Abstract
A high royal jelly-producing strain of honeybees (HRJHB) has been obtained by successive artificial selection of Italian honeybees (Apis mellifera ligustica) in China. The HRJHB can produce amounts of royal jelly that are dozens of times greater than their original counterparts, which has promoted China to be the largest producer of royal jelly in the world. In this study, we generated a chromosome-scale of the genome sequence for the HRJHB using PacBio long reads and Hi-C technique. The genome consists of 16 pseudo-chromosomes that contain 222 Mb of sequence, with a scaffold N50 of 13.6 Mb. BUSCO analysis yielded a completeness score of 99.3%. The genome has 12,288 predicted protein-coding genes and a rate of 8.11% of repetitive sequences. One chromosome inversion was identified between the HRJHB and the closely related Italian honeybees through whole-genome alignment analysis. The HRJHB’s genome sequence will be an important resource for understanding the genetic basis of high levels of royal jelly production, which may also shed light on the evolution of domesticated insects.
Subject terms: Genetics, Agriculture
| Measurement(s) | genome • DNA • transcriptome • sequence_assembly |
| Technology Type(s) | DNA sequencing • RNA sequencing • sequence assembly process |
| Sample Characteristic - Organism | Apis mellifera |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.16879888
Background & Summary
Royal jelly (RJ) is a proteinaceous secretion synthesized by the hypopharyngeal and mandibular glands of nurse worker bees and is used for feeding queen and larvae1. It also plays a critical role in the caste determination of honeybees2. Nowadays, RJ is widely used in medical products, health foods and cosmetics in many countries owing to the numerous biological activities it is known to perform including anti-bacterial, anti-oxidative, anti-inflammatory, immunomodulatory, anti-tumoral, and anti-aging activities3,4. China is now the largest producer and exporter of RJ in the world, which satisfies nearly all the global demand5. Since the 1980s, the yearly production of RJ in China has increased from 200 to around 3000 tons5. The rapidly increased production of RJ in China has been mainly attributed to the successful breeding of the high royal jelly-producing honeybees (HRJHB) (Fig. 1), and the effective utilization of corresponding production tools and techniques6.
Fig. 1.
High royal jelly-producing honeybees (HRJHB) in China. (a) Queen and workers in one colony. (b) Royal jelly in the queen’s cells.
HRJHB was derived from an Italian honeybee subspecies (Apis mellifera ligustica), which was chiefly introduced into China in the 1910s–1930s7. In 1960s, attempts were made by beekeepers in the Southeast region of China to select high RJ producing bee stocks to meet a high demand for RJ8. The colony that displayed a high rate of RJ production was selected for raising daughter queens and drones in each apiary8. Sometimes queens were also developed using larvae of high RJ producing colonies from different apiaries8. Queens then open-mated with local drones in the air8. After the aforementioned semi-controlled style of breeding, the annual RJ production per colony increased from 0.2–0.3 kg in the 1960s to 2–3 kg in the late 1980s and even reaching 6–8 kg in the 2000s8. This was perceived as a miracle and the HRJHB was rapidly introduced to other regions of China from the 1980s, onwards as well as other countries at a later date. At present, the annual production per HRJHB colony has reached more than 10 kg, which is dozens of times greater than that of common Italian honeybees (A. m. ligustica)5. RJ production has become a major income source for many beekeepers in China and the HRJHB has been certified as a new honeybee genetic resource by the Chinese government7.
Previous studies regarding isoenzymes, microsatellites and mitochondrial DNA have shown significant genetic differentiation between the HRJHB and the other common A. m. ligustica populations in China9–11. It was suggested that morphological markers, behavioural and physiological changes, and differently expressed proteins and genes, correlate to the high royal jelly-producing trait12–16. However, related research has so failed to develop an entirely clear picture of what causes the complex royal jelly-producing trait. In recent years, honeybee selection programs for high RJ production have also been implemented in Brazil and France beekeeping17,18. Additionally, further breeding of HRJHB for improving general resistance to disease is being carried out in China.
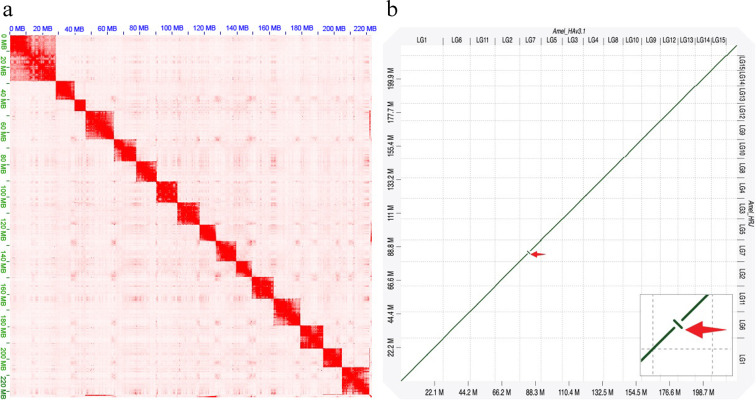
In this study, we generated a chromosome-scale of the genome assembly of the HRJHB using PacBio long-reads, Illumina short reads, and the Hi-C chromosome conformation capture technique (Table 1; Fig. 2a). The resultant genome has a total length of 222 Mb with 16 chromosomes, and the scaffold N50 was 13.6 Mb (Table 1). One chromosome inversion was identified between HRJHB and the closely related Italian honeybees via whole-genome alignment analysis (Fig. 2b). Moreover, through a combination of ab initio gene predictions, transcript evidence and homologous protein evidence, 12,288 protein coding genes were identified in this genome, therein 6,615 genes were assigned a GO term and 8,614 genes were assigned a protein domain (Table 2). Repetitive elements are made of 8.11% of the HRJHB genome sequence, but transposable elements (TEs) only occupy 2.15% (Table 2). Among those TEs, DNA transposons represented the most abundant TE class, which make up the majority of the total TE content (1.68% out of 2.15%). Furthermore, Tc1-mariner (TcMar) is the most abundant TE superfamily in the genome. The genome sequence provides a valuable resource for exploring the molecular basis of the high royal jelly-producing trait in honeybee and will facilitate further genetic improvements. The HRJHB may even represent a novel animal model for studying the effects of artificial selection on insects.
Table 1.
Sequencing data generated for the HRJHB genome assembly.
| Genome sequencing | |||
|---|---|---|---|
| Read number | Read_length(mean) | Total read length (Gb) | |
| PacBio long reads | 2,154,163 | 15,489 | 33.37 |
| Ilumina sequencing | 71786450 | 150 | 10.77 |
| RNA-seq | 130258000 | 150 | 18.05 |
| Hi-C sequencing | 218592996 | 150 | 32.79 |
| Genome assembly | |||
| Genome assembly size | 222 Mb | ||
| Number of scaffolds | 16 | ||
| Scaffold N50 | 13.6 Mb | ||
| BUSCO completeness | 99.30% | ||
Fig. 2.
Chromosome-scale assembly for HRJHB genome. (a) The HRJHB’s genome contig contact matrix using Hi-C data. (b) The HRJHB’s genome sequence was aligned with a closely related honeybee genome (NCBI assembly: Amel_HAv3). The red arrow indicates the chromosome inversion between the two genomes on LG7.
Table 2.
Annotation of protein-coding genes and repetitive sequences.
| Protein-coding genes | |||
| Total gene number | 12,288 | ||
| BUSCO completeness | 97% | ||
| Number of genes with a GO term | 6,615 | ||
| Number of genes with a protein domain | 8,614 | ||
| Repetitive sequences | |||
| TE superfamily | Length occupied (bp) | Percent of genome | |
| DNA transposons | TcMar | 3557056 | 1.67 |
| hAT | 14338 | 0.01 | |
| non-LTR retrotransposons | CR1 | 952652 | 0.45 |
| R2 | 49376 | 0.02 | |
| LTR retrotransposons | Copia | 6014 | 0.00 |
| Gypsy | 383280 | 0.18 | |
| Total TEs | 4962716 | 2.15 | |
| Other repeats | 13761592 | 5.96 | |
| Total repeats | 18724308 | 8.11 | |
Methods
Sample collection and genome sequencing
Samples of the HRJHB for genome and transcriptome sequencing were collected in 2019 from Zhejiang Province, China, where the HRJHB was originated and primarily distributed (Fig. 3).
Fig. 3.
Original area of HRJHB (red arrowhead).
Newly emerged drone bees (n = 20), that are descendants of the queen bee, were collected from a single colony (Fig. 1a). The thoraxes were pooled for PacBio single molecule real-time (SMRT) sequencing and Illumina HiSeq sequencing. Genomic DNA was extracted using the Gentra Puregene Tissue Kit (Qiagen) and was sequenced in accordance with the standard protocols. Newly emerged worker bees (n = 20) were collected from the same colony and their thoraxes were pooled for Hi-C sequencing. Hi-C library preparation was performed by Frasergen (http://www.frasergen.com/), which mainly followed a protocol described previously19. The obtained Hi-C sequencing libraries were sequenced on the Illumina HiSeq X Ten platform. Worker bees that were excreting royal jelly (n = 20) were collected from the same colony and their heads, thoraxes and abdomens (excluding the mid-gut tissues) were pooled for RNA-seq on the Illumina HiSeq X Ten platform.
De novo genome assembly for HRJHB
A total of 33.37 Gb of long reads were generated by the PacBio Sequel platform (Table 1), which were self-corrected and assembled into contigs using Canu v2.120, with default parameters. The obtained contigs were parsed by Purge Haplotigs v1.1.121 to get rid of the redundancies caused by the heterozygosity of the pooled honeybee samples. Then, the remaining non-redundant contigs were polished with Illumina HiSeq reads (Table 1) three times by utilizing software Pilon v1.2322. Finally, the Juicer tool23 was applied to map Hi-C reads (Table 1) against the polished contig sequences of HRJHB using the BWA algorithm24. The 3D-DNA pipeline25 was applied to scaffold the contig sequences in relation to the chromosome-scale of genome assembly.
Annotation of repeat sequences
TEs were de novo identified by RepeatModeler226, in line with default parameters. Using the obtained repeat library, each honeybee genome assembly was analyzed with RepeatMasker (http://www.repeatmasker.org) to yield a comprehensive summary of the TE landscape in each assembly. The annotation files produced by RepeatMasker were processed by in-house scripts to eliminate redundancies. In addition, refined annotation files were used to determine the TE diversity and abundance within each assembly and tandem repeats were identified with the Tandem Repeat Finder27, which was implemented in RepeatMasker.
Prediction and functional annotation of protein-coding genes
Annotation of protein-coding genes was based on ab initio gene predictions, transcript evidence, and homologous protein evidence, which were all applied in the MAKER computational pipeline28. Meanwhile, RNA-seq reads obtained in this study were assembled using Trinity29. The assembled RNA-seq transcripts, along with proteins from bees (superfamily Apoidea) that are available in the National Center for Biotechnology Information (NCBI) GenBank (last accessed on 01/28/2020), were imported into the MAKER pipeline to generate gene models. To obtain functional clues for the predicted gene models, protein sequences encoded by them were searched against the Uniprot-Swiss-Prot protein databases (last accessed on 01/28/2020) using the BLASTp algorithm implemented in BLAST suite v2.2830. In addition, protein domains and GO terms associated with gene models were identified by InterproScan-531.
Data Records
The raw data was submitted to the National Center for Biotechnology Information (NCBI) SRA database (Experiments for SRP300170) under BioProject accession number PRJNA68947432. The assembled genome has been deposited at DDBJ/ENA/GenBank under the accession GCA_019321825.133. Moreover, the genome annotation results have been deposited at the Figshare database34.
Technical Validation
Evaluation of the genome assembly
The completeness of the genome assembly was evaluated using a set of 4,415 hymenopteran benchmarking universal single-copy orthologs (BUSCOs) using software BUSCO v335. The results indicated that 99.3% of these BUSCOs were present in the genome assembly (Table 1), suggesting a remarkably complete assembly of the HRJHB genome.
Furthermore, the chromosome-level structural accuracy was assessed by performing whole-genome alignments between HRJHB genome and a closely related honeybee genome (GenBank assembly: Amel_HAv3) using software D-GENIES36. The alignment results revealed a highly conserved chromosome structure between the two genomes, indicating an accurate scaffolding of contigs in the HRJHB genome. Nevertheless, we did find one inversion on LG7 (Fig. 2b). The Hi-C heatmap revealed a well-organized interaction contact pattern along the diagonals within/around the chromosome inversion region in HRJHB (Fig. 4), which rules out the possibility that the structural variation was derived from unreliable Hi-C signals in the HRJHB assembly. In addition, as chromosome inversion has been found to be associated with honeybee adaptations37, the inversion identified in the HRJHB genome will guarantee that further analysis will be carried out to investigate its association with high royal jelly production.
Fig. 4.
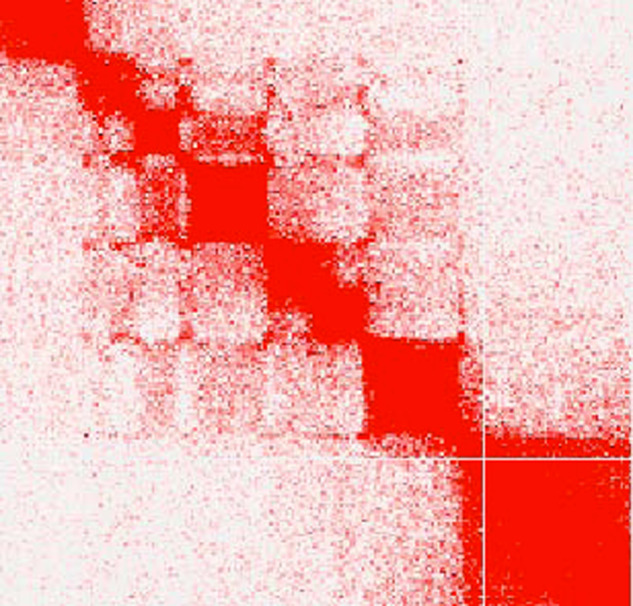
Hi-C heatmap around the identified chromosome inversion region in the HRJHB.
Acknowledgements
This work was funded by the Science and Technology Department of Zhejiang Province, China (2016C02054-11), the National Natural Science Foundation of China (31602014), and the Fundamental Research Funds of Chinese Academy of Agricultural Sciences (grant numbers: Y2019XK13, Y2021XK16).
Author contributions
L.C. and C.S. conceived the study. L.C. collected the samples. L.C. extracted the genomic DNA and conducted sequencing. C.S. and X.Z. performed bioinformatics analysis. C.S., L.C. and Y.C. wrote the manuscript. All authors read and approved the final manuscript.
Code availability
All software used in this work is in the public domain, with parameters being clearly described in Methods. If no detail parameters were mentioned for a software, default parameters were used as suggested by developer.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lianfei Cao, Email: caolf@mail.zaas.ac.cn.
Cheng Sun, Email: suncheng@caas.cn.
References
- 1.Knecht D, Kaatz HH. Patterns of larval food production by hypopharyngeal glands in adult worker honey bees. Apidologie. 1990;21:457–468. doi: 10.1051/apido:19900507. [DOI] [Google Scholar]
- 2.Kamakura M. Royalactin induces queen differentiation in honeybees. Nature. 2011;473:478–483. doi: 10.1038/nature10093. [DOI] [PubMed] [Google Scholar]
- 3.Ramadan MF, Al-Ghamdi A. Bioactive compounds and health-promoting properties of royal jelly: A review. J Funct Foods. 2012;4:39–52. doi: 10.1016/j.jff.2011.12.007. [DOI] [Google Scholar]
- 4.You MM, et al. Royal jelly alleviates cognitive deficits and b-amyloid accumulation in APP/PS1 mouse model via activation of the cAMP/PKA/CREB/BDNF pathway and inhibition of neuronal apoptosis. Front Aging Neurosci. 2019;10:428. doi: 10.3389/fnagi.2018.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng, H. Q., Cao, L. F., Huang, S. K., Neumann, P. & Hu, F. L. Current status of the beekeeping industry in China, In: Chantawannakul P., Williams G., Neumann P. (eds) Asian Beekeeping in the 21st Century. Springer Nature Singapore Pte Ltd., Singapore, 129–158 (2018).
- 6.Hu FL, et al. Standard methods for Apis mellifera royal jelly research. J Apic Sci. 2019;58:1–68. doi: 10.1080/00218839.2017.1286003. [DOI] [Google Scholar]
- 7.(CNCAGR) China National Commission of Animal Genetic Resources. Animal genetic resources in China –Bees. Chinese Agricultural Press, Beijing, China (2011).
- 8.Cao LF, Zheng HQ, Pirk CWW, Hu FL, Xu ZW. High royal jelly-producing honey bees (Apis mellifera ligustica) (Hymenoptera: Apidae) in China. J Econ Entomol. 2016;109:510–514. doi: 10.1093/jee/tow013. [DOI] [PubMed] [Google Scholar]
- 9.Sun LX, Chen ZY, Yuan JJ, Xie JJ. Genetic variability of MDHII in four lines of Apis mellifera ligustica. J Zhangzhou Teach Coll. 2004;17:54–59. [Google Scholar]
- 10.Chen SL, Li JK, Zhong BX, Su SK. Microsatellite analysis of royal jelly producing traits of Italian honeybee (Apis mellifera liguatica) Acta Genet Sin. 2005;32:1037–1044. [PubMed] [Google Scholar]
- 11.Cao LF, Zheng HQ, Shu QY, Hu FL, Xu ZW. Mitochondrial DNA characterization of high royal jelly-producing honeybees (Hymenoptera: Apidae) in China. J Apic Sci. 2017;61:217–222. doi: 10.1515/jas-2017-0016. [DOI] [Google Scholar]
- 12.Li JK, Feng M, Desalegn B, Fang Y, Zheng AJ. Proteome comparison of hypopharyngeal gland development between Italian and royal jelly producing worker honeybees (Apis mellifera L.) J Proteome Res. 2010;9:6578–6594. doi: 10.1021/pr100768t. [DOI] [PubMed] [Google Scholar]
- 13.Wu F, et al. Behavioural, physiological and molecular changes in alloparental caregivers may be responsible for selection response for female reproductive investment in honey bees. Mol Ecol. 2019;28:4212–4227. doi: 10.1111/mec.15207. [DOI] [PubMed] [Google Scholar]
- 14.Altaye SZ, Meng LF, Li JK. Molecular insights into the enhanced performance of royal jelly secretion by a stock of honeybee (Apis mellifera ligustica) selected for increasing royal jelly production. Apidologie. 2019;50:436–453. doi: 10.1007/s13592-019-00656-1. [DOI] [Google Scholar]
- 15.Nie HY, et al. Identification of genes related to high royal jelly production in the honey bee (Apis mellifera) using microarray analysis. Genet Mol Biol. 2017;789:781–789. doi: 10.1590/1678-4685-GMB-2017-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rizwan M, et al. Population genomics of honey bees reveals a selection signature indispensable for royal jelly production. Mol Cell Probes. 2020;52:101542. doi: 10.1016/j.mcp.2020.101542. [DOI] [PubMed] [Google Scholar]
- 17.Parpinelli RS, Ruvolo-Takasusuki MCC, Toledo VAA. MRJP microsatellite markers in Africanized Apis mellifera colonies selected on the basis of royal jelly production. Genet Mol Res. 2014;13:6724–6733. doi: 10.4238/2014.August.28.16. [DOI] [PubMed] [Google Scholar]
- 18.Wragg D, et al. Whole-genome resequencing of honeybee drones to detect genomic selection in a population managed for royal jelly. Sci Rep. 2016;6:27168. doi: 10.1038/srep27168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belton JM, et al. Hi–C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koren S, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roach MJ, Schmidt SA, Borneman AR. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 2018;19:460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durand NC, et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudchenko O, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flynn JM, et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci USA. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cantarel BL, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones PH, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.2021. NCBI Sequence Read Archive. SRP300170
- 33.2021. NCBI Assembly. GCA_019321825.1
- 34.Sun C. 2021. Genome annotation for high royal jelly-producing honeybee. figshare. [DOI]
- 35.Waterhouse RM, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2018;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cabanettes F, Klopp C. D-GENIES: Dot plot large genomes in an interactive, efficient and simple way. PeerJ. 2018;6:e4958. doi: 10.7717/peerj.4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christmas MJ, et al. Chromosomal inversions associated with environmental adaptation in honeybees. Mol Ecol. 2019;28:1358–1374. doi: 10.1111/mec.14944. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2021. NCBI Sequence Read Archive. SRP300170
- 2021. NCBI Assembly. GCA_019321825.1
- Sun C. 2021. Genome annotation for high royal jelly-producing honeybee. figshare. [DOI]
Data Availability Statement
All software used in this work is in the public domain, with parameters being clearly described in Methods. If no detail parameters were mentioned for a software, default parameters were used as suggested by developer.