Significance
The deubiquitinating enzyme CYLD specifically removes Lysine 63 (K63)–linked polyubiquitin chains from several substrates. Although it is highly expressed in the brain, its role in the central nervous system is far from being understood. A thorough behavioral, physiological, and morphological analysis revealed that CYLD-deficient mice exhibit major autism-like behaviors accompanied by deficits in the structure and function of hippocampal synapses. In addition, we show that CYLD is a modulator of mechanistic target of rapamycin (mTOR) signaling, synaptic α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunits, and autophagy in the hippocampus. We thereby introduce CYLD and K63-linked polyubiquitination processes at the synapse as molecular targets for both a better understanding of the pathomechanisms underlying autism spectrum disorder and the development of novel autism therapies.
Keywords: CYLD, autism spectrum disorder, synapse, autophagy, mTOR signaling
Abstract
Nondegradative ubiquitin chains attached to specific targets via Lysine 63 (K63) residues have emerged to play a fundamental role in synaptic function. The K63-specific deubiquitinase CYLD has been widely studied in immune cells and lately also in neurons. To better understand if CYLD plays a role in brain and synapse homeostasis, we analyzed the behavioral profile of CYLD-deficient mice. We found that the loss of CYLD results in major autism-like phenotypes including impaired social communication, increased repetitive behavior, and cognitive dysfunction. Furthermore, the absence of CYLD leads to a reduction in hippocampal network excitability, long-term potentiation, and pyramidal neuron spine numbers. By providing evidence that CYLD can modulate mechanistic target of rapamycin (mTOR) signaling and autophagy at the synapse, we propose that synaptic K63-linked ubiquitination processes could be fundamental in understanding the pathomechanisms underlying autism spectrum disorder.
CYLD is a deubiquitinating (DUB) enzyme first identified as being mutated in familial cylindromatosis, an autosomal dominant genetic predisposition to multiple tumors called cylindromas (1, 2). CYLD is located in the cytoplasm, and its C-terminal catalytic domain mediates the cleavage of tetra-ubiquitin to tri-, di-, and monoubiquitin with a preference for Lysine 63 (K63)– or Met1-linked polyubiquitin chains from several substrates (3). The N terminus comprises three cytoskeletal-associated protein–glycine-rich (CAP–Gly) domains, which can bind microtubules facilitating cytoskeleton formation (4).
Although CYLD is highly expressed in the brain (1, 5), surprisingly only few data are available on its role there. Specifically, CYLD was identified in the postsynaptic density (PSD) as detected by mass spectrometry analysis, immunoblotting, and immunoelectron microscopy (6, 7). In addition, immunogold labeling of dissociated hippocampal cultures under basal and depolarizing conditions showed that CYLD expression significantly increases at PSDs upon neuronal depolarization (7). It was further shown that CYLD recruitment to the PSD is dependent on the activation of Ca2+/calmodulin-dependent protein kinase II upon membrane depolarization and N-methyl-D-aspartate (NMDA) receptor activation (8). Thus, CYLD is recruited to synapses in an activity-dependent manner and involved in the regulation of several signaling pathways. In addition, CYLD was shown to control synapse organization by localizing and compartmentalizing specific synapse targets by deubiquitinating PSD-95, which functions as a major scaffold protein organizing the structure of the PSD (9). Moreover, CYLD was shown to positively regulate dendritic growth by promoting both α-tubulin acetylation in mouse hippocampal neurons and through the interaction of its first CAP–Gly domain with microtubules (10).
Recently, several neuropsychiatric disorders such as autism spectrum disorder (ASD) have been linked to molecular changes in synaptic connections. Interestingly, the proteome analysis of striatal synaptosomes from two mutant lines for Shank3, a major ASD candidate gene, revealed that the amount of synaptic CYLD is significantly increased and reduced in the striatum of Shank3 overexpressing and Shank3-deficient mice, respectively (11, 12).
In this study, we investigated the role of CYLD in synaptic dysfunction related to ASD. We show that Cyld−/− mice exhibit major autism-like phenotypes including impaired social communication, increased repetitive behavior, and cognitive dysfunction. In addition, we associate the behavioral phenotypes with reduced basal synaptic transmission, impaired network excitability, and reduced long-term potentiation (LTP) in Cyld−/− hippocampi. Furthermore, the presence of K63-linked polyubiquitin substrates at the synapse and the abundance of CYLD in the PSD fraction suggest that CYLD is a primary DUB for multiple synaptic proteins. Based on our data, we suggest that CYLD controls mechanistic target of rapamycin (mTOR) signaling and the regulation of autophagic processes. Indeed, the deletion of Cyld resulted in increased hippocampal mTOR activity correlating with decreased levels of the autophagy marker LC3B-II within hippocampal synaptosomes.
Materials and Methods
Mice.
Cyld−/− mice were generated with a targeting construct in which the ATG-containing exon 4 of the Cyld gene was disrupted with a lacZ reporter and a neomycin gene (13). Both Cyld+/− and Cyld−/− are fertile, have a normal life span, and do not spontaneously develop tumors (13).
Shank3b (Shank3tm2Gfng) mice were generated by replacing exons 13 through 16 of the Shank3 gene with a neomycin resistance (neo) cassette, leading to the deletion of major Shank3 isoforms (14). Shank3b mice were acquired from Jackson Laboratory (Stock No. 000664 - B6.129-Shank3tm2Gfng/J). Both Shank3b+/− and Shank3b−/− are viable and fertile.
Heterozygous Cyld+/− mice were crossed to obtain Cyld−/− and Cyld+/+ littermate controls. Homozygous Cyld−/− mice were crossed with homozygous Shank3b−/− mice in order to generate heterozygous mice for both Cyld and Shank3 (Cyld+/−Shank3b+/−). Cyld+/−Shank3b+/− mice were crossed to obtain double-mutant mice.
Only male mice of each line were used in all experiments performed. Since female mice were excluded, our conclusions might be somewhat limited regarding potential sex differences. Mice were housed and bred under specific pathogen-free conditions. Experiments were performed according to guidelines of the central animal facility institution (TARC, Mainz University Medical Center, under animal permit TVA G17-1-093) with mice of 5 to 14 wk of age on a C57BL/6J background who were housed under a 12-h light/dark schedule and had access to food and water. Behavioral experiments were performed at 8 to 14 wk; miniature excitatory postsynaptic current (mEPSC) recordings, morphological, synaptic, and Western blot analysis at 6 wk; and multielectrode array (MEA) recordings at 5 wk.
Behavioral Tests.
The mice tested were housed at four to five per cage in a room with a 12-h light/dark cycle (lights on at 0700 hours) and had free access to tap water and food. The mice were single housed for at least 10 d before performing the ultrasound vocalization test. Regarding Cyld−/− behavioral analysis, only one cohort of mice was analyzed compared to Cyld+/+ controls. In the case of Cyld−/−, Shank3−/−, and Cyld−/−Shank3−/− double-mutant mice, four different cohorts of animals were sequentially analyzed compared to Cyld+/+Shank3+/+ control littermates. All experiments were conducted in a blinded fashion. The order of behavioral tests performed is the following: open field (OF), elevated plus maze (EPM), three-chamber sociability and social memory, object recognition, rotarod, marble burying, grooming, Morris water maze (MWM), and vocalization.
OF.
Spontaneous activity in the OF was tested in a gray Perspex arena (120 cm in diameter, 25 cm high; illumination 120 lx) as described earlier (15). The mice were placed in the center and allowed to explore the OF for 10 min. The behavior was recorded by an overhead video camera and a personal computer (PC) equipped with “Ethovision XT 8.5” software (Noldus, Inc.) to calculate velocity, distance traveled, and time spent in central, intermediate, or peripheral zones of the OF.
EPM.
EPM analysis of anxiety-like behavior was performed as described earlier (15). The plus maze is made of gray plastic with a 5 × 5 cm central platform, 30 × 5 cm open arms and 30 × 5 × 15 cm closed arms; illumination 120 lx. The behavior was recorded for 5 min by an overhead video camera and a PC equipped with “Ethovision XT 8.5” software version 8.5.614 (Noldus, Inc.) to calculate the time spent in open or closed arms, distance traveled, and velocity.
Three-chamber sociability and social memory.
Sociability and social memory were evaluated as described earlier (15, 16). The social testing arena was a rectangular, three-chambered box, illumination <50 lx. Each chamber was 20 × 40 × 22 cm in size. Dividing walls were made from clear Plexiglas with rectangular openings (35 × 35 mm) allowing access into each chamber. Overhead cameras recorded the test and analysis made in a PC equipped with “Ethovision XT 8.5” (Noldus Inc.). There was no delay between the analysis of sociability and social memory: testing mice were removed from the testing arena, the object was replaced with a new stranger mouse, and the testing mice were subsequently placed back in the testing arena. The duration was the following: 5 min habituation to central chamber, 10 min sociability, and 10 min social memory. The stimulus mice were C57BL/6J males and 3 wk old. Based on the amount of time spent in each chamber, a “social index” and a “social memory index” (with a value of 50 meaning no preference) were calculated according to the following formulas:
| Social index=(timestranger 1/(timestranger 1+timeempty))⋅100 |
| Social memory index=(timestranger 2/(timestranger 2+timestranger 1))⋅100 |
Object recognition.
The object recognition test performed is an adaptation of the traditional novel object recognition test previously described (15). The object recognition test consists of three phases. In the first phase, the animal is presented to an empty test arena (white box with 40 × 40 × 40 cm dimensions) for 10 min in order to be familiarized with the test environment, which differs from the home cage. Following the habituation phase, the animal is returned to the home cage for a minimal time, which is necessary to place an object into the arena. In the second phase, the animal is returned to the test arena and presented to the object for 5 min. The time spent exploring the object (interaction time) is recorded by “Ethovision XT 8.5” (Noldus, Inc.). Exploration is defined as approaching the object with the nose closer than 2 cm. A total of 1 h after the first object exploration, the animal is returned to the testing arena where it is again presented to the same object for 5 min. The recognition memory readout is the reduction of exploration during the second time of exposure to the same object.
Rotarod.
The rotarod test was performed as described earlier (15) using a rotating drum (Ugo Basile) whose speed accelerates from 4 to 40 rpm over a 5-min time period. After 24 h of habituation on the stationary rotarod drum, the mice were placed individually on the accelerating drum, and the latency of falling off the drum was recorded using a stopwatch.
Marble burying.
The marble burying test examines repetitive motor behaviors (17). The mice are placed individually in a 45 × 25 × 20 cm box, which was previously filled with 5 cm of fresh bedding and 20 marbles evenly distributed on the bedding, for 30 min. Afterward, the number of marbles buried by the mice is measured. A marble is considered to be buried if at least 2/3 of the marble is covered with bedding.
Grooming.
Grooming is also a measure of repetitive motor activity (18). The mice were placed in a glass cylinder container 20 cm in diameter with no bedding. There was no habituation, and the measuring started 10 s after the mouse was placed in the container. Grooming behavior was measured for 15 min. Face wiping, scratching/rubbing of head and ears, and full-body grooming were counted as grooming behavior. The total duration of grooming was evaluated.
MWM.
The MWM test was performed as described earlier (19, 20). The swimming activity of each mouse was monitored via an overhead video recording by “Ethovision XT 8.5” (Noldus, Inc.), which relays information including latency to the platform, total distance traveled and trajectory of swimming, time and distance spent in each quadrant, and swim speed.
Ultrasound vocalization.
Rodent communication in the ultrasonic range was recorded as previously described (20) using the recording software Avisoft-SASLab Pro version 4.33 at a sampling frequency of 300 kHz. The microphone (UltraSoundGate CM16) was connected to a preamplifier (UltraSoundGate 116), which was connected to a computer (Avisoft Bioacoustics). For the test, male mice were first habituated to the test arena. Subsequently, an unfamiliar C57BL/6J female mouse in estrous was put into the same box, and the vocalization behavior of the male was recorded for 5 min. In Fig. 1, Cyld−/− and Cyld+/+ littermate controls were analyzed only once in the presence of the same C57BL/6J female. We then considered only the mice who emitted at least one call in the presence of the C57BL/6J female. In Fig. 2, we tested each single mouse four times in four different test phases in the presence of four different C57BL/6J females. We then averaged, for each mouse, the number of calls emitted in the presence of n females who induced the vocalization in that precise male. The latency until the first call of the male test mouse and the total number of calls were counted by manual scoring.
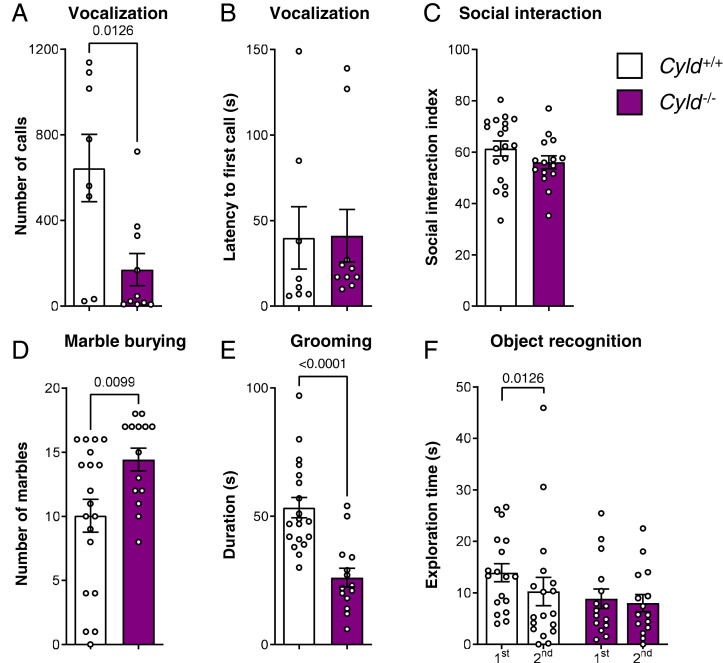
Fig. 1.
Cyld−/− mice display autism-like phenotypes including an impairment of social communication, increased repetitive behavior, and cognitive dysfunction. (A and B) Vocalization analysis of the number of calls emitted in the presence of a female mouse and latency to first call. (C) Social index of social interaction analysis in the three-chamber test. (D and E) Repetitive behavior assessment with both marble burying test and time of grooming. (F) Analysis of the exploratory behavior with the time of the first and the second exposure to the same object. For experiments in A and B, n = 8 for Cyld+/+ controls (11 excluded, no vocalization) and n = 10 for Cyld−/− (five excluded, no vocalization); for experiments in C through F, n = 19 for Cyld+/+ controls and n = 15 for Cyld−/−. All n values used for statistics refer to the number of mice used after identification of possible outliers with Grubbs' method. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.001 calculated by unpaired nonparametric Mann–Whitney U test. Graphs are mean ± SEM.
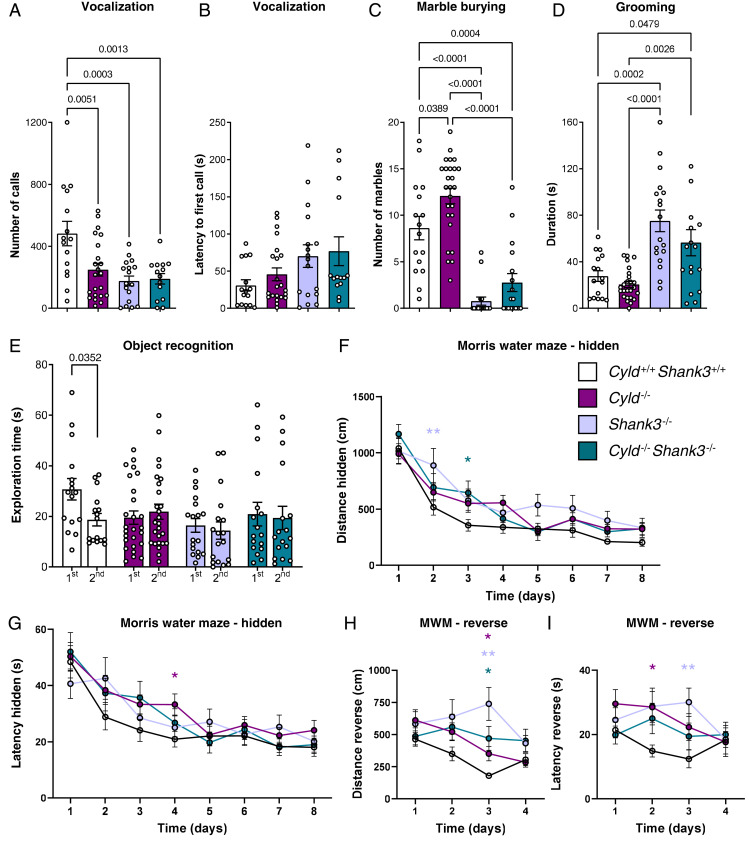
Fig. 2.
Behavioral analysis of Cyld−/−, Shank3−/−, and Cyld−/−Shank3−/− double-mutant mice. (A and B) The vocalization analysis of the number of calls emitted in the presence of a female mouse and latency to first call. (C and D) The repetitive behavior assessment with both marble burying test and time of grooming. (E) The analysis of the exploratory behavior with the time of the first and the second exposure to the same object. (F–I) The analysis of spatial memory by MWM test with measures of total distance traveled and latency to the platform during the hidden training phase and reverse phase. For experiments in A and B, n = 15 for Cyld+/+Shank3+/+ controls (one excluded, no vocalization), n = 22 for Cyld−/− mice (four excluded, no vocalization), n = 17 for Shank3−/− (one excluded, no vocalization), and n = 14 for Cyld−/−Shank3−/− mice (three excluded, no vocalization) from four independent cohorts. For experiments in C through E, n = 16 for Cyld+/+Shank3+/+ controls, n = 26 for Cyld−/− mice, n = 18 for Shank3−/−, and n = 17 for Cyld−/−Shank3−/− mice from four independent cohorts. For experiments in F through I, n = 15 for Cyld+/+Shank3+/+ controls, n = 22 for Cyld−/− mice, n = 17 for Shank3−/−, and n = 13 for Cyld−/−Shank3−/− mice from four independent cohorts. All n values used for statistics refer to the number of mice used after identification of possible outliers with Grubbs' method. Statistics were calculated by ordinary one-way ANOVA followed by Tukey’s multiple comparisons test (A–D) by unpaired nonparametric Mann–Whitney U test (E) or by two-way repeated-measures ANOVA with Dunnett’s multiple comparisons test (F–H). *P < 0.05, **P < 0.01, graphs are mean ± SEM.
Electrophysiology.
Acute brain slices.
The animals were deeply anesthetized with isoflurane, decapitated, and the brain quickly removed and transferred to a 4 °C cold choline-based cutting solution containing 87 mM NaCl, 25 mM NaHCO3, 37.5 mM choline chloride, 2.5 mM KCl, 1.25 mM NaH2PO4, 0.5 mM CaCl2, 7 MgCl2, and 25 mM glucose (pH 7.4) and oxygenated with carbogen (95% O2, 5% CO2). Next, the brain was cut into 200-µm thick coronal slices for patch-clamp recordings and 400-µm thick hippocampal slices for MEA recordings with a Leica VT1200 S vibratome. The slices recovered in the cutting solution for 20 min at 37 °C before they were transferred to standard artificial cerebrospinal fluid (ACSF) at 37 °C: 125 mM NaCl, 25 mM NaHCO3, 1.25 mM NaH2PO4, 2.5 mM KCl, 2 mM CaCl2, 1mM MgCl2, and 25 mM glucose (pH = 7.4).
Patch-clamp recordings.
For patch-clamp recordings, single slices were transferred to a submerged recording chamber mounted on top of an upright microscope (Olympus BX51WI, Olympus) and equipped with 4× (Plan N, numerical aperture [NA] 0.1, Olympus) and 40× (LUMPlan FI/IR, NA 0.8 w, Olympus) objectives. The recording electrode (borosilicate glass, outer diameter 2.0 mm; wall thickness 0.5 mm) was pulled using a micropipette puller (P1000, Sutter Instrument Company) and had a resistance of 5 to 6 MΩ. Electrical signals were acquired at 10 kHz by an EPC10 amplifier (HEKA) and recorded with Patchmaster software (HEKA). The data were analyzed semiautomatically using pClamp 10.4 software.
mEPSCs.
Cells were recorded in voltage-clamp mode (21). To measure mEPSCs, 50 μM (2R)-amino-5-phosphonovaleric acid (D-APV = NMDAR antagonist, NB-48–0302, Biotrend), 10 μM 4-(6-imino-3-(4-methoxyphenyl)pyridazin-1-yl) butanoic acid hydrobromide (gabazine = GABAAR-antagonist, 217808, Biotrend), and 1 μM tetrodotoxin (TTX = voltage gated sodium channel blocker, H1035, Hello Bio) were added to ACSF. The intracellular solution contained 120 mM Cs-gluconate, 10 mM CsCl, 8 mM NaCl, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), 10 mM phosphocreatine-Na, 0.3 mM guanosine triphosphate (GTP), 2 mM Mg2+-ATP, and 0.2 mM ethyleneglycol-bis(β-aminoethyl)-N,N,N′,N′-tetraacetic acid, pH 7.3 adjusted with CsOH. To study the morphology of the cells after recording, 0.1 to 0.5% biocytin (Sigma-Aldrich) was added to the intracellular recording solution and sonicated for 15 min. Cells were clamped to −70mV. The resting membrane potential was tested in current clamp mode directly after the membrane was ruptured. The access to the cell was constantly monitored by a hyperpolarizing the test pulse of 5 mV for 5 ms. The liquid junction potential was not corrected. The average of the amplitude, event frequency, and interevent interval was calculated from more than 30 mEPSCs events per cell.
MEA recordings.
Extracellular recordings of field excitatory postsynaptic potentials (fEPSPs) were acquired with a two-chamber MEA system (MEA2100 System, Multi Channel Systems MCS GmbH). MEA chips consisted of 60 electrodes, an electrode diameter of 30 μm, and an interelectrode distance of 200 μm (60MEA200/30iR; Multi Channel Systems MCS GmbH) and were continuously perfused with oxygenated (95% O2 and 5% CO2) ACSF. The temperature in the chambers was set to 32 °C. Acute hippocampal slices were placed on top of the MEA chips and incubated for 30 min prior to the recordings. fEPSPs were recorded using the Multi Channel Experimenter 2.16 (Multi Channel Systems MCS GmbH) with a 50-kHz sampling rate.
Input–Output (I/O) curves were generated by the application of a stimulus from 500 to 5,000 mV in 500-mV steps to the stratum radiatum close to the CA3 area. One recording electrode was selected in Cornu Ammonis 1 (CA1) (2). Prior to LTP induction, control fEPSPs were recorded at 30% of the maximum fEPSP amplitude for 10 min (one stimulus per minute). LTP was induced by a 1× 100-Hz high frequency stimulation (HFS). Next, control fEPSPs were evoked for 1 h. An independent control pathway was recorded using a second stimulation electrode located in the direction of the subiculum. LTP traces were normalized to the mean of 10 min of the baseline control recordings.
Events were detected and analyzed using the Multi Channel Analyzer 2.16 (Multi Channel Systems MCS GmbH), Microsoft Office Excel (Microsoft), and GraphPad Prism (GraphPad Software).
Morphological Analysis of Neurons and Dendritic Spines.
Immunofluorescence staining of biocytin-filled neurons.
Electrophyiologically patched neurons were filled with biocytin (see intracellular solution) for about 10 min and then transferred into 4% Histofix (Carl Roth) at 4 °C overnight. The slices were incubated for 1 to 2 h in 1× phosphate-buffered saline (PBS)/0.2% Triton. A total of 20% bovine serum albumin (Sigma-Aldrich) was added to prevent unspecific binding of the antibody. The slices were then incubated at 4 °C overnight in fresh blocking solution together with a streptavidin-coupled Alexa 594–conjugated antibody (1:1.000, S32356, Life Technologies). The slices were mounted onto a microscope slide (Thermo Scientific) in Prolong Gold Antifade (P10144, Life Technologies). The images were taken with an SP8 confocal microscope (Leica) at the Imaging Core Facility of Mainz University Medical Center using the Leica LAS Software (Leica). To detect the streptavidin-coupled Alexa 594–conjugated antibody, a 552-nm laser was used. Whole cell neurons were imaged with a 20× oil-immersion objective (Leica) and had the following voxel sizes: x/y = 0.314 μm and z = 1 μm. Medium spiny neuron dendrites (S4) were imaged with a 63× oil-immersion objective (Leica) and a voxel size of the following: x/y = 0.048 μm and z = 0.5 μm. The same parameters were used for pyramidal cells of the CA1 layer in the hippocampus. Here, we distinguished between apical and basal dendrites.
Dendritic tree analysis.
The images of biocytin-filled striatal medium spiny neurons and CA1 pyramidal neurons (PNs) were reconstructed through three-dimensional Imaris analysis. The stacks were opened with Imaris 8.4 and processed using the AutoPath (no loops) algorithm, which prevents looping back on neuronal processes. To quantify dendritic tree complexity, Sholl analysis and total dendritic length measurement from Imaris were used on reconstructed cells. The starting radius was defined as 1 µm from the center, with increments of 1 µm.
Dendritic spine analysis.
In order to yield a better resolution of the spines on the dendrite images, a blind deconvolution was performed using the Lightning tool of the LAS software (Leica). The spines were then analyzed using Neuronstudio (CNIC, Mount Sinai School of Medicine).
Immunofluorescence Staining of Striatal Synapses.
Tissue fixation and processing.
The mice were perfused intracardially with cold PBS and 4% paraformaldehyde (PFA). PFA-perfused central nervous system tissue was postfixed in 4% PFA overnight at 4 °C and embedded in paraffin. After antigen retrieval and unspecific binding blocking, PFA-fixed sections were incubated with primary antibodies. Bound antibodies were visualized with appropriate species-specific Cy2- or Cy3-conjugated secondary antibodies. The nuclei were stained with DAPI (Invitrogen). Immunostained sections were scanned using the confocal microscope LSM 800 (Zeiss), with objective magnification of 63×, sampling tiles from dorsal and ventral (bits 16, pixel size 0.06 mm).
Antibodies.
The Mouse anti-VGluT1 (Synaptic Systems, 135 011) was 1:500, the Rabbit anti-VGluT2 (Synaptic Systems, 135 408) was 1:500, the Chicken anti-Homer-1 (Synaptic Systems, 160 006) was 1:500, the Goat anti-chicken Alexa Fluor 647 (Thermo Fisher, A-21449) was 1:200, the Goat anti-mouse Alexa Fluor 555 (Thermo Fisher, A-21127) was 1:200, and the Goat anti-rabbit Alexa Fluor 488 (Thermo Fisher, A-11008) was 1:200.
Quantification of synaptic terminals.
To evaluate the number of excitatory presynaptic (VGluT1 and VGluT2) and postsynaptic terminals (Homer-1), positive signals were quantified using a custom-made script based on Cognition Network Language (Definiens Cognition Network Technology; Definiens Developer XD software). The total tissue area was normalized using DAPI+ signal to ensure similar cellular numbers in the field of view. White matter tracts within striatum were excluded during analysis.
Western Blotting.
Preparation of homogenates from brain tissue.
To obtain lysates, dissected tissue was homogenized with a manual homogenizer in cold lysis buffer (2% sodium dodecyl sulfate [SDS], 95 mM NaCl, 10 mM ethylenediaminetetraacetic acid, Protease Inhibitor Mixture Complete [Roche], and Phosphatase inhibitor mixtures PhosSTOP [Roche]), boiled for 5 min, centrifuged at 14,000 rpm at 16 °C, and the supernatant collected for protein estimation. The supernatants were then aliquoted and stored at −20 °C until used.
Isolation of crude synaptosomes from brain tissue.
Tissue was homogenized with 800 µL homogenization buffer (0.32 M sucrose in 5 mM Hepes buffer pH 7.4 ± Protease Inhibitor Mixture Complete [Roche] and Phosphatase inhibitor mixtures PhosSTOP [Roche], 1 tablet of each for 50 mL ± o-PA 5 mM ± N-ethylmaleimide 5 mM) per hippocampus or striatum using a manual homogenizer with appropriate vessel and pistil. After 12 strokes (12 times down and up), the homogenate was transferred to a 1.5-mL Eppendorf cup. Samples were subsequently spun at 1,000 × g for 10 min at 4 °C. The supernatant S1 was collected, and the pellet P1 was rehomogenized in 600 µL homogenization buffer in the Eppendorf cup with an Eppendorf pellet pestle. The sample was spun again at 1,000 × g for 10 min at 4 °C, and supernatants S1` and S1 were combined. Combined supernatants were centrifuged at 12,000 × g for 20 min at 4 °C. Supernatant S2 was collected; pellet P2 was resuspended in 800 µL homogenization buffer with an Eppendorf pellet pestle. The resuspended P2 was spun at 12,000 × g for 20 min at 4 °C. Supernatants S2` and S2 were combined. The remaining pellet P2 is the crude synaptosomal fraction, which can be resuspended in specific lysis buffer.
Sample preparation and immunoblotting procedure.
For brain tissue, protein concentrations were determined by the bicinchoninic acid (BCA) method (Pierce BCA Protein Assay Kit, Thermo Fisher Scientific, Inc.); samples (10 to 30 μg protein) were fractionated by SDS–polyacrylamide gel electrophoresis (PAGE) and blotted onto nitrocellulose using the Trans-Blot Turbo Transfer System (BioRad). Membranes were blocked in 4% milk in Tris-buffered saline with Tween-20 (TBST) 0.05%, Triton X-100 in Tris-buffered saline (24.2 g/L Trizma Base, 87.6 g/L NaCl in water, pH 7.5) for 1 h. Appropriate regions were excised and incubated with specific primary antibody at 4 °C overnight. Secondary fluorescent antibodies conjugated with 680 or 800 were incubated with membranes for 1 h at room temperature. The membranes were then washed three times in TBST 0.05% and two times in PBS 1× and developed with Odyssey.
Co-immunoprecipitation.
Precleared whole-cell lysate (300 µg) from a Cyld+/+ hippocampus was incubated with rabbit anti-CYLD (Cell Signaling, 8462) or control rabbit IgG (Sigma-Aldrich, 12 to 370) at 4 °C overnight with gentle agitation. Protein G agarose beads (Roche, 11719416001) (20 µL) were added, and the samples were incubated for 2 h at 4 °C. The immunocomplexes were then pelleted, washed multiple times at 4 °C, and subjected to SDS-PAGE and Western blotting.
Antibodies.
The Guinea pig anti-GluA2 (Synaptic Systems, 182 105) was 1:1,000, the Mouse anti-GAPDH (Invitrogen, AM4300) was 1:5,000, the Mouse anti-PSD-95 (Synaptic Systems, 124 011) was 1:1,000, the Mouse anti-rpS6 (Cell Signaling, 2317) was 1:1,000, the Rabbit anti-CYLD (Cell Signaling, 8462) was 1:1,000, the Rabbit anti-GluA1 (Synaptic Systems, 182 003) was 1:1,000, the Rabbit anti-Homer-1b/c (Synaptic Systems, 160 023) was 1:1,000, the Rabbit anti-LC3B (Sigma-Aldrich, L7543) was 1:1,000, the Rabbit anti-mTOR (Cell Signaling, 2983) was 1:1,000, the Rabbit anti-p-mTOR (Cell Signaling, 2971) was 1:1,000, the Rabbit anti-p-rpS6 (Cell Signaling, 2215) was 1:1,000, the Rabbit anti-p-S6K (Cell Signaling, 9234) was 1:1,000, the Rabbit anti-Shank3 (Sigma-Aldrich, HPA003446) was 1:1,000, the Rabbit anti-S6K (Cell Signaling, 2708) was 1:1,000, the Donkey anti-Guinea pig IgG (H + L) IRDye 800CW (Licor) was 1:10,000, the Goat anti-Mouse IgG (H + L) IRDye 680RD (Licor) was 1:10,000; and the Goat anti-Rabbit IgG (H + L) DyLight 800 4X PEG Conjugate (Cell Signaling, 5151) was 1:10,000.
Statistical Analysis.
Statistical analysis was performed with Prism (GraphPad, Version 9). Results are shown as the mean ± SEM. The number of experiments and the number of mice used for each experiment are indicated in the corresponding figure legends. P < 0.05 was taken as the limit of statistical significance. All behavioral tests, electrophysiological analysis, neuronal reconstruction, and analysis of dendritic tree and spine morphology were performed in a blinded manner.
Results
CYLD Deletion Leads to Social Deficits, Repetitive Stereotypic Movements, and Cognitive Impairment.
As CYLD is highly expressed in neurons and its synaptic protein levels depend on the dosage of the major ASD gene Shank3 (12), we set out to understand if it is involved in brain function by analyzing the autism-like behavior of Cyld−/− mice. Cyld−/− mice show normal motor function and anxiety behavior. In detail, Cyld−/− mice did not show any difference compared to controls in the latency to fall from the accelerating rod in the rotarod and in total distance traveled in both OF and EPM (SI Appendix, Fig. S1 A–C). Moreover, Cyld−/− mice did not exhibit any difference compared to the control animals in the time spent in the different OF arena areas and in the time spent in both closed and open arms of EPM (SI Appendix, Fig. S1 E–G). Within the social domain, one of the core diagnostic domains of ASD, we detected an impairment in social communication, which in rodents is reflected by calls in the ultrasonic range. We found that, in comparison to control mice, Cyld−/− male mice vocalize significantly less, without any difference in the latency to the first call produced (Fig. 1 A and B). However, Cyld−/− mice do not show any impairment in either social interaction or social memory in the three-chamber test (Fig. 1C and SI Appendix, Fig. S1D). Importantly, repetitive, stereotypic movements, which represent the second core diagnostic feature of ASD, are also affected in Cyld−/− mice. Cyld−/− mice show an increase in the number of buried marbles in the marble burying test (Fig. 1D) and a reduction in self-grooming behavior compared to controls (Fig. 1E). The third domain we found affected in Cyld−/− mice is cognition. Altered intellectual abilities are a major comorbid feature of ASD, and, to measure these alterations, we used the object recognition test. This test is based on the ability of the animal to memorize an object for a period of 60 min, leading to a reduction in exploration time during the second exposure of the same object. We found that indeed the control mice exhibit a significant reduction in exploration time between the first and second exploration (Fig. 1F). In contrast, Cyld−/− mice did not show any difference in exploration time among the two exposures to the same object (Fig. 1F), suggesting a deficit in recognition memory.
CYLD Deletion Does Not Modify Autism-Like Behavioral Phenotypes in Shank3-Deficient Mice.
Recently, it has been shown that synaptic CYLD is coregulated within the brain of Shank3 mutant mice (11, 12). Thus, we crossed Cyld−/− and Shank3−/− mice and analyzed the neurobehavioral phenotype of the resulting Cyld−/−Shank3−/− double-mutant mice in order to clarify if CYLD deletion can modify the autistic phenotype of Shank3−/− mice. We found that similarly to the Cyld−/− mice, the Shank3-deficient mice and the double-mutant animals also display an impairment in social communication, showing reduced numbers of calls in the presence of a female compared to controls (Fig. 2A). In contrast, the reduced number of calls was not associated with a change in the latency to the first call emitted (Fig. 2B). However, the occurrence of repetitive, stereotypic movements affects CYLD and Shank3-deficient mice in opposite ways. Cyld−/− mice show an increase in the number of buried marbles and a reduction in self-grooming behavior, and Shank3−/− mice show a decreased number of buried marbles and an increased duration of grooming (Fig. 2 C and D), while the double-mutant mice performed exactly like Shank3−/− mutant mice in both the marble burying test, with a reduction of buried marbles under cage bedding and in overgrooming (Fig. 2 C and D).
The analysis of cognitive functions with the object recognition test highlighted an impairment of memorizing an object in Cyld−/−, Shank3−/−, and Cyld−/−Shank3−/− mice. Differently from control littermates, the single-deficient or double-deficient mice did not show a reduction in the exploration time during the second exposure to the same object (Fig. 2E). To further explore cognitive features, we decided to extend our analysis using the MWM test. The analysis of both escape latency and distance traveled during hidden platform training in the MWM test showed a worse performance of Cyld−/−, Shank3−/−, and Cyld−/−Shank3−/− mice in reaching the platform compared to controls, with a significant difference in the distance at day 2 between control and Shank3−/− mice, at day 3 between control and Cyld−/−Shank3−/− mice, and in the latency at day 4 between control and Cyld−/− mice (Fig. 2 F and G). During the probe trial, we did not observe any difference in the percentage of time spent in the target quadrant among the four different genotypes (SI Appendix, Fig. S2A). At the end of the hidden phase training, Cyld−/−, Shank3−/−, Cyld−/−Shank3−/−, and controls reach the same level of performance. In the reversal learning phase, we found that the plasticity needed in order to localize the new place of the hidden platform within the pool was impaired in Cyld−/−, Shank3−/−, and Cyld−/−Shank3−/− mice compared to controls. A significant difference in the latency to reach the platform was detectable at day 2 between control and Cyld−/− mice and at day 3 between control and Shank3−/− mice. At day 3 of reversal learning, we could also detect a significant difference in the distance traveled between Cyld−/−, Shank3−/−, Cyld−/−Shank3−/−, and controls (Fig. 2 H and I), respectively. To sum up, Cyld−/− and Shank3−/− mutants show similar impairments in the social and cognitive domains but opposite phenotypes in the repetitive domain. Neither phenotype is restored nor exacerbated in Cyld−/−Shank3−/− double mutants whose repetitive behavioral profile rather resembles the one of single Shank3−/− mutants.
CYLD Function Modulates Hippocampal Plasticity.
Dysregulation of neuronal morphology and cytoarchitecture in the hippocampus are typical for ASD (22). Furthermore, a disruption of hippocampal circuits often underlies cognitive impairment, a major comorbid ASD feature present in Cyld−/− mice. To elucidate a possible dysfunction of CYLD-deficient hippocampal neurons, we performed an electrophysiological analysis. The evaluation of CA1 PNs showed that the mean amplitude of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor–mediated mEPSCs was significantly decreased in Cyld−/− mice when compared to controls, suggesting alterations of basic synaptic properties of CA1 PNs upon loss of CYLD (Fig. 3 A and B). No difference was detected in the mean of mEPSC frequency between Cyld−/− mice and controls (Fig. 3 C). An analysis of striatal medium spiny neurons (MSNs) was also performed since behavioral tests showed alterations of repetitive stereotypic movements in Cyld−/− mice. However, the analysis of mEPSCs of striatal MSNs did not show any difference in amplitude and frequency in both dorsal and ventral striatum among Cyld−/− mice and controls (SI Appendix, Fig. S3 A–F). The unaffected basal synaptic properties of striatal MSNs are in line with the unchanged numbers of corticostriatal and thalamostriatal connections, as we did not find any difference among genotypes in the density of presynaptic contacts stained with VGluT1 (corticostriatal inputs), VGluT2 (thalamostriatal inputs), or corresponding postsynaptic specializations on MSN dendrites stained with Homer-1 in both the dorsal and ventral striatum (SI Appendix, Fig. S4 A–L).
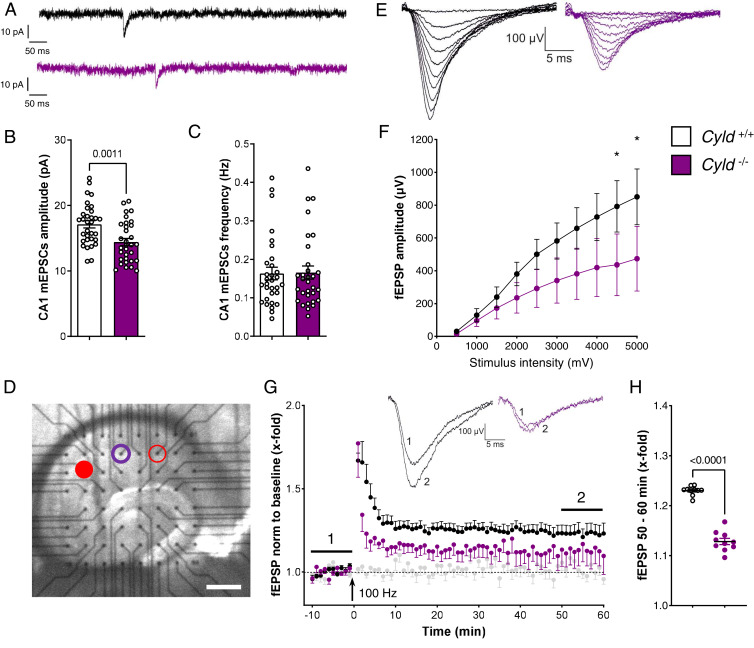
Fig. 3.
Electrophysiological analysis of CA1 mEPSCs and fEPSPs highlighted an impaired excitability in Cyld−/− mice. (A–C) Representative traces of AMPA-mediated mEPSCs and quantification of mEPSC frequency and amplitude in the hippocampal CA1 region of Cyld−/− mice and Cyld+/+ controls at P42. (D) Representative picture of the location of an acute hippocampal slice on top of the MEA chip. The red-filled dot indicates the stimulation electrode, the violet circle marks the recording electrode, and the empty red circle marks the second stimulation electrode of the independent control pathway. (E) Representative traces of fEPSP at gradually increasing stimulus intensity generate I/O curves in both experimental groups. (F) The I/O curves show decreased fEPSP amplitudes in Cyld −/− mice (*P < 0.05). (G) LTP induction of Schaffer collaterals using a 100 Hz HFS. Black dots correspond to the data from Cyld+/+ (10 slices of four mice), violet dot from Cyld −/− (11 slices of four mice), and the gray dots were recorded from the independent control pathway (11 slices). (H) The relative strength of the potentiation of fEPSPs 50 to 60 min after HFS was significantly lower in Cyld −/− (****P < 0.001). For experiments in A through C, n = 32 neurons from Cyld+/+ controls and n = 32 neurons from Cyld −/− mice. Statistics were calculated by unpaired Student’s t test (A–C). Nonparametric Mann–Whitney U tests were used to compare two groups displaying a nonnormal distribution. Two-way ANOVA was used to compare the I/O curve progression, and responses to single intensities were compared using Fisher’s Least Significant Difference. *P < 0.05, graphs are mean ± SEM.
In order to investigate further mechanisms linked to the autism-like phenotypes detected in CYLD-deficient mice, we extended our electrophysiological analysis by measuring neuronal network activity in acute hippocampal slices using MEA recordings. First, we evaluated the excitability of CA3 to CA1 synapses (Schaffer collaterals) in acute slices to understand if the lack of CYLD already implies changes in excitability. One stimulation electrode (filled red dot, Fig. 3D) was used to stimulate afferent fibers of the Schaffer collaterals to record an I/O relation of CA1 PNs. The amplitudes of fEPSPs (purple circle, Fig. 3D) showed an impaired excitability in Cyld−/− tissue compared to controls (Fig. 3 E and F), with a significant difference in the highest stimulation range (4,500 to 5,000 mV).
Next, we characterized neuronal long-term synaptic plasticity in Cyld−/− mice by performing classical LTP experiments in CA1. Baseline fEPSPs were electrically induced at an intensity that evoked 30% of the maximum response evoked by the I/O curve (1,500 mV). Input specificity was controlled by a second independent stimulation (control pathway, empty red circle, Fig. 3D, gray dots in Fig. 3G). Following 10 min of stable baseline recordings, HFS was applied for 1 s at a frequency of 100 Hz. The stimulation induced LTP at CA3 to CA1 synapses in control animals, while the level of LTP was much lower in slices from Cyld−/− mice, indicating an impaired hippocampal LTP in mice lacking CYLD (Fig. 3G). The mean amplitudes of fEPSPs recorded from Cyld−/− mice were significantly reduced 50 to 60 min after LTP induction when compared to control tissue (Fig. 3H).
CYLD Deletion Disrupts Neuroanatomical Properties of CA1 PNs.
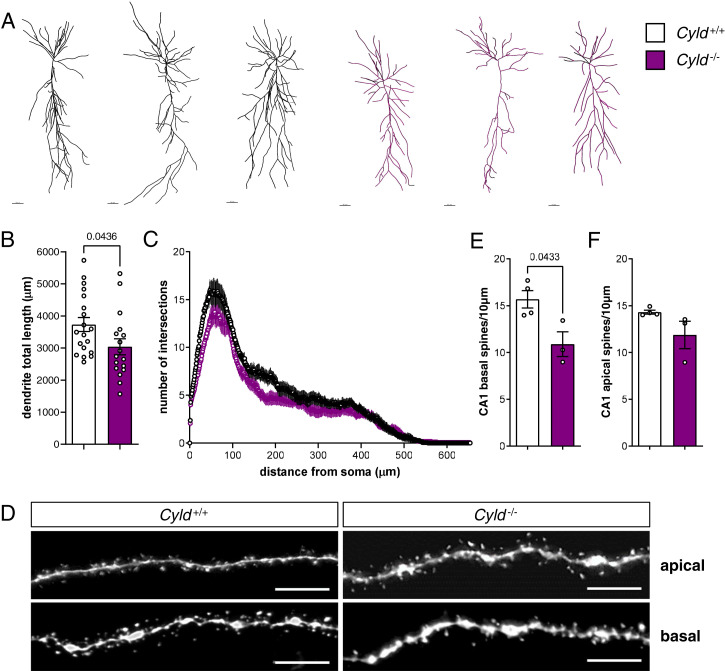
Next, we set out to investigate if CYLD deletion affects dendritic tree and spine changes in the young adult brain. To this end, we filled neurons with biocytin and analyzed neuronal and spine morphology. Using Imaris, we detected a decrease of dendrite total length in CYLD-deficient mice as compared to control animals (Fig. 4 A and B), while a Sholl analysis of branching complexity did not highlight any significant differences among genotypes (Fig. 4C). Moreover, we counted the number of spines at the basal and apical dendrites of CA1 PNs and found a significant decrease in the total number of basal spines (Fig. 4 D and E) and a trend toward a reduction of apical spines (Fig. 4F) in Cyld−/− mice compared to controls.
Fig. 4.
CYLD deletion results in reduced total dendrite length and basal spine number of CA1 hippocampal PNs. (A) Representative pictures of reconstructed biocytin-filled PNs by Imaris of Cyld−/− mice and Cyld+/+ controls at P42 in CA1. (B and C) Measurement of dendrite total length and Sholl analysis of CA1 PNs. (D) Representative pictures of apical and basal spines of Cyld−/− and Cyld+/+ PNs. (E and F) Quantification of basal and apical spines. For experiments in A through C, n = 19 neurons from Cyld+/+ controls and n = 17 neurons from Cyld−/− mice. For experiments in D through F, n = 4 for Cyld+/+ controls and n = 3 for Cyld−/− mice. The statistics were calculated by unpaired Student’s t test (B, E, and F) and two-way repeated ANOVA with Sidak’s (C) post hoc tests. Graphs are mean ± SEM. (Error bars in A: 40 µm; error bars in D: 5 µm.)
In contrast to hippocampal PNs, a morphological analysis of biocytin-filled striatal MSNs in the dorsal and ventral striatum did not highlight differences regarding neuronal dendritic complexity and total dendritic length (SI Appendix, Fig. S5 A–F). Moreover, the total spine number among MSN dendrites was also unchanged within the dorsal and ventral striatum of Cyld−/− and control mice (SI Appendix, Fig. S5 G–I).
CYLD Deletion Results in Altered Autophagy and mTOR Signaling.
To elucidate a possible molecular mechanism by which the deubiquitinating function of CYLD is able to regulate the normal protein homeostasis of synapses, we performed a biochemical characterization of major PSD proteins that have already been molecularly linked to CYLD (9, 11, 12). To isolate the crude synaptosome fraction, we followed a well-established protocol (SI Appendix, Fig. S6 A and B) which allows the enrichment of synapse-associated proteins (23). In particular, we analyzed by Western blotting Shank3 and PSD-95 protein levels in these fractions from both hippocampus and striatum and did not find any change between Cyld−/− mice compared to controls (SI Appendix, Fig. S7 A–F).
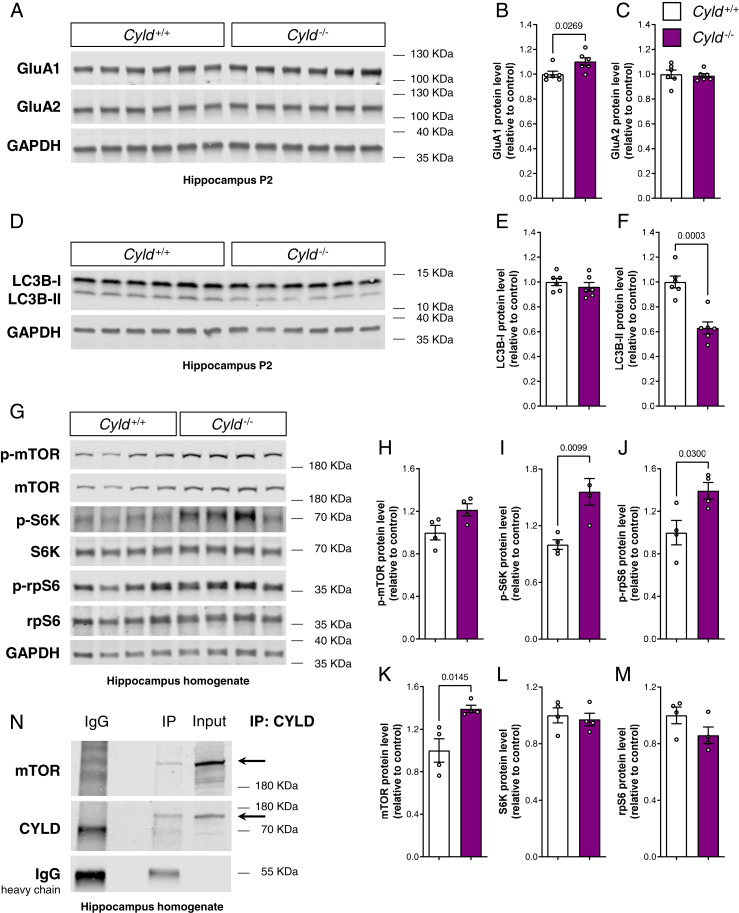
Given the electrophysiological impairment in hippocampal CA1 upon loss of CYLD, we further evaluated the levels of the two major subunits of the AMPA receptor GluA1 and GluA2, both crucial for functional plasticity (24). Interestingly, the amount of GluA1 in the hippocampal synaptosome fraction was significantly higher in Cyld−/− mice compared to controls, while GluA2 protein levels remain unchanged (Fig. 5 A–C). Because of recent evidence implicating CYLD in the autophagic–lysosomal pathway and the fact that autophagy significantly contributes to AMPA receptor internalization (25–27), we decided to analyze changes in autophagy-associated proteins in isolated synaptosomes. Indeed, the quantification of the autophagosome marker LC3B (microtubule-associated proteins 1A/1B light chain 3B) showed a significant reduction specifically for its lipidated form LC3B-II, but not its unlipidated form LCB-I, in Cyld−/− hippocampal synaptosomes under steady state conditions (Fig. 5 D–F), indicative for dysregulated autophagic activity. In striatal synaptosomes, we did not find any change in either AMPA receptor subunits GluA1 and GluA2 or in the autophagosome marker LC3B in line with both electrophysiological and morphological analyses performed (SI Appendix, Fig. S7 G–L).
Fig. 5.
The loss of CYLD leads to an increase of the AMPA receptor subunit GluA1, dysregulates autophagic flux, and causes mTOR signaling up-regulation within the hippocampus. (A–C) Western blot analysis of the AMPA receptor subunits GluA1 and GluA2, each normalized to GAPDH in the hippocampal P2 fraction. (D–F) Western blot analysis of LC3B-I and LC3B-II, each normalized to GAPDH in the hippocampal P2 fraction. (A–G) Western blot analysis of mTOR signaling cascade and relative quantification in hippocampus homogenate. p-mTOR, p-S6K, and p-rpS6 were normalized to total mTOR, S6K, and rpS6, respectively. Total mTOR, total S6K, and total rpS6 were normalized to GAPDH. Lysates (20 µg proteins for A and D and 30 µg proteins for G) of Cyld−/− mice and Cyld+/+ controls were run on a 4 to 15% gradient gel. (H) CYLD immunoprecipitation from wild-type hippocampus homogenate run on a 4 to 15% gradient gel and blotted for mTOR and CYLD. For experiments in A through F, n = 6 for Cyld+/+ controls and n = 6 for Cyld−/− mice at P42. For experiments in G through M, n = 4 for Cyld+/+ controls and n = 4 for Cyld−/− mice at P42. Statistics were calculated by unpaired Student’s t test and show a significant increase in GluA1, a significant decrease in LC3B-II protein levels, and a significant increase in total mTOR, p-S6K, and p-rpS6 in Cyld−/− hippocampi. Graphs are mean ± SEM.
Mechanistically, autophagosome formation is induced by an upstream blockage of mTOR activity (28). Thus, we investigated whether mTOR signaling was affected in the hippocampus of Cyld−/− mice. Strikingly, we detected a significant increase in total mTOR protein levels, resulting in a significant overactivation of the signaling cascade further downstream reflected by increased p-S6K and p-rpS6 in Cyld−/− mice compared to controls, while total levels of S6K and rpS6 did not change (Fig. 5 G–M). It has been shown that the K63-specific ubiquitination of mTOR leads to its activation and modulation of the autophagic flux (29). To address the potential function of CYLD in the regulation of mTOR signaling, we further analyzed whether CYLD interacted with the mTOR complex. For this, we specifically immunoprecipitated CYLD and detected mTOR in a wild-type hippocampus homogenate by immunoblotting (Fig. 5N), suggesting that CYLD functions as a DUB for mTOR.
Discussion
Evidence that CYLD is an important regulator of synapse homeostasis has increased over the last decade (30). Our behavioral analysis of CYLD-deficient mice shows that CYLD is essentially regulating three main behavioral domains impaired in ASD: social communication, repetitive behavior, and cognition. Since genotypes differed during the acquisition but not the recognition phase of the object recognition test, these findings might alternatively be explained by a differential response to novelty among genotypes. Nevertheless, the MWM data support the presence of cognitive impairment upon loss of CYLD.
Furthermore, by behaviorally investigating the recently proposed molecular relationship between Cyld and Shank3 gene dosage (11, 12), we found that deleting CYLD in mice deficient in Shank3 neither ameliorates nor exacerbates the behavioral profile of the latter. We therefore conclude that the decrease of synaptic CYLD in Shank3−/− animals is a consequence of imbalanced synaptic activity and corresponding molecular synaptic network composition caused by the loss of Shank3. Since CYLD is actively recruited to the PSD upon synaptic activity (7, 8), a possibility could be that CYLD cannot enter the synapse in Shank3-deficient mice anymore because of a disrupted postsynaptic scaffold.
We further characterized CYLD-deficient mice to delineate the circuits linked to the behavioral phenotype, focusing on regions relevant to ASD such as the striatum and the hippocampus. We found that CA1 PNs within the hippocampus of Cyld−/− mice exhibit impaired basal synaptic properties, disrupted network excitability, and reduced LTP. This dysfunction is accompanied by reduced total dendrite length and spine numbers, analogous to what has previously been described upon CYLD knockdown in primary hippocampal cultures (10). In contrast, we did not see any difference in several functional and morphological parameters of striatal MSNs, although CYLD is highly expressed in this region (1, 5). Furthermore, the number of corticostriatal and thalamostriatal inputs was not affected. These data suggest that the function of synaptic CYLD is region/circuit specific and affects only certain populations of neurons. CYLD deletion does not lead to a broad but rather specific cellular outcome. Particularly, we reckon that CYLD fine tunes synapse function by deubiquitinating specific synaptic target molecules and that the unbalanced regulation of these molecules finally leads to the functional, morphological, and behavioral phenotypes of Cyld−/− mice. It will be crucial in the future to obtain a detailed understanding of the neuronal populations and associated circuits underlying these phenotypes and to identify the possibly circuit-specific synaptic deubiquitination targets of CYLD.
Interestingly, the cellular hippocampal phenotypes we observed were accompanied by increased levels of the AMPA receptor subunit GluA1 in the synaptosomal fraction of Cyld−/− hippocampus. Decreased LC3B-II levels were also detected in the same synaptosomal fraction, suggesting that the removal of CYLD affects autophagic activity at hippocampal synapses. A limited number of studies with contrasting results have addressed how AMPA receptor endosomal trafficking converges with autophagy (26, 27). Shehata et al., for example, show that in hippocampal neurons, autophagy is involved in AMPA receptor degradation after chemical long-term depression (LTD) induced by stimulating neurons with brief low-dose NMDA (26). Thus, increased autophagy leads to LTD and correlates with increased LC3-II levels, mTOR dephosphorylation, and degradation of GluA1 (26). Instead, Shen et al. suggest that autophagy inhibition causes a reduction of endocytic recycling and is required for AMPA receptor internalization and degradation, promoting synaptic LTD (27). In our study, we also show that a dysregulated autophagic flux with reduced levels of the LC3B-II autophagosome marker correlates with an increase in GluA1 AMPA receptor subunit. Further elucidation on how exactly CYLD regulates autophagy and AMPA endosomal trafficking is crucial to be addressed in future studies.
We further noted the hippocampal up-regulation of mTOR signaling upon deletion of CYLD in mice. Recently, CYLD has been proposed to inhibit autophagic degradation at the stage of autolysosome efflux in cardiomyocytes by blocking mTOR reactivation (25). Controversially, in the same study of Qi et al., it has been shown that knockdown of CYLD in mouse embryonic fibroblasts leads to increased autophagic flux, while CYLD overexpression in cardiomyocyte leads to decreased mTOR signaling, measured via decreased pS6K levels, which also implies increased autophagosome formation (25). In addition, in vitro overexpression of a specific gain-of-function mutation of CYLD identified in amyotrophic lateral sclerosis (ALS)/frontotemporal dementia impairs autophagosome maturation (31). mTOR is known to contribute to synaptic plasticity and memory through the regulation of protein synthesis, and mutations in components of its pathway have been extensively associated with neurodevelopmental disorders (32–34). For example, peripheral blood mononuclear cells of individuals affected by mild and severe idiopathic autism show an increased activity of mTOR signaling, suggesting it as a molecular signature of clinical ASD severity (32). Furthermore, mTOR signaling is also one of the modulators of autophagy, and its activation inhibits autophagy at an early step in autophagosome formation (35). Tang et al. show that Tsc2+/− mice, in which mTOR is constitutively hyperactive, are characterized by postnatal spine pruning defects, ASD-like social behaviors, and decreased levels of the autophagic marker LC3-II (36). Yan et al. demonstrate that hippocampal neurons from Fmr1−/− mice modeling fragile X syndrome are characterized by reduced autophagy, down-regulation of LC3-II, and overactivation of mTOR (37). Furthermore, they show that Fmr1−/− neurons exhibit an accumulation of ubiquitinated protein aggregates, indicating that autophagy is involved here (37). Our analysis of hippocampal tissue and synaptosome preparations recapitulate the phenotype described in both Tsc2+/− mice and Fmr1−/− neurons, with an overactivation of mTOR signaling and a decrease in LC3B-II protein levels.
In addition to phosphorylation, K63 polyubiquitination is also a posttranslational modification regulating mTOR activity. Linares et al. described how the E3 ubiquitin ligase TRAF6 is required for mTOR activation via K63 polyubiquitination, and TRAF6 knockdown in cells leads to higher levels of LC3-II and enhanced autophagic flux (29). Our data now indicate that CYLD and mTOR interact in mouse hippocampi, which strongly suggests that CYLD is a DUB for mTOR. It has been previously proposed that TRAF6 and CYLD may contribute to synaptic homeostasis in a precise equilibrium in order to regulate the K63 polyubiquitination status of specific synaptic targets (9). Thus, if TRAF6 is the E3 ubiquitin ligase responsible for K63 polyubiquitination and activation of mTOR, CYLD might regulate K63 deubiquitination and inactivation of mTOR. One open question remains as to how K63 polyubiquitination and phosphorylation concert mTOR activity and how CYLD is specifically involved in these processes.
In summary, our study sheds light on the role of DUBs in the context of neurodevelopmental disorders. We provide strong evidence that CYLD is crucial to maintain synaptic homeostasis in the hippocampus and, consequently, cognitive function. It will be essential to identify the specific targets of CYLD at the synaptic level in the future to better clarify its precise molecular function within the neuronal circuitry underlying autism-like phenotypes. This will certainly pave a new way to target the K63 ubiquitination machinery at the synapse for the development of novel autism therapies.
Supplementary Material
Acknowledgments
M.J.S. was supported by the Volkswagen Foundation (“Experiment” Grant 95 235) and the German Research Foundation (DFG, Collaborative Research Center [CRC] 1080, Project B10). A.W. was supported by the DFG Grants TRR128 (Projects A03 and A07) and CRC1292 (Project TP15) and the Reinhart Koselleck Grant. A.M.C. and C.B. were supported by the DFG Grant CRC1177 (Project ID 259130777). D.M. was supported by the European Research Council (Grant 865026) and the Swiss NSF (310030_173010 and 310030_185321). T.M. was supported by the DFG (CRC 1080, project C02).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110755118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
Change History
January 3, 2022: The author footnote has been updated.
References
- 1.Bignell G. R., et al. , Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 25, 160–165 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Biggs P. J., et al. , Familial cylindromatosis (turban tumour syndrome) gene localised to chromosome 16q12-q13: evidence for its role as a tumour suppressor gene. Nat. Genet. 11, 441–443 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Sato Y., et al. , Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat. Struct. Mol. Biol. 22, 222–229 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Lork M., Verhelst K., Beyaert R., CYLD, A20 and OTULIN deubiquitinases in NF-κB signaling and cell death: so similar, yet so different. Cell Death Differ. 24, 1172–1183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazarei G., et al. , Expression analysis of novel striatal-enriched genes in Huntington disease. Hum. Mol. Genet. 19, 609–622 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dosemeci A., et al. , Composition of the synaptic PSD-95 complex. Mol. Cell. Proteomics 6, 1749–1760 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dosemeci A., Thein S., Yang Y., Reese T. S., Tao-Cheng J. H., CYLD, a deubiquitinase specific for lysine63-linked polyubiquitins, accumulates at the postsynaptic density in an activity-dependent manner. Biochem. Biophys. Res. Commun. 430, 245–249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thein S., et al. , CaMKII mediates recruitment and activation of the deubiquitinase CYLD at the postsynaptic density. PLoS One 9, e91312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Q., et al. , Proteasome-independent polyubiquitin linkage regulates synapse scaffolding, efficacy, and plasticity. Proc. Natl. Acad. Sci. U.S.A. 114, E8760–E8769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J., Sekine-Aizawa Y., Ebrahimi S., Tanaka S., Okabe S., Tumor suppressor protein CYLD regulates morphogenesis of dendrites and spines. Eur. J. Neurosci. 50, 2722–2739 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Reim D., et al. , Proteomic analysis of post-synaptic density fractions from Shank3 mutant mice reveals brain region specific changes relevant to autism spectrum disorder. Front. Mol. Neurosci. 10, 26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin C., et al. , Shank3 regulates striatal synaptic abundance of Cyld, a deubiquitinase specific for Lys63-linked polyubiquitin chains. J. Neurochem. 150, 776–786 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Massoumi R., Chmielarska K., Hennecke K., Pfeifer A., Fässler R., Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell 125, 665–677 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Peça J., et al. , Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 472, 437–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radyushkin K., et al. , Complexin2 null mutation requires a ‘second hit’ for induction of phenotypic changes relevant to schizophrenia. Genes Brain Behav. 9, 592–602 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Crawley J. N., Designing mouse behavioral tasks relevant to autistic-like behaviors. Ment. Retard. Dev. Disabil. Res. Rev. 10, 248–258 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Thomas A., et al. , Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology (Berl.) 204, 361–373 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalueff A. V., et al. , Neurobiology of rodent self-grooming and its value for translational neuroscience. Nat. Rev. Neurosci. 17, 45–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris R., Developments of a water-maze procedure for studying spatial learning in the rat. J. Neurosci. Methods 11, 47–60 (1984). [DOI] [PubMed] [Google Scholar]
- 20.Radyushkin K., et al. , Neuroligin-3-deficient mice: model of a monogenic heritable form of autism with an olfactory deficit. Genes Brain Behav. 8, 416–425 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Neher E., Sakmann B., Single-channel currents recorded from membrane of denervated frog muscle fibres. Nature 260, 799–802 (1976). [DOI] [PubMed] [Google Scholar]
- 22.Varghese M., et al. , Autism spectrum disorder: neuropathology and animal models. Acta Neuropathol. 134, 537–566 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smalla K.-H., Klemmer P., Wyneken U., “Isolation of the postsynaptic density: A specialization of the subsynaptic cytoskeleton” in The Cytoskeleton, Dermietzel R., Ed. (Humana Press, Totowa, NJ, 2013), pp. 265–280. [Google Scholar]
- 24.Henley J. M., Wilkinson K. A., Synaptic AMPA receptor composition in development, plasticity and disease. Nat. Rev. Neurosci. 17, 337–350 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Qi L., et al. , CYLD exaggerates pressure overload-induced cardiomyopathy via suppressing autolysosome efflux in cardiomyocytes. J. Mol. Cell. Cardiol. 145, 59–73 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shehata M., Matsumura H., Okubo-Suzuki R., Ohkawa N., Inokuchi K., Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J. Neurosci. 32, 10413–10422 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen H., Zhu H., Panja D., Gu Q., Li Z., Autophagy controls the induction and developmental decline of NMDAR-LTD through endocytic recycling. Nat. Commun. 11, 2979 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Zhang H., Regulation of autophagy by mTOR signaling pathway. Adv. Exp. Med. Biol. 1206, 67–83 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Linares J. F., et al. , K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol. Cell 51, 283–296 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zajicek A., Yao W. D., Remodeling without destruction: non-proteolytic ubiquitin chains in neural function and brain disorders. Mol. Psychiatry 26, 247–264 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobson-Stone C., et al. , CYLD is a causative gene for frontotemporal dementia - amyotrophic lateral sclerosis. Brain 143, 783–799 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosina E., et al. , Disruption of mTOR and MAPK pathways correlates with severity in idiopathic autism. Transl. Psychiatry 9, 50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borrie S. C., Brems H., Legius E., Bagni C., Cognitive sysfunctions in intellectual disabilities: The contributions of the Ras-MAPK and PI3K-AKT-mTOR pathways. Annu. Rev. Genomics Hum. Genet. 18, 115–142 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Winden K. D., Ebrahimi-Fakhari D., Sahin M., Abnormal mTOR activation in autism. Annu. Rev. Neurosci. 41, 1–23 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Kim J., Kundu M., Viollet B., Guan K. L., AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang G., et al. , Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 83, 1131–1143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan J., Porch M. W., Court-Vazquez B., Bennett M. V. L., Zukin R. S., Activation of autophagy rescues synaptic and cognitive deficits in fragile X mice. Proc. Natl. Acad. Sci. U.S.A. 115, E9707–E9716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.