Significance
RNA processing generally occurs as transcripts are being produced and the concomitant cotranscriptional processes are interconnected with chromatin regulation. These cotranscriptional mechanisms quantitatively influence transcriptional output. At the Arabidopsis gene FLC, repression involves alternative processing of FLC antisense transcripts linked to delivery of a local chromatin environment that determines FLC transcription initiation and elongation rate. Here, we show that AGO1, a factor known predominantly for its role in posttranscriptional gene silencing, is involved in this cotranscriptional repression mechanism. Conserved cotranscriptional regulators, the THO/TREX complex and NTC components of the activated spliceosome, physically associate with AGO1. Our analysis suggests that alternative interactions of cotranscriptional regulators with the RNA Pol II–spliceosome link RNA processing and chromatin modification to quantitatively regulate transcriptional output.
Keywords: RNA processing, ARGONAUTE1, THO/TREX complex, cotranscription
Abstract
Quantitative transcriptional control is essential for physiological and developmental processes in many organisms. Transcriptional output is influenced by cotranscriptional processes interconnected to chromatin regulation, but how the functions of different cotranscriptional regulators are integrated is poorly understood. The Arabidopsis floral repressor locus FLOWERING LOCUS C (FLC) is cotranscriptionally repressed by alternative processing of the antisense transcript COOLAIR. Proximal 3′-end processing of COOLAIR resolves a cotranscriptionally formed R-loop, and this process physically links to a histone-modifying complex FLD/SDG26/LD. This induces a chromatin environment locally that determines low transcription initiation and a slow elongation rate to both sense and antisense strands. Here, we show that ARGONAUTE1 (AGO1) genetically functions in this cotranscriptional repression mechanism. AGO1 associates with COOLAIR and influences COOLAIR splicing dynamics to promote proximal COOLAIR, R-loop resolution, and chromatin silencing. Proteomic analyses revealed physical associations between AGO1, subunits of RNA Polymerase II (Pol II), the splicing-related proteins—the spliceosome NineTeen Complex (NTC) and related proteins (NTR)—and the THO/TREX complex. We connect these activities by demonstrating that the THO/TREX complex activates FLC expression acting antagonistically to AGO1 in COOLAIR processing. Together these data reveal that antagonistic cotranscriptional regulation through AGO1 or THO/TREX influences COOLAIR processing to deliver a local chromatin environment that determines FLC transcriptional output. The involvement of these conserved cotranscriptional regulators suggests similar mechanisms may underpin quantitative transcriptional regulation generally.
RNA-mediated chromatin regulation has emerged as a key mechanism in gene regulation in eukaryotes (1). It is well-established how small RNAs (sRNAs) regulate chromatin silencing in heterochromatin formation in fission yeast, plants, and animal germlines (2). Long noncoding RNA (lncRNA) also plays pivotal roles in chromatin regulation. A classic example is the X inactive specific transcript (Xist)–mediated X chromosome inactivation in female mammals (3). One area, however, where our understanding is poor is how cotranscriptional processing of nascent transcripts can dynamically modulate transcriptional output through the regulation of chromatin.
Plants as sessile organisms have evolved to rapidly respond to environmental changes and thus provide an ideal system to dissect such mechanisms. In Arabidopsis thaliana, expression of the floral repressor locus FLOWERING LOCUS C (FLC) plays a central role in determining reproductive strategy—overwintering or rapid cycling—with considerable consequence on reproductive success in different climates (4–6). Regulators of FLC expression have been identified through genetic screens for early- and late-flowering plants. Remarkably, the majority of factors identified are general cotranscriptional regulatory factors involving RNA processing and chromatin regulation. For example, activators of FLC expression include homologs of yeast RNA polymerase II–associated factor 1 complex (Paf1-complex), SET-domain proteins, SWR1 chromatin-remodeling complex, and H2B monoubiquitination mediators (reviewed in ref. 7). Functioning antagonistically to these are repressors of FLC expression: conserved RNA binding proteins (FCA, FPA, and FLK), RNA 3′-end processing and splicing factors, and chromatin modifiers. These repressors were grouped into the autonomous pathway that regulates FLC through an RNA-mediated chromatin-silencing mechanism (reviewed in ref. 8). Our current understanding of this mechanism is that the RNA-binding protein FCA concentrates 3′-end processing factors in nuclear condensates and promotes proximal polyadenylation of a set of antisense long noncoding transcripts transcribed from FLC, called COOLAIR (9, 10). These 3′-end processing factors dynamically interact with a histone-modifying complex FLD/SDG26/LD, thereby removing H3K4me1 over the FLC gene body. The act of promoting proximal polyadenylation of COOLAIR resolves an R-loop, and this is essential to trigger the histone demethylation-induced chromatin silencing (11). This process creates a chromatin environment that reduces transcriptional initiation and slows the elongation rate, contributing to the quantitative repression of FLC expression (12, 13). In summary, the quantitative output of transcription at FLC is set by antagonistic functions of activators and repressors in a mechanism involving RNA/lncRNA processing and chromatin regulation. However, how these antagonistic functions converge and how quantitative output is modulated had not been previously addressed.
Argonaute proteins are well-known effectors in widely conserved RNA interference (RNAi) pathways. AGO proteins loaded with sRNAs form the core of RNA-induced silencing complexes that function in posttranscriptional gene silencing, either through cleavage of target RNA or translational inhibition, both of which mainly occur in the cytoplasm (14). AGO proteins also engage in transcriptional gene silencing in the nucleus, triggering epigenetic modifications on chromatin (15). In recent years, emerging evidence has demonstrated AGOs function more widely in gene regulation including transcriptional activation, RNA splicing, DNA repair, and chromatin topology (16–23). There are 10 AGOs in Arabidopsis, with diverse functions and association with different sRNA species. Arabidopsis AGO1 is conventionally considered to work predominantly with microRNAs (miRNAs) in RNAi (24), but recent evidence has shown AGO1 undergoes nucleocytosolic shuttling and associates with chromatin in response to ultraviolet light, plant hormones, and stress (25–28). These data suggest Arabidopsis AGO1 may play a much wider role in gene regulation.
Here, we report AGO1 as a player in the COOLAIR-mediated FLC repression mechanism. We find that AGO1 physically associates with the FLD/SDG26/LD complex. Genetic analysis confirmed the AGO1 involvement in FCA-mediated FLC repression, and we show AGO1 associates with antisense RNA COOLAIR and influences COOLAIR splicing. Proteomics analyses revealed a close relationship between AGO1, the THO/TREX and NTC/NTR complexes, and RNA Polymerase II (Pol II). We show the THO/TREX complex functions antagonistically with AGO1 on COOLAIR processing and thus FLC regulation. The detailed dissection of the FLC/COOLAIR transcriptional unit has enabled an understanding that differential assembly of cotranscriptional machineries with RNA Pol II results in changes in RNA processing that feeds back to alter local chromatin environment and thus influence subsequent transcriptional output.
Results
AGO1 Functions in the Same Genetic Pathway as FCA and FLD to Repress FLC.
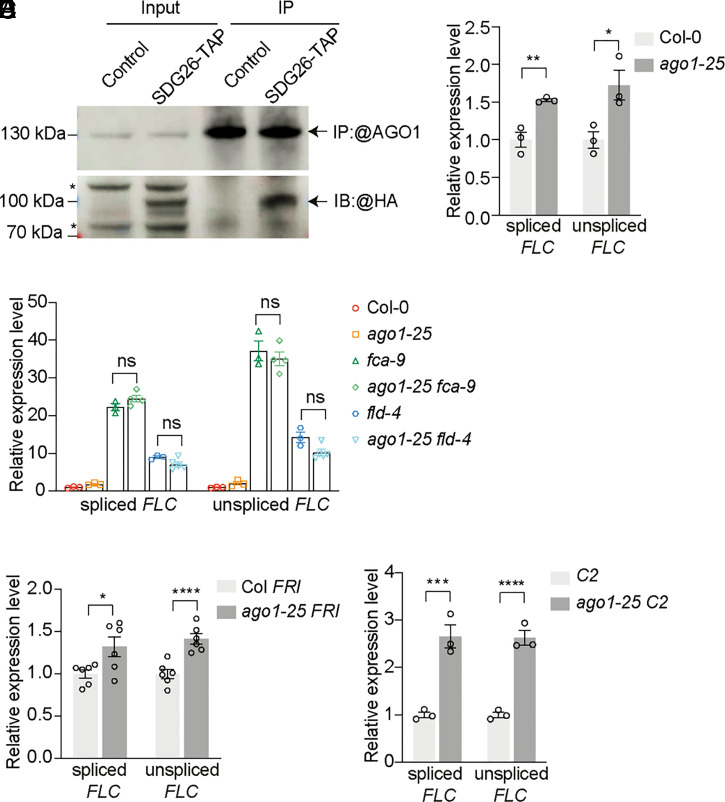
Interactors of the FLD/SDG26/LD complex have been identified using mass spectrometric analyses (12). A high peptide count for AGO1 was found in the SDG26-GFP affinity purification (SI Appendix, Fig. S1). Additionally, unique peptides of AGO1 were detected in cross-linked nuclear immunoprecipitation of SDG26-GFP (SI Appendix, Fig. S1). The interaction between AGO1 and SDG26 was further confirmed by coimmunoprecipitation (co-IP) using nuclear extracts from a stable transgenic line expressing SDG26 fused to a FLAG-HA TAP tag (SDG26-TAP) (Fig. 1A). The physical interaction between AGO1 and the FLC repressor complex FLD/SDG26/LD prompted us to investigate the role of AGO1 in FLC regulation. The FLD/SDG26/LD complex works downstream of FCA to repress FLC in the autonomous pathway. We therefore asked whether AGO1 functions in the same genetic pathway.
Fig. 1.
AGO1 genetically functions with FCA and FLD to repress FLC. (A) Co-IP in nuclear extracts from wild-type seedlings (Control) or seedlings expressing SDG26-TAP. IP: immunoprecipitation; IB: immunoblot. Asterisk indicates nonspecific signal. (B) Expression of spliced and unspliced FLC relative to UBC in Col-0 and ago1-25 seedlings. Data are normalized to wild-type Col-0. Data are presented as the mean ± SEM (n = 3). Asterisks indicate significant differences between the indicated plants (**P ≤ 0.007136, *P ≤ 0.031062, two-tailed t test). (C) Expression of spliced and unspliced FLC relative to UBC in various genotypes. Data are normalized to Col-0. Data are presented as the mean ± SEM (n = 3 to 5). Two-tailed P value from multiple t test corrected by Holm–Sidak method; ns, not significant. (D) Expression of spliced and unspliced FLC relative to UBC in Col FRI and ago1-25 FRI seedlings. Data are normalized to Col FRI. Data are presented as the mean ± SEM (n = 6). Asterisks indicate significant differences between the indicated plants (*P ≤ 0.029432, ****P ≤ 0.000551, two-tailed t test). (E) Expression of spliced and unspliced FLC relative to UBC in C2 and ago1-25 C2 seedlings. Data are normalized to C2. Data are presented as the mean ± SEM (n = 3). Asterisks indicate significant differences between the indicated plants (***P ≤ 0.002662, ****P ≤ 0.000588, two-tailed t test).
Since a complete loss of AGO1 function is sterile we exploited a weak allele ago1-25; this is slightly late-flowering (29) and confers increased FLC expression (Fig. 1B), similar to loss-of-function mutations of the autonomous pathway. ago1 fca and ago1 fld double mutants did not show any additive effect on FLC expression (Fig. 1C); however, an additive effect was observed when ago1 was combined with FRIGIDA (FRI), a strong activator of FLC (Fig. 1D). This indicates AGO1 functions in the same genetic pathway as FCA and FLD, but independently of FRI. To further investigate the role of AGO1 in FLC repression, we introduced ago1-25 into a genotype (named C2) carrying FRI and overexpressing FCAγ that we have used to successfully screen components required for FCA in FLC regulation (9, 10, 30, 31). FLC was significantly derepressed in C2 ago1-25 compared to C2 (Fig. 1E). Neither the native FCA protein level nor the overexpressed FCAγ was affected by the ago-25 mutation (SI Appendix, Fig. S2). These data collectively demonstrate AGO1 is required for FCA-mediated FLC repression. AGO1 harbors an Asp-Glu-Asp-His/Asp (DEDH/D) catalytic tetrad, which is commonly used for slicing target messenger RNA (mRNA) in Arabidopsis (24, 32). We therefore asked whether the slicer activity of AGO1 is required for repression of FLC. We analyzed FLC expression in a set of transgenic lines either expressing wild-type slicer or deficient slicer in ago1-25 (33). The derepression of FLC in ago1-25 is only complemented by introducing a wild-type slicer (SI Appendix, Fig. S3), suggesting that slicer activity or an intact slicer domain is required for AGO1-mediated FLC repression.
AGO1 Binds COOLAIR Transcripts and Influences COOLAIR Splicing.
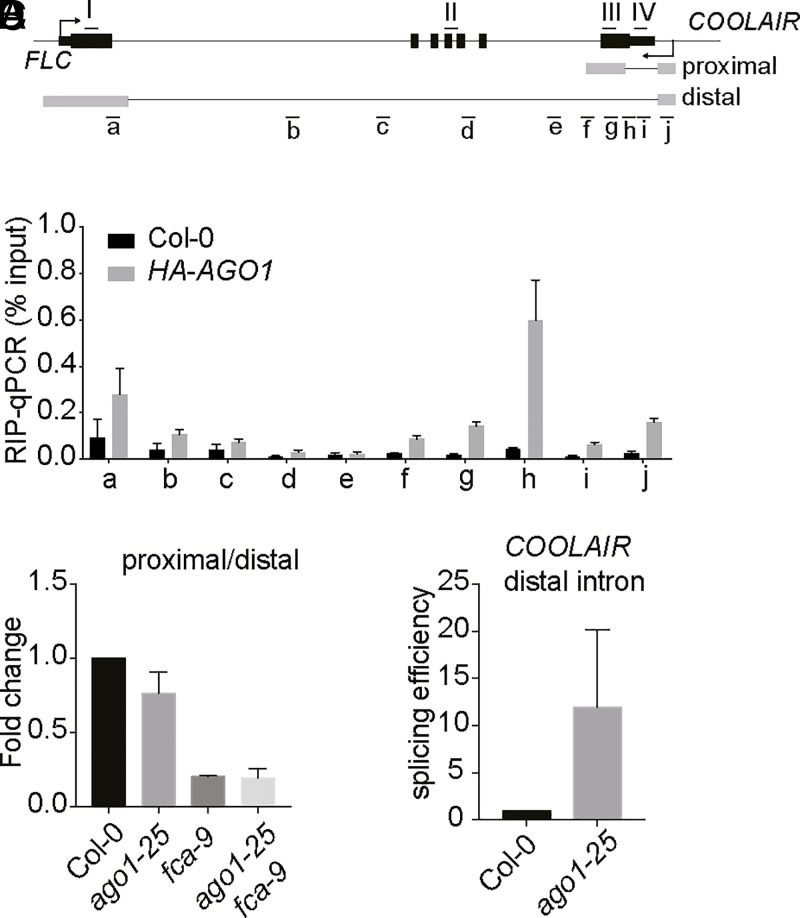
AGO1 was reported to directly bind to chromatin in the Arabidopsis genome to positively regulate gene expression, especially in response to hormone and stress stimuli (28). However, this does not seem to be the mechanism by which AGO1 regulates FLC for two reasons: First, FLC is repressed rather than activated by AGO1; second, chromatin immunoprecipitation (ChIP) sequencing data in this report did not suggest FLC chromatin is enriched for AGO1. Instead, the physical association and the genetic requirement of AGO1 in the autonomous pathway of FLC regulation prompts us to investigate a direct role of AGO1 in this antisense RNA-mediated chromatin silencing pathway. We performed RNA immunoprecipitation (RIP) in a transgenic line expressing hemagglutinin (HA)–tagged AGO1. Intriguingly, AGO1 associates with specific segments of COOLAIR, especially at a region spanning the exon–intron junction in the proximally polyadenylated COOLAIR (Fig. 2 A and B). This specific occupancy of AGO1 on COOLAIR prompted us to test whether AGO1 influences COOLAIR processing. We detected a reduced proximal-to-distal ratio of COOLAIR in ago1-25, which is epistatic to fca-9 (Fig. 2C). This suggested AGO1 works with FCA to promote proximal COOLAIR. Argonaute proteins have been documented to influence alternative splicing in other organisms (17–19, 34). We therefore hypothesized the shift to distal COOLAIR in ago1 mutant might be due to a shift in use of the distal splice acceptor site. By analyzing chromatin-bound RNA, we indeed detected a higher ratio of spliced to unspliced distal COOLAIR in ago1-25 (Fig. 2D). Taken together, these results suggest AGO1 targets COOLAIR transcript and influences splicing of COOLAIR.
Fig. 2.
AGO1 associates with COOLAIR and influences COOLAIR processing. (A) Schematic diagram showing FLC gene structure and COOLAIR transcripts. Black and gray boxes represent FLC and COOLAIR exons, respectively. Black and gray lines represent FLC and COOLAIR introns, respectively. The arrow indicates the transcription start site (TSS). Short black lines with letters underneath indicate positions of amplicons in qPCR amplification. (B) RIP–qPCR analyzing HA-AGO1 enrichment on nascent COOLAIR transcript. Wild-type Col-0 was used as background control. The x axis corresponds to the fragments shown in A. Data are mean ± SD (n = 3). (C) The ratio of proximal-to-distal isoforms of COOLAIR transcripts (refer to the schematic in A) in various genotypes relative to Col-0. Data are mean ± SEM (n = 3). (D) The splicing efficiency of distal intron (spliced/unspliced) determined through chromatin-bound RNA analysis. Data are normalized to Col-0. Data are mean ± SD (n = 3).
FCA forms liquid-like nuclear condensates in vivo that concentrate 3′-end processing factors to promote efficient polyadenylation (9). Thus, we asked whether AGO1 might influence the dynamics of FCA condensates. We compared FCA-mTurquoise2 condensates in wild type and ago1-25 mutant and found a small reduction of FCA condensate size in ago1-25 (SI Appendix, Fig. S4), the subtle difference likely due to ago1-25’s being a weak allele. Nevertheless, the slight AGO1 stabilization of the FCA condensates may increase dwell time over the locus sufficiently to promote splicing of the proximal intron linked to 3′-end processing to promote proximal COOLAIR formation.
AGO1 Promotes R-Loop Resolution and FLC Chromatin Silencing.
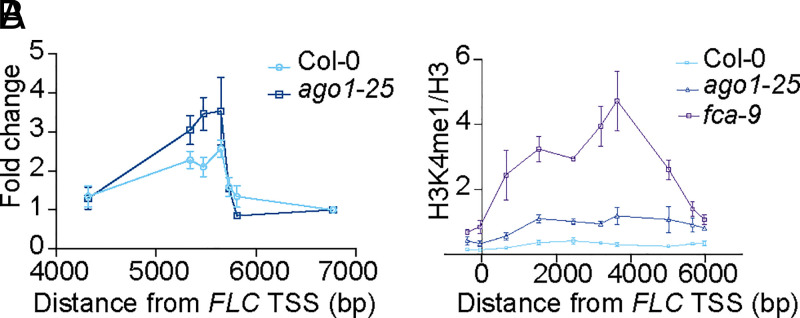
We had previously shown that processing of proximal COOLAIR promotes the resolution of a COOLAIR induced R-loop, which links to histone demethylation-induced chromatin silencing of FLC (11). Given AGO1 promoted proximal COOLAIR and repressed FLC, we speculated that AGO1 might also affect the R-loop dynamics. DNA/RNA immunoprecipitation (DRIP)-qPCR analysis showed the R-loop over the 3′ end of FLC indeed increases in ago1-25 (Fig. 3A). Consistent with this, H3K4me1 over FLC gene body was found to increase in ago1-25 (Fig. 3B). Taken together, AGO1 functions with FCA and FLD to promote FLC chromatin silencing, in a mechanism involving COOLAIR processing and R-loop resolution.
Fig. 3.
AGO1 promotes COOLAIR R-loop removal and FLC chromatic silencing. (A) DRIP–qPCR determining R-loop level in Col-0 and ago1-25. The number on the x axis is the distance to FLC TSS. Data are percentage of input normalized to a region without COOLAIR transcript (between 6,000 and 7,000 base pairs to FLC TSS). Data are presented as mean ± SEM (n = 3). (B) ChIP analysis of H3K4me1 level at FLC in various genotypes. The number on the x axis is the distance to FLC TSS. Data are mean ± SD (n = 3).
AGO1 canonically functions with miRNA and small interfering RNA (siRNA), which are mainly produced through DICER-LIKE (DCL) proteins in Arabidopsis. Previously, based on a T-DNA insertion mutant of DCL3 that largely released FLC silencing in 35S::FCA (overexpression of FCA ) background we reported that DCL3 is also involved in FCA/FLD-mediated FLC silencing (35). This particular dcl3 T-DNA allele suffers from posttranscriptional gene silencing due to a complex T-DNA insertion, which influences expression of other transgenes in that line (36). Introduction of an ethyl methanesulfonate–induced dcl3 allele into the 35S::FCA genotype did not result in released silencing of FLC, in contrast to the ago1-25 mutant (SI Appendix, Fig. S5). Therefore, we conclude AGO1 is involved in FCA-mediated FLC silencing independently of DCL3.
Physical Association of AGO1, RNA Pol II, and THO/TREX and NTC/NTR Complexes.
To understand the function of AGO1 in RNA processing, we investigated the proteins associated with AGO1 in the nucleus. A previous mass spectrometry analysis of nuclear AGO1 coimmunoprecipitated proteins identified SWI/SNF complex as an interactor that promotes transcription in response to stimuli (28). Other factors involved in transcription and RNA processing were also identified, including RNA Polymerase subunits, components of the NTC complex which couples RNA Pol II to splicing and the whole THO/TREX complex which links transcription with RNA processing and export (SI Appendix, Fig. S6A). We immunopurified HPR1/THO1, a component of the THO/TREX complex, from a transgenic line expressing HPR1 tagged with green fluorescent protein (GFP) (37). We were able to identify all the components of the THO/TREX complex, suggesting a successful IP. AGO1 was also identified as a coimmunoprecipitant of HPR1. Similar to the coimmunoprecipitated proteins of nuclear AGO1, RNA Pol II subunits and a plethora of factors in the NTC/NTR complex were identified, indicating a close relationship between these factors (SI Appendix, Fig. S6B and Dataset S1). We took advantage of the FLC/COOLAIR transcriptional unit to further understand the functional relevance of the physical associations between AGO1 and these conserved cotranscriptional regulators.
The THO/TREX Complex Is Required for FLC Activation and Antagonizes AGO1 and FCA.
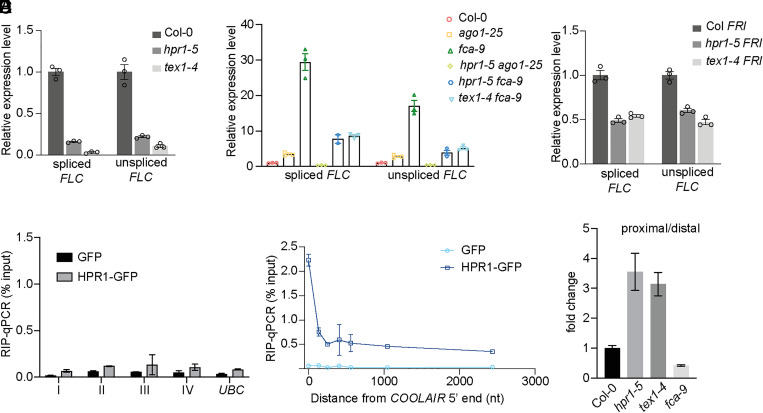
The conserved THO/TREX complex that links multiple steps of RNA processing with RNA export has been shown to promote messenger ribonucleoprotein assembly and prevent R-loop formation in yeast and mammals (38). Given the role of AGO1 in resolving COOLAIR-induced R-loop, we first tested the possibility that the THO/TREX complex may prevent COOLAIR-induced R-loop formation. Using DRIP-qPCR analysis, we did not observe an obvious R-loop level change in the THO/TREX complex mutants hpr1-5 and tex1-4 (SI Appendix, Fig. S7). We did, however, find a reduction of FLC expression in these mutants (Fig. 4A), suggesting the THO/TREX complex activates FLC. Combination of these mutations with ago1-25 or fca-9 gave double mutants with intermediate levels of FLC expression (Fig. 4B), indicating the THO/TREX complex is required for the derepression of FLC in ago1-25 or fca-9. We next asked whether this requirement is general for FLC activation, and therefore we introduced the THO/TREX mutants into the FRI genotype, where FLC is strongly activated. We found FLC activation is inhibited in the mutants (Fig. 4C), suggesting that the THO/TREX complex is also required for FRI-mediated FLC activation.
Fig. 4.
The THO/TREX complex up-regulates FLC and promotes distal COOLAIR. (A) Expression of spliced and unspliced FLC relative to UBC in various genotypes. Data are normalized to Col-0. Data are presented as the mean ± SEM (n = 3). (B) Expression of spliced and unspliced FLC relative to UBC in various genotypes. Data are normalized to Col-0. Data are presented as the mean ± SEM (n = 3). (C) Expression of spliced and unspliced FLC relative to UBC in various genotypes. Data are normalized to Col FRI. Data are presented as the mean ± SEM (n = 3). (D) RIP-qPCR analyzing HRP1-GFP enrichment on FLC mRNA in transgenic line expressing HPR1-GFP. Transgenic line expressing GFP alone was used as negative control. The x axis refers to the fragments shown in Fig. 2A. Data are mean ± SD (n = 3). (E) RIP-qPCR analyzing HRP1-GFP enrichment on nascent COOLAIR. Transgenic line expressing GFP was used as negative control. The x axis is the distance to COOLAIR 5′. Data are mean ± SD (n = 3). (F) The ratio of proximal-to-distal isoforms of COOLAIR transcripts (refer to the schematic in Fig. 2A) in various genotypes relative to Col-0. Data are mean ± SEM (n = 3).
The THO/TREX Complex Binds to Nascent COOLAIR and Promotes Production of Distal COOLAIR.
The general requirement of the THO/TREX complex to activate FLC through different pathways supports a direct cotranscriptional effect of this complex at the FLC locus. Therefore, we performed RIP-qPCR analysis to check the occupancy of this complex on the transcripts using a transgenic line expressing HPR1 tagged with GFP. There was relatively little enrichment of the HPR1-GFP protein with FLC mRNA (Fig. 4D), suggesting that FLC mRNA export is less likely to be affected. In contrast, HPR1-GFP was highly enriched with the COOLAIR nascent transcript (Fig. 4E). This suggests the THO/TREX complex mainly functions through cotranscriptional regulation of COOLAIR, which in turn influences FLC transcriptional activity in the FLC/COOLAIR circuit. We therefore analyzed COOLAIR processing and found low levels of distal COOLAIR in hpr1-5 and tex1-4 (Fig. 4F). The proximal COOLAIR was not affected in these mutants, consistent with no change in R-loop levels. In yeast, the THO/TREX complex was shown to promote transcription elongation and we speculate that this complex plays a similar mechanism at FLC promoting COOLAIR transcription elongation, and thus enhancing distal COOLAIR production.
Discussion
The interconnection between RNA Pol II, cotranscriptional RNA processing, and local chromatin is poorly understood, in part due to the complex feedbacks between the different steps. We have been analyzing these interconnections through analysis of Arabidopsis FLC regulation. Genetic screens uncovered FLC activators and repressors that identified cotranscriptional regulators involved in RNA processing and chromatin modification. The FLC repression mechanism links RNA 3′-end processing with chromatin regulators, and the SET-domain protein SDG26 was shown to play an important role in bridging these two processes. In the work reported here, we show that AGO1 is a factor in this repression mechanism through physical interaction with FLD/SDG26/LD complex and the genetic relationship with FCA and FLD. We also find THO/TREX acts antagonistically with AGO1, thus identifying one of the convergence points of FLC activators and repressors. AGO1 and THO/TREX function antagonistically on COOLAIR processing and FLC regulation and both interact with the NTC components. Thus, our analysis of a developmentally important regulator has shed light on antagonistic cotranscriptional regulation that potentially underlies quantitative regulation of transcription generally.
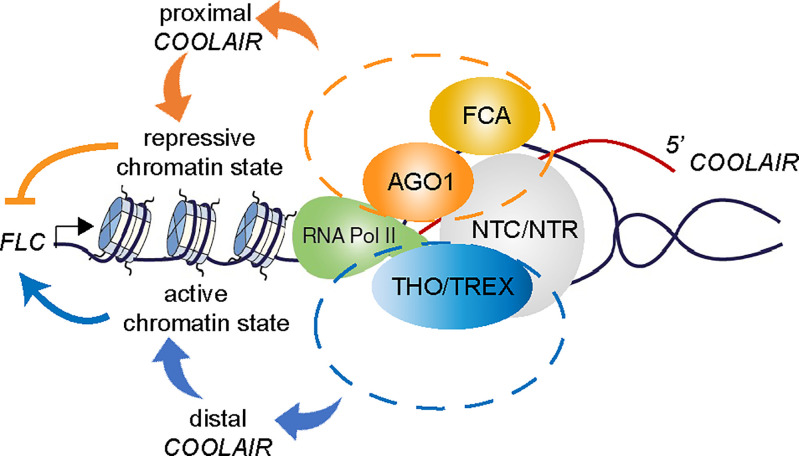
The multicomponent NTC/NTR complex forms part of the catalytic spliceosome and associates closely with both AGO1 and the THO/TREX complex. The physical association between AGO1 and the NTC/NTR was also revealed in other organisms. A conserved NTC/NTR component MAC7/Aquarius/EMB-4 was reported to interact with nuclear Argonaute HRDE-1 to mediate RNAi-dependent cotranscriptional silencing in a Caenorhabditis elegans germline (39, 40). We also find MAC7 may function together with AGO1 in FLC repression as FLC expression is derepressed in a mac7-1 mutant (SI Appendix, Fig. S8). Our data show AGO1 and the THO/TREX complex function antagonistically on COOLAIR processing and FLC regulation. In Arabidopsis, the isolation of loss-of-function mutations of the NTC/NTR complex has revealed differential regulation by different components even on the same target FLC. For example, a mutation on PRP8 causes elevated FLC level (30), while a mutation on BRR2 reduces FLC expression (41). One could argue this is through indirect effects of the complex; however, we propose that the NTC/NTR complex functions as the nexus determining which cotranscriptional complexes associate with RNA Pol II state (with feedback between these steps). We propose that AGO1 and the THO/TREX complex are differentially recruited by the NTC/NTR to influence the cotranscriptional coupling of the spliceosome to RNA Pol II. At FLC/COOLAIR this links to delivery of a local chromatin environment that determines FLC transcriptional initiation and elongation rate (Fig. 5). In this respect, the antagonistic cotranscriptional complexes conferring quantitative transcriptional regulation are like a tug of war between RNA Pol II-associating complexes.
Fig. 5.
Model for how modulation of antisense RNA processing determines sense transcriptional output. Antagonistic assembly of cotranscriptional machinery with RNA Pol II influences transcript processing. During COOLAIR transcription, FCA, AGO1, and components of NTC/NTR link to RNA Pol II and promote proximal COOLAIR. THO/TREX dynamically competes for RNA Pol II to promote distal COOLAIR. The differential composition of the cotranscriptional regulators with the Pol II complex influences the chromatin state locally, which in turn influences the transcriptional output of the whole locus.
The mechanistic basis for this tug of war between AGO1 and THO/TREX requires additional analysis. AGO1 and the THO/TREX physically interact with NTC components in the nucleus and may associate with different Pol II states, which determines the differential processing of RNA. Nevertheless, we do not rule out the possibility that they function together at some targets as shown in trans-acting siRNA biogenesis (42, 43). In regard to the cotranscriptional silencing mechanism, a classic Argonaute protein’s involvement is in an RNAi-mediated heterochromatin-silencing mechanism, elegantly elaborated in fission yeast, worms, and plants (1). An important question, therefore, is whether there are sRNAs involved in FLC regulation. So far, only one group of Pol IV/RDR2/DCL3-dependent sRNA hybridizing to the FLC 3′ region has been reported, and these are highly enriched in mature siliques rather than seedlings (44). Analysis of flowering time and FLC expression in a range of mutants defective in sRNA production showed a dcl1 dcl3 double mutant exhibited significantly higher FLC expression. Although DCL3 is not required for FCA-mediated FLC repression (SI Appendix, Fig. S5) and DCL1 is mainly required for producing miRNA (45) (FLC is not a miRNA target), redundant DICER activity could produce sRNA. Therefore, we searched sRNA sequence databases (http://ipf.sustech.edu.cn/pub/asrd/) (46) and found a low number of sRNA reads at the FLC locus, including low-abundance sRNAs complementary to the intron/exon junction region of proximal COOLAIR (SI Appendix, Fig. S9). This location was intriguing given the observed change in COOLAIR splicing in ago1. We therefore analyzed RNAs coimmunoprecipitated with AGO1 and found sRNA fragments complementary to COOLAIR over the proximal intron/exon junction region (SI Appendix, Fig. S10 A–C), where AGO1 was enriched on COOLAIR (Fig. 2B). A previous report in fission yeast had defined a class of Dicer-independent sRNA called primal sRNA (priRNA) generated from degradation products of transcripts that associate with Ago1 and target antisense transcripts (47). The priRNAs mediate an Argonaute-involved transcriptome surveillance mechanism that may be relevant to our findings. We therefore suggest a close connection between cotranscriptional RNA processing and turnover of aberrant transcripts by RNA-silencing factors. It is also possible that Argonaute proteins are recruited directly to nascent RNA or via protein–protein interactions, independently of sRNA (18, 23, 48). What recruits AGO1 to the COOLAIR transcript is an important question to be addressed in the future.
Materials and Methods
Plant Materials and Growth Conditions.
Mutant alleles ago1-25 (29), fca-9 (49), fld-4 (35), hpr1-5, tex1-4 (37), and mac7-1 (50) were described previously. Transgenic lines C2 (35) and FCA-mTurquoise2 (11) were described previously. Transgenic line HPR1-GFP was described previously (37) and was provided by Chi-Kuang Wen, CAS Center for Excellence in Molecular Plant Science, Shanghai, China. AGO-HA, AGO1(DDH), AGO1(DDD), AGO1(DAH), and AGO1(ADH) were transgenic lines in ago1-25 background, which were generously provided by James C. Carrington, Donald Danforth Plant Science Center, Olivette, MO, and were described previously (33). dcl3 allele was provided by David C. Baulcombe, University of Cambridge, Cambridge, UK.
Seeds were surface-sterilized and sown on standard half-strength Murashige and Skoog (1/2 MS) medium plate without glucose. Plates were stratified at 4 °C for 3 d before transferred to long-day conditions (16-h light at 20 °C, 8-h darkness at 16 °).
Expression Analysis.
Ten-day-old seedlings were harvested, and RNA was extracted. TURBO DNase (Ambion) was used to remove DNA contamination before reverse transcription. RNA was reverse-transcribed by SuperScript IV Reverse Transcriptase (Invitrogen) using gene-specific primers. qPCR analysis was performed, and data were normalized to UBC. Primers are described in SI Appendix, Table S1.
Western Blot Analysis.
Total protein extracts from 10-d-old seedlings were prepared and immunoblot was performed as described previously (9). Anti-FCA (1:8,000 dilution, homemade) antibody and rabbit immunoglobulin G horseradish peroxidase-linked whole antibody (1:10,000 dilution, NA934; GE Healthcare) were used as primary and secondary antibody, respectively, before using chemiluminescence for detection.
RIP.
For HPR1-GFP RIP, 3 g of 14-d-old seedlings were cross-linked in 1% formaldehyde. Nuclei were enriched using Honda buffer (20 mM Hepes, 0.44 M sucrose, 1.25% Ficoll, 2.5% Dextran T40, 10 mM MgCl2, 0.5% Triton X-100, 5 mM dithiothreitol [DTT], and 1× protease inhibitor mixture [Roche]) in the presence of 10 U/mL RNase inhibitor (RNaseOUT; Ambion). After resuspending the pellet in 1 mL Nuclear Lysis Buffer (50 mM Tris·HCl pH 7.5, 100 mM NaCl, 1% Triton X-100, 1 mM MgCl2, 0.1 mM CaCl2, 1× protease inhibitor mixture, and 100 U/mL RNase inhibitor), 40 U/mL DNase (Invitrogen) was added for incubation for 10 min at 37 °C. The lysate was incubated 1 h at 4 °C after adding 10 μL 10% sodium dodecyl sulfate (SDS) and 5 M NaCl. The lysate was then sonicated with Diagenode Bioruptor 10 times, 30 s on/30 s off at high setting. Soluble chromatin extracts were collected after centrifugation. GFP-trap magnetic agarose beads (ChromoTek) were prewashed with low-salt wash buffer and blocked with 0.1% bovine serum albumin and 1 mg/mL transfer RNA (tRNA) in low-salt wash buffer for 1 h before adding them into IP solution. After 2 h incubation at 4 °C, the beads were washed through high-salt washing buffer and low-salt washing buffer. The beads were suspended in RNA elution buffer (100 mM NaCl, 50 mM Tris·HCl, pH 7.0, 1 mM ethylenediaminetetraacetic acid [EDTA], and 1% SDS) at 95 °C for 15 min then were reverse cross-linked at 65 °C for 1 h in the presence of Protease K and RNase inhibitor. RNA was extracted using TRIzol and subjected to reverse transcription using gene-specific primers. qPCR was performed and data were presented as IP/1% input. UBC was used as a negative control. Primers are listed in SI Appendix, Table S1.
HA-AGO1 RIP was performed as described previously (9). Nuclei were enriched through cross-linked seedlings. After sonication, nuclear extract was incubated with anti-HA magnetic beads (88836; Thermo Scientific) for 2 h at 4 °C. After wash, the immunoprecipitates were eluted and reverse cross-linked. RNA was extracted using TRIzol and subsequent RT-qPCR was conducted. Primers are listed in SI Appendix, Table S1.
Co-IP.
Four grams of 14-d-old seedlings were cross-linked in 1% formaldehyde. Nuclei were prepared using Honda buffer (described above) and suspended in RIPA buffer (50 mM Tris·HCl, 150 mM NaCl, 1% Nonidet P-40, 0.5% Sodium deoxycholate, 0.1% SDS, 1× protease inhibitor mixture). Nuclear extracts were sonicated and incubated with Benzonase for 15 min. Four micrograms of AGO1 antibody (AS09527; Agrisera) was added into the soluble extracts incubated for 2 h before adding 25 μL Dynabeads Protein A (Invitrogen) for another 1.5 h. Beads were washed three times before eluted in 1x LDS buffer.
DRIP.
DRIP was performed as previously described (11). Briefly, 2 g 10-d-old seedlings were used to extract DNA. DNA was treated with Proteinase K overnight and extracted again using phenol/chloroform. The DNA pellet were dissolved in water and quantified with Qubit DNA quantification kit (Invitrogen). 1 μg of DNA were subjected to sonication, which was used for IP with 5 μg of S9.6 antibody (ENH001; Kerafast) overnight at 4 °C, before adding Protein G Agarose (Invitrogen) into IP solution for another 2 h. The immunoprecipitants were washed, eluted and precipitated, which then was subjected to qPCR analysis. The data were represented as normalized to 1% of input. Primers were listed in SI Appendix, Table S1.
Histone ChIP.
Histone ChIP was performed as previously described with modifications (11). Protein A magnetic beads (10002D; Invitrogen), anti-H3 (ab176842; Abcam), and anti-H3K4me1 (ab176877; Abcam) were used. After immunoprecipitation, the eluted DNA was quantified by qPCR with primers listed in SI Appendix, Table S1. Data were normalized to 1% of input and presented as the ratio of H3K4me1 to H3.
Chromatin-Bound RNA Preparation.
Nuclei from 2 g of 10-d-old seedlings were prepared as described before in RIP. The nuclei pellet was weighed and resuspended in an equal volume of resuspension buffer (50% glycerol, 0.5 mM EDTA, 1 mM DTT, 25 mM Tris·HCl, pH 7.5, and 100 mM NaCl). After washing twice with two volumes of UREA wash buffer (25 mM Tris·HCl, pH 7.5, 300 mM NaCl, 1 M urea, 0.5 mM EDTA, 1 mM DTT, and 1% Tween 20) via pipetting up and down, the chromatin pellet was spun down, which then was rinsed gently with TES. The RNA was extracted using TRIzol.
Microscopy.
The seeds carrying heterozygous ago1-25 and homozygous FCA-mTurquoise2 were sown and the 7-d-old seedlings were segregated into two populations. One population which developed normally was referred to genotype carrying wild-type AGO1 and the other with developmental defects (unexpanded dark green cotyledons) was referred to genotype carrying ago1-25 homozygous mutation. Root tips were imaged as described previously (11). The Analyze Particles tool was applied to obtain “area” data for each condensate after manual thresholding by ImageJ software. The same settings were used on all images.
Mass Spectrometry.
Fine powder from 14-d-old seedling was dissolved in IP buffer (50 mM Tris·HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, and 1× protease inhibitor mixture). Extracts were immunoprecipitated with GFP-trap magnetic agarose beads (ChromoTek). The affinity-purified protein samples were separated on gel. The gel slices were washed with 50 mM TEAB buffer, pH 8 (Sigma) and incubated with 10 mM DTT for 30 min at 65 °C followed by incubation with 30 mM iodoacetamide at room temperature (both in 50 mM TEAB). After washing and dehydration with acetonitrile, the gels were soaked with 50 mM TEAB containing 10 ng/µL Sequencing Grade Trypsin (Promega) and incubated at 50 °C for 8 h. The eluted peptide solution was dried down, and the peptides were dissolved in 0.1%TFA/3% acetonitrile. Aliquots were analyzed by nanoLC-MS/MS on an Orbitrap Fusion Tribrid mass spectrometer coupled to an UltiMate 3000 RSLCnano LC system (Thermo Fisher Scientific). Recalibrated peaklists were generated with MaxQuant 1.6.0.16 REF in LFQ mode using the TAIR10_pep_20101214 A. thaliana protein sequence database (https://www.arabidopsis.org/, 35,386 entries) plus the MaxQuant contaminants database (245 entries). The quantitative LFQ results from MaxQuant with default parameters were used together with search results from an in-house Mascot Server 2.4.1 (Matrixscience, London) on the same databases. The Mascot search results were imported into Scaffold 4.11 (https://www.proteomesoftware.com/) using identification probabilities of 99% for proteins and 95% for peptides.
sRNA Fragments Detection.
One gram of Col-0 and HA-AGO1 seedlings were ground into fine powder and subjected to immunoprecipitation using anti-HA magnetic beads (88836; Thermo). The beads were then extracted with TRIzol agent (Invitrogen) to retrieve the RNAs bound to AGO1. RNAs were reverse-transcribed using a Mir-XTM miRNA First Strand Synthesis Kit (Takara) according to the manufacturer’s instructions. The complementary DNA was amplified using sRNA sequences as forward primers (designed multiple primers complementary to COOLAIR sequence across the proximal COOLAIR transcript) and the adaptor as the reverse primer. miR159 was included as a positive control. qPCR was performed using the Mir-XTM miRNA qRT-PCR SYBR Kit (Takara) on LightCycler480 II (ROCHE) and qPCR data were normalized to tRNA. Primers are listed in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. C. Wen, Prof. Sir D. Baulcombe, and Dr. J.Carrington for providing seeds. We thank Gerhard Saalbach in the Proteomics Facility of the John Innes Center. We thank all the members of the C.D. group for discussions. Work in the C.D. group is supported by the European Research Council grant EPISWITCH (833254), a Wellcome Senior Investigator grant (210654), a Royal Society Professorship (RP\R1\180002), and the Biotechnology and Biological Sciences Research Council Institute Strategic Programme GEN (BB/P013511/1).
Footnotes
Reviewers: A.R.K., Universidad de Buenos Aires; E.A.M., University of Cambridge.
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113757118/-/DCSupplemental.
Data Availability
Previously published data were used for this work (12, 28). All other study data are included in the article and/or supporting information.
References
- 1.Holoch D., Moazed D., RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 16, 71–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martienssen R., Moazed D., RNAi and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 7, a019323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penny G. D., Kay G. F., Sheardown S. A., Rastan S., Brockdorff N., Requirement for Xist in X chromosome inactivation. Nature 379, 131–137 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Sheldon C. C., et al. , The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michaels S. D., Amasino R. M., FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson G. G., Dean C., Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289 (2002). [DOI] [PubMed] [Google Scholar]
- 7.He Y., Control of the transition to flowering by chromatin modifications. Mol. Plant 2, 554–564 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Wu Z., Fang X., Zhu D., Dean C., Autonomous Pathway: FLOWERING LOCUS C repression through an antisense-mediated chromatin-silencing mechanism. Plant Physiol. 182, 27–37 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang X., et al. , Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature 569, 265–269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu F., Marquardt S., Lister C., Swiezewski S., Dean C., Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94–97 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Xu C., et al. , R-loop resolution promotes co-transcriptional chromatin silencing. Nat. Commun. 12, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang X., et al. , The 3′ processing of antisense RNAs physically links to chromatin-based transcriptional control. Proc. Natl. Acad. Sci. U.S.A. 117, 15316–15321 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z., et al. , Quantitative regulation of FLC via coordinated transcriptional initiation and elongation. Proc. Natl. Acad. Sci. U.S.A. 113, 218–223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Höck J., Meister G., The Argonaute protein family. Genome Biol. 9, 1–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castel S. E., Martienssen R. A., RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14, 100–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alló M., et al. , Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol. 16, 717–724 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Ameyar-Zazoua M., et al. , Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 19, 998–1004 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Taliaferro J. M., et al. , Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: Alternative pre-mRNA splicing and transcriptional repression. Genes Dev. 27, 378–389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarallo R., et al. , The nuclear receptor ERβ engages AGO2 in regulation of gene transcription, RNA splicing and RISC loading. Genome Biol. 18, 1–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cernilogar F. M., et al. , Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480, 391–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen T. B., et al. , Argonaute-associated short introns are a novel class of gene regulators. Nat. Commun. 7, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shuaib M., et al. , Nuclear AGO1 regulates gene expression by affecting chromatin architecture in human cells. Cell Syst. 9, 446–458 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Gómez Acuña L. I., et al. , Nuclear role for human Argonaute-1 as an estrogen-dependent transcription coactivator. J. Cell Biol. 219, e201908097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baumberger N., Baulcombe D. C., Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 102, 11928–11933 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolata J., et al. , Salt stress reveals a new role for ARGONAUTE1 in miRNA biogenesis at the transcriptional and posttranscriptional levels. Plant Physiol. 172, 297–312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schalk C., et al. , Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proc. Natl. Acad. Sci. U.S.A. 114, E2965–E2974 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bologna N. G., et al. , Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant MicroRNA pathway. Mol. Cell 69, 709–719 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Liu C., et al. , Arabidopsis ARGONAUTE 1 binds chromatin to promote gene transcription in response to hormones and stresses. Dev. Cell 44, 348–361 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Morel J. B., et al. , Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14, 629–639 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marquardt S., et al. , Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell 54, 156–165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z. W., Wu Z., Raitskin O., Sun Q., Dean C., Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc. Natl. Acad. Sci. U.S.A. 111, 7468–7473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arribas-Hernández L., et al. , The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell 28, 1563–1580 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carbonell A., et al. , Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24, 3613–3629 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu Y., et al. , Argonaute binding within human nuclear RNA and its impact on alternative splicing. RNA 27, 991–1003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu F., et al. , The Arabidopsis RNA-binding protein FCA requires a Lysine-Specific Demethylase 1 homolog to downregulate FLC. Mol. Cell 28, 398–407 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Mlotshwa S., et al. , Transcriptional silencing induced by Arabidopsis T-DNA mutants is associated with 35S promoter siRNAs and requires genes involved in siRNA-mediated chromatin silencing. Plant J. 64, 699–704 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu C., Zhou X., Wen C. K., HYPER RECOMBINATION1 of the THO/TREX complex plays a role in controlling transcription of the REVERSION-TO-ETHYLENE SENSITIVITY1 gene in Arabidopsis. PLoS Genet. 11, e1004956 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luna R., Rondón A. G., Pérez-Calero C., Salas-Armenteros I., Aguilera A., The THO complex as a paradigm for the prevention of cotranscriptional R-Loops. Cold Spring Harb. Symp. Quant. Biol. 84, 105–114 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Tyc K. M., et al. , The conserved intron binding protein EMB-4 plays differential roles in germline small RNA pathways of C. elegans. Dev. Cell 42, 256–270 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Akay A., et al. , The Helicase Aquarius/EMB-4 is required to overcome intronic barriers to allow nuclear RNAi pathways to heritably silence transcription. Dev. Cell 42, 241–255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahrez W., et al. , BRR2a affects flowering time via FLC splicing. PLoS Genet. 12, e1005924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jauvion V., Elmayan T., Vaucheret H., The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell 22, 2697–2709 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yelina N. E., et al. , Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 13948–13953 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swiezewski S., et al. , Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc. Natl. Acad. Sci. U.S.A. 104, 3633–3638 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurihara Y., Watanabe Y., Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U.S.A. 101, 12753–12758 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng L., et al. , An online database for exploring over 2,000 Arabidopsis small RNA libraries. Plant Physiol. 182, 685–691 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halic M., Moazed D., Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell 140, 504–516 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leung A. K., et al. , Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 18, 237–244 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henderson I. R., Liu F., Drea S., Simpson G. G., Dean C., An allelic series reveals essential roles for FY in plant development in addition to flowering-time control. Development 132, 3597–3607 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Jia T., et al. , The Arabidopsis MOS4-associated complex promotes MicroRNA biogenesis and precursor messenger RNA splicing. Plant Cell 29, 2626–2643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Previously published data were used for this work (12, 28). All other study data are included in the article and/or supporting information.