Significance
Soil organic matter contains a large fraction of global C whose fate is largely governed by soil microbial communities. Global C-cycle models can be improved using information about microbial processes in soil, but the inner workings of soil communities remain largely hidden from view. We used stable isotopes to characterize the guild structure of soil bacteria based on how they assimilated C from different sources. Guild structure corresponded with growth dynamics and rrn copy number. We also show that this guild structure explains microbial biogeographical patterns at continental and global scales. These results suggest that microbial contributions to soil C cycling are largely governed by an anabolic process that governs energy allocation to growth, competition, and survival.
Keywords: DNA-SIP, bacterial, soil, C cycle, ecology
Abstract
Soil microorganisms determine the fate of soil organic matter (SOM), and their activities compose a major component of the global carbon (C) cycle. We employed a multisubstrate, DNA-stable isotope probing experiment to track bacterial assimilation of C derived from distinct sources that varied in bioavailability. This approach allowed us to measure microbial contributions to SOM processing by measuring the C assimilation dynamics of diverse microorganisms as they interacted within soil. We identified and tracked 1,286 bacterial taxa that assimilated 13C in an agricultural soil over a period of 48 d. Overall 13C-assimilation dynamics of bacterial taxa, defined by the source and timing of the 13C they assimilated, exhibited low phylogenetic conservation. We identified bacterial guilds composed of taxa that had similar 13C assimilation dynamics. We show that C-source bioavailability explained significant variation in both C mineralization dynamics and guild structure, and that the growth dynamics of bacterial guilds differed significantly in response to C addition. We also demonstrate that the guild structure explains significant variation in the biogeographical distribution of bacteria at continental and global scales. These results suggest that an understanding of in situ growth dynamics is essential for understanding microbial contributions to soil C cycling. We interpret these findings in the context of bacterial life history strategies and their relationship to terrestrial C cycling.
Soil organic matter (SOM) represents the largest terrestrial pool of organic carbon (C) on Earth (1), and SOM dynamics have major impacts on the global C cycle. SOM is sensitive to land management (2–4) and climate change (5), and SOM pools influence soil health (6, 7), agricultural productivity (8), and climate stability (6, 9). However, the mechanisms that promote SOM persistence and loss remain uncertain, and this introduces variability into global C-cycle models (10–13). Microorganisms are dominant drivers of SOM decomposition, stabilization, and mineralization (14), and C-cycle models increasingly seek to incorporate microbial traits to improve predictions of SOM dynamics (15, 16). For example, microbially explicit C-cycle models, such as MEND (17), MIMICS (18), and CORPSE (19), improve prediction of C-cycle dynamics when compared to models that ignore microbial processing (20).
Soil microbes play a critical role in governing global C flux, but the microbial mechanisms that determine SOM dynamics remain poorly characterized (13). Microbial contributions to soil C cycling are typically determined in the aggregate (e.g., soil respiration) without providing mechanistic insight into processes of individual microbes. Determining the characteristics of individual microbes typically requires laboratory cultivation, but most soil microbes remain uncultivated and poorly characterized (21–23). In the absence of cultivation, the characteristics of uncultivated microbes are often inferred by assuming phylogenetic conservation with cultivated representatives (24–26). However, phylogenetic conservation varies dramatically among microbial traits and functionalities (27), and many soil microbes lack closely related cultivated isolates (28). Microbial characteristics are also often inferred through metagenomic analyses that indicate the frequency of specific gene families and metabolic pathways in the environment (29, 30).
Inferences based on phylogenetic conservatism and metagenomic analyses predict “potential metabolism,” but this information cannot currently predict the ecological interactions that govern realized activity in the environment. Microbe–microbe and microbe–environment interactions alter patterns of gene expression, activity, and growth, and so potential metabolism tells us little of the actual microbial processes that drive soil C cycling at any particular place and time. The “realized activity” of a microbe in situ depends on a complex pattern of biotic and abiotic interactions. Therefore, realized activity in the C cycle might correlate only weakly with the metabolic potential of the community as inferred from phylogenetic conservation and metagenomic analyses (31, 32).
DNA-stable isotope probing (DNA-SIP) can assess the in situ activity of microorganisms within complex habitats based on patterns of C assimilation by identifying taxa that assimilate 13C from labeled substrates (33). DNA-SIP can track the assimilation of C from multiple substrates into thousands of microbial taxa in soil (34–36) with high specificity and sensitivity (36). We used multiple-window high-resolution DNA-SIP (MW-HR-SIP) to track 13C from nine distinct C sources through the soil bacterial food web over a period of 48 d (Fig. 1). These nine sources of C were selected because they varied in bioavailability and are common in soils. We initially defined bioavailability on the basis of solubility and hydrophobicity, which predicts sorption to particulate organic matter (37) and availability for transport across the cell membrane. The C sources we used were cellulose (a component of plant cell walls), glucose and xylose (monomers of cellulose and hemicellulose respectively), vanillin (a phenolic acid derived from lignin), glycerol and palmitic acid (components of plant lipids), an amino acid mixture (monomers of protein), and lactate and oxalate (major fermentation products from soil bacteria and fungi).
Fig. 1.
This experiment employed soil microcosms, each amended with all nine C sources (added at 0.4 mg C per gram of soil), only one of which was >99% 13C-labeled in treatment microcosms. Control microcosms had all nine C sources added but none were isotopically labeled. Microcosms were destructively sampled at multiple time points (black bars) based on mineralization rates from preliminary experiment. Headspace samples were taken every 1 to 7 d. DNA extracted from microcosm soil was used both for DNA-SIP and whole bacterial community sequencing (unfractionated).
Our multisubstrate design, where only the identity of the 13C substrate was varied (Fig. 1), allowed us to systematically investigate C assimilation dynamics as a function of C source while maintaining identical conditions across samples. In addition, by sampling over time we focused on the ultimate fate of C within the bacterial community and not just the bacteria responsible for initial substrate assimilation. That is, our goal was not to differentiate primary assimilation from later stages but rather to map bacterial guild structure by examining how C from diverse sources infiltrates the community over time. Guilds are groups of organisms that access resources in a similar manner (38). We used C assimilation dynamics to infer guild structure based on the source and timing of the 13C assimilated by bacterial taxa, such that bacteria within a guild exhibit similar patterns of C assimilation over time. We predicted that variation in C assimilation dynamics across bacterial taxa would be poorly explained by phylogenetic relatedness but that bacteria within guilds would have similar growth dynamics linked to shared life history strategies.
Results and Discussion
C Source Bioavailability Explains C Mineralization Rates.
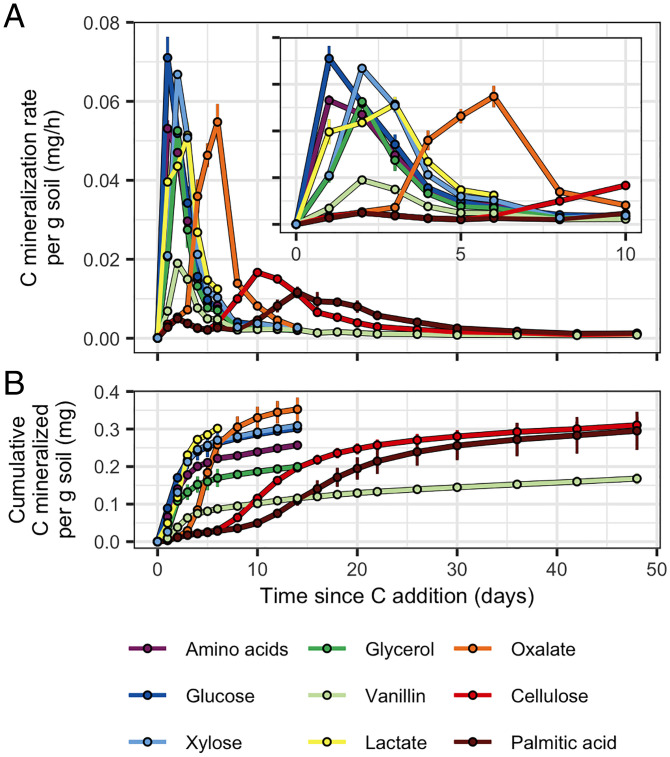
We tracked C mineralization dynamics from each C source for up to 48 d by measuring 13CO2 production. Peak 13C mineralization rates occurred on day 1 for glucose and amino acids; day 2 for xylose, glycerol, and vanillin; day 3 for lactate; day 7 for oxalate; day 10 for cellulose; and day 14 for palmitic acid (Fig. 2A, SI Appendix, Fig. S1). Cumulative 13C mineralization varied substantially between sources, being lowest for vanillin (42% C mineralized) and highest for oxalate (88% C mineralized; Fig. 2B and SI Appendix, Fig. S2). As expected, total C mineralization (13C + 12C) was similar across all treatments (SI Appendix, Fig. S1) since all microcosms received the same set of substrates with only the identity of the 13C-labeled substrate varied. These mineralization dynamics were confirmed in an independent experiment with soils from the same site (SI Appendix, Fig. S2).
Fig. 2.
C source mineralization dynamics varied based on bioavailability. (A) 13C mineralization rates and (B) cumulative amounts of 13C mineralized per gram dry weight of soil. (A, Inset) A finer-scale representation of the 13C mineralization rates over days 0 to 10. Error bars represent ± SD among microcosm replicates (n = 3).
Substrate bioavailability predicts degradation rate in soil (39), and so we selected C sources that differed in their solubility and hydrophobicity [as defined by octanol–water partition coefficients predicted using the XLogP3 model (40)] as described in SI Appendix, Supplemental Materials and Methods and indicated in Fig. 1. We found a significant and strong positive correlation between LogP and the day of peak C mineralization across C sources (Pearson’s r = 0.932, P = 0.001; SI Appendix, Fig. S3A) and a significant negative correlation between LogP and the maximum C mineralization rate (Pearson’s r = −0.855, P = 0.007; SI Appendix, Fig. S3B). These findings suggest that C sources with low bioavailability, such as cellulose (an insoluble polymer) and palmitic acid (a waxy fatty acid), are degraded slowly, while those with high bioavailability, such as glucose, xylose, amino acids, and lactate are degraded rapidly. Vanillin (an aromatic molecule) had intermediate bioavailability, allowing it to be mineralized rapidly; however, its hydrophobicity favors adsorption to organic surfaces in soil (41, 42). Hence, while vanillin in the aqueous phase degrades rapidly, its sorption to organic matter may limit microbial access over time. Highly oxidized compounds such as oxalate favor allocation of C to catabolic processes, resulting in greater cumulative mineralization (43, 44). Given that LogP cannot be determined for cellulose (because of its insolubility), and that there was a strong relationship between LogP and day of peak mineralization (SI Appendix, Fig. S3A), we subsequently applied an operational definition of bioavailability based on the observed day of peak mineralization. This approach allowed quantification of bioavailability for both soluble and insoluble substrates.
Uncultivated Bacteria Are Major Players in Soil C Cycling.
We assessed the temporal dynamics of bacterial C assimilation by performing MW-HR-SIP for each of the nine 13C-labeled C sources over 48 d. Sampling times were selected based on the 13C mineralization dynamics for each substrate. A total of 12,394 unique bacterial operational taxonomic units (OTUs) were observed and 6,613 of these passed independent filtering on the basis of sparsity. From this set, MW-HR-SIP identified 1,286 “incorporators” (i.e., OTUs with significant evidence of 13C incorporation into DNA; Fig. 3 and Dataset S1). The vast majority of these incorporators (86.4%) represented uncultivated taxa, and only 13.6% matched cultivated strains (i.e., > 97% 16S ribosomal RNA (rRNA) gene sequence identity to cultivated isolates). In addition, 26.8% of the incorporators had less than 90% 16S rRNA gene sequence identity to any cultivated strains. C from low and intermediate bioavailability sources (i.e., cellulose, palmitic acid, and vanillin) was assimilated primarily by uncultivated OTUs (SI Appendix, Fig. S4). In contrast, C from high bioavailability sources (i.e., glucose, xylose, amino acids, glycerol, oxalate, and lactate) was assimilated primarily by OTUs matching known isolates (SI Appendix, Fig. S4). Microbes that access C from low bioavailability sources are likely to have traits (e.g., slow growth, metabolic dependency, and surface attachment) that diminish their tolerance for laboratory growth media and may limit their representation in culture collections (45, 46).
Fig. 3.
The dynamics of C assimilation varied across the 1,286 OTUs that exhibited significant 13C labeling. OTUs are ordered by their phylogeny with phylum, or class for Proteobacteria, indicated by different colors in the phylogenetic tree. Only phyla/classes with 10 or more 13C-labeled OTUs are colored, with others colored gray. Each vertical block indicates 13C-labeling results for a single day as specified at the top. Each column within a block indicates results for a given substrate as specified along the bottom. Rows display 13C-labeling dynamics for each OTU as follows: red bars indicate 13C-labeling, light gray bars indicate OTU detection in the gradient but no evidence of labeling, white indicates the OTU was not detected in the gradient. Phylum/class abbreviations: Bact. = Bacteroidetes, Acido. = Acidobacteria, Gemm. = Gemmatimonadetes, Chl-flx. = Chloroflexi, Firmi. = Firmicutes, Actino. = Actinobacteria, Verruc. = Verrucomicrobia, Plancto. = Planctomycetes, Beta. = Betaproteobacteria, Gamma. = Gammaproteobacteria, Delta. = Deltaproteobacteria, Alpha. = Alphaproteobacteria.
We also examined unfractionated DNA (i.e., DNA extracted directly from soil and not subjected to MW-HR-SIP) in order to evaluate overall change of microbial community composition over time. The majority of incorporators were also detected in unfractionated DNA (1,153, or about 90%), though 133 incorporators (∼10%) were not observed in unfractionated DNA. This result indicates that rare OTUs, not readily detected in 16S rRNA gene surveys, are active participants in soil C cycling. Density gradient centrifugation fractionates DNA by buoyant density with sequencing performed across many gradient fractions, and this approach translates into far greater sequencing depth than is typical in microbial community analyses, allowing for the detection of rare taxa not readily observed when sequencing unfractionated soil DNA.
Phylogenetic Conservation of C Assimilation Dynamics.
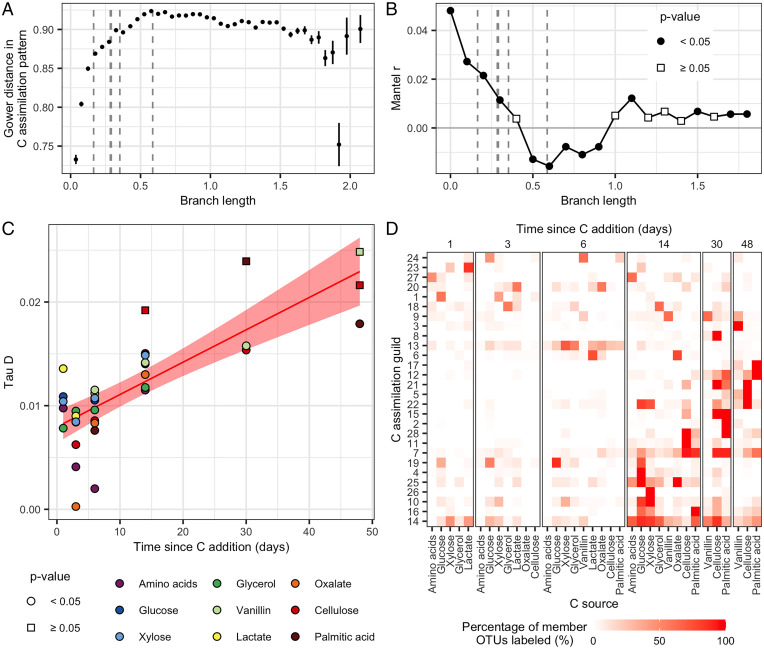
The ecophysiology of bacterial OTUs is often inferred from their phylogenetic relatedness (47–50). We therefore sought to determine if C assimilation activity could be predicted by phylogenetic similarity. We used two methods to test for the phylogenetic conservation in C assimilation dynamics. First, we calculated dissimilarity in assimilation dynamics between OTUs. Dissimilarity in C assimilation was calculated as Gower distance, which has values between 0 (identity) and 1 (complete dissimilarity). Phylogenetic signal is strong when closely related taxa have highly similar assimilation patterns (Gower distance = 0), absent when closely related taxa have completely dissimilar assimilation patterns (Gower distance = 1), and weak when closely related taxa have highly dissimilar assimilation patterns (e.g., >0.7). Patterns of C assimilation were weakly conserved among closely related taxa and dissimilarity increased rapidly with phylogenetic distance (Fig. 4 A and B). Most variation was explained at the species level with minimal phylogenetic signal present at taxonomic ranks above the genus level. Even when analyzing C assimilation dynamics independently for each C source, phylogeny was a weak predictor of realized activity (SI Appendix, Fig. S5). Second, we used consenTRAIT (49) to measure the phylogenetic depth (i.e., 16S rRNA gene evolutionary distance; τD) at which patterns of C assimilation were conserved. We found τD values ranging from ∼0.001 to 0.025 across the nine C sources (Fig. 4C), consistent with τD values of C substrate utilization measured by Martiny et al. (49). Phylogenetic depth varied with respect to C source, being highest for taxa that assimilated C from vanillin, cellulose, and palmitic acid, though this phylogenetic clustering was not statistically significant when assessed across all time points (consenTRAIT; cellulose day 14, P = 0.153; cellulose day 48, P = 0.647; palmitic acid day 30, P = 0.906; vanillin day 48, P = 0.940; P values adjusted for multiple comparisons; Fig. 4C). However, we also observed significant correlation between τD and time of C assimilation (Pearson’s r = 0.791, P < 0.001; Fig. 4C). For example, phylogenetic clustering of taxa that accessed C from cellulose, palmitic acid, and vanillin was higher for days 14 to 48 than it was for days 3 to 6 (Fig. 4C).
Fig. 4.
(A) The Gower’s distance in C assimilation dynamics between OTUs increases rapidly in relation to their phylogenetic distance (branch length). The vertical dashed lines indicate distance values that correspond to estimates of taxonomic ranks for genus, family, order, class, and phylum (from left to right). OTU pairs are grouped into phylogenetic distance bins with error bars indicating ± SE. (B) There is a weak correlation between Gower’s distance in C assimilation dynamics and phylogenetic distance (branch length) between OTUs, with more variance explained among closely related OTUs (Mantel test, points colored by significance of correlation, note very low Mantel r). (C) Phylogenetic depth (Tau D, calculated with consenTRAIT) of incorporators with respect to C source and time of labeling. All P values are globally adjusted with the Benjamini–Hochberg correction. There is a positive correlation between Tau D and time (Pearson’s r = 0.79, P < 0.001). This linear relationship is represented by the red line with the ribbon representing the SE. (D) OTUs were grouped into 28 guilds on the basis of their 13C-labeling patterns. Color intensity indicates the percentage of member OTUs who have detectable 13C labeling from each substrate on each day.
These results suggest a link between the bioavailability of C sources and the phylogenetic conservation of assimilation dynamics. For example, the assimilation of C from low bioavailability sources (such as cellulose or palmitic acid) requires metabolic or ecological traits that are more deeply conserved than those required to assimilate C from high bioavailability sources (such as glucose, xylose, or amino acids). However, the results also showed that the phylogenetic conservation of traits that governed access to soil C were highly variable. Even within a single substrate such as cellulose, the degree of phylogenetic clustering depended on time of assimilation. The fact that the phylogenetic clustering of incorporators varied depending on the time of assimilation suggests that patterns of C assimilation were not produced solely by the presence or absence of specific catabolic pathways. Rather, we expect that shared ecological characteristics (e.g., growth dynamics, predation, cross-feeding, motility, attachment, secondary metabolite production, etc.) explain considerable variation in access to soil C. As a consequence, we expect traits that govern access to soil C to exhibit considerable variation in their phylogenetic conservation (27).
C Assimilation Dynamics Define Bacterial Guild Structure.
Since phylogenetic relatedness was a weak and inconsistent predictor of C assimilation dynamics in soil, we sought an alternative means to group bacteria into ecologically relevant clusters. We used unsupervised clustering to group incorporators into operationally defined guilds based on their 13C-labeling dynamics. This approach generated 28 guilds (Fig. 4D). Each guild contained between 15 and 122 incorporators and represented 3 to 13 different phyla (Dataset S1). We then examined the growth and 13C assimilation dynamics of guilds and related these characteristics with rRNA operon (rrn) copy number. The rrn copy number of bacteria grown in culture correlates with maximal growth rate as well as C use efficiency and has been proposed as a component of microbial life-history strategies (51, 52). In all cases, values were averaged across all OTU within a guild (Dataset S2).
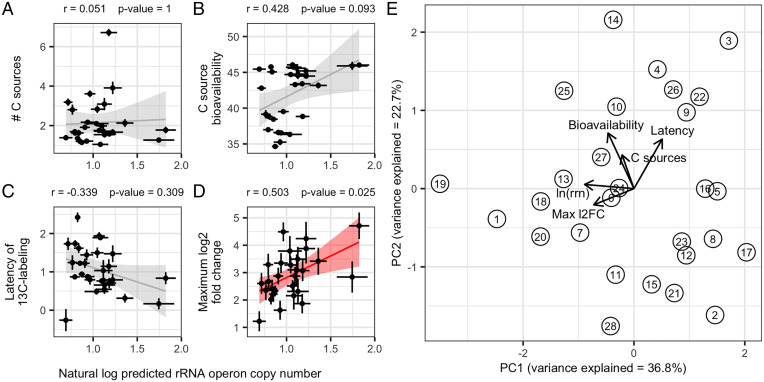
After adjusting for multiple comparisons (n = 4), we found a positive correlation only between rrn copy number and the maximum log2 fold change in normalized abundance (max L2FC; Pearson’s r = 0.503, P = 0.025; Fig. 5D). Max L2FC was defined as the maximum change in differential abundance before and after C sources were added to soil. High-copy-number guilds exhibited large changes in normalized abundance in response to C input, while low-copy-number guilds varied much less over time. In other words, guilds with high copy number exhibited highly dynamic populations while guilds with low copy number were less dynamic. Since sequencing is compositional (53) and since several of the bacterial groups with the largest growth response had high copy number (Dataset S1), relative abundance was normalized both by predicted rrn copy number and DNA yield for each sample. Our observation links the rrn copy number of guilds to their in situ population dynamics in response to C addition. Two guilds (guilds 1 and 19) had considerably higher rrn copy numbers than the other guilds, and these two guilds could be driving much of the relationship between rrn copy number and max L2FC. When these two guilds were removed from the analysis the relationship with maximum log2 fold change remained marginally significant (Pearson’s r = 0.458, P = 0.074; SI Appendix, Fig. S6).
Fig. 5.
We tested whether the average rrn copy number of guilds was correlated with other guild characteristics. (A) The average number of C sources from which C was assimilated by each guild. (B) The average bioavailability of the sources from which C was assimilated by each guild. Bioavailability is defined here as a unitless value determined operationally based on mineralization dynamics for each substrate, with higher values for more rapidly mineralized substrates. (C) The average latency of C assimilation for each guild. Latency is defined operationally and it indicates the time delay between peak 13C mineralization and the time of 13C assimilation into DNA for each C source. (D) The dynamic growth response (max L2FC) of each guild as measured by increase in normalized abundance in response to C addition. The values provided are for Pearson’s correlation with lines indicating linear regression and shading indicating SE (significant results are in red). All P values were corrected for multiple comparisons with Benjamini–Hochberg procedure (n = 4). Error bars for points indicate ± SE across OTUs within each guild. (E) PCA differentiates guild structure on the basis of rrn copy number [ln(rrn)], C source bioavailability, latency of C assimilation, dynamic growth response (Max l2FC), and number of C sources (C sources). Circles represent guilds with numbers indicating guild identity and arrows represent loadings of each variable onto the principal component axes (PC1 and PC2).
We did not find any correlation between rrn copy number of guilds and the number of sources from which they derived their C (Pearson’s r = 0.051, P = 0.797; Fig. 5A), the average bioavailability of these C sources (Pearson’s r = 0.428, P = 0.093; Fig. 5B), or the “latency” of 13C labeling (Pearson’s r = −0.339, P = 0.077; Fig. 5C). We defined the latency of an OTU as the delay between the time of peak 13C mineralization and the time that 13C labeling was first detected. A lower value for latency indicates that C assimilation took place when C mineralization was maximal, and hence it is more likely that assimilation is coupled closely to the process of mineralization. A higher value for latency indicates that C assimilation took place long after rates of C mineralization began to decline. Hence, a high latency value makes it more likely that an organism was assimilating C that had been transformed by prior microbial processing.
To evaluate the ecological relevance of the observed guild structure, we examined how the phylogenetic structure of guilds varied with respect to their growth and 13C-assimilation dynamics. Variation in selection pressure should alter the phylogenetic structure of guilds, producing phylogenetic clustering when selection pressure is low and overdispersion when selection pressure is high (54). We predicted that competitive pressure should vary with C source bioavailability such that competition is greatest for less bioavailable C sources. This expectation is based on the assumption that competition is maximal when resources are limiting and that bacterial access to resources depends upon diffusive transport in the aqueous phase, which in turn is affected by resource bioavailability. We determined the degree of phylogenetic clustering by using the nearest taxon index (NTI). After adjusting for multiple comparisons (n = 4), we found positive correlations between guild NTI and both C source bioavailability (Pearson’s r = 0.64, P = 0.001; SI Appendix, Fig. S7B) and max L2FC (Pearson’s r = 0.51, P = 0.022; SI Appendix, Fig. S7D). No correlation was observed between guild NTI and either the number of C sources (Pearson’s r = 0.453, P = 0.062; SI Appendix, Fig. S7A) or C assimilation latency (Pearson’s r = −0.226, P = 0.994; SI Appendix, Fig. S7C).
These results indicate that competitive interactions within guilds are linked to the bioavailability of source C and the growth dynamics of guild members (as measured by max L2FC). More specifically, phylogenetic overdispersion, indicating that guilds were structured by competitive interactions, was found in guilds that assimilated C more slowly, changed little in normalized abundance, and accessed C from sources of low bioavailability. In contrast, phylogenetic clustering, indicating that guilds were structured by environmental factors (which in this experiment is driven by the sudden overabundance of a suitable resource), was found in guilds that assimilated C rapidly, changed dramatically in normalized relative abundance, and accessed C from sources of high bioavailabilty.
While rrn copy number is one trait that can inform a guild’s ecology and relationship to SOC, traits often impose trade-offs, causing interactions that can confound simple correlations. We therefore used a principal component analysis (PCA) to explore variation in guild responses as a function of their growth and assimilation dynamics (Fig. 5E). The first principal component (PC1) explained 36.8% of variation in guild responses and this axis corresponded with variation in rrn copy number (43.1% loading), growth dynamics (max L2FC; 28.5% loading), latency (14.1% loading), C source bioavailability (11.9% loading), and number of C sources (2.48% loading). The second principal component (PC2) explained 22.7% of the variation in guild responses. This second axis largely corresponded with variation in C source bioavailability (44.5% loading), latency (35.2% loading), number of C sources (16.3% loading), growth dynamics (max L2FC; 3.8% loading), and rrn copy number (0.3% loading).
The overall guild structure suggests three extremes in guild characteristics, with certain characteristics maximized at each vertex (Fig. 5E). For example, guilds with the highest rrn copy number and the highest max L2FC (i.e., the most dynamic growth response) had low PC1 but intermediate PC2. Guilds assimilating C from the least bioavailable and fewest C sources had high PC1 and low PC2. Guilds with the highest latency in C assimilation and greatest diversity of C assimilation patterns (i.e., those that access C from the most C sources) had high PC1 and high PC2. These vertices represent the characteristic extremes of the bacterial guild structure, though many intermediate guilds exist.
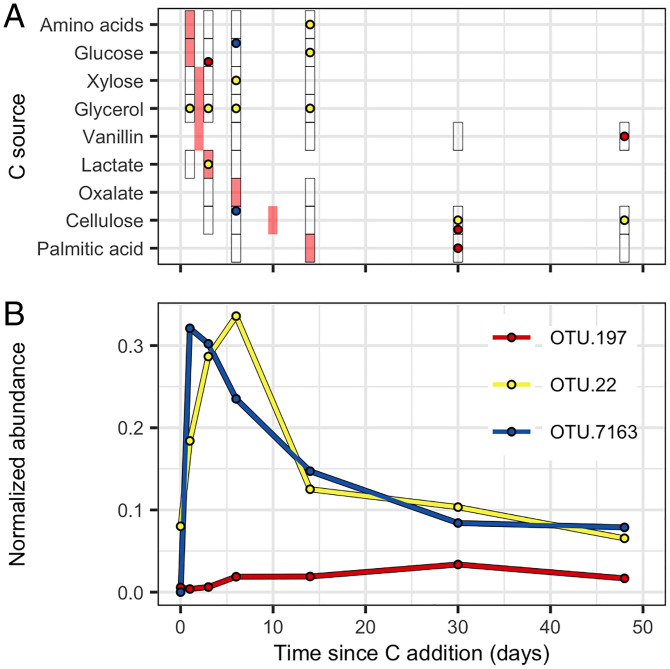
We selected the most abundant OTUs (based on the summed normalized abundance across time) that represented these three vertices to illustrate the maximal differences between guilds. The most abundant OTU from guild 19 was OTU.7163, classified as a Pseudomonas species. This pseudomonad was estimated to have 5 rrn operons, it had very low initial abundance, increased rapidly after C input reaching maximal abundance within 1 d followed by a dramatic decline over time, and it assimilated C at early time points (Fig. 6). Correspondingly, guild 19 had a high max L2FC, intermediate C source bioavailability and low latency (Fig. 5). The most abundant OTU from guild 14 was OTU.22, classified as an Agromyces species. This microbacterium was estimated to have 1 rrn operon, it had high initial abundance, increased more slowly after C input reaching maximal abundance after 6 d followed by a rapid decline over time, and it was consistently 13C-labeled throughout the experiment assimilating C from diverse sources (Fig. 6). Correspondingly, guild 14 had an intermediate max L2FC, many C sources, and high latency (Fig. 5). The most abundant OTU from guild 2 was OTU.197, classified as a species of Verrucomicrobia. This verrucomicrobium was estimated to have 2 rrn operons, it increased in abundance very slowly reaching maximal abundance after 30 d, remaining at relatively low abundance throughout, and it assimilated C from glucose, vanillin, and cellulose at mostly later time points (Fig. 6). Correspondingly, guild 2 had a low max L2FC, low C source bioavailability and low latency (Fig. 5). Generalized growth responses for all guilds are provided in SI Appendix, Fig. S8. It is clear from Figs. 5 and 6 and SI Appendix, Fig. S8 that guilds differ in their growth dynamics (max L2FC), and access to C (latency, C sources, and bioavailability), and these differences suggest trade-offs between growth dynamics and the ability to access C in the soil environment over time.
Fig. 6.
Growth and 13C-labeling dynamics of the most abundant OTUs representing guilds 2 (OTU.197), 14 (OTU.22), and 19 (OTU.7163), which are the guilds most differentiated by PCA (Fig. 5E). (A) The 13C-labeling pattern differs between the three exemplar OTUs. Circles show times when 13C labeling was detected for each substrate and OTU responses are identified by circle color as defined in B. Black rectangles indicate the times when DNA-SIP was performed for each 13C-labeled substrate and red rectangles indicate the time of peak C mineralization for each C source (Fig. 2). (B) Growth dynamics differ between the three exemplar OTUs. Normalized abundance (expressed as micrograms of DNA) is calculated from relative abundance values normalized by rrn copy number and DNA yield.
The guild structure we identified can be interpreted in the context of life-history theory, specifically Grime’s C-S-R framework (55). While Grime’s framework was conceived to describe plant life-history strategies, it has recently been applied to microbial systems (56–58). Ruderal (R) strategists of the C-S-R framework have little investment in resource acquisition, being adapted for rapid growth in response to a sudden influx of nutrients. From our guild structure, ruderal bacteria are exemplified by the pseudomonads and bacilli of guild 19. These taxa had high rrn copy number, highly dynamic growth response (max L2FC), and low latency, and 13C labeling tends to occur immediately following nutrient addition, all together suggesting the ability to grow rapidly as a strategy for exploiting episodic nutrient pulses. Competitive (C) strategists are adapted for resource acquisition, likely allocating energy to features such as mycelia, biofilms, extracellular enzymes, siderophores, and antimicrobials that require substantial investment of energy but increase fitness under conditions defined by prolonged competition for resources. Thus, competitors should grow more slowly (i.e., lower maximum growth rate) with less-dynamic swings in population size. In our guild structure, competitors were exemplified by the Agromyces from guild 14. These taxa had higher latency suggesting the ability to access 13C following metabolic processing by other microbes, and they accessed C from a greater diversity of sources and over longer periods of time than ruderals. Both ruderals and competitors can be considered copiotrophs because of their ability to quickly respond to nutrient inputs (56), but these strategies would manifest differently in their population dynamics and sensitivity to nonequilibrium states (e.g., disturbance). Stress tolerators (S) were defined by Grime on the basis of abiotic stress (55), but we propose that for soil bacteria these organisms are adapted for life at low energy flux (i.e., conditions that would be stressful and result in starvation or dormancy for copiotrophs). We therefore propose that for bacteria in soil, stress tolerators are synonymous with oligotrophs. These stress-tolerant soil bacteria are adapted to access low-bioavailability C sources that would be less competitive resources for growth-adapted copiotrophic organisms. Thus, in our guild structure, stress tolerant (S) strategists were those exemplified by the Verrucomicrobia of guild 2. These taxa had low rrn copy number and very slow growth response but also low latency and a tendency to access C from low bioavailability sources. While we focus on guilds 19, 14, and 2 to illustrate the presence of trade-offs, Fig. 5 illustrates that there is a wide diversity of strategies for accessing C from soil, including intermediate strategies not well binned strictly as ruderals, competitors, or stress tolerators (Fig. 5 and SI Appendix, Fig. S8). Similarly, members from the same taxonomic group may display dissimilar strategies. For example, while some Pseudomonas species exemplify the ruderal strategy others can be found in guilds representing competitors or stress tolerators (Dataset S1), demonstrating the physiological diversity and versatility of this genus.
Finally, we emphasize that the ecological trade-offs that define fitness in soil are surely different from those that define fitness in other habitats (e.g., aquatic environments). To illustrate this idea, consider the copiotroph–oligotroph framework, which was first used to describe the life history strategies of aquatic bacteria. Aquatic oligotrophs thrive at low substrate concentrations where substrate flux across the membrane is low and this imposes selection for high metabolic efficiency (59–66). Aquatic oligotrophic habitats are typically characterized by substrates that have low concentration at steady state (67). Under such equilibrium conditions, oligotroph growth rates are governed by substrate affinity as described by Michaelis–Menton kinetics (59). Soils, however, represent a highly dynamic, nonequilibrium system in which substrate availability varies over time and space in response to nutrient inputs, water availability, and sorption/desorption processes (68). Hence, the characteristics of “soil oligotrophs” are likely to differ from those of “aquatic oligotrophs.” While both groups seek to optimize metabolic efficiency, the path to efficiency is different in soil and aquatic habitats.
Substrate bioavailability is likely a major determinant of microbial access to soil C. Insoluble substrates, and sorbed substrates, cannot cross the cytoplasmic membrane. Growth on these substrates is limited by the diffusion of enzymes and enzyme products between cells and their substrates. As a result, we predict that access to C from low bioavailability substrates favors oligotrophs in soil, because limitations on diffusion and transport should select for metabolic efficiency. Furthermore, diffusive fluxes in soil environments are favored by intimate contact between organism and substrate. Such close contacts persist in water films that coat soil aggregates and remain in place as soils dry and diffusion is limited by increasing tortuosity. Hence, we predict that oligotrophic activity persists over a wide range of moisture availability in soils and that such activity is mediated by diverse collections of interacting organisms. In addition, we predict that access to highly soluble substrates in soil favors copiotrophs with ruderal or competitive strategies. These substrates are ephemeral in soils outside of the rhizosphere, being transiently available at high concentration following wetting events or nutrient input. The pulsed nature of soluble organic matter in soil pore water should favor rapid utilization of the C when nutrients are transiently present in excess, because the benefits of high affinity should matter most when resources are present at low concentration and the benefits of high efficiency should matter most when resources are present at steady state.
High Turnover of Bacterial Biomass.
In terms of understanding microbial contributions to soil C cycling, much focus has been placed on primary microbial degradation of C derived from plants, and the degradation of plant biomass and exudates is typically considered in relation to microbial catabolism (24–26, 32, 42). However, recent evidence suggests that persistent SOM is largely composed from the products of microbial anabolic reactions (69). Hence, microbial turnover and mortality is likely of considerable importance in soil C cycling. We observed rapid growth in response to C inputs, CO2 flux peaked at 2 d, and soil DNA increased over 6 d (a 52% increase), but there was also evidence of substantial turnover and mortality. For example, soil DNA declined considerably from day 6 through day 48 (a 20% decrease), and the normalized abundance of many taxa decreased over time. Of the 13C-labeled taxa, 387 were observed at high frequency over time (detection in six of seven time points) in unfractionated soil DNA. Of these high-frequency OTUs only 59 (15%) were observed to have a maximum in normalized relative abundance at the end of the experiment (i.e., no evidence of mortality), while 221 (57%) decreased by more than 50% in normalized abundance from an early peak in abundance. Decreases in normalized abundance were often dramatic and rapid (Fig. 6), particularly for guilds having high max L2FC (Fig. 5). In addition, among the frequently observed 13C-labeled taxa, a total of 202 (52%) were 13C-labeled at an early time point but were unlabeled by that same source at later time points. Loss of 13C label can result from growth and isotope dilution over time, but when coupled to evidence of decreases in mineralization rate, decreases in DNA yield, and decreases in normalized abundance of most taxa over time, this result suggests mortality and turnover of 13C DNA. Finally, the high latency for many 13C-labeled taxa (Fig. 5) indicates that many taxa became 13C-labeled indirectly by consuming microbial products rather than by direct assimilation of added C. The operation of a viral shunt in soil (70) would explain well these growth and assimilation dynamics. It also seems likely that fungi, which are not measured here, contribute significantly to C mineralization dynamics, and hence bacteria with high latency are likely to be assimilating the products of both fungal and bacterial metabolism. Collectively these results argue that secondary processing, fueled by the products of microbial anabolic metabolism, comprises a major component of the C that cycles through soil communities. Based on our guild structure we would predict that much of this microbial biomass C ultimately flows through taxa having “competitive” life-history strategies (those with high latency in Fig. 5).
Guild Structure Explains Bacterial Biogeography.
To test for a linkage between guild structure and biogeography, we mapped incorporators to global bacterial surveys to determine whether guild membership explained variation in biogeography of soil bacteria. We employed RLQ analysis, a multivariate approach that finds correspondence between organism properties (R) and environmental parameters at various sites (Q; e.g., habitat or soil pH) based on organismal abundance within sites (L) (71, 72). We mapped incorporator OTUs and their guild membership to two 16S rRNA gene surveys of soil bacterial diversity spanning continental (QIITA study 619) and global scales (QIITA study 928) (73). Guild membership explained significant variation in microbial relative abundance across environmental parameters (SI Appendix, Table S1 and Fig. S9). For simplicity, guilds were divided into three groups based on C assimilation profile. Group D assimilated C primarily from dissolved substrates of high bioavailability (glucose, xylose, amino acids, glycerol, lactate, and oxalate). Group P assimilated C primarily from particulate substrates of low bioavailability (cellulose and palmitic acid). Group V assimilated C from vanillin; this group was treated separately as it was likely present in both aqueous and nonaqueous forms with transition between these two states changing over time (owing to its insolubility and tendency to sorb to SOM). Guilds were further grouped by time of labeling: early (E) and late (L). At continental scales, guilds that primarily used dissolved substrates at late time points were associated with forest soils and soils having high organic C and N (DL; SI Appendix, Fig. S9A). At global scales, these guilds were positively associated with latitude (DL; SI Appendix, Fig. S9B), likely as a result of the northern concentration of forest ecosystems and SOM at global scales. The guilds most strongly associated with tropical and subtropical broadleaf forests were those that assimilated C from particulate substrates early (PE; SI Appendix, Fig. S9B). The guilds that were most strongly associated with temperate grasslands, savannas, and shrubland were those that assimilated C from diverse substrates (DL, PL, and VL; SI Appendix, Fig. S9B). Guilds that used C from particulate substrates and vanillin at later timepoints were most closely associated with desert biomes (PL and VL; SI Appendix, Fig. S9A). These results show that patterns of microbial C assimilation can be linked to patterns of microbial biogeography and that guild structure has relevance across large biogeographical scales.
Conclusions
We conducted a multisubstrate DNA-SIP experiment to measure the in situ activities of bacteria in the soil C cycle. We show that degradation dynamics and patterns of bacterial C assimilation depended on the bioavailability of C inputs. In total, 1,286 bacteria were found to incorporate C derived from nine different sources, and most of these bacteria were distantly related to cultivated isolates. We found that C assimilation dynamics in soil had low phylogenetic conservation among bacteria. Instead, C assimilation dynamics allowed us to group bacteria into guilds whose properties were generally consistent with well-known life-history strategies. We also show that this guild structure can explain variation in bacterial biogeography. Our results demonstrate that life-history strategies are likely linked to C assimilation dynamics. SOM is largely derived from the products of anabolic microbial metabolism (14), and therefore microbial assimilation and turnover of C is a primary determinant of C fate in soil. Based on our results, we hypothesize that C fate in soil is not determined solely, or even primarily, by the metabolic pathways that enable its catalysis but rather by the life history traits that govern microbial growth and C acquisition in situ. As a result, life-history theory could be useful in predicting the C-cycle activity of microbial communities (18). While more research is necessary to better understand the genetic and ecological underpinnings of life history strategies in soil bacteria, our results add to the growing body of knowledge indicating the importance of life-history theory to bacterially mediated terrestrial C cycling.
Materials and Methods
Soil Microcosm Experiments.
Soil microcosms were designed and incubated with 13C-labeled and unlabeled substrates as previously described (35) except that C amendment consisted of nine substrates: cellulose, xylose, glucose, glycerol, vanillin, palmitic acid, amino acid mixture, lactate, and oxalate (SI Appendix, Table S2). For most substrates, octanol–water partition coefficient (LogP) was determined using XLogP3 through PubChem (40). Since amino acids were added as a mixture, this substrate LogP was averaged across the 20 common amino acids. LogP is not available for cellulose as it is insoluble. Each substrate was added at 0.4 mg C⋅g−1 soil, corresponding to a total addition of 3.6 mg C⋅g−1 soil. The soils had 1.22% total soil C (35, 74), and so the total C addition represents about 30% of native soil C. Only one of the nine substrates was 13C-labeled in each treatment and the remainder were unlabeled (Fig. 1). Soil was from Penn Yan, NY, with site and collection methods previously described (35, 74) and outlined in SI Appendix. 13C and 12C mineralization dynamics were determined based on concentrations of 12CO2 and 13CO2 in microcosm headspace measured over time as detailed in SI Appendix. Soil from microcosms were destructively sampled depending on substrate mineralization dynamics over days 1, 3, 6, 14, 30, and 48 after substrate addition.
DNA Extraction and Isopycnic Centrifugation.
DNA extraction and isopycnic centrifugation was conducted as previously described (35) and detailed in SI Appendix. In short, for each treatment, size-selected DNA (≥4 kb) (75) was centrifuged in a CsCl density gradient and 100-µL fractions were collected. An aliquot of DNA taken before isopycnic centrifugation was used for whole-microcosm bacterial community sequencing (i.e., unfractionated DNA).
16S rRNA Gene Amplification and Sequencing.
We amplified and sequenced the V4 region of the 16S rRNA gene as previously described (76) and detailed in SI Appendix using dual indexed primers (515f and 806r) developed by Kozich et al. (77). The 16S rRNA amplicon libraries were processed as described previously (35) and detailed in SI Appendix. OTUs were clustered at 97% sequence similarity. Raw sequencing reads can be accessed at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (accession no. PRJNA668741).
Identifying Incorporators.
OTUs that incorporated 13C into their DNA (i.e., incorporators) were identified with MW-HR-SIP (36) using development code for the HTSSIP R package (78). In short, we used DESeq2 (79) to identify OTUs significantly enriched in the “heavy” gradient fractions of the 13C-labeled treatments compared to the “heavy” gradient fractions in corresponding 12C control. Incorporators were defined as OTUs with a log2 fold enrichment greater than 0.25 and a Benjamini–Hochberg adjusted P value less than 0.1. We used the overlapping BD windows 1.70 to 1.73, 1.72 to 1.75, and 1.74 to 1.77 g/mL, which were shown via simulations to have the highest sensitivity across all tested scenarios while not losing specificity (36). Independent sparsity filtering was conducted for each BD window to minimize the number of comparisons performed.
The taxonomic novelty of incorporators was determined by aligning OTU representative sequences to the SILVA All-Species Living Tree project small subunit database version 123, representing all sequenced type strains curated at that time (80). Alignment was performed using nucleotide BLAST (81) with an expected value cutoff of 1E-20. Incorporators with no hits ≥97% sequence identity were considered to have no closely related isolates, while those with no hits ≥90% were considered to have no related isolates. Statistical analyses were conducted in R version 3.1.2 (82).
Analyses of Phylogenetic Conservation.
Fasttree v2.1.9 (83) was used to infer a maximum likelihood phylogeny from the 16S rRNA sequence alignment of representative OTUs. The phylogeny was rooted with Sulfolobus solfataricus DSM1616 (GenBank accession no. X90478) as the outgroup. Phylogenetic conservation of realized function was examined both by comparing functional distance with phylogenetic distance and testing for a phylogenetic signal with a modified version of consenTRAIT (49).
Analysis of Guild Structure.
Incorporators were grouped into guilds based on their 13C-labeling patterns as determined by log2 fold change in 13C-labeled treatment fractions versus corresponding unlabeled control fractions. Specifically, clustering was performed by 1) calculating pairwise Gower distances for each OTU pair, 2) hierarchically clustering (UPGMA agglomeration method) based on the resulting distance matrix, 3) generating a dendrogram of the hierarchical associations, and 4) detecting clusters in the dendrogram using the cutreeHybrid function in the DynamicTreeCut R package (84). The rrn copy number of each OTU was predicted as described previously (85). Briefly, pplacer v1.1 (86) was used to insert OTU representative sequences into a 16S rRNA gene reference phylogeny of bacteria with known rrn copy number. The resulting phylogeny was used by guppy v1.1 to predict rrn copy numbers of each OTU based on its relatedness to the reference taxa. The natural log of the copy number was used for all comparisons (51). NTI of guilds was calculated using the R package picante (87).
Four guild characteristics were calculated with respect to C assimilation and growth dynamics. All characteristics were calculated for individual OTUs and then averaged by guild. The number of C sources from which C was assimilated was simply the number of C sources from which an OTU was 13C-labeled at any time point. Since logP is not available for cellulose, C source bioavailability was defined operationally based on the day of maximal 13C mineralization rate for each substrate (as detailed in SI Appendix). The C source bioavailability was determined as the average of all sources from which 13C was assimilated. Latency of C assimilation was determined by the natural log of the ratio between the first day of 13C labeling and the day of peak C mineralization for each C source. This value was then averaged across all sources from which C was assimilated. The maximum log2 fold change in OTU abundance in the unfractionated DNA was calculated following abundance normalization to minimize bias due to compositional data. Normalization included two steps: normalizing for predicted rrn copy number and normalizing by sample DNA yield. OTU relative abundances were first divided by their predicted rRNA operon copy number, then by the estimate copy number for the entire community. Within each timepoint, normalized abundances were then multiplied by the average DNA yield across the replicate microcosms. The extracted DNA yield (micrograms of DNA per gram of dry weight soil) was quantified with the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific). We recognize that this calculation does not take into account inflation in DNA yield due to the growth of fungi and other organisms not accounted for in our study; however, we believe that these effects will be small compared to the effects of bacterial growth, particularly as we are simply comparing between bacterial OTUs within this study. Untreated bulk soils were used as our baseline abundance, with OTUs undetected in these soils assigned the lowest abundance measured. For each OTU, the maximum log2 fold change in abundance was calculated as the difference between this baseline and the maximum abundance that was recorded after C addition. If abundance at all time points was less than the baseline or undetected, the maximum abundance was assigned the baseline abundance, making the maximum log2 fold change = 0 for such OTUs. An OTU must be growing if 13C labeling is detected; hence, a decrease in abundance at times when taxa are labeled indicates either that the rate of mortality for the population exceeds the division rate (deaths > births), that labeling occurred prior to growth decline, or that normalization was not entirely successful at eliminating compositional sequencing bias.
Mapping Incorporators to Biogeography Datasets.
Detailed descriptions of the independent datasets and their analysis for this study are found in SI Appendix. In short, OTU count tables, representative 16S rRNA gene sequences, and metadata was downloaded from Qiita (88) for QIITA studies 619 (continental dataset) and 928 (global dataset). Incorporators were mapped to external dataset OTUs separately for each study with the mothur alignment tool (89) using the SILVA reference alignment as a template. To examine the biogeography of guilds at continental (QIITA 619) and global (QIITA 928) scales, guilds were grouped by assimilation dynamics (as described in Results and Discussion). Then, for each QIITA dataset, guild designations (R) of mapped OTUs were used in combination with their OTU count tables (L) and sample metadata (Q) for RLQ and forth corner analyses (72, 90) as detailed in SI Appendix.
Supplementary Material
Acknowledgments
We thank Chuck Pepe-Ranney, Ashley Campbell, Marquessa Henry, and Braulio Castillo, Anay Hindupur, and Jillian Waters for their help with experimental design and implementation. This material is based on work supported by the Department of Energy Office of Science, Office of Biological & Environmental Research Genomic Science Program under Awards DE-SC0004486 and DE-SC0010558.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115292118/-/DCSupplemental.
Data Availability
DNA sequence data have been deposited in NCBI Sequence Read Archive (BioProject accession no. PRJNA668741).
References
- 1.Jobbágy E. G., Jackson R. B., The vertical distribution of soil organic carbon and its relation to climate and vegetation. Ecol. Appl. 10, 423–436 (2000). [Google Scholar]
- 2.Brovkin V., et al. , Effect of anthropogenic land-use and land-cover changes on climate and land carbon storage in CMIP5 projections for the twenty-first century. J. Clim. 26, 6859–6881 (2013). [Google Scholar]
- 3.Scharlemann J. P. W., Tanner E. V. J., Hiederer R., Kapos V., Global soil carbon: Understanding and managing the largest terrestrial carbon pool. Carbon Manag. 5, 81–91 (2014). [Google Scholar]
- 4.Luo Z., Wang G., Wang E., Global subsoil organic carbon turnover times dominantly controlled by soil properties rather than climate. Nat. Commun. 10, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M. A., et al. , Managing uncertainty in soil carbon feedbacks to climate change. Nat. Clim. Chang. 6, 751–758 (2016). [Google Scholar]
- 6.Lal R., Soil carbon sequestration impacts on global climate change and food security. Science 304, 1623–1627 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Lal R., Soil health and carbon management. Food Energy Secur. 5, 212–222 (2016). [Google Scholar]
- 8.Reeves D. W., The role of soil organic matter in maintaining soil quality in continuous cropping systems. Soil Tillage Res. 43, 131–167 (1997). [Google Scholar]
- 9.Smith P., Soils as carbon sinks: The global context. Soil Use Manage. 20, 212–218 (2004). [Google Scholar]
- 10.Heimann M., Reichstein M., Terrestrial ecosystem carbon dynamics and climate feedbacks. Nature 451, 289–292 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Todd-Brown K. E. O., et al. , Changes in soil organic carbon storage predicted by Earth system models during the 21st century. Biogeosciences 11, 2341–2356 (2014). [Google Scholar]
- 12.Luo Y., Keenan T. F., Smith M., Predictability of the terrestrial carbon cycle. Glob. Change Biol. 21, 1737–1751 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Wieder W. R., et al. , Carbon cycle confidence and uncertainty: Exploring variation among soil biogeochemical models. Glob. Change Biol. 24, 1563–1579 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Schimel J. P., Schaeffer S. M., Microbial control over carbon cycling in soil. Front. Microbiol. 3, 10.3389/fmicb.2012.00348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Todd-Brown K. E. O., et al. , Causes of variation in soil carbon simulations from CMIP5 Earth system models and comparison with observations. Biogeosciences 10, 1717–1736 (2013). [Google Scholar]
- 16.Wieder W. R., et al. , Explicitly representing soil microbial processes in Earth system models. Global Biogeochem. Cycles 29, 1782–1800 (2015). [Google Scholar]
- 17.Wang G., Post W. M., Mayes M. A., Development of microbial-enzyme-mediated decomposition model parameters through steady-state and dynamic analyses. Ecol. Appl. 23, 255–272 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Wieder W. R., Grandy A. S., Kallenbach C. M., Bonan G. B., Integrating microbial physiology and physio-chemical principles in soils with the MIcrobial-MIneral Carbon Stabilization (MIMICS) model. Biogeosciences 11, 3899–3917 (2014). [Google Scholar]
- 19.Sulman B. N., Phillips R. P., Oishi A. C., Shevliakova E., Pacala S. W., Microbe-driven turnover offsets mineral-mediated storage of soil carbon under elevated CO2. Nat. Clim. Chang. 4, 1099–1102 (2014). [Google Scholar]
- 20.Wieder W. R., Bonan G. B., Allison S. D., Global soil carbon projections are improved by modelling microbial processes. Nat. Clim. Chang. 3, 909–912 (2013). [Google Scholar]
- 21.Hug L. A., et al. , A new view of the tree of life. Nat. Microbiol. 1, 1–6 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Lloyd K. G., Steen A. D., Ladau J., Yin J., Crosby L., Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems 3, e00055–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steen A. D., et al. , High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 13, 3126–3130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langille M. G. I., et al. , Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aßhauer K. P., Wemheuer B., Daniel R., Meinicke P., Tax4Fun: Predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31, 2882–2884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brbić M., et al. , The landscape of microbial phenotypic traits and associated genes. Nucleic Acids Res. 44, 10074–10090 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martiny J. B. H., Jones S. E., Lennon J. T., Martiny A. C., Microbiomes in light of traits: A phylogenetic perspective. Science 350, 10.1126/science.aac9323 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Choi J., et al. , Strategies to improve reference databases for soil microbiomes. ISME J. 11, 829–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fierer N., et al. , Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. U.S.A. 109, 21390–21395 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alneberg J., et al. , Genomes from uncultivated prokaryotes: A comparison of metagenome-assembled and single-amplified genomes. Microbiome 6, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wohl D. L., Arora S., Gladstone J. R., Functional redundancy supports biodiversity and ecosystem function in a closed and constant environment. Ecology 85, 1534–1540 (2004). [Google Scholar]
- 32.Evans R., et al. , Defining the functional traits that drive bacterial decomposer community productivity. ISME J. 11, 1680–1687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dumont M. G., Murrell J. C., Stable isotope probing—linking microbial identity to function. Nat. Rev. Microbiol. 3, 499–504 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Hungate B. A., et al. , Quantitative microbial ecology through stable isotope probing. Appl. Environ. Microbiol. 81, 7570–7581 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pepe-Ranney C., Campbell A. N., Koechli C. N., Berthrong S., Buckley D. H., Unearthing the ecology of soil microorganisms using a high resolution DNA-SIP approach to explore cellulose and xylose metabolism in soil. Front. Microbiol. 7, 10.3389/fmicb.2016.00703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youngblut N. D., Barnett S. E., Buckley D. H., SIPSim: A modeling toolkit to predict accuracy and aid design of DNA-SIP experiments. Front. Microbiol. 9, 10.3389/fmicb.2018.00570 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sijm D., Kraaij R., Belfroid A., Bioavailability in soil or sediment: Exposure of different organisms and approaches to study it. Environ. Pollut. 108, 113–119 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Simberloff D., Dayan T., The guild concept and the structure of ecological communities. Annu. Rev. Ecol. Syst. 22, 115–143 (1991). [Google Scholar]
- 39.Berg B., Hannus K., Popoff T., Theander O., Changes in organic chemical components of needle litter during decomposition. Long-term decomposition in a Scots pine forest. Can. J. Bot. 60, 1310–1319 (1982). [Google Scholar]
- 40.Cheng T., et al. , Computation of octanol-water partition coefficients by guiding an additive model with knowledge. J. Chem. Inf. Model. 47, 2140–2148 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Polubesova T., Chen Y., Navon R., Chefetz B., Interactions of hydrophobic fractions of dissolved organic matter with Fe(3+) - and Cu(2+)-montmorillonite. Environ. Sci. Technol. 42, 4797–4803 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Keiluweit M., Kleber M., Molecular-level interactions in soils and sediments: The role of aromatic π-systems. Environ. Sci. Technol. 43, 3421–3429 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Brant J. B., Sulzman E. W., Myrold D. D., Microbial community utilization of added carbon substrates in response to long-term carbon input manipulation. Soil Biol. Biochem. 38, 2219–2232 (2006). [Google Scholar]
- 44.Manzoni S., Taylor P., Richter A., Porporato A., Ågren G. I., Environmental and stoichiometric controls on microbial carbon-use efficiency in soils. New Phytol. 196, 79–91 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Alain K., Querellou J., Cultivating the uncultured: Limits, advances and future challenges. Extremophiles 13, 583–594 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Pham V. H. T., Kim J., Cultivation of unculturable soil bacteria. Trends Biotechnol. 30, 475–484 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Lennon J. T., Aanderud Z. T., Lehmkuhl B. K., Schoolmaster D. R. Jr., Mapping the niche space of soil microorganisms using taxonomy and traits. Ecology 93, 1867–1879 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Placella S. A., Brodie E. L., Firestone M. K., Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc. Natl. Acad. Sci. U.S.A. 109, 10931–10936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martiny A. C., Treseder K., Pusch G., Phylogenetic conservatism of functional traits in microorganisms. ISME J. 7, 830–838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dolan K. L., Peña J., Allison S. D., Martiny J. B. H., Phylogenetic conservation of substrate use specialization in leaf litter bacteria. PLoS One 12, e0174472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roller B. R., Schmidt T. M., The physiology and ecological implications of efficient growth. ISME J. 9, 1481–1487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fierer N., Bradford M. A., Jackson R. B., Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Gloor G. B., Macklaim J. M., Pawlowsky-Glahn V., Egozcue J. J., Microbiome datasets are compositional: And this is not optional. Front. Microbiol. 8, 2224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horner-Devine M. C., Bohannan B. J. M., Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87, S100–S108 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Grime J. P., Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Am. Nat. 111, 1169–1194 (1977). [Google Scholar]
- 56.Prosser J. I., et al. , The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5, 384–392 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Ho A., et al. , Conceptualizing functional traits and ecological characteristics of methane-oxidizing bacteria as life strategies. Environ. Microbiol. Rep. 5, 335–345 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Fierer N., Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Button D. K., Biochemical basis for whole-cell uptake kinetics: Specific affinity, oligotrophic capacity, and the meaning of the michaelis constant. Appl. Environ. Microbiol. 57, 2033–2038 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Button D. K., Nutrient-limited microbial growth kinetics: Overview and recent advances. Antonie van Leeuwenhoek 63, 225–235 (1993). [DOI] [PubMed] [Google Scholar]
- 61.Zobell C. E., Grant C. W., Bacterial utilization of low concentrations of organic matter. J. Bacteriol. 45, 555–564 (1943). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Postgate J. R., Hunter J. R., The survival of starved bacteria. J. Gen. Microbiol. 29, 233–263 (1962). [DOI] [PubMed] [Google Scholar]
- 63.Jannasch H. W., Growth of marine bacteria at limiting concentrations of organic carbon in seawater. Limnol. Oceanogr. 12, 264–271 (1967). [Google Scholar]
- 64.Kuznetsov S. I., Dubinina G. A., Lapteva N. A., Biology of oligotrophic bacteria. Annu. Rev. Microbiol. 33, 377–387 (1979). [DOI] [PubMed] [Google Scholar]
- 65.Akagi Y., Simidu U., Taga N., Growth responses of oligotrophic and heterotrophic marine bacteria in various substrate concentrations, and taxonomic studies on them. Can. J. Microbiol. 26, 800–806 (1980). [Google Scholar]
- 66.Button D. K., Nutrient uptake by microorganisms according to kinetic parameters from theory as related to cytoarchitecture. Microbiol. Mol. Biol. Rev. 62, 636–645 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arnosti C., Microbial extracellular enzymes and the marine carbon cycle. Annu. Rev. Mar. Sci. 3, 401–425 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Sollins P., Homann P., Caldwell B. A., Stabilization and destabilization of soil organic matter: Mechanisms and controls. Geoderma 74, 65–105 (1996). [Google Scholar]
- 69.Liang C., Amelung W., Lehmann J., Kästner M., Quantitative assessment of microbial necromass contribution to soil organic matter. Glob. Change Biol. 25, 3578–3590 (2019). [DOI] [PubMed] [Google Scholar]
- 70.Kuzyakov Y., Mason-Jones K., Viruses in soil: Nano-scale undead drivers of microbial life, biogeochemical turnover and ecosystem functions. Soil Biol. Biochem. 127, 305–317 (2018). [Google Scholar]
- 71.Dolédec S., Chessel D., ter Braak C. J. F., Champely S., Matching species traits to environmental variables: A new three-table ordination method. Environ. Ecol. Stat. 3, 143–166 (1996). [Google Scholar]
- 72.Dray S., et al. , Combining the fourth-corner and the RLQ methods for assessing trait responses to environmental variation. Ecology 95, 14–21 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Bates S. T., et al. , Examining the global distribution of dominant archaeal populations in soil. ISME J. 5, 908–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berthrong S. T., Buckley D. H., Drinkwater L. E., Agricultural management and labile carbon additions affect soil microbial community structure and interact with carbon and nitrogen cycling. Microb. Ecol. 66, 158–170 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Youngblut N. D., Buckley D. H., Intra-genomic variation in G + C content and its implications for DNA stable isotope probing. Environ. Microbiol. Rep. 6, 767–775 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Barnett S. E., Youngblut N. D., Buckley D. H., Soil characteristics and land-use drive bacterial community assembly patterns. FEMS Microbiol. Ecol. 96, fiz194 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., Schloss P. D., Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Youngblut N. D., Barnett S. E., Buckley D. H., HTSSIP: An R package for analysis of high throughput sequencing data from nucleic acid stable isotope probing (SIP) experiments. PLoS One 13, e0189616 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yarza P., et al. , The All-Species Living Tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31, 241–250 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 82.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2018). [Google Scholar]
- 83.Price M. N., Dehal P. S., Arkin A. P., FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langfelder P., Zhang B., Horvath S., Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics 24, 719–720 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Kembel S. W., Wu M., Eisen J. A., Green J. L., Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLOS Comput. Biol. 8, e1002743 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsen F. A., Kodner R. B., Armbrust E. V., pplacer: Linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics 11, 1–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kembel S. W., et al. , Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464 (2010). [DOI] [PubMed] [Google Scholar]
- 88.A. Gonzalezet al., . Qiita: rapid, web-enabled microbiome meta-analysis. Nat. Methods. 15, 796–798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schloss P. D., et al. , Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dray S., Chessel D., Thioulouse J., Co-inertia analysis and the linking of ecological data tables. Ecology 84, 3078–3089 (2003). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequence data have been deposited in NCBI Sequence Read Archive (BioProject accession no. PRJNA668741).