Abstract
During ovarian follicular development, granulosa cells proliferate and progressively differentiate to support oocyte maturation and ovulation. To determine the underlying links between proliferation and differentiation in granulosa cells, we determined changes in 1) the expression of genes regulating DNA methylation and 2) DNA methylation patterns, histone acetylation levels and genomic DNA structure. In response to equine chorionic gonadotropin (eCG), granulosa cell proliferation increased, DNA methyltransferase (DNMT1) significantly decreased and Tet methylcytosine dioxygenase 2 (TET2) significantly increased in S-phase granulosa cells. Comprehensive MeDIP-seq analyses documented that eCG treatment decreased methylation of promoter regions in approximately 40% of the genes in granulosa cells. The expression of specific demethylated genes was significantly increased in association with specific histone modifications and changes in DNA structure. These epigenetic processes were suppressed by a cell cycle inhibitor. Based on these results, we propose that the timing of sequential epigenetic events is essential for progressive, stepwise changes in granulosa cell differentiation.
Subject terms: Reproductive biology, DNA methylation
Tomoko Kawai et al. investigate DNA methylation patterns during granulosa cell differentiation and proliferation. Their results suggest that sequential epigenetic events are essential for progressive changes in granulosa cell differentiation.
Introduction
Several epigenetic mechanisms have been identified in cells as they differentiate, including stem cell and embryonic specification, epithelial to mesenchymal changes in cancer cells, and immune cell lineages1–3. Specific factors that are known to control epigenetic processes include DNA methyltransferases (DNMTs), which act to methylate specific regions of the genome, especially CpG-rich promoter regions, and hence block and impair transcription4,5. Once the DNA methylation patterns in a specific cell type are established, they are copied to a daughter cell by the functions of the DNMT1/PCNA/UHRF1 complexes6. Methylation events are reversed and regulated by TET enzymes (Tet methylcytosine dioxygenases) that convert DNA methylcytosine to 5-hydroxymethylcytosine7,8. Collectively, these pathways control or modify the methylated CpG islands found frequently in promoters of key regulatory genes to induce specific gene expression patterns.
The development of ovarian follicles capable of ovulating and releasing a fertilizable oocyte is a complex process that is regulated by intrafollicular factors as well as peripheral hormones, including follicle stimulating hormone (FSH), which binds to the FSH receptor and regulates granulosa cell proliferation and functions that are essential supporters/regulators of oocyte growth and maturation9–11. Specifically, during the transition of secondary follicles (SFs) to antral follicles, granulosa cells express constant levels of the FSH receptor and produce estrogen but little to no progesterone, a marker of differentiated granulosa cells that have terminally differentiated into luteal cells9. During the growth of antral follicles to the preovulatory stage, proliferating granulosa cells differentiate in response to FSH and acquire the ability to respond to the ovulatory surge of luteinizing hormone (LH)9. The FSH-mediated differentiation process includes the induction of ~500 genes12,13, including Lhcgr, which encodes the LH receptor that is essential for LH-mediated ovulation14,15.
FSH-stimulated granulosa cell proliferation is dependent on the expression of the cell cycle regulator cyclin D216,17. Mutant mice lacking the Ccnd2 gene that encodes cyclin D2 exhibit female infertile phenotypes characterized by small follicles, depletion of granulosa cells, and altered gene expression patterns, including the lack of Lhcgr18, suggesting that cell proliferation might be involved in the differentiation of granulosa cells and the large-scale epigenetic changes that occur during follicular development and ovulation. Specifically, DNA methylation of the promoter region of Lhcgr in granulosa cells is ~50% in small follicles; however, methylation decreases to <20% in proliferating granulosa cells of preovulatory follicles (PFs)19. In contrast, Lhcgr is constitutively expressed in theca cells of growing and PFs, and the DNA methylation rate is low19, indicating that the DNA methylation pattern might be altered in granulosa cells but not in theca cells during the marked transition of granulosa functions in PFs. Canon et al.20 reported that proliferating B cells have much lower rates of DNA methylation in marker genes of plasma cells than nonproliferating B cells. However, the relationship between cell proliferation and epigenetic regulation of cell differentiation remains unclear.
The reduction of DNA methylation in the Lhcgr promoter region of granulosa cells is dependent on not only FSH but also the de novo synthesis of retinoic acid (RA) and SMAD pathways19. RA and SMADs are known factors involved in cell fate determination due to their impact on epigenetic modifications21,22. Therefore, the studies described herein were undertaken to analyze granulosa cells as a model to determine the underlying mechanisms by which DNA methylation changes dramatically in highly proliferative cells.
Results
Temporal changes occur in the expression of DNMT1 and other epigenetic regulatory factors in proliferating granulosa cells at specific stages of follicular development
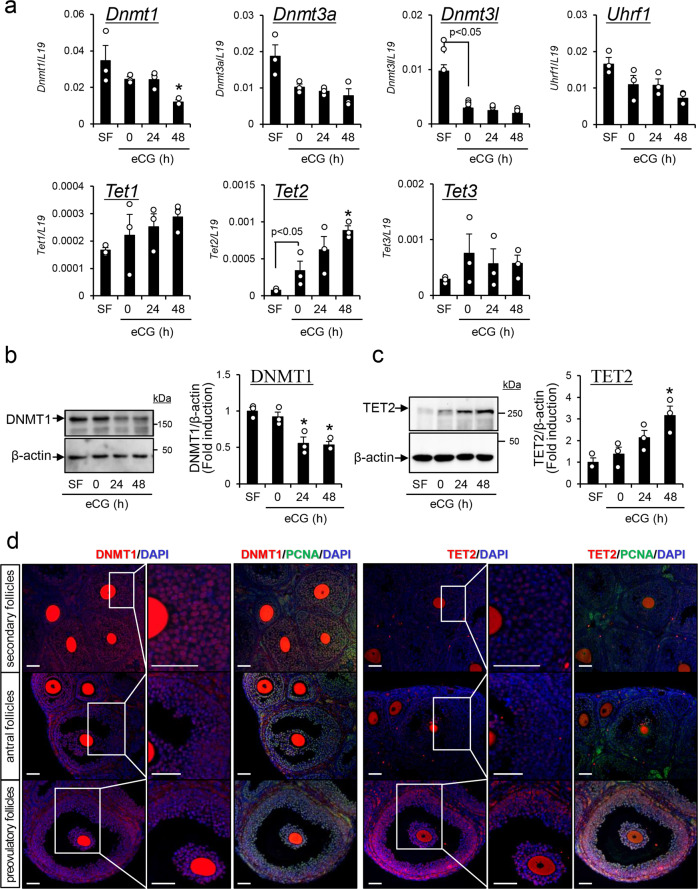
The induction of follicle development by equine chorionic gonadotropin (eCG) was used as a general treatment for superovulation in mice. The experimental design in detail was mentioned in Supplemental Notes 1–6. In granulosa cells of PFs isolated from mouse ovaries 48 h after eCG injection, the expression levels of Dnmt1 were significantly decreased compared with those in antral follicles isolated from mice before eCG injection (Fig. 1a; 0 h). In contrast, the expression of Dnmt3l was significantly decreased in granulosa cells in antral follicles isolated from mouse ovaries before eCG injection compared with that in SFs. Tet2 expression was significantly induced in antral follicles (eCG 0 h) from SFs; a further significant induction was observed in PFs at 48 h after eCG injection (Fig. 1a). The expression levels of Dnmt3a, Uhrf1, Tet1, and Tet3 did not change significantly during PF growth (Fig. 1a). Changes in DNMT1 and Tet methylcytosine dioxygenase 2 (TET2) in granulosa cells during follicular development were also analyzed by western blotting (Fig. 1b, c). DNMT1 significantly decreased in granulosa cells at 24 and 48 h after eCG injection compared with that before eCG injection (eCG 0 h). The level of TET2 was dramatically higher in granulosa cells at 48 h after eCG injection than before eCG injection (eCG 0 h).
Fig. 1. Kinetic changes in the expression levels of genes involved in epigenetic regulatory mechanisms in mouse granulosa cells during follicular development.
a The expression levels of epigenetic-related genes in granulosa cells. Levels of mRNA were normalized to that of L19. Data represents the mean ± SEM (n = 3 biological replicates). Significant differences were observed between SF and eCG at 0 h (p < 0.05). *Significant differences were induced by eCG injection compared with before eCG injection (0 h) (p < 0.05). SF, secondary follicles. b, c Temporal changes of DNMT1 (b) and TET2 (c) in granulosa cells of secondary follicles (SF) with multilayered granulosa cells and antral follicles before or after eCG stimulation. β-Actin was used as a loading control. The results are representative of three independent experiments. Immunoblots shown are representative of three independent experiments. In each experiment, the different protein samples were used (n = 3 biological replicates). Quantitative expression of DNMT1 and TET2 relative to the expression of β-actin (control), as determined by western blotting. The SF value was set as 1 and the data are expressed as fold induction. The values are the mean ± SEM (n = 3 biological replicates). Significant differences were observed between SF and eCG at 0 h (p < 0.05). *Significant differences were induced by eCG injection compared with before eCG injection (0 h) (p < 0.05). Full-length blotting images of DNMT1, TET2, and β-actin are shown in Supplementary Fig. 8. d The localization of DNMT1, TET2, and PCNA in secondary follicles (SFs) and antral follicles (AFs) of 3-week-old immature mice or preovulatory follicles (PFs) of ovaries collected 48 h after eCG injection. DNMT1 and TET2 were visualized with Cy3, PCNA was visualized with FITC, and nuclei were stained with DAPI in the ovaries of 3-week-old mice treated with or without eCG. All of different length scale bars show 50 μm.
To observe the cell-specific localization of DNMT1, TET2, and PCNA (a marker of proliferation) in granulosa cells during the S-phase of the mitotic cell cycle, immunofluorescence analyses were performed. Most of the cells surrounding oocytes (granulosa cells in SFs and cumulus cells in antral follicles and PFs) were DNMT1 positive (Fig. 1d and Supplementary Fig. 1). A high level of DNMT1 was also detected in granulosa cells of antral follicles, whereas DNMT1 was significantly decreased in granulosa cells of PFs (48 h after eCG injection). In contrast, the number of PCNA-positive cells was significantly increased in both granulosa cells and cumulus cells in response to eCG treatment (Fig. 1d and Supplementary Fig. 1). Most PCNA-positive cells were also DNMT1 positive in cumulus cells and in granulosa cells of SFs. However, the number of DNMT1-positive cells relative to PCNA-positive cells significantly decreased in a stage-dependent manner in granulosa cells as they differentiated (Fig. 1d and Supplementary Fig. 1). TET2-positive cells were localized in follicles from the secondary to preovulatory stages. However, TET2-positive cells were significantly increased in both cumulus cells and granulosa cells at 48 h after eCG injection (Fig. 1d and Supplementary Fig. 1). TET2 expression was observed in both proliferating (PCNA+) and nonproliferating (PCNA−) cells.
The induction of DNMT1 is significantly decreased in S-phase granulosa cells during follicular development
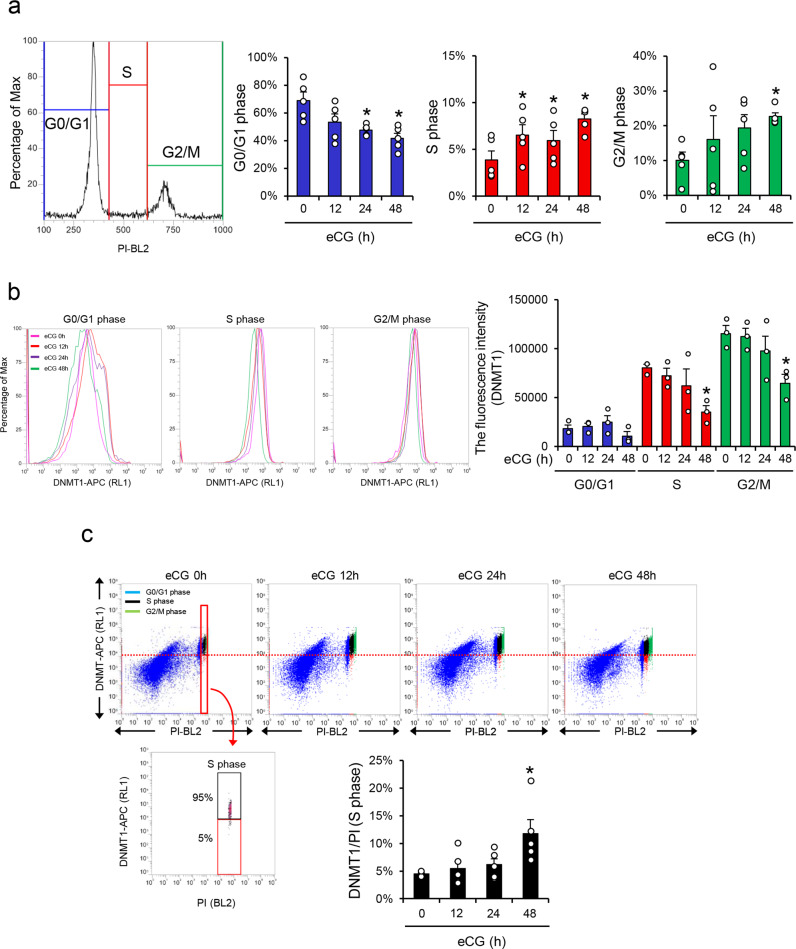
To determine the relationship between cell cycle progression and the expression level of DNMT1 in more detail, the stage of the cell cycle was analyzed by propidium iodide (PI) staining, and the expression level of DNMT1 was analyzed by flow cytometry in granulosa cells collected at specific time points. Dot blot graphs were made on the basis of FSC-A (forward scatter area) and SSC-A (side scatter area), and 20,000 granulosa cells were gated (Supplementary Fig. 2a, b). After eCG injection, the proliferative activity increased in granulosa cells; the percentage of cells in the G0/G1 phase was significantly decreased, and the percentages of cells in S-phase and G2/M-phase were significantly increased (Fig. 2a). In each cell cycle stage, the fluorescence intensity of DNMT1 was detected by flow cytometric analyses (Fig. 2b). DNMT1 increased from the G0/G1 stage to the S-phase. However, the fluorescence intensity of DNMT1 was significantly decreased in S-phase granulosa cells collected from large antral follicles (PFs) at 48 h after eCG injection (Fig. 2b). The lower threshold of the DNMT1 level at S-phase was set as 5% fewer expressed cells at eCG 0 h, as a statistically significant change (Fig. 2c, red dashed line). The number of cells expressing low levels of DNMT1 (red collar) at S-phase was significantly increased at 48 h after eCG injection (Fig. 2c).
Fig. 2. Relationship between the level of DNMT1 and cell cycle stages in granulosa cells during the follicular development process.
a, b Positive fluorescence signal of PI (a) or DNMT1 (b) in granulosa cells collected from ovaries before or after eCG injection. Granulosa cells were collected from 3-week-old mice and 3-week-old mice at 12, 24, or 48 h after eCG injection. The granulosa cells were incubated with Cy3-tagged anti-DNMT1 antibody and PI (propidium iodide), and then the fluorescence signals were detected by flow cytometry analysis. a Each percentage of G0/G1 phase cells, S-phase cells, or G2/M-phase cells is shown in the bar graph. b The mean fluorescence intensity was analyzed in DNMT1-positive cells. The unstained sample was used to set a region for the cells that were not stained (negative). All of experiments are represented as means ± SEM, n = 3 (Fig. 2b) or n = 5 (Fig. 2a) biological replicates. *Significant differences were observed between eCG at 0 h and eCG-primed mice (p < 0.05). Significant differences in percentage values were transformed into normally distributed numbers by angle transformation and then analyzed by one-way ANOVA. Tukey–Kramer was used as post hoc test. c Variations in positive fluorescence signals of DNMT1 in granulosa cells in each cell cycle stage. The dot plot graph of double-positive granulosa cells with DNMT1 and PI. The percentage of cells that expressed lower levels of DNMT1 at S-phase is shown in the bar graph. The lower threshold of DNMT1 level at S-phase was set as 5% of lower expressed cells at eCG 0 h (red line). The cells that expressed lower levels of DNMT1 are shown in the red box and cells with higher DNMT1 expression are shown in the black box at S-phase. Experiment is represented as means ± SEM, n = 5 biological replicates. *Significant differences were observed between eCG at 0 h and eCG-primed mice (p < 0.05). Significant differences in percentage values were transformed into normally distributed numbers by angle transformation and then analyzed by one-way ANOVA. Tukey–Kramer was used as post hoc test.
Genome-wide DNA demethylation occurred in the promoter regions of genes in granulosa cells of PFs
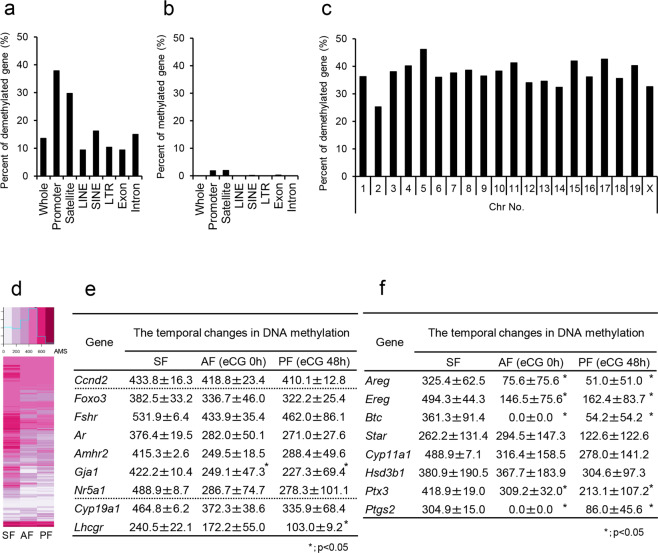
The comprehensive changes in the DNA methylation status in granulosa cells during follicular development were analyzed by methylated DNA immunoprecipitation coupled with next-generation sequencing (MeDIP-Seq) analyses. The absolute methylation signals (AMS) were used for the analysis of the percentage of DNA methylation of the promoter region in each gene23,24. The significantly demethylated genes in granulosa cells of PFs (48 h after eCG injection) compared with those in SFs represented 13.6% of the whole genome. The most dramatic changes in methylation status occurred in promoter regions (37.87%) and satellite regions (29.73%) of the genome (Fig. 3a). In contrast, increases in methylated regions in the whole genome of granulosa cells during follicular development were minor (Fig. 3b). The genes in which their promoter regions were demethylated accounted for more than 30% of all chromosomes, including the X chromosome (Fig. 3c). Cluster analysis also showed that as an overall trend, promoter regions were demethylated in a stage-dependent manner in granulosa cells during follicular development (Fig. 3d). The changes in the methylation status of specific genes highly expressed in granulosa cells during follicular development (Fig. 3e) and during the ovulation process (Fig. 3f) are listed. The methylation status did not change dramatically in the genes Ccnd2, Foxo3, Fshr, and Ar, which are highly expressed in SFs and maintained in PFs25,26. However, the methylation levels dramatically decreased to approximately half that in the Amhr2, Gja1, Nr5a1, and Lhcgr genes, in which transcription is known to be increased after eCG injection25,27,28. The Areg, Ereg, Btc, Star, Cyp11a1, Hsd3b, Ptx3, and Ptgs2 genes, which regulate ovulation and were induced after hCG injection29,30, were also dramatically demethylated in granulosa cells of PFs. Annotation analysis of demethylated genes was performed to identify the predicted functions in biological processes and the results are shown in Supplementary Tables 1 and 2.
Fig. 3. Comprehensive analysis of genome-wide DNA methylation status in granulosa cells during follicular development.
a, b The percentage of genes that were significantly demethylated (a) or methylated (b) during follicular development in each genomic region. c The percentage of significantly demethylated genes in promoter regions of each chromosome. d Clustering analysis of demethylated genes in promoter regions of granulosa cells during follicular development. SF, granulosa cells in secondary follicles with multilayered granulosa cells; AF, granulosa cells in antral follicles (3-week mice not treated with eCG); PF, granulosa cells in preovulatory follicles (at 48 h after eCG injection). e, f Temporal changes in DNA methylation of marker genes (e) and LH-targeting genes (f) in granulosa cells during the follicle development process. The values are represented as the mean ± SEM (n = 3 biological replicates). The mean ± SEM values from MeDIP-seq analysis are absolute methylation signals (AMS) in specific promoter regions containing CpG sites. For genes with multiple specific promoter regions, including CpG sites, the average value was calculated. The significant differences between values in antral follicles (3-week mice not treated with eCG) or preovulatory follicles (at 48 h after eCG injection) and that in granulosa cells of secondary follicles (SF) were analyzed (p < 0.05).
Validation of epigenetic modifications (DNA methylation status, H3K27 acetylation, and opened chromatin structure) in candidate genes identified by MeDIP-seq analysis in granulosa cells
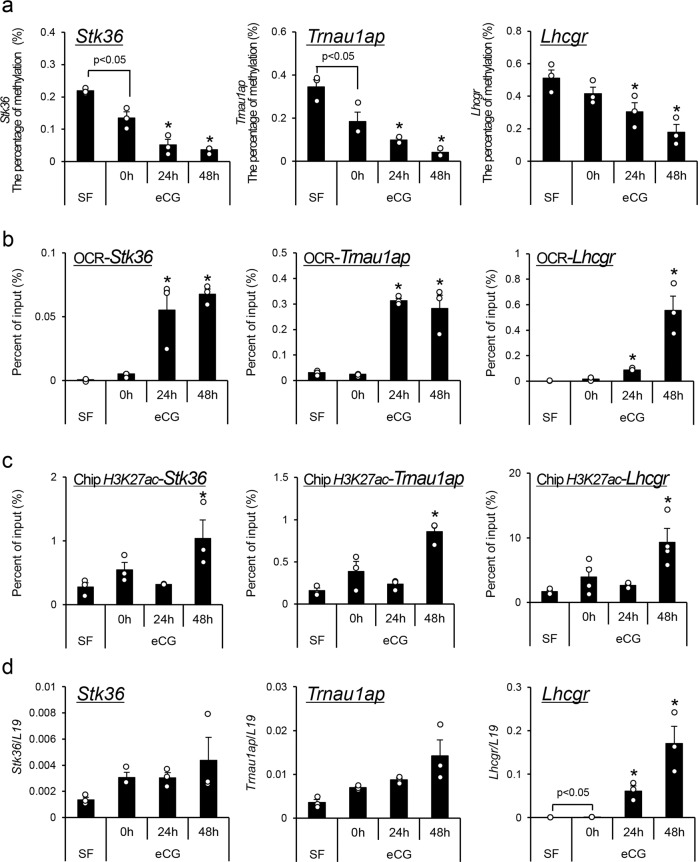
From the top nine most highly demethylated genes listed in Supplementary Table 3, Stk36 and Trnau1ap, as well as Lhcgr, were selected for validation of their promoter methylation status by bisulfite sequence analyses. In the promoter regions of the Stk36, Trnau1ap, and Lhcgr genes, the percentage of methylated CpG sites was significantly decreased in a time-dependent manner after eCG injection (Fig. 4a). Promoter demethylation of these genes was associated with significantly increased PCR products by formaldehyde-assisted isolation of regulatory elements-quantitative PCR (FAIRE-qPCR) (opened chromatin status of promoter regions, OCRs) (Fig. 4b). The acetylation of K27 in histone H3 (H3K27ac) was also increased in the promoter regions of the Stk36, Trnau1ap, and Lhcgr genes (Fig. 4c). Furthermore, the expression of mRNA for each gene, especially Lhcgr, was also significantly increased in granulosa cells after eCG treatment (Fig. 4d). Changes in DNA structure, histone acetylation and transcription were also significantly induced in the other seven demethylated genes listed in Supplementary Table 3 (Supplementary Fig. 3a–c).
Fig. 4. Changes in epigenetic regulation of the demethylated genes Stk36, Trnau1ap, and Lhcgr in granulosa cells during follicular development.
a The temporal changes in methylation of promoter regions were analyzed by bisulfite sequencing. Granulosa cells were collected from secondary follicles with multilayered granulosa cells or antral follicles (3-week mice not treated with eCG) before or after eCG injection. Values are represented as the mean ± SEM (n = 3 biological replicates). Significant differences were observed between SF and eCG at 0 h (p < 0.05). *Significant differences were induced by eCG injection compared with before eCG injection (0 h) (p < 0.05). SF, secondary follicles. b, c The kinetic changes of the opened chromatin region (OCR) detected by FAIRE-qPCR (b) or the acetylated H3K27 detected by ChIP assay (c) in the promoter region of each gene in granulosa cells of secondary follicles with multilayered granulosa cells or antral follicles (3-week mice not treated with eCG) before or after eCG injection. The SF value was set as 1 and the data are expressed as fold induction. Values are represented as the mean ± SEM (n = 3 biological replicates). Significant differences in percentage values were transformed into normally distributed numbers by angle transformation and then analyzed by one-way ANOVA. Tukey–Kramer was used as post hoc test. *Significant differences were induced by eCG injection compared with before eCG injection (0 h) (p < 0.05). d Kinetic changes of the expression level of each gene in granulosa cells. Levels of mRNA were normalized to that of L19. Values are represented as the mean ± SEM (n = 3 biological replicates). Significant differences were observed between SF and eCG at 0 h (p < 0.05). *Significant differences were induced by eCG injection compared with before eCG injection (0 h) (p < 0.05).
Histone modification and enhanced gene expression occurred only after hCG injection following eCG-induced demethylation and altered chromatin structure in LH target genes
The promoter regions of LH target genes (Areg, Ereg, Btc, Star, Cyp11a1, and Ptgs2) were demethylated by eCG stimulation in granulosa cells (Fig. 3f). The chromatin structure examined by FAIRE-qPCR assays was also opened in the promoter regions of these genes in granulosa cells 48 h after eCG injection (just before hCG injection) (Supplementary Fig. 4a). However, the levels of H3K27 acetylation of these LH target genes in granulosa cells were not significantly changed by eCG injection (Supplementary Fig. 4b). The injection of hCG following 48 h after eCG stimulation was required to increase the levels of H3K27 acetylation in the promoter regions of LH target genes (Supplementary Fig. 4b). The higher total level of H3K27 acetylated histone H3 was maintained by 4 h after hCG injection (Supplementary Fig. 5). Increased acetylation occurred simultaneously with significantly increased expression of the LH target genes in eCG-primed granulosa cells exposed to an ovulatory dose of hCG (Supplementary Fig. 4c).
A cell cycle inhibitor significantly suppressed DNA demethylation in FSH- and testosterone-stimulated granulosa cells
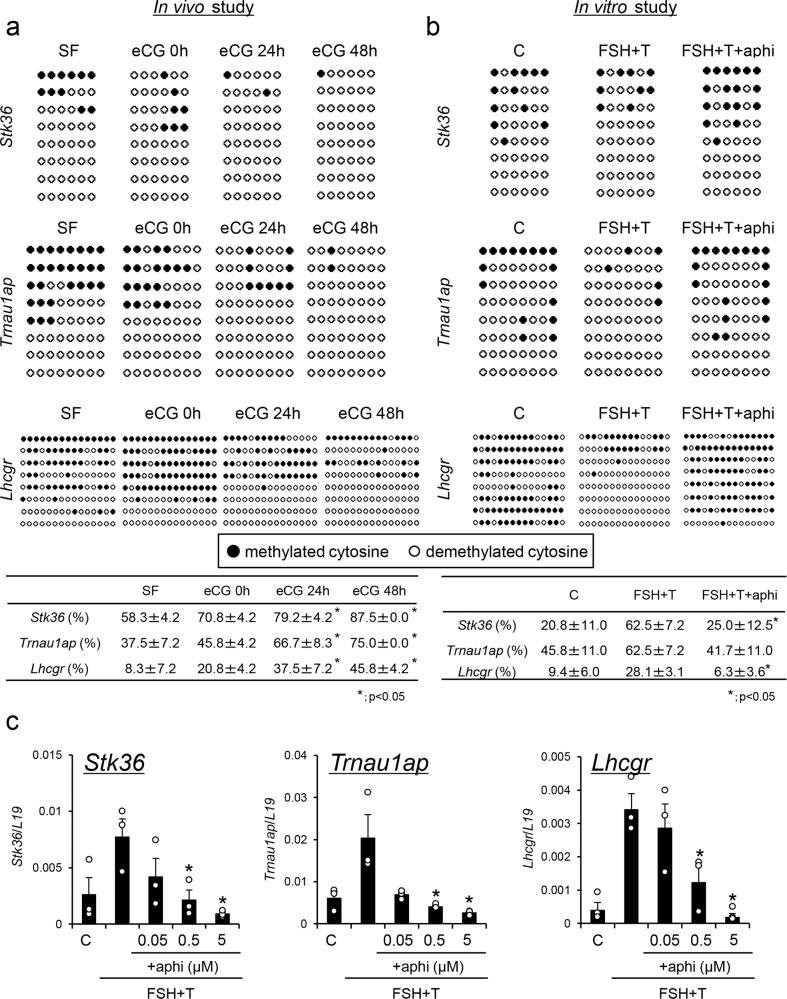
To determine the mechanisms that mediate promoter demethylation of the Stk36, Trnau1ap, and Lhcgr genes in granulosa cells, the percentage of CpG islands in which all CpG sites were not methylated in each promoter region was calculated based on bisulfite sequence analysis. The percentage of nonmethylated CpG islands of Stk36, Trnau1ap, and Lhcgr significantly increased in granulosa cells obtained from SFs to PFs in vivo (Fig. 5a). When granulosa cells collected from immature mice were cultured with FSH and testosterone, which are known as inducers of granulosa cell differentiation in vitro19, an increase in nonmethylated CpG islands was also observed (Fig. 5b). The ratio of demethylated CpG islands was significantly reduced by the addition of aphidicolin, a cell division inhibitor (Fig. 5b). Likewise, the expression of Stk36, Trnau1ap, and Lhcgr was increased in granulosa cells cultured with FSH and testosterone (FSH + T), whereas the induction of these genes was significantly suppressed by aphidicolin in a dose-dependent manner (Fig. 5c). These results indicated that demethylation was linked to cell cycle progression and was possibly related to changes in the cellular levels of DNMT1 and TET2 (Fig. 1).
Fig. 5. The effect of cell cycle inhibitor on the demethylation of Stk36, Trnau1ap, and Lhcgr, and on their expression in granulosa cells during follicular development.
a The percentage of nonmethylated CpG islands in the promoter regions of Stk36, Trnau1ap, and Lhcgr in granulosa cells during the follicular development process. ●, Methylated cytosine. ○, Unmethylated cytosine. SF, secondary follicles. Values are represented as the mean ± SEM (n = 3 biological replicates). *There were significant differences compared to SF (p < 0.05). Significant differences in percentage values were transformed into normally distributed numbers by angle transformation and then analyzed by one-way ANOVA. Tukey–Kramer was used as post hoc test. b The effects of the cell cycle inhibitor aphidicolin (aphi) on the appearance of nonmethylated CpG islands in the promoter regions of Stk36, Trnau1ap, and Lhcgr in granulosa cells cultured with FSH and testosterone (FSH + T) in the presence of 1% FBS. ●, Methylated cytosine. ○, Unmethylated cytosine. C, without any hormones (control). Values are represented as the mean ± SEM (n = 3 biological replicates). *Significant differences were observed between the control group and the aphidicolin treatment group in the presence of FSH + testosterone and 1% FBS (p < 0.05). Significant differences in percentage values were transformed into normally distributed numbers by angle transformation and then analyzed by one-way ANOVA. Tukey–Kramer was used as post hoc test. c The effects of aphidicolin on the expression of Stk36, Trnau1ap, and Lhcgr in granulosa cells. Granulosa cells were cultured without any hormones (control C) or with FSH (100 ng/ml) and testosterone (T; 10 ng/ml) without or with different concentrations of aphidicolin (aphi: 0.05, 0.5, 5 μM) in the presence of 1% FBS. Levels of mRNA were normalized to that of L19. Values are represented as the mean ± SEM (n = 3 biological replicates). *Significant differences were observed between the control group and the aphidicolin treatment group in the presence of FSH + testosterone and 1% FBS (p < 0.05).
Oocyte-secreted factors blocked the reduction of Dnmt1 expression and the induction of Tet2, whereas RA overcame the negative effects
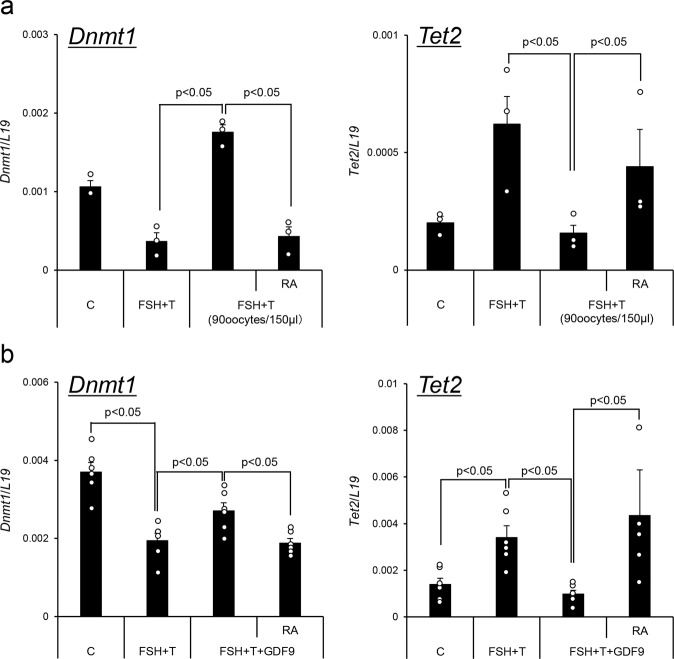
To determine factors that impact epigenetic events mediated by DNMT1 and TET2 during granulosa cell differentiation, granulosa cells were cultured with FSH and testosterone (FSH + T) in the presence of serum. The expression of Dnmt1 mRNA was significantly decreased, whereas the expression of Tet2 mRNA was significantly increased (Fig. 6a, b). However, when granulosa cells were cocultured with denuded oocytes, the effects of FSH + T were dramatically reduced (Fig. 6a). The reduced expression of Dnmt1 and the induction of Tet2 were also regulated by additional treatment with GDF9 (Fig. 6b). However, the addition of RA to the medium overcame the negative effects mediated by coculture with denuded oocytes or treatment with GDF9 (Fig. 6a, b).
Fig. 6. Oocytes impact the expression of Dnmt1 and Tet2 in granulosa cells and cumulus cells.
a The effect of coculture with denuded oocytes on the expression of Dnmt1 and Tet2 in granulosa cells. Granulosa cells were collected from ovaries of 3-week-old mice after treatment with eCG for 6 h and were cultured in medium supplemented with FSH plus T in the presence of 1% FBS. GV-stage oocytes were collected from COCs. None or 90 GV-stage oocytes/150 μl were cocultured with granulosa cells. Values are represented as the mean ± SEM (n = 3 biological replicates). Levels of mRNA were normalized to that of L19. FSH + T. Granulosa cells were cultured with FSH (100 ng/ml) and testosterone (10 ng/ml) for 48 h. RA, RA (1 μM) was added to medium in which granulosa cells were cultured with 90 GV-stage oocytes. Significant differences were observed compared with cells treated with no oocytes or cells treated with 90 oocytes per 150 μl of medium (p < 0.05). b The effects of GDF9 on the expression of Dnmt1 and Tet2 in granulosa cells. Granulosa cells were collected from ovaries of 3-week-old mice after treatment with eCG for 6 h and were cultured in each treatment group in the presence of 1% (v/v) FBS. control (C); granulosa cells were cultured without any hormones for 48 h. FSH + T, granulosa cells were cultured with FSH (100 ng/ml) and testosterone (10 ng/ml) for 48 h. FSH + T + GDF9, granulosa cells were cultured with FSH + T and mouse recombinant GDF9 (100 ng/ml). FSH + T + GDF9 + RA, Granulosa cells were cultured with FSH + T + GDF9 and RA (1 μM) for 48 h. Levels of mRNA were normalized to that of L19. Values are represented as the mean ± SEM (n = 6 biological replicates). Significant differences were observed among the treatment groups (p < 0.05).
RA is a key regulator of DNMT1 expression in S-phase granulosa cells
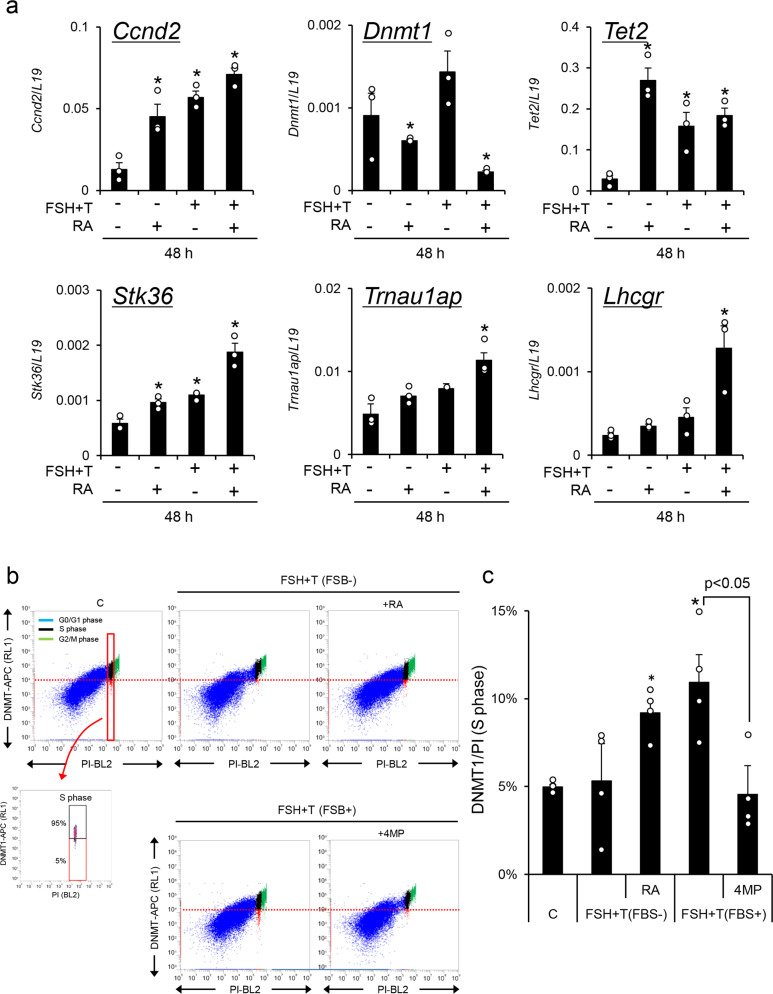
To determine how the expression levels of Ccnd2, Dnmt1, Tet2, and Lhcgr were regulated in granulosa cells during follicular development, granulosa cells were cultured with FSH + testosterone (FSH + T) and/or RA in the absence of serum (which contains retinol and an RA precursor). Treatment with RA alone significantly decreased Dnmt1 expression and increased Tet2 expression (Fig. 7a). The DNA methylation ratios in CpG islands in the promoter regions of Stk36 and Trnau1ap were slightly but not significantly decreased; however, further significant reduction was induced by the addition of FSH + T to the RA-containing medium (Supplementary Fig. 6). On the other hand, FSH + T alone (without RA) did not induce the suppression of Dnmt1 or demethylation of CpG islands in the promoter regions of Stk36 and Trnau1ap when granulosa cells were cultured without fetal beef serum. The expression patterns of Stk36 and Trnau1ap were similar to those in Lhcgr (Fig. 7a). The percentage of cells that expressed low levels of DNMT1 at S-phase was significantly increased by the addition of RA to FSH + T-containing serum-free medium. Using serum-containing medium, FSH + T significantly increased the percentage of cells that expressed low levels of DNMT1. The induction was significantly suppressed by the additional 4MP (ADH inhibitor), which reduced the cellular levels of RA (Fig. 7b, c). Collectively, these results indicate that RA exerts potent regulatory effects on the expression of DNMT1 in granulosa cells.
Fig. 7. The roles of retinoic acid (RA) in epigenetic regulation in granulosa cells during follicular development.
a The expression of Ccnd2, Dnmt1, Tet2, Lhcgr, Stk36, and Trnau1ap in cultured granulosa cells. Granulosa cells were collected from ovaries of 3-week-old mice after treatment with eCG for 6 h and were cultured in the absence of serum. FSH (100 ng/ml). T, testosterone (10 ng/ml). RA (1 μM). Levels of mRNA were normalized to that of L19. Values are represented as the mean ± SEM (n = 3 biological replicates). The value of the control (without any factors) was set as 1 and the data are presented as fold induction. *Significant differences were observed between the control group and the RA and/or FSH + T treatment group (p < 0.05). b, c The percentage of cells that expressed lower levels of DNMT1 was significantly increased by RA but decreased by the alcohol dehydrogenase (ADH) inhibitor (4MP). b The dot plot graph of double-positive granulosa cells with DNMT1 and PI during follicular development. c The percentage of cells that expressed lower levels of DNMT1 at S-phase. The lower threshold of DNMT1 levels at S-phase was set as 5% of cells with lower DNMT1 expression in the control group (C; without any factors) (red line). The cells that expressed lower levels of DNMT1 are shown in the red box, and cells with higher DNMT1 expression are shown in the black box at S-phase. Values are represented as the mean ± SEM (n = 4 biological replicates). *Significant differences were observed between the control group and the RA treatment group in the presence of FSH + T or the FSH + T treatment group in the presence of 1% FBS (p < 0.05). Significant differences were observed compared with those treated with no 4MP (p < 0.05). Significant differences in percentage values were transformed into normally distributed numbers by angle transformation and then analyzed by one-way ANOVA. Tukey–Kramer was used as post hoc test.
Discussion
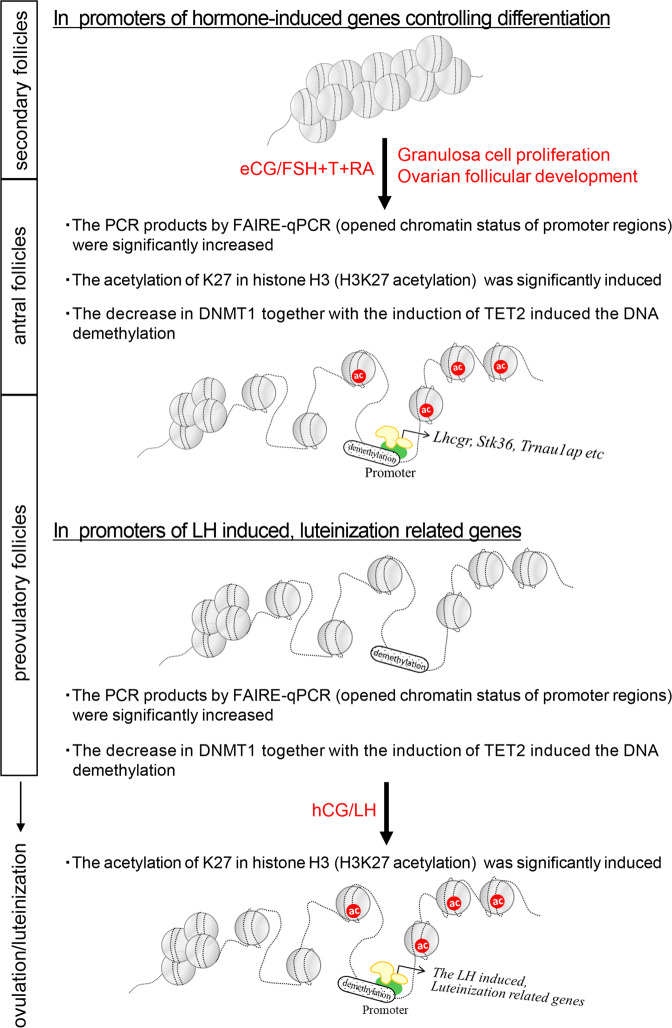
Development-related changes in tissue specification depend on processes that tightly regulate cell proliferation and differentiation. Although transcription factors play a key role in gene activation, gene expression patterns in each cell also depend on changes in cell-specific chromatin structure, notably the open chromatin structure in the S-phase of the cell cycle and in promoter and enhancer regions of activated genes31–33. Genomic structural variations are regulated by the differences in DNA methylation and histone modifications34 that are controlled by hormones and signaling pathways that impact cell proliferation and differentiation20,35. In this study, we document that hormone-regulated proliferation and differentiation of granulosa cells during follicular development from the secondary to preovulatory stage are dependent on specific temporal changes in DNA methylation and histone acetylation (Fig. 8).
Fig. 8. A schematic of epigenetic-related mechanisms in granulosa cells during follicular development process.
DNA demethylation and histone modification contribute to changes in gene expression as proliferating granulosa cells undergo progressive differentiation during follicular maturation.
In proliferating cells, DNA replication points are formed at G1 phase, in which genomic AT-rich regions are open35. At S-phase, proteins involved in DNA replication, such as DNA helicase and PCNA, are recruited to these regions36. DNMT1 is also observed in replication sites and interacts with PCNA to ensure inheritance of the existing DNA methylation patterns at S-phase and faithful daughter cell identity37. During S-phase, UHRF1 and DNMT3a interact with DNMT1 on the N-terminal domain and convert unmethylated cytosine to 5-methylated cytosine38. When TET factors are recruited to methylated regions of DNA during S-phase, TET converts 5-methylated cytosine to 5-hydroxymethylcytosine via oxidative demethylation mechanisms, leading to demethylation of methylated DNA regions39. In the present study, DNMT1 was increased in PCNA-positive S-phase granulosa cells compared with that in cells in the G0/G1 phase, thereby participating in replication accuracy. However, the level of DNMT1 in S-phase decreased in granulosa cells exposed to eCG, to induce follicular development, indicating that the DNA methylation status associated with the induction of genes controlling differentiation would be related to both the loss of the maintenance of 5-methylated cytosine due to decreased levels of DNMT1 and the conversion from 5-methylated cytosine to 5-hydroxymethylcytosine due to an increase in TET2 in S-phase. Strikingly, CpG islands in which all CpG sites were not methylated in each promoter were newly observed in granulosa cells after eCG injection in the promoter regions of Lhcgr, Stk36, and Trnau1ap (which included the top nine demethylated genes in granulosa cells). The findings supported our hypothesis that the methylation pattern would not be copied in daughter cells where the expression level of DNMT1 was low. This model by which granulosa cell functions change with proliferation is compatible with the mechanisms by which B cells and epithelial cells acquire new functions in a cell proliferation-dependent manner20,35.
To identify the promoters of genes that were selectively demethylated in S-phase during granulosa cell differentiation, MeDIP-Seq analyses were performed. Highly demethylated promoters accounted for ~40% of the genome, including not only well-known granulosa cell markers but also promoters of other genes. Annotation analysis predicted changes in metabolic processes, phosphorylation/signaling pathways, cytoskeletal organization, transmembrane transport, catabolic processes, and chromatin modification. The factors involved in transcription and DNA modification were also significantly changed. During the follicular development process, mitochondrial ATP production is dominant and increased in granulosa cells after eCG injection, and this process is required for cell proliferation40. Multiple signaling pathways are activated in granulosa cells during the follicular development process, including the serine/threonine kinase or PI3K/AKT pathways41. In the top nine demethylated genes, Stk36 (serine/threonine kinase 36) inhibits the activation of Gli factors that are activated by the hedgehog signal transduction cascade and regulate cell proliferation and tumorigenesis42. Tranu1ap (tRNA selenocysteine 1 associated protein 1) has been shown to inhibit proliferation in cancer cells by acting through the PI3K/AKT pathway43. Moreover, it is also well known that in granulosa cells, changes in the cytoskeleton and cell membrane transport occur during the follicular development process44, suggesting that dynamic DNA demethylation in the promoter region of the genome is essential for granulosa cell proliferation and differentiation. However, the functions of most demethylated genes detected in this study, including the top nine genes, such as Stk36 and Trnau1ap, remain to be determined.
Interestingly, in the genes that were induced in granulosa cells during the ovulation process (after hCG stimulation), the demethylation of promoter regions had already occurred before hCG stimulation. In the promoters of LH target genes, Star and Cyp11a1, histone-H4 acetylation (Ac-H4) and trimethylation of histone H3 lysine-4 (H3K4me3) are increased, whereas H3K9me3 and H3K27me3 are decreased in luteinized granulosa cells after hCG injection45. Of critical importance, granulosa cells isolated from small follicles are not capable of undergoing luteinization when cultured with LH or forskolin + phorbol 12-myristate 13-acetate (PMA), which are compounds that mimic LH signaling in preovulatory granulosa cells46. In contrast, granulosa cells isolated from PFs not only express high levels of the LH receptor but also rapidly differentiate into luteinized cells by both treatments46, suggesting that histone modification is induced by the LH-stimulating signaling pathway in promoter regions where DNA demethylation and changes in DNA structure have already occurred. The histone modifications and the expression of LH target genes critical for ovulation were induced by only hCG in this study, as shown in previous reports47. On the other hand, in FSH-target genes that were expressed in granulosa cells by eCG injection, DNA demethylation, changes in DNA structure and histone modification were induced at the same time points. The timing of histone modification is dependent on the types of transcription factors that are activated by different signaling pathways and recruited to histone acetylation factors48,49. Thus, the order of DNA demethylation, changes in DNA structure, and histone acetylation appears to be strictly determined in each promoter region in granulosa cells.
Granulosa cells and cumulus cells undergo differentiation in antral follicles; however, their functional differences appear to be related, in part, to factors produced either specifically by the oocyte (GDF9 and BMP15) that activate SMAD signaling or by the granulosa cells (RA) that enhance differentiation19,50. Cumulus cells directly surround oocyte and transfer energy sources via gap junctional communication to oocyte during the follicular development process51,52. After the LH surge of ovulational stimuli, cumulus cells are not luteinized but accumulate hyaluronan-rich matrix, which impacts ovulation and fertilization processes53. Granulosa cells are luteinized to form the corpus luteum, which produces progesterone to induce and maintain pregnancy54. Herein, we show that coculture of granulosa cells with denuded oocytes maintained the expression of Dnmt1 but suppressed the expression of Tet2, thus favoring promoter methylation in granulosa cells. These effects on oocytes were also induced by treatment with GDF9 and their negative effects were completely overcome by exogenous RA. RA is produced from retinol in two steps55, and the reactions are dependent on the expression of ADH and ALDH in FSH-stimulated granulosa cells during follicular development56. Importantly, the induction of both enzymes is suppressed in granulosa cells by coculture with oocytes19. Thus, the proliferation of granulosa cells that leads to an increase in follicle diameter might be required to allow RA synthesis by reducing the local concentration of oocyte-secreted factors in follicular fluid.
In antral follicles, follicular fluid accumulates within the follicle that separates granulosa cells from the enclosed cumulus cell–oocyte complex. As cumulus cells are strongly regulated by the oocyte via oocyte-secreted factors that activate SMAD pathways57, RA production is limited19, and a high level of Dnmt1 was observed in cumulus cells of PFs. Thus, the distance from the oocyte determines the epigenetic status in follicular somatic cells and their fate as cumulus cells or granulosa cells. In other words, cell proliferation first indirectly weakens the mechanisms of precise copying of the DNA methylation status due to the distance from oocyte and second, cell proliferation directly changes the methylation status and transcriptome in granulosa cells during the follicular development process. Collectively, these results indicate that the epigenetic regulation of granulosa cell differentiation mediated by cell proliferation, RA, and oocyte-secreted factors is one of the most highly orchestrated processes in female reproduction.
The maturation and developmental competence of oocytes decrease with increasing age in not only female mice but also women58,59. One of the reasons has been reported to be that the level of oocyte-secreted factors is lower in oocytes recovered from aged mice than in oocytes recovered from younger mice60. In older infertility patients, abnormal luteinization and low quality of oocytes in small antral follicles have been observed61,62. The decreasing ovarian functions in older women/female mice would be involved in abnormal promoter DNA methylation of critical genes in granulosa cells and cumulus cells, based on our evidence that oocyte-secreted factors strongly regulate the epigenetic changes in both cells. Moreover, bacterial infections of the female genital tract result in pelvic inflammatory disease, which causes infertility63. The injection of lipopolysaccharide, which is a component of gram-negative bacteria, alters the DNA methylation status in the Lhcgr promoter region in granulosa cells of the mouse ovary64. Based on the present study that focused on the changes in epigenetic status in granulosa cells during follicular development in immature mice, it is expected that analyses of the epigenetic status in granulosa cells will make it possible to identify potential causes of reproductive disorders and infertility. In particular, the analysis of DNA methylation in granulosa cells may lead to the development of new treatments and preventions for infertility or the development of contraceptives, because DNA methylation status is an index (or measure) of whether gene expression is possible or not.
In conclusion, during follicular development, granulosa cells progressively proliferate and differentiate to acquire the ability to respond to LH, promote ovulation and undergo luteinization. Importantly, we show herein that this process involves progressive changes in the DNA methylation patterns that are regulated by the induction of TET2 and the decrease of DNMT1 in S-phase in granulosa cells of eCG-stimulated antral follicles. The induction of TET2 and the decrease of DNMT1 are regulated not only by FSH and testosterone but also by increased levels of RA signaling and decreased oocyte-secreted factors such as GDF9. Furthermore, genome-wide DNA demethylation leads to opened genome structures that, when coupled with specific histone modifications (acetylation), promote the activation of the transcription of specific genes at specific time points. We propose that the timing of sequential molecular and epigenetic events is essential for the stepwise changes in granulosa cell differentiation during follicular development and ovulation and that the RA signaling pathway plays a key role in modifying the effects of gonadotropins and oocyte-secreted factors.
Methods
Materials
Dulbecco’s modified Eagle medium (DMEM)/F12 and penicillin–streptomycin were purchased from Invitrogen (Carlsbad, CA, USA), fetal bovine serum was purchased from Life Technologies, Inc. (Grand Island, NY, USA), oligonucleotide poly-(dT) was purchased from Invitrogen, and AMV reverse transcriptase was purchased from Promega Corp. (Madison, WI, USA). Routine chemicals and reagents were obtained from Nacalai Chemical, Co. (Osaka, Japan) or Sigma Chemical, Co. (St. Louis, MO, USA).
Animals
Immature female (3-week-old) C57BL/6 mice were obtained from Charles River Laboratories Japan (Yokohama, Japan). Twenty-two-day-old female mice were injected intraperitoneally with 4 IU of eCG to stimulate follicular growth; after 48 h, they were injected with 5 IU of hCG to stimulate ovulation and luteinization. In our experiment, granulosa cells or ovaries were collected from one mouse in each treatment group for real-time PCR analysis, western blotting, and immunofluorescence staining. For low cytometry, granulosa cells were collected from two mice in each treatment group and four mice were used for bisulfite sequence assay, MeDIP-Seq analysis, chromatin immunoprecipitation (ChIP) assay, and FAIRE-qPCR analysis in each treatment group. The animals were housed under a 12 h light/12 h dark schedule in the Experimental Animal Center at Hiroshima University and provided with food and water ad libitum. The animal study was approved by the Hiroshima University Animal Committee (Permit Number: C18–34) and the mice were maintained in accordance with the Hiroshima University Guidelines for the Care and Use of Laboratory Animals.
Isolation of granulosa cells
SFs with multilayered granulosa cells (Type 5b65) were isolated from mouse (2-week-old) ovaries by puncture with a 26 G 1/2 needle under a stereomicroscope. Granulosa cells were collected from antral follicles in ovaries of immature (3-week-old) mice at 0, 12, 24, or 48 h after eCG injection or 4 or 8 h after hCG injection following 48 h eCG injection by puncture with a 26 G 1/2 needle under a stereomicroscope (Supplementary Fig. 7).
RNA extraction and real-time PCR
Total RNA was obtained from mouse granulosa cells of SFs with multilayered granulosa cells (Type 5b), mouse granulosa cells or cumulus-oocyte complexes (COCs) using RNAeasy Mini Kit (Qiagen Sciences, Germantown, MD, USA) according to the manufacturer’s instructions. The RNA samples were treated with DNase (Qiagen) and 50 ng of total RNA was reverse transcribed in 20 μl reaction buffer. Three microliters of cDNA was used for real-time PCR. Real-time PCR was performed on a StepOne Real-Time PCR System (Applied Biosystems, Warrington, UK) with power SYBR Green PCR Master Mix (Applied Biosystems) at 60 °C for annealing temperature and 40 cycles of amplification. The primer sets are shown in Supplementary Table 5. L19 was used as a control for reaction efficiency and variations in concentrations of mRNA in the original reverse transcription (RT) reaction.
Genomic DNA extraction
Genomic DNA was obtained from granulosa cells of SFs with multilayered granulosa cells (Type 5b), granulosa cells at 0, 24, and 48 h after eCG injection or cultured granulosa cells of mouse ovaries using QIAamp DNA Blood Mini Kit (Qiagen Sciences) according to the manufacturer’s instructions. One milligram of genomic DNA was prepared for the bisulfite sequence assay.
MeDIP-Seq analysis
One microgram of DNA was used as an input sample and treated with MethylMiner Methylated DNA Enrichment kit (Invitrogen). Three biological replicates were used for each group. The DNA was sonicated by Covaris S2 (Covaris, Woburn, MA) under the following conditions (setting: value, duty cycle: 10%, intensity: 5, cycles per burst: 200, time: 2 cycles of 60 s each). The average size of the fragment after sonication was 350 bp. The fragmented DNA was dissociated into single strands, and the methylated region was enriched by the methyl-CpG binding domain of the human MBD2 protein using MethylMiner Methylated DNA Enrichment Kit (Invitrogen66,67). Quantification of DNA methylation at multiple false discovery rates (FDRs) was adjusted to the p-value (p < 0.05) cutoff. TruSeq DNA HT Sample Prep kit (Illumina, Tokyo, Japan) was used for the paired end library method. The sequence of the collected sample was decoded using an Illumina HiSeq 2000 and the methylated region was identified by MEDIPS software. AMS were used to judge the DNA methylation level of the promoter region in each gene. The prediction of gene function was analyzed using Functional Annotation Bioinformatics Microarray Analysis (https://david-d.ncifcrf.gov/tools.jsp). The accession number for all sequence data shown in this study is DNA Data Bank of Japan Sequence Read Archive: DRA010809 (http://trace.ddbj.nig.ac.jp/dra/index_e.html). The analysis of methylation of imprinted genes is shown in Supplementary Table 4.
Bisulfite sequence assay
One milligram of genomic DNA and 0.3 M NaOH (Nacalai) were added to the sodium bisulfite reaction mix using EpiTect Bisulfite Kit (Qiagen Sciences). Following the reaction, the bisulfite-converted DNA was purified and then used for PCR using specific primer sets to amplify the sequences containing CpG islands in the promoter region of each gene. The primer sets are shown in Supplementary Table 6. Sequence analysis of PCR products was performed on more than eight colonies for each treatment. The methylation of CpG islands in the promoter region was analyzed by QUantification tool for Methylation Analysis (http://quma.cdb.riken.jp/). The percentage of nonmethylated CpG islands in the promoter region of each gene was calculated as the percentage of colonies with all nonmethylated CpG islands in the eight colonies.
ChIP assay
The DNA–protein complexes were collected from SFs with multilayered granulosa cells (Type 5b) or granulosa cells of mouse (3-week-old) ovaries at 0, 24, and 48 h after eCG injection or at 4 and 8 h after hCG injection. The ChIP assay was performed using SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) (Cell Signaling, MA, USA)) according to the manufacturer’s instructions. Three or four biological replicates were performed for each group. Then, 100–1000 bp of the sheared chromatin was obtained by ultrasonic disruptor (TOMY UD-200). ChIP was performed using anti-histone H3 containing the acetylated lysine 27 (H3K27ac) antibody (1 : 100) (Cell Signaling). Normal rabbit IgG antibody (1 : 100) (Cell Signaling) was used as a negative control and histone H3 (D2B12) XP® rabbit mAb (ChIP formula) (1 : 100) (Cell Signaling) was used as a positive control. Three microliters of the ChIP DNA or total input DNA (diluted to 1/50) were used for real-time PCR. Real-time PCR was performed on a StepOne Real-Time PCR System (Applied Biosystems) with power SYBR Green PCR Master Mix (Applied Biosystems) at 60 °C for annealing temperature and 40 cycles of amplification. The primer sets used for the detection of each specific region are shown in Supplementary Table 6.
FAIRE-qPCR analysis
SFs with multilayered granulosa cells (Type 5b) were isolated from mouse (2-week-old) ovaries, and granulosa cells were isolated from mouse (3-week-old) ovaries at 0, 24, and 48 h after eCG injection or at 4 and 8 h after hCG injection. Recovery of the genome regions of open chromatin was performed according to an established procedure68,69. Three microliters of the opened chromatin DNA or total input DNA (diluted to 1/10) were used for real-time PCR. Real-time PCR was performed on a StepOne Real-Time PCR System (Applied Biosystems) with power SYBR Green PCR Master Mix (Applied Biosystems) at 60 °C for annealing temperature and 40 cycles of amplification. The primer sets used for the detection of each specific region are shown in Supplementary Table 6.
In vitro culture of granulosa cells
Granulosa cells were collected from ovaries of immature (3-week-old) mice at 6 h following injections of eCG19. Cells were seeded onto serum-recoated 24- or 96-well plate. Granulosa cells were treated with 100 ng/ml FSH (NIDDK, Torrance, CA)/DMEM/F12 medium, testosterone (10 ng/ml, Sigma)/ethanol, and/or RA (1 μM, Sigma)/dimethyl sulfoxide (DMSO) in the presence or absence of 1% (v/v) fetal bovine serum. Some granulosa cells were treated with 0.05, 0.5, or 5 μM aphidicolin (Sigma)/DMSO or 100 ng/ml GDF9 (mouse recombinant GDF9, R&D Systems, Inc., Minneapolis, MN) in the presence of FSH, testosterone, and 1% (v/v) fetal bovine serum.
Isolation and coculture of oocytes with granulosa cells
Granulosa cells were collected from ovaries of immature (3-week-old) mice as described previously in the “Methods” section. COCs isolated from each ovarian antral follicle by needle puncture. The cumulus cells surrounding oocyte were removed by repeating pipetting using a glass pipette under the stereomicroscope. The denuded oocytes at germinal vesicle (GV) stage (GV-stage oocytes) were used for the coculture study. Granulosa cells and GV-stage oocytes were cocultured in a 96-well plate (HTS Transwell-96 Permeable Support with 8.0 µm Pore Polyester Membrane no. 3374; Corning Incorporated, Kennebunk, ME) for 48 h and treated with 100 ng/ml FSH and 10 ng/ml testosterone or 1 μM RA in the presence of 1% fetal bovine serum and 0.2 mM isobutylmethylxanthine (IBMX). IBMX was added to suppress spontaneous oocyte maturation, because GV-stage oocytes strongly suppressed Lhcgr expression in granulosa cells.
Immunofluorescence staining
Ovarian tissues were embedded in paraffin wax (ThermoFisher Scientific, Inc., Waltham, USA) after dehydration. Paraffin-embedded tissue sections (5 μm) were deparaffinized. The sections were blocked with the M.O.M kit (VECTOR) and then incubated with primary antibody (1 : 100 anti-PCNA antibody, Cell Signaling#2586S, 1 : 100 anti-DNMT1 antibody, GeneTex#GTX116011, 1 : 100 anti-TET2 antibody, Abcam#ab124297) in 0.3% (v/v) Triton X-100 in phosphate-buffered saline (PBS) (−) overnight at 4 °C. After washing with 0.3% (v/v) Triton X-100 in PBS (−), the ovaries were visualized with Alexa Fluor 568 goat anti-rabbit secondary antibody (1 : 100) (Sigma) or fluorescein isothiocyanate-conjugated goat anti-mouse IgG secondary antibody (1 : 100) (Sigma) and DAPI (Vector Laboratories, Inc., CA, USA). Digital images were captured using Keyence BZ-9000 microscope (Keyence, Co., Osaka, Japan).
Western blotting
Proteins (20 μg) were separated by SDS-polyacrylamide gel (7.5%, 10%, or 12.5%) electrophoresis and transferred to polyvinylidene difluoride membranes (GE Bioscience, USA). The membranes were blocked with 5% (w/v) bovine serum albumin (Nacalai Chemical, Co.). The blots were incubated with primary antibody (1 : 1000 anti-H3K27ac antibody, Cell Signaling#8173S, 1 : 500 anti-DNMT1 antibody, GeneTex#GTX116011, 1 : 500 anti-TET2 antibody, Cell Signaling#36449, or 1 : 1000 anti-β-actin antibody, Sigma#A5316, Supplementary Table 7) overnight at 4 °C. After washing in TBST, enhanced chemiluminescence (ECL) detection was performed by using the ECL system (GE Bioscience) and appropriately exposing the blots of DNMT1 to Fuji X-ray film (Fujifilm, Tokyo, Japan). The images for western blottings of H3K27ac and TET2 proteins were detected by ChemiDoc Touch MP imaging system (BIO-RAD). The expression of each protein was calculated by the Gel Analyzer of ImageJ (ImageJ.exe).
Flow cytometry
For detection of DNMT1, granulosa cells were fixed with 70% (v/v) methanol in PBS (−) for 10 min at 4 °C. After washing, these cells were treated with 0.1% (v/v) Triton X-100 in DMEM/F12 at room temperature for 15 min and then further treated with the M.O.M kit (VECTOR) in DMEM/F12 at room temperature for 30 min. The primary antibody (1 : 50 anti-DNMT1 antibody, Novus Biologicals#NB100–56519APC, Supplementary Table 7) was added at 37 °C for 30 min. For detection of PI signal, fixed granulosa cells were treated with PI staining reagents (Cellstain®-PI solution, P378, 1 : 100) at 37 °C for 30 min. The fluorescence intensity of 20,000 cell-specific events was detected using Attune NxT Acoustic Focusing Cytometer Ver 2.6 (ThermoFisher). PI fluorescence was detected by BL2 channel and different stages of the cell cycle were determined by specific pick as G0/G1, S and G2/M phases indicated by first, second, and third picks, respectively. DNMT1 fluorescence was detected by RL1 channel.
Statistics and reproducibility
All statistics are described in figure legends. In the comparison between the SF and the follicular follicle before eCG injection, statistical analyses of data (n > 3) for comparison were carried out by Student’s t-test (Statview; Abacus Concepts, Inc., Berkeley, CA). In the comparison of multiple treatment groups, statistical analyses of data (n > 3) for comparison were carried out by one-way analysis of variance (ANOVA). Tukey–Kramer was used as post hoc test. In the statistical analysis of % data, percentage values were transformed into normally distributed numbers by angle transformation and then analyzed by one-way ANOVA. Tukey–Kramer was used as post hoc test. Student’s t-test and one-way ANOVA were performed if the data were normally distributed. Data are presented as mean ± SEM (n > 3 biological replicates) and p < 0.05 was considered significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
Ovine FSH was kindly provided by Dr. A.F. Parlow, the National Hormone and Pituitary Program, the National Institute of Diabetes and Digestive and Kidney Disease, USA. This work was supported in part by JSPS KAKENHI Grant Number JP 19H03108 (to M.S.).
Author contributions
T.K. performed the experiments and analyzed the data. M.S. and J.S.R. designed the study, and wrote and revised the manuscript.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and Supplementary Data 1, and will be available from the corresponding author upon reasonable request. The accession number for all sequence data shown in this study is DNA Data Bank of Japan (DDBJ) Sequence Read Archive: DRA010809.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Communications Biology thanks Changjiu He, Fatima Smagulova, and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editor: George Inglis. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02849-w.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, et al. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Shipony Z, et al. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature. 2014;513:115–119. doi: 10.1038/nature13458. [DOI] [PubMed] [Google Scholar]
- 4.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long HK, King HW, Patient RK, Odom DT, Klose RJ. Protection of CpG islands from DNA methylation is DNA-encoded and evolutionarily conserved. Nucleic Acids Res. 2016;44:6693–6706. doi: 10.1093/nar/gkw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T, et al. Structural and mechanistic insights into UHRF1-mediated DNMT1 activation in the maintenance DNA methylation. Nucleic Acids Res. 2018;46:3218–3231. doi: 10.1093/nar/gky104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–750. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen H, Xu W, Lan F. Histone lysine demethylases in mammalian embryonic development. Exp. Mol. Med. 2017;49:e325. doi: 10.1038/emm.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards JS. Hormonal control of gene expression in the ovary. Endocr. Rev. 1994;15:725–751. doi: 10.1210/edrv-15-6-725. [DOI] [PubMed] [Google Scholar]
- 10.Edson MA, Nagaraja AK, Matzuk MM. The mammalian ovary from genesis to revelation. Endocr. Rev. 2009;30:624–712. doi: 10.1210/er.2009-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva-Buttkus PD, et al. Effect of cell shape and packing density on granulosa cell proliferation and formation of multiple layers during early follicle development in the ovary. J. Cell Sci. 2008;121:3890–3900. doi: 10.1242/jcs.036400. [DOI] [PubMed] [Google Scholar]
- 12.McRae RS, Johnston HM, Mihm M, O’Shaughnessy PJ. Changes in mouse granulosa cell gene expression during early luteinization. Endocrinology. 2005;146:309–317. doi: 10.1210/en.2004-0999. [DOI] [PubMed] [Google Scholar]
- 13.Wigglesworth K, Lee KB, Emori C, Sugiura K, Eppig JJ. Transcriptomic diversification of developing cumulus and mural granulosa cells in mouse ovarian follicles. Biol. Reprod. 2015;92:1–14. doi: 10.1095/biolreprod.114.121756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narayan Prema. Genetic models for the study of luteinizing hormone receptor function. Front. Endocrinol. 2015;6:152. doi: 10.3389/fendo.2015.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binder AK, et al. The absence of ER-β results in altered gene expression in ovarian granulosa cells isolated from in vivo preovulatory follicles. Endocrinology. 2013;154:2174–2187. doi: 10.1210/en.2012-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robker RL, Richards JS. Hormone-induced proliferation and differentiation of granulosa cells: a coordinated balance of the cell cycle regulators cyclin D2 and p27Kip1. Mol. Endocrinol. 1998;12:924–940. doi: 10.1210/mend.12.7.0138. [DOI] [PubMed] [Google Scholar]
- 17.Cannon JD, Cherian-Shaw M, Lovekamp-Swan T, Chaffin CL. Granulosa cell expression of G1/S phase cyclins and cyclin-dependent kinases in PMSG-induced follicle growth. Mol. Cell Endocrinol. 2007;264:6–15. doi: 10.1016/j.mce.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 18.Sicinski P, et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature. 1996;384:470–474. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- 19.Kawai T, Richards JS, Shimada M. The cell type-specific expression of Lhcgr in mouse ovarian cells: evidence for a DNA-demethylation-dependent mechanism. Endocrinology. 2018;159:2062–2074. doi: 10.1210/en.2018-00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caron G, et al. Cell-cycle-dependent reconfiguration of the DNA methylome during terminal differentiation of human B cells into plasma cells. Cell Rep. 2015;13:1059–1071. doi: 10.1016/j.celrep.2015.09.051. [DOI] [PubMed] [Google Scholar]
- 21.Das S, et al. MicroRNA mediates DNA demethylation events triggered by retinoic acid during neuroblastoma cell differentiation. Cancer Res. 2010;70:7874–7881. doi: 10.1158/0008-5472.CAN-10-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakashima H, et al. MeCP2 controls neural stem cell fate specification through miR-199a-mediated inhibition of BMP-Smad signaling. Cell Rep. 2021;35:109124. doi: 10.1016/j.celrep.2021.109124. [DOI] [PubMed] [Google Scholar]
- 23.Bell JT, et al. Differential methylation of the TRPA1 promoter in pain sensitivity. Nat. Commun. 2014;5:2978. doi: 10.1038/ncomms3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao Y, et al. MeSiC: a model-based method for estimating 5mC levels at single-CpG resolution from MeDIP-seq. Sci. Rep. 2015;5:14699. doi: 10.1038/srep14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards JS, Pangas SA. The ovary: basic biology and clinical implications. J. Clin. Invest. 2010;120:963–972. doi: 10.1172/JCI41350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abedel-Majed MA, Romereim SM, Davis JS, Cupp AS. Perturbations in lineage specification of granulosa and theca cells may alter corpus luteum formation and function. Front. Endocrinol. 2019;10:832. doi: 10.3389/fendo.2019.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayek El S, Clarke HJ. Follicle-stimulating hormone increases gap junctional communication between somatic and germ-line follicular compartments during murine oogenesis. Biol. Reprod. 2015;93:47. doi: 10.1095/biolreprod.115.129569. [DOI] [PubMed] [Google Scholar]
- 28.Claes A, et al. Molecular changes in the equine follicle in relation to variations in antral follicle count and anti-Müllerian hormone concentrations. Equine Vet. J. 2016;48:741–748. doi: 10.1111/evj.12514. [DOI] [PubMed] [Google Scholar]
- 29.Park JY, et al. EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science. 2004;303:682–684. doi: 10.1126/science.1092463. [DOI] [PubMed] [Google Scholar]
- 30.Richards JS, Ascoli M. Endocrine, paracrine, and autocrine signaling pathways that regulate ovulation. Trends Endocrinol. Metab. 2018;29:313–325. doi: 10.1016/j.tem.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Shimada M, Hernandez-Gonzalez I, Gonzalez-Robayna I, Richards JS. Paracrine and autocrine regulation of epidermal growth factor-like factors in cumulus oocyte complexes and granulosa cells: key roles for prostaglandin synthase 2 and progesterone receptor. Mol. Endocrinol. 2006;20:1352–1365. doi: 10.1210/me.2005-0504. [DOI] [PubMed] [Google Scholar]
- 32.Shimada M, et al. Hyaluronan fragments generated by sperm-secreted hyaluronidase stimulate cytokine/chemokine production via the TLR2 and TLR4 pathway in cumulus cells of ovulated COCs, which may enhance fertilization. Development. 2008;135:2001–2011. doi: 10.1242/dev.020461. [DOI] [PubMed] [Google Scholar]
- 33.Rodríguez-Gil A, Riedlinger T, Ritter O, Saul VV, Schmitz LM. Formaldehyde-assisted isolation of regulatory elements to measure chromatin accessibility in mammalian cells. J. Vis. Exp. 2018;134:57272. doi: 10.3791/57272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xi H, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HJ, et al. Capturing cell type-specific chromatin compartment patterns by applying topic modeling to single-cell Hi-C data. PLoS Comput. Biol. 2020;16:e1008173. doi: 10.1371/journal.pcbi.1008173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Jia ST, Jia S. New insights into the regulation of heterochromatin. Trends Genet. 2016;32:284–294. doi: 10.1016/j.tig.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 38.Schröder C, et al. Regions of common inter-individual DNA methylation differences in human monocytes: genetic basis and potential function. Epigenet. Chromatin. 2017;10:37. doi: 10.1186/s13072-017-0144-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai YP, et al. TET1 regulates hypoxia-induced epithelial-mesenchymal transition by acting as a co-activator. Genome Biol. 2014;15:513. doi: 10.1186/s13059-014-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoque SAM, Kawai T, Zhu Z, Shimada M. Mitochondrial protein turnover is critical for granulosa cell proliferation and differentiation in antral follicles. J. Endocr. Soc. 2019;3:324–339. doi: 10.1210/js.2018-00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez-Robayna IJ, et al. Follicle-stimulating hormone (FSH) stimulates phosphorylation and activation of protein kinase B (PKB/Akt) and serum and glucocorticoid-lnduced kinase (Sgk): evidence for A kinase-independent signaling by FSH in granulosa cells. Mol. Endocrinol. 2000;14:1283–1300. doi: 10.1210/mend.14.8.0500. [DOI] [PubMed] [Google Scholar]
- 42.Maloveryan A, Finta C, Osterlund T, Kogerman P. A possible role of mouse Fused (STK36) in Hedgehog signaling and Gli transcription factor regulation. J. Cell Commun. Signal. 2007;1:165–173. doi: 10.1007/s12079-007-0014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu Xiaoyan, et al. Knockdown of Trnau1ap inhibits the proliferation and migration of NIH3T3, JEG-3 and Bewo cells via the PI3K/Akt signaling pathway. Biochem. Biophys. Res. Commun. 2018;503:521–527. doi: 10.1016/j.bbrc.2018.05.065. [DOI] [PubMed] [Google Scholar]
- 44.Elzaiat M, et al. High-throughput exploration of the network dependent on AKT1 in mouse ovarian granulosa cells. Mol. Cell Proteomics. 2019;18:1307–1319. doi: 10.1074/mcp.RA119.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maki O, et al. Epigenetic changes of the Cyp11a1 promoter region in granulosa cells undergoing luteinization during ovulation in female rats. Endocrinology. 2016;157:3344–3354. doi: 10.1210/en.2016-1264. [DOI] [PubMed] [Google Scholar]
- 46.Jo M, Curry TE. Luteinizing hormone-induced RUNX1 regulates the expression of genes in granulosa cells of rat periovulatory follicles. Mol. Endocrinol. 2006;20:2156–2172. doi: 10.1210/me.2005-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang YL, et al. CBP-CITED4 is required for luteinizing hormone-triggered target gene expression during ovulation. Mol. Hum. Reprod. 2014;20:850–860. doi: 10.1093/molehr/gau040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cosma MP. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. cell. 2002;10:227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 49.Emerson BM. Specificity of gene regulation. Cell. 2002;109:267–270. doi: 10.1016/s0092-8674(02)00740-7. [DOI] [PubMed] [Google Scholar]
- 50.Sanfins A, Rodrigues P, Albertini DF. GDF-9 and BMP-15 direct the follicle symphony. J. Assist Reprod. Genet. 2018;35:1741–1750. doi: 10.1007/s10815-018-1268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su YQ, Sugiura K, Eppig JJ. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: energy metabolism. Semin. Reprod. Med. 2009;27:32–42. [Google Scholar]
- 52.Sugiura K, Pendola FL, Eppig JJ. Mouse oocyte control of granulosa cell development and function: paracrine regulation of cumulus cell metabolism. Dev. Biol. 2005;279:20–30. doi: 10.1016/j.ydbio.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 53.Richards JS, Russell DL, Robker RL, Dajee M, Alliston TN. Molecular mechanisms of ovulation and luteinization. Mol. Cell Endocrinol. 1998;145:47–54. doi: 10.1016/s0303-7207(98)00168-3. [DOI] [PubMed] [Google Scholar]
- 54.Stocco C, Telleria C, Gibori G. The molecular control of corpus luteum formation, function, and regression. Endocr. Rev. 2007;28:117–149. doi: 10.1210/er.2006-0022. [DOI] [PubMed] [Google Scholar]
- 55.Kumar S, Sandell LL, Trainor PA, Koentgen F, Duester G. Alcohol and aldehyde dehydrogenases: retinoid metabolic effects in mouse knockout models. Biochim. Biophys. Acta. 2012;1821:198–205. doi: 10.1016/j.bbalip.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kawai T, Yanaka N, Richards JS, Shimada M. De novo-synthesized retinoic acid in ovarian antral follicles enhances FSH-mediated ovarian follicular cell differentiation and female fertility. Endocrinology. 2016;157:2160–2172. doi: 10.1210/en.2015-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su YQ, et al. Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte-cumulus cell complex in mice: genetic evidence for an oocyte-granulosa cell regulatory loop. Dev. Biol. 2004;276:64–73. doi: 10.1016/j.ydbio.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 58.Shen M, et al. Observation of the influences of diosgenin on aging ovarian reserve and function in a mouse model. Eur. J. Med. Res. 2017;22:42. doi: 10.1186/s40001-017-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Navot D, et al. Poor oocyte quality rather than implantation failure as a cause of age-related decline in female fertility. Lancet. 1991;337:1375–1377. doi: 10.1016/0140-6736(91)93060-m. [DOI] [PubMed] [Google Scholar]
- 60.Gong Y, et al. Age-related decline in the expression of GDF9 and BMP15 genes in follicle fluid and granulosa cells derived from poor ovarian responders. J. Ovarian Res. 2021;14:1. doi: 10.1186/s13048-020-00757-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franks S, Stark J, Hardy K. Follicle dynamics and anovulation in polycystic ovary syndrome. Hum. Reprod. Update. 2008;14:367–378. doi: 10.1093/humupd/dmn015. [DOI] [PubMed] [Google Scholar]
- 62.Dumesic DA, Abbott DH. Implications of polycystic ovary syndrome (PCOS) on oocyte development. Semin. Reprod. Med. 2008;26:53–61. doi: 10.1055/s-2007-992925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ross JDC. An update on pelvic inflammatory disease. Sex. Transm. Infect. 2002;78:18–19. doi: 10.1136/sti.78.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adetunji AO, Kawai T, Shimada M. Impact of lipopolysaccharide administration on luteinizing hormone/choriogonadotropin receptor (Lhcgr) expression in mouse ovaries. J. Reprod. Immunol. 2020;142:103193. doi: 10.1016/j.jri.2020.103193. [DOI] [PubMed] [Google Scholar]
- 65.Pedersen T, Peters H. Proposal for a classification of oocytes and follicles in the mouse ovary. J. Reprod. Fertil. 1968;17:555–557. doi: 10.1530/jrf.0.0170555. [DOI] [PubMed] [Google Scholar]
- 66.Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab. Investig. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- 67.Gebhard C, et al. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006;66:6118–6128. doi: 10.1158/0008-5472.CAN-06-0376. [DOI] [PubMed] [Google Scholar]
- 68.Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gil AR, Riedlinger T, Ritter O, Saul VV, Schmitz LM. Formaldehyde-assisted isolation of regulatory elements to measure chromatin accessibility in mammalian cells. J. Vis. Exp. 2018;134:57272. doi: 10.3791/57272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and Supplementary Data 1, and will be available from the corresponding author upon reasonable request. The accession number for all sequence data shown in this study is DNA Data Bank of Japan (DDBJ) Sequence Read Archive: DRA010809.