. 1996;48:60–67. doi: 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 21.Lim M Y, Fry K, Yun A, Chong S, Linnen J, Fung K, Kim J P. Sequence variation and phylogenetic analysis of envelope glycoprotein of hepatitis G virus. J Gen Virol. 1997;78:2771–2777. doi: 10.1099/0022-1317-78-11-2771. [DOI] [PubMed] [Google Scholar]
- 22.Linnen J, Wages J, Jr, Zhang-Keck Z-Y, Fry K E, Krawczynski K Z, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih J W-K, Young L, Piatak M, Jr, Hoover C, Fernandez J, Chen S, Zou J-C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 23.Masuko K, Mitsui T, Iwano K, Yamazaki C, Okuda K, Meguro T, Murayama N, Inoue T, Tsuda F, Okamoto H, Miyakawa Y, Mayumi M. Infection with hepatitis GB virus C in patients on maintenance hemodialysis. N Engl J Med. 1996;334:1485–1490. doi: 10.1056/NEJM199606063342301. [DOI] [PubMed] [Google Scholar]
- 24.Mellor J, Holmes E C, Jarvis L M, Yap P L, Simmonds P The International HCV Collaborative Study Group. Investigation of the pattern of hepatitis C sequence diversity in different geographical regions: implications for virus classification. J Gen Virol. 1995;76:2493–2507. doi: 10.1099/0022-1317-76-10-2493. [DOI] [PubMed] [Google Scholar]
- 25.Muerhoff A S, Simons J N, Leary T P, Erker J C, Chalmers M L, Pilot-Matias T J, Dawson G J, Desai M D, Mushahwar I K. Sequence heterogeneity within the 5′-terminal region of the hepatitis GB virus C genome and evidence for genotypes. J Hepatol. 1996;25:379–384. doi: 10.1016/s0168-8278(96)80125-5. [DOI] [PubMed] [Google Scholar]
- 26.Muerhoff A S, Smith D B, Leary T P, Erker J C, Desai S M, Mushahwar I K. Identification of GB virus C variants by phylogenetic analysis of 5′-untranslated region sequences. J Virol. 1997;71:6501–6508. doi: 10.1128/jvi.71.9.6501-6508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naito H, Win K M, Abe K. Identification of a novel genotype of hepatitis G virus in Southeast Asia. J Clin Microbiol. 1999;37:1217–1220. doi: 10.1128/jcm.37.4.1217-1220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakatsuji Y, Shih J W K, Tanaka E, Kiyosawa K, Wages J, Jr, Kim J P, Alter H J. Prevalence and disease association of hepatitis G virus infection in Japan. J Viral Hepatitis. 1996;3:307–316. doi: 10.1111/j.1365-2893.1996.tb00103.x. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto H, Nakao H, Inoue T, Fukuda M, Kishimoto J, Iizuka H, Tsuda F, Miyakawa Y, Mayumi M. The entire nucleotide sequences of two GB virus C/hepatitis G virus isolates of distinct genotypes from Japan. J Gen Virol. 1997;78:737–745. doi: 10.1099/0022-1317-78-4-737. [DOI] [PubMed] [Google Scholar]
- 30.Pickering J M, Thomas H C, Karayannis P. Genetic diversity between hepatitis G virus isolates: analysis of nucleotide variation in the NS-3 and putative ‘core’ peptide genes. J Gen Virol. 1997;78:53–60. doi: 10.1099/0022-1317-78-1-53. [DOI] [PubMed] [Google Scholar]
- 31.Sallie R, Shaw J, Mutimer D. GBV-C virus and fulminant hepatic failure. Lancet. 1996;347:1552. doi: 10.1016/s0140-6736(96)90704-7. [DOI] [PubMed] [Google Scholar]
- 32.Sampietro M, Badalamenti S, Lunghi G. Hepatitis GB virus C. N Engl J Med. 1996;335:1392. doi: 10.1056/NEJM199610313351811. [DOI] [PubMed] [Google Scholar]
- 33.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoff A S, Schlauder G G, Desai S M, Mushahwar I K. Isolation of novel virus like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 34.Smith D B, Cuceanu N, Davidson F, Jarvis L M, Mokili J L K, Hamid S, Ludlam C A, Simmonds P. Discrimination of hepatitis G virus/GBV-C geographical variants by analysis of the 5′ non-coding region. J Gen Virol. 1997;78:1533–1542. doi: 10.1099/0022-1317-78-7-1533. [DOI] [PubMed] [Google Scholar]
- 35.Soetjipto R, Handajani, Lusida M I, Darmadi S, Adi P, Soemarto, Ishido S, Katayama Y, Hotta H. Differential prevalence of hepatitis C virus subtypes in healthy blood donors, patients on maintenance hemodialysis, and patients with hepatocellular carcinoma in Surabaya, Indonesia. J Clin Microbiol. 1996;34:2875–2880. doi: 10.1128/jcm.34.12.2875-2880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K, Hijikata M, Aoyama K, Hoshino H, Hino K, Mishiro S. Characterization of GBV-C/HGV viral genome: comparison among different isolates for a ∼2kb-sequence that covers entire E1 and most of 5′UTR and E2′. Int Hepatol Commun. 1997;6:253–263. [Google Scholar]
- 37.Tsuda F, Hadiwandowo S, Sawada N, Fukuda M, Tanaka T, Okamoto H, Miyakawa Y, Mayumi M. Infection with GB virus C (GBV C) in patients with chronic liver diseases or on maintenance hemodialysis in Indonesia. J Med Virol. 1996;49:248–252. doi: 10.1002/(SICI)1096-9071(199607)49:3<248::AID-JMV15>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 38.Wang H L, Hou Y-D, Jin D-Y. Identification of a single genotype of hepatitis G virus by comparison of one complete genome from a healthy carrier with eight from patients with hepatitis. J Gen Virol. 1997;78:3247–3253. doi: 10.1099/0022-1317-78-12-3247. [DOI] [PubMed] [Google Scholar]
- 39.Wang J-T, Chen P-J, Liu D-P, Sheu J-C, Wang T-H, Chen D-S. Prevalence and infectivity of hepatitis G virus and its strain variant, the GB agent, in volunteer blood donors in Taiwan. Transfusion. 1998;38:290–295. doi: 10.1046/j.1537-2995.1998.38398222874.x. [DOI] [PubMed] [Google Scholar]
- 40.Wu R-R, Mizokami M, Cao K, Nakano T, Ge X-M, Wang S-S, Orito E, Ohba K, Mukaide M, Hikiji K, Lau J Y N, Iino S. GB virus C/hepatitis G virus infection in southern China. J Infect Dis. 1997;175:168–171. doi: 10.1093/infdis/175.1.168. [DOI] [PubMed] [Google Scholar]
- 41.Yashina T L, Favorov M O, Khudyakov Y E, Fields H A, Znoiko O O, Shkurko T V, Bonafonte T, Sevall J S, Agopian M S, Peter J B. Detection of hepatitis G virus (HGV) RNA: clinical characteristics of acute HGV infection. J Infect Dis. 1997;175:1302–1307. doi: 10.1086/516460. [DOI] [PubMed] [Google Scholar]
- 42.Yoshiba M, Okamoto H, Mishiro S. Detection of the GBV-C hepatitis virus genome in serum from patients with fulminant hepatitis of unknown aetiology. Lancet. 1995;346:1131–1132. doi: 10.1016/s0140-6736(95)91802-7. [DOI] [PubMed] [Google Scholar]