Significance
Heme is essential for respiration. As a cofactor of cytochromes, heme functions as a main electron carrier in all respiratory electron transport chains. Therefore, it is natural to expect all respiring and free-living microorganisms to make heme. Against this expectation, here we show that the acI lineage, one of the most abundant bacterial groups in freshwater environments, is unable to biosynthesize heme and requires exogenous heme. Furthermore, we provide genomic evidence for putative heme auxotrophy among many not-yet-cultured aquatic microbial groups. Heme should be the focus of future research on the metabolic dependency among microorganisms and the role of exchangeable metabolites in structuring diverse ecosystems, and would also be a media component that must be considered when cultivating novel microbes.
Keywords: acI lineage, heme, auxotrophy, riboflavin, Black Queen Hypothesis
Abstract
Heme, a porphyrin ring complexed with iron, is a metalloprosthetic group of numerous proteins involved in diverse metabolic and respiratory processes across all domains of life, and is thus considered essential for respiring organisms. Several microbial groups are known to lack the de novo heme biosynthetic pathway and therefore require exogenous heme from the environment. These heme auxotroph groups are largely limited to pathogens, symbionts, or microorganisms living in nutrient-replete conditions, whereas the complete absence of heme biosynthesis is extremely rare in free-living organisms. Here, we show that the acI lineage, a predominant and ubiquitous free-living bacterial group in freshwater habitats, is auxotrophic for heme, based on the experimental or genomic evidence. We found that two recently cultivated acI isolates require exogenous heme for their growth. One of the cultured acI isolates also exhibited auxotrophy for riboflavin. According to whole-genome analyses, all (n = 20) isolated acI strains lacked essential enzymes necessary for heme biosynthesis, indicating that heme auxotrophy is a conserved trait in this lineage. Analyses of >24,000 representative genomes for species clusters of the Genome Taxonomy Database revealed that heme auxotrophy is widespread across abundant but not-yet-cultivated microbial groups, including Patescibacteria, Marinisomatota (SAR406), Actinomarinales (OM1), and Marine groups IIb and III of Euryarchaeota. Our findings indicate that heme auxotrophy is a more common phenomenon than previously thought, and may lead to use of heme as a growth factor to increase the cultured microbial diversity.
The actinobacterial acI lineage is the most abundant bacterioplankton group in freshwater environments (1–4). They have streamlined genomes that suggest auxotrophy for various essential compounds (such as vitamins and amino acids) and have diversified into multiple subgroups that display spatiotemporal patterning typical of niche specialization (5–8). Laboratory cultivation is crucial for the experimental validation of genome-based inferences on the characteristics of environmentally relevant microbial groups (9, 10), as exemplified by studies on the marine SAR11 group (11). However, efforts to cultivate the acI lineage using genome predictions and by inferring ecophysiological traits from the habitats of these cells were unsuccessful (6, 12), until we recently reported an approach for the robust cultivation of two acI strains, thus establishing the first stable cultures of the acI lineage (13).
Stable axenic cultivation of two acI strains was achieved by supplying catalase as a critical growth supplement (13), after which diverse subclades of the acI lineage were isolated with this approach (14). In this previous work (13), the two strains, “Candidatus Planktophila rubra” IMCC25003 (hereinafter referred to as IMCC25003) and “Candidatus Planktophila aquatilis” IMCC26103 (hereinafter referred to as IMCC26103), grew robustly and reproducibly when bovine liver catalase was added to growth media. The growth of these strains coincided with decreases in H2O2 concentrations caused by the addition of catalase, suggesting that the acI group might be sensitive to oxidative stress and therefore proliferated due to the lowered H2O2 concentrations in the culture medium. However, two experimental observations could not be easily reconciled with this interpretation. First, when H2O2 concentration was lowered by the addition of pyruvate, a chemical scavenger of H2O2, the enhancement of acI growth was marginal. Second, the H2O2 concentration that allowed the growth of the two acI strains in the catalase-supplemented laboratory experiment (∼10 nM) was much lower than those measured in the lake from which the strains were isolated (∼60 nM). Therefore, we conjectured that “catalase is essential for the growth of acI strains via mechanisms involving, but not restricted to, H2O2 decomposition” (13).
While further investigating this unsolved discrepancy, we discovered that all genomes of the acI lineage were deficient in the heme biosynthetic pathway (Fig. 1) (discussed in more detail below). Hemes are iron-containing porphyrins that are present as cofactors in a diverse range of bacterial proteins, including respiratory cytochromes, nonrespiratory-chain cytochromes, noncytochrome hemoproteins, gas sensor proteins, peroxidases, and catalases (15). Although several microbial groups with pathogenic, symbiotic, or commensal lifestyles are known to lack the de novo heme biosynthetic pathway (16–21), the complete absence of heme biosynthesis has rarely been reported in free-living organisms (15). Therefore, the deficiency in the heme biosynthetic pathway of the acI lineage was unexpected, inescapably leading us to speculate that acI bacteria require heme, an essential cofactor of catalase, for their growth, rather than the H2O2 scavenging function of catalase.
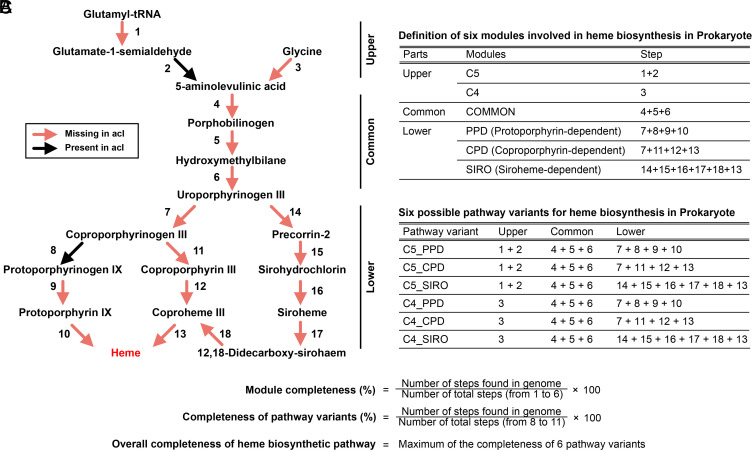
Fig. 1.
Overview of heme biosynthetic pathways in prokaryotes and the acI lineage. (A) Diagram showing the heme biosynthetic pathway steps. The presence (black) or absence (red) of each step in the 20 acI genomes are indicated by colored arrows. (B) Definition of modules and heme biosynthetic pathway variants used in this study. (C) Formulae for the calculation of completeness of modules and pathway variants. The maximum value among the completeness of six pathway variants of each genome was taken as the overall completeness of the genome. Refer to SI Appendix, Table S1 for a list of enzyme names and corresponding KO IDs of the steps defined in the diagram.
In this study, based on genome predictions and experimental evidence, we report that the acI lineage is auxotrophic for heme. To our knowledge, these experiments with acI bacteria are unique in showing that major and abundant lineages of free-living microorganisms inhabiting natural aquatic environments can be heme auxotrophs. We further demonstrate through comparative genomics that heme auxotrophy may be widespread among many abundant prokaryotic groups.
Results
Genomic Evidence of Heme Auxotrophy in the acI Lineage.
All of 20 complete genome sequences from isolated acI strains (5, 6) were found to lack essential heme biosynthesis genes. The heme biosynthetic pathway is complex and consists of three parts (15) (Fig. 1). In the upper part, 5-aminolevulinic acid is synthesized via two different modules known as C4 and C5. In the essential middle part, 5-aminolevulinic acid is converted to uroporphyrinogen III (a common precursor of all heme compounds), via a three-step process common to all organisms, designated as “COMMON.” In the lower part, heme synthesis from uroporphyrinogen III is mediated by three different modules, each designated PPD (protoporphyrin-dependent), CPD (coproporphyrinogen-dependent), and SIRO (siroheme-dependent). Therefore, six variant pathways, combinations of one of the two upper modules with one of the three lower modules, are possible in theory for heme biosynthesis. All acI genomes lack genes for the essential middle part of the pathway and also lack most genes for the upper and lower parts (Fig. 1), which strongly indicates that these strains cannot synthesize heme and therefore require exogenous heme for survival. In accordance with our analyses, the lack of the heme biosynthetic pathway in the acI lineage was also suggested in another recent study (22). Given that heme-containing cytochromes are essential for respiration (23, 24), it was surprising to find that the acI lineage, which is comprised of free-living and obligately aerobic bacteria, does not possess complete sets of heme biosynthesis coding sequences.
Experimental Evidence of Heme Auxotrophy in the Cultured acI Bacteria.
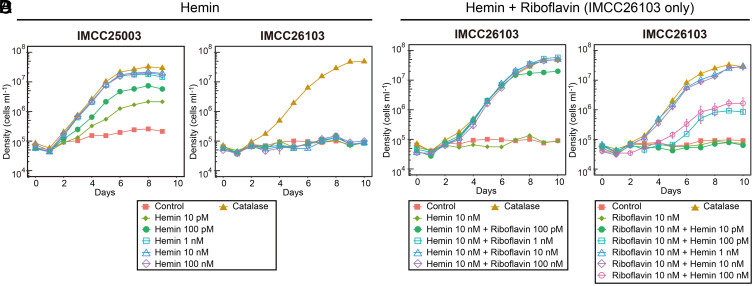
As the first step to test heme auxotrophy of acI bacteria, we investigated the effect of hemin (the oxidized form of heme b) on the growth of the two acI strains, IMCC25003 and IMCC26103, belonging to the acI-A1 and acI-A4 tribes, respectively. IMCC25003 growth was dependent on hemin in a catalase-free medium. Growth was enhanced by hemin at concentrations as low as 10 pM, and enhancement was saturated at hemin concentrations of 1 nM or higher, with maximum cell densities (1.8 to 2.5 × 107 cells mL−1) comparable to that obtained from bovine liver catalase addition (10 U mL−1) (Fig. 2A). Given that the heme concentration in 10 U mL−1 catalase is equivalent to ∼50 nM, this amount of added catalase would be predicted to provide sufficient heme to support full IMCC25003 culture growth. On the other hand, hemin showed no effect on the growth of IMCC26103 (Fig. 2B).
Fig. 2.
Hemin-dependent growth of the two acI strains. (A and B) Effect of various concentrations (0 to 100 nM) of hemin on the growth of “Ca. P. rubra” IMCC25003 (A) and “Ca. P. aquatilis” IMCC26103 (B). (C) Effect of various concentrations (0 to 100 nM) of riboflavin on the growth of IMCC26103 in the presence of hemin (10 nM). (D) Effect of various concentrations (0 to 100 nM) of hemin on the growth of IMCC26103 in the presence of riboflavin (10 nM). The culture medium without hemin and riboflavin was designated as “Control” and the medium with 10 U mL−1 of bovine liver catalase was designated as “Catalase.” All experiments were performed in triplicate. Error bars indicate the SE. Note that error bars smaller than the size of the symbols are hidden.
Since hemin enhanced the growth of IMCC25003, but not IMCC26103, we hypothesized that IMCC26103 requires additional growth factors that are present in catalase preparations and we predicted that, similar to IMCC25003, IMCC26103 would exhibit hemin-dependent growth if this additional factor was provided. A reinvestigation of genome-inferred metabolic potential (6) identified riboflavin, a precursor of FMN and FAD, as the most likely additional growth factor required by IMCC26103 but not IMCC25003. The full gene set for de novo biosynthesis of riboflavin was identified only in IMCC25003. IMCC26103 instead had a gene for the riboflavin transporter PnuX, preceded by the FMN riboswitch (SI Appendix, Fig. S1). Furthermore, the vitamin mixture used in the experiments did not contain riboflavin (SI Appendix, Table S2). Consistent with our genome-based predictions, IMCC26103 grew when riboflavin (≥100 pM) was provided together with hemin (10 nM), and its maximum cell densities in cultures supplemented with 1 to 100 nM of riboflavin reached to levels that were comparable to those of cultures supplied with catalase (Fig. 2C).
Given that these results indicated that the catalase used in this study contained riboflavin, we directly measured the riboflavin concentration in the catalase solution. HPLC analysis of the catalase stock solution (105 U mL−1) demonstrated that 10 U mL−1 of catalase contained ∼690 pM of riboflavin (including FMN and FAD), which explains the growth enhancement of IMCC26103 in the catalase-supplemented cultures. Riboflavin concentrations at picomolar ranges have been reported from various natural waters, including the Northeast Pacific Ocean (1.3 to 9 pM) (25), a coastal region (up to 4.4 pM) (26), and an estuary (45 to 128 pM) (27). Moreover, a recent study reported 14 to 40 pM of riboflavin and 93 to 300 pM of FMN in coastal water samples (28). Considering that the minimum riboflavin concentration required for the growth of acI bacteria would be lower than 100 pM based on the growth curves of IMCC26103 and that freshwater is generally more productive than marine waters, riboflavin supply (including FMN and FAD) in freshwater environments would be sufficient to support the growth of acI bacteria that are auxotrophic for riboflavin.
Upon demonstrating that IMCC26103 requires riboflavin for growth, the effect of hemin on the growth of the strain was reexamined in the presence of riboflavin (10 nM). IMCC26103 exhibited hemin-dependent growth (Fig. 2D), as shown for IMCC25003 (Fig. 2A), providing clear evidence of heme auxotrophy for both of the cultivated acI strains tested. Both acI strains grew well without catalase if the essential component of catalase, heme, and the impurity in the catalase stock, riboflavin, were supplied. It is worth noting that the relationships between growth and hemin concentration were different for the two strains. For example, a hemin supply between 1 and 10 nM maximized the growth of both strains; however, a higher hemin concentration (≥100 pM) was required to grow IMCC26103 (in contrast to 10 pM for IMCC25003), and growth was reduced at 100 nM with IMCC26103, but not IMCC25003 (Fig. 2 A and D). The requirement of higher hemin concentration by IMCC26103 might be ascribed to a difference in putative heme transporters (29). Upon searching for heme uptake systems in the acI genomes, gene clusters similar to the dppABCDF system (30, 31) were found in both strains, but a gene cluster similar to hemTUV (32, 33) was found only in IMCC25003, suggesting that acI bacteria may have a varied repertoire of heme uptake systems that result in differences in the minimal heme concentrations required for growth (see SI Appendix, Table S3 for the distribution of heme uptake systems in 20 complete acI genome sequences). We speculate that the reduced growth enhancement of IMCC26103 at higher hemin concentrations (100 nM) may be due to sensitivity to oxidative stress caused by excess heme (e.g., via the Fenton reaction) (34, 35), as only IMCC25003, but not IMCC26103, possesses the katG gene (13).
Concentrations of dissolved iron–porphyrin-like complexes (up to 11.5 nM) (36) and heme b (47.3 to 70.8 pM) (37) in natural waters are similar to or higher than 10 to 100 pM, the minimum heme concentrations that supported the growth of acI strains, and therefore it seems likely that acI bacterioplankton survive in their habitats through uptake of exogenous heme. In natural systems, heme could originate from many sources (e.g., hemoproteins such as cytochromes), liberated from co-occurring organisms by diverse mechanisms, including viral lysis (38, 39). The concentration of dissolved heme is thus expected to fluctuate spatiotemporally in response to environmental changes associated with microbial dynamics. This fluctuation in heme concentration might be linked to taxon-specific distribution of diverse acI clades and tribes (5, 40), as the affinity of acI bacteria toward heme may vary depending on different uptake systems (SI Appendix, Table S3). Additional measurements of dissolved heme concentration in aquatic habitats and the further characterization of heme uptake systems of the acI lineage are required to better understand the effect of variable heme concentrations on acI populations.
Growth Response of acI Strains to Hydrogen Peroxide.
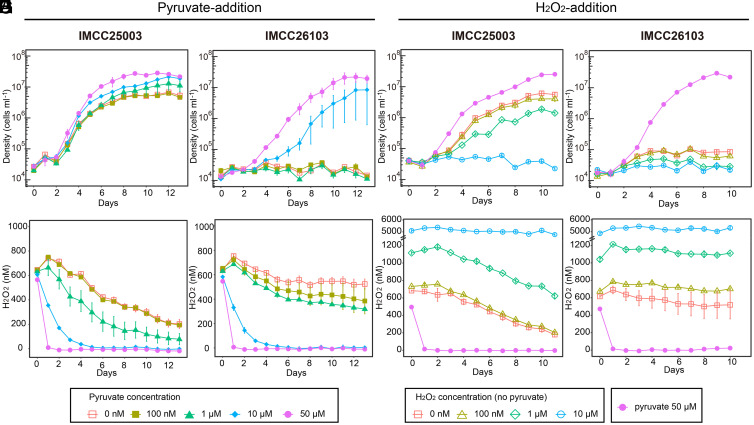
With the establishment of robust growth conditions for acI strains by supplying heme and riboflavin, which do not degrade H2O2 (unlike catalase), we were able to reinvestigate the extreme growth sensitivity of the acI strains to H2O2 reported in our previous study (13). First, we used pyruvate (100 nM to 50 µM) to reduce H2O2 concentrations in our basal medium (filtered and autoclaved lake water; ∼600 nM of H2O2), as pyruvate is a H2O2-scavenging chemical and is not utilized by the acI strains as a carbon source (13). IMCC25003 grew in all conditions, with a slight inhibition dependent on H2O2 concentration (Fig. 3A). Even at H2O2 concentrations above 200 nM throughout the incubation period (pyruvate: 0 or 100 nM), maximum cell densities reached ∼6 × 106 cells mL−1 (Fig. 3 A and E). On the other hand, IMCC26103 exhibited growth only when ≥10 µM pyruvate was added, reducing the H2O2 concentration far below 200 nM, thus indicating that IMCC26103 is more sensitive to H2O2 than IMCC25003 (Fig. 3 B and F). To assess the growth of the acI strains at higher H2O2 concentrations, the two strains were cultured in media supplemented with H2O2 (100 nM to 10 μM). Although inhibited, IMCC25003 grew at initial H2O2 concentrations of nearly 1 μM, and complete inhibition was observed only when H2O2 remained above 5 μM (Fig. 3 C and G). As expected, IMCC26103 failed to grow in any of the H2O2-supplemented conditions (Fig. 3 D and H).
Fig. 3.
Effects of H2O2 on the growth of acI strains. Various concentrations of pyruvate (0 to 50 μM) (A, B, E, and F) and H2O2 (0 to 10 μM) (C, D, G, and H) were added to the basal media containing hemin (10 nM) and riboflavin (1 nM) to adjust the hydrogen peroxide concentration of the media. (A–D) Growth of IMCC25003 (A and C) and IMCC26103 (B and D). (E–H) Changes in H2O2 concentration during cultivation of IMCC25003 (E and G) and IMCC26103 (F and H). All experiments were performed in triplicate. Error bars indicate the SE. Note that error bars shorter than the size of the symbols are hidden.
These observations using culture media supplemented with heme, riboflavin, and varying H2O2 concentrations demonstrate that the acI strains investigated herein are not as sensitive to H2O2 as previously thought. We postulate that these differences in H2O2 sensitivity between the two acI strains are due to the presence of katG (a gene for bifunctional catalase-peroxidase) in IMCC25003, but not in IMCC26103. The KatG protein of IMCC25003 displayed catalase activity in our previous work (13), and therefore its presence in IMCC25003 likely explains why this strain can defend itself against H2O2 stress better than IMCC26103. This difference could lead to niche partitioning among the acI strains, as large spatiotemporal variation in H2O2 concentrations has been reported in freshwater habitats (41). When confronted with high concentrations of H2O2, katG-possessing acI bacteria would gain an advantage despite having to invest resources for KatG production. In contrast, low H2O2 concentrations would favor katG-deficient acI cells, as these conditions would allow for these cells to save the resources required for KatG production. It is also worth noting that the putative fitness advantage gained by possessing the katG gene would be affected also by the availability and concentration of heme, in addition to H2O2 concentrations, as KatG cannot function without heme as pointed out in a recent study (22).
Genomic Evidence of Heme Auxotrophy Across the Uncultured Microbial Majority.
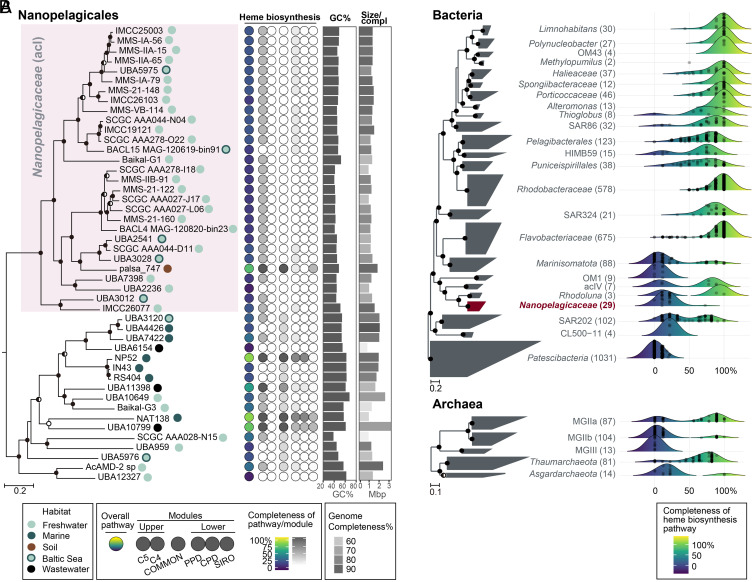
To gain insights into the mechanisms by which the acI lineage lost the heme biosynthetic pathway, we analyzed the distribution of the pathway in the genomes of Nanopelagicales. In the Genome Taxonomy Database (GTDB) (42), the acI lineage is classified as Nanopelagicaceae within the order Nanopelagicales constituting several families. This analysis was performed by taking the maximum value of the completeness of six variants of heme biosynthetic pathways found in each representative genome for species clusters within Nanopelagicales available in the GTDB (release 89) (Fig. 1 and Dataset S1). The pathway was almost completely deficient in all freshwater Nanopelagicaceae genomes, including single-amplified genomes (SAGs) and metagenome-assembled genomes (MAGs), whereas only one MAG from permafrost soil (palsa_747) contained genes encoding early steps in the pathway (Fig. 4A). Other Nanopelagicales genomes not affiliated with the acI lineage exhibited more variable but generally higher completeness than the acI genomes (SI Appendix, Fig. S2). Unlike the acI lineage in which all genomes (except for palsa_747) exhibited <25% completeness, the completeness of non-acI lineage ranged 0 to 89% (SI Appendix, Fig. S2). Most genomes (15 of 17) had at least one gene for the essential (COMMON) part of the heme biosynthetic pathway, and three MAGs retrieved from the ocean (NP52 and NAT138) or wastewater (UBA10799) had nearly complete pathways (Fig. 4A). Given that most of the phylum Actinobacteria genomes exhibited the complete heme biosynthetic pathway (SI Appendix, Fig. S3), it seemed plausible that the loss of the heme biosynthetic pathway occurred in some members of the order Nanopelagicales and that the acI lineage had completely lost the pathway while adapting to freshwater environments through genome streamlining (43, 44).
Fig. 4.
Heme biosynthetic pathway completeness of Nanopelagicales and major aquatic prokaryotic groups. (A) Heme biosynthetic pathway completeness of representative genomes for species clusters belonging to Nanopelagicales. A phylogenomic tree of Nanopelagicales constructed using a concatenated alignment of conserved marker proteins is shown on the left. The circles on the right side of genome names indicate isolation sources (habitats). The Nanopelagicaceae (acI) genomes are shaded in pink. Rhodoluna lacicola MWH-Ta8 (not shown here) was set as an outgroup. The middle column illustrates the overall (leftmost circle) and per-module (the remaining six circles) completeness of the heme biosynthetic pathway among the Nanopelagicales genomes. Refer to Fig. 1 for the heme biosynthetic pathway completeness calculations. The circle colors indicate the pathway completeness according to the color gradients in the bottom legend. The GC contents and genome sizes of the genomes are shown with bar graphs on the right. Genome completeness is indicated by the darkness of the genome size bars, as indicated by the scale at the bottom. (B) Distribution of the completeness of the heme biosynthetic pathway in major aquatic prokaryotic groups. Phylogenomic trees of representative genomes for species clusters affiliated with major aquatic bacterial and archaeal groups (SI Appendix, Table S4 and Dataset S1) are shown on the left. The number of genomes of the groups are indicated at the right of the group names (within parentheses). The Nanopelagicaceae family (acI lineage) is indicated in red. The phylum Patescibacteria and phyla Thaumarchaeota and Asgardarchaeota were set as the outgroups in the bacterial and archaeal trees, respectively. On the right, ridgeline density plots were used to illustrate the distribution of the completeness of heme biosynthetic pathways in the prokaryotic groups. The colors under the ridgelines indicate the completeness following a color gradient shown at the bottom. The points in the plots indicate the completeness of each genome of the groups. For the trees in this figure, bootstrap values (A; from 100 resamplings) and local support values (B; from 1,000 resamplings) are shown at the nodes as filled circles (≥90%), half-filled circles (≥70%), and empty circles (≥50%).
Having shown the absence of the heme biosynthetic pathway in the freshwater acI bacteria, we expanded the scope of our investigation to assess genomic evidence for this trait among all bacteria and archaea. This analysis targeted a total of 24,706 bacterial and archaeal genomes representing species clusters of the GTDB database (Dataset S1). We found that many phyla consisted exclusively of members that lacked heme biosynthesis genes (e.g., Dependentiae, Patescibacteria, and DPANN containing Nanoarchaeota) or contained substantial proportions of predicted heme auxotrophs (e.g., Marinisomatota [SAR406], Chloroflexota, and Asgardarchaeota) (SI Appendix, Figs. S3 and S4). Many of the phyla in which heme biosynthetic pathway gaps were frequent consisted of not-yet or rarely cultured organisms, the so-called microbial dark matter (45), suggesting that heme auxotrophy is a plausible explanation for the challenges of culturing the uncultured microbial majority. Alternatively, considering the enormous metabolic diversity of prokaryotes, unknown heme biosynthesis pathways are a possibility, and several of the groups predicted to be heme-auxotrophs (e.g., Patescibacteria and DPANN) are inferred from their genomes to lack respiration and rely strictly on fermentative metabolism (46, 47); these cells could conceivably replicate with little to no exogenous heme.
The apparent heme auxotrophy in several abundant planktonic clades, such as SAR406 and Marine Group II archaea (MG-II), led us to analyze aquatic microorganisms in more detail (Fig. 4B). A total of 3,233 representative genomes for species clusters of 29 major phylogenetic groups residing in marine, freshwater, and groundwater environments were analyzed (4, 48). The selected groups, their GTDB taxonomy, and related literatures are listed in SI Appendix, Table S4. Evidence of deficiencies in heme biosynthetic pathways was widespread, although varying in degree, in many not-yet-cultivated groups, such as SAR406, Actinomarinales (OM1), CL500-11, Patescibacteria, MG-IIb, MG-III, and Asgardarchaeota (Fig. 4B). Inspection of phylogenomic trees indicated that heme auxotrophy is relegated to specific subclades within each of these groups (SI Appendix, Figs. S5–S11), which indicates that the loss of heme biosynthetic pathways likely occurred independently many times during evolutionary adaptation to specific environmental niches.
We investigated the relationship between genome completeness and the heme biosynthetic pathway completeness (hereinafter, heme completeness) to ascertain whether or not heme auxotrophy could be an artifact caused by the incompleteness of SAGs and MAGs. For this analysis, the 24,706 genomes were divided into two groups according to the GTDB metadata: Isolate and MAG/SAG. Most of the isolates had almost complete genomes with complete or nearly complete heme biosynthetic pathways, with a small number of putative heme auxotrophs, which we defined as heme completeness of <50% (SI Appendix, Fig. S12A). Genome completeness of MAG/SAGs ranged from 50 to 100%, with an evident bias toward higher completeness, whereas heme completeness of MAG/SAGs showed a bimodal distribution with maxima at the complete and incomplete limits of the spectrum (SI Appendix, Fig. S12A), indicating that a substantial number of the taxa represented by MAG/SAGs are heme auxotrophs. When the MAG/SAGs were partitioned into five intervals according to genome completeness, genomes with low heme completeness were a sizable proportion across all intervals, including the most complete one (≥90%) (SI Appendix, Fig. S12B). The substantial proportion of heme incomplete genomes among the most complete MAG/SAGs indicates that the conclusion of widespread heme auxotrophy among not-yet-cultured taxa is not an artifact caused by incompleteness of MAG/SAGs. As exemplified by the two cultured acI strains that showed no growth when heme was not available, heme auxotrophs might have escaped general cultivation efforts better than heme prototrophs. Because genomes of not-yet-cultured microbial groups can be obtained only in the form of MAG/SAGs in most cases, it is likely that most heme auxotroph genomes are MAG/SAGs, thus leading to higher proportions of heme auxotrophs among MAG/SAGs than among isolate genomes regardless of genome completeness.
Having demonstrated that the bimodal distribution of heme completeness in MAG/SAGs was not related to genome completeness, we next sought to determine whether heme auxotrophs across diverse phylogenetic groups share genomic features that might be associated with the loss of heme biosynthesis, with a focus on heme-requiring proteins. We examined the number of proteins assigned to the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) groups that are associated with heme-requiring enzymes, such as cytochrome c oxidase, succinate dehydrogenase, and catalase (Materials and Methods). The number of heme-requiring proteins in the acI lineage was smaller than that in the other genomes belonging to the same class, phyla, or domain. However, the acI lineage had more heme proteins per unit genome length (SI Appendix, Fig. S13). When all bacterial and archaeal genomes were segregated into two groups according to heme completeness (threshold value of 50%), the genomes with lower heme completeness (<50%) had a smaller number of heme-requiring proteins than those with higher completeness (≥50%), irrespective of normalization by genome size (SI Appendix, Fig. S14). These results suggest that the number of heme-requiring proteins may have generally decreased in heme auxotrophs. However, the essential role of some heme-requiring proteins in respiring microbes (e.g., the acI) may have limited the loss of the heme-requiring proteins more strictly than other proteins in streamlined genomes, leading to the increase in the relative proportion of heme-requiring proteins in those genomes.
Discussion
Our findings confirmed heme auxotrophy in the predominant freshwater microbe acI and uncovered genomic evidence indicating that heme auxotrophy has evolved repeatedly among microorganisms from many environments, particularly among cosmopolitan aquatic plankton. The previously reported effect of catalase on the growth of the two acI strains (13) was found to be due to heme, an essential cofactor of catalase, rather than the enzymatic activity of catalase. Discarding the heme biosynthetic pathway could, in principle, be a risky strategy for free-living aerobes because many essential biochemical reactions rely on heme-requiring proteins (e.g., oxidative phosphorylation). Therefore, cells lacking the heme biosynthetic pathway would be destined to depend on exogenous heme, the supply of which could be variable. However, the few studies that have measured free dissolved heme or heme-like molecules in aquatic habitats have detected concentrations that would be sufficient to support the growth of the acI strains studied herein (36, 37). Furthermore, a recent study suggested that, despite lacking the heme biosynthetic pathway, the acI lineage can be successful in freshwater habitats by forming functional cohorts with other diverse microbes where many exometabolites, including heme, play important roles as metabolic currencies (22).
Fitness benefits from abandoning heme biosynthesis likely favor auxotrophic dependency (43). Heme is synthesized through complex biochemical pathways involving 8 to 11 steps (Fig. 1). The total length of the proteins involved in these pathways has been estimated at ∼5,800 to 7,200 amino acids (22), which may be regarded to be quite expensive, especially for microbes inhabiting resource-limited environments. Aside from saving the resources that would be needed to maintain and express the heme biosynthetic machinery, auxotrophic cells might also be able to avoid costs associated with toxicity from heme and heme biosynthesis. Several mechanisms of heme toxicity have been described: generation of reactive oxygen species (ROS) by the Fenton reaction, catalyzed by iron liberated from heme (34, 35), and ROS production by intermediates of heme biosynthesis. H2O2 is produced by several enzyme-dependent reactions, including the oxidation of coproporphyrinogen III to coproporphyrin III (Fig. 1: step 11 in CPD module) and the oxygen-dependent conversion of coproporphyrinogen III to protoporphyrin IX via protoporphyrinogen IX (Fig. 1: steps 8 and 9 in PPD module) (15). The final step of CPD module produces superoxide (49) and nonmetallated intermediates in heme biosynthesis (e.g., protoporphyrin IX) generate ROS when exposed to light and oxygen (35, 49–51). Ostensibly to counter this toxicity and ROS-mediated stress, cells are known to finely regulate heme biosynthesis, have strategies for heme detoxification, such as efflux systems (52, 53), and combat oxidative stress with multiple strategies. In addition to the direct costs of heme biosynthesis, which include acquiring the necessary iron, metabolic burdens associated with regulation and ROS detoxification might be diminished by abandoning heme biosynthesis.
In addition to heme, we also experimentally demonstrated auxotrophy for riboflavin (i.e., vitamin B2) in one of the two acI strains studied here. Genomic analyses further indicated that three strains among the 20 genome-sequenced acI strains are auxotrophic for riboflavin (SI Appendix, Fig. S1). Although flavin cofactors are highly versatile and involved in numerous enzymatic reactions (54, 55), the distribution of flavin cofactors in aquatic habitats and riboflavin auxotrophy among ecologically relevant microbial groups remain largely underexplored compared to other vitamin B compounds, such as thiamine, biotin, and cobalamin (56–58). To the best our knowledge, clear demonstrations of riboflavin auxotrophy among abundant and free-living prokaryotic groups are rare. Therefore, our discovery of riboflavin auxotrophy in the acI lineage is remarkable and expands the scope of studies on vitamin-mediated microbial interaction in the environment.
Collectively, our findings contribute to a growing list of examples in which cells dispense with expensive metabolic pathways in favor of increased reliance on compounds that are fundamentally important to their metabolism but are synthesized by members of surrounding microbial communities, thus providing another case of the Black Queen hypothesis (BQH) (43, 59). Since the first proposal based on the finding of cross-protection of Prochlorococcus from oxidative stress by other co-occurring microbes (43), the BQH has served as a conceptual framework to understand microbial interactions and community evolution. Auxotrophies for so-called exometabolites, such as amino acids and vitamins (22, 58–60), are acknowledged to be the consequences of adaptive gene loss. Furthermore, it has been experimentally demonstrated that the loss of biosynthetic genes can provide selective advantages when their final products are available in the environment (61, 62). In this study, heme is established as a previously unknown kind of exchangeable metabolite that allows microbes to discard the biosynthetic pathway following the BQH.
Two important themes in microbiome science converge in the findings we report: the connectivity of microbiomes and the challenge of culturing a broader range of the uncultured microbial majority. The motivations for advancing the knowledge on these topics are the need to expand the range of cell types that can be studied using experimental microbial cell biology approaches, and also the need to understand the rules that govern interactions among the members of microbial communities (63). The discovery of widespread heme reliance in microbial ecosystems will likely stimulate future investigations of heme concentrations across biomes and the influence of heme traffic between cells on the structuring of microbial communities.
Materials and Methods
Cultivation of the acI Strains.
The basal culture medium used in this study was prepared by adding carbon compounds, nitrogen and phosphorus sources, an amino acid mixture, a vitamin mixture, and trace metals (SI Appendix, Table S2) into 0.2-μm-filtered and autoclaved lake water (13). The lake water used to prepare the medium was collected from the surface (1 m) of Lake Soyang in January 2020. Strains IMCC25003 and IMCC26103 were maintained aerobically in the basal medium supplemented with catalase (10 U mL−1) at 20 °C. A catalase stock solution (105 U mL−1 in 10 mM PBS, pH 7.4) was prepared using bovine liver catalase (C9322, Sigma-Aldrich). For the growth experiments, stock solutions of hemin (5 mM in 5 mM NaOH; 51280, Sigma-Aldrich) and riboflavin (500 μM in Milli-Q water; R9504, Sigma-Aldrich) were prepared and sterilized with filtration through a 0.1-μm pore-size membrane filter. A H2O2 solution (H1009, Sigma-Aldrich) was diluted at a 1:10,000 ratio with sterile water. A pyruvic acid stock solution (50 mM) was also prepared and sterilized with a 0.1-μm pore-size membrane filter prior to use. All culture experiments were performed at 20 °C in the dark using acid-washed polycarbonate flasks. Actively growing cells in the late exponential phase were used as inocula. Cell counts were obtained using a flow cytometer (Guava easyCyte Plus, Millipore) after staining with SYBR Green I (final concentration 5×; Invitrogen) for 1 h.
Determination of Riboflavin Concentration.
HPLC analysis was conducted to quantify the amount of riboflavin in the catalase stock solution. A total of 5 mL of catalase stock solution was diluted to 50 mL with Milli-Q water and heated at 80 °C in a water bath for 20 min, then analyzed with an HPLC system coupled with a fluorescence detector (Agilent) and a CAPCELL PAK C18 MG (250 × 4.6 mm, 5 μm; Shiseido) column. The detector was set to 445-nm excitation and 530-nm emission wavelengths. The mobile phase was prepared with 10 mM NaH2PO4 (pH 5.5) and methanol (76:24 [vol/vol]), and the analysis was conducted with 10 μL of injection volume at a 0.9 mL min−1 flow rate for 30 min. The amount of riboflavin, FMN, and FAD in the sample was calculated based on the peak area of the chromatogram, using standard curves with a high R2 value (0.999) obtained when FAD, FMN, and riboflavin standards were separated under the same conditions. The riboflavin, FMN, and FAD used for the determination of standard curves were purchased from Sigma-Aldrich.
Measurement of Hydrogen Peroxide Concentration.
Hydrogen peroxide concentration was analyzed using the Amplex Red hydrogen peroxide/peroxidase assay kit (A22188, Invitrogen) according to the manufacturer’s instructions. For the assay, 100 μL of Amplex Red working solution (prepared according to the manufacturer’s instructions) was added to each assay tube containing 100 μL of sample. After incubation for 5 min at room temperature in the dark, the fluorescence of resorufin (the oxidized form of the Amplex Red reagent) was measured at excitation and emission wavelengths of 470 nm and 665 to 720 nm, respectively, using a Qubit 3 fluorometer (Thermo Fisher Scientific). To quantify H2O2 concentrations, a standard curve (0.01 to 25 μM of H2O2) was generated by diluting 3.6% (vol/vol) of H2O2 solution with sodium phosphate buffer supplied with the kit.
Analysis on the Completeness of the Heme Biosynthetic Pathway in Prokaryotic Genomes.
A total of 24,706 representative genome sequences for species clusters in the GTDB (Release 89; 23,458 Bacteria and 1,248 Archaea) were downloaded from the GTDB server. This genome set comprises 14,406 isolate genomes, 9,861 MAGs, and 439 SAGs (42). According to the GTDB metadata (available at https://data.gtdb.ecogenomic.org/releases/release89/89.0/), a total of 17,763 genomes (71.9%) satisfied the criteria for high-quality in terms of completeness (≥90%) and contamination (≤5%), and >99.8% of genomes satisfied the criteria for medium quality (completeness of ≥50% and contamination of <10%) (SI Appendix, Table S5) (64). Additionally, the genome sequences of the 20 acI strains isolated to date (5, 6) were downloaded from GenBank. Annotation of the genome sequences was performed using Prokka (v1.14.6), with no rRNA, no tRNA, and no annotation options (65). Predicted protein sequences were searched against the KofamKOALA database (accessed on February 19, 2020) using KofamScan 1.2.0 (66). To reduce run time, the KofamScan was executed using a custom-built hal file containing only 25 KO IDs involved in heme biosynthetic pathways (Fig. 1 and SI Appendix, Table S1), with the “-f mapper” option to retain only the hits above thresholds specific to KO IDs (67). The 25 KO IDs were curated based on a KEGG pathway map titled “Porphyrin and chlorophyll metabolism (ko00860),” KEGG modules M00121 (heme biosynthesis, plants and bacteria, glutamate → heme), M00868 (heme biosynthesis, animals and fungi, glycine → heme), M00846 (siroheme biosynthesis, glutamyl-tRNA → siroheme), and M00847 (heme biosynthesis, archaea, siroheme → heme), in addition to several previous studies (15, 68). The KofamScan output files were then merged for the next step.
To calculate the completeness of the heme biosynthetic pathway using the KofamScan output, we modified “KEGG_decoder.py” script (69) using custom-built definitions for the six modules (C5, C4, COMMON, PPD, CPD, and SIRO) and the six variants (C5_PPD, C5_CPD, C5_SIRO, C4_PPD, C4_CPD, and C4_SIRO) of the pathway (Fig. 1). These pathway definitions are based on the curated pathways and the 25 associated KOs presented in Fig. 1 and SI Appendix, Table S1. The modified Python script (available at https://github.com/SuhyunInha/Heme) was executed with the merged KofamScan output file as an input, which produced completeness values for each of the six modules and six variants for all genomes. For each genome, the maximum completeness value among the six variants was taken as the overall heme biosynthetic pathway completeness.
To estimate the number of heme-requiring proteins encoded in the genomes, we searched and downloaded the list of enzymes that contain heme or cytochrome in the cofactor field from the BRENDA enzyme database (Release 2021.1) (70). From the list, several enzymes that are involved in the biosynthesis of heme were removed. The EC numbers of the remaining 322 enzymes were used to extract the corresponding KO IDs, based on the htext file (“ko00001.keg”) downloaded from the KEGG database (https://www.genome.jp/brite/ko00001; accessed in April 2021). A custom hal file was constructed from the 477 KO IDs that had been extracted, and further used as a profile database for running KofamScan. The protein sequences predicted in the genomes were used as input. The number of heme-requiring proteins (i.e., proteins assigned to the KO IDs associated with heme-requiring enzymes) per genome was calculated from the KofamScan output files. The list of analyzed KO IDs and EC numbers, the custom-built hal file, and summarized results are available at the above Github repository, together with the R script used for the analysis.
Reconstruction of Phylogenomic Trees and Visualization of the Heme Biosynthetic Pathway Completeness.
To visualize the distribution of the heme biosynthetic pathway completeness in prokaryotic genomes, phylogenomic trees were constructed using the trimmed and concatenated multiple sequence alignment of 120 bacterial and 122 archaeal marker proteins downloaded from the GTDB (Release 89). Two phylum-level trees (SI Appendix, Figs. S3 and S4) were inferred by FastTree2 with the default options (71) using all the representative genomes for species clusters. Phyla containing 10 or fewer genomes were not included in the tree building for visualization. The tree in Fig. 4B was also built using FastTree2, using the genomes affiliated with the phylogenetic groups listed in SI Appendix, Table S4. All other trees were built using RAxML 8.2.7 with the PROTGAMMAAUTO option (72). Genome statistics including completeness and contamination calculated by CheckM, size, and GC content were downloaded from the GTDB. Visualization of pathway completeness (modules and variants) was performed using the tidyverse (73), ggridges (74), and viridis packages in R environment (v4.0.2) (75).
Supplementary Material
Acknowledgments
This study was supported by Mid-Career Research Program NRF-2019-R1A2B5B02070538 (to J.-C.C.) and Science Research Center Grant of the National Research Foundation NRF-2018R1A5A1025077 (to J.-W.L., C.O.J., J.-C.C.) through the National Research Foundation funded by the Ministry of Sciences and Information and Communications Technology; and by the Basic Science Research Program funded by the Ministry of Education, Republic of Korea NRF-2020R1A6A3A01100626 (to S.K.) and NRF-2019R1I1A1A01063401 (to I.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102750118/-/DCSupplemental.
Data and Code Availability.
All study data are included in the article and supporting information. The scripts used in this paper are available in GitHub at https://github.com/SuhyunInha/Heme.
References
- 1.Glöckner F. O., et al. , Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66, 5053–5065 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zwart G., Crump B. C., Agterveld M. P. K. V., Hagen F., Han S. K., Typical freshwater bacteria: An analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28, 141–155 (2002). [Google Scholar]
- 3.Warnecke F., Amann R., Pernthaler J., Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6, 242–253 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Newton R. J., Jones S. E., Eiler A., McMahon K. D., Bertilsson S., A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 75, 14–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuenschwander S. M., Ghai R., Pernthaler J., Salcher M. M., Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J. 12, 185–198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang I., Kim S., Islam M. R., Cho J.-C., The first complete genome sequences of the acI lineage, the most abundant freshwater Actinobacteria, obtained by whole-genome-amplification of dilution-to-extinction cultures. Sci. Rep. 7, 42252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton J. J., et al. , Metabolic network analysis and metatranscriptomics reveal auxotrophies and nutrient sources of the cosmopolitan freshwater microbial lineage acI. mSystems 2, e00091-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia S. L., et al. , Model communities hint at promiscuous metabolic linkages between ubiquitous free-living freshwater bacteria. MSphere 3, e00202-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thrash J. C., Culturing the uncultured: Risk versus reward. mSystems 4, e00130-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carini P., A “cultural” renaissance: Genomics breathes new life into an old craft. mSystems 4, e00092-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovannoni S. J., SAR11 bacteria: The most abundant plankton in the oceans. Annu. Rev. Mar. Sci. 9, 231–255 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Garcia S. L., McMahon K. D., Grossart H. P., Warnecke F., Successful enrichment of the ubiquitous freshwater acI Actinobacteria. Environ. Microbiol. Rep. 6, 21–27 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Kim S., Kang I., Seo J.-H., Cho J.-C., Culturing the ubiquitous freshwater actinobacterial acI lineage by supplying a biochemical ‘helper’ catalase. ISME J. 13, 2252–2263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S., Park M. S., Song J., Kang I., Cho J.-C., High-throughput cultivation based on dilution-to-extinction with catalase supplementation and a case study of cultivating acI bacteria from Lake Soyang. J. Microbiol. 58, 893–905 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Dailey H. A., et al. , Prokaryotic heme biosynthesis: Multiple pathways to a common essential product. Microbiol. Mol. Biol. Rev. 81, e00048-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruss A., Borezée-Durant E., Lechardeur D., Environmental heme utilization by heme-auxotrophic bacteria. Adv. Microb. Physiol. 61, 69–124 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Granick S., Gilder H., The porphyrin requirements of Haemophilus influenzae and some functions of the vinyl and propionic acid side chains of heme. J. Gen. Physiol. 30, 1–13 (1946). [PMC free article] [PubMed] [Google Scholar]
- 18.Roden J. A., Wells D. H., Chomel B. B., Kasten R. W., Koehler J. E., Hemin binding protein C is found in outer membrane vesicles and protects Bartonella henselae against toxic concentrations of hemin. Infect. Immun. 80, 929–942 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Ham R. C. H. J., et al. , Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. U.S.A. 100, 581–586 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rollat-Farnier P.-A., et al. , Two host clades, two bacterial arsenals: Evolution through gene losses in facultative endosymbionts. Genome Biol. Evol. 7, 839–855 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baureder M., Hederstedt L., Heme proteins in lactic acid bacteria. Adv. Microb. Physiol. 62, 1–43 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Mondav R., et al. , Streamlined and abundant bacterioplankton thrive in functional cohorts. mSystems 5, e00316-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morel F. M. M., Price N. M., The biogeochemical cycles of trace metals in the oceans. Science 300, 944–947 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Hogle S. L., Barbeau K. A., Gledhill M., Heme in the marine environment: From cells to the iron cycle. Metallomics 6, 1107–1120 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Cohen N. R., et al. , Iron and vitamin interactions in marine diatom isolates and natural assemblages of the Northeast Pacific Ocean. Limnol. Oceanogr. 62, 2076–2096 (2017). [Google Scholar]
- 26.Sañudo-Wilhelmy S. A., et al. , Multiple B-vitamin depletion in large areas of the coastal ocean. Proc. Natl. Acad. Sci. U.S.A. 109, 14041–14045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heal K. R., et al. , Determination of four forms of vitamin B12 and other B vitamins in seawater by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 28, 2398–2404 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Monteverde D. R., et al. , Distribution of extracellular flavins in a coastal marine basin and their relationship to redox gradients and microbial community members. Environ. Sci. Technol. 52, 12265–12274 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Tong Y., Guo M., Bacterial heme-transport proteins and their heme-coordination modes. Arch. Biochem. Biophys. 481, 1–15 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Létoffé S., Delepelaire P., Wandersman C., The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc. Natl. Acad. Sci. U.S.A. 103, 12891–12896 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitra A., Ko Y. H., Cingolani G., Niederweis M., Heme and hemoglobin utilization by Mycobacterium tuberculosis. Nat. Commun. 10, 4260 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stojiljkovic I., Hantke K., Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13, 719–732 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Létoffé S., Delepelaire P., Wandersman C., Functional differences between heme permeases: Serratia marcescens HemTUV permease exhibits a narrower substrate specificity (restricted to heme) than the Escherichia coli DppABCDF peptide-heme permease. J. Bacteriol. 190, 1866–1870 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Everse J., Hsia N., The toxicities of native and modified hemoglobins. Free Radic. Biol. Med. 22, 1075–1099 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Choby J. E., Skaar E. P., Heme synthesis and acquisition in bacterial pathogens. J. Mol. Biol. 428, 3408–3428 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vong L., Laës A., Blain S., Determination of iron-porphyrin-like complexes at nanomolar levels in seawater. Anal. Chim. Acta 588, 237–244 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Isaji Y., Ogawa N. O., Takano Y., Ohkouchi N., Quantification and carbon and nitrogen isotopic measurements of heme B in environmental samples. Anal. Chem. 92, 11213–11222 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Zheng Q., et al. , Highly enriched N-containing organic molecules of Synechococcus lysates and their rapid transformation by heterotrophic bacteria. Limnol. Oceanogr. 66, 335–348 (2020). [Google Scholar]
- 39.Bellworthy J., Gledhill M., Esposito M., Achterberg E. P., Abundance of the iron containing biomolecule, heme b, during the progression of a spring phytoplankton bloom in a mesocosm experiment. PLoS One 12, e0176268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paver S. F., Newton R. J., Coleman M. L., Microbial communities of the Laurentian Great Lakes reflect connectivity and local biogeochemistry. Environ. Microbiol. 22, 433–446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry M. A., et al. , Cyanobacterial harmful algal blooms are a biological disturbance to Western Lake Erie bacterial communities. Environ. Microbiol. 19, 1149–1162 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Parks D. H., et al. , A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 38, 1079–1086 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Morris J. J., Lenski R. E., Zinser E. R., The Black Queen Hypothesis: Evolution of dependencies through adaptive gene loss. MBio 3, e00036-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giovannoni S. J., Cameron Thrash J., Temperton B., Implications of streamlining theory for microbial ecology. ISME J. 8, 1553–1565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rinke C., et al. , Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Brown C. T., et al. , Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Beam J. P., et al. , Ancestral absence of electron transport chains in Patescibacteria and DPANN. Front. Microbiol. 11, 1848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuhrman J. A., Hagström Å., “Bacterial and archaeal community structure and its patterns.” in Microbial Ecology of the Oceans, Kirchman D. L., Ed. (Wiley, 2008), pp. 45–90. [Google Scholar]
- 49.Hobbs C., Dailey H. A., Shepherd M., The HemQ coprohaem decarboxylase generates reactive oxygen species: Implications for the evolution of classical haem biosynthesis. Biochem. J. 473, 3997–4009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stojiljkovic I., Evavold B. D., Kumar V., Antimicrobial properties of porphyrins. Expert Opin. Investig. Drugs 10, 309–320 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Hamblin M. R., et al. , Helicobacter pylori accumulates photoactive porphyrins and is killed by visible light. Antimicrob. Agents Chemother. 49, 2822–2827 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernandez A., et al. , Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae. PLoS Pathog. 6, e1000860 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stauff D. L., et al. , Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J. Bacteriol. 190, 3588–3596 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Macheroux P., Kappes B., Ealick S. E., Flavogenomics—A genomic and structural view of flavin-dependent proteins. FEBS J. 278, 2625–2634 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Leys D., Scrutton N. S., Sweating the assets of flavin cofactors: New insight of chemical versatility from knowledge of structure and mechanism. Curr. Opin. Struct. Biol. 41, 19–26 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Sañudo-Wilhelmy S. A., Gómez-Consarnau L., Suffridge C., Webb E. A., The role of B vitamins in marine biogeochemistry. Annu. Rev. Mar. Sci. 6, 339–367 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Suffridge C. P., et al. , B vitamins and their congeners as potential drivers of microbial community composition in an oligotrophic marine ecosystem. J. Geophys. Res. Biogeosci. 123, 2890–2907 (2018). [Google Scholar]
- 58.Paerl R. W., et al. , Prevalent reliance of bacterioplankton on exogenous vitamin B1 and precursor availability. Proc. Natl. Acad. Sci. U.S.A. 115, E10447–E10456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson W. M., et al. , Auxotrophic interactions: A stabilizing attribute of aquatic microbial communities? FEMS Microbiol. Ecol. 96, fiaa115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia S. L., et al. , Auxotrophy and intrapopulation complementary in the ‘interactome’ of a cultivated freshwater model community. Mol. Ecol. 24, 4449–4459 (2015). [DOI] [PubMed] [Google Scholar]
- 61.D’Souza G., et al. , Less is more: Selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution 68, 2559–2570 (2014). [DOI] [PubMed] [Google Scholar]
- 62.D’Souza G., Kost C., Experimental evolution of metabolic dependency in bacteria. PLoS Genet. 12, e1006364 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zengler K., Zaramela L. S., The social network of microorganisms - how auxotrophies shape complex communities. Nat. Rev. Microbiol. 16, 383–390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bowers R. M., et al. ; Genome Standards Consortium, Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seemann T., Prokka: Rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Aramaki T., et al. , KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanehisa M., Goto S., KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cavallaro G., Decaria L., Rosato A., Genome-based analysis of heme biosynthesis and uptake in prokaryotic systems. J. Proteome Res. 7, 4946–4954 (2008). [DOI] [PubMed] [Google Scholar]
- 69.Graham E. D., Heidelberg J. F., Tully B. J., Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J. 12, 1861–1866 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang A., et al. , BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 49 (D1), D498–D508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Price M. N., Dehal P. S., Arkin A. P., FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wickham H., et al. , Welcome to the Tidyverse. J. Open Source Softw. 4, 1686 (2019). [Google Scholar]
- 74.Wilke C. O., ggridges: Ridgeline plots in ‘ggplot2’. R package version 0.5.1. https://cran.r-project.org/web/packages/ggridges/index.html (2018).
- 75.Team R Core, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information. The scripts used in this paper are available in GitHub at https://github.com/SuhyunInha/Heme.