Significance
This manuscript identifies a mechanism by which sleep can sometimes occur at the wrong circadian time of day. Sleep is regulated by two processes, the circadian system, which confers 24-h rhythms on sleep, and the homeostatic system, which ensures an organism sleeps sufficiently. Sleep occurs at night because of circadian timing cues and also homeostatic sleep drive triggered by prolonged daytime wakefulness. However, the homeostatic system can sometimes override the circadian system; for instance, following a night of sleep deprivation. We identify a mechanism by which the homeostatic system suppresses circadian signals, allowing sleep to occur against the circadian clock. We find connections between homeostatic and circadian circuits and show that homeostatic sleep drive decreases circadian neuronal activity.
Keywords: circadian rhythms, sleep homeostasis, neuropeptides, circuit, Drosophila
Abstract
Sleep is controlled by homeostatic mechanisms, which drive sleep after wakefulness, and a circadian clock, which confers the 24-h rhythm of sleep. These processes interact with each other to control the timing of sleep in a daily cycle as well as following sleep deprivation. However, the mechanisms by which they interact are poorly understood. We show here that hugin+ neurons, previously identified as neurons that function downstream of the clock to regulate rhythms of locomotor activity, are also targets of the sleep homeostat. Sleep deprivation decreases activity of hugin+ neurons, likely to suppress circadian-driven activity during recovery sleep, and ablation of hugin+ neurons promotes sleep increases generated by activation of the homeostatic sleep locus, the dorsal fan-shaped body (dFB). Also, mutations in peptides produced by the hugin+ locus increase recovery sleep following deprivation. Transsynaptic mapping reveals that hugin+ neurons feed back onto central clock neurons, which also show decreased activity upon sleep loss, in a Hugin peptide–dependent fashion. We propose that hugin+ neurons integrate circadian and sleep signals to modulate circadian circuitry and regulate the timing of sleep.
Sleep is regulated by two processes, circadian and homeostatic (1). The endogenous circadian clock, together with its downstream pathways, is synchronized to external day–night cycles and determines the timing of sleep to produce 24-h rhythms in sleep and wake. The homeostatic process tracks sleep:wake history and generates sleep drive, based on the extent of wakefulness. Overtly, sleep homeostasis can be seen as an increase in sleep duration and depth after prolonged wakefulness. Generally, circadian and homeostatic processes are studied as separate mechanisms that regulate sleep, but they clearly intersect and are coordinated in a daily cycle to promote the onset and maintenance of sleep at night. Following sleep deprivation, the homeostatic system can drive sleep at the wrong time of day, but even under these conditions, interactions between the two systems determine the timing and duration of sleep. However, the neuronal mechanisms by which circadian and homeostatic pathways signal to each other are largely unknown.
The functions and regulation of sleep are extensively studied in model organisms, such as Drosophila melanogaster (2). In the Drosophila brain, the circadian clock is expressed in ∼150 clock neurons that are organized into neuroanatomical groups, of which the ventral and dorsal lateral neurons are the most important for driving rhythms of locomotor activity (3, 4). The small ventrolateral neurons (s-LNvs) link to other brain regions through different circuits. One of those circuits connects the s-LNvs to the site of the motor ganglion, the thoracic nerve cord: s-LNvs → DN1s → Dh44+ neurons → hugin+ neurons→ ventral nerve cord (5). Dh44-expressing neurons in the pars intercerebralis regulate locomotor activity rhythms in part through the signaling of DH44 neuropeptide to hugin-expressing neurons in the subesophageal zone (SEZ) (6, 7). Dh44+ and hugin+ circadian output neurons do not contain canonical molecular clocks, but display cycling in neuronal activity or peptide release, likely under control of upstream circadian signals (7–9). Links between these neurons and loci regulating sleep homeostasis have not been identified yet.
Regulation of sleep homeostasis in flies involves the central complex and mushroom body (10–13). Of particular importance is a group of sleep-promoting neurons labeled by the 23E10-GAL4 driver that projects to the dorsal fan-shaped body (dFB) neuropil in the central complex. These dFB neurons promote sleep when activated (14, 15), and they are required for normal sleep rebound after deprivation (16). 23E10+ neurons receive input signals from R5 ellipsoid body neurons, which track sleep need (17).
As hugin+ neurons are significantly downstream of central clock neurons and close to behavioral outputs, we asked whether they also have a role in sleep. We find that hugin+ neurons are dispensable for determining daily sleep amount, but they show decreases in activity following sleep deprivation. They also receive projections from the dFB and counter sleep-promoting effects of 23E10+ neurons, such that ablation of hugin+ neurons enhances sleep driven by 23E10+ cells. Further supporting a role in sleep, mutations in Hugin peptides affect recovery sleep after deprivation and enhance sleep driven by 23E10+ cells. hugin+ neurons target PDF+ s-LNv clock neurons, which also show decreases in intracellular Ca2+ levels following sleep deprivation. Thus hugin+ neurons serve as an integrations site of signals from both the sleep homeostat and the circadian clock.
Results
Silencing hugin+ Neurons Does Not Affect Sleep.
As noted above, because hugin+ neurons mediate circadian output and are just upstream of the motor ganglion (7), we asked whether they also regulate sleep. To test this, we expressed the temperature-sensitive TrpA1 channel (18) in hugin+ neurons and activated them with high temperature while measuring sleep behavior. In other experiments, we expressed temperature-sensitive shibirets, a dominant-negative dynamin gene (19), to inhibit synaptic transmission from hugin+ neurons at high temperature. Unlike 23E10+ neurons that increase sleep when activated with TrpA1 (14, 15, 20), neither activation of hugin+ neurons with TrpA1 (SI Appendix, Fig. S1A) nor inhibition with shibirets (SI Appendix, Fig. S1B) produced any changes in sleep. Despite unchanged sleep levels, hugin > shibirets flies were less active than control flies, as measured by number of beam crossings per day (SI Appendix, Fig. S1C), which confirms our previous findings that hugin+ neurons regulate locomotor activity (7).
Since mechanisms that participate in baseline and sleep recovery may be different, we asked whether hugin+ neurons play a role in regulating sleep homeostasis. We used the same thermogenetic approaches to activate or inhibit the hugin+ neurons while simultaneously sleep depriving the flies at night using a mechanical method. After sleep deprivation, recovery sleep was monitored in the flies during the daytime. We found no significant difference in recovery sleep between the experimental and control genotypes when hugin+ neurons were activated or inhibited (SI Appendix, Fig. S1D). Given that mechanical sleep deprivation can recruit multiple pathways to elicit rebound (21), it is possible that disrupting the activity of hugin+ neurons alone does not affect sleep amount or homeostasis.
Sleep Deprivation Decreases Ca2+ Levels in hugin+ Neurons.
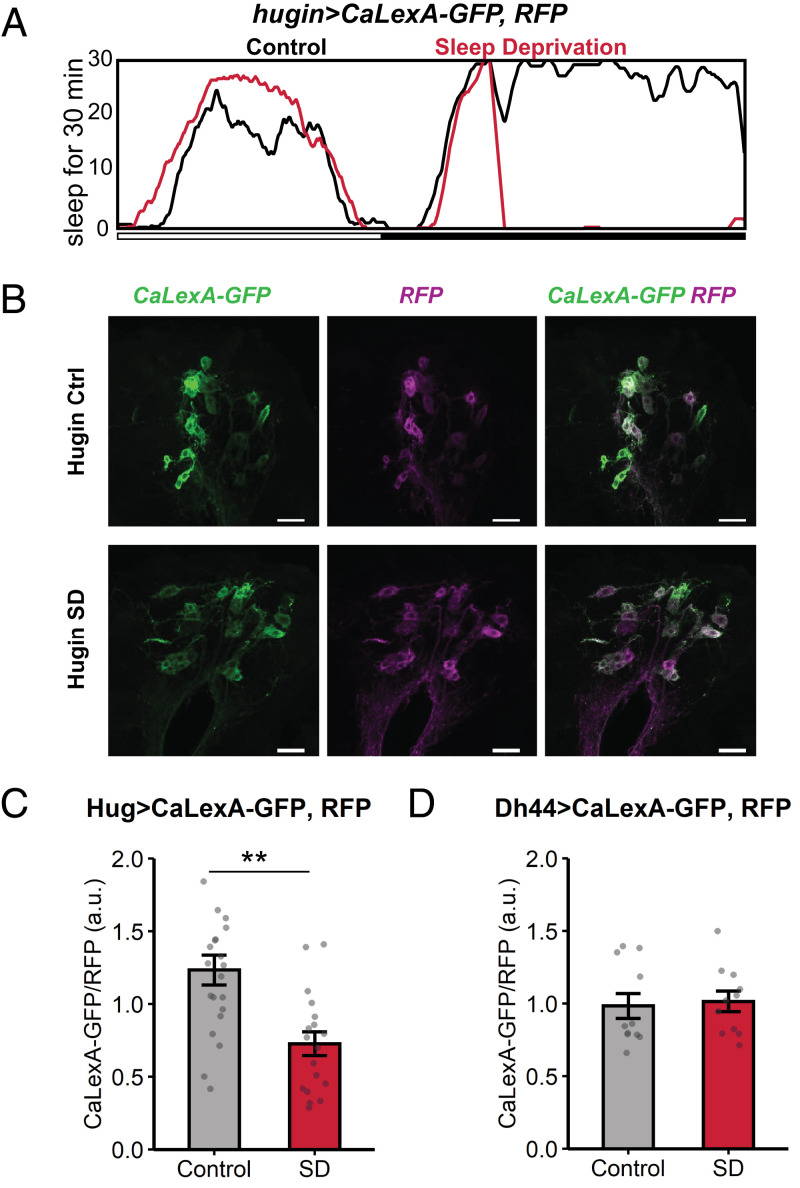
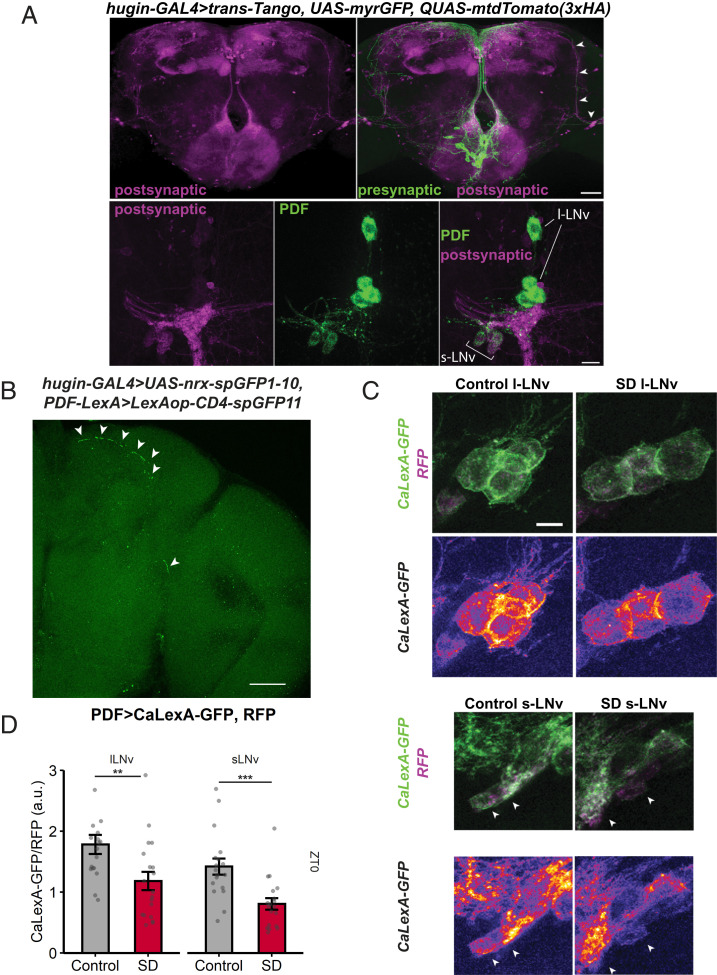
Although hugin+ neurons did not affect sleep, we considered the possibility that they were affected by sleep loss. Sleep is correlated with changes in neuronal activity in sleep-regulatory circuits, including the mushroom body, dFB, and R5 ellipsoid body (12, 17, 22, 23). For example, sleep-promoting dFB neurons tend to be more electrically active after sleep deprivation, when sleep pressure is high, than in rested flies (24). To determine whether sleep loss alters neuronal activity in hugin+ neurons, we measured intracellular Ca+ using CaLexA (calcium-dependent nuclear import of LexA) (25), which drives expression of GFP in response to sustained increases in Ca2+ levels. We used hugin-GAL4 to express CaLexA-GFP transgenes and UAS-CD8:RFP for normalizing the GFP signal. hugin > CaLexA-GFP; RFP flies were deprived of sleep for 9 h at the end of the night (zeitgeber time [ZT] 15 to 24) and subsequently collected for GFP measurements (Fig. 1A). A control group, flies that were not sleep deprived, was assayed at the same time of day as the deprived group. In the sleep-deprived flies, the CaLexA-dependent GFP signal in hugin+ neurons was lower than in control flies (Fig. 1 B–C). To rule out a general effect of sleep deprivation on Ca2+, we also tested whether sleep deprivation affects Ca2+ levels in Dh44+ neurons, another group of circadian output neurons (6). However, the CaLexA-GFP signal in Dh44+ neurons was not significantly different between the sleep-deprived and control flies (Fig. 1D). Thus, sleep deprivation specifically affects Ca2+ levels of hugin+ neurons, suggesting that the homeostat inhibits hugin+ neurons.
Fig. 1.
Ca2+ levels of hugin+ neurons are suppressed with sleep deprivation. (A) Sleep profiles of hugin > CaLexA-GFP; RFP flies subjected to no sleep deprivation (control, black, n = 8 flies) or 9-h sleep deprivation (SD, red, n = 8 flies). Sleep graphed as minutes per 30-min bin over 21 h. (B) Representative images show GFP reporting Ca2+ levels via the CaLexA system and RFP normalizer signals in hugin > CaLexA-GFP; RFP flies from control or SD groups. Maximum intensity projection images show hugin+ neurons in the subesophageal zone. (Scale bar, 25 µm.) (C) Levels of GFP signal normalized to RFP signal in hugin+ cell bodies from control (n = 21 flies) and SD (n = 18 flies) groups. **P = 0.000449, Welch’s t test. (D) Levels of GFP signal normalized to RFP signal in Dh44+ cell bodies from control (n = 11 flies) and SD (n = 11 flies) groups. n.s., P = 0.782 by Welch’s t test. Summary statistics are displayed as mean ± SEM.
To verify that the sleep-loss–induced decrease in Ca2+ in hugin+ neurons is not exclusive to mechanical sleep deprivation, we tested whether heat-induced sleep loss affects the activity of hugin+ neurons. We used the CaLexA system to measure Ca2+ in hugin > CaLexA-GFP; RFP flies after a full day at 30 °C. A control group, flies that were kept at a constant low temperature, was assayed at the same time of day as the heat-exposed group. As is typical for Drosophila at high temperatures, hugin > CaLexA-GFP; RFP flies slept less overall at 30 °C, exhibiting a slight sleep increase during the day with dramatic sleep loss (208 min) at night (26) (SI Appendix, Fig. S2A). We assayed calcium in the morning following the loss of sleep at night and found a decreased CaLexA-GFP signal (normalized by RFP) in hugin+ neurons (SI Appendix, Fig. S2B), showing that sleep loss due to heat also inhibits the activity of hugin+ neurons.
hugin+ Neurons Receive Projections from the Sleep-Promoting dFB.
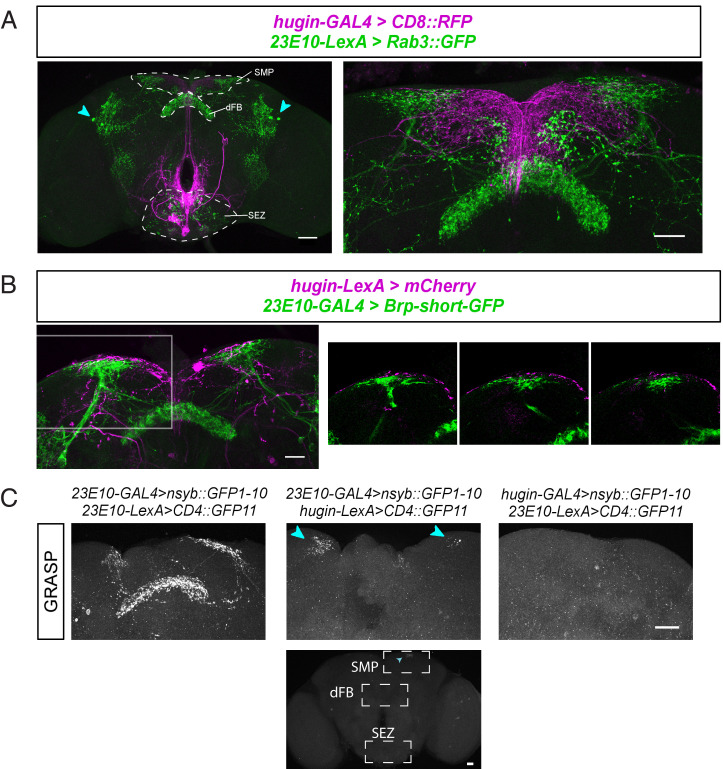
Given that hugin+ neurons are affected by sleep loss, we sought to determine whether they are connected to circuitry of the sleep homeostat. Based upon our previous findings that hugin+ neurons project back to the dorsal part of the fly brain (7), including the superior medial protocerebrum (SMP) in the vicinity of the dFB, we asked whether hugin+ neurons contact the dFB. Using the sleep-promoting 23E10-GAL4 driver (16, 24, 27, 28) to label dFB neuron dendrites and simultaneously using the LexA system to mark presynaptic sites of hugin+ neurons, we found that both sets of projections localize to the SMP (SI Appendix, Fig. S3). Published images also suggest presynaptic sites of dFB neurons in the SMP, although these are primarily localized in a single dorsal layer of the fan-shaped body (20, 28–30). Using 23E10-LexA to express Rab3::GFP, we confirmed the presence of presynaptic projections of 23E10+ neurons in the dFB and SMP, albeit with a weaker signal in the SMP (Fig. 2A). Additionally, we expressed brp-shortGFP, a nonfunctional 754-residue portion of bruchpilot (BRP) that localizes to presynaptic active zones (31, 32), in 23E10+ neurons. This presynaptic marker labeled projections in both the dFB and SMP (Fig. 2B).
Fig. 2.
Sleep-promoting dFB neurons contact hugin+ circadian output neurons. (A) Colabeling of hugin+ neurons with membrane marker (magenta) and 23E10+ dFB neurons with RAB3::GFP, a presynaptic marker (green). The Left image shows colabeling of neurons in the whole fly brain; arrowheads indicate 23E10+ cell bodies. SMP, dFB, and SEZ regions are labeled. The Right image shows the dorsal protocerebrum, where hugin+ projections intermingle with 23E10+ projections in the SMP. (B) Colabeling of hugin+ neurons with membrane marker (magenta) and 23E10+ dFB neurons with BRP-shortGFP, a presynaptic marker (green). The Left image shows colabeling of neurons in the dorsal brain. The Right series of images shows single confocal sections of the region indicated by a white box, where hugin+ projections intermingle with 23E10+ projections in the SMP. (C) Synaptic nSyb::spGFP1-10 is expressed in presynaptic neurons and complementary spGFP11 expressed in putative postsynaptic neurons. GFP reconstitution occurs only if synaptic connectivity exists. (C, Left) When both nSyb::spGFP1-10 and spGFP11 are expressed in 23E10+ dFB neurons, GFP reconstitution occurs in the dFB and SMP. (C, Middle, Top) Cyan arrowheads point to the GFP reconstitution in the SMP when nSyb::spGFP1-10 is expressed in 23E10+ dFB neurons and spGFP11 is expressed in hugin+ neurons. (C, Middle, Bottom) GFP is only reconstituted in the SMP (not dFB or SEZ) when nSyb::spGFP1-10 is expressed in 23E10+ dFB neurons and spGFP11 is expressed in hugin+ neurons. (C, Right) No GFP reconstitution when nSyb::spGFP1-10 is expressed in hugin+ neurons and spGFP11 is expressed in 23E10+ dFB neurons. (Scale bars: A, Left is 50 µm; A, Right, B, and C are 25 µm.)
To look for a possible synaptic connection between 23E10+ and hugin+ neurons, we used an assay called GFP Reconstitution Across Synaptic Partners (GRASP). This system uses the expression of a split GFP, one part tethered to neuronal Synaptobrevin (nSyb::spGFP1-10) in the putative presynaptic cells and the complement tethered to the membrane (CD4::spGFP11) of the putative postsynaptic neurons (33). As opposed to the original GRASP that did not label synapses specifically (34), trafficking of nSyb to the presynaptic vesicle membrane ensures that nSyb-GRASP identifies membrane contacts specifically at synapses and also indicates directionality of the contact. We first tested the nSyb-GRASP tool by coexpressing both presynaptic nSyb::spGFP1-10 and complementary CD4::spGFP11 in 23E10+ dFB neurons. In these flies, GFP reconstituted in both the dFB and SMP (Fig. 2 C, Left), confirming that 23E10+ dFB neurons have presynaptic sites in both these regions. In flies with the presynaptic nSyb::spGFP1-10 expressed in 23E10+ dFB neurons and complementary CD4::spGFP11 expressed in hugin+ neurons, fluorescent GFP reconstituted in the SMP, but not in the dFB or in the SEZ (Fig. 2 C, Top, Middle, and Bottom). We also performed the reciprocal experiment, with nSyb::spGFP1-10 expressed in the hugin+ neurons and complementary CD4::spGFP11 expressed in 23E10+ dFB neurons, but did not observe any GFP fluorescence in the brain (Fig. 2 C, Right), suggesting that hugin+ neurons do not signal to dFB cells. As GRASP signals can be difficult to detect, we cannot entirely exclude the possibility of a reciprocal connection, but these data clearly demonstrate that 23E10+ dFB neurons are presynaptic to hugin+ neurons in the SMP.
hugin+ Neurons Modulate Output of 23E10+ Sleep-Promoting dFB Neurons.
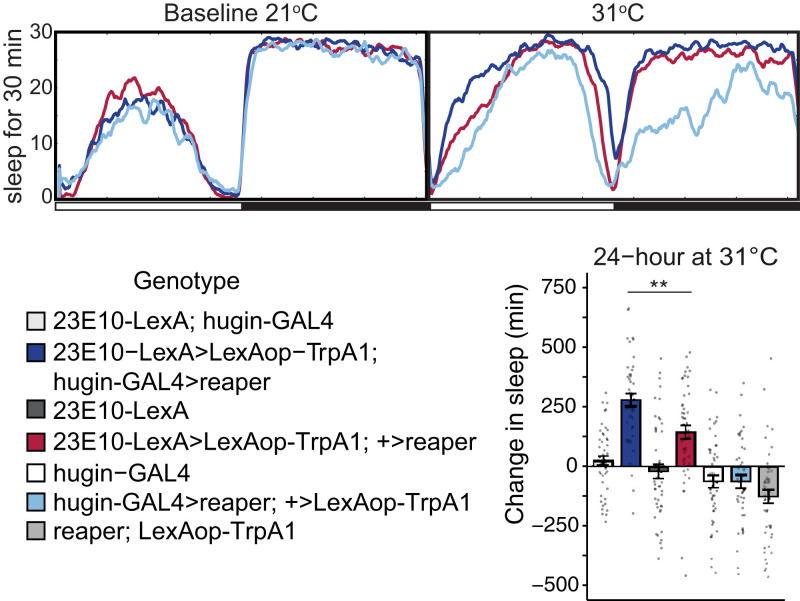
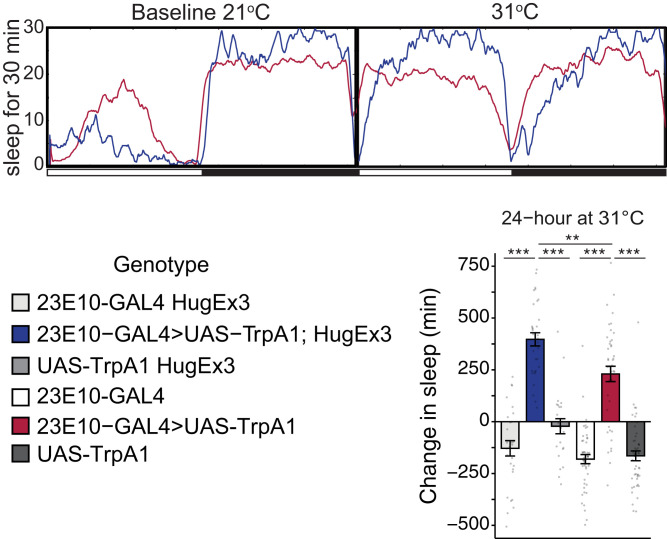
We next tested whether hugin+ neurons affect the sleep-promoting output of 23E10+ neurons, by activating 23E10+ dFB neurons in flies where hugin+ neurons are ablated (Fig. 3). We confirmed that expression of Reaper in hugin+ neurons ablated these neurons by the loss of coexpressing GFP signal (SI Appendix, Fig. S4A) and by monitoring behavioral phenotypes. Similar to our previous work where we inhibited hugin+ neurons (7), we found that ablation of hugin+ neurons significantly decreased morning activity and also resulted in a trend toward lower activity in the evening (ZT 6 to 12) (SI Appendix, Fig. S4B). Thermogenetic activation of the 23E10+ neurons using the LexA/LexAop system to drive two copies of TrpA1 (23E10-LexA > LexAop-TrpA1(2x); +> UAS-reaper) led to sleep increase during the day as reported previously (14, 15, 20) (Fig. 3). Activation of 23E10+ neurons in flies lacking hugin+ neurons (23E10-LexA > LexAop-TrpA1(2x); hugin-GAL4 > UAS-reaper) enhanced the typical increase in sleep, mostly during the daytime (Fig. 3 and SI Appendix, Fig. S5A). This result indicates that hugin+ neurons counter the output of 23E10+ neurons.
Fig. 3.
hugin+ neurons are modulators of 23E10+ sleep-promoting dFB neurons. Ablating hugin+ neurons leads to increased sleep after dFB activation. Sleep profiles and quantification of sleep during 23E10+ dFB neuron activation with TrpA1 in flies where hugin+ neurons were ablated using reaper (n = 42 to 47). The bar graphs quantify sleep changes with temperature-dependent (31 °C) activation of 23E10+ neurons, relative to sleep levels on the preactivation day. Key groups are shown in the sleep profiles (Top). Note that unless 23E10 neurons are activated, flies lose sleep at night at 31 °C (e.g., light blue profile). Sleep was significantly higher (**P = 0.0095) when hugin+ neurons were ablated (dark blue) compared to just activating 23E10+ neurons of the dFB (red). Experimental groups (dark blue, red) were also statistically different from all other controls shown. Circles are individual fly data points, and summary statistics are displayed as mean ± SEM. Means compared with one-way ANOVA and Tukey’s test.
Mutations in the hugin Locus Affect Sleep Rebound and the Sleep-Promoting Output of 23E10+ dFB Neurons.
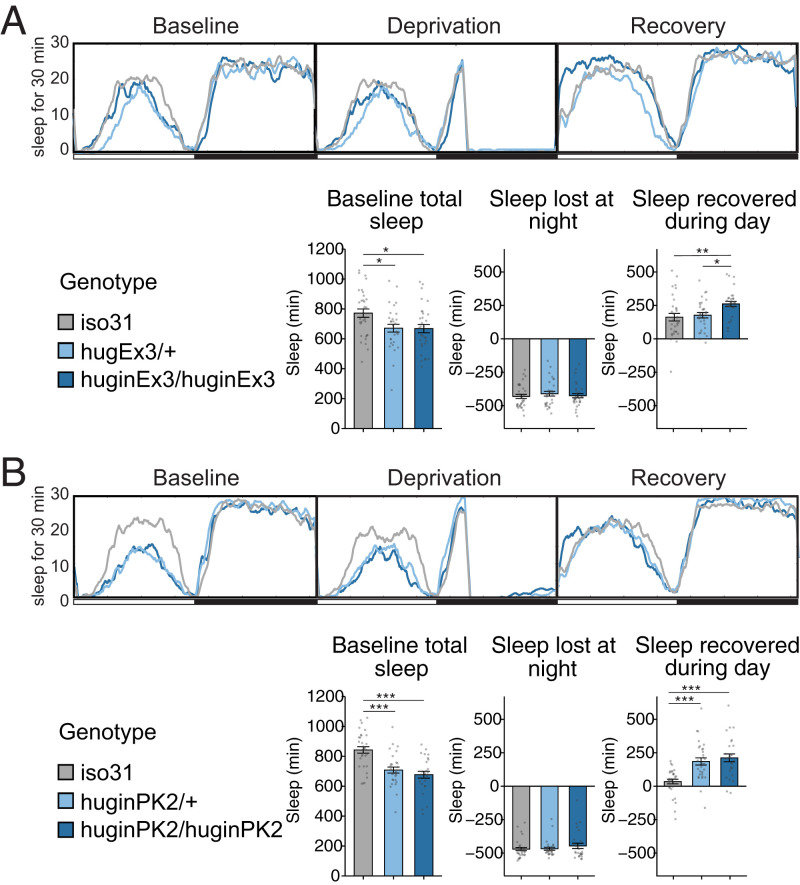
To determine whether the ability of hugin+ neurons to affect the sleep homeostat is dependent on Hugin peptide signaling, we used CRISPR-CAS9 to produce mutant alleles of hugin that affect expression of one or both of its encoded neuropeptides, Hugin-γ and Pyrokinin 2 (PK2) (35). The huginPK mutant contains a 1-base pair (bp) deletion that truncates the PK2 peptide, while the huginΔEx3 mutant lacks the majority of exon 3 and is predicted to eliminate both neuropeptides. We backcrossed each CRISPR line for five generations to exclude potential off-target mutations and then tested the flies for circadian and sleep behavior. The huginPK2 mutants showed weaker circadian rhythms, while the huginΔEx3 flies showed a small lengthening of circadian period (SI Appendix, Table S1); thus, period effects may arise from the Hugin peptide, and it is possible that weaker rhythms of PK2 flies reflect dominant negative effects of the truncated peptide predicted for this allele. Interestingly, both hugin mutations produced a dominant but small decrease in baseline sleep, which is likely a developmental effect (Fig. 4 A and B). Since our data support the idea that hugin+ neurons respond to the sleep homeostat, we measured rebound sleep in both mutants after mechanical deprivation of sleep for 9 h at the end of the night (ZT 15 to 24). We found that both mutants exhibited increased rebound sleep (Fig. 4 A and B), indicating that outputs of hugin+ neurons modulate recovery sleep. Increased rebound sleep in huginΔEx3 mutants manifested as a recessive trait, which is typical of loss-of-function rebound mutants (24, 28). However, huginPK2 flies showed a dominant effect, again, perhaps because of dominant negative effects of this allele.
Fig. 4.
Hugin peptide signaling regulates baseline sleep and circadian rhythms. Hugin CRISPR mutants, huginΔEx3 (A) and huginPK (B), were assayed for baseline sleep as well as during and after mechanical sleep deprivation from ZT 15 to 24. Recovery sleep is plotted as the difference in 12 h sleep on the day after mechanical sleep deprivation relative to sleep amount on the predeprivation day. For both panels: n = 27 to 32 flies. Circles are individual fly data points, and summary statistics are displayed as mean ± SEM. Means were compared using one-way ANOVA and Tukey’s test. *P < 0.05, **P < 0.01, ***P < 0.001.
We next tested whether the Hugin peptide affects the sleep-promoting output of 23E10+ neurons, by activating 23E10+ dFB neurons in Hugin CRISPR mutant flies. Thermogenetic activation of the 23E10+ neurons using the GAL4/UAS system to drive TrpA1 (23E10-GAL4 > UAS-TrpA1) led to sleep increase (Fig. 5) as seen previously in Fig. 3. Activation of 23E10+ neurons in flies lacking Hugin peptide (23E10-GAL4 > UAS-TrpA1 Ex3) enhanced the typical increase in sleep, primarily during the day (Fig. 5 and SI Appendix, Fig. S5B), as seen also with ablation of hugin+ neurons. This result indicates that Hugin peptide signaling counters the output of 23E10+ neurons.
Fig. 5.
hugin CRISPR mutation affects sleep promotion by 23E10+ dFB neurons. Activating the dFB in Hugin CRISPR mutants, huginΔEx3 leads to increased sleep. Sleep profiles and quantification of sleep during 23E10+ dFB neuron activation with TrpA1 in flies with homozygous CRISPR mutations at the hugin locus (n = 28 to 48). The bar graphs quantify sleep changes with temperature-dependent (31 °C) activation of 23E10+ neurons, relative to sleep levels on the preactivation day. Sleep was significantly higher (P = 0.0023) in Hugin CRISPR mutants (dark blue) compared to just activating the dFB (red). Experimental groups (dark blue, red) were statistically different from all other controls shown. Circles are individual fly data points, and summary statistics are displayed as mean ± SEM. Means compared with one-way ANOVA and Tukey’s test. **P < 0.01, ***P < 0.001. HugEx3 in genotypes denotes a homozygous CRISPR deletion.
hugin Mediates Responses of Pdf+ Clock Neurons to Sleep Loss.
We previously showed that hugin+ neurons project into the ventral nerve cord (VNC) (7). To identify additional targets of these neurons, we used trans-Tango, a pan-neuronal transsynaptic labeling system in which a tethered ligand at synapses activates tdTomato expression in postsynaptic partners (36). Presynaptic neurons are simultaneously labeled with myr::GFP, a different fluorescent protein. We expressed the trans-Tango ligand in hugin+ neurons and observed trans-Tango–dependent signal in many brain regions, including the pars intercerebralis, mushroom body lobes, mushroom body calyx and pedunculus, SMP, subesophageal zone, and accessory medulla (Fig. 6 A, Top). The postsynaptic neurons with cell bodies in the accessory medulla and projections to the superior medial protocerebrum were reminiscent of small ventrolateral clock neurons (s-LNvs) (Fig. 6 A, Top Right, magenta, arrowheads). s-LNvs secrete the peptide Pigment Dispersing Factor (PDF), prompting us to label for PDF peptide and confirm that a subset of the postsynaptic partners observed in hugin > trans-Tango flies is PDF positive. Pdf+ neurons include the large ventrolateral neurons (l-LNv), which were also positive for the trans-Tango-dependent signal, but showed weaker signal than the s-LNvs (Fig. 5 A, Bottom), indicating that s-LNvs are primary targets of hugin+ neurons. To confirm the synaptic connection between hugin+ and PDF+ neurons, we used GRASP as above. Expression of synaptic nrx::spGFP1-10 in hugin+ neurons and spGFP11 in PDF+ neurons resulted in GFP reconstitution in the SMP, superior lateral protocerebrum (SLP), and posterior lateral protocerebrum (PLP) (Fig. 6B). Thus, two independent techniques identified a synaptic connection localized to the same part of the brain, indicating that hugin+ neurons indeed project to Pdf+ neurons.
Fig. 6.
hugin+ neurons target PDF-expressing clock neurons, which show decreased Ca2+ levels upon sleep deprivation. (A) Use of trans-Tango to map projections of hugin+ neurons (green). Trans-Tango system reveals the synaptic partners (magenta) of hugin+ neurons in the brain. (A, Top) Image is a maximum intensity projection from the anterior side, and arrowheads indicate postsynaptic signal that resembles the projections of PDF+ neurons. (A, Bottom) Colabeling of PDF peptide (green) and postsynaptic signal (magenta) in flies with trans-Tango ligand expressed in hugin+ neurons. PDF+ s-LNvs are postsynaptic to hugin+ neurons. s-LNv, small ventrolateral neurons; l-LNv, large ventrolateral neurons. (Scale bars in A: Top, 50 μm; Bottom, 15 μm.) (B) GRASP to assay connectivity between hugin+ neurons and LNvs. Synaptic nrx::spGFP1-10 is expressed in presynaptic neurons and complementary spGFP11 is expressed in putative postsynaptic neurons. GFP reconstitution occurs only if synaptic connectivity exists. Expression of nrx::spGFP1-10 in hugin+ neurons and spGFP11 in PDF+ neurons resulted in GFP reconstitution (white arrows) in the SMP, SLP, and PLP. (Scale bar, 50 μm.) (C) Representative images of l-LNvs or s-LNvs from a Pdf > CaLexA-GFP; RFP fly in control or SD group. Top row shows merged images of GFP signal reporting Ca2+ levels with the CaLexA system and RFP normalizer signal. Bottom row shows “fire” pseudocolor image of CaLexA-GFP signal (blue/purple = low intensity and yellow/white = high intensity). (Scale bar, 10 µm for all images in this panel.) (D) Pdf > CaLexA-GFP; RFP flies were subjected to no sleep deprivation (control, gray) or 9-h sleep deprivation (SD, red). Graph shows GFP levels normalized to RFP levels in Pdf+ l-LNv or s-LNv from control (n = 18 flies) and SD (n = 19 flies) groups. Summary statistics are displayed as mean ± SEM **P = 0.00910, ***P = 0.000655 by Welch’s t test.
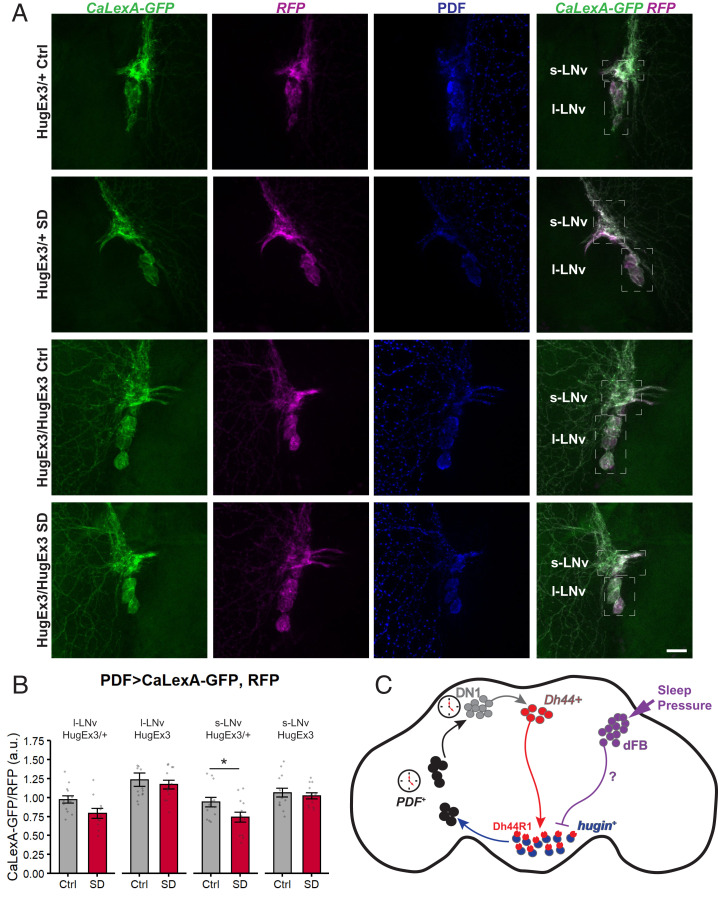
Our data identify a circuit that links sleep homeostasis centers to circadian clock neurons (23E10+ dFB → hugin+ SEZ → Pdf+ s-LNvs), and suggest a potential mechanism for homeostatic components to regulate circadian clock outputs. To test whether the activity of Pdf+ neurons themselves is altered with sleep deprivation, we used CaLexA to measure Ca2+ level changes in Pdf+ neurons during sleep deprivation. With mechanical sleep deprivation, the CaLexA-GFP signal in both Pdf+ s-LNv and l-LNv cell bodies was lower in the sleep-deprived flies as compared to controls (Fig. 6 C and D). We next tested whether the Ca2+ decrease in Pdf+ neurons after deprivation is dependent on Hugin peptide signaling. By comparing the activity of Pdf+ neurons in control and sleep deprivation conditions, we found that in s-LNvs, sleep deprivation resulted in a decrease in Ca2+ levels in the control huginΔEx3/+ group, but not in huginΔEx3/ΔEx3 mutants (Fig. 7 A and B). This result is of particular interest since the trans-Tango experiment suggests that s-LNvs are primary targets of hugin+ neurons. While sleep deprivation did not significantly affect Ca2+ levels in l-LNvs of hugin mutants or huginΔEx3/+ controls, we suspect that the large area of the large neurons dilutes changes in calcium, so we cannot definitively say that only s-LNvs show consistent responses to sleep loss. Together these data indicate that sleep deprivation acts through Hugin peptides to suppress the activity of LNvs.
Fig. 7.
Hugin peptide mediates the decrease in Ca2+ levels of Pdf-expressing clock neurons after sleep deprivation. (A) Representative images of l-LNvs and s-LNvs from a Pdf > CaLexA-GFP; RFP flies in control or SD group dissected at ZT 0. The fourth column shows merged images of GFP signal reporting Ca2+ levels with the CaLexA system and RFP normalizer signal. Note the lower green signal in the huginΔEx3/+ heterozygous fly subjected to SD (second row) compared to the huginΔEx3/+ heterozygous control fly (Top row). (Scale bar, 25 µm for all images in this panel.) (B) Ca2+ levels in Pdf+ neurons were measured with Pdf > CaLexA-GFP; RFP reporter in huginΔEx3/+ heterozygous or huginΔEx3/ΔEx3 mutant flies that were unperturbed (control, gray) or subjected to 9-h sleep deprivation (SD, red). Graph shows GFP levels normalized to RFP levels in Pdf+ l-LNvs or s-LNvs (n = 13 or 14 flies per group). l-LNv: Two-way ANOVA (genotype × condition) revealed a significant main effect of genotype [F(1, 47) = 23.29, P < 0.0001], a nonsignificant effect of condition [F(1, 47) = 3.68, P = 0.061], and a nonsignificant interaction between factors [F(1, 47) = 0.604, P = 0.441]. s-LNv: Two-way ANOVA revealed a significant main effect of genotype [F(1, 49) = 12.47, P = 0.0009], a significant effect of condition [F(1, 49) = 4.36, P = 0.042], and a nonsignificant interaction between factors [F(1, 49) = 1.852, P = 0.180]. *P = 0.0385, significantly different by Sidak’s multiple comparison test. Summary statistics are displayed as mean ± SEM. (C) Proposed model for regulation of a circadian output circuit by sleep homeostatic drive. Sleep drive triggered by prolonged wakefulness suppresses locomotor activity by decreasing firing of hugin+ neurons (blue) and, through them, also firing of PDF+ clock neurons (black). We suggest that effects of the sleep homeostat on hugin+ neurons are mediated by projections of 23E10+ dFB neurons (purple).
Discussion
The circadian clock and homeostat both regulate sleep, but it is not clear how the two processes functionally interact. We show here that circadian and sleep circuits intersect at circadian output neurons that secrete the Hugin peptide. Homeostatic sleep drive signals through hugin+ neurons to suppress circadian outputs, thereby allowing for sleep at times when the circadian system typically promotes wake. We also find that hugin+ circadian output neurons feedback to s-LNvs, the central clock neurons. Thus, the sleep homeostat influences outputs of the circadian clock by modulating the activity of circadian output neurons and clock neurons (Fig. 7C).
Previously, we showed that a circuit from s-LNvs → DN1 → Dh44+ neurons → hugin+ neurons controls rhythms of locomotor activity. hugin+ neurons promote locomotion in the morning and evening, but especially in the evening during the day-to-night transition (6, 7). Activation or inhibition of these neurons does not affect sleep, but Hugin peptides affect the homeostatic response to sleep deprivation. As for Hugin peptides, the major phenotype of the dFB 23E10+ neurons is in the context of sleep loss. Given that 23E10+ neurons project to hugin+ neurons and are activated by sleep loss while hugin+ neurons show decreased activity (24), we suggest that 23E10+ neurons inhibit hugin+ neurons. This would also explain why ablation of hugin+ neurons enhances sleep-promoting effects of 23E10+ activation. We were unable to reliably detect inhibition of hugin+ neurons by 23E10+ neurons through optogenetic or other stimulus–response experiments, but there are several reasons why this is technically challenging. Based on the GRASP data (Fig. 2C) we know that dFB presynaptic sites synapse onto hugin+ neurons in the SMP, which likely represents only a subset of hugin+ neurons as the population is heterogeneous and also projects to other regions such as the VNC. hugin+ neuron presynaptic sites, rather than dendrites, are located in the SMP region (7), suggesting that the connection is an inhibitory axo-axonic synapse. Axo-axonic synapses are more common than previously reported, according to the recently published Drosophila connectome (37), and inhibitory neuromodulation at such synapses may control neuromodulator/neuropeptide release locally without concomitant changes in the cell body (38). So, the fact that we did not immediately detect hyperpolarization in hugin cell bodies using a P2X2/GCaMP approach does not preclude the possibility of inhibition. On the other hand, the CaLexA signal we report in hugin neurons of sleep-deprived brains represents integration of calcium levels over time. Regardless of whether the dFB is their major source of homeostatic input, it is clear that hugin+ neurons respond to sleep loss. Our data suggest that during sleep deprivation, the homeostat not only generates sleep drive but also actively disengages activity-promoting circuits. As noted above, both the dFSB and hugin+ neurons are most relevant in the context of sleep loss when homeostatic need is high.
Our work demonstrates that sleep homeostasis also influences the activity of circadian clock neurons, LNvs. The LNvs are considered wake promoting, with the predominant effect being light dependent and deriving from the l-LNv subset (39–41). While s-LNvs alone are not sufficient to promote wake, down-regulation of PDF receptor in s-LNvs increases sleep, suggesting that PDF signaling to s-LNvs modulates wake-promoting effects of l-LNvs (40, 41). In addition, the down-regulation of short Neuropeptide F signaling between s-LNvs and l-LNvs decreases nighttime sleep, suggesting that some outputs of the LNvs also promote sleep (42). Notably, both s-LNvs and l-LNvs show more depolarized resting membrane potentials during the day than during the night, supporting the idea that LNvs are more active during times of increased arousal (43, 44). Targets of the s-LNvs, the DN1 neurons are another example of circadian neurons that affect sleep through the central complex, in this case the ellipsoid body (45, 46). Additionally, another subset of clock neurons, LPNAstA neurons, projects to the dFB to promote sleep and could serve as a mechanism of circadian control of the sleep homeostat (20).
An increase in sleep drive caused by social enrichment was previously associated with an increased number of synapses in the LNv projections into the medulla, a brain region that processes visual information from the eyes (47). Here, we report Ca2+ levels in LNvs decrease with sleep deprivation, which we hypothesize dampens the wake-promoting effects of LNvs to allow for recovery sleep. It is possible that decreased Ca2+ levels in LNvs with sleep deprivation precede synaptic downscaling that is thought to occur with recovery sleep (47). Bushey et al. suggested potentiation of s-LNv activity by sleep loss, based upon increased vesicles and PDF expression in s-LNV axon terminals (48); however, this study did not demonstrate changes in synaptic release. Given that the number of total vesicles at the axon terminal is not directly correlated with the number of primed vesicles or the probability of release (49), those experiments were not necessarily indicative of high calcium. hugin mutants do not exhibit a decrease in s-LNv Ca2+ levels with sleep deprivation, which suggests that Hugin peptide signaling is involved in the suppression of s-LNv activity with sleep deprivation. While the hugin mutant phenotype of increased s-LNv calcium in conjunction with increased rebound sleep might seem antithetical, higher calcium in the s-LNvs likely does not translate into increased locomotor activity in these mutants as hugin+ neurons are a conduit for transmission of s-LNV signals (7). In addition to the s-LNv-hugin-dFB link we report here, other sleep homeostat pathways may also modulate hugin+ neurons and/or LNvs. Notably, GABA and myoinhibitory peptide signal to LNvs to regulate sleep, although the source of these neuromodulators is not known yet (41, 50, 51).
The mammalian ortholog of Hugin, Neuromedin U (NMU) (35), is also implicated in sleep regulation. While NMU injection in rats does not change total sleep time, it changes sleep architecture (52). NMU overexpression decreases sleep in zebrafish (53), which is consistent with our findings that hugin+ neurons promote wake. Chiu et al. (53) did not report alterations in baseline sleep levels in their NMU knockout fish, and, as noted above, we believe that decreased daily sleep in hugin mutants represents a developmental effect. Following sleep deprivation at night, hugin mutants show increased rebound in the morning, supporting the idea that Hugin opposes increases in sleep at this time of day. Interestingly, NMU overexpression in zebrafish increases the duration of stimulus-evoked arousal during sleep. To the extent that increased arousal reflects decreased sleep drive, this indicates a conserved role for Hugin/NMU in regulating sleep.
In Drosophila, influences of the sleep homeostat on the circadian system have previously not been demonstrated. In rodents, sleep deprivation dampens electrical activity in the suprachiasmatic nucleus (SCN) for up to 7 h and reduces the ability of the circadian clock to phase shift by light (1, 54–56). We find a similar effect in flies, where neuronal activity is depressed in LNv central clock neurons. In the rodent model, the mechanism mediating the reduced SCN activity is not clear, but may involve serotonin signaling from the raphe dorsalis (57). We show here that effects of sleep deprivation on LNvs are mediated by Hugin signaling from output neurons that connect to a major sleep homeostatic locus. Importantly, sleep deprivation does not appear to shift rest:activity patterns in flies (58), nor does it affect clock gene expression in the rodent SCN (59), suggesting that timekeeping by the clock is relatively unperturbed. Therefore, sleep homeostasis appears to primarily influence clock outputs.
Methods
D. melanogaster.
Flies were maintained on cornmeal–molasses medium. For thermogenetic and trans-Tango experiments, flies were raised at 18 °C, and all other flies were maintained at 25 °C. w1118 iso31 strain was used as the wild-type strain. For sleep behavior experiments, transgenic lines were backcrossed into the iso31 genetic background. For controls, UAS and GAL4 fly lines were tested as heterozygotes after crossing to iso31. The following flies were from the Bloomington Drosophila Stock Center (BDSC): 23E10-GAL4 (#49032) (60), 23E10-LexA (#52693) (61), Hugin-GAL4 (#58769) (62), Hugin-LexA (#52715), Dh44-GAL4 (#39347), UAS-CD8::RFP (#32219), LexAop-Rab3::GFP (#52239) (63), LexAop-6xmCherry-HA (#52271), UAS-nSyb::GFP1-10, LexAop-CD4::GFP11 (#64314), and UAS-reaper (#5773) (64). LexAop-CD4-spGFP11; UAS-nrx-spGFP1-10 was a gift from N. Shah (65). Trans-Tango fly was a gift from G. Barnea (36). CaLexA fly was a gift from J. W. Wang (25). PDF-GAL4 was a gift from J. Hall (66). UAS-TrpA1 was a gift from L. C. Griffith (18). UAS-shibirets (20XUAS-IVS-Shibire[ts1]-p10-INS) and LexAop-TrpA1 (chromosome 2) were gifts from G. Rubin (67). LexAop-TrpA1 (chromosome 3) was a gift from S. Waddell (68). The LexAop-TrpA1; LexAop-TrpA1 fly line was created by H. Toda (A.S. laboratory).
Generation of hugin Mutants.
huginΔEx3 and huginPK2 mutants were generated with the CRISPR-CAS9 system. The guide RNA sequences for generating huginΔEx3 were 5′-GGGAGCCCGCTTATCGCGTG-3′ and 5′-GGAGGACGGAGGACGAGCCC-3′, and the guide RNAs for huginPK2 were 5′-GTGCCGTTCAAGCCACGCCT-3′ and 5′-GGCAAACGTGCTCAAGTGTG-3′. Guide RNAs were cloned into the pCFD4 plasmid (69), and the plasmids encoding the guide RNAs were injected into vasa-Cas9 flies (BDSC #51323) (70) at Rainbow Transgenic Flies, Inc. Mutations in the F1 generation were identified with PCR screening and confirmed with Sanger sequencing. huginΔEx3 is a 260-bp deletion (dm6/chr3R: 12,528,601 to 12,528,860), and huginPK2 is a 1-bp deletion (dm6/chr3R: 12,528,750). The mutant alleles were backcrossed for five generations into the iso31 background.
Immunohistochemistry.
For polarity labeling, CaLexA experiments, and reaper confirmation ∼7-d-old females raised at 25 °C were used. For trans-Tango experiments, ∼15- to 20-d-old females raised at 18 °C were used, as previously described (36). All fly brains were dissected in phosphate-buffered saline with 0.1% Triton-X (PBST) and fixed with 4% formaldehyde in PBS for 20 min at room temperature. Brains were rinsed 3 times, 10 min per wash, with PBST, blocked in 5% normal goat serum in PBST (NGST) for 60 min, and incubated in primary antibody diluted in NGST for >16 h at 4 °C. Brains were rinsed 3 × 10 min in PBST, incubated 2 h in secondary antibody diluted in NGST, rinsed 3 times,10 min per wash, in PBST, and mounted with Vectashield media (Vector Laboratories Inc.). For the reaper confirmation experiment, both primary and secondary antibody incubation was for two nights at 4 °C. Primary antibodies used were: rabbit anti-GFP at 2 µg/mL (Thermo Fisher Scientific Inc. A-11122), rat anti-RFP at 1 µg/mL (ChromoTek 5F8), mouse anti-BRP at 1:100 (Developmental Studies Hybridoma Bank nc82), rat anti-HA at 1 µg/mL (Roche clone 3F10), and mouse anti-PDF at 0.3 µg/mL (Developmental Studies Hybridoma Bank c7-c). Secondary antibodies were from Thermo Fisher Scientific and used at 1:1,000: Alexa Fluor 488 goat anti-rabbit, Alexa Fluor 555 goat anti-rat, Alexa Fluor 647 goat anti-rat, and Alexa Fluor 647 goat anti-mouse.
GRASP.
nSyb-GRASP flies were dissected in extracellular saline [103 mM NaCl, 3 mM KCl, 1 mM NaH2PO4, 4 mM MgCl2, 10 mM D-(+)-trehalose dehydrate, 10 mM D-(+)-glucose, 5 mM N-Tris(hydroxymethyl) methyl-2-aminoethane sulfonic acid, 26 mM NaHCO3, pH 7.4]. Dissected brains were exposed to a high concentration of KCl to increase GRASP signal, as previously described (33). Dissected brains were incubated in 1 mL 70 mM KCl in saline three times (∼5 s per KCl incubation), alternating with 1 mL saline (∼5 s per wash), and then transferred to 1 mL saline to incubate for 10 min. Brains were fixed with 4% formaldehyde in PBS for 20 min at room temperature, rinsed 3 times, 10 min per wash, in PBST, and mounted with Vectashield media. nrx-GRASP brains were processed using the same protocol except without the KCl washes. Endogenous GRASP signal without antibody labeling was imaged.
Confocal Microscopy.
Eight-bit images were acquired using a Leica TCS SP5 laser scanning confocal microscope with a 40×/1.3 numerical aperture (NA) or 20×/0.7 NA objective and a 1-μm or 2-μm z-step size. Maximum intensity z-projection and sum intensity z-projection (reaper confirmation only) images were generated in Fiji, a distribution of ImageJ software (71).
Sleep Behavior Assay.
Individual ∼7-d-old female flies were loaded into glass tubes containing 5% sucrose and 2% agar. Locomotor activity was monitored with the Drosophila activity monitoring system (DAMS) (Trikinetics). Flies were monitored for sleep in a 12 h:12 h (12:12) light:dark cycle at 25 °C for CaLexA experiments or at 21 °C for thermogenetic experiments. Incubator temperature shifts occurred at lights-on, ZT 0. For mechanical sleep deprivation experiments, flies were loaded into the DAMS and sleep deprived during the night by shaking on an adapted vortex for 2 s randomly within every 20-s interval. Sleep was defined as 5 consecutive minutes of inactivity. Sleep analysis was performed with PySolo software (72). Data from flies that survived the duration of the experiments were pooled and analyzed. Behavioral data were analyzed with one-way analysis of variance (ANOVA) with Tukey’s test as the post hoc test to compare means between groups. Differences between groups were considered significant if P < 0.05 by the post hoc test.
Circadian Rest:Activity Rhythms Behavior Assay.
Rest:activity rhythm assays were performed and analyzed as described by King et al. in ref. 7. Flies were entrained to a 12 h light:12 h dark (LD) cycle from birth and put in total darkness for analysis at ∼7 d old. Data from days 3 to 9 for flies that survived the duration of the experiment were analyzed. Period and 24 h fast Fourier transform (FFT) were analyzed by one-way ANOVA with Tukey’s test in Prism 8 for Mac Os X.
CaLexA Analysis.
Fluorescence intensity measurement was performed in Fiji. Regions of interest (ROIs) were manually drawn to encompass individual RFP-positive cell bodies, and mean pixel intensities of RFP and GFP were measured from the ROI. For each cell, the CaLexA-GFP/RFP signal (arbitrary unit [a.u.]) was calculated as a ratio between the mean pixel intensities of GFP and RFP. For each brain, the average CaLexA-GFP/RFP signal of a cell is a sample point. Welch’s t test was used to compare differences in CaLexA-GFP/RFP signal between sleep-deprived and control groups. The two-way ANOVA was used to compare differences between groups that were split into genotype and conditions, and the Sidak’s multiple comparison post hoc test was used for pairwise comparisons.
Statistical Analysis.
The statistical details of experiments can be found in the figure legends. All statistical tests were performed in R (version 3.3.1) expect for the circadian data and activity traces, which were performed in Prism 8 for Mac Os X. Graphs were generated in R using the ggplot2 package, except for sleep profiles, which were generated in Pysolo.
Supplementary Material
Acknowledgments
We thank Drs. Gilad Barnea (Brown University, Providence, RI), Leslie Griffith (Brandeis University, Waltham, MA), Gerald Rubin (Janelia Research Campus, Ashburn, VA), Jeffery Hall (Brandeis University, Waltham, MA), Nirao Shah (Stanford University, Stanford, CA), Scott Waddell (Oxford University, Oxford, UK), and Jing Wang (UC San Diego, La Jolla, CA) for generously providing fly lines. Also, we used stocks from the Bloomington Drosophila Stock Center (NIH P40OD018537). We thank Zhifeng Yue, Kiet Luu, and Juliana T. Choi for assistance with experiments and Michael Nusbaum for helpful discussions. Parts of this manuscript were included in the doctoral dissertation of A.N.K. (73). The work was supported by NIH grant R37NS048471 (to A.S.). J.E.S. was supported by a training grant in neuroscience (NIH T32-NS105607), an NIH diversity supplement (NIH NS48471), and by a grant to the University of Pennsylvania from the HHMI through the James H. Gilliam Fellowship for Advanced Study program. A.N.K. was supported in part by a training grant in genetics (NIH T32GM008216) and a National Science Research Award fellowship (NIH F31NS100395). C.T.H. was supported by a training grant in age-related neurodegenerative diseases (NIH T32AG00255).
Footnotes
Reviewers: R.A., Northwestern University; and M.S.-H., Columbia University Irving Medical Center.
Competing interest statement: The principal investigator, A.S., has co-authored two publications with reviewer Ravi Allada: The first was a book chapter, which was submitted in January 2017 and published in August 2017; thus, it was more than 48 m ago (https://pubmed.ncbi.nlm.nih.gov/28432135/). The second was a paper led by John Hogenesch that laid out guidelines for circadian rhythms. This paper had >90 co-authors, and Ravi Allada and A.S. had no direct contact regarding this paper, which was published in October 2017 (https://pubmed.ncbi.nlm.nih.gov/29098954/). Importantly, A.S. and Ravi Allada have never collaborated on a research project.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111183118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Borbély A. A., Daan S., Wirz-Justice A., Deboer T., The two-process model of sleep regulation: A reappraisal. J. Sleep Res. 25, 131–143 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Dubowy C., Sehgal A., Circadian rhythms and sleep in Drosophila melanogaster. Genetics 205, 1373–1397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlichting M., Díaz M. M., Xin J., Rosbash M., Neuron-specific knockouts indicate the importance of network communication to Drosophila rhythmicity. eLife 8, e48301 10.7554/eLife.48301. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delventhal R., et al. , Dissection of central clock function in Drosophila through cell-specific CRISPR-mediated clock gene disruption. eLife 8, e48308 10.7554/eLife.48308. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King A. N., Sehgal A., Molecular and circuit mechanisms mediating circadian clock output in the Drosophila brain. Eur. J. Neurosci. 51, 268–281 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavanaugh D. J., et al. , Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell 157, 689–701 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King A. N., et al. , A peptidergic circuit links the circadian clock to locomotor activity. Curr. Biol. 27, 1915–1927 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai L., et al. , A conserved circadian function for the neurofibromatosis 1 gene. Cell Rep. 22, 3416–3426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavey M., Collins B., Bertet C., Blau J., Circadian rhythms in neuronal activity propagate through output circuits. Nat. Neurosci. 19, 587–595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joiner W. J., Crocker A., White B. H., Sehgal A., Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757–760 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Pitman J. L., McGill J. J., Keegan K. P., Allada R., A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441, 753–756 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Sitaraman D., et al. , Propagation of homeostatic sleep signals by segregated synaptic microcircuits of the Drosophila mushroom body. Curr. Biol. 25, 2915–2927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donlea J. M., Neuronal and molecular mechanisms of sleep homeostasis. Curr. Opin. Insect Sci. 24, 51–57 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Donlea J. M., Thimgan M. S., Suzuki Y., Gottschalk L., Shaw P. J., Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332, 1571–1576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueno T., et al. , Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat. Neurosci. 15, 1516–1523 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Qian Y., et al. , Sleep homeostasis regulated by 5HT2b receptor in a small subset of neurons in the dorsal fan-shaped body of drosophila. eLife 6, e26519 10.7554/eLife.26519. (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S., Liu Q., Tabuchi M., Wu M. N., Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165, 1347–1360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pulver S. R., Pashkovski S. L., Hornstein N. J., Garrity P. A., Griffith L. C., Temporal dynamics of neuronal activation by Channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J. Neurophysiol. 101, 3075–3088 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitamoto T., Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J. Neurobiol. 47, 81–92 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Ni J. D., et al. , Differential regulation of the Drosophila sleep homeostat by circadian and arousal inputs. eLife 8, e40487 10.7554/eLife.40487. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubowy C., et al. , Genetic dissociation of daily sleep and sleep following thermogenetic sleep deprivation in Drosophila. Sleep (Basel) 39, 1083–1095 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bushey D., Tononi G., Cirelli C., Sleep- and wake-dependent changes in neuronal activity and reactivity demonstrated in fly neurons using in vivo calcium imaging. Proc. Natl. Acad. Sci. U.S.A. 112, 4785–4790 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap M. H. W., et al. , Oscillatory brain activity in spontaneous and induced sleep stages in flies. Nat. Commun. 8, 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donlea J. M., Pimentel D., Miesenböck G., Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81, 860–872 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuyama K., Zhang Y., Rao Y., Wang J. W., Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J. Neurogenet. 26, 89–102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parisky K. M., Agosto Rivera J. L., Donelson N. C., Kotecha S., Griffith L. C., Reorganization of sleep by temperature in Drosophila requires light, the homeostat, and the circadian clock. Curr. Biol. 26, 882–892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pimentel D., et al. , Operation of a homeostatic sleep switch. Nature 536, 333–337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donlea J. M., et al. , Recurrent circuitry for balancing sleep need and sleep. Neuron 97, 378–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W., et al. , Morphological characterization of single fan-shaped body neurons in Drosophila melanogaster. Cell Tissue Res. 336, 509–519 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Cavanaugh D. J., Vigderman A. S., Dean T., Garbe D. S., Sehgal A., The Drosophila circadian clock gates sleep through time-of-day dependent modulation of sleep-promoting neurons. Sleep (Basel) 39, 345–356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid A., et al. , Activity-dependent site-specific changes of glutamate receptor composition in vivo. Nat. Neurosci. 11, 659–666 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Fouquet W., et al. , Maturation of active zone assembly by Drosophila Bruchpilot. J. Cell Biol. 186, 129–145 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macpherson L. J., et al. , Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat. Commun. 6, 10024 10.1038/ncomms10024. (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feinberg E. H., et al. , GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Melcher C., Bader R., Walther S., Simakov O., Pankratz M. J., Neuromedin U and its putative Drosophila homolog hugin. PLoS Biol. 4, e68 10.1371/journal.pbio.0040068. (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talay M., et al. , Transsynaptic mapping of second-order taste neurons in flies by trans-Tango. Neuron 96, 783–795 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheffer L., et al. , A connectome and analysis of the adult drosophila central brain. bioRxiv [Preprint] 1–83 (2020). 10.1101/2020.04.07.030213. [DOI] [PMC free article] [PubMed]
- 38.Cover K. K., Mathur B. N., Axo-axonic synapses: Diversity in neural circuit function. J. Comp. Neurol. 529, 2391–2401 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheeba V., et al. , Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr. Biol. 18, 1537–1545 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang Y., Griffith L. C., Rosbash M., Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc. Natl. Acad. Sci. U.S.A. 105, 19587–19594 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parisky K. M., et al. , PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60, 672–682 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang Y., et al. , Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron 80, 171–183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao G., Nitabach M. N., Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J. Neurosci. 28, 6493–6501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheeba V., Gu H., Sharma V. K., O’Dowd D. K., Holmes T. C., Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J. Neurophysiol. 99, 976–988 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamaze A., Krätschmer P., Chen K. F., Lowe S., Jepson J. E. C., A wake-promoting circadian output circuit in Drosophila. Curr. Biol. 28, 3098–3105 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Guo F., Holla M., Díaz M. M., Rosbash M., A circadian output circuit controls sleep-wake arousal in Drosophila. Neuron 100, 624–635 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Donlea J. M., Ramanan N., Shaw P. J., Use-dependent plasticity in clock neurons regulates sleep need in drosophila. Science 324, 105–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bushey D., Tononi G., Cirelli C., Sleep and synaptic homeostasis: Structural evidence in Drosophila. Science 332, 1576–1581 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung J. H., Kirk L. M., Bourne J. N., Harris K. M., Shortened tethering filaments stabilize presynaptic vesicles in support of elevated release probability during LTP in rat hippocampus. Proc. Natl. Acad. Sci. U.S.A. 118, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung B. Y., Kilman V. L., Keath J. R., Pitman J. L., Allada R., The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr. Biol. 19, 386–390 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oh Y., et al. , A homeostatic sleep-stabilizing pathway in Drosophila composed of the sex peptide receptor and its ligand, the myoinhibitory peptide. PLoS Biol. 12, e1001974 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahnaou A., Drinkenburg W. H. I. M., Neuromedin U(2) receptor signaling mediates alteration of sleep-wake architecture in rats. Neuropeptides 45, 165–174 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Chiu C. N., et al. , A zebrafish genetic screen identifies neuromedin U as a regulator of sleep/wake states. Neuron 89, 842–856 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mistlberger R. E., Landry G. J., Marchant E. G., Sleep deprivation can attenuate light-induced phase shifts of circadian rhythms in hamsters. Neurosci. Lett. 238, 5–8 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Challet E., Turek F. W., Laute M., Van Reeth O., Sleep deprivation decreases phase-shift responses of circadian rhythms to light in the mouse: Role of serotonergic and metabolic signals. Brain Res. 909, 81–91 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Deboer T., Détári L., Meijer J. H., Long term effects of sleep deprivation on the mammalian circadian pacemaker. Sleep 30, 257–262 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Deboer T., Sleep homeostasis and the circadian clock: Do the circadian pacemaker and the sleep homeostat influence each other’s functioning? Neurobiol. Sleep Circadian Rhythms 5, 68–77 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hendricks J. C., et al. , Rest in Drosophila is a sleep-like state. Neuron 25, 129–138 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Curie T., Maret S., Emmenegger Y., Franken P., In vivo imaging of the central and peripheral effects of sleep deprivation and suprachiasmatic nuclei lesion on PERIOD-2 protein in mice. Sleep (Basel) 38, 1381–1394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jenett A., et al. , A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfeiffer B. D., et al. , Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melcher C., Pankratz M. J., Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 3, e305 10.1371/journal.pbio.0030305. (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shearin H. K., Dvarishkis A. R., Kozeluh C. D., Stowers R. S., Expansion of the gateway multisite recombination cloning toolkit. PLoS One 8, e77724 10.1371/journal.pone.0077724. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White K., Tahaoglu E., Steller H., Cell killing by the Drosophila gene reaper. Science 271, 805–807 (1996). [DOI] [PubMed] [Google Scholar]
- 65.Fan P., et al. , Genetic and neural mechanisms that inhibit Drosophila from mating with other species. Cell 154, 89–102 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park J. H., et al. , Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 97, 3608–3613 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfeiffer B. D., Truman J. W., Rubin G. M., Using translational enhancers to increase transgene expression in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 109, 6626–6631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burke C. J., et al. , Layered reward signalling through octopamine and dopamine in Drosophila. Nature 492, 433–437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Port F., Chen H. M., Lee T., Bullock S. L., Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, E2967–E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gratz S. J., et al. , Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilestro G. F., Cirelli C., pySolo: A complete suite for sleep analysis in Drosophila. Bioinformatics 25, 1466–1467 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.A. King, . Neural circuits controlling circadian rhythms. Publicly Accessible Penn Dissertations, 2842. University of Pennsylvania, Philadelphia, PA. https://repository.upenn.edu/edissertations/2842 (2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.