Significance
Bacterial populations that transition to stationary phase can efficiently maintain their competitive fitness potential even when subject to various stressors that damage cellular macromolecules. However, the mechanism remains unclear. Here, we show that stationary phase staphylococci that harbor damaged proteins in their cytoplasm undergo a regulated form of cell death to maintain the competitive fitness potential of the bacterial population. The activation of cell death is achieved by the down-regulation or degradation of cellular antioxidant proteins by the ATP-dependent ClpXP protease, which usually functions to maintain protein quality control in cells.
Keywords: Staphylococcus aureus, clpP, stationary phase fitness, clpX, sodA
Abstract
The transition from growth to stationary phase is a natural response of bacteria to starvation and stress. When stress is alleviated and more favorable growth conditions return, bacteria resume proliferation without a significant loss in fitness. Although specific adaptations that enhance the persistence and survival of bacteria in stationary phase have been identified, mechanisms that help maintain the competitive fitness potential of nondividing bacterial populations have remained obscure. Here, we demonstrate that staphylococci that enter stationary phase following growth in media supplemented with excess glucose, undergo regulated cell death to maintain the competitive fitness potential of the population. Upon a decrease in extracellular pH, the acetate generated as a byproduct of glucose metabolism induces cytoplasmic acidification and extensive protein damage in nondividing cells. Although cell death ensues, it does not occur as a passive consequence of protein damage. Instead, we demonstrate that the expression and activity of the ClpXP protease is induced, resulting in the degeneration of cellular antioxidant capacity and, ultimately, cell death. Under these conditions, inactivation of either clpX or clpP resulted in the extended survival of unfit cells in stationary phase, but at the cost of maintaining population fitness. Finally, we show that cell death from antibiotics that interfere with bacterial protein synthesis can also be partly ascribed to the corresponding increase in clpP expression and activity. The functional conservation of ClpP in eukaryotes and bacteria suggests that ClpP-dependent cell death and fitness maintenance may be a widespread phenomenon in these domains of life.
Bacterial pathogens can efficiently switch between growth and nongrowing stationary states in response to changes in nutrient status and environmental challenges (1, 2). Often, the entry into stationary phase is triggered by nutrient deprivation imposed by the host or due to competition with other microbial cohabitants (3, 4). Multiple stresses, including changes in pH, temperature, and osmolarity, can also limit active growth (5). The physiological and adaptive responses to stress that accompany the transition of bacterial cells to a nongrowing state are generally optimized for long-term survival (6). These changes enhance persistence in pathogens and make them tolerant to various stressors, including antibiotics and the host immune system (2, 6). However, the fate of bacterial cells that undergo stress-induced cellular damage in stationary phase remains unclear. Such cells may become unfit for subsequent growth and may need to be eliminated. Their removal would allow competition for limiting nutrients to be confined to healthier populations and would also ensure that the surviving population is fit for reentry into the growth cycle under more favorable conditions (7).
We have previously utilized Staphylococcus aureus as a model bacterial pathogen to study stationary phase survival dynamics (8, 9). When grown under conditions of excess glucose, S. aureus prematurely enters the stationary phase before glucose is completely exhausted from the media (9). Interestingly, the bacterial cells continue to consume glucose and excrete acetate as a byproduct, even though growth is arrested, suggesting that growth and carbon catabolism are uncoupled at this stage. The early entry into the stationary phase under these conditions results from the weak acid properties of acetate. As the pH of the culture shifts toward the pKa of acetate (pH, 4.8) during growth, a net movement of protonated acetate into cells is observed, which results in cytoplasmic acidification (9). This process initiates a series of events during stationary phase that is not seen when cells are grown in lower glucose concentrations. Notably, the cells entering stationary phase following growth in excess glucose have a lower respiratory potential, produce significant levels of endogenous reactive oxygen species (ROS), and accumulate chromosomal damage. These events culminate in cell death. Both the observed decrease in cell viability and the associated hallmarks of cell death can be avoided if acetate production is decreased by the genetic inactivation of cidC (pyruvate: menaquinone oxidoreductase) or if the entry of protonated acetate into cells is limited by controlling the rate of acidification with 4-morpholinepropanesulfonic acid (MOPS) buffer (pH, 7.4) (9). Thus, acetate-mediated cytoplasmic acidification directly potentiates cell death in stationary phase.
Proteolysis is a conserved process that takes place in all organisms to enforce protein quality control and modulate the intracellular levels of native proteins (10). In bacteria, the ClpP proteases comprise one of the important AAA+ proteolytic machines. S. aureus encodes the ClpXP and ClpCP proteases (11). The ClpP subunits assemble as two homo-heptameric rings to form a barrel structure with peptidase activity. The ClpX and ClpC subunits are ATPases associated with the ClpP barrel as hexameric rings and provide substrate specificity through direct interaction with substrates or specific adaptor proteins (12). Staphylococci that encounter toxic acetate levels during growth are reported to increase expression of the clp genes, presumably to maintain protein quality in these cells (13).
Here, we explore the mechanisms involved in maintaining the competitive fitness of stationary bacterial populations under stress. We demonstrate that acetate-mediated intracellular acidification increases the susceptibility of proteins to oxidation and aggregation in stationary-phase cells. Although the accumulation of oxidized protein aggregates in cells can be cytotoxic (14, 15), the observed cell death in stationary phase cells does not directly result from passive toxicity due to protein damage. Instead, we demonstrate that damaged cells die from the increased expression and activity of the ATP-dependent ClpXP proteolytic machinery, which targets the destruction of superoxide dismutase (SodA), a critical determinant of cell survival. The active process of eliminating cells with extensive protein damage through ClpXP-dependent proteolysis ensures the survival of the fittest cells in stationary phase. This activation of cell death is not unique to acetate-mediated weak acid stress. Antimicrobials that interfere with protein biosynthesis can also variably induce clpP expression and promote ClpXP-dependent cell death in nongrowing staphylococci suggesting a broader, more conserved, and beneficial role for ATP-powered cell death processes in bacteria.
Results
Acetate Increases Protein Oxidation and Aggregation under Acidic Conditions.
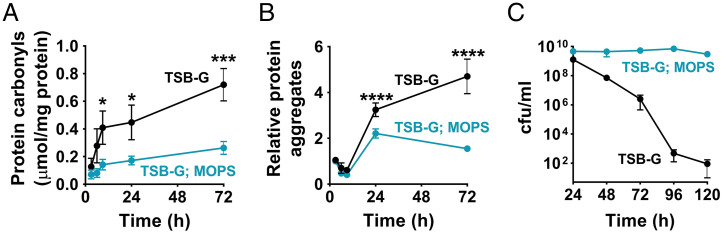
Since cytoplasmic proteins are sensitive to the oxidation status and pH of their environment, we initially predicted that stationary-phase cell death following acetate-mediated intracellular acidification might result from protein damage. To test this hypothesis, we measured the levels of protein oxidation and aggregation under conditions that caused cytoplasmic acidification (9). S. aureus JE2 was cultured in media supplemented with excess glucose (45 mM), where acetate accumulated to approximately 37 mM and the extracellular pH remained close to 4.8 throughout the stationary phase (SI Appendix, Fig. S1A). The protein oxidation status was determined by measuring the carbonyl content of the total cytoplasmic protein fraction and protein aggregation was measured as the SDS-insoluble protein fraction from the total cellular protein pools. We observed that the levels of carbonylated proteins and protein-aggregates increased throughout staphylococcal growth as the media acidified (Fig. 1 A and B and SI Appendix, Fig. S1A). Additionally, cell viability decreased over 7-logs during stationary phase (Fig. 1C and SI Appendix, Table S1). Notably, the levels of protein damage and the rate of cell death were lower when cells were grown in media that was buffered to a pH 7.4 with 100 mM MOPS (Fig. 1 and SI Appendix, Table S1), a condition that prevented prolonged exposure of cells to the toxic effects of acetate (SI Appendix, Fig. S1B), despite the high levels of glucose in the media (9). Acetate-mediated cell death in stationary phase could also be prevented by inactivating ackA, the major contributor of acetate in S. aureus (SI Appendix, Fig. S1 C and D). However, further analysis of the ackA mutant was not attempted due to the pleiotropic effects of this mutation on cell physiology. Together, these observations highlight protein oxidation and protein aggregation as important correlates of stationary phase cell death following acetate stress.
Fig. 1.
Acetic acid induces protein damage and cell death. The toxicity of acetic acid on staphylococci was distinguished by growth in MOPS-buffered (100 mM, pH 7.3) and -unbuffered TSB. Acetic acid remains as charged anions in MOPS-buffered media, preventing its entry into cells. (A) Protein oxidation was determined at different stages of growth by measuring the carbonyl content of intracellular proteins (n = 3, mean ± SD). (B) Protein aggregates were measured as the detergent-insoluble fraction of total intracellular proteins. The aggregates were isolated, resolved by SDS/PAGE, and quantified using ImageJ software. The fold-differences in protein aggregate levels are relative to those observed at 3 h of growth (midexponential phase). (n = 5, mean ± SD) (C) Cell viability in stationary phase was determined by enumeration of total bacterial colony forming units following dilution plating. TSB-G, TSB supplemented with 45 mM glucose. Two-way ANOVA with Sidak’s multiple comparisons posttest; *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001.
ClpXP Catalyzed Proteolysis Is Essential for Acetate-Mediated Cell Death.
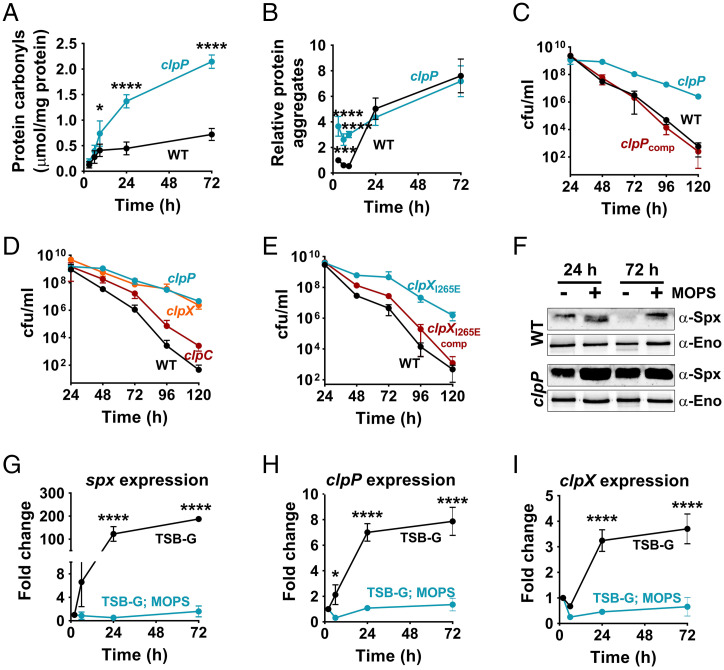
The accumulation of oxidized proteins and protein-aggregates can themselves have toxic sequelae resulting in the organism’s death (14). In S. aureus, ClpP has been described as the major intracellular protease involved in protein quality control and can recycle damaged or unwanted proteins (11, 16). Accordingly, we reasoned that the inactivation of clpP should increase the rate of cell death due to the accumulation of damaged proteins. Upon evaluating the cytoplasmic protein aggregates and oxidized proteins in the clpP mutant, we confirmed an increase in the level of protein damage relative to the parental strain (Fig. 2 A and B). The increase in protein aggregates over the wild-type (WT) strain was primarily observed in the exponential phase (Fig. 2B). In comparison, we detected a more notable increase in oxidized proteins in the clpP mutant over time and upon entry of cells into stationary phase (Fig. 2A). To determine whether the elevated levels of damaged proteins following clpP mutation increased the rate of staphylococcal cell death, we assessed the survival of a clpP mutant over stationary phase. Unexpectedly, the rate of stationary phase cell death was significantly lower in the clpP mutant than the WT strain (Fig. 2C and SI Appendix, Table S1), which suggested that cell death did not result from protein damage per se, but was a consequence of the ClpP activation that followed. The complementation of the clpP mutant by introducing the clpP allele under the control of its native promoter within the SaPI chromosomal locus fully restored the rate of cell death in the clpP mutant to WT levels (Fig. 2C and SI Appendix, Table S1), which confirmed a role for ClpP in promoting cell death.
Fig. 2.
The ClpXP proteolytic activity enhances cell death. (A) Protein oxidation (n = 3, mean ± SD), (B) protein aggregation (n = 5, mean ± SD), and (C) viability of the WT and clpP mutants (n = 3, mean ± SD). (D) The cell viabilities of the clpX, clpC (n = 4, mean ± SD) and (E) clpXI265E mutants were determined in stationary phase relative to the WT strain (n = 3, mean ± SD). (F) The intracellular abundance of Spx was used as an indicator of ClpXP proteolytic activity following growth in MOPS-buffered (100 mM, pH 7.3) and unbuffered media. S. aureus Spx was detected using cross-reactive polyclonal antibodies raised against B. subtilis Spx. Enolase was detected as a loading control. (G) The fold-change in the expression of spx, (H) clpP, and (I) clpX was determined over time during different growth phases by qRT-PCR (n = 3, mean ± SD). Fold-change expression values are relative to 3 h of growth (midexponential phase). Two-way ANOVA with Sidak’s multiple comparisons posttest; *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001.
The observed decrease in cell death of the clpP mutant did not result from a reduction in extracellular acidification as the concentration of acetate and pH in stationary phase closely matched those of the WT under all tested culture conditions (SI Appendix; compare SI Appendix, Figs. S1 A and B and S2 A and B). Nor were there any specific extracellular factors excreted by the WT that could induce cell death in the clpP mutant, as cross-substitution of cell-free stationary phase culture supernatants between the WT and the clpP mutant did not alter the kinetics of cell death of either strain (SI Appendix, Fig. S2C). This suggested that the ClpP-dependent cell death in the WT strain resulted from endogenous factors.
The proteolytic function of ClpP is dependent on Clp ATPase subunits. In S. aureus, the ClpX and ClpC ATPases interact with ClpP (12). Accordingly, we determined the contribution of ClpX and ClpC in acetate-mediated cell death. Although independent mutations of either ATPase subunits exhibited a decreased rate of cell death compared to the WT strain, the clpX mutant more fully phenocopied the clpP mutant (Fig. 2D and SI Appendix, Table S1), which suggested that cell death was primarily under the control of the ClpXP protease. This conclusion was confirmed by our observation that acetate-mediated cell death was decreased to a similar extent in a strain expressing a mutant variant of ClpX, ClpXI265E (Fig. 2E and SI Appendix, Table S1). The ClpXI265E variant retained the ClpX chaperone activity, but not the ClpXP protease function due to a single amino acid substitution in the ClpP recognition motif of ClpX (11). Additionally, chromosomal complementation of native clpX in the clpXI265E mutant restored the rate of cell death to WT levels (Fig. 2E and SI Appendix, Table S1). These observations strongly suggested that the ClpXP proteolytic function was solely responsible for increasing cell death following acetate-mediated protein damage.
Next, we determined how acetate-mediated intracellular acidification affected ClpXP activity. Previous studies have concluded that Spx is an exclusive substrate of the ClpXP protease (17, 18). Accordingly, the intracellular levels of Spx were used as a proxy for gauging ClpXP activation. In contrast to the Spx levels observed at 24 h when WT survival was at its peak, immunoblotting using anti-Spx antibodies revealed that Spx was depleted in the late stationary phase (72 h) when cell viability was actively decreasing (Fig. 2F). The depletion of Spx at 72 h was only observed in cultures grown in the absence of MOPS buffer that was subject to prolonged acidification (Fig. 2F) and was not due to a decrease in spx transcription (Fig. 2G). Indeed, Spx levels underwent depletion even though its transcription had dramatically increased (Fig. 2 F and G). Additionally, we also confirmed that the reduction of Spx at 72 h was dependent on ClpP activity, as its inactivation restored Spx levels (Fig. 2F). These observations suggest that ClpXP activity is enhanced following acetate-mediated intracellular acidification. The increased ClpXP activity was also matched with increased expression of both clpP and clpX following acidification (Fig. 2 H and I). These observations suggest that acetate-mediated protein damage at low pH increases the expression and proteolytic activity of ClpXP, resulting in cell death.
ClpP Enhances Cell Death by Diminishing the Cellular Antioxidant Capacity.
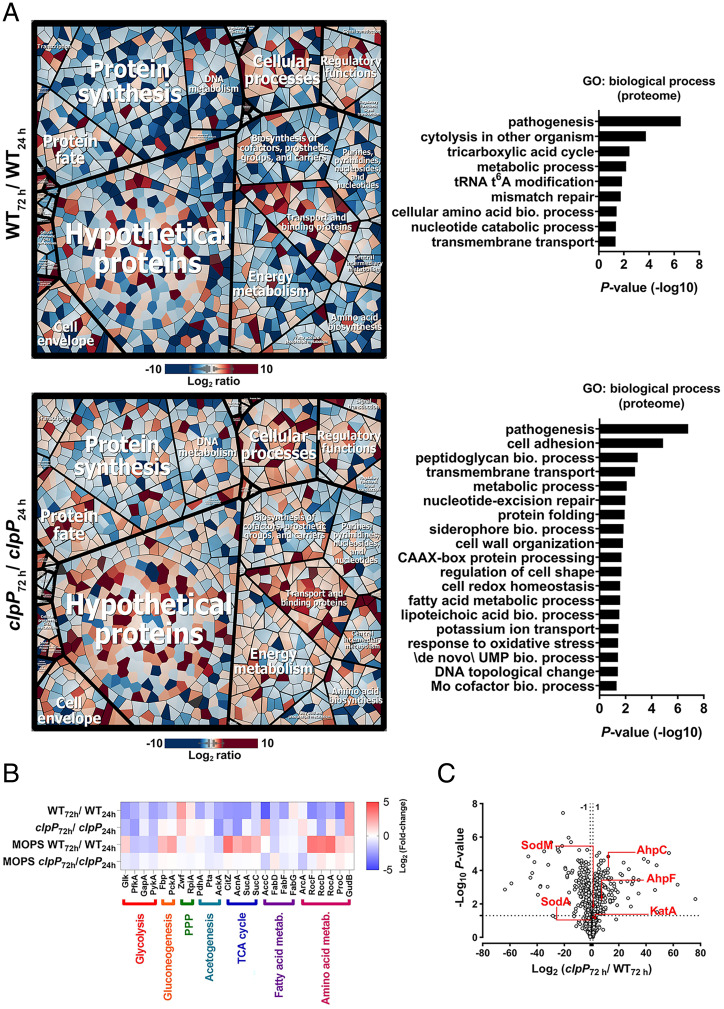
Thus far, our analyses suggested that ClpXP targeted the degradation of proteins that are important for cell survival in the late stationary phase. To identify potential targets of ClpXP, we determined proteomic changes in the WT and the clpP mutant at 24 h and 72 h in stationary phase by mass spectrometry (Datasets S1–S6). The identified proteins were organized hierarchically into functional classes based on their TIGRFAM annotation and the differences in protein abundance were visualized using Voronoi Treemaps (Fig. 3A). Furthermore, proteins associated with cell metabolism were grouped according to metabolic pathways (19) (Dataset S7). A critical subset of these proteins involved in central metabolism and amino acid metabolism was visualized as a heatmap (Fig. 3B). We observed notable differences between the WT and clpP proteomes during the transition from 24 h to 72 h in stationary phase (Fig. 3 A and B and SI Appendix, Fig. S3). At 72 h, most of the proteins in the WT strain associated with cell metabolism and biosynthesis of nucleotides, proteins, amino acids, and cofactors had decreased in abundance relative to levels observed at 24 h.
Fig. 3.
Altered proteome of stationary phase cells undergoing acetate stress. (A) Voronoi Treemaps and GO-term enrichment analysis of WT and clpP mutant are depicted. The Voronoi Treemaps were generated from log2 ratios of their respective intracellular proteins at 72 h and 24 h of growth in TSB-G. Proteins with altered ratios were clustered based on the TIGRFAM annotations and depicted as functional categories. Additional subcategories and gene ID annotations are indicated in SI Appendix, Fig. S3. (B) Heatmap representing the intracellular changes in select metabolic enzymes of WT and clpP at 72 h relative to 24 h. The corresponding reaction level differences are detailed in Dataset S7. (C) Volcano plot of the intracellular proteins in stationary phase cells. Each data point represents individual proteins organized according to their mean clpP/WT log2 fold-change ratios (y axis) following 72 h of growth in TSB-G. The horizontal dotted line indicates the cutoff for proteins that showed significantly altered abundance (P ≤ 0.05).
In contrast, the protein levels were relatively more stable in the clpP mutant during the same time-transition in unbuffered media (compare clpP72h/24h vs. WT72h/24h in Fig. 3 A and B and Dataset S7). This trend was partially reversed when bacterial cells were grown in MOPS-buffered media (Fig. 3B and SI Appendix, Figs. S4A and S5). Furthermore, proteins associated with gene ontology (GO) terms, including “redox homeostasis” and “response to oxidative stress” were uniquely enriched in the clpP mutant (clpP72h/24h) compared to the WT strain (WT72h/24h), which suggested that ClpP may be able to target proteins that are important for detoxification of endogenous ROS in the WT strain (Fig. 3A). Indeed, a direct comparison of the protein abundance between the WT and clpP mutant at 72 h confirmed increased levels of antioxidant proteins in the latter mutant following acetate stress (Fig. 3C) but not when the media was buffered with MOPS (SI Appendix, Fig. S4B). Overall, these findings are consistent with the hypothesis that the clpP mutant is more metabolically active than the WT strain in stationary phase and survives acetate stress by modulating intracellular ROS.
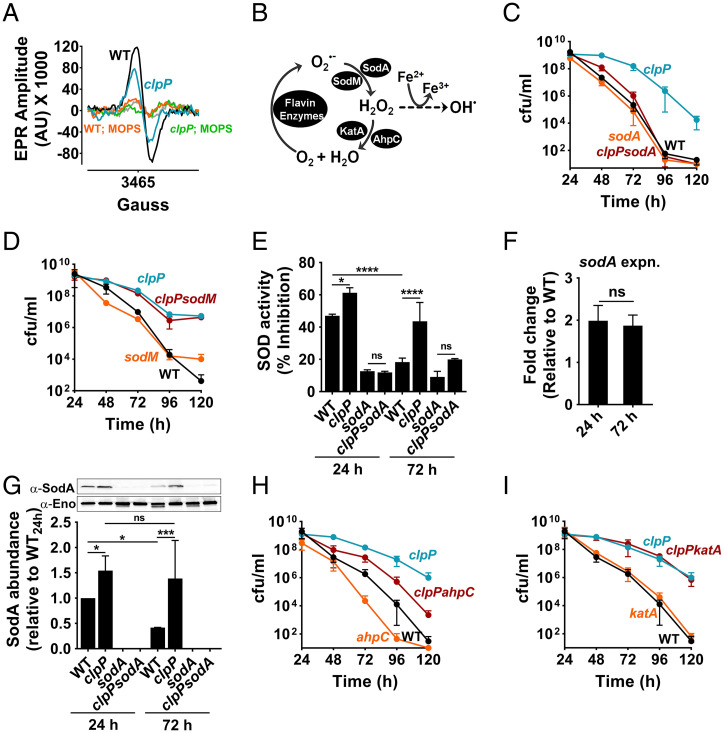
To determine whether inactivation of clpP affected ROS production in the stationary phase, we performed electron paramagnetic resonance (EPR) spectroscopy on WT and clpP mutant samples at different times during growth (SI Appendix, Fig. S6A) using CMH as a ROS-specific spin probe (20). EPR spectroscopy demonstrated that the clpP mutant produced lower ROS levels than the WT strain at 72 h (Fig. 4A). However, ROS production in stationary phase was not detected when WT and the clpP mutant were grown in media buffered with MOPS, pH 7.4 (Fig. 4A). S. aureus has multiple enzymes that control the endogenous production of ROS: specifically, SodA and SodM dismutate superoxide (O2•-) to hydrogen peroxide (H2O2) (Fig. 4B). Subsequently, catalase and AhpC convert H2O2 to water and limits the formation of toxic hydroxyl radical (OH•) (Fig. 4B). Given that the abundance of these antioxidant enzymes was increased in the clpP mutant (Fig. 3C), we hypothesized that they might individually or collectively aid cell survival by enhancing endogenous ROS detoxification.
Fig. 4.
ClpXP targets SodA to subvert the antioxidant capacity of cells. (A) Representative EPR spectroscopic trace of ROS derived from whole cells in stationary phase (72 h). ROS was detected using the spin probe, CMH. (B) Schematic of the antioxidant enzymes of S. aureus and their target ROS. AhpC, alkylhydroperoxidase subunit; KatA, catalase; SodA/SodM, Mn-dependent superoxide dismutases. (C) Stationary phase viability of sodA and (D) sodM mutants relative to WT and clpP mutant strains (n = 3, mean ± SD). (E) SOD activity of cell extracts at 24 h and 72 h were determined as the percent inhibition of WST-1 reduction by superoxide using a commercially available kit (Sigma). WST-1, water-soluble tetrazolium salt (n = 3, mean ± SD; one-way ANOVA with Tukey's multiple comparisons test, *P ≤ 0.05, ****P ≤ 0.0001). (F) The fold-change in the expression of sodA was determined at 24 h and 72 h in the clpP mutant. The fold-change expression values were calculated relative to the WT strain at identical time-points (n = 3, mean ± SD; two-tailed unpaired t test, ns: not significant). (G) The intracellular levels of SodA were detected in various mutants by Western blot analysis using cross-reactive polyclonal antibodies raised against Bacillus anthracis SodA (Upper). Enolase was detected as a loading control. Densitometric quantification was performed by ImageJ (Lower) (n = 3, mean ± SD; one-way ANOVA with uncorrected Fisher's LSD, *P ≤ 0.05, ***P ≤ 0.001). (H) Cell viability of ahpC and (I) katA mutants in stationary phase (n = 3, mean ± SD).
To test this hypothesis, we determined the contribution of each enzyme to improving the survival of the clpP mutant. Although mutation of sodA did not change the kinetics of cell death in the WT strain, its mutation in the clpP background (clpPsodA strain) completely abrogated survival of the clpP mutant to WT levels (Fig. 4C and SI Appendix, Table S1). Separately, inactivation of sodM had no noticeable effect on the death kinetics in either the WT or clpP mutant backgrounds (Fig. 4D and SI Appendix, Table S1), despite increased expression in the clpP mutant over time (SI Appendix, Fig. S6B). Consistent with a role for sodA in the survival of the clpP mutant, we observed that the total superoxide dismutase activity was elevated in the clpP mutant relative to the WT at 24 h and 72 h of growth (Fig. 4E).
Conversely, SodA activity decreased in the WT strain in a ClpP-dependent manner and correlated with increased cell death as cells transitioned from 24 h to 72 h in stationary phase (Fig. 4E). A similar decrease in SodA activity did not occur when the WT strain was grown in MOPS-buffered tryptic soy broth (TSB)-G (SI Appendix, Fig. S7). Although sodA expression had maximized during exponential growth (SI Appendix, Fig. S6B), the observed differences in SodA enzyme activity in unbuffered growth conditions did not result from differences in sodA expression during the transition from 24 h to 72 h in stationary phase (Fig. 4F), which suggested that SodA is likely to be a substrate of the ClpXP protease under these conditions. Indeed, Western blot analysis using anti-SodA antibodies revealed a ClpP-dependent decrease in SodA levels (by ∼44%) in the WT strain at 72 h relative to 24 h in stationary phase (Fig. 4G). The decrease in SodA abundance (Fig. 4G) is consistent with the observed reduction in SOD activity (>40%) when WT transitioned from 24 h to 72 h (Fig. 4E). The transcription of ahpC in the clpP mutant was threefold higher than the WT at 72 h (SI Appendix, Fig. S6B). The clpPahpC double mutant exhibited a decreased survival compared to the clpP mutant (Fig. 4H and SI Appendix, Table S1). However, the ahpC mutant itself exhibited an increased rate of cell death relative to the WT strain (Fig. 4H and SI Appendix, Table S1). This suggested that although AhpC activity is essential for stationary phase survival, its contribution toward the survival of the clpP mutant is likely to be secondary to SodA activity. We did not observe a role for katA in increasing the survival of the clpP mutant (Fig. 4I and SI Appendix, Table S1), despite increased expression over time (SI Appendix, Fig. S6B). Together, these observations suggest that under acetic acid stress, the targeted degradation of SodA and the reduction of ahpC expression by ClpP in the stationary phase could trigger cell death.
ClpP-Dependent Cell Death Enhances the Competitive Fitness Potential of Staphylococci.
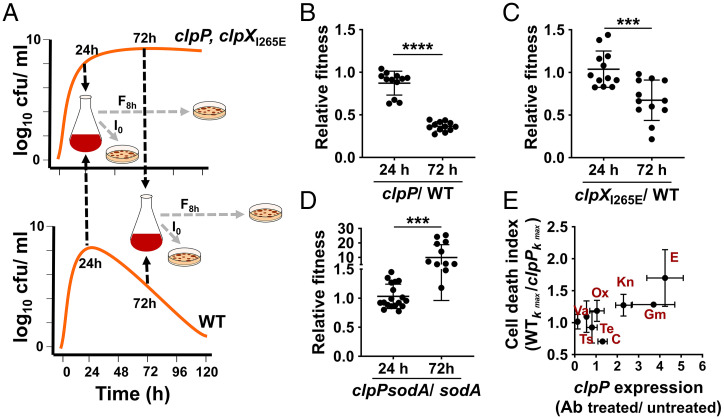
The necessity of mechanisms that activate cell death in single-cell organisms like S. aureus is counterintuitive. Hence, we suspected that the benefits of ClpP-dependent cell death might exist at the population rather than the single-cell level. Since the accumulation of damaged proteins was likely to affect growth, we hypothesized that the enhanced survival of the clpP mutant during the stationary phase might come at the cost of maintaining the overall competitive fitness potential of the population. To test this hypothesis, we harvested the WT and clpP mutant before the onset of cell death when cell viability for both strains was highest (24-h growth) and at a subsequent time point when cell viability of the WT (but not the clpP mutant) was declining (72 h) (Fig. 5A). We performed coculture competition assays of the clpP mutant and WT strains derived from each time point. We assessed their mean competitive fitness (w) as a ratio of their Malthusian parameters (exponential growth rate) over an 8-h growth period (21). Although the clpP mutant isolated from 24-h-old cultures exhibited only a modest decrease in fitness relative to the WT strain (w24 = 0.87), the mean competitive fitness of the clpP mutant was significantly diminished when the competition was initiated from 72-h-old cultures (w72 = 0.365) (Fig. 5B). We observed a similar trend when the competition was conducted between clpXI265E and the WT strain (w24 = 1.03 vs. w72 = 0.673) (Fig. 5C). The poor competitive outcome of the clpP and clpXI265E mutants occurred despite the increased survival of these mutants relative to the WT strain at 72 h. To test whether the observed competitive fitness defect of the clpP mutant resulted from increased SodA activity rather than pleiotropic effects of the clpP mutation, we performed coculture competition of the clpPsodA double mutant with its isogenic sodA parent strain following 72-h growth. We observed that the competitive defect of the clpP mutant was reversed if sodA was inactivated in that background (clpPsodA mutant) (compare Fig. 5D with Fig. 5B). These results suggest that the ClpXP-dependent decrease in SodA activity not only initiates cell death but also is essential for maintaining the overall competitive fitness potential of staphylococcal populations undergoing stress in stationary phase.
Fig. 5.
ClpXP activity eliminates fitness-compromised cells in stationary phase. (A) The competitive fitness of the clpP and clpXI265E mutants that grew to stationary phase in TSB-G (24 h or 72 h) were determined relative to the WT strain harvested at identical time points in the same growth media (TSB-G). For the determination of relative fitness, coculture competitions were carried out for 8 h in fresh TSB. Initial bacterial counts (I0) and final counts after 8 h of growth (F8h) were used to measure the relative fitness (w). Relative fitness ratios of the (B) clpP to WT, (C) clpXI265E to WT, and (D) sodA to clpPsodA are depicted here. (n = 12, mean ± SD; two-tailed unpaired t test, ***P ≤ 0.001, ****P ≤ 0.0001). (E) Correlation of clpP expression and ClpP-dependent cell death. The fold-change in clpP expression was determined at 24 h after challenging WT with various antibiotics at the postexponential phase (6 h). The fold-change expression values were calculated relative to the WT strain grown to 24 h without antibiotic treatment. The cell death index reflects a measure of the ClpP-dependent cell death observed in stationary phase following antibiotic challenge. The cell death index was calculated as the ratio of the death rates (kmax) of WT and clpP mutant. A value above 1 indicates a greater rate of cell death in the WT relative to the clpP mutant. Measures of clpP expression and cell death index were determined following the growth of S. aureus in TSB. C, chloramphenicol; E, erythromycin: Gm, gentamicin: Kn, kanamycin; Ox, oxacillin; Te, tetracycline; Ts, trimethoprim; Va, vancomycin.
We next asked if the evolutionary pressure to maintain competitive fitness through regulated cell death could be triggered by antimicrobial compounds that interfere with protein homeostasis and induce clpP expression. To test this hypothesis, we treated the WT strain with different classes of antibiotics at the postexponential phase (25× MIC). We also assessed their ability to induce clpP expression (Fig. 5E) and induce ClpP-dependent cell death (SI Appendix, Table S2). Our analysis revealed that different antibiotics were able to differentially activate clpP expression by 24 h relative to untreated control at the same time-point; notably, aminoglycosides that interfered with protein synthesis were most prominent in this regard (Fig. 5E). Importantly, we observed a strong positive correlation between the ability of antibiotics that induce clpP expression to also promote clpP-dependent cell death [Pearson coefficient (r) = 0.7612, r2 = 0.5794, P = 0.0282] (Fig. 5E and SI Appendix, Table S2). Collectively, these findings strongly suggest that nondividing populations with irreparable protein damage are targeted for elimination in a ClpXP-dependent manner to sustain population fitness.
Discussion
The evolutionary selection of energy-dependent cell death processes in bacteria may have origins in niches where interspecies competition and antimicrobial warfare are commonly observed. Antimicrobial compounds and metabolic byproducts, such as weak acids that interfere with protein homeostasis, are frequently produced by competing bacterial species (22, 23). Such interactions could adversely bias fitness outcomes of target bacterial populations that are affected by the harmful effects of these metabolites (24). While bacteria may transition to nongrowing states under these conditions and become more tolerant, the continued stress from these antimicrobials is likely to increase damaged cells in the population (25). The elimination of unfit cells through an active self-destruction program could improve the competitive fitness of the bacterial population when cells re-enter growth under more favorable conditions.
How are unfit cells eliminated? Studies have shown that stress-induced oxidized protein aggregates in the cytoplasm can be lethal (14). However, our findings demonstrate that this is not the case in staphylococci that enter stationary phase following acetate stress. Indeed, the clpP mutant accumulated more oxidized proteins and protein aggregates than the WT strain but still survived significantly better in the stationary phase.
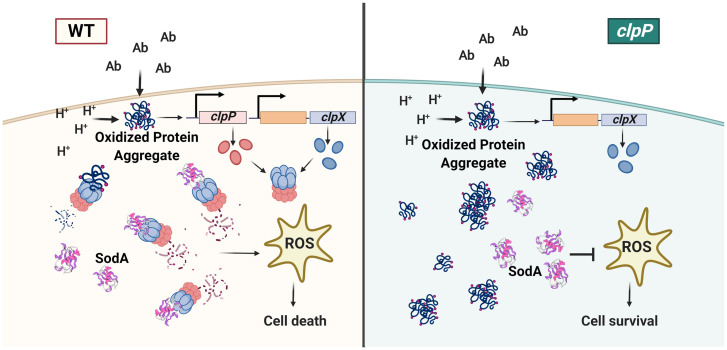
According to our model (Fig. 6), cells that accumulate damaged proteins in stationary phase induce ClpXP expression and activity in response. The extent of intracellular protein damage following stress may dictate the fate of cells. When cell division halts and cells enter stationary phase, protein quality control is critical because cells can neither dilute damage through cell division nor divert all energy to protein repair as resources are scarce during stationary phase. If the damage is beyond repair and affects the competitive fitness of the population, the fitness-compromised cells must undergo cell death. Thus, regulated cell death may represent a means to dilute damage in nondividing bacterial populations, just as rapid cell division in dividing populations.
Fig. 6.
Model of the ClpXP mediated cell death pathway in S. aureus. S. aureus accumulates oxidized protein aggregates and turns on the expression of clpX and clpP when they encounter antibiotic (Ab) stress or undergo intracellular acidification (H+). In rapidly dividing cells that can dilute the effect of damaged proteins through cell division, the ClpXP protease may be sufficient to recycle protein aggregates and restore the fitness of cells. However, nondividing populations that contain protein aggregates undergo cell death to maintain population fitness (Left). To initiate cell death, the ClpXP protease targets the degradation of SodA, which results in lethal oxidative damage due to sustained superoxide production. In the clpP mutant (Right), the lack of protein turnover results in the retention of proteins that are highly susceptible to oxidation and aggregation. However, the increased levels and activity of SodA promote cell survival due to decreased ROS-mediated killing. This image was created with BioRender.com.
Our findings suggest that ClpXP mediates cell death by targeting the degradation of SodA. The resulting inability to efficiently detoxify superoxide triggers cell death. Several lines of evidence cumulatively support this model. First, acetate-mediated cytoplasmic acidification increased the expression of both clpX and clpP in stationary phase. Consistent with this phenotype, an increase in ClpXP-dependent proteolytic activity could be deduced from the degradation of Spx, a ClpXP-specific substrate, and more broadly from the ClpP-dependent changes in the intracellular proteome of WT strain during the late stationary phase. Second, the observed cell death was not related to the chaperone function of ClpX in S. aureus (26) as the clpXI265E mutant was unable to initiate cell death in response to acetic acid stress. This reconfirmed the role of the ClpXP protease as the primary mediator of cell death. Finally, although the abundance of multiple intracellular proteins decreased upon ClpP activation, those involved in countering oxidative stress and redox balance were prominent. SodA activity was significantly reduced in the late stationary phase in a ClpP-dependent manner. Furthermore, inactivation of sodA in the clpP mutant restored the kinetics of cell death to WT levels, thus highlighting SodA as a target of ClpXP for the induction of cell death in stationary phase. It is important to note that SodA was previously identified as a critical factor for long-term survival of S. aureus in stationary phase (27). Thus, ClpXP induces cell death by targeting bona fide proteins involved in maintaining cell viability. Interestingly, the magnitude of rescue from cell death is more when buffering the media with MOPS than when clpP is inactivated. This may in part be due to the residual production of ROS in the clpP mutant that prevents it from complete rescue. Alternatively, it is possible that a ClpP-independent mechanism is at play. For example, acetate may still exert significant toxicity in the clpP mutant by accumulating in the cytoplasm as anions.
The toxicity of superoxide and its downstream derivatives, such as hydrogen peroxide and hydroxyl radicals, is well documented. It involves damage to macromolecules and metal centers in enzymes critical for cell viability (28). Irreparable DNA double-stranded breaks due to hydroxyl radicals arising from Fenton chemistry and oxidation of essential proteins could result in permanent loss of replicative ability, resulting in cell death (29). Damage within cells has been proposed to induce a programmed cell death process mediated by the Cid and Lrg system (30, 31). Although recent evidence suggests that the Cid/Lrg proteins are involved in the transport of metabolites across the cell membrane (32), oxidative stress may trigger their oligomerization and pore formation in the cell membrane resulting in cell death (33). Proteomic analysis revealed altered levels of several proteins involved in diverse cellular functions when cell death is triggered following stress. Thus, the involvement of additional pathways in coordination with or independent of SodA in the regulation of cell death cannot be ruled out.
While ClpXP can turnover native proteins and control their intracellular abundance (34, 35), the specific signals that mark SodA for degradation by ClpXP in unfit cells are unknown. Similarly, the sources of cellular ROS in stress-induced damaged cells remain to be identified. However, the ability of ClpXP to regulate cell death appears to be conserved in prokaryotes and eukaryotes. In a recent study, the Bacillus subtilis ClpXP protease was shown to eliminate defective sporulating cells with misassembled envelopes by degrading SpoIVA and inducing lysis (36). Such a mechanism in B. subtilis was proposed to select against the loss of a sporulation program (36, 37). In Escherichia coli, it was demonstrated that the ClpPA protease ultimately promotes survival during starvation and phage infection by inducing toxin–antitoxin (MazEF)-mediated programmed cell death in a subpopulation of cells. Accordingly, it was shown that the ClpPA targets the antitoxin MazE for degradation, thus freeing the MazF toxin to inhibit the translation of essential proteins by cleaving mRNA (38). In eukaryotes, the mitochondrial ClpXP protease was found to restrict the lifespan of the fungus Podospora anserina (39). Interestingly, the human ortholog of clpP was also able to reduce the extended lifespan of the P. anserina clpP mutant when heterologously expressed in that organism (39), suggesting functional conservation of ClpP in controlling cell death and lifespan across the kingdoms of life.
Like the clpX mutation, the inactivation of clpC also increased the survival of stationary phase cells that had undergone protein damage, albeit this was a modest increase in our model. The importance of the staphylococcal ClpCP protease in promoting cell death in stationary phase has been described previously (40). The increased survival of the clpC mutant was attributed to a greatly reduced tricarboxylic acid (TCA) cycle activity resulting from poor aconitase function in this mutant. Consequently, the lower cellular energy available to the clpC mutant was hypothesized to prevent cell entry into the energy-dependent death phase (41, 42). Independently, the reduced TCA cycle activity and low respiratory potential in the clpC mutant could also result in less ROS production and increased stationary phase survival (40, 43). More recently, the ClpCP protease was shown to modulate intracellular survival by targeting the MazEF toxin–antitoxin system in S. aureus (44). The degradation of the MazE antitoxin by the ClpCP protease resulted in MazF toxin-mediated bacterial stasis (44).
Previously, Dwyer et al. (45) demonstrated that antibiotic stress induced several hallmarks of apoptosis in E. coli and enabled the destruction of cells similar to programmed cell death in eukaryotes. Remarkably, the ClpXP protease was shown to play a significant role in the antibiotic-induced bacterial cell death in addition to RecA and members of the SOS response regulon (45). Our findings also highlight a link between certain classes of antibiotics and ClpXP-dependent cell death. Notably, we observed increased clpP expression and ClpP-dependent cell death when antibiotics that target bacterial ribosomes—like gentamicin, kanamycin, and erythromycin—were used against S. aureus. These antibiotics primarily disrupt protein homeostasis by blocking protein synthesis (46, 47). Although some of these antibiotics are thought to be bacteriostatic (48, 49) at high concentrations, most of these antibiotics may become bactericidal due to indirect effects from protein damage (46, 50). Our finding that antibiotic-mediated killing may be alleviated by inactivating clpP indicates a significant contribution for ClpP proteolysis in cell death during antibiotic stress.
In conclusion, our findings suggest that stationary phase staphylococci can activate a ClpXP-regulated cell death program in response to stress and eliminate unfit cells from the population. We propose that such a regulated cell death mechanism is evolutionarily selected in bacteria to maintain competitive fitness during growth transitions.
Materials and Methods
DNA Manipulation and Strain Construction.
S. aureus JE2 and isogenic mutants were obtained from the Nebraska Transposon Mutant Library (51). The transposon mutants were transduced into strain JE2 using ϕ11. Full-length genes and their native promoters were inserted into the chromosome for complementation of mutant strains using the SaPI integration vector. Additional details on strain construction and growth conditions are described in SI Appendix, Supplemental Experimental Procedures.
Protein Aggregation and Carbonylation.
The protein aggregates were isolated from bacterial cells as described by Engman et al. (52), To measure protein carbonylation, bacterial cells were initially suspended in 1.5 mL of PBS containing 50 mM dithiothreitol (DTT) and mechanically disrupted using the Precellys bead homogenizer. Proteins in the supernatant were precipitated by adding 10% trichloroacetic acid (TCA). Carbonyl-containing proteins were derivatized by incubating with 500 µL of 10 mM 2,4-dinitrophenylhydrazine (DNPH) dissolved in 2 M HCl. The derivatized proteins were reprecipitated with 500 µL of 20% TCA. The protein pellets were washed three times in 1 mL of 1:1 ethanol-ethyl acetate, and finally dissolved in 6 M guanidine in 20 mM potassium phosphate buffer adjusted to pH 2.3 with trifluoracetic acid. The absorbance was measured at 370 nm, and carbonyl content was determined using an extinction coefficient of 22,000 M−1 cm−1 (53).
Measurement of the Competitive Fitness.
Cultures of JE2 and its isogenic clpP or clpXI265E grown in TSB 45 mM glucose were used to assess competitive fitness. Following 24 h of growth in media containing excess glucose, we used ∼107 bacteria per milliliter of each strain for competition experiments. Due to the increased death of WT at 72 h into stationary phase, competition experiments were initiated from ∼105 to ∼106 cfu/mL. The bacterial colony forming units (cfu) were enumerated on TSA plates with and without 5 µg/mL erythromycin immediately after initiation of competition and at 8 h between tested strains. The competitive fitness was calculated using the Malthusian parameter for competitors using the following formula: w = ln (Mf/Mi)/ln (Wf/Wi), where f and i represent cfu counts at final (8 h) and initial (time 0) of competition assay, respectively (21). M and W refer to mutant and WT, respectively. Coculture competitions between sodA and clpPsodA were similarly carried out.
EPR spectroscopy.
The bacterial cultures (OD600 = 10) were harvested at various time points during growth, pelleted, and resuspended in 1 mL KDD buffer (Krebs-Hepes buffer, pH 7.4; 99 mM NaCl, 4.69 mM KCl, 2.5 mM CaCl2, 1.2 mM MgSO4, 25 mM NaHCO3, 1.03 mM KH2PO4, 5.6 mM D-glucose, 20 mM Hepes, 5 µM DETC, and 25 µM deferoxamine) for determination of ROS production. The bacterial suspensions were mixed with the ROS-sensitive spin probe CMH (working stock ∼4 mM prepared in KDD buffer) to a final concentration of 200 µM and incubated at room temperature for 15 min. A Bruker e-scan EPR spectrometer was used with the following acquisition parameters: field sweep width, 60.0 G; microwave frequency, 9.75 kHz; microwave power, 21.90 mW; modulation amplitude, 2.37 G; conversion time, 10.24 ms; time constant, 40.96 ms (20).
SOD Activity.
For SOD activity, WT and mutants were grown in TSB 45 mM glucose and 2-mL culture was harvested at 24 h and 72 h. The cultures were centrifuged at 16,000 × g for 5 min. The cell pellets were initially washed with saline before finally being resuspended in 0.5 mM KPO4 buffer pH 7.3 with 1× EDTA free protease inhibitor (Roche). Following mechanical lysis of cells using a bead beater, the clear lysate was retrieved by centrifuging crude extract at 16,000 × g for 5 min at 4 °C. The protein concentration was estimated using Bradford protein assay (Bio-Rad) as per the manufacturer’s instructions and 10 µg of protein was used to evaluate SOD activity using an SOD assay kit (Sigma-Aldrich).
LC-MS/MS Analysis.
Mass spectrometry analysis was performed by the University of Nebraska Medical Center Mass-Spectrometry and Proteomics Core Facility. Method details are described in more detail in SI Appendix, Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Prof. Peter Zuber, Oregon Health & Science University, for generously providing us with anti-Spx antibodies. This work was financially supported by research grants from the National Institute of Allergy and Infectious Diseases P01 AI83211 (to K.W.B. and V.C.T.), R01-AI125589 (to K.W.B.), and R01 AI125588 (to V.C.T.); Nebraska Collaboration Initiative Seed Grant 21-1106-6009 (to R.S., K.W.B., and V.C.T.); and Qassim University postgraduate scholarship No. 167637 (to A.A.A.). The University of Nebraska Medical Center Mass Spectrometry and Proteomics Core Facility is administrated through the Office of the Vice-Chancellor for Research and supported by state funds from the Nebraska Research Initiative. The electron paramagnetic resonance spectroscopy core is supported, in part, by NIH Center of Biomedical Research Excellence Grant 1P30GM103335 awarded to the University of Nebraska’s Redox Biology Center. The funders had no role in study design, data collection, interpretation, and decision to submit this work for publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
1Present address: Unaizah College of Medicine and Medical Sciences, Qassim University, Unaizah 56219, Saudi Arabia.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109671118/-/DCSupplemental.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (54) partner repository with the dataset identifier PXD029457. The source data for proteomics is provided in Datasets S1–S6. All other study data are included in the article and SI Appendix.
References
- 1.Kolter R., Siegele D. A., Tormo A., The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47, 855–874 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Lewis K., Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Rohmer L., Hocquet D., Miller S. I., Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19, 341–348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y. J., Rubin E. J., Feast or famine: The host-pathogen battle over amino acids. Cell. Microbiol. 15, 1079–1087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaishankar J., Srivastava P., Molecular basis of stationary phase survival and applications. Front. Microbiol. 8, 2000 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nyström T., Stationary-phase physiology. Annu. Rev. Microbiol. 58, 161–181 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Allocati N., Masulli M., Di Ilio C., De Laurenzi V., Die for the community: An overview of programmed cell death in bacteria. Cell Death Dis. 6, e1609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhari S. S., et al. , The LysR-type transcriptional regulator, CidR, regulates stationary phase cell death in Staphylococcus aureus. Mol. Microbiol. 101, 942–953 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas V. C., et al. , A central role for carbon-overflow pathways in the modulation of bacterial cell death. PLoS Pathog. 10, e1004205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauer R. T., et al. , Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119, 9–18 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stahlhut S. G., et al. , The ClpXP protease is dispensable for degradation of unfolded proteins in Staphylococcus aureus. Sci. Rep. 7, 11739– (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frees D., Savijoki K., Varmanen P., Ingmer H., Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria. Mol. Microbiol. 63, 1285–1295 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Rode T. M., et al. , Responses of Staphylococcus aureus exposed to HCl and organic acid stress. Can. J. Microbiol. 56, 777–792 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Maisonneuve E., Ezraty B., Dukan S., Protein aggregates: An aging factor involved in cell death. J. Bacteriol. 190, 6070–6075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyström T., Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 24, 1311–1317 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frees D., et al. , Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54, 1445–1462 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Feng J., et al. , Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J. Proteome Res. 12, 547–558 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Pamp S. J., Frees D., Engelmann S., Hecker M., Ingmer H., Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188, 4861–4870 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazharul Islam M., et al. , An integrated computational and experimental study to investigate Staphylococcus aureus metabolism. NPJ Syst. Biol. Appl. 6, 3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas V. C., Chaudhari S. S., Jones J., Zimmerman M. C., Bayles K. W., Electron paramagnetic resonance (EPR) spectroscopy to detect reactive oxygen species in Staphylococcus aureus. Bio Protoc. 5, e1586 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiser M. J., Lenski R. E., A comparison of methods to measure fitness in Escherichia coli. PLoS One 10, e0126210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hibbing M. E., Fuqua C., Parsek M. R., Peterson S. B., Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer M. A., Kainz K., Carmona-Gutierrez D., Madeo F., Microbial wars: Competition in ecological niches and within the microbiome. Microb. Cell 5, 215–219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghoul M., Mitri S., The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Cornforth D. M., Foster K. R., Competition sensing: The social side of bacterial stress responses. Nat. Rev. Microbiol. 11, 285–293 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Jelsbak L., et al. , The chaperone ClpX stimulates expression of Staphylococcus aureus protein A by Rot dependent and independent pathways. PLoS One 5, e12752 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clements M. O., Watson S. P., Foster S. J., Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J. Bacteriol. 181, 3898–3903 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imlay J. A., The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keyer K., Imlay J. A., Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. U.S.A. 93, 13635–13640 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayles K. W., The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol. 5, 721–726 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Bayles K. W., Bacterial programmed cell death: Making sense of a paradox. Nat. Rev. Microbiol. 12, 63–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van den Esker M. H., Kovács A. T., Kuipers O. P., YsbA and LytST are essential for pyruvate utilization in Bacillus subtilis. Environ. Microbiol. 19, 83–94 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Ranjit D. K., Endres J. L., Bayles K. W., Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J. Bacteriol. 193, 2468–2476 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker T. A., Sauer R. T., ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta 1823, 15–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaBreck C. J., May S., Viola M. G., Conti J., Camberg J. L., The protein chaperone ClpX targets native and non-native aggregated substrates for remodeling, disassembly, and degradation with ClpP. Front. Mol. Biosci. 4, 26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan I. S., Weiss C. A., Popham D. L., Ramamurthi K. S., A quality-control mechanism removes unfit cells from a population of sporulating bacteria. Dev. Cell 34, 682–693 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decker A. R., Ramamurthi K. S., Cell death pathway that monitors spore morphogenesis. Trends Microbiol. 25, 637–647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aizenman E., Engelberg-Kulka H., Glaser G., An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3',5'-bispyrophosphate: A model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U.S.A. 93, 6059–6063 (1996). Correction in: Proc. Natl. Acad. Sci. U.S.A. 93, 9991 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer F., Weil A., Hamann A., Osiewacz H. D., Human CLPP reverts the longevity phenotype of a fungal ClpP deletion strain. Nat. Commun. 4, 1397 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatterjee I., Neumayer D., Herrmann M., Senescence of staphylococci: Using functional genomics to unravel the roles of ClpC ATPase during late stationary phase. Int. J. Med. Microbiol. 300, 130–136 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Chatterjee I., Maisonneuve E., Ezraty B., Herrmann M., Dukan S., Staphylococcus aureus ClpC is involved in protection of carbon-metabolizing enzymes from carbonylation during stationary growth phase. Int. J. Med. Microbiol. 301, 341–346 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Chatterjee I., et al. , Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J. Bacteriol. 187, 4488–4496 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatterjee I., et al. , Staphylococcus aureus ClpC ATPase is a late growth phase effector of metabolism and persistence. Proteomics 9, 1152–1176 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Gunaratnam G., et al. , ClpC affects the intracellular survival capacity of Staphylococcus aureus in non-professional phagocytic cells. Sci. Rep. 9, 16267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dwyer D. J., Camacho D. M., Kohanski M. A., Callura J. M., Collins J. J., Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol. Cell 46, 561–572 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis B. D., Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51, 341–350 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kannan K., et al. , The general mode of translation inhibition by macrolide antibiotics. Proc. Natl. Acad. Sci. U.S.A. 111, 15958–15963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Unger L., Kisch A., Observations on bacteriostatic and bactericidal action of erythromycin. Proc. Soc. Exp. Biol. Med. 98, 176–178 (1958). [DOI] [PubMed] [Google Scholar]
- 49.Heifets L. B., Lindholm-Levy P. J., Comstock R. D., Bacteriostatic and bactericidal activities of gentamicin alone and in combination with clarithromycin against Mycobacterium avium. Antimicrob. Agents Chemother. 36, 1695–1698 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ling J., et al. , Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol. Cell 48, 713–722 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fey P. D., et al. , A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4, e00537-e12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engman J., von Wachenfeldt C., Regulated protein aggregation: A mechanism to control the activity of the ClpXP adaptor protein YjbH. Mol. Microbiol. 95, 51–63 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Levine R. L., et al. , Determination of carbonyl content in oxidatively modified proteins. Methods Enzymol. 186, 464–478 (1990). [DOI] [PubMed] [Google Scholar]
- 54.Y. Perez-Riverol, The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (54) partner repository with the dataset identifier PXD029457. The source data for proteomics is provided in Datasets S1–S6. All other study data are included in the article and SI Appendix.