Significance
Understanding the factors that sculpt gut microbial communities in mammals is of great interest. Here, we studied a diverse clade of herbivorous rodents (woodrats, Neotoma) with variable but well-characterized diets and habitats to quantify the relative contributions of host genetics, geography, and diet, alongside neutral processes, in structuring the gut microbiome under natural and controlled conditions. While diet and geography made significant contributions to microbiome structure, host phylogeny explained the greatest proportion of observed variation. Provision of a common diet in captivity altered natural microbial communities, with communities from different species varying in their resistance to this perturbation. Captivity increased the amount of variation explained by host phylogeny, further emphasizing the extent to which host genetics structure mammalian microbiomes.
Keywords: neutral model, DNA metabarcoding, phylosymbiosis, Neotoma, 16S rRNA
Abstract
The microbiome is critical for host survival and fitness, but gaps remain in our understanding of how this symbiotic community is structured. Despite evidence that related hosts often harbor similar bacterial communities, it is unclear whether this pattern is due to genetic similarities between hosts or to common ecological selection pressures. Here, using herbivorous rodents in the genus Neotoma, we quantify how geography, diet, and host genetics, alongside neutral processes, influence microbiome structure and stability under natural and captive conditions. Using bacterial and plant metabarcoding, we first characterized dietary and microbiome compositions for animals from 25 populations, representing seven species from 19 sites across the southwestern United States. We then brought wild animals into captivity, reducing the influence of environmental variation. In nature, geography, diet, and phylogeny collectively explained ∼50% of observed microbiome variation. Diet and microbiome diversity were correlated, with different toxin-enriched diets selecting for distinct microbial symbionts. Although diet and geography influenced natural microbiome structure, the effects of host phylogeny were stronger for both wild and captive animals. In captivity, gut microbiomes were altered; however, responses were species specific, indicating again that host genetic background is the most significant predictor of microbiome composition and stability. In captivity, diet effects declined and the effects of host genetic similarity increased. By bridging a critical divide between studies in wild and captive animals, this work underscores the extent to which genetics shape microbiome structure and stability in closely related hosts.
Gut microbial communities assist in nutrition, pathogen defense, and toxin metabolism and are critical for animal development, survival, and fitness (1). Although dysbiotic and low-diversity microbiomes are often associated with disease (2), healthy hosts also exhibit substantial temporal and interindividual variation in their gut microbial communities (3, 4). While it is widely recognized that host genetic background, diet, and geographic location can influence microbiome composition, there is substantial interest in understanding the relative importance of these factors (5, 6). Moreover, previous studies have largely ignored the role of neutral processes, such as passive dispersal and ecological drift, known to significantly influence community assembly (7). To advance our understanding of how these factors explain microbiome structure and stability, we quantified the contributions of geography, diet, and phylogenetic relatedness (hereon “phylogeny”) in a mammalian genus (Neotoma) with conserved morphology and life history across diverse diets and geographic locales.
Closely related animals often host similar bacterial communities, producing a pattern described as “phylosymbiosis” (5). Despite evidence that phylosymbiosis is common in nature and linked to host health (8), the mechanisms promoting these patterns remain poorly understood (5, 9). Host genetic similarity impacts microbiome composition within species (10, 11), suggesting that accumulated genetic differences between host species should also contribute to divergence in microbial communities. Studies involving closely related hosts provide more mechanistic insight into microbiome structure (12, 13). However, in cross-species comparisons, genetic differences often covary with other traits that also significantly influence microbiome composition (14).
While host genetics consistently influence microbiome structure, distinct animal diets can exert selective pressures that eclipse the effects of host genetics (15, 16). Diet strongly influences microbiome structure and function in controlled laboratory studies (e.g., refs. 17, 18). Although wild animal diets are difficult to manipulate, seasonal changes in microbiome composition are often attributed to changes in diet (19, 20). Further adding to the notion that diet strongly influences microbial community composition, divergent diets among closely related species are often associated with dissimilar microbiomes (13, 21). Conversely, across distantly related hosts, convergent diets can select for similar communities (15, 22, 23). However, these multispecies studies often rely on captive hosts, have limited intraspecific replication, and use coarse diet classifications (e.g., herbivore and carnivore). Since these broad dietary shifts are associated with substantial morphological changes, the influence of diet alone is difficult to quantify. Such effects could be disentangled by analyzing diet differences among more recently diverged, morphologically similar populations. Furthermore, with advances in diet characterization, most notably the use of DNA metabarcoding, wild animals can be subject to fine-scale analyses of the relative abundance and diversity of their dietary components (24). Such high-resolution characterization permits exploration of how diet diversity and composition shape the gut microbiome in wild animal populations.
Another important factor that merits consideration is that, in natural systems, diets are often site specific, making it difficult to separate diet effects from the effects of uncharacterized environmental differences and other neutral processes (e.g., ref. 25). Bacterial symbionts are often dispersal limited, with geographic structuring common in host-associated microbiomes (26, 27). These environmental effects can be controlled by sampling at one site (e.g., refs. 16, 24, 28) or quantified by replicated sampling at multiple sites. Additionally, neutral processes such as random dispersal, speciation, and extinction, or random changes in the abundances of species (termed “ecological drift”) are hallmarks of community ecology (7). While understanding the specific processes behind ecological stochasticity is challenging, neutral models provide a means to explore the extent of neutral and selective forces in shaping the assembly of microbial communities (29). Despite this importance, studies of the gut microbiome have only recently begun to investigate neutral processes (e.g., refs. 30, 31). Overall, moving toward a comprehensive understanding of microbiome structure requires teasing apart the interacting effects of geography, diet, and host phylogeny in wild animals, while accounting for both neutral and selective processes.
To advance our understanding of forces shaping mammalian microbiomes, we leveraged a natural system consisting of many species in the genus Neotoma (“woodrats”). These herbivorous, solitary rodents occur throughout North America, reaching their highest diversity in the southwestern United States, where multiple species often occur in sympatry (32). Behavior and morphology are highly conserved within the genus, but diet composition and specialization vary among species within the same habitat and among populations within a species (33). Different diets expose woodrats to different nutrients and plant secondary compounds (PSCs), and their specialized gut microbiomes assist in both digestion and PSC detoxification (34, 35). To quantify how differences in host phylogeny, diet, and geography predict Neotoma gut microbiome composition, we characterized diet and bacterial communities in seven woodrat species from multiple populations across the southwestern United States. After sampling wild animals, we brought the same individuals into captivity and placed them on the same diet. Similar captive conditions were expected to reduce differences in exposure; however, as wild-caught animals retain at least part of their natural microbiota in captivity (36), these experimental conditions were not expected to expose animals to identical microbial pools. Captivity was anticipated to alter microbiome composition, with dietary specialists predicted to be most sensitive to this perturbation. If ongoing exposure to different diets and environments substantially structured wild gut microbiomes, we expected captivity to reduce variation in microbiome composition between hosts. Alternatively, if host genetics were the dominant structuring factor, we expected that species would retain distinct microbiomes in captivity, and that reducing environmental variation would increase our ability to detect patterns of phylosymbiosis (8). Furthermore, we predicted that phylosymbiosis would be strongest for “selected” microbiota that deviated from patterns predicted under neutral assembly processes. By using a unique paired longitudinal sampling of wild and captive animals, we analyzed how host relatedness, diet, environment, and neutral processes predicted the structure and stability of naturally assembled mammalian microbiomes.
Results
To quantify how phylogeny, geography, and diet predicted mammalian gut microbiome structure, we sampled 25 woodrat populations representing seven species from 19 sites (Fig. 1 A and B). Trapping yielded samples from 2 to 10 individuals per population (SI Appendix, Table S1). While smaller sample sizes from some populations may reduce our ability to detect variation in these populations, animals from the same population generally hosted similar microbiomes, supporting the notion that samples from two to three individuals can provide insight into local microbial communities (SI Appendix, Fig. S1). For each individual, we characterized the natural gut microbiome and diet using bacterial and plant DNA metabarcoding (SI Appendix, Text S1). As we found that standard plant primers poorly amplified cacti, where cacti were abundant, we used stable isotopes to estimate cactus consumption and adjust individual diet proportions (SI Appendix, Text S1). We grouped plant taxa by family and calculated diet diversity as both richness (number of observed plant families) and Shannon diversity, which combines richness and relative abundance (SI Appendix, Fig. S2). As these metrics were correlated (Pearson correlation, r = 0.83, df = 145, t = 17.7, P < 0.001), we used richness in subsequent analyses. Diet richness varied among populations (ANOVA, F24,130 = 4.1, P < 0.001), with some exhibiting high specialization while others consumed a variety of plant taxa. Diet composition also varied among populations, both when calculated using plant occurrence (PERMANOVA, Jaccard, R2 = 0.52, P = 0.001) or relative abundance (Bray–Curtis, R2 = 0.62, P = 0.001). All wild diets included plants known to contain substantial levels of toxic secondary compounds (SI Appendix, Table S2); however, the presence and abundance of these plants varied between populations (Fig. 1C). In particular, multiple populations consumed diets of at least 10% cactus (n = 6), creosote bush (n = 5), and conifers (n = 6), creating natural diet replicates across species and sites.
Fig. 1.
Phylogeny, geography, and diet all contributed to microbiome structure in wild woodrats (Neotoma). (A) Sampled populations represented seven species (phylogeny and branch lengths from ref. 62) from (B) 19 sites (SI Appendix, Table S1). (C) Wild populations consumed different diets and exhibited different levels of dietary specialization. In the bipartite plot, lines connecting woodrat populations to the plants they feed on are scaled based on the relative abundance of each plant in the population’s diet. Dietary specialists (diet >60% one taxa) (63) are indicated with an asterisk. (D) In wild woodrat populations, microbiome composition was structured by host evolutionary history, (E) geography, and (F) diet (animals feeding on plant families known for substantial amounts of PSCs are colored by plant family as per C).
As predicted, diet influenced microbiome composition and diversity. Populations feeding on similar plants shared distinct sets of differentially abundant bacteria, with diets comprising at least 10% cactus, creosote, or conifer associated with 47, 34, and 54 enriched bacterial amplicon sequence variants (ASVs), respectively (SI Appendix, Figs. S3 and S4). Diet and microbiome richness were correlated, with each added plant family corresponding to ∼7 additional bacterial ASVs (Generalized Linear Mixed Model [GLMM]; β = 0.032, SE = 0.0097, z value = 3.32, P = 0.001, Fig. 2). Populations where individuals fed on more plant taxa also exhibited higher microbiome dispersion, but only when microbiome dissimilarity was measured with metrics that incorporated bacterial phylogeny (GLMM; UniFrac: β = 0.07, SE = 0.02, z value = 2.99, P = 0.003; weighted UniFrac: β = 0.08, SE = 0.03, z value = 2.68, P = 0.007; P > 0.05 for Jaccard and Bray–Curtis). Finally, using hierarchical clustering of population-averaged microbiome and diet data from all 25 populations, we found that diet and microbiome composition were more congruent than expected by chance (normalized clustering information distance: diet vs. microbiome = 0.32; random pairs = 0.15, P value <0.001, Fig. 1F).
Fig. 2.
Wild woodrat populations varied in dietary specialization, and for individuals, diet richness (measured as number of plant families) positively correlated with microbial richness (number of ASVs). Analyses were conducted using mixed effects models that included DNA concentration as a fixed effect and host species, sequencing run, and population as random factors, using all individuals (n = 147) with both wild microbiome and diet data. The displayed regression line depicts population-level predictions calculated using the ggpredict function in the ggeffects package v1.1.1 (64).
Naturally assembled microbiomes also showed evidence of geographic structuring, driven by strong site effects. Bacterial communities differed among sites for all dissimilarity metrics (PERMANOVA, all distance metrics P = 0.001) and, when controlling for host relatedness, increasing geographic distance among individuals significantly increased microbiome dissimilarity (partial Mantel test, r = 0.24, P = 0.0001). These patterns likely reflect high interindividual similarity within sites, as geographic effects were no longer significant when microbiomes were aggregated by host population (partial Mantel test, r = 0.10, P = 0.13, Fig. 1D). Local environmental effects were also detectable at sites where multiple species occurred in sympatry. Using species pairs where both sympatric and allopatric individuals were sampled, we found that heterospecific animals from the same site had more similar microbiomes than matched species pairs from different sites (Welch’s t test, Jaccard dissimilarity, all three pairs P < 0.02, SI Appendix, Fig. S5). Furthermore, although sympatric and allopatric Neotoma lepida and Neotoma macrotis populations shared a similar percentage of ASVs (9.4 vs. 10.4%), sympatry increased shared ASVs for N. lepida and Neotoma bryanti (39.4 vs. 26.0%) and N. bryanti and N. macrotis (28.1 vs. 12.4%), suggesting that sympatric animals share a microbial source pool.
Across all sites, different host species harbored distinct microbial communities (PERMANOVA, all distance metrics, P = 0.001) with more closely related individuals hosting more similar microbiomes (partial Mantel test controlling for geographic distance, r = 0.39, P = 0.0001; analyses based on normalized clustering information distance produced similar results and are included in SI Appendix, Text S1). This pattern of phylosymbiosis was even stronger when microbiome data were aggregated at the population level (r = 0.63, P = 0.0001), but was no longer detectable when data were aggregated by host species (Mantel test, all distance metrics: P > 0.15), suggesting that congruence in individual and population level analyses resulted from strong similarity within species. Alternatively, the nonsignificant results at the species level may simply reflect the smaller sample size (e.g., trees with 7 vs. 25 tips).
When the relative contributions of geography, phylogeny, and diet were analyzed together in multiple regression on distance matrices (MRM) models, all three factors were significant (P = 0.001) and together explained 49% of microbiome variance in wild hosts (Fig. 3). Phylogeny, geography, and diet each explained 35, 20, and 16% of observed variance, respectively (Bray–Curtis dissimilarities, Fig. 3A). As expected for a natural system, some variance was attributed to multiple factors (depicted as overlapping Venn diagram regions, Fig. 3A), with phylogeny, geography, and diet each uniquely explaining 21, 5, and 6% of the variance, respectively. Analyses with Jaccard dissimilarities produced similar results, while models with phylogenetically weighted distances explained less variance (Fig. 3B and SI Appendix, Table S3), consistent with related populations hosting distinct, but closely related taxa.
Fig. 3.
Diet, geography, and phylogenetic relatedness all contributed to wild microbiome structure, but host phylogeny was the most influential factor (A). The relative structuring effects of geographic origin (site), phylogeny, and diet differed between wild and captive animals, and Bray–Curtis (BC), Jaccard (J), weighted UniFrac (WU), and unweighted UniFrac (U) microbiome dissimilarity metrics (B). In captivity, the effects of wild diet decreased, but phylogeny effects increased, resulting in more explained variance (C).
If community assembly were driven primarily by neutral processes such as bacterial dispersal, the distribution of ASVs should follow frequency and occurrence patterns predicted under the prokaryote neutral model (PNM) (29, 30). As deviations from this model provide evidence for nonneutral processes such as host selection, we fit the PNM to ASVs from each site with more than seven animals to test whether increased host species diversity reduced model fit. As predicted, model fit decreased with increasing Neotoma diversity at a site (linear regression, R2 = 0.27, P = 0.048, SI Appendix, Fig. S6), further emphasizing that different host species preferentially selected different ASVs from shared environments. We also used the prokaryote neutral model to test whether observed patterns in microbiome composition were driven by the subset of ASVs exhibiting evidence of selection. Using the PNM, we classified ASVs from all wild woodrats as “neutral” or selected (over or under the frequency expected under neutral assembly) (Fig. 4A) (30). As expected, compared to MRM models with neutral taxa, models with selected taxa explained more total variance (R2 = 0.45 vs. 0.38). Furthermore, the extent to which host phylogeny, diet, and geography explained microbiome variance differed between neutral, over-, and underrepresented taxa (Fig. 4B). Notably, while phylogenetic effects were weaker for neutral compared to over- and underrepresented bacterial taxa (R2 = 0.21 vs. 0.26 vs. 0.26, respectively), geographic effects were stronger (R2 = 0.22 vs. 0.16 vs. 0.15). Neutral and overrepresented ASVs occurred in most bacterial families detected in woodrats. In contrast, underrepresented taxa were found only in the families Ruminococcaceae, Lactobacillaceae, Lachnospiraceae, Prevotellaceae, Muribaculaceae, Bifidobacteriaceae, and Desulfovibrionaceae (Fig. 4C).
Fig. 4.
We fit the neutral model to ASVs from all wild woodrats. (A) ASVs that occur more or less frequently than predicted by the model are shown in blue and orange, respectively. (B) Neutral, over-, and underrepresented taxa explained different components of microbiome variance, calculated using multiple regression on distance matrix models with different microbial groups. (C) Neutral, over-, and underrepresented taxa differed in taxonomic distributions. Each point represents an ASV, showing all ASVs with at least 10 reads, sized by total abundance, and colored with respect to neutral model fit. For “Coriobacteriales,” the asterisk indicates incertae sedis, and for phyla, Actinobacteria is abbreviated as “Actino.”
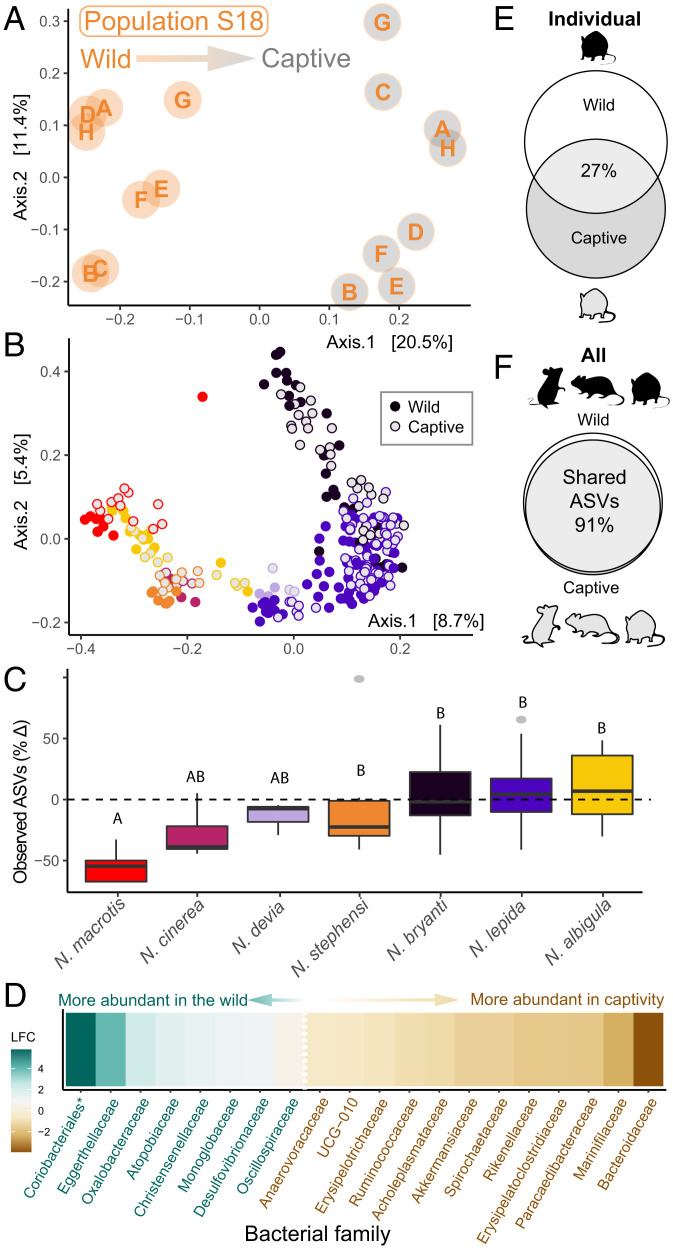
To reduce diet and environmental influences, wild woodrats were brought into the same captive facility for 1 mo and fed the same nontoxic alfalfa chow (Harlan Teklad, formula 2031). Captivity did not reduce variation among individuals for most distance metrics (Bray–Curtis, Jaccard, UniFrac: P > 0.1; weighted UniFrac; distance to median: captive = 0.13, wild = 0.15, permutation test P = 0.001), suggesting that homogenous conditions did not generally homogenize gut microbiomes. After a month in captivity, microbiomes still differed between populations (PERMANOVA, R2 = 0.43, P = 0.001), but individuals changed significantly from their state at capture (PERMANOVA, R2 = 0.02, P = 0.001, Fig. 5 A and B). Although microbiomes in dietary specialists were expected to be more sensitive to perturbation (37), we found no evidence that the degree of dietary specialization predicted changes in microbiome richness or composition (all models, P > 0.1, SI Appendix, Table S4). The extent to which captivity altered microbiome richness and composition differed among host species (SI Appendix, Table S4 and Fig. 5C) and was not conserved among closely related species (Moran’s I, padj > 0.10 for all metrics). Captivity induced the largest microbiome changes in N. macrotis. Although microbial communities from captive N. macrotis were still most similar to their wild counterparts (Fig. 5B), individuals exhibited a 55 ± 11% decline in ASV richness (Fig. 5C), with significant declines in ASVs from Muribaculaceae, Lachnospiraceae, and 11 other bacterial families. While these changes were most pronounced in N. macrotis, for each woodrat population, captivity significantly altered the abundance of 27.8 ± 22 ASVs (increasing 12 ± 9 and decreasing 16 ± 17, respectively), with differential abundance analyses identifying a distinct set of ASVs that changed within each population. Most of these differentially abundant ASVs were from Muribaculaceae (48%) or Lachnospiraceae (31%), families which also comprised 43 and 26% of ASVs detected in woodrats (SI Appendix, Fig. S7 A and B).
Fig. 5.
Captivity altered gut microbiomes. (A) Within a population (S18 shown), individuals (represented by letters) showed consistent community-specific shifts. (B) Despite changes, captive microbiomes still clustered with their counterparts from wild hosts (points represent individuals, colored by species as in C). (C) Animals varied in their response to captivity, with different species exhibiting different changes in ASV richness (letter codes indicate significant differences in percent change of observed ASVs). (D) Captivity also altered the abundance of 20 bacterial families (asterisks indicate incertae sedis), reducing taxa known to degrade PSCs. (E) In captivity, individuals retained <50% of their wild ASVs; (F) however, across all animals, >90% of wild ASVs were detected in captivity.
Animals with less diverse wild microbiomes maintained less diverse communities in captivity (LM, β = 0.52, SE = 0.09, t value = 5.83, P < 0.001), with no evidence for invasion by novel microbes from the captive facility or diet (SI Appendix, Text S2). While collectively most ASVs (>90%) occurred in woodrats under both wild and captive conditions, for individuals, 58 ± 11% of ASVs detected in the wild were not found in subsequent captive samples (Fig. 5 E and F), with low abundance taxa the least likely to be detected again (GLMM, β = 0.97, SE = 0.02, z value = 39.17, P < 0.001). In captivity, the bacterial ASVs that were enriched in natural cactus, conifer, and creosote diets became less abundant [mean log2 fold change (LFC), wild to captive = 0.30, t(131) = 6.6, P < 0.001]. Furthermore, across all hosts, captivity significantly altered the relative abundance of 20 bacterial families and 36 genera, with the greatest increases seen in Bacteriodaceae and the greatest reductions seen in Oxalobacteraceae, Eggerthellaceae, and Coriobacteriales (Fig. 5D and SI Appendix, Tables S5 and S6).
Captivity reduced natural diet and environmental differences, and the resulting changes to animal microbiomes increased patterns of phylosymbiosis. Compared to MRM models for wild hosts, models for captive hosts explained more total variance (Fig. 3B), with geography, phylogeny, and diet together explaining 62% of microbiome composition (Fig. 3C). Consistent with the increased variance explained by captive MRM models, neutral model fit was also lower for captive microbiomes (wild R2 = 0.37, captive R2 = 0.27), potentially due to either the reinforcement of structuring effects of host phylogeny or fewer opportunities for animals to acquire neutral ASVs in the relatively clean captive environment. In captivity, the effects of geographic origin remained similar, while variance explained by natural diet declined from 16 to 9% and variance explained by phylogeny increased from 35 to 56% (Fig. 3C and SI Appendix, Fig. S8). Together, these data suggest that host genetics are a dominant factor structuring gut microbial communities in woodrats.
Discussion
Mammalian microbiomes are frequently observed to exhibit phylosymbiosis, with closely related hosts maintaining more similar microbiomes (8). This pattern is thought to be mediated by multiple factors, ranging from stochastic influences such as the availability and ability of microbes to disperse in shared environments, to the adaptive pressures of shared ecological niche, diet, and genetics (5, 9). However, the relative contributions of these factors are not well understood because few groups of closely related animal species experience the diet and environmental variation required for accurate study. To quantify how these factors predict the structure of mammalian microbiomes, we used woodrats—a clade of closely related, morphologically similar rodents with extensive natural diet and geographic diversity.
In wild woodrats, host relatedness proved to be the strongest predictor of microbiome structure, with diet and local environment also playing an important role in determining composition and diversity. While host and bacterial cospeciation can yield phylosymbiosis (38), woodrats originated too recently (<10 Ma) (39) and the 16S rRNA gene mutates too slowly (1 to 2% per 100 Ma) (40) for cospeciation to be detectable in this dataset. This suggests that closely related woodrats assembled similar communities due to similarities in host filtering. Host filtering, due to differences in physiology or selective pressures, has been proposed as an important driver of phylosymbiosis (14). Despite extensive differences in diet and local environment, closely related wild woodrats hosted more similar microbiomes, suggesting that similar filtering is primarily mediated by host genetics. The specific genes producing these patterns in woodrats are unknown; however, as genes mediating innate immunity are linked to microbiome variation in model organisms (41), similar pathways might also contribute to phylosymbsiosis in wild mammals.
Different woodrat populations consumed distinct diets, which significantly affected microbiome composition and diversity. While it is often unclear whether diet effects are due to differences in diet components or gut morphology, gut morphology in all woodrats is similar (42), suggesting the observed microbiome variation was due to different diet components. Natural woodrat diets differed in PSCs and nutrients, both of which can influence gut microbial communities. In woodrats, PSCs are primarily metabolized in the foregut and act as both antimicrobials and microbial substrates (33). Changing exposure to PSCs alters the woodrat gut microbiome in controlled laboratory settings (43) and should also exert strong selection on microbial communities in wild animals. Microbes also play an important role in metabolizing dietary fiber and regulating energy homeostasis, with different microbes specializing on different polysaccharides (44). Although all woodrats consume relatively high fiber diets, the type and quantity of these fermentable compounds differ among plants. For hindgut fermenters like woodrats, differences in dietary fiber are expected to particularly impact microbes in the cecum, where fiber is degraded. As microbiota in the feces are often most similar to those in the hindgut (45, but see ref. 42), studies such as ours, that rely on noninvasive fecal samples, are likely to be particularly sensitive to changes in dietary fiber. While PSCs can also be metabolized as an energy source (46), it seems likely that their energetic contribution is minor, and that their effects arise from their ability to act as antimicrobials and of course, to sicken the host. In our dataset, diet likely impacts the microbiome via differences in both toxins and nutrients, particularly dietary fiber.
Our dataset included populations that varied in degree of dietary specialization, presenting an opportunity to examine the extent to which diet diversity predicts microbiome diversity. Populations that fed on more diverse diets maintained more diversity in their gut microbial communities, consistent with expectations (47) and likely driven by a selective pressure to harbor a diverse array of polysaccharide lyases. It should also be noted that increased diet diversity naturally increases opportunities for animals to acquire novel microbes. Approximately one quarter of the microbes in woodrat feces also occur on vegetation (36), indicating that plant-associated microbes likely contribute substantially to the woodrat gut microbiome. Many of these plant-associated microbes are anticipated to be transient in the animal gut (48), but if some are adapted to metabolize plant polysaccharides and/or toxins, then the maintenance of these bacteria, or the transfer of their genetic material to gut microflora (49), could enhance an herbivore’s ability to feed on novel diets.
While wild individuals retained a substantial part of their microbiome when brought into captivity, captivity altered woodrat gut microbiomes in ways similar to those seen in other vertebrates (37, 50). These changes could be explained by a range of factors, including physiological responses to stress, exposure to new microbes, loss of natural microbial sources, and novel diets (51). In our study, all animals were provided with the same captive conditions, creating an opportunity to examine the extent to which host factors such as genotype and dietary specialization predict microbiome stability. Although past work found that the microbiome of a dietary specialist, Neotoma stephensi, was more sensitive to captivity than that of a dietary generalist, Neotoma albigula (37), here we found that natural diet breadth had no effect on changes in microbiome richness or composition. For woodrats, microbiome responses were most strongly predicted by host species identity, with one species (N. macrotis) showing particularly large declines in diversity and pronounced community changes when brought into captivity. These species-specific responses to the same disturbance suggest that host genetics control not only microbiome composition, but also the stability of these communities.
Novel diets were likely the primary driver of observed microbiome changes in captivity (18). For captive animals, we replaced diverse PSC-rich natural diets with simple, nontoxic, alfalfa-based chow. On this chow, microbiome variation attributed to past natural diets declined, as did the abundance of taxa in families known to degrade PSCs (e.g., Coriobacteriales, Eggerthellaceae, and Oxalobacteriaceae) (35, 52). This suggests that these taxa are lost in the absence of direct selection from PSCs and other natural diet components. However, it should be noted that these taxa could be maintained at low levels (i.e., below our detection threshold) and recover when preferred resources become available (17, 53). Such storage effects play a key role in maintaining diverse communities of free-living species (54), and might also help maintain diverse symbiont assemblages that allow hosts to recover from perturbation, undergo niche expansion, and acclimate to changing environments (44).
While the structuring effects of diet declined in captivity, geographic effects persisted. Geographic structuring is commonly seen in vertebrate microbiomes (e.g., (refs. 27, 55, 56), potentially due to drift, in situ diversification, or different local selective pressures (26). Geographic effects could reflect exposure to transient environmental microbes through activities such as foraging or grooming (56). However, persistence of geographic patterns in captive woodrats indicates that animals retained site-specific microbes, suggesting that these symbionts contribute to core microbiome functions. Alternatively, some retained geographic effects might reflect population-level genetic differences that influenced bacterial filtering. Although efforts were taken to minimize genetic differences within species (e.g., N. lepida were all derived from the same clade (2A) (57), genetic differences between populations, and even single genes (58), can alter microbiome structure (11). While studies in wild house mice suggest that the effects of geography are stronger than those of host population structure (27), as patterns observed in captive woodrats are likely influenced by the microbial communities that animals assembled in the wild, woodrats from each population would need to be reared under conditions where microbial inputs are fully controlled to differentiate the effects of exposure and host filtering.
Although captivity altered microbial communities, woodrat populations still retained distinct microbiomes under these conditions. Removing animals from natural habitats reduced environmental and dietary variation, revealing the extent to which host genetics mediate microbiome structure (8). In captivity, congruence between the host and microbial community dendrograms increased, underscoring the fact that environmental differences can obscure patterns of phylosymbiosis, perhaps explaining why this pattern is observed in many, but not all, mammalian clades (5, 9, 59). In general, congruence between host phylogeny and microbial community structure will only establish and persist if selection favors certain community members enough to overcome the alternative selective and stochastic effects of diet and environment, as well as other neutral processes. For some systems, the structuring effects of host genetics might only be detectable when other sources of microbiome variation are reduced, highlighting a potential utility of captive studies.
While differences in host phylogeny, diet, and geography explain substantial amounts of microbiome variation, neutral processes like dispersal and ecological drift also contribute to the observed differences across hosts. Stochastic processes alone explain a substantial amount of microbiome structure for model organisms under controlled conditions (30), as well as for a population of wild house mice (31). Across wild woodrats, ∼55% of ASVs occurred at frequencies predicted by neutral assembly processes, suggesting that for these neutral taxa, all woodrats are equally accessible and colonizable. Thus, neutral processes should be considered when investigating the forces that sculpt host-associated microbiomes (31). However, in contrast to more homogenous systems, the neutral model was a relatively poor fit for microbiota samples from wild woodrats with diverse diets and genotypes.
Neutral models also allow recognition of microbial ASVs that are more or less widespread than predicted. A number of taxa were identified as “overrepresented.” These taxa may be beneficial and, due to their ability to provide important metabolic functions, are potentially maintained by deterministic processes, such as host selection. Alternatively, higher prevalence in overrepresented taxa may reflect greater dispersion capabilities, or association with dietary plants. For example, in our study, the majority of spore-forming, anaerobic Clostridia were found to be overrepresented, consistent with the hypothesis that spore-forming bacteria are more transmissible. A much smaller, more taxonomically restricted set of “underrepresented” ASVs occurred less frequently than expected, showing distribution patterns consistent with being selected against (e.g., pathogens). Alternatively, these underrepresented ASVs could be associated with specific sites, diets, or host genotypes, because they provide distinct, beneficial metabolic functions to specific populations of animals. For example, several of the microbial families that were underrepresented in our analysis are known to contribute to fiber digestion [Ruminococcaceae, Prevotellaceae (44, 60) and associated hydrogen metabolism Desulfovibrionaceae (61)]. Increasing our understanding of the influence of deterministic processes in the maintenance of microbes should enhance our ability to identify functionally important members of microbial communities.
Determining how selective and stochastic factors contribute to mammalian microbiome structure has proven difficult as few natural systems allow replicated sampling of closely related host species that also exhibit dietary and geographic variation. Using the extensive natural replication found in wild woodrats, our work shows that diet, geography, and other stochastic processes influence microbiome structure. However, host relatedness is the most important and persistent predictor of microbial community composition and stability.
Materials and Methods
To examine factors influencing mammalian microbiome structure, we sampled 2 to 10 individuals from 25 woodrat populations across the southwestern United States between 2017 and 2019 (Fig. 1 A and B and SI Appendix, Table S1). To assay natural diets and gut microbiomes, we collected a “wild” fecal sample from each animal at the time of capture, before transporting animals to the University of Utah’s School of Biological Sciences animal facility. We fed individually housed woodrats commercial high-fiber chow and collected a second “captive” fecal sample after approximately 1 mo. We used Illumina sequencing to generate microbiome and diet data. As plant primers poorly amplified cactus, we used stable-isotope ratios to adjust dietary cactus proportions. Using a woodrat phylogeny based on the Bayesian inference analyses in refs. 62 and 57, we examined how geography, phylogeny, and diet contributed to wild and captive microbiome structure. We first examined factors individually using PERMANOVAs, tests for tree congruence, and differential abundance analyses. We then examined the combined effects of all three factors using multiple regression on distance matrices and variance partitioning. We also examined the extent to which communities fit the prokaryote neutral model and whether neutral and nonneutral taxa differed in taxonomy and explained variance, following methods from ref. 30. Finally, we tested whether individual responses to captivity differed based on host and microbiome factors, as well as how captivity altered the abundance of specific bacterial taxa. Detailed methods are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Kaylene Yamada, James Patton, James Malcolm, Granger Cocke, Hayden Christensen, Lane Mulvey, Thomas Eiting, Sydney Stephens, Madeline Nelson, and Margaret Doolin for assistance with sample collection and animal husbandry; Marjorie Matocq and Daniel Nelson for assistance with microsatellite analyses; and the reviewers for their thoughtful feedback. Funding was provided by NSF Dimensions DEB Award 1342615, NSF IOS Award 1656497; Ruth L. Kirschstein National Research Service Award NIH T32AI055434; Genetics Training Grant NIH 5T32GM007464-38; and a Smithsonian-Mpala fellowship. Animal use was approved by the University of Utah Institutional Animal Care and Use Committee (16-02011) and conducted under permits from California (SC-8123), Utah (1COLL5194-1, 2), Nevada (333663), and Arizona (SP773078).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108787118/-/DCSupplemental.
Data Availability
Amplicon sequences (16S rRNA and chloroplast trnL) have been deposited in the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/PRJNA722312). Code is available in GitHub at https://github.com/SBWeinstein/Neotoma2021 and https://github.com/robertgreenhalgh/stand.
References
- 1.Moran N. A., Ochman H., Hammer T. J., Evolutionary and ecological consequences of gut microbial communities. Annu. Rev. Ecol. Evol. Syst. 50, 451–475 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kriss M., Hazleton K. Z., Nusbacher N. M., Martin C. G., Lozupone C. A., Low diversity gut microbiota dysbiosis: Drivers, functional implications and recovery. Curr. Opin. Microbiol. 44, 34–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lozupone C. A., Stombaugh J. I., Gordon J. I., Jansson J. K., Knight R., Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores G. E., et al. , Temporal variability is a personalized feature of the human microbiome. Genome Biol. 15, 531 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim S. J., Bordenstein S. R., An introduction to phylosymbiosis. Proc. Biol. Sci. 287, 20192900 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colston T. J., Jackson C. R., Microbiome evolution along divergent branches of the vertebrate tree of life: What is known and unknown. Mol. Ecol. 25, 3776–3800 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Zhou J., Ning D., Stochastic community assembly: Does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81, e00002–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks A. W., Kohl K. D., Brucker R. M., van Opstal E. J., Bordenstein S. R., Phylosymbiosis: Relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14, e2000225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohl K. D., Ecological and evolutionary mechanisms underlying patterns of phylosymbiosis in host-associated microbial communities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benson A. K., et al. , Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. U.S.A. 107, 18933–18938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki T. A., et al. , Host genetic determinants of the gut microbiota of wild mice. Mol. Ecol. 28, 3197–3207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochman H., et al. , Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 8, e1000546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrillo-Araujo M., et al. , Phyllostomid bat microbiome composition is associated to host phylogeny and feeding strategies. Front. Microbiol. 6, 447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazel F., et al. , Is host filtering the main driver of phylosymbiosis across the tree of life? mSystems 3, e00097-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muegge B. D., et al. , Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perofsky A. C., Lewis R. J., Meyers L. A., Terrestriality and bacterial transfer: A comparative study of gut microbiomes in sympatric Malagasy mammals. ISME J. 13, 50–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmody R. N., et al. , Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Mota R., Kohl K. D., Orr T. J., Denise Dearing M., Natural diets promote retention of the native gut microbiota in captive rodents. ISME J. 14, 67–78 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurice C. F., et al. , Marked seasonal variation in the wild mouse gut microbiota. ISME J. 9, 2423–2434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergmann G. T., Craine J. M., Robeson M. S. 2nd, Fierer N., Seasonal shifts in diet and gut microbiota of the American bison (Bison bison). PLoS One 10, e0142409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amato K. R., et al. , Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 13, 576–587 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delsuc F., et al. , Convergence of gut microbiomes in myrmecophagous mammals. Mol. Ecol. 23, 1301–1317 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Henderson G., et al. , Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5, 14567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kartzinel T. R., Hsing J. C., Musili P. M., Brown B. R. P., Pringle R. M., Covariation of diet and gut microbiome in African megafauna. Proc. Natl. Acad. Sci. U.S.A. 116, 23588–23593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moeller A. H., et al. , Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res. 23, 1715–1720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moeller A. H., et al. , Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc. Natl. Acad. Sci. U.S.A. 114, 13768–13773 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linnenbrink M., et al. , The role of biogeography in shaping diversity of the intestinal microbiota in house mice. Mol. Ecol. 22, 1904–1916 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Baxter N. T., et al. , Intra- and interindividual variations mask interspecies variation in the microbiota of sympatric peromyscus populations. Appl. Environ. Microbiol. 81, 396–404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloan W. T., et al. , Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 8, 732–740 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Burns A. R., et al. , Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 10, 655–664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sieber M., et al. , Neutrality in the metaorganism. PLoS Biol. 17, e3000298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reid F. A., A Field Guide to Mammals of North America (Peterson Field Guides, Houghton Mifflin Company, New York, 4th ed., 2006). [Google Scholar]
- 33.Kohl K. D., Dearing M. D., The woodrat gut microbiota as an experimental system for understanding microbial metabolism of dietary toxins. Front. Microbiol. 7, 1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohl K. D., Weiss R. B., Cox J., Dale C., Dearing M. D., Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol. Lett. 17, 1238–1246 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Miller A. W., Oakeson K. F., Dale C., Dearing M. D., Microbial community transplant results in increased and long-term oxalate degradation. Microb. Ecol. 72, 470–478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohl K. D., Dearing M. D., Wild-caught rodents retain a majority of their natural gut microbiota upon entrance into captivity. Environ. Microbiol. Rep. 6, 191–195 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Kohl K. D., Skopec M. M., Dearing M. D., Captivity results in disparate loss of gut microbial diversity in closely related hosts. Conserv. Physiol. 2, cou009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders J. G., et al. , Stability and phylogenetic correlation in gut microbiota: Lessons from ants and apes. Mol. Ecol. 23, 1268–1283 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Martin R. A., Zakrzewski R. J., On the ancestry of woodrats. J. Mammal. 100, 1564–1582 (2019). [Google Scholar]
- 40.Kuo C.-H., Ochman H., Inferring clocks when lacking rocks: The variable rates of molecular evolution in bacteria. Biol. Direct 4, 35 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurilshikov A., Wijmenga C., Fu J., Zhernakova A., Host genetics and gut microbiome: Challenges and perspectives. Trends Immunol. 38, 633–647 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Kohl K. D., Miller A. W., Marvin J. E., Mackie R., Dearing M. D., Herbivorous rodents (Neotoma spp.) harbour abundant and active foregut microbiota. Environ. Microbiol. 16, 2869–2878 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Miller A. W., Oakeson K. F., Dale C., Dearing M. D., Effect of dietary oxalate on the gut microbiota of the mammalian herbivore Neotoma albigula. Appl. Environ. Microbiol. 82, 2669–2675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martens E. C., Kelly A. G., Tauzin A. S., Brumer H., The devil lies in the details: How variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J. Mol. Biol. 426, 3851–3865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dill-McFarland K. A., Weimer P. J., Pauli J. N., Peery M. Z., Suen G., Diet specialization selects for an unusual and simplified gut microbiota in two- and three-toed sloths. Environ. Microbiol. 18, 1391–1402 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Musilova L., Ridl J., Polivkova M., Macek T., Uhlik O., Effects of secondary plant metabolites on microbial populations: Changes in community structure and metabolic activity in contaminated environments. Int. J. Mol. Sci. 17, 1205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reese A. T., Dunn R. R., Drivers of microbiome biodiversity: A review of general rules, feces, and ignorance. MBio 9, e01294-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammer T. J., Janzen D. H., Hallwachs W., Jaffe S. P., Fierer N., Caterpillars lack a resident gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 114, 9641–9646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hehemann J.-H., et al. , Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912 (2010). [DOI] [PubMed] [Google Scholar]
- 50.McKenzie V. J., et al. , The effects of captivity on the mammalian gut microbiome. Integr. Comp. Biol. 57, 690–704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trevelline B. K., Fontaine S. S., Hartup B. K., Kohl K. D., Conservation biology needs a microbial renaissance: A call for the consideration of host-associated microbiota in wildlife management practices. Proc. Biol. Sci. 286, 20182448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maruo T., Sakamoto M., Ito C., Toda T., Benno Y., Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int. J. Syst. Evol. Microbiol. 58, 1221–1227 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Sonnenburg E. D., et al. , Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chesson P., Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Evol. Syst. 31, 343–366 (2000). [Google Scholar]
- 55.Lankau E. W., Hong P. Y., Mackie R. I., Ecological drift and local exposures drive enteric bacterial community differences within species of Galápagos iguanas. Mol. Ecol. 21, 1779–1788 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Grieneisen L. E., et al. , Genes, geology and germs: Gut microbiota across a primate hybrid zone are explained by site soil properties, not host species. Proc. Biol. Sci. 286, 20190431 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patton J. L., Huckaby D. G., Álvarez-Castañeda S. T., The Evolutionary History and a Systematic Revision of Woodrats of the Neotoma lepida Group (University of California Press, 2007), vol. 135. [Google Scholar]
- 58.Goodrich J. K., Davenport E. R., Clark A. G., Ley R. E., The relationship between the human genome and microbiome comes into view. Annu. Rev. Genet. 51, 413–433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grond K., et al. , No evidence for phylosymbiosis in western chipmunk species. FEMS Microbiol. Ecol. 96, fiz182 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Avgustin G., Flint H. J., Whitehead T. R., Distribution of xylanase genes and enzymes among strains of Prevotella (Bacteroides) ruminicola from the rumen. FEMS Microbiol. Lett. 78, 137–143 (1992). [DOI] [PubMed] [Google Scholar]
- 61.Rey F. E., et al. , Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc. Natl. Acad. Sci. U.S.A. 110, 13582–13587 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matocq M. D., Shurtliff Q. R., Feldman C. R., Phylogenetics of the woodrat genus Neotoma (Rodentia: Muridae): Implications for the evolution of phenotypic variation in male external genitalia. Mol. Phylogenet. Evol. 42, 637–652 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Shipley L. A., Forbey J. S., Moore B. D., Revisiting the dietary niche: When is a mammalian herbivore a specialist? Integr. Comp. Biol. 49, 274–290 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Lüdecke D., ggeffects: Tidy data frames of marginal effects from regression models. J. Open Source Softw. 3, 772 (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Amplicon sequences (16S rRNA and chloroplast trnL) have been deposited in the National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/PRJNA722312). Code is available in GitHub at https://github.com/SBWeinstein/Neotoma2021 and https://github.com/robertgreenhalgh/stand.