ABSTRACT
Oncogenic Ras mutations are highly prevalent in hematopoietic malignancies. However, it is difficult to directly target oncogenic RAS proteins for therapeutic intervention. We have developed a Drosophila acute myeloid leukemia model induced by human KRASG12V, which exhibits a dramatic increase in myeloid-like leukemia cells. We performed both genetic and drug screens using this model. The genetic screen identified 24 candidate genes able to attenuate the oncogenic RAS-induced phenotype, including two key hypoxia pathway genes HIF1A and ARNT (HIF1B). The drug screen revealed that echinomycin, an inhibitor of HIF1A, can effectively attenuate the leukemia phenotype caused by KRASG12V. Furthermore, we showed that echinomycin treatment can effectively suppress oncogenic RAS-driven leukemia cell proliferation, using both human leukemia cell lines and a mouse xenograft model. These data suggest that inhibiting the hypoxia pathway could be an effective treatment approach and that echinomycin is a promising targeted drug to attenuate oncogenic RAS-induced cancer phenotypes.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Drosophila, Mouse xenografts, Leukemia, Oncogenic RAS, Echinomycin, Hypoxia pathway, HIF1A
Summary: Hypoxia pathway inhibition, either genetically or pharmacologically, rescues RAS-induced oncogenesis in a Drosophila acute myeloid leukemia model, mouse xenograft model and human leukemia cells.
INTRODUCTION
Mutations of the RAS family genes, consisting of HRAS, KRAS and NRAS, are involved in over 30% of all human cancers (Downward, 2015; Pylayeva-Gupta et al., 2011). KRAS is an especially prevalent contributor to human cancers affecting the pancreas, colon and lung (Downward, 2015; Pylayeva-Gupta et al., 2011). RAS mutations have also been associated with different types of leukemias, including acute myeloid leukemia (AML) (Beaupre and Kurzrock, 1999a,b; Braun and Shannon, 2008; Liu et al., 2019).
RAS genes encode highly conserved GTP-binding proteins involved in eukaryotic cell proliferation and differentiation (McCormick, 1994), which cycle between active GTP-bound and inactive GDP-bound states. Activated RAS, in turn, activates downstream effector pathways. Many oncogenic RAS mutations lock RAS in a constitutively activated state, which triggers the activation of multiple interacting signaling pathways (Bos, 1989; Downward, 2003). Activated RAS promotes cellular transformation through molecular signaling that stimulates cell proliferation, inhibits differentiation, blocks cell death and promotes angiogenesis (Hanahan and Weinberg, 2000). To date, efforts selectively and directly targeting oncogenic RAS have been extremely challenging. Although a few direct inhibitors for oncogenic RAS have been developed recently, they have shown variable responsiveness. Thus, invigorating the call for combinatorial strategies to optimize treatment of RAS-mediated cancers has been proposed (Hallin et al., 2020).
The KRASG12V allele is one of the most prevalent oncogenes found in human cancers, and has been the focus of many previous studies to identify candidate genes or pathways representing potential therapeutic targets (Barbie et al., 2009; Corcoran et al., 2013; Kim et al., 2013; Luo et al., 2009; Sarthy et al., 2007; Scholl et al., 2009; Singh et al., 2012; Steckel et al., 2012; Vicent et al., 2010). However, seemingly promising candidates identified through in vitro genetic screens have failed to translate into clinically promising therapeutic targets. They often stumble at the in vivo validation phase, with the in vitro findings unable to be replicated in animal models and human patients (Downward, 2015). The translation hurdle urges the need for oncogenic RAS genetic screens and drug screens using in vivo lower-animal models that accommodate large-scale screens, such as Drosophila, followed by validation studies in human cells and mammalian animal models, such as mice.
Studies in Drosophila have proven extremely valuable for elucidating key features of RAS cellular physiology, and pathways affected by RAS protein activation are highly conserved from flies to humans (Asha et al., 2003; Brumby and Richardson, 2003, 2005; Gonzalez, 2013; Lusk et al., 2017; Ma et al., 2017; Pagliarini and Xu, 2003; Vidal and Cagan, 2006; Wu et al., 2010). Drosophila possesses a single Ras85D gene, in contrast to the human KRAS, NRAS and HRAS family members. Although genetic screens have been conducted previously for fly cancer models with oncogenic mutations in the fly Ras85D gene (Asha et al., 2003; Chabu and Xu, 2014; Gladstone and Su, 2011; Ma et al., 2017; Pagliarini and Xu, 2003), no previous studies have been described that use a fly cancer model induced by a human oncogenic RAS mutation.
In this study, we established a new Drosophila cancer model induced by expressing human KRASG12V in fly hemocytes. This model exhibits a dramatic increase in hemocyte numbers caused by overproliferation, which provides an ideal phenotype for genetic and drug screens. Using this model in a genetic screen, we identified 24 promising hits that, when silenced in the hemocytes, attenuated the leukemia phenotype of the KRASG12V model, including two key components of the hypoxia pathway, HIF1A and ARNT (HIF1B), which are involved in the development of both solid tumors and hematological malignancies (Deynoux et al., 2016; Jun et al., 2017; Semenza, 2003; Szymczak et al., 2018). A drug screen using the same model revealed that echinomycin, an inhibitor of HIF1A, could antagonize the KRASG12V-induced leukemia phenotype. We validated this finding in human leukemia cell lines as well as a mouse xenograft model, demonstrating the promising translational value of this finding and expanding its application to a broad range of oncogenic RAS mutations.
RESULTS
Generation of a new KRASG12V-induced Drosophila leukemia model
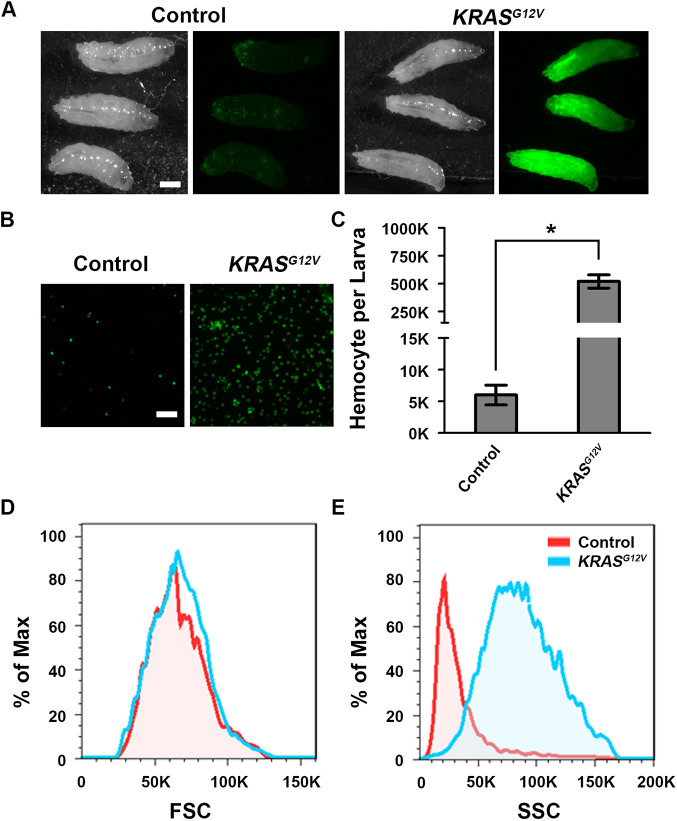
We generated a new UAS-KRASG12V transgenic line to express the oncogenic human KRASG12V allele in different types of tissues. When we used Hml-Gal4 to express KRASG12V (alone with UAS-GFP as a marker) in the Drosophila hemocytes, we noticed a dramatic increase in GFP+ hemocytes (Fig. 1A). At 25°C, third-instar larvae with Hml-Gal4-driven UAS-KRASG12V (genotype Hml-Gal4, UAS-GFP; UAS-KRASG12V) contained nearly 100-fold more circulating hemocytes than the control (genotype Hml-Gal4, UAS-GFP) flies (Fig. 1B,C). Using a flow-cytometry equipment with high-throughput imaging function, we analyzed the size and granularity of hemocytes in the control and KRASG12V flies (Fig. 1D,E). We found that the size of KRASG12V hemocytes did not change significantly (Fig. 1D); however, the hemocytes exhibited significantly increased intracellular granularity (Fig. 1E).
Fig. 1.
Human KRASG12V mutant transgene drives abnormal hemocyte proliferation in a Drosophila leukemia model. (A) Transgenic third-instar larvae carrying hemocyte-specific Hml-Gal4 driver directing expression of UAS-GFP (Control), and UAS-GFP plus UAS-HsKRASG12V (KRASG12V). Panels show stereo micrographs of larvae (left) and GFP fluorescence micrographs (right). Scale bar: 0.3 mm. (B) Hemocytes in hemolymph samples of equal volume extracted from control and KRASG12V third-instar larvae. Scale bar: 50 μm. (C) Quantification of total hemocytes per control and KRASG12V third-instar larvae (n=6; results are presented as mean±s.d.; *P<0.05; unpaired Student's t-test). (D) Frequency plots comparing control and KRASG12V third-instar larval hemocyte cell size. Forward scatter (FSC) is a measurement of the amount of the laser beam that passes around the cell, which gives a relative size of a cell. (E) Frequency plots comparing control and KRASG12V third-instar larval hemocyte cell structure. Side scatter (SSC) is a measurement of the amount of the laser beam that bounces off particulates inside the cell, which is an indicator of granularity in a cell.
KRASG12V increases the percentage of Wg-expressing and Lz-expressing hemocytes, and decreases P1-labeled matured hemocytes
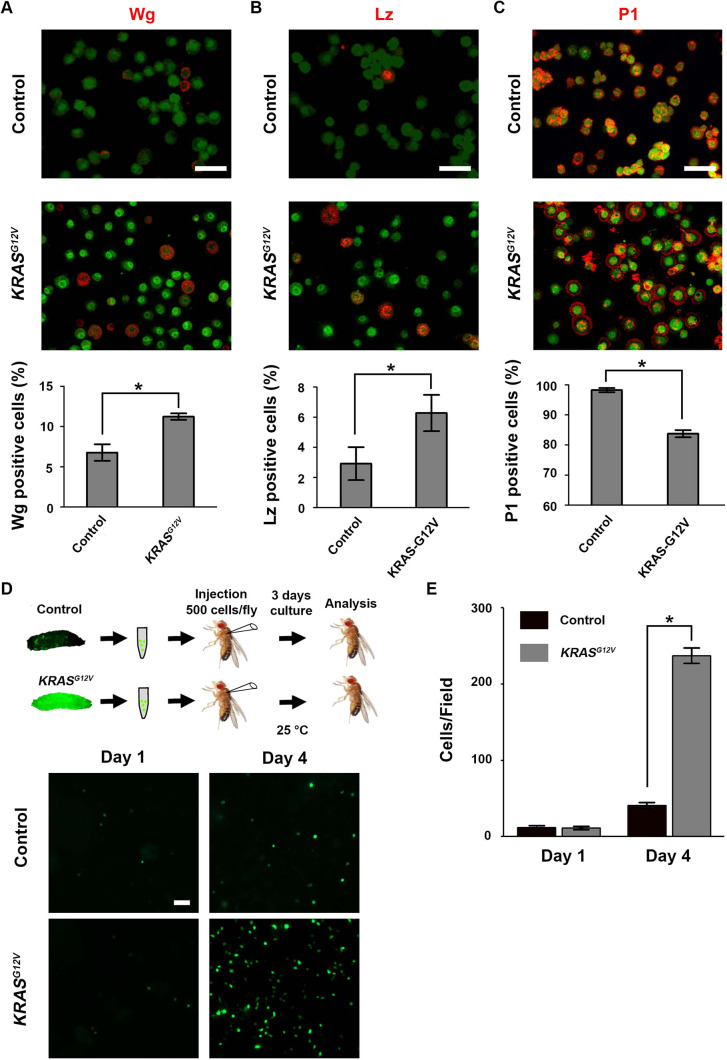
The Drosophila hematopoietic system consists of different cell types that undergo different stages of differentiation (Evans et al., 2014; Fu et al., 2020). We analyzed the relative proportions of hemocyte subtypes that express signature differentiation markers, such as Wingless (Wg), Lozenge (Lz) and Nimrod C1 (Fig. 2), to determine the effects of KRASG12V on hemocyte differentiation. We found that KRASG12V induced increased numbers of Wg-expressing hemocytes (Fig. 2A), which has been associated with stem-like hemocyte precursors because Wg expression withdraws as hemocytes differentiate (Sinenko et al., 2009). We found that Wg-expressing hemocytes also exhibited weaker Hml-Gal4-driven GFP expression (Fig. 2A). We also found that crystal cells with Lz expression increased their proportion in KRASG12V flies (Fig. 2B). In contrast, plasmatocyte expression of Nimrod C1, a terminally differentiated plasmatocyte marker that is recognized by the anti-P1 antibody (Kurucz et al., 2007), was significantly reduced in proportion in the KRASG12V flies (Fig. 2C).
Fig. 2.
Human KRASG12V transgene expression alters the distribution of hemocyte subtypes and drives abnormal cell population expansion after transplantation. (A) Pro-hemocytes were selectively immunolabeled with anti-Wingless (Wg) antibody. The percentage of pro-hemocytes in total hemocyte is increased ∼1.5- to 2-fold in KRASG12V larvae (quantification shown below; n=6; results are presented as mean±s.d.; *P<0.05; unpaired Student's t-test). Scale bar: 30 μm. (B) Crystal cells were selectively immunolabeled with anti-Lozenge (Lz) antibody. The percentage of crystal cells in total hemocytes is increased ∼2-fold in KRASG12V larvae (quantification shown below; n=6; results are presented as mean±s.d.; *P<0.05; unpaired Student's t-test). Scale bar: 30 μm. (C) Plasmatocytes were selectively immunolabeled with anti-P1 antibody. The percentage of plasmatocytes in total hemocytes is reduced ∼10% in hemocytes KRASG12V larvae (quantification shown below; n=6; results are presented as mean±s.d.; *P<0.05; unpaired Student's t-test). Scale bar: 30 μm. (D) Schematic showing larval hemocyte transplantation into adult flies. Hemocytes were collected from control (Hml-Gal4, UAS-GFP) or KRASG12V (Hml-Gal4, UAS-GFP; UAS-KRASG12V) third-instar larvae, then injected (500 hemocytes) into adult flies. Injected flies were maintained for 4 days, at the end of which time the hemolymph was collected and analyzed microscopically for hemocyte density. Lower panels show fluorescence micrographs of control compared to KRASG12V hemocytes (expressing GFP) 1 day after transplantation (day 1) and day 4 post-transplantation into adult flies. Scale bar: 50 μm. (E) Quantification of transplanted hemocyte numbers at day 1 and day 4 post-transplantation (results are presented as mean±s.d.; *P<0.05; unpaired Student's t-test).
KRASG12V-induced hemocyte overproliferation is cell autonomous
We next tested whether KRASG12V-induced overproliferation of hemocytes was cell autonomous using a leukemia cell transplant assay. We collected GFP+ hemocytes from the KRASG12V leukemia fly model donor, in parallel with the control donor (Hml-Gal4, UAS-GFP). After careful titrating, we injected ∼500 GFP+ donor hemocytes into wild-type (w1118) flies and followed up by counting GFP+ hemocytes in transplant recipients at two successive time points. The number of GFP+ hemocytes did not change significantly in the first few hours after transplantation (Fig. 2D, Day 1). However, after 3 days (Day 4 in Fig. 2D), GFP+ hemocytes from the KRASG12V donors were ∼20-fold more abundant than GFP+ hemocytes from the control donors (Fig. 2D,E), suggesting that KRASG12V-induced hemocyte overproliferation is cell autonomous.
KRASG12V compromises the immune function of hemocytes in leukemia flies
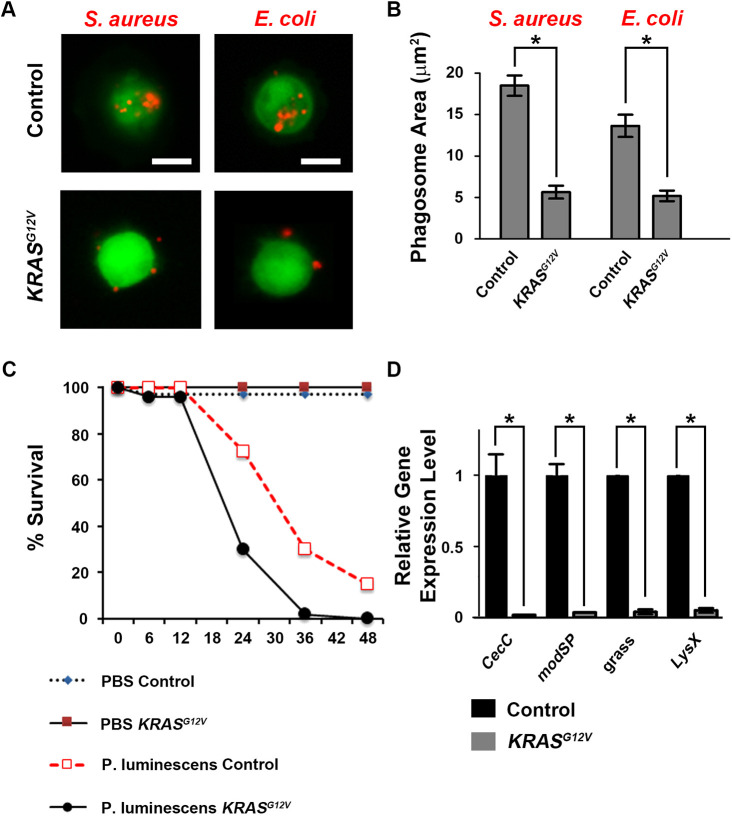
Human leukemia patients are at increased risk of suffering mortality as a result of infection due, at least in part, to immune cell dysfunction associated with transformation (Lamble and Lind, 2018). Hemocytes are critical components of the Drosophila innate immune system, and we observed that KRASG12V expression led to reduced numbers of the P1-expressing plasmatocytes that are largely responsible for phagocytosis of bacteria (Govind, 2008). We therefore analyzed the immune functional capabilities of KRASG12V-expressing hemocytes. Drosophila immunity can distinguish between Gram+ and Gram− bacteria through different pathways. First, we tested the ability of larval hemocytes to phagocytose these two different types of bacteria: Gram+ bacteria Staphylococcus aureus and Gram− bacteria Escherichia coli, each tagged with the fluorescent label pHrodo Red. Whereas normal hemocytes readily phagocytosed both bacterial species, KRASG12V hemocytes showed significantly reduced phagocytic activity for both the Gram+ bacteria S. aureus and the Gram− bacteria E. coli (Fig. 3A,B), even though the percentage of KRASG12V hemocytes displaying phagocytic activity was not reduced (Fig. S1).
Fig. 3.
Human KRASG12V transgene expression is associated with Drosophila hemocyte immune function deficits. (A) Hemocytes (green) from control (Hml-Gal4, UAS-GFP) and KRASG12V (Hml-Gal4, UAS-GFP; UAS-KRASG12V) third-instar larvae co-incubated with fluorescent pHrodo Red-tagged S. aureus or E. coli (red). Scale bars: 10 μm. (B) Quantification of phagosome area in control and KRASG12V hemocytes co-incubated with fluorescent pHrodo Red-tagged S. aureus or E. coli (n=100; results presented as mean±s.d.; *P<0.05; unpaired Student's t-test). (C) Survival curves of control and KRASG12V adult flies infected with pathogenic P. luminescens bacteria or sterile PBS (vehicle) at 18°C. Forty flies were analyzed for each group. (D) Quantitative RT-PCR showing the relative expression level of genes involved in antibacterial defense in hemocytes from control compared to KRASG12V flies (n=3 replicates in each group; results presented as mean±s.d.; *P<0.05; unpaired Student's t-test). CecC, Cecropin C; modSP, modular serine protease; grass, Gram+-specific serine protease; LysX, Lysozyme X.
We also tested overall immune function by infecting adult flies with pathogenic Photorhabdus luminescens bacteria, which are known to suppress the insect immune response and even cause lethality to healthy flies. Because flies expressing KRASG12V showed lethality at the pupal stage at 25°C, we maintained the Hml-Gal4, UAS-GFP; UAS-KRASG12V leukemia model at 18°C to obtain adult flies for injection. We found that KRASG12V still induced significant overproliferation of hemocytes at 18°C, but not as dramatically as at 25°C (Fig. S2). The mortality rates of the control and KRASG12V flies were not affected by injection of phosphate buffered saline (1× PBS), demonstrating that injection alone did not harm the flies. When injected with same volume of P. luminescens in 1× PBS, the KRASG12V flies exhibited significantly decreased resistance to the infection. At 24 h after infection, only ∼30% of KRASG12V flies remained alive compared to over 70% of control flies (Fig. 3C). Finally, using RT-PCR, we examined the hemocyte expression levels of four genes (CecC, modSP, grass and LysX) that have been previously associated with defense against both Gram− and Gram+ bacteria in Drosophila (Arefin et al., 2017). We found that KRASG12V expression was associated with dramatically reduced expression of these antibacterial response genes in hemocytes (Fig. 3D). Overall, our data suggested that KRASG12V-induced overproliferation of hemocytes leads to significantly compromised immunity and increased susceptibility to infection.
A genetic screen in Drosophila to identify modifiers of the KRASG12V-induced hemocyte overproliferation phenotype
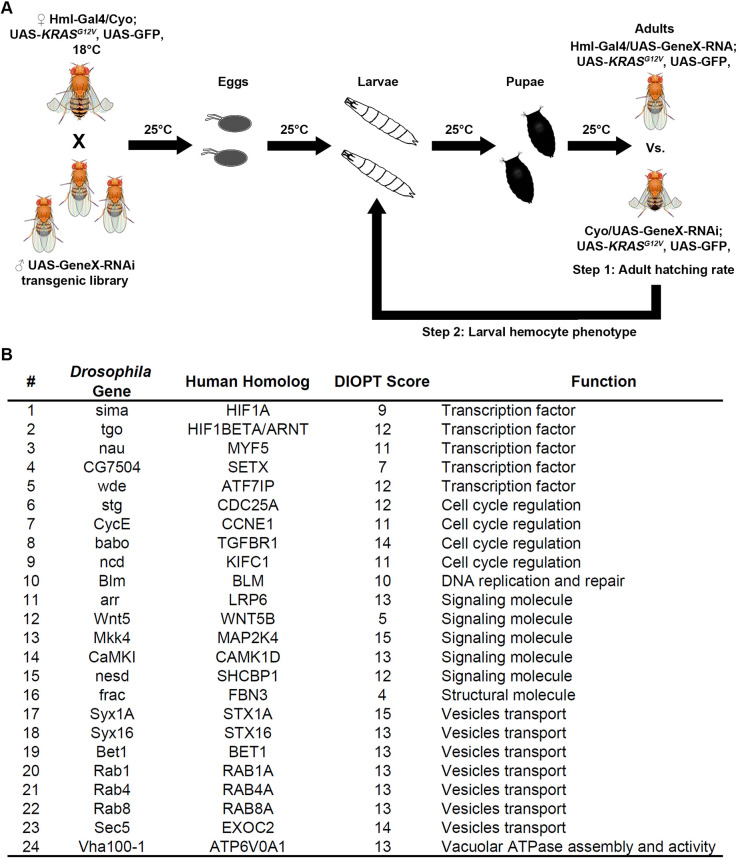
The discovery that our KRASG12V fly leukemia model can survive to adult stage at 18°C, but not at 25°C (owing to Gal4 being more stable at 25°C and hence driving higher levels of KRASG12V expression), allowed us to design a high-throughput genetic screen to identify the genes that, when silenced in the hemocytes, attenuate the adult lethality of the KRASG12V fly leukemia model at 25°C (Fig. 4A). We designed this screen to be performed in two steps. Step 1 is a high-throughput one-step cross to compare the number of adult fly progenies with straight-wing (Hml-Gal4; UAS-KRASG12V. UAS-GeneX-RNAi) versus curly-wing (control, with only the UAS-KRASG12V. UAS-GeneX-RNAi but not Hml-Gal4) phenotypes. The ratio will be 1:1 if silencing of GeneX prevents KRASG12V-induced hemocyte overproliferation. The ratio will be 0:1 if silencing of GeneX is completely lethal to the flies or has no effect on KRASG12V-induced lethality. In step 2, we evaluated and confirmed the positive hits from step 1 by examining the hemocyte number at the third-instar larval stage (Fig. 4A).
Fig. 4.
Drosophila KRASG12V genetic screen and the hits that rescue the hemocyte overproliferation and developmental lethality phenotypes. (A) Schematic illustration of the approach used for the genetic screen (see Materials and Methods section for a detailed description of rationale, strategy and procedure). (B) The 24 Drosophila genes that, when silenced in hemocytes, completely rescued KRASG12V-induced hemocyte overproliferation and developmental lethality phenotypes. Also listed are the human homologs of these genes, their DIOPT score (indicating the degree of conservedness) and the predicted function of the proteins encoded by these genes.
After screening ∼3000 genes that are expressed in the hemocytes based on our unpublished hemocyte RNA sequencing (RNA-seq) data, we identified 24 genes that could completely rescue the lethality and hemocyte overproliferation of our KRASG12V leukemia model at 25°C (Fig. 4B). All of these 24 genes are evolutionarily conserved from flies to humans [based on Drosophila RNAi Screening Center (DRSC) Integrative Ortholog Prediction Tool (DIOPT) score]. These genes encode proteins that are involved in transcription, cell cycle regulation, DNA replication and repair, signaling transduction, vesicle transport and vacuolar ATPase assembly (Fig. 4B). Numbers of hemocytes per third-instar larva were counted for gene silencing of each of these 24 hits, by itself and within the genetic background of the KRASG12V leukemia fly model. Data were obtained for two independent RNA interference (RNAi) lines for each of the genes to ensure that none of the findings were due to off-target effects (Table S1). Consistent results were obtained for all 24 genes across both strains. We found that silencing any of these 24 genes could completely rescue the KRASG12V-induced hemocyte overproliferation (Table S2).
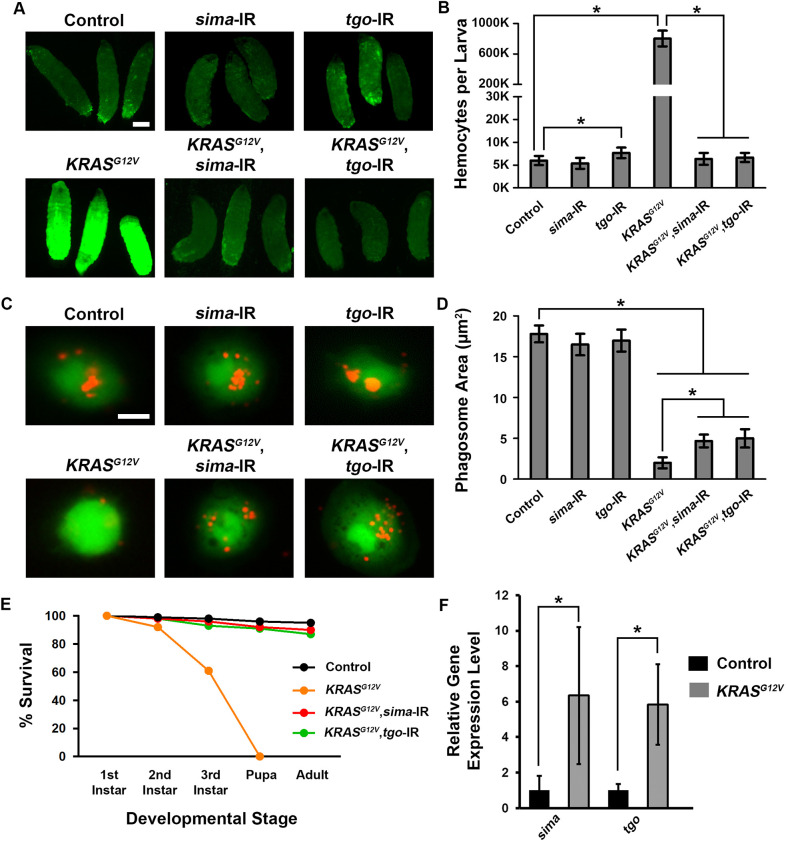
Drosophila HIF1A and ARNT orthologs sima and tgo are required for the survival of KRASG12V-induced leukemia cells
Among the 24 hits identified from the KRASG12V genetic screen, sima (also called HIF-1A) and tango (tgo) are highly conserved fly orthologs of mammalian hypoxia-inducible factor 1 subunit alpha (HIF1A) and aryl hydrocarbon receptor nuclear translocator (ARNT, also called HIF1B), respectively. HIF1A and ARNT are transcription factors critically involved in the response to hypoxia (Dengler et al., 2014). Hemocyte-specific silencing of either sima or tgo by itself did not affect hemocyte proliferation, but silencing sima or tgo in the KRASG12V leukemia model background completely restored the number of hemocytes down to wild-type levels (Fig. 5A,B). Next, we tested whether silencing sima or tgo could also rescue hemocyte phagocytosis function. Indeed, both were able to rescue the KRASG12V-induced hemocyte phagocytosis defect, while silencing of sima or tgo alone did not affect phagocytosis (Fig. 5C,D). Further, we found that silencing of sima or tgo could completely rescue the lethality caused by Hml-Gal4>KRASG12V at 25°C at each developmental stage compared to control flies (Fig. 5E). Concurrently, we observed significantly increased gene expression levels for sima and tgo in fly hemocytes carrying human KRASG12V (Fig. 5F).
Fig. 5.
Silencing Drosophila HIF1 complex gene homologs sima and tgo rescues KRASG12V-induced hemocyte overproliferation, immune deficiency and pupal-stage developmental lethality. (A) Left upper and lower panels show control (Hml-Gal4, UAS-GFP) and KRASG12V (Hml-Gal4, UAS-GFP; UAS-KRASG12V) third-instar larvae, respectively, in which all hemocytes express GFP (green fluorescence). Middle and right panels illustrate the effects of silencing HIF1A homolog sima (sima-IR) and ARNT homolog tgo (tgo-IR), respectively, in control (upper) and KRASG12V (lower) third-instar larvae. Scale bar: 0.3 mm. (B) Quantification of total circulating hemocyte numbers with and without silencing of sima or tgo gene expression in control and KRASG12V third-instar larvae (n=6; results are presented as mean±s.d.; *P<0.05; Kruskal–Wallis H-test). (C) Left upper and lower panels show hemocytes (green) extracted from control and KRASG12V third-instar larvae, respectively, co-incubated with fluorescent Dextran-tagged S. aureus (red). Middle and right panels illustrate the effects of silencing HIF1A homolog sima (sima-IR) and ARNT homolog tgo (tgo-IR), respectively. Scale bar: 10 μm. (D) Quantification of phagosome area with and without silencing of sima or tgo gene expression in control and KRASG12V hemocytes (n=100; results are presented as mean±s.d.; *P<0.05; Kruskal–Wallis H-test). (E) Survival during development of control, KRASG12V and KRASG12V flies in which sima (sima-IR) or tgo (tgo-IR) were silenced in hemocytes. (F) Quantitative RT-PCR showing the relative expression level of sima and tgo in hemocytes from control compared to KRASG12V flies (n=3 replicates in each group; results are presented as mean±s.d.; *P<0.05; unpaired Student's t-test).
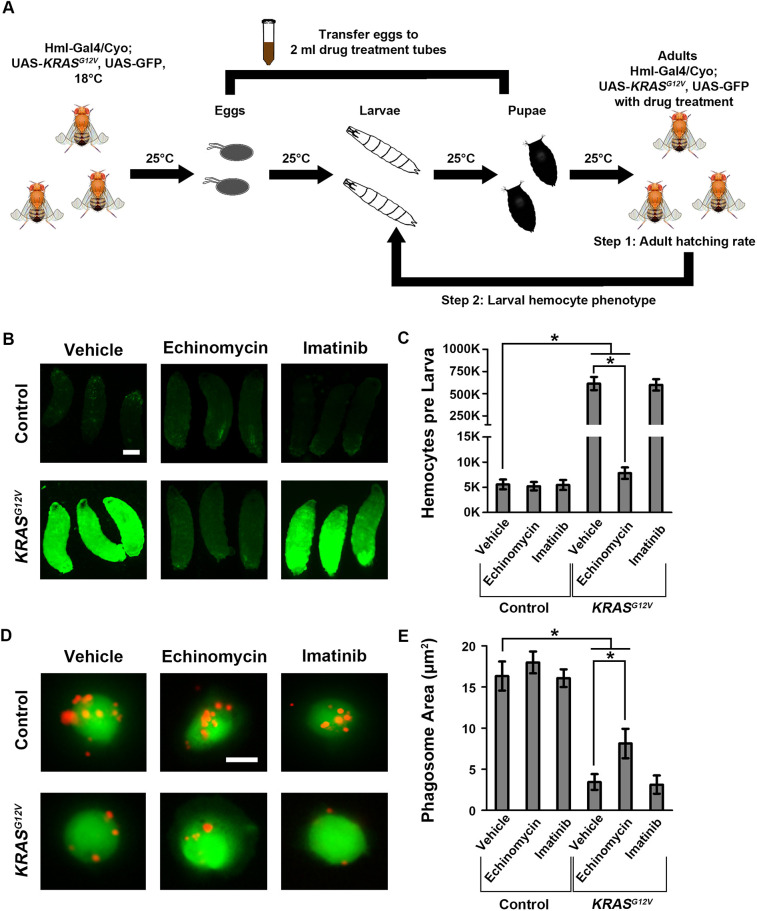
A drug screen using the KRASG12V leukemia fly model identifies echinomycin as an effective hit
In parallel with the large-scale genetic screen, we designed and performed a small-scale drug screen to identify compounds capable of rescuing the lethality of the KRASG12V leukemia model at 25°C (Fig. 6A). We first collected eggs from the Hml-Gal4>KRASG12V leukemia model and transferred ten fertilized eggs to each drug treatment vial, which contained a concentration of 0.5 μM drug in semi-liquid soft-gel food. When the fertilized eggs had hatched into larvae, they were fed on the drug-containing food. The drug treatment vials were kept at 25°C for 10 days to count the number of adult flies that eclosed in each vial. If a drug is completely lethal to the flies or has no effect on inhibiting KRASG12V-induced hemocyte overproliferation, all flies would die before eclosion to adults. If a drug inhibits the KRASG12V-induced hemocyte overproliferation, this will be evident in the number of eclosed adult flies.
Fig. 6.
Echinomycin reverses KRASG12V-induced hemocyte overproliferation and immune deficiency. (A) Schematic illustration of the approach used for the KRASG12V Drosophila leukemia model drug screen (see Materials and Methods section for a detailed description of rationale, strategy and procedure). (B) Left upper and lower panels show control (Hml-Gal4, UAS-GFP) and KRASG12V (Hml-Gal4, UAS-GFP; UAS-KRASG12V) third-instar larvae, respectively, in which all hemocytes express GFP (green fluorescence). Middle and right panels show the effects of treating larvae with echinomycin and imatinib, respectively. Scale bar: 0.3 mm. (C) Quantification of total circulating hemocyte numbers without (vehicle) or with echinomycin or imatinib treatment in control and KRASG12V third-instar larvae (n=6; results are presented as mean±s.d.; *P<0.05; Kruskal–Wallis H-test). (D) Left upper and lower panels show hemocytes (green) extracted from control and KRASG12V third-instar larvae, respectively, co-incubated with fluorescent Dextran-tagged S. aureus (red). Middle and right panels illustrate the effects of treatment with echinomycin and imatinib, respectively. Scale bar: 10 μm. (E) Quantification of phagosome area in control and KRASG12V hemocytes with vehicle, echinomycin or imatinib (n=100; results are presented as mean±s.d.; *P<0.05; Kruskal–Wallis H-test).
We tested a collection of 34 chemical inhibitors (Table S3) available at our laboratory. This drug screen identified echinomycin to be the only drug among the 34 compounds that could completely rescue the lethality of the KRASG12V leukemia fly model at 25°C (Fig. 6B,C). We also found that echinomycin could partially rescue the reduced phagocytic activity of KRASG12V hemocytes (Fig. S3). Unexpectedly, the protein-tyrosine kinase inhibitor imatinib, inhibiting BCR-ABL, which can induce constitutive active downstream targets of oncogenic RAS (Cilloni and Saglio, 2012), failed to rescue both the KRASG12V leukemia model adult-stage mortality (Fig. S3) and the reduced phagocytic activity of KRASG12V hemocytes (Fig. 6B-E).
Echinomycin is a known inhibitor of the DNA-binding activity of HIF1A and HIF1B proteins (Kong et al., 2005). The identification of echinomycin as an effective drug for our KRASG12V leukemia model further demonstrates the importance of the hypoxia signaling pathway in the KRASG12V-induced leukemia phenotype. By identifying both the genetic factors and their inhibitors from parallel genetic and drug screens, our findings also show how effective the Drosophila KRASG12V leukemia model can be in both types of screening platform in RAS-associated cancer studies.
Echinomycin reduces the viability of human leukemia cells carrying oncogenic RAS mutations
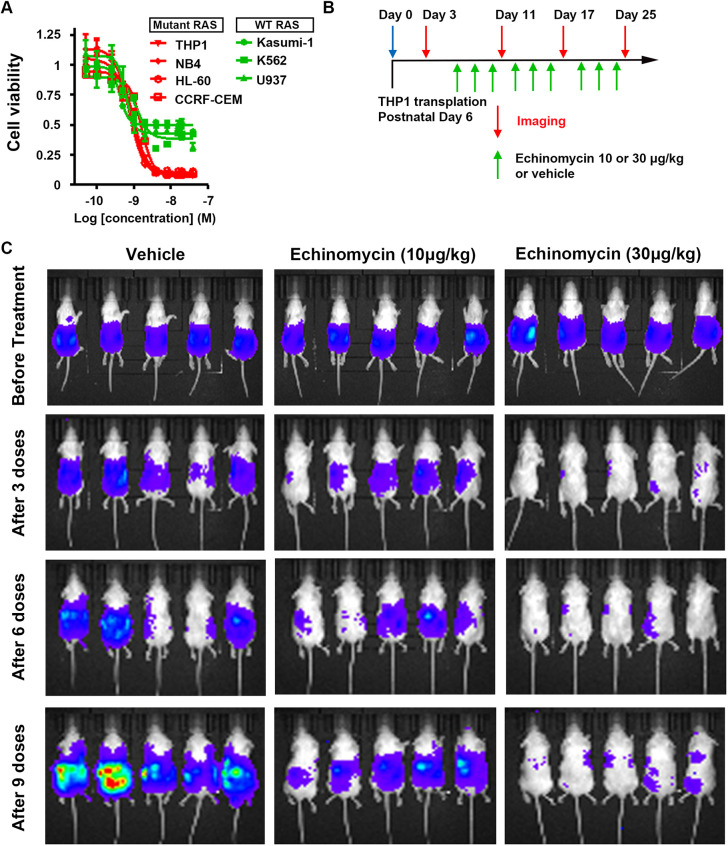
To validate our findings from the Drosophila KRASG12V leukemia model, we tested the effectiveness of echinomycin treatment in a panel of human leukemia cell lines, some of which carried oncogenic RAS mutations and other non-RAS mutations (Fig. 7A). We found that echinomycin treatment more potently reduced cell viability in the four oncogenic RAS cell lines [THP1 (NRASG12D), NB4 (KRASA18D), HL-60 (NRASQ61L) and CCRF-CEM (KRASG12D); Fig. 7A, red] than in the three leukemia cell lines without oncogenic RAS mutations (Kasumi-1, K562 and U937 leukemia cell line; Fig. 7A, green). Only ∼10% of RAS-driven leukemia cells remained viable after exposure to 4 nM echinomycin for 48 h, whereas ∼50% of non-RAS involved leukemia cells remained viable (Fig. 7A). This finding both validated the effect of echinomycin on attenuating the Drosophila KRASG12V leukemia model in human cells and expanded the effective range of echinomycin to a broader range of oncogenic RAS mutations.
Fig. 7.
Echinomycin reduces human leukemia cell viability and proliferation. (A) Cell viability of human leukemia cell lines following echinomycin treatment. Human leukemia cell lines were treated with echinomycin for 48 h at the indicated concentrations. Cell viability was then determined by incubating cells overnight with MTT tetrazolium salt (Sigma-Aldrich). Experiments were performed in triplicate. Red, leukemia cell lines carrying RAS mutations; green, leukemia cell lines in which RAS is not mutated. (B) Schematic illustration of THP1 cell (human leukemia cell line carrying NRASG12D) intrahepatic transplantation to generate mouse xenograft models, and time course of echinomycin treatments and diagnostic bioluminescence imaging to monitor THP1 cell transplant growth. (C) Bioluminescence imaging of mice engrafted with human THP1 leukemia cells bearing NRASG12D mutation. Mice were treated every other day with 10 μg/kg or 30 μg/kg echinomycin (as indicated) or vehicle control and were imaged after every third dose.
Echinomycin inhibits human NRASG12D leukemia cell proliferation in a mouse xenograft model
To further validate the effectiveness of echinomycin on leukemia cells induced by oncogenic RAS mutations in a mammalian in vivo model, we examined the effect of echinomycin administration on the growth of transplanted THP1 leukemia cells carrying oncogenic NRASG12D (alone with a luciferase expression vector) in a xenograft mouse model (Fig. 7B). At day 3 post-transplantation and prior to drug administration, THP1 cell growth was readily apparent using live bioluminescence imaging of luciferase (Fig. 7C). Echinomycin treatment involving doses of either 10 μg/kg body weight or 30 μg/kg body weight effectively suppressed in vivo growth of human THP1 leukemia cells, with a clearly dose-dependent pattern, because higher dose treatment almost completely suppressed leukemia cell growth (Fig. 7C). These observations further validated that echinomycin can effectively suppress oncogenic RAS-induced leukemia cell growth in mammals, and further illustrated the value of the Drosophila leukemia model to identify this promising targeted drug to treat oncogenic RAS-induced cancer phenotypes.
DISCUSSION
In this study, we generated a new KRASG12V leukemia model in Drosophila by expressing human oncogenic KRASG12V in fly hemocytes, which led to a nearly 100-fold increase in hemocyte proliferation and adult fly lethality. Using this easy-to-score phenotype, we designed highly efficient genetic and drug screens to identify genes and compounds able to inhibit oncogenic KRAS. Fortuitously, the parallel genetic and drug screens converged on the same pathways, as they identified both the key genes involved in the hypoxia pathway (sima and tgo), as well as the hypoxia inhibitor echinomycin as potent oncogenic KRAS antagonists. This discovery has provided strong evidence supporting an essential role for the hypoxia pathway in the oncogenic KRAS-induced cancer phenotype and has identified echinomycin as a promising hypoxia-targeting drug.
The genetic screen identified sima and tgo as mediators of the KRASG12V pathomechanism. The two genes encode the fly homologs of human HIF1A and ARNT, respectively. HIF activation is a well-established feature of cancer cell survival in solid tumors, as cells positioned in the tumor interior are subject to severe oxygen deprivation, i.e. hypoxia. Activated HIFs lead to changes in the microenvironment that support tumor survival and growth, such as angiogenesis (Bousquet et al., 2016; Talks et al., 2000). In addition to the role of HIFs in solid tumors, HIF-mediated signaling has been found to play a crucial role in leukemia (Deynoux et al., 2016; Szymczak et al., 2018; Wang et al., 2011b). In addition, activated KRAS has been shown to induce HIF1A and ARNT target gene expression in human colon cancer cells (Chun et al., 2010). Echinomycin treatment has demonstrated therapeutic effect on acute myeloid leukemia cells with TP53 mutation in xenograft mouse models (Wang et al., 2020), but our study has shown, for the first time, that echinomycin is effective for oncogenic RAS in Drosophila, mouse and human cell systems. Our data also indicated significantly increased expression of sima and tgo in human KRASG12V hemocytes (Fig. 5F), suggesting that uncontrolled proliferation of hemocytes within the larval hemocoel (body cavity), in our in vivo screen, could produce a hypoxic environment in which HIF gene function and HIF pathway activation are essential for cancer cell survival. In this scenario, HIF gene function would sustain progression and maintenance of leukemia in the fly, ultimately leading to death during the pupal stage. Consistent with this interpretation, HIF1A or ARNT gene silencing in hemocytes completely rescued adult fly development (Fig. 5E). To our knowledge, this is the first time HIF1 gene(s) have been identified in a KRAS genetic screen and illustrates the value of an in vivo Drosophila model expressing a human oncogene for genetic screening purposes.
Furthermore, our fly leukemia model can be directly applied in an in vivo drug screen (Aritakula and Ramasamy, 2008; Dar et al., 2012; Gladstone and Su, 2011; Markstein et al., 2014; Willoughby et al., 2013). Testing the effect of multiple drugs at multiple concentrations in fly larvae revealed echinomycin as a strong inhibitor of KRASG12V-induced hemocyte overproliferation. Interestingly, echinomycin is an inhibitor of the HIF1 pathway (Kong et al., 2005; Wang et al., 2011a, 2014). It prevents HIF1 binding to DNA target sites and thus disrupts HIF1 activation of target genes in response to hypoxic stress. Remarkably, echinomycin treatment was as effective as silencing either of the sima or tgo HIF pathway genes (Fig. 6). Moreover, echinomycin showed no toxicity to the fly at a concentration that completely reversed the leukemia phenotype (Fig. S3). Thus, these results have demonstrated another unique advantage of in vivo drug testing in Drosophila to identify potentially beneficial anticancer compounds of low organismal toxicity. Furthermore, we showed that echinomycin was particularly effective in multiple oncogenic RAS human leukemia lines compared to non-RAS cell lines and demonstrated its effect in a mammalian in vivo model for oncogenic RAS (xenograft mouse model using human THP1 cells with oncogenic NRASG12D; Fig. 7). In addition, our recent data showed that echinomycin also effectively inhibited HIF1A oncoprotein and regressed lung tumor cell growth based on multiple RAS cell lines, including NCI-H727 (KRASG12V), NCI-H1944 (KRASG13D) and Calu-1 (KRASG12C) (Huang et al., 2021). Taken together, these findings demonstrate the potential of echinomycin in treating RAS-induced cancer phenotypes. These findings are in line with previous studies that showed that echinomycin suppressed the growth of multiple AML cell lines and T-lymphoblastic leukemia cell lines (Yonekura et al., 2013) and effectively treated a mouse Mll-Flt3 AML model (Wang et al., 2011b, 2014). However, our study provides the first in vivo evidence that echinomycin treatment can attenuate oncogenic KRAS-induced leukemia in both Drosophila and mouse xenograft models.
Functionally, the KRASG12V-expressing hemocytes showed excessive proliferation, which ultimately resulted in complete mortality at the pupal stage (i.e. no adult flies emerged). Furthermore, they displayed impaired innate immune capabilities as evidenced by significantly reduced phagocytic activity (which could be partially rescued by HIF1 silencing; Fig. 5C,D), increased sensitivity to bacterial infection and reduced immune-related gene expression (Fig. 3D). These observations stand in contrast with previous reports that found that Drosophila strains carrying activated fly Ras85DG12V in hemocytes displayed no changes in phagocytosis activity compared to wild-type flies (Arefin et al., 2017). This possibly reflects the differential effects of human KRASG12V versus fly Ras85DG12V when expressed in hemocytes. Furthermore, a closer look at the expanded number of hemocytes caused by KRASG12V-induced overproliferation revealed a substantially altered composition of circulating hemocytes. Composition was notable for an expanded population of hemocytes that expressed Wg, an early hemocyte lineage surface marker (Fig. 2A), and a reduced P1+ plasmatocyte population, which is typically the most abundant and shows phagocytic activity (Fig. 2C). Interestingly, increased HIF1A has been shown to mediate cancer stem cell maintenance in leukemia, a highly proliferative cell type (Wang et al., 2011b, 2014). KRASG12V expression also drove expansion of the normally very small population of Lz+ crystal cells (Fig. 2B), which contribute to melanization reactions linked to innate immunity (Stofanko et al., 2010; Williams, 2007). It is possible that a subgroup of the Wg+ early hemocyte population also expresses the more mature crystal cell and/or plasmatocyte markers, as it has been previously reported that oncogenic RAS can lead to reprogramming and dedifferentiation (Ischenko et al., 2013). Owing to antibody limitations (all used here are mouse monoclonal), we cannot rule out this possibility at this time. In a recent study of single-cell profiling of Drosophila blood, we identified new hemocyte cell types (Fu et al., 2020). We are currently using this information to investigate how KRASG12V expression changes the composition of the hemocyte population.
Future mechanistic studies to elucidate the pathways underlying the differential susceptibility of oncogenic RAS leukemic cells to HIF inhibition by echinomycin will be of great interest. These studies could identify additional drug targets to inhibit HIF pathway signaling. Testing the efficacy of these and additional drug candidates, such as those shown to inhibit oncogenic KRAS and HIF pathways in a colorectal cancer screen (Bousquet et al., 2016), in treating oncogenic RAS in model systems like our fly are urgently needed to identify valuable treatment options in the clinic. In conclusion, the data presented here demonstrate the significant role of hypoxia signaling in oncogenic RAS leukemias and identify echinomycin as a viable candidate for treatment. They also establish the potential of our Drosophila model for use in conducting genetic screens and drug screens for human cancers.
MATERIALS AND METHODS
Fly stocks
Flies were reared on standard food at room temperature or at 18°C or 25°C (as indicated). Drosophila lines were obtained from the Bloomington Drosophila Stock Center or Vienna Drosophila Resource Center unless otherwise indicated (Table S3). In order to generate transgenic fly lines carrying UAS-HsKRASG12V, we cloned HsKRASG12V into the pUAST-attB vector, and the transgenes were introduced into a fixed chromosomal docking site by germ line transformation, to ensure that the RAS alleles were transcribed at precisely equivalent levels in hemocytes.
RASG12V leukemia model
The Drosophila model was generated through genetic crosses that ultimately yielded flies of genotype Hml-Gal4, UAS-GFP (second chromosome); UAS-HsKRASG12V (third chromosome). In such flies, Hml-Gal4 drove oncogenic UAS-Ras and UAS-GFP transgene expression specifically in hemocytes.
Drosophila genetic screen
A genetic screen was used to identify genes required for hemocyte cell viability/proliferation in a background of human KRASG12V expression. The approach involved simultaneous Hml-Gal4-driven hemocyte-specific expression of UAS-KRASG12V with a UAS-RNAi gene-silencing construct targeting a fly gene that is expressed in the hemocyte based on our unpublished RNA-seq data. Lethality for a given UAS-RNAi-targeted gene was determined by crossing homozygous UAS-RNAi transgenic male flies to females carrying the Hml-Gal4 driver balanced over CyO (‘Curly O’) and homozygous for UAS-KRASG12V and UAS-GFP (females were raised at 18°C, at which temperature the yeast-derived Gal4 protein is unstable and therefore ineffective at driving UAS-KRASG12V expression to levels inducing pupal-stage lethality; adult females were shifted to 25°C for genetic crosses and all subsequent screening steps were performed at 25°C). Progeny were first screened for emergence of straight-winged adult flies, indicating that silencing of the given gene by RNAi expression in hemocytes had prevented KRASG12V-induced pupal-stage death (curly-winged adult flies inherited the CyO balancer chromosome rather than the Hml-Gal4 driver and therefore expressed neither KRASG12V nor RNAi in hemocytes). In cases of straight-winged adult fly emergence, larval-stage progeny were then examined to determine the number of circulating hemocytes (all of which expressed GFP).
Hemocyte counting
Circulating hemocytes from single wandering third-instar larvae grown at 25°C were collected in 20 μl Schneider's Drosophila medium (Gibco). Larvae were carefully opened from the rear dorsal aspect and circulating hemocytes were gently transferred into the medium without disturbing the lymph gland. To count hemocytes, 10 μl of this mix was added to a hemocytometer, and GFP+ cells were counted using fluorescent microscopy. Hemocytes from six individual larvae of each genotype were counted using a hemocytometer.
Quantitative RT-PCR
Circulating hemocytes were collected from wandering third-instar larvae. Total RNA was extracted using TRIzol LS Reagent (Thermo Fisher Scientific) according to the manufacturer's instruction. Briefly, 250 µl hemocytes were lysed with 750 µl TRIzol LS Reagent, then RNA was extracted with chloroform and precipitated with isopropanol. RNA was washed with 75% ethanol and finally solubilized in TE buffer. The concentration and quality (A260/A280) of RNA was checked by a Nanodrop 2000 spectrophotometer (Thermo Scientific). Complementary DNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative RT-PCR of immune response genes was performed with Power SYBR Green PCR Master Mix (Applied Biosystems), and data were analyzed using the 2-ΔΔCT method. Gene rp49 (also called RpL32) was used as an internal control for normalization of gene expression levels.
Flow cytometry
Circulating hemocytes were collected from ten and two wandering third-instar Hml-Gal4, UAS-GFP and Hml-Gal4, UAS-GFP; UAS-KRASG12V larvae grown at 25°C, respectively. The size and granularity of collected live cells were immediately analyzed by BD FACS Calibur flow cytometry, and results were analyzed using FlowJo software (BD Biosciences).
Hemocyte labeling
Circulating hemocytes were collected from wandering third-instar larvae. Cells were attached to slides at 29°C for 60 min. Cells were then fixed for 10 min in 4% paraformaldehyde in PBS, blocked with 0.5% Triton X-100 plus 5% bovine serum albumin (in 1× PBS) for 1 h, incubated with primary antibodies anti-P1 (a kind gift from Dr I. Ando, Biological Research Center, Szeged, Hungary; 1:1000), anti-4D4 (Developmental Studies Hybridoma Bank; 1:1000) or anti-Lz (Developmental Studies Hybridoma Bank; 1:1000) at 4°C overnight. The cells were then washed three times with 1× PBS, incubated with Cy3-conjugated goat anti-mouse antibody (Jackson ImmunoResearch; 1:2000), washed three times with 1× PBS, mounted and imaged using a Zeiss Axio Imager with ApoTome2.
Hemocyte phagocytosis assay
Circulating hemocytes were collected from ten wandering third-instar larvae and incubated for 20 min in Schneider's Drosophila medium with 1 mg/ml S. aureus or E. coli bacterial cells labeled with pHrodo Red (Thermo Fisher Scientific). Hemocytes were then fixed for 10 min in 4% paraformaldehyde in PBS. Confocal imaging was performed using a Zeiss ApoTome.2 microscope and a Plan-Apochromat 20×/0.8 NA air objective. For quantitative comparisons of fluorescence intensity between samples, we selected an imaging setting that avoided oversaturation and used it across all image collection for comparative analysis. ImageJ Software Version 1.49 was used for image processing.
Drosophila drug treatment
A total of 34 compounds (both commercially available and proprietary; Table S3) were included in the pilot drug screen, including those targeting mTOR, AKT, AMPK and hypoxia signaling pathways. Drosophila food was ground to a powder-like consistency, and ∼2 mg was dispensed into a 2 ml tube. Drug was added to the food to a concentration of 0.5 μM. Ten eggs were transferred to each tube and maintained on drug-supplemented food at 25°C until growth and development to adult flies. Hemocytes were then collected for analysis in wandering third-instar larvae of those hatched flies.
Transplantation of larval hemocytes into adult flies
Hemolymph samples containing circulating hemocytes were collected from third-instar larvae of control flies. Hemocytes from control (Hml-Gal4, UAS-GFP) and KRASG12V (Hml-Gal4, UAS-GFP; UAS-KRASG12V) larvae were collected in ice-cold Schneider's Drosophila medium. Hemocytes were centrifuged and resuspended in the same medium at 2.5×106 cell/ml. A newly emerged wild-type male fly was anesthetized with CO2, then 200 nl of hemocyte cell suspension was injected into the abdomen using a Nanoject II injector (Drummond Scientific) fitted with a glass capillary prepared on a micropipette puller (Narishige PN-30). Hemocytes from each larval genotype were transplanted into 50-100 adult flies of wild-type genetic background. Transplant-bearing flies were then transferred to fresh food and maintained for 2 h (day 1) or 4 days prior to analysis. For analysis, hemolymph samples collected from three files were pooled in 20 µl Schneider's Drosophila medium, and GFP+ hemocytes were counted using a hemocytometer.
Infection and survival assays
Experiments were performed as described previously (Shokal and Eleftherianos, 2017). P. luminescens was cultured in sterile Luria–Bertani (LB) broth for 18-22 h at 30°C on a shaker (220 rpm). The bacteria were then pelleted by centrifugation, washed and re-suspended in 1× sterile PBS. The concentration of bacteria for fly infections was adjusted to optical density (OD, 600 nm) 0.1 using a spectrophotometer. Seven- to 10-day-old adult male flies were anesthetized with CO2, then 18.4 nl bacterial suspension or sterile 1× PBS (injury control) was injected into the thorax using a Nanoject II apparatus (Drummond Scientific) equipped with a glass capillary prepared on a micropipette puller. Forty flies of each genotype were infected and subsequently maintained at 25°C. Survival was monitored for 48 h following injection.
Human leukemia cell viability assay
The human leukemia cell line CCRF-CEM was purchased from American Type Culture Collection (ATCC), and all other cell lines were maintained in Y.L.’s laboratory. The status of KRAS of U937 (KRAS wild type) and NB4 (KRASA18D) was obtained from Babij et al. (2011). Information on the other cell lines was obtained from ATCC, including Kasumi-1 and K562 (both KRAS wild type), THP1 (NRASG12D), HL-60 (NRASQ61L) and CCRF-CEM (KRASG12D). In total, four human cell lines with KRAS mutations and three lines with wild-type KRAS were assayed. Cell viability was determined using Cell Proliferation Kit I (MTT; Sigma-Aldrich) according to the manufacturer's instructions. Briefly, cells were seeded at a density of 8×104 cells/well in a 96-well plate and maintained at 37°C for 48 h in the presence of vehicle (control) or echinomycin at increasing concentrations. Next, 10 μl MTT was added to each well and the plate incubated for a further 4 h in a humidified incubator. Then, 100 μl of solubilization solution was added to each well and the plate was kept at 37°C overnight. Dye was quantitated using a scanning multi-well spectrophotometer (BioTek Instruments).
Human leukemia xenograft
Initially, 21 Nod.Scid.Il2rg0 [NOD scid gamma (NSG)] mice, postnatal day 6, received intrahepatic transplants of 106 lentiviral luciferase transduced human THP1 NRAS mutant leukemia cells in ice-cold PBS as previously reported (Wu et al., 2016). Human leukemia cell engraftment was confirmed 3 days later by live bioluminescence imaging using a Xenogen IVIS Spectrum Imaging System (Caliper Life Sciences). Beginning on postnatal day 6, mice received intraperitoneal injections of vehicle, or 10 μg/kg or 30 μg/kg echinomycin (seven mice in each treatment group) every other day for a total of nine injections. Two mice in the 30 μg/kg echinomycin group did not survive the first three doses of treatment; therefore, results for only five mice in each group were included in further analysis. Bioluminescence imaging was performed after every third dose. For imaging, mice received IP d-luciferin (K+ salt, 150 mg/kg, Caliper Life Sciences) and 10 min later were anesthetized with isoflurane. Imaging results were analyzed using Living Image software (PerkinElmer). Signal values (photons/s/cm2/sr) were obtained for regions of interest, and quantitative comparisons across control and treatment groups were based on mean bioluminescence values.
Statistical analysis
Statistical tests were performed using PAST.exe software. Data were first tested for normality by using the Shapiro–Wilk test (a=0.05). Normally distributed data were analyzed either by unpaired Student's t-test (two groups) and Bonferroni comparison to adjust the P-value or by a one-way ANOVA followed by a Tukey–Kramer post-test for comparing multiple groups. Data without a normal distribution were analyzed by either a Mann–Whitney test (two groups) and Bonferroni comparison to adjust P-value or a Kruskal–Wallis H-test followed by a Dunn's test for comparisons between multiple groups. Statistical significance was defined as P<0.05.
Ethics approval
This study was approved by the University of Maryland School of Medicine Institution Review Board.
This article is part of a collection ‘The RAS Pathway: Diseases, Therapeutics and Beyond’, which was launched in a dedicated Special Issue guest edited by Donita Brady and Arvin Dar. See related articles in this collection at https://journals.biologists.com/dmm/collection/5089/The-RAS-Pathway.
Supplementary Material
Acknowledgements
The authors thank the Bloomington Drosophila Stock Center and the Vienna Drosophila Resource Center for the transgenic RNAi fly stocks. We thank Dr I. Ando (Biological Research Center, Szeged, Hungary; primary anti-P1 antibody) and the Developmental Studies Hybridoma Bank at the University of Iowa for antibodies. Further, we thank Dr Joyce van de Leemput for editing the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Y.L., Z.H.; Methodology: J.-y.Z., X.H., Y.F., Y.W., P.Z., Y.L., Z.H.; Validation: J.-y.Z., X.H., Y.W.; Formal analysis: J.-y.Z., X.H., Y.F., Y.W., Z.H.; Investigation: J.-y.Z., X.H., Y.F., Y.W., Z.H.; Resources: Z.H.; Data curation: J.-y.Z., X.H., Y.W., P.Z., Y.L., Z.H.; Writing - original draft: Z.H.; Writing - review & editing: Z.H.; Visualization: J.-y.Z., X.H., Y.W.; Supervision: P.Z., Y.L., Z.H.; Project administration: Y.L., Z.H.; Funding acquisition: Y.L., Z.H.
Funding
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (R01-DK098410 to Z.H.), the National Heart, Lung, and Blood Institute (R01-HL134940 to Z.H.) and the National Institute of Allergy and Infectious Diseases (R01-AI064350 to Y.L. and P.Z.).
References
- Arefin, B., Kunc, M., Krautz, R. and Theopold, U. (2017). The immune phenotype of three Drosophila leukemia models. G3 7, 2139-2149. 10.1534/g3.117.039487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aritakula, A. and Ramasamy, A. (2008). Drosophila-based in vivo assay for the validation of inhibitors of the epidermal growth factor receptor/Ras pathway. J. Biosci. 33, 731-742. 10.1007/s12038-008-0093-9 [DOI] [PubMed] [Google Scholar]
- Asha, H., Nagy, I., Kovacs, G., Stetson, D., Ando, I. and Dearolf, C. R. (2003). Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163, 203-215. 10.1093/genetics/163.1.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babij, C., Zhang, Y., Kurzeja, R. J., Munzli, A., Shehabeldin, A., Fernando, M., Quon, K., Kassner, P. D., Ruefli-Brasse, A. A., Watson, V. J.et al. (2011). STK33 kinase activity is nonessential in KRAS-dependent cancer cells. Cancer Res. 71, 5818-5826. 10.1158/0008-5472.CAN-11-0778 [DOI] [PubMed] [Google Scholar]
- Barbie, D. A., Tamayo, P., Boehm, J. S., Kim, S. Y., Moody, S. E., Dunn, I. F., Schinzel, A. C., Sandy, P., Meylan, E., Scholl, C.et al. (2009). Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108-112. 10.1038/nature08460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaupre, D. M. and Kurzrock, R. (1999a). RAS and leukemia: from basic mechanisms to gene-directed therapy. J. Clin. Oncol. 17, 1071-1079. 10.1200/JCO.1999.17.3.1071 [DOI] [PubMed] [Google Scholar]
- Beaupre, D. M. and Kurzrock, R. (1999b). RAS inhibitors in hematologic cancers: biologic considerations and clinical applications. Invest. New Drugs 17, 137-143. 10.1023/A:1006319116226 [DOI] [PubMed] [Google Scholar]
- Bos, J. L. (1989). RAS oncogenes in human cancer: a review. Cancer Res. 49, 4682-4689. [PubMed] [Google Scholar]
- Bousquet, M. S., Ma, J. J., Ratnayake, R., Havre, P. A., Yao, J., Dang, N. H., Paul, V. J., Carney, T. J., Dang, L. H. and Luesch, H. (2016). Multidimensional screening platform for simultaneously targeting oncogenic KRAS and hypoxia-inducible factors pathways in colorectal cancer. ACS Chem. Biol. 11, 1322-1331. 10.1021/acschembio.5b00860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, B. S. and Shannon, K. (2008). Targeting Ras in myeloid leukemias. Clin. Cancer Res. 14, 2249-2252. 10.1158/1078-0432.CCR-07-1005 [DOI] [PubMed] [Google Scholar]
- Brumby, A. M. and Richardson, H. E. (2003). scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 22, 5769-5779. 10.1093/emboj/cdg548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby, A. M. and Richardson, H. E. (2005). Using Drosophila melanogaster to map human cancer pathways. Nat. Rev. Cancer 5, 626-639. 10.1038/nrc1671 [DOI] [PubMed] [Google Scholar]
- Chabu, C. and Xu, T. (2014). Oncogenic Ras stimulates Eiger/TNF exocytosis to promote growth. Development 141, 4729-4739. 10.1242/dev.108092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun, S. Y., Johnson, C., Washburn, J. G., Cruz-Correa, M. R., Dang, D. T. and Dang, L. H. (2010). Oncogenic KRAS modulates mitochondrial metabolism in human colon cancer cells by inducing HIF-1alpha and HIF-2alpha target genes. Mol. Cancer 9, 293. 10.1186/1476-4598-9-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilloni, D. and Saglio, G. (2012). Molecular pathways: BCR-ABL. Clin. Cancer Res. 18, 930-937. 10.1158/1078-0432.CCR-10-1613 [DOI] [PubMed] [Google Scholar]
- Corcoran, R. B., Cheng, K. A., Hata, A. N., Faber, A. C., Ebi, H., Coffee, E. M., Greninger, P., Brown, R. D., Godfrey, J. T., Cohoon, T. J.et al. (2013). Synthetic lethal interaction of combined BCL-XL and MEK inhibition promotes tumor regressions in KRAS mutant cancer models. Cancer Cell 23, 121-128. 10.1016/j.ccr.2012.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar, A. C., Das, T. K., Shokat, K. M. and Cagan, R. L. (2012). Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature 486, 80-84. 10.1038/nature11127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler, V. L., Galbraith, M. and Espinosa, J. M. (2014). Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 49, 1-15. 10.3109/10409238.2013.838205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deynoux, M., Sunter, N., Herault, O. and Mazurier, F. (2016). Hypoxia and hypoxia-inducible factors in leukemias. Front. Oncol. 6, 41. 10.3389/fonc.2016.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward, J. (2003). Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3, 11-22. 10.1038/nrc969 [DOI] [PubMed] [Google Scholar]
- Downward, J. (2015). RAS synthetic lethal screens revisited: still seeking the elusive prize? Clin. Cancer Res. 21, 1802-1809. 10.1158/1078-0432.CCR-14-2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, C. J., Liu, T. and Banerjee, U. (2014). Drosophila hematopoiesis: markers and methods for molecular genetic analysis. Methods 68, 242-251. 10.1016/j.ymeth.2014.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Huang, X., Zhang, P., van de Leemput, J. and Han, Z. (2020). Single-cell RNA sequencing identifies novel cell types in Drosophila blood. J. Genet. Genomics 47, 175-186. 10.1016/j.jgg.2020.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladstone, M. and Su, T. T. (2011). Chemical genetics and drug screening in Drosophila cancer models. J. Genet. Genomics 38, 497-504. 10.1016/j.jgg.2011.09.003 [DOI] [PubMed] [Google Scholar]
- Gonzalez, C. (2013). Drosophila melanogaster: a model and a tool to investigate malignancy and identify new therapeutics. Nat. Rev. Cancer 13, 172-183. 10.1038/nrc3461 [DOI] [PubMed] [Google Scholar]
- Govind, S. (2008). Innate immunity in Drosophila: pathogens and pathways. Insect. Sci. 15, 29-43. 10.1111/j.1744-7917.2008.00185.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallin, J., Engstrom, L. D., Hargis, L., Calinisan, A., Aranda, R., Briere, D. M., Sudhakar, N., Bowcut, V., Baer, B. R., Ballard, J. A.et al. (2020). The KRAS(G12C) inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 10, 54-71. 10.1158/2159-8290.CD-19-1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. and Weinberg, R. A. (2000). The hallmarks of cancer. Cell 100, 57-70. 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Huang, X., Liu, Y., Wang, Y., Bailey, C., Zheng, P. and Liu, Y. (2021). Dual targeting oncoproteins MYC and HIF1alpha regresses tumor growth of lung cancer and lymphoma. Cancers 13, 694. 10.3390/cancers13040694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischenko, I., Zhi, J., Moll, U. M., Nemajerova, A. and Petrenko, O. (2013). Direct reprogramming by oncogenic Ras and Myc. Proc. Natl. Acad. Sci. USA 110, 3937-3942. 10.1073/pnas.1219592110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun, J. C., Rathore, A., Younas, H., Gilkes, D. and Polotsky, V. Y. (2017). Hypoxia-inducible factors and cancer. Curr. Sleep Med. Rep. 3, 1-10. 10.1007/s40675-017-0062-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. S., Mendiratta, S., Kim, J., Pecot, C. V., Larsen, J. E., Zubovych, I., Seo, B. Y., Kim, J., Eskiocak, B., Chung, H.et al. (2013). Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell 155, 552-566. 10.1016/j.cell.2013.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, D., Park, E. J., Stephen, A. G., Calvani, M., Cardellina, J. H., Monks, A., Fisher, R. J., Shoemaker, R. H. and Melillo, G. (2005). Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res. 65, 9047-9055. 10.1158/0008-5472.CAN-05-1235 [DOI] [PubMed] [Google Scholar]
- Kurucz, E., Markus, R., Zsamboki, J., Folkl-Medzihradszky, K., Darula, Z., Vilmos, P., Udvardy, A., Krausz, I., Lukacsovich, T., Gateff, E.et al. (2007). Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr. Biol. 17, 649-654. 10.1016/j.cub.2007.02.041 [DOI] [PubMed] [Google Scholar]
- Lamble, A. J. and Lind, E. F. (2018). Targeting the immune microenvironment in acute myeloid leukemia: a focus on T cell immunity. Front Oncol 8, 213. 10.3389/fonc.2018.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Ye, Q., Zhao, X.-P., Zhang, P.-B., Li, S., Li, R.-Q. and Zhao, X.-L. (2019). RAS mutations in acute myeloid leukaemia patients: A review and meta-analysis. Clin. Chim. Acta 489, 254-260. 10.1016/j.cca.2018.08.040 [DOI] [PubMed] [Google Scholar]
- Luo, J., Emanuele, M. J., Li, D., Creighton, C. J., Schlabach, M. R., Westbrook, T. F., Wong, K. K. and Elledge, S. J. (2009). A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835-848. 10.1016/j.cell.2009.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk, J. B., Lam, V. Y. and Tolwinski, N. S. (2017). Epidermal growth factor pathway signaling in Drosophila embryogenesis: tools for understanding cancer. Cancers 9, 16. 10.3390/cancers9020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X., Lu, J.-Y., Dong, Y., Li, D., Malagon, J. N. and Xu, T. (2017). PP6 disruption synergizes with oncogenic Ras to promote JNK-dependent tumor growth and invasion. Cell Rep 19, 2657-2664. 10.1016/j.celrep.2017.05.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein, M., Dettorre, S., Cho, J., Neumuller, R. A., Craig-Muller, S. and Perrimon, N. (2014). Systematic screen of chemotherapeutics in Drosophila stem cell tumors. Proc. Natl. Acad. Sci. USA 111, 4530-4535. 10.1073/pnas.1401160111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, F. (1994). Raf: the holy grail of Ras biology? Trends Cell Biol. 4, 347-350. 10.1016/0962-8924(94)90075-2 [DOI] [PubMed] [Google Scholar]
- Pagliarini, R. A. and Xu, T. (2003). A genetic screen in Drosophila for metastatic behavior. Science 302, 1227-1231. 10.1126/science.1088474 [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta, Y., Grabocka, E. and Bar-Sagi, D. (2011). RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer 11, 761-774. 10.1038/nrc3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarthy, A. V., Morgan-Lappe, S. E., Zakula, D., Vernetti, L., Schurdak, M., Packer, J. C., Anderson, M. G., Shirasawa, S., Sasazuki, T. and Fesik, S. W. (2007). Survivin depletion preferentially reduces the survival of activated K-Ras-transformed cells. Mol. Cancer Ther. 6, 269-276. 10.1158/1535-7163.MCT-06-0560 [DOI] [PubMed] [Google Scholar]
- Scholl, C., Frohling, S., Dunn, I. F., Schinzel, A. C., Barbie, D. A., Kim, S. Y., Silver, S. J., Tamayo, P., Wadlow, R. C., Ramaswamy, S.et al. (2009). Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell 137, 821-834. 10.1016/j.cell.2009.03.017 [DOI] [PubMed] [Google Scholar]
- Semenza, G. L. (2003). Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721-732. 10.1038/nrc1187 [DOI] [PubMed] [Google Scholar]
- Shokal, U. and Eleftherianos, I. (2017). Thioester-containing protein-4 regulates the Drosophila immune signaling and function against the pathogen Photorhabdus. J. Innate. Immun. 9, 83-93. 10.1159/000450610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinenko, S. A., Mandal, L., Martinez-Agosto, J. A. and Banerjee, U. (2009). Dual role of wingless signaling in stem-like hematopoietic precursor maintenance in Drosophila. Dev. Cell 16, 756-763. 10.1016/j.devcel.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A., Sweeney, M. F., Yu, M., Burger, A., Greninger, P., Benes, C., Haber, D. A. and Settleman, J. (2012). TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell 148, 639-650. 10.1016/j.cell.2011.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckel, M., Molina-Arcas, M., Weigelt, B., Marani, M., Warne, P. H., Kuznetsov, H., Kelly, G., Saunders, B., Howell, M., Downward, J.et al. (2012). Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res. 22, 1227-1245. 10.1038/cr.2012.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stofanko, M., Kwon, S. Y. and Badenhorst, P. (2010). Lineage tracing of lamellocytes demonstrates Drosophila macrophage plasticity. PLoS ONE 5, e14051. 10.1371/journal.pone.0014051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak, D., Dybko, J. and Kuliczkowski, K. (2018). The role of hypoxia-inducible factors in leukemias. Adv. Clin. Exp. Med. 27, 271-275. 10.17219/acem/69261 [DOI] [PubMed] [Google Scholar]
- Talks, K. L., Turley, H., Gatter, K. C., Maxwell, P. H., Pugh, C. W., Ratcliffe, P. J. and Harris, A. L. (2000). The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol. 157, 411-421. 10.1016/S0002-9440(10)64554-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent, S., Chen, R., Sayles, L. C., Lin, C., Walker, R. G., Gillespie, A. K., Subramanian, A., Hinkle, G., Yang, X., Saif, S.et al. (2010). Wilms tumor 1 (WT1) regulates KRAS-driven oncogenesis and senescence in mouse and human models. J. Clin. Invest. 120, 3940-3952. 10.1172/JCI44165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal, M. and Cagan, R. L. (2006). Drosophila models for cancer research. Curr. Opin. Genet. Dev. 16, 10-16. 10.1016/j.gde.2005.12.004 [DOI] [PubMed] [Google Scholar]
- Wang, R., Zhou, S. and Li, S. (2011a). Cancer therapeutic agents targeting hypoxia-inducible factor-1. Curr. Med. Chem. 18, 3168-3189. 10.2174/092986711796391606 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Liu, Y., Malek, S. N., Zheng, P. and Liu, Y. (2011b). Targeting HIF1alpha eliminates cancer stem cells in hematological malignancies. Cell Stem Cell 8, 399-411. 10.1016/j.stem.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Liu, Y., Tang, F., Bernot, K. M., Schore, R., Marcucci, G., Caligiuri, M. A., Zheng, P. and Liu, Y. (2014). Echinomycin protects mice against relapsed acute myeloid leukemia without adverse effect on hematopoietic stem cells. Blood 124, 1127-1135. 10.1182/blood-2013-12-544221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Liu, Y., Bailey, C., Zhang, H., He, M., Sun, D., Zhang, P., Parkin, B., Baer, M. R., Zheng, P.et al. (2020). Therapeutic targeting of TP53-mutated acute myeloid leukemia by inhibiting HIF-1alpha with echinomycin. Oncogene 39, 3015-3027. 10.1038/s41388-020-1201-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M. J. (2007). Drosophila hemopoiesis and cellular immunity. J. Immunol. 178, 4711-4716. 10.4049/jimmunol.178.8.4711 [DOI] [PubMed] [Google Scholar]
- Willoughby, L. F., Schlosser, T., Manning, S. A., Parisot, J. P., Street, I. P., Richardson, H. E., Humbert, P. O. and Brumby, A. M. (2013). An in vivo large-scale chemical screening platform using Drosophila for anti-cancer drug discovery. Dis. Model. Mech. 6, 521-529. 10.1242/dmm.009985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T., Heuillard, E., Lindner, V., Bou About, G., Ignat, M., Dillenseger, J. P., Anton, N., Dalimier, E., Gosse, F., Foure, G.et al. (2016). Multimodal imaging of a humanized orthotopic model of hepatocellular carcinoma in immunodeficient mice. Sci. Rep. 6, 35230. 10.1038/srep35230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M., Pastor-Pareja, J. C. and Xu, T. (2010). Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature 463, 545-548. 10.1038/nature08702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura, S., Itoh, M., Okuhashi, Y., Takahashi, Y., Ono, A., Nara, N. and Tohda, S. (2013). Effects of the HIF1 inhibitor, echinomycin, on growth and NOTCH signalling in leukaemia cells. Anticancer Res. 33, 3099-3103. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.