Significance
CRISPR-SID was established in the diploid frog Xenopus tropicalis for in vivo elucidation of cancer cell vulnerabilities. CRISPR-SID uses deep-learning predictions and binomial theory to identify genes under positive or negative selection during autochthonous tumor development. Using CRISPR-SID in a genetic model for desmoid tumors, treatment-recalcitrant mesenchymal tumors driven by hyperactivation of the Wnt signaling pathway, we identified EZH2 and SUZ12, both encoding critical components of the polycomb repressive complex 2, as dependency genes for desmoid tumors. Finally, we demonstrate the promise of EZH2 inhibition as a therapeutic strategy for desmoid tumors. With the simplicity of CRISPR sgRNA multiplexing in Xenopus embryos, the CRISPR-SID method may be applicable to reveal vulnerabilities in other tumor types.
Keywords: cancer dependency, Xenopus, desmoid tumor, APC, EZH2
Abstract
Cancer precision medicine implies identification of tumor-specific vulnerabilities associated with defined oncogenic pathways. Desmoid tumors are soft-tissue neoplasms strictly driven by Wnt signaling network hyperactivation. Despite this clearly defined genetic etiology and the strict and unique implication of the Wnt/β-catenin pathway, no specific molecular targets for these tumors have been identified. To address this caveat, we developed fast, efficient, and penetrant genetic Xenopus tropicalis desmoid tumor models to identify and characterize drug targets. We used multiplexed CRISPR/Cas9 genome editing in these models to simultaneously target a tumor suppressor gene (apc) and candidate dependency genes. Our methodology CRISPR/Cas9 selection–mediated identification of dependencies (CRISPR-SID) uses calculated deviations between experimentally observed gene editing outcomes and deep-learning–predicted double-strand break repair patterns to identify genes under negative selection during tumorigenesis. This revealed EZH2 and SUZ12, both encoding polycomb repressive complex 2 components, and the transcription factor CREB3L1 as genetic dependencies for desmoid tumors. In vivo EZH2 inhibition by Tazemetostat induced partial regression of established autochthonous tumors. In vitro models of patient desmoid tumor cells revealed a direct effect of Tazemetostat on Wnt pathway activity. CRISPR-SID represents a potent approach for in vivo mapping of tumor vulnerabilities and drug target identification.
The identification of novel cancer vulnerabilities is paramount to enable therapeutic discoveries and to improve patient prospects. Recent technological advances allow genetic dependency mapping across a wide array of cancer cell lines at an unprecedented speed (1). At the same time, the introduction of transcription activator-like effector nucleases (TALEN) and CRISPR has empowered straightforward genome engineering in a large number of animal models previously recalcitrant to genetic studies. A limiting experimental factor for mammalian organisms is the large-scale introduction of the genome-editing reagents in early embryos. However, for aquatic vertebrates such as fish and amphibians with external development and large-size embryos, simple microinjections allow fast, straightforward, and multiplexed introduction of these genome editing compounds. Over the past years, we harnessed TALENs and CRISPR/Cas9 in the true diploid model Xenopus tropicalis empowering this amphibian species as a cost-efficient platform for cancer biology (2–4). We further proposed the use of CRISPR/Cas9 to validate cancer cell vulnerabilities in vivo by inactivating suspected genetic dependencies in developing tumors (5). Clonal cancer growth is an inherently Darwinian process, where genetic mutations that increase fitness are under positive selection and mutations disfavoring fitness or growth are under negative selection pressure (6, 7). By monitoring the selection for specific patterns of CRISPR/Cas9 editing outcomes during autochthonous tumor development, we here establish an experimental pipeline for in vivo elucidation of cancer cell vulnerabilities. In brief, we use deep learning to forecast CRISPR/Cas9 gene editing outcomes and subsequently employ binomial statistics to ascertain deviations between expected and observed editing patterns as a measure of tumoral selection mechanisms, thereby identifying genetic vulnerabilities.
Desmoid tumors are orphan-type soft-tissue neoplasms driven strictly by Wnt signaling network hyperactivation (8, 9). Desmoid tumors can occur either sporadically and harbor activating missense mutations in the CTNNB1 gene or can be associated with germline loss-of-function mutations in the tumor suppressor gene APC (associated with familial adenomatous polyposis [FAP] or Gardner’s syndrome). Desmoid tumors do not metastasize and pose a low risk of death in sporadic disease, but they confer substantial clinical complications and, dependent on the location of the tumor, cause severe morbidity or lead to amputation of one of the extremities. However, in FAP patients, who routinely undergo colectomy, desmoid tumors are the leading cause of morbidity and mortality (10). Despite the clearly defined genetic etiology and the strict and unique implication of the Wnt/β-catenin pathway, no systemic specific molecular therapy for the tumors exists. A number of agents such as the tyrosine kinase inhibitor Sorafenib and the gamma-secretase inhibitor Nirogacestat have been shown to exert activity against desmoid tumors (11, 12). However, in both cases, the actual drug target and the underlying molecular mode of action in the desmoid tumor cells remain unknown (13).
Here, using multiplexed CRISPR/Cas9-based genome editing, we performed desmoid tumor genetic dependency mapping directly in vivo using X. tropicalis. We show genetic dependencies during autochthonous desmoid tumorigenesis and report preclinical data revealing Ezh2 as a druggable vulnerability in desmoid tumors.
Results
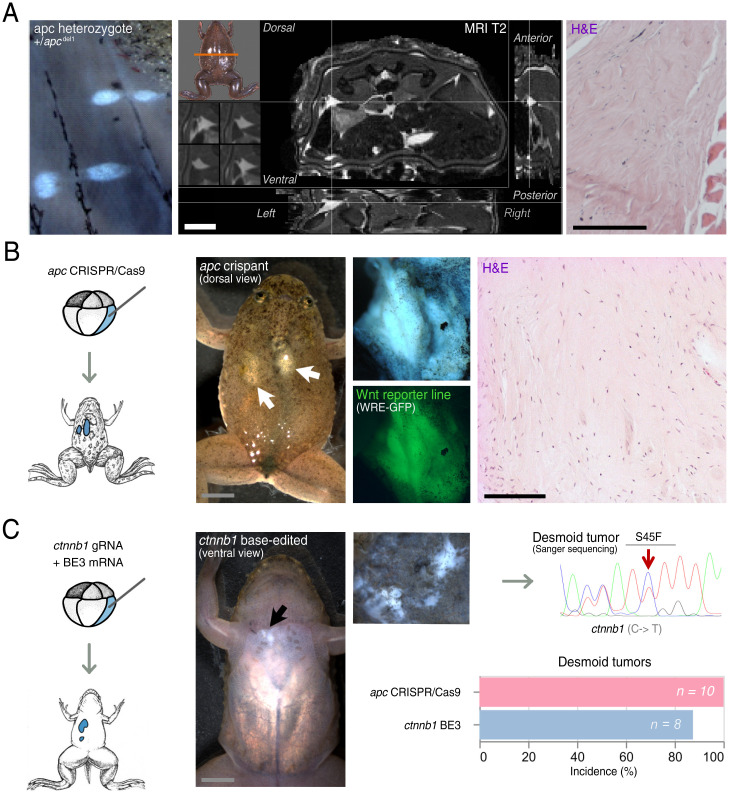
Wnt Signaling Network Modulation Initiates Desmoid Tumorigenesis in X. tropicalis.
Previously, we reported penetrant induction of desmoid tumors in F0 X. tropicalis by TALENs-mediated genome engineering of the tumor suppressor gene apc (4). From these, we generated a stable F1 X. tropicalis line containing a germline heterozygous 1–base pair deletion in the mutation cluster region (MCR) of the apc gene (c.4120del, apcMCR-Δ1/+) (Dataset S1A). In phenocopy to Gardner syndrome patients, apcMCR-Δ1/+ animals develop desmoid tumors, showing 100% penetrance (n = 7) when monitored at the age of 5 y (Fig. 1A). Desmoid tumors present on MRI as collagen-rich neoplasms, appreciable by T2-weighted hyperintensity, and exhibit conventional key histopathological hallmarks of long-sweeping fascicles composed of uniform and slender fibroblasts/myofibroblasts (Fig. 1A) (14). In the meantime, we similarly induced desmoid tumors by CRISPR/Cas9-mediated targeting of apc, which further facilitates downstream multiplexed gene inactivation studies. For this, we similarly targeted the MCR of apc with CRISPR/Cas9 and performed genome editing (Dataset S1B). Upon injection of 8-cell stage embryos carrying a Wnt/β-catenin reporter construct (15) in the two dorsal-vegetal blastomeres, which contribute primarily to the anterior endoderm and dorsal mesoderm, subsequent postmetamorphic X. tropicalis presented with fully penetrant development of multiple desmoid tumors (n = 10) at 3 mo of age (Fig. 1B). Epifluorescence exposed active Wnt signaling, and histopathology demonstrated the presence of all relevant conventional hallmarks of desmoid tumors (Fig. 1B). Deep PCR amplicon sequencing of a desmoid tumor revealed the presence of biallelic apc frameshifting mutations in the MCR, encoded by the final exon, with as a predicted consequence expression of a truncated Apc protein (Dataset S1C). Finally, desmoid tumors could also be induced by CRISPR-base editing (BE3) mediating a targeted C-to-T conversion in the ctnnb1 gene (Fig. 1C) (16, 17). As such, we modulate the conserved S45 phosphorylation site of β-catenin in X. tropicalis, recapitulating a clinically relevant human S45F missense mutation (Fig. 1C) (18). The S45 residue is normally phosphorylated by the casein kinase 1, which primes the β-catenin protein for subsequent phosphorylation by the glycogen synthase kinase 3-β in the destruction complex, and thereby earmarks it for ubiquitination and degradation by the proteasome. The stabilized S45F mutant β-catenin protein drove desmoid tumorigenesis in 87.5% of F0 animals (n = 8) via monoclonal expansion of C-to-T edited cells at 3 mo of age (Fig. 1C). Collectively, we describe three genetic models for desmoid tumors in X. tropicalis by modulating the Wnt signaling pathway, which can now be exploited for genetic dependency mapping and preclinical drug studies.
Fig. 1.
Hyperactivation of the Wnt/β-catenin signaling pathway via truncation of Apc or constitutive activation of β-catenin induces desmoid tumors in X. tropicalis. (A) X. tropicalis carrying a germline heterozygous 1–base pair deletion in the MCR of the apc gene (c.4120del, apcMCR-Δ1/+) develop desmoid tumors with a 100% penetrance at 3 y of age (Left). The desmoid tumors present as T2 hyperintense foci on MRI (Middle, intersection of the crosshairs) and are characterized by classical desmoid tumor histopathology of long sweeping fascicles composed of uniform and slender fibroblasts/myofibroblasts (Right). (B) apc CRISPR/Cas9 injections in the vegetal-dorsal blastomere leads to mosaic mutant animals that manifest several desmoid tumors. These desmoid tumors exhibit active and high Wnt signaling activity as demonstrated by GFP expression using a Wnt reporter line (WntREs:dEGFP) (15) (Middle). Classical histopathology of desmoid tumors with long sweeping fascicles of bland fibroblasts/myofibroblasts exhibiting, in this case, local invasion into musculature (Right). (C) Injection of a BE3 cytosine base editor with a ctnnb1 gRNA leads to development of desmoid tumors. Sanger sequencing of a dissected desmoid tumor demonstrates base pair edits culminating in amino acid changes corresponding the human S45F mutation in CTNNB1 associated with sporadic desmoid tumorigenesis. Penetrance of desmoid tumors by 3 mo of age is 100% for apc CRISPR/Cas9-injected animals and 87.5% for ctnnb1 base-edited animals (Scale bar in histology picture, 100 µm).
CRISPR-SID: Quantitative Assessments of Positive and Negative Selection Pressure on Gene Inactivation during Tumorigenesis.
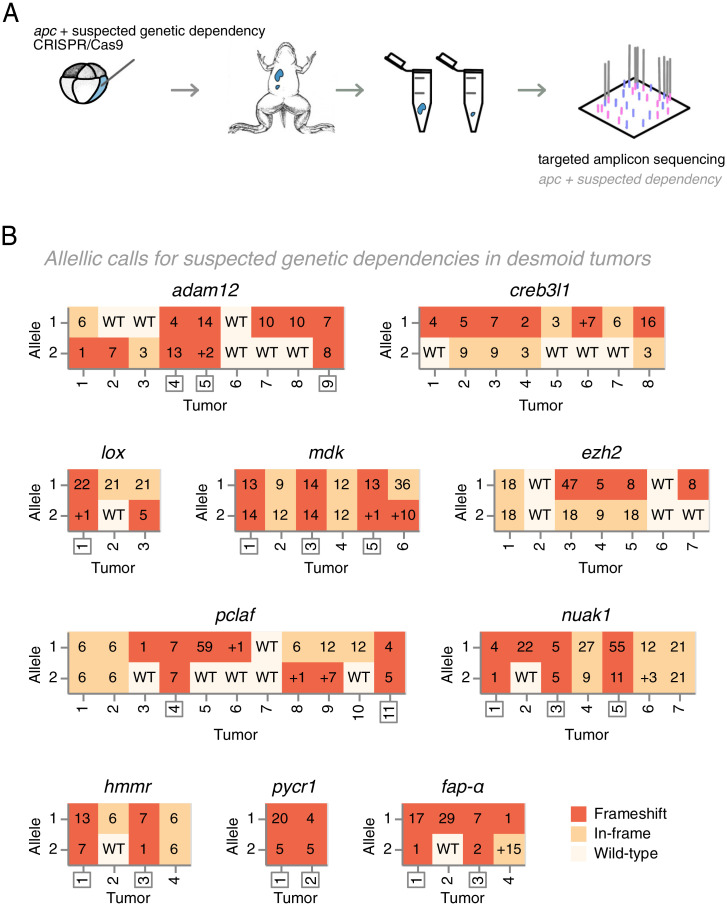
To further explore the use of our CRISPR/Cas9-mediated apc desmoid tumor model for genetic dependency mapping, we first selected suspected genetic dependencies. We reanalyzed available transcriptomics studies and identified genes which are consistently up-regulated in desmoid tumors compared with other fibrotic lesions (Dataset S2) (19, 20). In addition, we integrated protein druggability, via DGIdb (67), and mined published desmoid tumor literature (Dataset S3). As such, we generated a shortlist of 10 suspected genetic dependencies in desmoid tumors: hmmr, adam12, fap-α, mdk, nuak1, lox, pycr1, creb3l1, pclaf, and ezh2 (21–27). For these suspected dependencies, we validated high-efficiency guide RNAs (gRNAs) by injection in X. tropicalis embryos followed by deep amplicon sequencing of the targeted regions (Dataset S1D).
Next we developed a methodology, CRISPR-SID (CRISPR/Cas9 selection–mediated identification of dependencies), to ascertain selection pressure toward specific CRISPR/Cas9-induced mutational spectra enriched during autochthonous genetic tumor development. This selection pressure can manifest itself during repair of the CRISPR/Cas9-induced double strand break as positive, via selection for biallelic frameshift mutations in tumor suppressors, or as negative, via counterselection of biallelic frameshift mutations in genetic dependencies. This results from Darwinian natural selection mechanisms occurring in vivo within the mosaic CRISPR/Cas9-mutated Xenopus during tumor initiation and development. We wanted to quantitatively assess these selection mechanisms at play by comparing CRISPR/Cas9 editing outcomes in a previously reported machine-learning prediction algorithm and via deep amplicon sequencing of Xenopus tissues under different degrees of selection pressure.
We first deployed the deep learning algorithm InDelphi, which accurately predicts CRISPR/Cas9 editing outcomes. While InDelphi was trained in human cell lines and in mouse embryonic stem cells (28), we recently reported that the algorithm is also highly predictive for double strand break repair outcomes in Xenopus and zebrafish embryos (29). The model provides us with high-accuracy prediction of genome editing outcomes for our gRNAs in the absence of tumoral Darwinian selection mechanisms. We coinjected gRNAs targeting each suspected genetic dependency together with apc gRNA and Cas9 protein in blastomeres contributing to the anterior endoderm and dorsal mesoderm in X. tropicalis (Dataset S1E). Postmetamorphosis, each developing desmoid tumor was subjected to deep amplicon sequencing, and its mutational spectrum at each CRISPR/Cas9 target site was classified (Fig. 2 A and B) (Dataset S4). We next employed binomial statistics to ascertain whether the classification of the mutation spectrum occurring in desmoid tumors is binomially likely or unlikely given the InDelphi deep-learning predictions (SI Appendix, Extended Information). A significant deviation in the tumor mutation spectrum, compared with InDelphi model predictions, is indicative for the presence of Darwinian selection mechanisms that either favor or disfavor inactivation of specific genes during desmoid tumor development.
Fig. 2.
Assessing and classifying editing outcomes at CRISPR/Cas9 target sites of candidate dependency genes in apc mutant desmoid tumors. (A) X. tropicalis embryos are cotargeted at apc and, respectively, one suspected genetic dependency (e.g., adam12). Desmoid tumors were dissected from 3-mo-old postmetamorphic animals, and both CRISPR/Cas9 target sites were subjected to targeted amplicon sequencing to determine gene editing outcomes. (B) Indels and allelic status at the suspected dependency CRISPR/Cas9 target sites in dissected desmoid tumors. Frameshift mutations (FS) are indicated in dark-red, in-frame (IF) mutations in ochre, while wild-type allelic calls are indicated in off-white. Numbers in boxes show specific INDEL mutations, where Δn indicates a deletion of n nucleotides and +n indicates insertions of n nucleotides. For eight suspected genetic dependencies, desmoid tumors were retrieved with biallelic frameshift mutations (demarcated by boxed numbers). In contrast, for ezh2 and creb3l1, biallelic frameshift mutations were never retrieved.
CRISPR-SID Ascertains Positive Selection for Biallelic apc Frameshift Mutations in Desmoid Tumors.
Illustrating CRISPR-SID by example, classifying the mutational spectrum of apc in desmoid tumors, we observed a high rate of biallelic apc frameshift mutations (SI Appendix, Fig. S1A). Using InDelphi CRISPR/Cas9 gene editing outcome predictions for biallelic frameshift mutations in a given desmoid tumor (P = 0.81) (SI Appendix, Fig. S1B), we employed binomial statistics to calculate the probability that 100% of the tumors would carry biallelic frameshifts in absence of selection (x = 61, n = 61). This probability is extremely low and indicative of positive selection pressure for biallelic apc mutations in desmoid tumors (P[X = 61] < 0.01) (SI Appendix, Fig. S1B). Using CRISPR-SID, we thus certify the known role of apc as a tumor suppressor in desmoid tumor biology, based solely on deviations between tumor-sampled and InDelphi-predicted apc editing outcomes. Evidently, this experimental observation is in direct agreement with the well-described biology of desmoid tumors and the role of apc as a tumor suppressor gene (8, 30).
To validate, we used the same rationale to compare the apc gene editing outcomes sampled from whole early embryos and analyzed by deep amplicon sequencing with apc gene editing outcomes in the desmoid tumors. In line with InDelphi model predictions, binomial statistics revealed the probability of the desmoid tumors to have this specific classification of apc gene editing outcomes as unlikely to occur by random chance (P[X = 61] < 0.01) (SI Appendix, Fig. S1B).
CRISPR-SID Ascertains Negative Selection Pressure and Identifies ezh2 and creb3l1 as Genetic Dependencies for Desmoid Tumors.
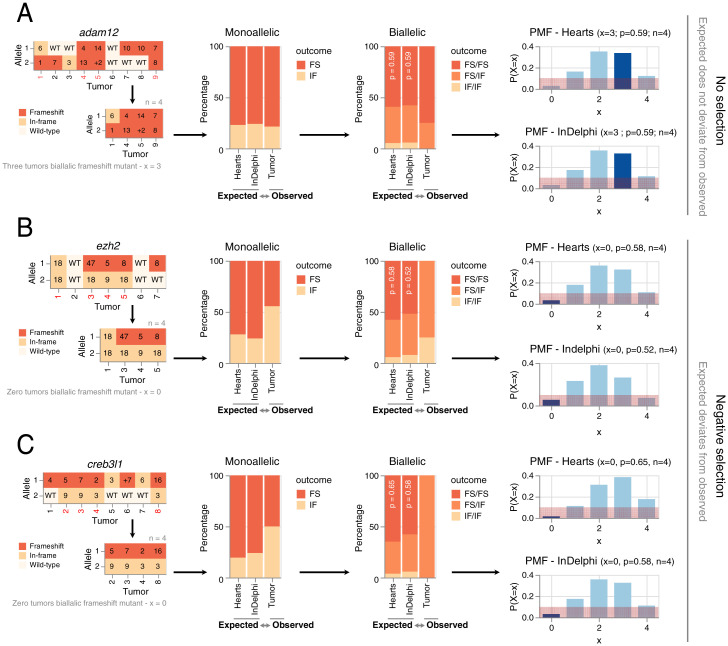
Classifying the mutation spectrum of each of the 10 suspected genetic dependencies (Fig. 2A), we observed for eight genes no significant deviations between InDelphi CRISPR/Cas9 editing outcome predictions and the experimental editing outcomes obtained in the desmoid tumors (Fig. 2 A and B and SI Appendix, Fig. S2). Importantly for this statistical analysis, only the tumors in which both alleles of the targeted dependency are mutated are taken into account. As such, this analysis in se is independent of cutting efficiency of a particular gRNA. As an example, using InDelphi predictions of the probability on adam12 biallelic frameshift mutations in a given desmoid tumor (P = 0.58), we employed binomial statistics to calculate the chance that, as observed, 75% of those tumors that were edited in both alleles would harbor biallelic frameshift mutations (x = 3, n = 4). This probability is high (P[X = 3] = 0.37) and indicative of lack of any selection pressure for a specific adam12 CRISPR/Cas9 editing outcome (Fig. 3A and SI Appendix, Extended Information). Biologically, the lack of deviation implies that for these eight genes, desmoid tumors can develop with biallelic inactivation of said genes, excluding them as genetic dependencies for desmoid tumorigenesis.
Fig. 3.
Statistical probability of ezh2 or creb3l1 biallelic frameshift mutations in desmoid tumors in relation to deep-learning predictions of gene editing outcomes. (A) Lack of selection mechanisms toward specific adam12 gene editing outcomes during desmoid tumorigenesis. The CRISPR/Cas9 editing outcomes for adam12 in desmoid tumors containing biallelic mutations (in-frame + frameshift) are likely and do not deviate from what is predicted by InDelphi and experimentally sampled in heart tissue. Given the adam12 editing outcomes sampled in hearts (i.e., in absence of selective pressure), the probability of a single biallelic mutant desmoid tumor to have biallelic frameshift editing outcomes is 59%. The probability of sampling 60% (3/5 tumors) with biallelic frameshift adam12 allelic status is likely (probability is 35%). Similarly, using InDelphi predictions, the probability of sampling this proportion of desmoid tumors with adam12 biallelic frameshift mutations is likely given binomial theory (probability is 34%). Red demarcation represents a 10% probability interval. PMF, probability mass function. (B and C) Editing outcomes for ezh2 and creb3l1 remain similar between InDelphi predictions and experimental observations in the heart of X. tropicalis. Significant deviations from the two former values are seen in the gene editing outcomes occurring in desmoid tumors. In line, the probabilities of never sampling biallelic frameshift mutations in ezh2 and creb3l1 within desmoid tumors is binomially unlikely (probability is 5% or smaller) given the editing outcomes in absence of tumor selection (heart tissue and InDelphi).
In direct contrast, we observed for two suspected dependencies, ezh2 and creb3l1, significant deviations between the InDelphi CRISPR/Cas9 editing outcome predictions and the actual experimental mutation patterns obtained by targeted deep amplicon sequencing of sampled desmoid tumors. For both genes, only tumors were obtained where at least one allele of the dependency gene was either wild type or had an in-frame INDEL mutation. For ezh2, the probability that 0% of those tumors that have both alleles mutated have in fact no biallelic frameshift mutations (x = 0, n = 4) is unlikely (P[X = 0]=0.053) given InDelphi predicted biallelic editing outcome probabilities (P = 0.52) (Fig. 3B and Dataset S5). Similarly, the probability that 0% of the tumors would have biallelic creb3l1 frameshift mutations is unlikely (P[X = 0) = 0.03) (x = 0, n = 4, P = 0.58) (Fig. 3C). We ruled out biases due to aberrant InDelphi predictions by determining the CRISPR/Cas9 editing outcomes in the hearts of animals from this respective injection setup. As the heart receives major contribution from the injected blastomeres, the editing outcome patterns for ezh2 and creb3l1 in their respective setup can also be used to determine editing outcomes in absence of tumoral selection pressure. Given these, the chance that 0% of the tumors in the ezh2 (Fig. 3B) and creb3l1 (Fig. 3C, Dataset S5) setups develop without biallelic frameshift mutations is considered unlikely (respectively, P[X = 0] = 0.03 and P[X = 0] = 0.02) (Dataset S5).
Taken together, our CRISPR-SID in vivo dependency mapping revealed that genetic disruption of lox, adam-12, mdk, hmmr, nuak1, fap-α, pclaf, and pycr1 does not prevent desmoid tumor development. In contrast, we observed significant deviations from the expected CRISPR/Cas9 editing outcomes in tumors for the ezh2 and creb3l1 genes. This negative selection indicates that ezh2 and creb3l1 are genetic dependencies for desmoid tumors.
Desmoid Tumor Development Critically Depends on the Activity of the Polycomb Repressive Complex 2.
EZH2 functions as the catalytic subunit in the polycomb repressive complex 2 (PRC2), mediating epigenetic alterations via its carboxyl-terminal histone methyl transferase SET domain (31). EZH2 inhibition is rapidly gaining traction as a therapeutic strategy, and an EZH2 inhibitor, Tazemetostat (Tazverik), has recently been approved by the Food and Drug Administration(FDA) via orphan drug designation for treatment of refractory follicular lymphoma and for epithelioid sarcoma, a rare soft tissue cancer (32–34). Given the well-tolerated clinical profile of Tazemetostat (35), we set out to validate EZH2 as a dependency in desmoid tumors. Encouragingly, we observed profound H3K27me3 immunoreactivity in Xenopus desmoid tumors indicative of high PRC2 activity, in line with human desmoid tumor biopsies (SI Appendix, Fig. S3A) (36).
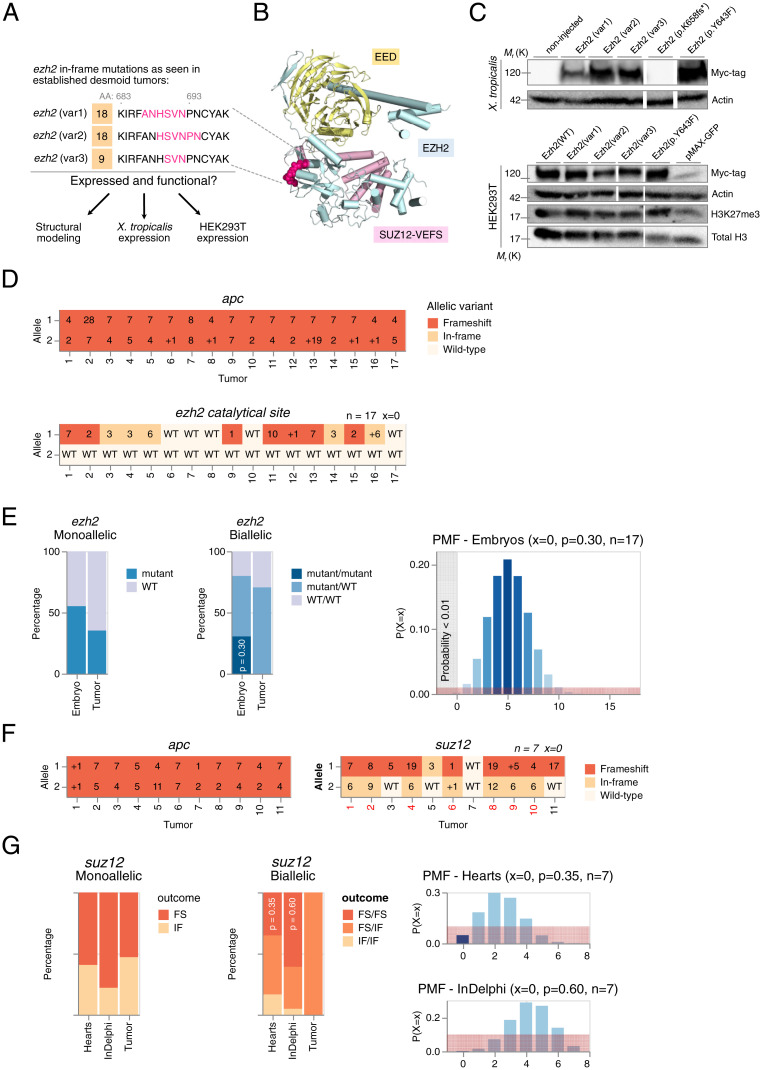
We further validated that the ezh2 editing outcomes in tumors are indicative for negative selection toward ezh2 biallelic frameshift mutations. As outlined above in the section CRISPR-SID Ascertains Negative Selection Pressure and Identifies ezh2 and creb3l1 as Genetic Dependencies for Desmoid Tumors, we found that in line with in vitro CRISPR/Cas9 screens (37), selection pressure for functional Ezh2 protein manifests itself via a higher than expected rate of in-frame mutations. Inherent to the selection model, this evidently imposes the condition that the observed ezh2 variants indeed encode functional proteins. Exploiting the conservation of sequence identity between human, Anolis carolinensis, and X. tropicalis EZH2 protein (38) (SI Appendix, Fig. S4), we generated a structural scaffold to map EZH2 protein variants retrieved from desmoid tumor deep amplicon sequencing (Fig. 4A). These three in-frame deletion protein variants map within a loop region at the periphery of the EZH2 subunit, which we determine likely to be structurally tolerated (Fig. 4B). Furthermore, we found that these protein variants express in vivo using Xenopus embryo expression assays and retained both expression and H3K27 trimethyltransferase function upon overexpression in HEK293T cells (Fig. 4C). This illustrates, next to negative selection against biallelic ezh2 frameshift mutations in tumors, a tolerance for in-frame variants encoding mutant but functional Ezh2 protein. In brief, under strong selection pressure due to CRISPR/Cas9 editing at the ezh2 target locus, we show that desmoid tumors can escape imposed selection pressure by harboring functional in-frame ezh2 mutations.
Fig. 4.
Assessing the role of the PRC2 in desmoid tumorigenesis. (A) In-frame ezh2 variants observed in desmoid tumors (Fig. 3B). (B) Mapping of identified deletion mutations in the EZH2 subunit of the PCR2. The deletion variants occur in a loop (magenta) at the periphery of the Ezh2 subunit and are expected to be well tolerated structurally. (C) Expression studies of Ezh2 protein variants in X. tropicalis and HEK293T cells. EZH2(p.Y643F) (corresponds to EZH2(p.Y642F) in human) and EZH2(p.K658fs*) variants function as positive and negative control, respectively. Actin and Total H3 are shown as loading controls. Myc-tagged Ezh2 protein variants express in X. tropicalis (Top) and in HEK293T cells (Bottom) and remain functional as they increase the levels of H3K27me3, when compared with the negative control (pMAX-GFP). Immunoblots were cut and pasted (white spaces) for clarity. Unaltered original blots are shown in SI Appendix, Fig. S7. (D) INDELs and allelic status at the suspected dependency CRISPR/Cas9 target sites in desmoid tumors. Frameshift mutations (FS) are indicated in dark orange, in-frame mutations (IF) in ochre, and wild-type (WT) in off-white. (E) Gene editing outcomes at the ezh2 exon encoding the Ezh2 catalytical pocket reveals selection toward retention of at least one wild-type ezh2 allele. Since each mutation (frameshift or in-frame) at this location can be considered as loss-of-function (LOF), we calculate the probability of a single desmoid tumor to have biallelic LOF ezh2 editing to be 30%, given ezh2 editing efficiencies in embryos from the same injection setup. Therefore, the probability of sampling 0% (17 tumors) without biallelic LOF allelic status is very unlikely (probability < 0.01%) according to binomial theory. Red demarcation represents a 1% probability interval. (F and G) The suz12 CRISPR/Cas9 editing outcomes, sampled in desmoid tumors, are unlikely and deviate from the expected. Given the suz12 editing outcomes sampled in hearts, the probability of a single biallelic mutant desmoid tumor to have biallelic frameshift editing outcomes is 35%. The probability of sampling 0% (0/7 tumors) with biallelic frameshift suz12 allelic status is unlikely (probability is <5%). Similarly, using InDelphi predictions, the probability of sampling this proportion of desmoid tumors with suz12 biallelic frameshift mutations is unlikely given binomial theory (probability is <0.01%). This demonstrates selection mechanisms toward specific suz12 gene editing outcomes during desmoid tumorigenesis. Red demarcation represents a 10% probability interval.
Previous studies have demonstrated that negative selection pressure in CRISPR screens increases dramatically when targeting specific protein domains driving the dependency since small in-frame deletions are in most cases not tolerated in critical functional domains (37, 39). Thus, each such mutation, whether in-frame or frameshift, can be considered a loss-of-function mutation. To investigate the importance of the histone methyltransferase activity of Ezh2 in its function as a dependency factor, we designed a gRNA targeting the SET domain at valine 659 (ezh2catal. site). With ezh2catal. site, we observed strong selection pressure on retaining at least one wild-type nonedited allele in all desmoid tumors sampled (Fig. 4D). Based on editing efficiencies of the ezh2catal. site gRNA in embryos from the same injection setup, the chance of recovering 17 tumors with none showing biallelic ezh2 editing events is binomially highly unlikely (P < 0.01) (Fig. 4E) (Dataset S6B). This confirms that the methyltransferase activity of Ezh2 is indeed crucial toward its activity as a dependency factor for desmoid tumor formation.
Importantly, there is growing evidence that histone methyltransferases like EZH2 can have methyltransferase-dependent functions not associated with PRC2 (40). Hence, we investigated whether suz12, encoding an essential structural subunit of PRC2, could be similarly identified as a genetic dependency for desmoid tumor development. For this, we designed a gRNA targeting suz12 at asparagine 338 (suz12N338). We performed CRISPR-SID revealing negative selection on biallelic suz12 frameshift mutations using a priori experimental data in the heart (P = 0.048; n = 7) and InDelphi predictions (P < 0.01; n = 7) (Fig. 4 F and G).
Collectively, these experiments demonstrate that both ezh2 and suz12 are genetic dependencies during desmoid tumor development with selection pressure specifically on the SET domain of the Ezh2 protein. This makes a strong case for a dependency on functional PCR2, mediating H3K27me3 marks, during desmoid tumorigenesis.
Chemical Inhibition of Ezh2, via Tazemetostat, Elicits a Therapeutic Response in Established Desmoid Tumors.
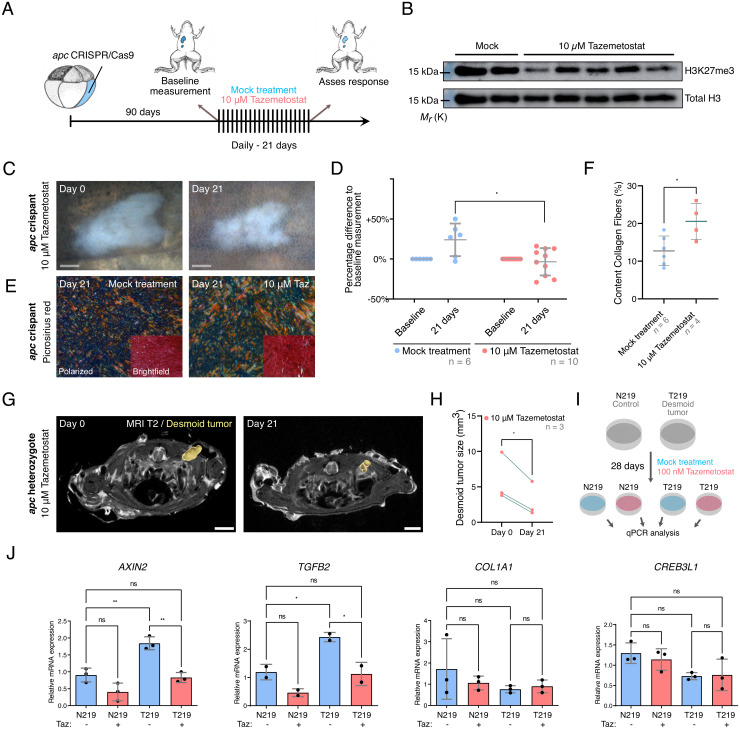
To investigate whether the genetic dependency of desmoid tumorigenesis for PRC2 could be harnessed toward a molecular therapy, we investigated whether EZH2 inhibition could elicit a therapeutic response in animals with established desmoid tumors. To exploit our apc mosaic mutant CRISPR/Cas9 X. tropicalis desmoid tumor model for preclinical drug studies, we exposed desmoid tumor–bearing froglets via the rearing water to the EZH2-inhibitor Tazemetostat (Tazverik/EZP-6438) (Fig. 5A). A critical issue when administrating a compound via the rearing water of the animals is to ascertain active absorption and biodistribution. Using a dose of 10 µM Tazemetostat, we demonstrated a reduction but not complete elimination of H3K27me3 levels in the livers of drug-exposed animals, indicative of moderate in vivo Ezh2 inhibition (Fig. 5B). With this concentration, no adverse effects were detected on the well-being of the exposed animals. A 3-wk Tazemetostat exposure resulted in a reduction or stasis in desmoid tumor size, whereas sustained tumor growth was observed in the control arm exposed to 0.1% dimethyl sulfoxide (DMSO) (P < 0.05) (Fig. 5 C and D). While the externally visible desmoid tumors normally had a cloudy milky appearance with smooth borders, after Tazemetostat treatment, they appeared more nodular and compacted, with irregular margins (Fig. 5C). Hence, we assessed the collagen content and organization via picrosirius red staining and polarization microscopy (Fig. 5E). This revealed that Tazemetostat-treated desmoid tumors have a significantly higher proportion of collagen organized in fibers when compared with mock-treated animals (P < 0.05) (Fig. 5F).
Fig. 5.
Ezh2 inhibition via Tazemetostat elicits a therapeutic response in desmoid tumors. (A and B) Tazemetostat-treated F0 apc crispants show a reduction in H3K27me3 levels in the liver (unaltered original scans of blots SI Appendix, Fig. S7). (C and D) Quantitatively, desmoid tumor size reveals stable or progressive disease in the control arm, while in Tazemetostat-exposed animals regression or stasis of tumors could be observed. Each data point represents one desmoid tumor; note that some animals developed multiple tumors (two-way ANOVA with repeated measurements; Dataset S6D; *P < 0.05). (E and F) Picrosirius Red staining with polarized light detection. The degree of alignment of collagen fibers is significantly increased in Tazemetostat-treated tumors when compared with mock-treated tumors (Student’s t test; Dataset S6D; *P < 0.05). (G and H) Desmoid tumors in apcMCR-Δ1/+ heterozygotes respond treatment with Tazemetostat (paired Student’s t test; Dataset S6D; *P < 0.05). (I) Treatment scheme of primary human desmoid tumor culture (T219) and paired normal cells derived from the fascia (N219) with Tazemetostat (100 nM) for 4 wk. (J) qRT-PCR analysis of AXIN2, TGFB2, COL1A1, and CREB3L1 expression in DMSO- (blue bars) or Tazemetostat (red bars)-treated cells (one-way ANOVA; Dataset S6D; *P < 0.05 **P < 0.01) (Gray scale bar, 1 mm; white scale bar, 3 mm).
While the above-mentioned apc crispant model is very penetrant and fast, it has the potential caveat, inherent to the method, that the two alleles of the tumor suppressor gene apc are already disrupted at an early developmental stage. However, FAP and Gardner’s syndrome patients harbor an inherited heterozygous APC mutation, and desmoid tumor development occurs upon a second hit in the APC gene or upon loss-of-heterozygosity. Similarly, for sporadic desmoid tumorigenesis, somatic monoallelic activating mutations in the CTNNB1 gene are acquired at adulthood or juvenile age. In order to investigate whether Tazemetostat could as well affect established desmoid tumors in the context of FAP or Gardner’s syndrome, we first performed MRI on apcMCR-Δ1/+ animals to detect the possible presence of desmoid tumors. MRI scans of apc heterozygotes (aged >3 y; n = 3) revealed hyperintense T2 signals indicative of desmoid tumors (41). These animals were exposed for 4 wk to Tazemetostat (added at a concentration of 10 µM to the rearing water), after which another T2 scan was taken. Dissection and histopathology confirmed the presence of several desmoid tumors, especially in between the dorsal back muscles and on the abdominal wall. Upon necropsy, we retrospectively localized the most prominent desmoid tumors, present between the back muscles, on the MRI scan images. This revealed a significant shrinkage of all three desmoid tumors under observation after Tazemetostat treatment (P < 0.05) (Fig. 5 G and H and SI Appendix, Fig. S5).
In sum, the drug treatment experiments show that established desmoid tumors in X. tropicalis are dependent on the methyl transferase activity of Ezh2, which can be modulated by Tazemetostat treatment.
Tazemetostat Inhibits the Wnt/β-Catenin Transcriptional Response in Human CTNNB1 Mutant Primary Tumor Cells.
In order to expand our identified dependency on functional PRC2 in desmoid tumors toward the human situation, we initially confirmed the presence of strong nuclear EZH2 immunoreactivity in human desmoid tumor specimens (SI Appendix, Fig. S3 B and C). In line with this, H3K27me3 immunoreactivity had previously been reported as a histopathological characteristic of desmoid tumors (36). Interestingly, tying back to our initial dependency mapping of creb3l1 in this study, we also found nuclear immunostaining of the N-terminally cleaved fragment of CREB3L1, indicative of an active role as transcription factor in clinical desmoid tumors (21) (SI Appendix, Fig. S3C).
Finally, in order to investigate whether Tazemetostat has an effect on the growth of human desmoid tumor cells cultured in vitro, we compared three primary cell lines obtained from clinical desmoid tumors and also included an equal number of normal fibroblasts (42). A dose–response curve with different concentrations of Tazemetostat (5 µM to 0.1 nM) showed absence of tumor-specific sensitivity for growth inhibition (SI Appendix, Fig. S6).
Next, in order to investigate a potential effect at the level of Wnt signaling responses, we selected a primary desmoid tumor culture (T219) harboring the CTNNB1S45F mutation, a mutation frequently found in sporadic desmoid tumors (43), for which paired normal fibroblasts (N219) were available. The desmoid tumor and normal cells were treated for 4 wk with 100 nM Tazemetostat. At the end of the treatment, qRT-PCR was performed to evaluate expression of genes highly transcribed in desmoid tumors (COL1A1, COL1A2, CREB3L1, and TGFB2) and of the AXIN2 gene, which is a general direct Wnt response gene (44, 45). Interestingly, Tazemetostat reduced AXIN2 and TGFB2 expression levels in desmoid tumor cells to the level observed in DMSO-treated normal cells. The expression of COL1A1, COL1A2, and CREB3L1 was not affected. Taken together, we conclude that EZH2 inhibition by Tazemetostat has a direct effect downstream or at the level of the β-catenin protein and normalizes Wnt signaling responses in human desmoid tumor cells.
Discussion
Desmoid tumors are uniquely driven by Wnt signaling hyperactivation and lack current standard of care (11). Here, we describe how CRISPR/Cas9-mediated desmoid tumor models in X. tropicalis were leveraged to identify Ezh2 as a druggable dependency.
We modulated the Wnt signaling pathway using both classical CRISPR/Cas9 for introducing truncating INDEL mutations in the apc gene, thereby mimicking familial adenomatous polyposis, and CRISPR base editing, for establishing an activating missense mutation (S45F) in beta-catenin, associated with sporadic desmoid tumors (16, 17). We also established a stable X. tropicalis line (apcMCR-Δ1/+) phenocopying presentation of desmoid tumors as seen in FAP and Gardner syndrome (4). Importantly, our models all converge on the induction of desmoid tumorigenesis, independent of the specific lesion in the Wnt signaling network. This further enforces the view that desmoid tumors are a consequence of Wnt signaling network hyperactivation in the broad sense (8, 9, 43).
Exploiting our models, we developed a methodology, CRISPR-SID, to identify cancer genetic dependencies. Dependency mapping allows to identify genetic perturbations that negatively affect the potential for either tumor initiation, tumor cell growth, or tumor cell survival, as those perturbations will be depleted via Darwinian selection (46). However, ascertaining negative selection in any process is challenging without high-confidence knowledge of that process outcome in absence of selection pressure. Therefore, for in vivo dependency mapping, which relies on the absence of biallelic gene disruption in a candidate dependency gene, one could always argue that biallelic frameshift mutations in this gene could still be observed if a larger sample size would be taken. As, while substantial evidence may prove the absence of biallelic frameshifts in a gene within tumors, there is no direct evidence that denies that this state could exist. This is especially the case for CRISPR/Cas9 experiments where there is never full experimental control on the cutting efficiency of the introduced gRNA, which can be dependent on sequence context and the chromatin status of the target region (47–49).
We believe our current study addresses this conundrum. We provide the framework to ascertain that the lack of sampling biallelic frameshift mutations in a given gene during tumorigenesis is not due to random chance but rather the result of selection processes. We show that the ever-increasing accuracy by which gene editing outcomes can be predicted by deep convolutional network learning approaches (a priori) can be used to demonstrate the deviation from expected gene editing outcomes in processes under selection pressure, such as tumorigenesis (a posteriori) (28, 29). Using this, we unveil selection processes at play during autochthonous desmoid tumorigenesis revealing genetic dependency on ezh2, suz12, and creb3l1. This selection pressure can be ascertained by substantial deviations between predicted (x% frameshift double strand break [DSB] repairs) and actual experimentally observed CRISPR/Cas9 editing outcomes (0% frameshift DSB repairs), a fact which we harness to identify cancer cell vulnerabilities. This in se turns a negative depletion signal into a positive enrichment signal. As most gRNAs will elicit editing outcomes with a higher frequency of frameshift mutations than in-frame mutations (28), the consistent observation of in-frame editing outcomes, due to selection pressure, can yield high statistical certainties with modest sample sizes of tumors. This provides the basis of the CRISPR-SID method. Interestingly, for in vitro CRISPR dependency screens, the retention of functional in-frame variants is still considered a nuisance that increases noise within negative selection screens. However, we postulate that if accurate a priori predictions of gene editing outcomes are available for the cell system under scrutiny, selection for in-frame mutants can be harnessed as a positive enrichment signal.
With the potential of our CRISPR/Cas9-based apc mosaic mutant desmoid tumor model, which combines high penetrance (100%) with short latency (10 to 12 wk) of tumor induction, we performed in-depth investigation into the nature of the ezh2 dependency. This is important since EZH2 is best known for its canonical role as a histone methyltransferase in the PRC2 but has also been documented to show noncanonical functions independent of PRC2, either via methylation of other substrates or even as a transcriptional activator (40). While in-frame DSB repairs could be observed in the initially targeted CXC domain of ezh2 (Fig. 3B), targeting its catalytic SET domain ascertained manifest selection pressure against either frameshift or in-frame DSB repairs (Fig. 4 D and E). Furthermore, we also identified suz12, encoding another component of PRC2 (38, 50), as a dependency gene. This clearly shows a strict dependency on canonical PRC2 activity. In line with our observations, clinical desmoid tumor samples have prominent nuclear localized expression of EZH2 and show strong immunoreactivity for H3K27me3, reflecting high PRC2 catalytic activity (36).
Exploiting our X. tropicalis model for preclinical compound validation, we exposed animals carrying established autochthonous desmoid tumors, either F0 apc crispants or apcΔMCR/+ heterozygotes, to the EZH2-inhibitor Tazemetostat. We found consistent reduction or stasis of the desmoid tumors, already after a 3- or 4-wk treatment. Given the fact that Tazemetostat is well tolerated and FDA approved for the treatment of follicular lymphoma and epithelioid sarcoma (32, 33), the drug could hold great promise for repurposing to treat patients with desmoid tumors. Importantly, Tazemetostat treatment in the Xenopus model induced a rapid measurable response. This contrasts with the timelines toward clinical responses of the tyrosine kinase inhibitor sorafenib (median time 9.6 mo) and the gamma-secretase inhibitor Nirogacestat (>8 mo) currently available to patients afflicted with desmoid tumors (11, 51). Also, for both latter cases, the actual drug target involved in the beneficial clinical outcome in desmoid tumors remains unknown. It is also worth noting that both those therapeutic agents elicit no major effects on desmoid tumor cells under laboratory culture conditions, underlining the need to identify novel drugs directly in animal models (13).
Despite their limitations, such as the absence of a tumor microenvironment, human T219 desmoid tumor cell cultures provided insight into the possible mechanism by which EZH2 inhibition may affect the tumors. We found that Tazemetostat significantly reduced the high expression of the AXIN2 and TGFB2 genes to levels observed in control-treated normal fibroblasts. The AXIN2 gene is a direct and the least tissue-specific transcriptional target of the Wnt/β-catenin pathway (44). The TGFB2 gene is not known as a direct Wnt target but has been found to be up-regulated by Wnt stimulation, likely in a tissue-dependent way (52–54). As mentioned, we also find the TGFB2 gene up-regulated in human desmoid tumors (Dataset S2).
Interestingly and importantly, the human T219 desmoid tumor cells carry activating missense mutations in the CTNNB1 gene that increase the stability of the β-catenin protein by interfering with its casein kinase 1 and glycogen synthase kinase 3-β–mediated phosphorylation and subsequent ubiquitination (42, 55). Hence, reduced expression of the AXIN2 Wnt/β-catenin response gene by Tazemetostat is indicative of a reduced transcriptional response of the β-catenin protein. Since the drug must thereby act either at the level of, or downstream from, the β-catenin protein, this implies that the drug can be effective in the context of both FAP/Gardner syndrome, associated with germline APC mutations, and in the context of sporadic desmoid tumors, originating from somatic CTNNB1 mutations.
At the moment, it is still unclear what is the exact nature of the clinical response of the desmoid tumors to the Tazemetostat treatment. One enticing possibility is that the desmoid tumors are addicted to the high signaling activity of the Wnt/β-catenin pathway, which makes them particularly sensitive to the inhibition of the pathway response. However, we failed to detect manifest cell proliferation or cell death responses, neither in vitro on human desmoid tumor cells nor in vivo in the Xenopus desmoid tumors. An alternative mode of action, possibly related to the role of PRC2 in the maintenance of stem cell characteristics, could be that EZH2 inhibition drives the tumor cells into a condition of terminal differentiation. In this context, we found that in the Xenopus desmoid tumors, Tazemetostat induced changes in the organization of the collagen fibers, which are excessive in desmoid tumors. Interestingly, it was previously shown that collagen content and cellularity in desmoid tumors are inversely correlated (56). Hence, increased presence of matured collagen fibers may be indicative of a differentiation response in desmoid tumor cells. Intriguingly, CREB3L1, which we also identified as a dependency factor for desmoid tumors, is known as a regulator of collagen synthesis and secretion (21, 57). This may suggest that desmoid tumors critically depend on a deregulated collagen synthesis program and that modulation of the latter has potential as a therapeutic strategy. A final possibility explaining the activity of Tazemetostat in an in vivo context and lack of a tumor-specific response in vitro may point to the involvement of the tumor microenvironment or the immune system, either innate or adaptive, in the drug response (58). Experiments addressing these different scenarios are currently ongoing.
This study further empowers the use of X. tropicalis for modeling human cancer. At the moment, genetic Xenopus cancer models are not familiar to many cancer researchers. This is for a large extent due to the fact that they have only recently emerged, thanks to the application of TALEN and CRISPR-mediated genome editing. It is reassuring that all studies up until now have shown that the Xenopus models accurately recapitulate human clinical data and genetically engineered mouse models. Examples are next to apc [this work and ref. 4, rb1/rbl1 (2), and tp53 (3)]. Although we have meanwhile modeled cancer by multiplexing different pathways [e.g., rb1, rbl1, pten, and tp53 (3)], CRISPR-SID would likely be most useful for cancers with a low number of genetic insults, such as retinoblastoma (RB1) and neurofibromatosis (NF1 or NF2).
A major advantage of the X. tropicalis CRISPR-SID model for dependency mapping is evidently the speed and the straightforwardness of the experimental approach. Microinjections are technically uncomplicated to perform, and multiplexed CRISPR/Cas9 genome editing is well-established (3). Also, since CRISPR-SID is based on positive and negative selection, high-efficiency genome editing is not per se required. This can be important for candidate dependency genes that are essential for early developmental processes, as in fact is the case for ezh2 and suz12 (59, 60). In this context, it is also very convenient that in Xenopus early embryos the different blastomeres, contributing to discrete developmental lineages, can be readily distinguished by their pigmentation characteristics and are strictly separated (61). Hence, it is possible to target genome editing to specific lineages and to titrate down gRNA/Cas9 ribonucleoprotein (RNP) complexes, thereby reducing interference of possible developmental phenotypes (62).
We expect CRISPR-SID to be applicable for multiple cancers and to be further adaptable to other model systems where CRISPR/Cas9 multiplexing can be easily achieved, such as zebrafish, tumor organoid culture systems, or cancer cell lines. The CRISPR-SID methodology will be uniquely positioned for in vivo validation of genetic dependencies identified in negative selection CRISPR/Cas9 screens in cancer cell cultures or in vivo xenografts of the latter. In addition, the multiplexing methodology could be used for the identification of codriver mutations (subject to positive selection) or synthetic lethality (via presence of negative selection), possibly in combination with established molecular therapies. The throughput of the technology could be further improved by the (adapted) application of the recently described MIC-Drop approach (multiplexed intermixed CRISPR droplets), where sgRNA/Cas9 ribonucleotide complexes and barcodes are packed into microdroplets and injected into fertilized eggs (63).
In conclusion, we demonstrate an experimental approach, CRISPR-SID, to ascertain negative selection in in vivo CRISPR/Cas9 experiments using deep learning predictions and binomial theory. We harness this approach to identify genetic dependencies on ezh2, suz12, and creb3l1 during autochthonous desmoid tumorigenesis. Finally, we demonstrate the promise of EZH2 inhibition as a possible therapeutic strategy for desmoid tumors.
Materials and Methods
Generating the apcMCR-Δ1/+ Line.
F0 apc mosaics generated by apc TALENs injection (4) were outcrossed with WT X. tropicalis, and offspring was genotyped to identify heterozygotes via heteroduplex mobility assay. One distinct band pattern on heteroduplex mobility analysis was determined by deep amplicon sequencing and BATCH-GE (64) to be indicative of a 1–base pair deletion in apc, and these animals were grouped and further raised for analysis.
CRISPR/Cas9 Genome Editing.
The apc gRNA was designed with the Doench, Hartenian algorithm (65). Mdk, wisp-1, lox, adam-12, hmmr, nuak1, creb3l1, FAP-α, pclafs8, pclafG10, pycr1, Ezh2R49, Ezh2S692, Ezh2V659, tp53, and suz12 gRNAs were designed with the CRISPRScan software package (66). In vitro transcription, purification, and quality control of injection-ready gRNA was performed as described before (Dataset S7A) (29).
DNA Extraction and Targeted Deep Sequencing.
In order to analyze CRISPR/Cas9 genome editing efficiencies, embryos, tissue, or tumors were incubated overnight at 55 °C in lysis buffer (50 mM Tris pH 8.8, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.5% Tween-20, 200 μg/mL proteinase K). For assessing total genome editing efficiency for a specific injection setup, we pooled nine stage-46 tadpoles before performing lysis. CRISPR/Cas9-target-site–containing amplicons were obtained by PCR with the primer pairs shown in Dataset S7B. Deep amplicon sequencing was performed with a validated pipeline, INDEL frequency data and variant calling for all sequencing was performed with BATCH-GE bioinformatics analysis (64).
Generation of X. tropicalis Mosaic Mutants by CRISPR/Cas9.
Wild-type X. tropicalis females and males were primed with 20 U and 10 U PREGNYL© human chorionic gonadotropine (hCG) (Merck), respectively. Natural matings were set-up 2 d later after boosting the female and male with 150 and 100 U hCG, respectively. Embryos were injected with the concentrations of precomplexed RNP and in the blastomeres described in Dataset S7C. For all experiments, we used NLS-Cas9-NLS (VIB Protein Service Facility, UGent), generated as described before (2). For the experiments with BE3 cytosine base editor (17), 100 pg synthetic BE3 mRNA en 40 pg sgRNA were injected per blastomere. All experiments were approved by the Ethical Committee for Animal Experimentation from Ghent University, Faculty of Science and VIB-Site Ghent (License EC2018-079). All methods were carried out in accordance with the relevant guidelines set out by this committee.
In Silico Differential Expression Analysis.
In order to identify genes that are consistently up-regulated in desmoid tumor fibromatosis compared with other fibrotic lesions, we reanalyzed the data from two previously published studies. The first dataset was generated by gene expression microarrays from 8 desmoid tumors, 10 dermatofibrosarcoma protuberans, and 13 solitary fibrous tumors (20). The second dataset comprised seven desmoid tumors compared with nine different types of fibrous lesions that were profiled by 3'-end sequencing for expression quantification (3SEQ) (19, 67). We performed multiclass differential expression analyses using sequence alignment maps (SAM) for microarray data and SAMseq for 3SEQ data. We identified 251 genes (Dataset S2) that were up-regulated in desmoid tumors in both datasets with contrast ≥ 2 and false detection rate ≤ 0.01. We performed functional annotation of these 251 genes using gene set enrichment analysis (MSigDB, Molecular Signatures Database version 6.2). Druggability was assessed via the DGIdb database (67).
CRISPR-SID.
For assessment of the gRNA efficiencies for the candidate dependency genes, each gRNA was injected together with Cas9 recombinant protein as precomplexed RNP, either unilaterally or bilaterally, in two-cell X. tropicalis embryos (Dataset S7C). Genome editing efficiencies were determined as described in the section DNA Extraction and Targeted Deep Sequencing. CRISPR-SID was performed by multiplex CRISPR/Cas9, thus generating double crispants for both apc and, respectively, each of the 11 putative dependency genes under scrutiny. Genome editing efficiencies were determined as described in the section DNA Extraction and Targeted Deep Sequencing. Animals were further raised until postmetamorphosis. Please note that, considering the fluctuating apc editing efficiencies across setups, direct comparison of tumor incidence rates to delineate impact of the candidate dependency gene knockout on desmoid tumorigenesis is impossible. For each animal developing one or multiple desmoid tumors, targeted PCR amplicon sequencing was performed for the apc and the respective candidate dependency gene on the desmoid tumors and, in most cases, the heart. Noteworthy, the heart is a tissue receiving significant contribution from the vegetal-dorsal lineage and as such will serve as a proxy to determine the probability of frameshift mutations occurring for a specific gRNA in absence of selection pressure. In practice, each isolated desmoid tumor will have a varying content of nonneoplastic cells, which will be edited at basal efficiencies similar to the vegetal-dorsal lineage of the animal. In order to perform a CRISPR-SID analysis, we require monoclonal tumors, and as such all tumors which showed more than two frequent mutation read variants in the apc gene were excluded from further analysis. Genes were excluded as genetic dependencies when they demonstrated biallelic frameshift mutations in desmoid tumors. The probability of y biallelic frameshifts was calculated using the binomial probability distribution, which takes the form . The calculated probability of k biallelic frameshift mutations is then function of the number of biallelic frameshifts, k, the number of samples, n, which is known, and the probability of the occurrence of biallelic frameshifts, p, which has been determined either experimentally by genetic analysis of the ezh2S692 and creb3l1L207 INDEL patterns in the heart, or as predicted by inDelphi software (28). For a more detailed method, see SI Appendix, Extended Information.
Imaging, Histology, and Immunohistochemistry.
Pictures of desmoid-tumor–bearing animals, including the active Wnt signaling within desmoid tumors by destabilized enhanced green fluorescent protein (dEGFP) fluorescence, were taken on a Carl Zeiss StereoLUMAR.V12 stereomicroscope. X. tropicalis desmoid tumor tissue samples were harvested from benzocaine-euthanized animals, fixed overnight at 4 °C in 4% paraformaldehyde (PFA), dehydrated, and embedded in paraffin. Tissue sections were cut (5 µm), stained, and imaged as described before (3). Clinical human desmoid tumor samples were identified by D.C., and anonymized tissue sections were provided to perform immunofluorescence. Antigen retrieval was performed using a PickCell 2100-Retriever (ProteoGenix) in citrate buffer (10 mM citric acid, 0.1% Tween-20, pH 6). Slides were subsequently blocked with blocking buffer (3% natural serum, 1% bovine serum albumin (BSA), and 0.1% Tween-20) and incubated overnight with anti-EZH2 antibody (3748; Abcam), anti-CREB3L1 (AF4080; R&Dsystems), or anti-H3K27me3 (07-449, Merck). Detection was performed with goat anti-rabbit DyLight-594, donkey anti-goat DyLight-594, and goat anti-rabbit DyLight-633, respectively. Counterstaining was performed with Hoechst-33342, and sections were imaged using a Leica TCS LSI zoom confocal microscope. Picrosirius red-polarization detection of collagen fibers in desmoid tumor tissue sections was performed as previously described (68). Images were acquired on a Axio Observer.Z1 inverted microscope attached to a monochrome Axiocam 506 camera (Carl Zeiss Microscopy GmbH) and analyzed using Fiji software. For quantification of Picrosirius red staining, following image processing was performed. The red channel was isolated from RGB input and masked as a binary. Mask was filtered for elongated collagen fibers based on size exceeding background threshold (>10 pixels) and circularity between 0.4, and 1 (representing elongated polygon blobs). Remaining mask was normalized to the total area of microphotograph containing desmoid tumor and described as percentage content collagen fibers.
Expression of EZH2 Protein Variants.
Carboxyl-terminal Myc-epitope–tagged expression plasmid pCS2-MT-EZH2 was generated by cloning X. tropicalis ezh2 cDNA, excluding the stop codon, in-frame into the BamHI-ClaI sites of the pCS2-MT vector. Specific mutations, NM_00101793.2:c.2163_2180del; NM_001017293.2:c.2165_2182del; NM_001017293.2:c.2171_2179del; NM_001017293.2:c.2068_2074del; NM_001017293.2:c.2127A > T, were introduced in full-length ezh2 cDNA by the Q5 Site-Directed Mutagenesis Kit (NEB; E0544S). Resulting plasmids were validated by Sanger sequencing, digested with NotI, and employed for SP6 in vitro transcription of ezh2 variant mRNA. Two-cell X. tropicalis embryos were injected with 500 pg capped mRNA and allowed to develop to stage 19, and cell extract was prepared in E1A lysis buffer (250 mM NaCl, 50 mM Tris pH 7.4, 0.1% Nonidet P-40) containing a complete protease inhibitor mixture (1:25) (Roche) and centrifuged for 10 min at 14,000 rpm in a microcentrifuge at 4 °C. Cleared supernatant, without yolk contamination, was further used. For transfection experiments, wells were seeded with 200,000 HEK293T cells and transfected the next day using CaCl2 transformation with 100 ng each of the pCS2-MT-EZH2 plasmids or 100 ng pMAX-GFP plasmid transfection control together with 900 ng pCS2-MT carrier plasmid. For two replicates, cell extracts were prepared in RIPA buffer (25mM Tris⋅HCl pH 7.6, 150mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS)) containing complete HALT protease and phosphatase inhibitor mixture (Pierce). For the third replica, cells were harvested in 1× phosphate-buffered saline (PBS), centrifuged, resuspended, and split in two. One part was used for cell extract preparation by RIPA buffer lysis as described before. The other part was used for histone extraction. Histone extracts were prepared as described before (69); the nucleus was first purified by centrifugation (1× PBS, 0.5% Triton X-100, and 5 mM sodium butyrate) followed by acid extraction (0.4 M HCl) for 1 h at 4 °C, and finally, the extract was neutralized with 0.5M Na2HPO4. All protein extracts were denatured in Laemmli buffer, separated by SDS-polyacrylamide gel electrophoresis (20 µg for total protein extracts and 1 µg for histone extracts) and transferred to a polyvinylidene fluoride (PVDF) membrane. Following antibodies were used for immunodetection: anti-Myc tag (9106; Abcam), anti-β-actin-HRP (47778; Santa Cruz), anti-H3K27me3 (39155; Active-motif), and anti-total H3 (1791; Abcam).
In Vivo EZH2 Inhibition.
Desmoid tumor–bearing apc crispants were generated as described before. Ninety-day-old animals were randomly assigned to either the treatment or the control arm. Animals were kept in 0.1× Marc's Modified Ringer's (MMR) containing either 10 µM Tazemetostat (HY-13803; MedChem Express) / 0.11% DMSO or 0.11% DMSO. Every day, one-half of the 0.1× MMR, containing either Tazemetostat or DMSO, was refreshed. A baseline tumor photography was made using a Carl Zeiss StereoLUMAR.V12 stereomicroscope, and a second photograph was taken after 21 d of treatment. Tumor sizes were quantified using ImageJ, and statistical analysis was performed using a two-way ANOVA with repeated measures. For Tazemetostat treatment in the apcMCR-Δ1/+ line, adult animals were kept in water supplemented with 10 µM Tazemetostat for 4 wk, with refreshment of one-half of Tazemetostat containing water every second day. Before and after the treatment, a whole-body MRI scan was taken. For MRI, adult animals were sedated by submersion in 0.2% MS222 (Tricaine) dissolved in water buffered to pH 7.0 with Sodium Bicarbonate. Upon disappearance of moving reflexes, animals were briefly submerged in fresh water and wrapped in a wet paper towel and plastic foil. MRI scans were acquired on a 7-Tesla preclinical MRI scanner (Bruker PharmaScan 70/16) using a 40-mm quadrature volume transmit/receive radiofrequency coil (Bruker). Anesthetized animals were positioned on the scanner’s animal bed. A localizer scan was acquired followed by a T1- and T2-weighted Rapid Acquisition with Relaxation Enhancement sequence to collect anatomical information of thorax and abdomen. The total acquisition time for each scan approximated 9 min. After imaging, animals were placed in a tilted container with fresh water, making sure to keep the head in the air. Fresh water was regularly pipetted on the partially submerged animal until breathing and movements became visible after which they were placed back in the original water tanks.
Cell Culture Drug Treatment and qRT-PCR Expression Analysis.
A primary human primary desmoid tumor cell culture (T219) and a patient matched population of normal fibroblasts (N219) (42) were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum (FCS) and seeded in 6-well plates. Cells were treated for 4 wk with 100 nM Tazemetostat or with vehicle (0.1% DMSO) with refreshment of one-half of the medium every second day. Total RNA was isolated from human desmoid tumor cell cultures using TRIzol reagent (Invitrogen) according to the manufacturer instructions. First-strand cDNA synthesis was performed using the SensiFAST cDNA Synthesis Kit (Bioline), and RNA concentration levels and purity were determined using a NanoDrop spectrophotometer (Thermo-Scientific). Samples were deidentified and a separate researcher performed qRT-PCR using the SensiFAST SYBR No-ROX Kit (Bioline) combined with a LightCycler 480 Real-Time PCR System (Roche). Expression was normalized to housekeeping genes Actb, B2m, and Gapd. Data were analyzed using the qBase+ software (Biogazelle). All primers including sequences used in this study can be found in Dataset S7D.
Supplementary Material
Acknowledgments
T.N. was funded by “Kom op tegen Kanker” (Stand up to Cancer), the Flemish Cancer Society, and a PhD fellowship with Vlaams Agenschap Innoveren en Ondernemen – HERMES (VLAIO-HERMES) (Agentschap voor Innovatie door Wetenschap en Technologie - Strategisch Basisonderzoek (IWT-SB)). Research in our laboratory is supported by the Research Foundation—Flanders (FWO-Vlaanderen; Grant Nos. G0A1515N and G029413N) and by grants from the Concerted Research Actions from Ghent University (BOF15/GOA/011 and BOF20/GOA/023). Further support was obtained from the Hercules Foundation, Flanders (Grant No. AUGE/11/14), the Desmoid Tumor Research Foundation, the Desmoid Tumour Foundation of Canada, and SOS Desmoïde. We thank Arne Martens, Nicolas Skrypek, and Marja Kreike for helpful suggestions regarding the cell culture experiments. We are indebted to Marnik Vuylsteke for consulting on statistical analysis. We acknowledge Marjolein Carron and Lana Hellebaut for proofreading and scientific discussions leading to new relevant insights. We thank Dr. Yonglong Chen for his kind gift of the BE3 base editor plasmid.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2115116118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Dempster J. M., et al. , Agreement between two large pan-cancer CRISPR-Cas9 gene dependency data sets. Nat. Commun. 10, 5817 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naert T., et al. , CRISPR/Cas9 mediated knockout of rb1 and rbl1 leads to rapid and penetrant retinoblastoma development in Xenopus tropicalis. Sci. Rep. 6, 35264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naert T., et al. , RBL1 (p107) functions as tumor suppressor in glioblastoma and small-cell pancreatic neuroendocrine carcinoma in Xenopus tropicalis. Oncogene 39, 2692–2706 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Van Nieuwenhuysen T., et al. , TALEN-mediated apc mutation in Xenopus tropicalis phenocopies familial adenomatous polyposis. Oncoscience 2, 555–566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naert T., Vleminckx K., CRISPR/Cas9 disease models in zebrafish and Xenopus: The genetic renaissance of fish and frogs. Drug Discov. Today. Technol. 28, 41–52 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Maresch R., et al. , Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat. Commun. 7, 10770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber J., et al. , CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc. Natl. Acad. Sci. U.S.A. 112, 13982–13987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alman B. A., Li C., Pajerski M. E., Diaz-Cano S., Wolfe H. J., Increased beta-catenin protein and somatic APC mutations in sporadic aggressive fibromatoses (desmoid tumors). Am. J. Pathol. 151, 329–334 (1997). [PMC free article] [PubMed] [Google Scholar]
- 9.Tejpar S., et al. , Predominance of beta-catenin mutations and beta-catenin dysregulation in sporadic aggressive fibromatosis (desmoid tumor). Oncogene 18, 6615–6620 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Nugent K. P., Spigelman A. D., Phillips R. K., Life expectancy after colectomy and ileorectal anastomosis for familial adenomatous polyposis. Dis. Colon Rectum 36, 1059–1062 (1993). [DOI] [PubMed] [Google Scholar]
- 11.Gounder M. M., et al. , Sorafenib for advanced and refractory desmoid tumors. N. Engl. J. Med. 379, 2417–2428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kummar S., et al. , Clinical activity of the γ-secretase inhibitor PF-03084014 in adults with desmoid tumors (aggressive fibromatosis). J. Clin. Oncol. 35, 1561–1569 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gounder M. M., Notch inhibition in desmoids: “Sure it works in practice, but does it work in theory?”. Cancer 121, 3933–3937 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Zreik R. T., Fritchie K. J., Morphologic spectrum of desmoid-type fibromatosis. Am. J. Clin. Pathol. 145, 332–340 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Tran H. T., Sekkali B., Van Imschoot G., Janssens S., Vleminckx K., Wnt/beta-catenin signaling is involved in the induction and maintenance of primitive hematopoiesis in the vertebrate embryo. Proc. Natl. Acad. Sci. U.S.A. 107, 16160–16165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komor A. C., Kim Y. B., Packer M. S., Zuris J. A., Liu D. R., Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Z., et al. , Modeling human point mutation diseases in Xenopus tropicalis with a modified CRISPR/Cas9 system. FASEB J. 33, 6962–6968 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Penel N., Chibon F., Salas S., Adult desmoid tumors: Biology, management and ongoing trials. Curr. Opin. Oncol. 29, 268–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo X., Zhu S. X., Brunner A. L., van de Rijn M., West R. B., Next generation sequencing-based expression profiling identifies signatures from benign stromal proliferations that define stromal components of breast cancer. Breast Cancer Res. 15, R117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West R. B., et al. , Determination of stromal signatures in breast carcinoma. PLoS Biol. 3, e187 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cotto K. C., et al. , DGIdb 3.0: A redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 46, D1068–D1073 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Q., Lee C. E., Denard B., Ye J., Sustained induction of collagen synthesis by TGF-β requires regulated intramembrane proteolysis of CREB3L1. PLoS One 9, e108528 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colombo C., et al. , Increased midkine expression correlates with desmoid tumour recurrence: A potential biomarker and therapeutic target. J. Pathol. 225, 574–582 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Jung H. Y., et al. , PAF and EZH2 induce Wnt/β-catenin signaling hyperactivation. Mol. Cell 52, 193–205 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misemer B. S., et al. , Expression of FAP, ADAM12, WISP1, and SOX11 is heterogeneous in aggressive fibromatosis and spatially relates to the histologic features of tumor activity. Cancer Med. 3, 81–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reversade B., et al. , Mutations in PYCR1 cause cutis laxa with progeroid features. Nat. Genet. 41, 1016–1021 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Skubitz K. M., Skubitz A. P., Gene expression in aggressive fibromatosis. J. Lab. Clin. Med. 143, 89–98 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Tolg C., Poon R., Fodde R., Turley E. A., Alman B. A., Genetic deletion of receptor for hyaluronan-mediated motility (Rhamm) attenuates the formation of aggressive fibromatosis (desmoid tumor). Oncogene 22, 6873–6882 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Shen M. W., et al. , Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naert T., et al. , Maximizing CRISPR/Cas9 phenotype penetrance applying predictive modeling of editing outcomes in Xenopus and zebrafish embryos. Sci. Rep. 10, 14662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latchford A., et al. , APC mutations in FAP-associated desmoid tumours are non-random but not ‘just right’. Hum. Mol. Genet. 16, 78–82 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Kim K. H., Roberts C. W., Targeting EZH2 in cancer. Nat. Med. 22, 128–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gounder M., et al. , Tazemetostat in advanced epithelioid sarcoma with loss of INI1/SMARCB1: An international, open-label, phase 2 basket study. Lancet Oncol. 21, 1423–1432 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Hoy S. M., Tazemetostat: First approval. Drugs 80, 513–521 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Girard J., et al. , Targeted therapies for follicular lymphoma. Curr. Hematol. Malig. Rep. 16, 25–31 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Duan R., Du W., Guo W., EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 13, 104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mito J. K., Qian X., Doyle L. A., Hornick J. L., Jo V. Y., Role of histone H3K27 trimethylation loss as a marker for malignant peripheral nerve sheath tumor in fine-needle aspiration and small biopsy specimens. Am. J. Clin. Pathol. 148, 179–189 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Shi J., et al. , Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 33, 661–667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brooun A., et al. , Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat. Commun. 7, 11384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michlits G., et al. , Multilayered VBC score predicts sgRNAs that efficiently generate loss-of-function alleles. Nat. Methods 17, 708–716 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Wang J., Wang G. G., No easy way out for EZH2: Its pleiotropic, noncanonical effects on gene regulation and cellular function. Int. J. Mol. Sci. 21, 9501 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cassidy M. R., et al. , Association of MRI T2 signal intensity with desmoid tumor progression during active observation: A retrospective cohort study. Ann. Surg. 271, 748–755 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mercier K. A., Al-Jazrawe M., Poon R., Acuff Z., Alman B., A metabolomics pilot study on desmoid tumors and novel drug candidates. Sci. Rep. 8, 584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trautmann M., et al. , Novel pathogenic alterations in pediatric and adult desmoid-type fibromatosis - A systematic analysis of 204 cases. Sci. Rep. 10, 3368 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jho E. H., et al. , Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22, 1172–1183 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timbergen M. J. M., et al. , Activated signaling pathways and targeted therapies in desmoid-type fibromatosis: A literature review. Front. Oncol. 9, 397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boehm J. S., et al. , Cancer research needs a better map. Nature 589, 514–516 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Jensen K. T., et al. , Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett. 591, 1892–1901 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Jain S., et al. , TALEN outperforms Cas9 in editing heterochromatin target sites. Nat. Commun. 12, 606 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X., et al. , Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci. Rep. 6, 19675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L., et al. , CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2. J. Clin. Invest. 128, 446–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi T., Prensner J. R., Robson C. D., Janeway K. A., Weigel B. J., Safety and efficacy of gamma-secretase inhibitor nirogacestat (PF-03084014) in desmoid tumor: Report of four pediatric/young adult cases. Pediatr. Blood Cancer 67, e28636 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Rudolf A., et al. , β-catenin activation in muscle progenitor cells regulates tissue repair. Cell Rep. 15, 1277–1290 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Biressi S., Miyabara E. H., Gopinath S. D., Carlig P. M., Rando T. A., A Wnt-TGFβ2 axis induces a fibrogenic program in muscle stem cells from dystrophic mice. Sci. Transl. Med. 6, 267ra176 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bridgewater D., et al. , β-catenin causes renal dysplasia via upregulation of Tgfβ2 and Dkk1. J. Am. Soc. Nephrol. 22, 718–731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clevers H., Nusse R., Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 (2012). [DOI] [PubMed] [Google Scholar]
- 57.McCarville M. B., et al. , MRI and biologic behavior of desmoid tumors in children. AJR Am. J. Roentgenol. 189, 633–640 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Guillemyn B., et al. , A homozygous pathogenic missense variant broadens the phenotypic and mutational spectrum of CREB3L1-related osteogenesis imperfecta. Hum. Mol. Genet. 28, 1801–1809 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Eich M. L., Athar M., Ferguson J. E. III, Varambally S., EZH2-targeted therapies in cancer: Hype or a reality. Cancer Res. 80, 5449–5458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Carroll D., et al. , The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21, 4330–4336 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E., Helin K., Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 23, 4061–4071 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moody S. A., Lineage tracing and fate mapping in Xenopus embryos. Cold Spring Harb. Protoc. 2018, 10.1101/pdb.prot097253 (2018). [DOI] [PubMed] [Google Scholar]
- 63.DeLay B. D., et al. , Tissue-specific gene inactivation in Xenopus laevis: Knockout of lhx1 in the kidney with CRISPR/Cas9. Genetics 208, 673–686 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parvez S., et al. , MIC-Drop: A platform for large-scale in vivo CRISPR screens. Science 373, 1146–1151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boel A., et al. , BATCH-GE: Batch analysis of next-generation sequencing data for genome editing assessment. Sci. Rep. 6, 30330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doench J. G., et al. , Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moreno-Mateos M. A., et al. , CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rittié L., Method for picrosirius red-polarization detection of collagen fibers in tissue sections. Methods Mol. Biol. 1627, 395–407 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Skrypek N., et al. , ZEB2 stably represses RAB25 expression through epigenetic regulation by SIRT1 and DNMTs during epithelial-to-mesenchymal transition. Epigenetics Chromatin 11, 70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.