Abstract
Lymphocyte development and differentiation are regulated by the basic helix-loop-helix (bHLH) transcription factors encoded by the E2A and HEB genes. These bHLH proteins bind to E-box enhancers in the form of homodimers or heterodimers and, consequently, activate transcription of the target genes. E2A homodimers are the predominant bHLH proteins present in B-lineage cells and are shown genetically to play critical roles in B-cell development. E2A-HEB heterodimers, the major bHLH dimers found in thymocyte extracts, are thought to play a similar role in T-cell development. However, disruption of either the E2A or HEB gene led to only partial blocks in T-cell development. The exact role of E2A-HEB heterodimers and possibly the E2A and HEB homodimers in T-cell development cannot be distinguished in simple disruption analysis due to a functional compensation from the residual bHLH homodimers. To further define the function of E2A-HEB heterodimers, we generated and analyzed a dominant negative allele of HEB, which produces a physiological amount of HEB proteins capable of forming nonfunctional heterodimers with E2A proteins. Mice carrying this mutation show a stronger and earlier block in T-cell development than HEB complete knockout mice. The developmental block is specific to the α/β T-cell lineage at a stage before the completion of V(D)J recombination at the TCRβ gene locus. This defect is intrinsic to the T-cell lineage and cannot be rescued by expression of a functional T-cell receptor transgene. These results indicate that E2A-HEB heterodimers play obligatory roles both before and after TCRβ gene rearrangement during the α/β lineage T-cell development.
T lymphocytes are derived in the thymus following a stepwise developmental pathway. Each T cell acquires a unique T-cell receptor (TCR), composed of either an α/β or γ/δ heterodimer, on the cell surface after the V(D)J recombination at the corresponding TCR gene loci. The α/β cell lineage development in the thymus has been conveniently divided into several stages based on the expression of TCR and its coreceptor CD4 and CD8 surface molecules. The most immature population is negative for TCR, CD4, and CD8 expression (double negative, or DN). These cells progress and expand to the TCRlow CD4+ CD8+ (double positive, or DP) stage, which makes up 70 to 80% of total cell mass of the thymus (11). DP cells are then subject to major histocompatibility complex-mediated positive and negative selection before maturing into either TCR+ CD4− CD8+ cytotoxic T cells or TCR+ CD4+ CD8− helper T cells (single positive, or SP). These cytotoxic and helper T cells exit the thymus to peripheral lymph organs, where they provide and mediate antigen-specific immune responses, respectively.
Lineage commitment and initiation of V(D)J recombination occur in the DN population, which is composed of less than 2% of total thymocytes in young adult mice. With additional markers such as CD44 and CD25, the DN cells can be further divided into precommitment (CD44+ CD25−, or DN1) and postcommitment (CD44+ CD25+, or DN2) T-lineage cells (13). Following lineage commitment, the DN2 cells become DN3 cells by down-regulating CD44 but maintaining CD25 expression. While a small number of committed cells develop into the γ/δ T-cell lineage, most DN3 cells in the late fetal and postnatal thymus enter the α/β T-cell lineage and initiate V(D)J recombination at the TCRβ gene locus (10). The formation of the pre-TCR complex, composed of a functional TCRβ chain paired with pre-Tα, is necessary for generating a signal driving the DN cells to the DP stage (11, 33). The nonreceptor tyrosine kinase p56lck and several other tyrosine kinases have been mapped immediately downstream of the pre-TCR signal (22, 23, 39). Downstream of the tyrosine kinases, the linker molecules SLP76 and LAT also play essential roles in mediating the pre-TCR signals (28, 44). It is less clear how these membrane proximal signals are transduced by nuclear transcription factors leading to gene expression and ultimately to cellular differentiation.
Several transcription factors have been identified by gene targeting analysis to play important roles during early T-cell development. These include the Ikaros, GATA3, TCF1, E2A, HEB, and a number of other structurally and functionally related genes. Ikaros encodes a lymphoid cell-restricted zinc finger DNA-binding protein which functions as a protein dimer (12). Ikaros together with several related zinc finger proteins seem to play complex and sustaining roles throughout lymphopoiesis (42). GATA3 is a member of the GATA zinc finger protein family. A Rag2-reconstitution test showed that disruption of the GATA3 gene leads to a complete elimination of the T-cell lineage while having little or no effect on the B-cell and other hematopoietic lineages (36). TCF1 is a T-cell-specific high-mobility group transcription factor important for TCRα gene expression (29). Disruption of the TCF1 gene leads to an accumulation of an intermediate cell type, TCR− CD4− CD8+ (immature single positive, or ISP), which is in transition from DN to DP stages of T-cell development (40). This function of TCF1 is partially compensated for by LEF1, a structurally related high-mobility group transcription factor which plays a much broader role in embryonic and tissue development (27).
E2A and HEB encode transcription factors that belong to the basic helix-loop-helix (bHLH) protein family. The basic region and the HLH domain of bHLH proteins mediate DNA binding and protein dimerization, respectively. Proteins containing the HLH domain but not the basic region (6) are effective dominant negative inhibitors of bHLH proteins. bHLH genes are evolutionarily conserved and found to play important roles in lineage specification and differentiation of many tissue types, including skeletal muscle and lymphocytes (17, 43). The E-protein class of the bHLH family, including the gene products of E2A, HEB, and E2-2, has been shown to participate in lymphopoiesis (3, 5, 45, 46). The bHLH domain of E-proteins mediates the formation of both homodimers and heterodimers, which recognize the canonical CANNTG E-box sequence found in many lymphoid cell-specific genes, including the immunoglobulin (Ig) heavy and light chain genes (25, 26). E2A plays an essential role in B-cell lineage development (2, 45), whereas HEB and E2-2 play facilitating roles by modulating E2A protein activity through two possible mechanisms (46). First, HEB and E2-2 may help increase the availability of E2A proteins by dimerizing with the Id proteins, which are the common inhibitory molecules of all E-proteins (35, 46). Second, HEB and E2-2 may also form heterodimers with the E2A proteins and participate directly in B-cell-specific gene transcription (1, 15). Although functional specificities among different E-protein dimers still remain to be defined (47), many studies have indicated that E2A homodimers are unique to B-lineage cells (7, 32) and are capable of activating directly B-cell- and lymphoid cell-specific genes such as Ig heavy chain Iμ, λ5, EBF, TdT, and Rag1 (3, 9, 19, 31, 34).
In contrast, T-cell development does not seem to be heavily dependent on any single E-protein gene. Among all three E-protein gene knockouts, disruption of HEB induces the most severe defect in T-cell development. Mice lacking the HEB gene show an accumulation of ISP cells and a roughly 5- to 10-fold reduction of total thymocytes. The ISP cells in HEBko mice are CD4lo/− CD8+ CD5lo HSA+ TCRlo/− and are in noncycling state (5). These characteristics are reminiscent of the TCFko mice discussed above (40). However, mature T cells are found in the thymus and peripheral lymphoid organs of HEB knockout mice, indicating a functional compensation for HEB by other genes (5). A partial block at the DN1 stage and normal thymocyte development were reported for the E2A and E2-2 knockout mice, respectively (3, 46). Two crucial pieces of evidence indicated that E2A and HEB play overlapping roles in T-cell development. First, Sawada and Littman (30) showed in their analysis of CD4 gene enhancers that the CD4-3 E-box site was predominantly occupied by the E2A-HEB heterodimers in cloned T-cell and thymocyte nuclear extracts. Second, a genetic interaction between HEB and E2A genes was revealed by analyzing E2A and HEB compound heterozygous mice, which displayed a thymic defect similar to that in HEB knockout mice (46). These observations raised a possibility that E2A-HEB heterodimers could directly instruct T-cell-specific gene expression in a way parallel to the function of E2A homodimers in B-cell development.
In this study, we find that the T-cell-specific E2A-HEB heterodimers are replaced by E2A homodimers in the HEB knockout mice and by HEB homodimers in the E2A knockout mice. This observation substantiates the conclusion made from genetic studies (46) that E2A and HEB are able to compensate for each other in T-cell development. To test the function of E2A-HEB heterodimers, we generated and analyzed mice carrying a dominant negative allele of HEB, named HEBbm. We show that HEBbm produces physiological levels of a mutant HEB protein capable of forming nonfunctional heterodimers with E2A proteins. Although several minor defects are found in other cell types, the most severe dominant negative effects of HEBbm are restricted to the T-cell lineage, where HEB is highly expressed. Overall, we find that HEBbm induces a stronger and earlier block in T-cell development than that in HEB knockout (HEBko) mice. Comparing HEBbm mice with HEBko mice, HEBbm mice show several distinct phenotypes. First, the thymic cellularity in HEBbm mice is reduced 3- to 10-fold from that in HEBko mice or 15- to 100-fold from that in the wild-type control. Second, T-cell development in HEBbm mice is blocked within the DN stage, one stage earlier than the ISP stage block seen in HEBko mice. Third, a defect in V(D)J recombination at the TCRβ gene locus is found in DN3 cells from HEBbm mice but not from HEBko mice reported earlier (5). Finally, we show that the developmental block in the DN stage cannot be rescued by forced expression of a functional TCR gene. These studies indicate that E2A-HEB heterodimers play essential roles both before and after the assembly of functional TCRβ genes.
MATERIALS AND METHODS
Construction of the HEBbm allele.
The basic region mutation was introduced by PCR-based mutagenesis on a 0.7-kb BamHI fragment of genomic DNA covering the entire bHLH encoding exon and adjacent intron sequences. The basic DNA-binding domain was changed from RRMANNARERLRV to RRMANNAREGHGV with an NcoI site incorporated in the DNA sequence. The mutated DNA fragment was verified by sequencing analysis before it was introduced into the targeting vector.
Southern blotting and PCR genotyping of the HEBbm allele.
Southern blotting was performed by probing the NcoI-digested mouse tail DNA with an HEB genomic DNA derived from the intron sequence upstream of the bHLH-encoding exon. The wild-type and the HEBbm alleles were revealed as 5.8- and 3.8-kb fragments, respectively. PCR genotyping was conducted by first amplifying toe DNA with a sense primer, YZ-119 (5′GACATCAAGGTCTCATCTAGG3′), and an antisense primer, YZ-122 (5′TCTCACTTGCTGTTCTAGACT3′). The PCR products were digested by NcoI and then separated on agarose gels. The wild-type and the HEBbm alleles were detected as 2.0-kb and 1.8-kb products, respectively, after NcoI digestion.
Western and EMSA analyses.
Nuclear extracts were prepared from total thymocytes according to the protocol previously described (5). Protein concentrations were determined by the Bio-Rad protein assay. A sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis minigel was loaded with 10 μg of nuclear extract, blotted to nitrocellulose with a Trans-Blot SD electrophoretic transfer cell (Bio-Rad), and blocked with 10% nonfat dried milk in 1× Tris-buffered saline–0.5% Tween 20. Primary antibodies were anti-HEB rabbit polyclonal antiserum purchased from Santa Cruz Biotech. The secondary donkey anti-rabbit antibody conjugated to horseradish peroxidase was used at a 1:5,000 dilution, followed by enhanced chemiluminescence treatment as suggested by the manufacturer (ECL kit; Amersham, Little Chalfont, Buckinghamshire, England). Electrophoretic mobility shift assay (EMSA) analysis was performed according to Sawada and Littman (30).
Protein synthesis.
Thymocyte RNA from an HEBbm heterozygous mouse was used in reverse transcription reaction with random hexamers. The cDNA was used in a PCR to clone the carboxyl-terminal 220-amino-acid coding region of the wild-type and the mutant HEB transcripts. The E47 subclone (pB4X) used in the assay contains the carboxyl-terminal 280-amino-acid coding region of the human E47 transcript. These cDNA fragments were cloned in the pBluescript expression vector. RNA transcripts were prepared using T3 or T7 polymerase and were quantified before they were used in translation. The rabbit reticulocyte lysate system with or without [35S]methionine was used for protein synthesis. The 35S-containing samples were used in sodium dodecyl sulfate gel for determining protein concentrations, whereas the nonradioactive samples were used in EMSAs.
PCR analysis of TCRβ gene V(D)J rearrangements.
DNA was isolated from total thymocytes (106) or cell-sorted DN3 thymocytes (2 × 104 to 10 × 104). Cells were lysed in 10 mM Tris-HCl (pH 8.0)–1 mM EDTA (pH 8.0)–0.2 μg of proteinase K/ml–0.2% Triton X-100 for 30 min at 55°C and then 10 min at 94°C. The rearrangement assay and PCR primers were used as previously described (5, 38). Briefly, PCRs were performed for 30 cycles of 45 s at 94°C, 45 s at 57°C, and 1 min at 72°C. Products were run on a 1.3% agarose gel, blotted onto nitrocellulose, and probed with a Jβ2-specific probe (1.4 kb HincII-SacII DNA fragment covering the entire Jβ2 region).
Flow cytometry.
Bone marrow, spleen, lymph node, and fetal thymus cells were isolated from age-matched mice in phosphate-buffered saline supplemented with 5% bovine calf serum and were used immediately for fluorescence-activated cell sorting (FACS) analysis. Cell suspensions were stained with a combination of a fluorescein isothiocyanate-conjugated antibody and a phycoerythrin (PE)-conjugated antibody plus 7-aminoactinomycin D (7AAD; Molecular Probes) and analyzed on a FACScan (Becton Dickinson). CD4-PE-cy5, CD8-PE-cy5, and TCR-PE-cy5 (clone H57-597) were also used together with 7AAD in the analysis of DN thymocytes. Cells from the adoptive transfer experiment were analyzed on a FACS Calibur with allophycocyanin (apc) included as the fourth color. Two-parameter dot plots were shown after gating populations by size and viability. Antibodies were purchased from Caltag and Pharmingen.
Irradiation and stem cell reconstitution.
C57BL/6 mice congenic for the Ly5.1 allotype marker were originally purchased from Jackson Laboratories and then bred in-house. Host mice at 8 to 12 weeks of age were irradiated with 1,100 rads 1 day before stem cell transfusion and were maintained thereafter in sterile bedding with antibiotics added in drinking water. Donor cells were prepared either from frozen stocks or from freshly isolated fetal liver and neonatal bone marrow cells. Either 0.1 × 106 to 0.5 × 106 total fetal liver cells or 0.5 × 105 to 2 × 105 total bone marrow cells were delivered to the host in 0.2 ml of phosphate-buffered saline through tail vein injection. For each donor type, three to five recipients were used in the test. Mice were sacrificed 1 or 2 months after irradiation for FACS analysis.
RESULTS
Functional compensation among individual E-protein dimers.
To understand further the functional relationship between HEB and E2A proteins in T-cell development, we performed EMSA to determine the DNA-binding ability of individual E-protein dimers present in thymocyte extracts of various mutant strains (Fig. 1). Functional E-protein dimers were detected via their ability to bind to the CD4-3 E-box DNA (30). Antibody supershift analysis was used to identify the components of DNA bound dimers. In agreement with Sawada and Littman's observation, we find that the E2A-HEB heterodimers account for the majority of E-box DNA-binding activities in wild-type thymocyte extract. The identity of E2A-HEB heterodimers (arrow A in lane 1) was determined by the supershifts generated with anti-HEB antibodies (arrow C in lane 2) and anti-E2A antibodies (arrow B in lane 3). The presumed E2A and HEB homodimers (complexes resistant to antibody supershift in lanes 2 and 3, respectively) account for only a small but detectable amount of E-box binding activity. Deletion of one and two copies of HEB resulted in a proportional increase in the amount of E2A homodimers determined by the anti-HEB antibody-resistant complexes (arrow A in lanes 2, 5, and 8) and the anti-E2A antibody-dependent supershift (arrow B in lane 9). Similarly, the amount of HEB homodimers, determined by the anti-E2A antibody-resistant complexes (arrow A in lanes 3, 12, and 15) and the anti-HEB antibody-dependent supershift (arrow C in lane 14), is proportionally increased due to deletion of one and two copies of E2A. These results indicate that homodimers of E2A or HEB are capable of replacing the DNA-binding activity of E2A-HEB heterodimers and therefore provide a molecular basis for the functional compensation between the HEB and E2A gene.
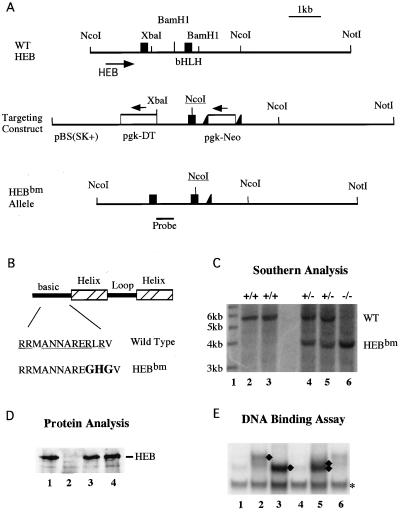
FIG. 1.
EMSA analysis of E-protein dimer formation on the CD4-3 E-box DNA. Thymocyte extracts from wild-type (WT) (lanes 1 to 3), HEBko heterozygous (lanes 4 to 6), HEBko homozygous (lanes 7 to 9), E2Ako heterozygous (lanes 10 to 12), and E2Ako homozygous (lanes 13 to 14) mice were incubated with 32P-labeled CD4-3 DNA in the absence of antibodies (lanes 1, 4, 7, 10, and 13), in the presence of anti-HEB antiserum (lanes 2, 5, 8, 11, and 14), or in the presence in anti-E2A antibody Yae (lanes 3, 6, 9, 12, and 15). Arrows on the left (A, B, and C) indicate CD4-3-bound E-protein dimer complexes, the E2A-dependent supershift complexes, and the HEB-dependent supershift complexes, respectively.
Construction of a dominant negative HEB allele.
The compensation among different E-protein dimers renders a leaky thymic phenotype and obscures the functional analysis of the native E-protein dimers in thymopoiesis. It is technically difficult to generate mice lacking both E2A and HEB by interbreeding of the single knockout mice because the double mutation leads to early embryonic death (46). We therefore undertook a dominant negative approach to investigate the function of E2A-HEB heterodimers and to create a better genetic model for understanding the mechanism of T-cell-specific gene regulation by E-protein dimers. It has been shown that an E-protein with altered DNA-binding domain can form nonfunctional heterodimers with other bHLH proteins (41). We hypothesized that disruption of the DNA-binding domain of HEB will suppress the function of both HEB homodimers and E2A-HEB heterodimers.
A two-step knockin approach was used to generate such a dominant negative HEB allele in mice. As illustrated in Fig. 2, a genomic HEB DNA construct carrying a point mutation (marked by NcoI) in the basic region of HEB (Fig. 2A and B) and a pgk-Neo selection marker (flanked by LoxP sites, or floxed) was introduced into mouse embryonic stem cells. The diphtheria toxin gene provided a negative selection against the nonhomologous recombination events. The floxed pgk-Neo was subsequently deleted in the targeted embryonic stem cell clones following transient transfection with the CMVCre plasmid. The final knockin clone with Neo deleted was verified by Southern blotting and was subsequently introduced into mouse embryos. Germ line transmission was established and confirmed by Southern blotting (Fig. 2C). This allele is referred to hereafter as HEBbm, for basic region mutation.
FIG. 2.
Generation of the HEBbm allele. (A) Schematic diagrams of the bHLH region of the HEB gene (top), the gene targeting construct (middle), and the final knockin HEBbm allele. Filled boxes and open boxes represent exons and selection markers, respectively. Intron and vector sequences are shown as single lines. LoxP insertions on each side of the Neo cassette are indicated by filled triangles. The positions of restriction sites and the probe relevant to the Southern analysis are indicated. The basic region mutation was introduced along with a NcoI site (underlined) into the bHLH encoding exon. WT, wild type. (B) The sequence of the basic region of HEB. Underlines indicate amino acids conserved among all E-proteins. Sequences mutated are shown in boldface. (C) Southern blot analysis of tail DNA digested with NcoI. The wild-type and HEBbm alleles are identified as 5.8- and 3.8-kb fragments, respectively, in the blot. Lane 1 contains an end-labeled 1-kb ladder as size markers. The size of each marker band is shown on the left. Two wild-type samples (lanes 2 and 3), two HEBbm heterozygous samples (lanes 4 and 5), and one HEBbm homozygous sample (lane 6) are shown in the blot. (D) Western analysis of thymocyte nuclear extracts for HEB and HEBbm proteins. Lanes: 1, wild type; 2, HEBko homozygote; 3, HEBko/bm compound heterozygote; 4, HEBbm heterozygote. Ten micrograms of proteins was loaded in each lane. (E) EMSA of HEBbm proteins. Proteins synthesized from programmed reticulocyte lysates were incubated with end-labeled CD4-3 probe. Lanes contain the following: lane 1, 3 μl of reticulocyte lysates (RL) without any RNA added in protein synthesis; lane 2, 3 μl of E47 proteins; lane 3, 3 μl of HEB proteins; lane 4, 3 μl of HEBbm proteins; lane 5, 3 μl of E47 plus 3 μl of HEB proteins; and lane 6, 3 μl of E47 plus 2 μl of HEBbm proteins. The E47, HEB, and HEBbm proteins synthesized in the programmed RL were in equal concentrations determined by a 35S protein gel (data not shown). Diamonds indicate bound E47 homodimers, HEB homodimers, and E47-HEB heterodimers; the asterisk indicates an unrelated RL-dependent shift.
Western analysis was performed to evaluate the expression level of the HEBbm protein. Thymocyte extracts from HEBbm/ko transheterozygous mice were used to determine the mobility and expression level of proteins produced from a single copy HEBbm allele. As shown in previous studies (5), HEBko homozygous mice show no detectable HEB protein (Fig. 2D, lane 2). In contrast, the single copy HEBbm allele on the HEB null background produces a protein (Fig. 2D, lane 3) indistinguishable from the HEB protein detected in wild-type mice (Fig. 2D, lane 1). To evaluate the dimerization and DNA-binding activity of HEBbm proteins, we subcloned the wild-type and the mutant HEB cDNAs from thymocytes of an HEBbm heterozygous mouse. Proteins were synthesized in reticulocyte lysate and tested in EMSAs with radiolabeled CD4-3 E-box DNA. E47, a bHLH protein encoded by the E2A gene, was also synthesized. As shown in Fig. 2E, E47 homodimers (lane 2), HEB homodimers (lane 3), and E47-HEB heterodimers (lane 5) are readily detected (marked by diamonds). However, under the same assay conditions, the HEBbm protein is incapable of binding to the DNA (lane 4) but is capable of reducing the DNA-binding activity of E47 homodimers (lane 6). This test confirms that HEBbm proteins are capable of interfering with the DNA-binding activity of E2A proteins.
Gross phenotypic comparison of HEBbm and HEBko mice.
HEBbm heterozygous mice are grossly indistinguishable from their wild-type littermates and are completely fertile. However, after 6 to 12 months of age, approximately 10 to 20% of the mice showed episodes of seizures upon handling by the tail, a phenomenon not seen in the wild-type and HEBko heterozygous controls. This phenotype underscores the importance of an earlier observation that HEB is highly expressed in the brain (21). HEBbm homozygotes were recovered from heterozygous interbreeding at full-term gestation at the expected Mendelian frequency (9 HEBbm/bm in a total of 34 genotyped E17.5 to E18.5 fetuses). Some HEBbm homozygous fetuses displayed exencephaly, a low-penitrant phenotype that has previously been observed in the HEBko and E2Ako homozygous embryos. Similar to HEBko homozygous mice, HEBbm homozygous neonates displayed severe growth retardation and often died within 2 weeks of birth (only 1.4% were HEBbm/bm of 785 genotyped 2-week-old F2 offspring).
A severe and specific defect in thymopoiesis.
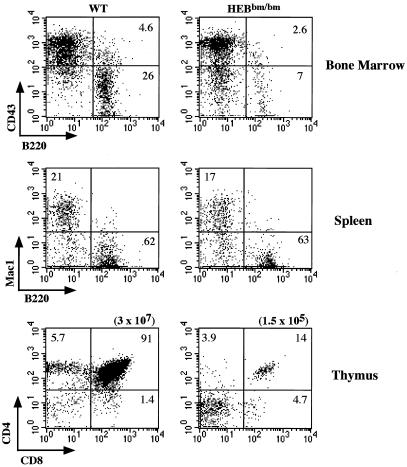
The effect of HEBbm on hematopoiesis was evaluated by flow cytometry analysis with hematopoietic lineage and stage-specific markers. Similar to HEBko homozygous mice, no obvious defect was found in the erythroid (determined by ter119 marker) and myeloid (determined by Mac1 and Gr1 markers) cell lineages in HEBbm homozygous mice (data not shown). B-cell development was normal in HEBbm heterozygous mice but impaired in HEBbm homozygous mice. Numbers of pro-B cells (CD43+ B220+), pre-B and immature B cells (CD43low B220+), and mature B cells (CD43− B220high) in HEBbm homozygous mice were found reduced compared to littermate controls (Fig. 3A, top panel). However, an analysis of spleen and lymph nodes showed no gross abnormality of B cells in the peripheral lymph organs, suggesting that the effect of HEBbm may be restricted to bone marrow B-cell development (Fig. 3A, bottom panel, and data not shown).
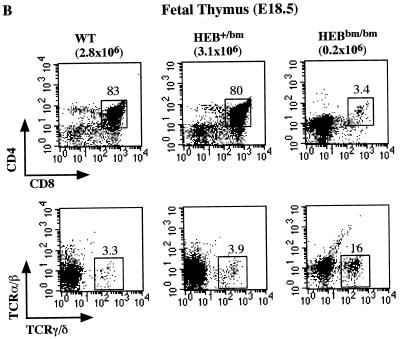
FIG. 3.
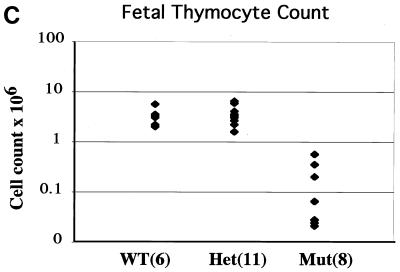
FACS analysis of B- and T-cell development in HEBbm mice. (A) Analysis of bone marrow (top) and lymph node (bottom) cells from adult HEBbm/+ and HEBbm/bm mice. CD43 and B220 markers were used to separate pro-B cells (CD43+ B220+), pre- and immature B cells (CD43low B220+), and mature B cells (CD43− B220high) in the bone marrow. TCRβ and B220 markers were used to separate T and B cells, respectively, in the lymph nodes. The relative percentage for each population is shown in the plots. Inguinal lymph nodes were collected from each animal, with the total cell numbers given on top of the plots. (B) E18.5 fetal thymus of wild-type, HEBbm/+, and HEBbm/bm thymocytes were analyzed in three separate stainings for CD4 and CD8 (top) and TCRα/β and TCRγ/δ (bottom) markers. CD4-, CD8-, and TCR-positive cells were gated out in the bottom panel. Cell counts of total thymocytes and individual populations are shown in the plots. Data are representative of multiple tests (n = 7 for HEBbm/bm). Events displayed for all plots in A and B are after size and 7AAD gating, which eliminates nonlymphoid and dead cells, respectively. (C) Cell count of E18.5 fetal thymus collected from five litters of timed mating between HEBbm heterozygous mice. Numbers of fetuses for each genotype included in the analysis are shown next to the genotype name in the chart. Means and standard deviations (in parentheses) for wild type, HEBbm/+, and HEBbm/bm are 3.3 (1.4) × 106, 3.6 (1.5) × 106, and 0.2 (0.2) × 106, respectively. Two-tailed t test shows a statistical difference between wild type and HEBbm/bm (P = 1.7 × 10−5) and no significant difference between wild type and HEBbm/+ (P = 0.7).
In contrast to B-cell development, T-cell development in HEBbm/bm was severely disrupted in both peripheral and central lymph organs (Fig. 3A, B, and C). The defect in T-cell development begins in fetal thymus. HEBbm/bm fetal thymus showed an approximately 15-fold reduction in the thymic cellularity, an accumulation of DN cells, and a near absence of DP and SP cells. This developmental defect seems specific to the α/β T-cell lineage since the γ/δ T cells were detected in HEBbm/bm mice, albeit with a slightly reduced number per thymus.
Multiple roles of HEB in T-cell development revealed by gene dosage effect.
A similar defect was observed in postnatal thymus (Fig. 4). A direct comparison of age-matched HEBbm mice and HEBko mice showed that the HEBbm mutation causes a much tighter and earlier block in T-cell development than the complete disruption of the HEB gene. T-cell development in the HEBbm homozygous mice was almost completely blocked at the DN stage rather than the ISP stage as seen in the HEBko/ko mice (5). This severe defect in thymopoiesis was accompanied by about a 10-fold reduction of total thymocytes relative to the HEBko/ko background and about a 100-fold reduction relative to levels in the wild-type controls among 2- to 3-week-old mice (Fig. 4 and data not shown). Interestingly, T-cell development in the HEBbm/ko transheterozygous mice can proceed to the ISP stage but not to the SP stage, a phenotype intermediate of HEBko homozygous mice and HEBbm homozygous mice. This result indicates that HEBbm exerts its dominant negative effects at several discrete developmental stages in a dosage-dependent manner.
FIG. 4.
FACS analysis of thymocytes from mice carrying various HEB alleles. (A) Two- to three-week-old mice were used in the analysis, with genotype and thymocyte count shown on the top. CD4 and CD8 plots were drawn after eliminating nonlymphocytes and dead cells in the size scatter and 7AAD plots, respectively. The relative percentage of cells in each quadrant was given in the plots. Data are representative of multiple tests including 8 wild-type and HEB+/ko mice, 8 HEBko/ko mice, 3 HEBko/bm mice, and 4 HEBbm/bm mice.
T-cell developmental defect is intrinsic to the T-cell lineage.
In addition to their high-level expression in the thymus, HEB transcripts were also detected in many other tissues, such as the brain. The low recovery rate of HEBbm/bm neonates indicates that HEB must play essential roles in cell types other than the T-cell lineage. Therefore, the dominant negative effect of the HEBbm mutation on T-cell development could be due to a disruption of T-cell intrinsic differentiation program or due to a perturbation of the environment needed for proper T-cell development. To distinguish these two possibilities, we performed an adoptive transfer experiment. Bone marrow cells from wild-type or HEBbm/bm neonatal animals were transfused into irradiated wild-type hosts. The ability of donor stem cells to provide radioprotection and to give rise to T-lineage cells was evaluated 1 or 2 months after transfusion. We find that HEBbm/bm bone marrow cells are able to protect the hosts from lethal irradiation, to produce myeloid cells, and with a reduced efficiency to produce B cells (Fig. 5). However, T-cell development from HEBbm/bm donors is severely impaired. The quantitative defect in B-cell development and the strong block in T-cell development recapitulate the phenotypes of HEBbm/bm mice. This result shows that the dominant negative effect of the HEBbm allele is due to disruption of normal HEB function within the lymphoid lineages.
FIG. 5.
Adoptive transfer test of wild-type (left) and HEBbm/bm (right) stem cells into wild-type hosts. Bone marrow cells (1 to 2 × 105) from wild-type or HEBbm homozygous neonates were transferred into C57BL/6 Ly5A congenic mice irradiated with 1,100 rads. Bone marrow cells, splenocytes, and thymocytes were collected and analyzed 1 month after adoptive transfer. A four-color flow cytometry test was carried out with CD45.2-fluorescein isothiocyanate (donor-specific marker) and 7AAD included in all experiments. The CD45.2+ 7AAD− cells were analyzed with CD43-pe and B220-apc markers for bone marrow cells, Mac1-pe and B220-apc for splenocytes, and CD8-pe and CD4-apc for thymocytes. Total thymocytes recovered from the wild type transfer and HEBbm/bm mutant transfer were indicated on the top of the plots. Data are representative of three separate sets of transfer experiment.
Impairments in V(D)J recombination at the TCRβ gene locus.
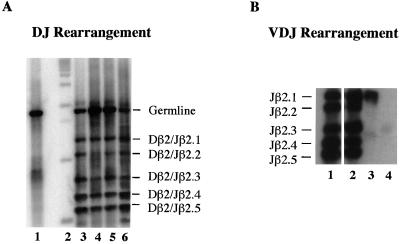
It has been well established that cells at the DN3 stage undergo V(D)J recombination at the TCRβ gene locus (10). A sequential assembly of the V, D, and J segments (D to J and then V to DJ) and the expression of a functional β chain paired with the pre-Tα chain is necessary for the DN3 cells to proceed to the next developmental stage. An analysis of the rearrangement status at the TCRβ gene locus showed a severe impairment in V to DJ rearrangements (Fig. 6B) in HEBbm/bm mice. While wild-type mice show efficient and random V(D)J rearrangements in both total thymocytes and sorted DN3 cells, HEBbm/bm mice show very inefficient and sporadic V(D)J rearrangements in total thymocytes and no VDJ rearrangements in sorted DN3 cells (Fig. 6B and data not shown for Vβ5 to Jβ2 rearrangements). This phenotype is in sharp contrast to that of HEBko/ko mice, which show relatively normal V(D)J rearrangements at the TCRβ gene locus (5). These results indicate that E-protein dimers, most likely the E2A-HEB heterodimers, are required for V(D)J rearrangements at the TCRβ gene locus.
FIG. 6.
(A) PCR analysis of TCRβ gene DJ rearrangement. DNA samples were prepared from either total thymocytes or sorted CD25+ CD44− DN3 cells of 2- to 3-week-old mice. DJ rearrangement products from the Dβ2 and Jβ2 region were analyzed by PCR with Dβ2 and Jβ2.6 primers. PCR products were blotted and hybridized with a J segment-specific probe. The expected DJ rearrangement products are indicated on the right. Samples in the blot are toe DNA (lane 1), a 1-kb size ladder (lane 2), wild-type total thymocytes (lane 3), HEBbm/bm total thymocytes (lane 4), HEBbm/bm DN3 cells (lane 5), and wild-type DN3 cells (lane 6). (B) PCR analysis of TCRβ gene VDJ rearrangement. DNA used in the DJ rearrangement assay were PCR amplified with Vβ8 and Jβ2.6 primers. PCR products with expected Jβ usage are indicated on the left. Samples in the blot are wild-type total thymocytes (lane 1), wild-type DN3 cells (lane 2), HEBbm/bm total thymocytes (lane 3), and HEBbm/bm DN3 cells (lane 4). Similar results were obtained with Vβ5 and Jβ2.6 primers.
DN cells in HEBbm/bm mice cannot be rescued to the DP stage by forced expression of a functional TCR transgene.
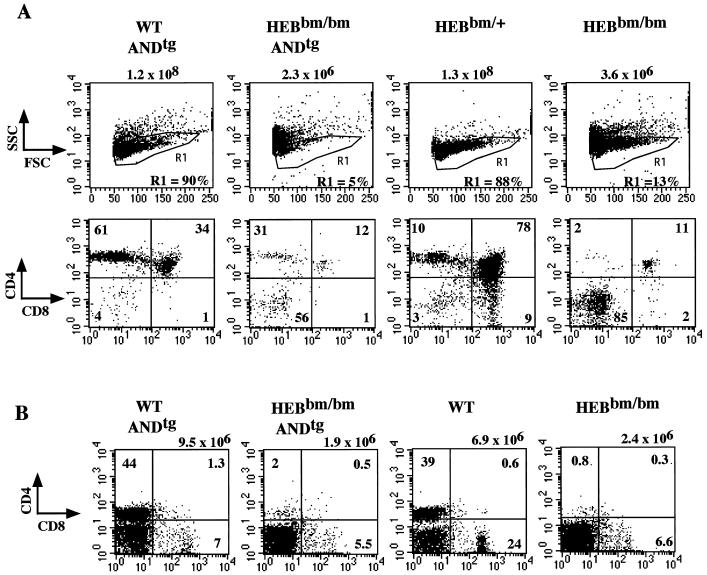
Expression of a functional TCR is necessary and sometimes sufficient for DN3 cells to progress to the DP stage and later from the DP to the SP stage (33). We therefore attempted to rescue the V(D)J recombination defect in HEBbm/bm mice by introducing a functional TCRα/β transgene. AND is a functional α/β TCR transgene capable of overcoming the block imposed by the RAG2 gene knockout, which arrests T-cell development at the DN3 stage. As previously reported (18), expression of this transgene in both H-2b and H-2k backgrounds with endogenous peptides will drive major histocompatibility complex class II-dependent positive selection, resulting in an increased CD4 SP population (Fig. 7A). When AND was bred to the HEBbm/bm background, we saw no increase in DP population and thymic cellularity. However, an increase in the percentage of CD4 SP cells is clearly visible, indicating that AND is expressed and capable of driving differentiation from the DP to the SP stage (Fig. 7A). An analysis of peripheral T cells from adult double transgenic mice also confirmed that the AND transgene failed to rescue the developmental block imposed by HEBbm. No substantial increase in numbers or percentage of total lymph node T cells was seen due to the addition of the AND transgene in HEBbm/bm mice (Fig. 7B). This study indicates that impaired V(D)J recombination is not sufficient to explain the developmental arrest in the HEBbm/bm mice. It implies that E2A-HEB heterodimers are also involved in controlling other genes, which function downstream or parallel to the TCR signaling pathway.
FIG. 7.
The AND TCR transgene cannot rescue the HEBbm mutation. (A) E18.5 fetal thymuses were analyzed by costaining with CD8, CD4, and TCR antibodies. Size plots are shown with genotypes indicated on the top. Cells highlighted in the R1 gate in the size plots (top panel) are displayed for CD4/CD8 staining (bottom panel). Data are representative of multiple litters. (B) Lymph nodes of single and double transgenic mice were analyzed with CD8, CD4, and 7AAD. The percentages of CD4 single positive and CD8 single positive populations are indicated in the CD4/CD8 plots. The total cell number of inguinal lymph nodes for each genotype analyzed is given on the top. Similar results were obtained from the analysis of splenocytes.
Although no significant increase in thymic cellularity is seen after introduction of AND into HEBbm mice, a substantial increase in the ratio of CD4 SP to DP cells is detected in the thymus. These CD4 SP cells express a normal level of AND TCR on their cell surface, suggesting that they are resulting from AND-mediated positive selection. This observation indicates that E2A-HEB heterodimers are dispensable during differentiation from the DP to the CD4 SP stage. Because of a persistent block in the transition from DN to DP, the absolute number of CD4 SP cells remains small in both thymus and peripheral lymphoid organs. The appearance of phenotypically normal CD4 SP cells in thymus also indicates that the CD4 expression is not absolutely dependent on E2A-HEB heterodimers at this stage of development.
DISCUSSION
In summary, we have shown that the dominant negative HEBbm allele blocks T-cell development at an earlier stage than the previously reported HEB knockout allele. This observation provides the strongest genetic evidence to date to support the notion that E2A-HEB heterodimers are the major bHLH dimers involved in early T-cell development. Results from our study of HEBbm mice suggest that E2A-HEB heterodimers may play a role in T cells similar to that of E2A homodimers in B cells. In both cell lineages, E-proteins may be directly involved in regulating the lymphoid cell-specific genes, such as TCR genes in T cells and Ig genes in B cells. The use of different E-protein dimers in B- and T-cell lineage development may reflect a need for lineage-restricted gene expression.
The dominant negative mutation reveals multiple roles of HEB in T-cell development.
Earlier studies have indicated that E2A homodimers directly control B-cell development. A search for the counterpart of E2A dimers involved in T-cell development has led us to investigate HEB function. HEB is highly homologous to E2A and is highly expressed in thymocytes (15). Disruption of the HEB gene leads to impaired T-cell development with a developmental block at the transition from the DN to the DP stage (5). However, TCRβ gene expression and rearrangement seem unaffected by HEB disruption. Moreover, T cells can develop to maturity in HEB knockout mice, albeit at a reduced frequency and with delayed timing. We now show that a mutation in the DNA-binding domain of HEB induces a tighter and earlier block in T-cell development. Therefore, the dominant negative approach reveals a function of HEB which could not be seen by conventional gene knockout studies.
Studies by Heemskerk et al. have shown that retrovirus-mediated expression of Id3 in CD34+ progenitors can promote NK cell development at the expense of the T-cell lineage in fetal thymic organ culture (14). Further, ectopic expression of the same Id3 retroviral construct in pro-T cells leads to specific inhibition of α/β but not γ/δ T-cell lineage development (8). Works from two other groups have also shown that a strong block in T-cell development is induced in transgenic animals overexpressing Id1 or Id2 under the control of the lck proximal promoter (20, 24). Our observations from HEBbm studies are generally in agreement with these reports. The fact that HEBbm inhibits α/β but not γ/δ T-cell lineage development indicates a highly restricted role for HEB in lymphoid lineage cells after their commitment to the T-cell lineage. We have also carried out a preliminary study to evaluate the potential role of HEB in NK cell development in the thymus. Analysis of fetal thymi from three HEBbm/bm and four wild-type and HEBbm/+ littermate controls showed that the number of NK1.1-positive cells per thymus is decreased approximately 10-fold in HEBbm/bm. We do not know at the present time if this inhibition on thymic NK cell development is intrinsic to the NK cell lineage or regulated by other cells in the thymus.
One caveat in Id overexpression studies is that one cannot distinguish which E-protein dimer is specifically inhibited by Id. In addition, the postulated interaction between Id2 and Rb proteins (16) further complicated the interpretation of the results. The use of the knockin rather than the transgenic approach renders protein expression at a physiological level. Thus, the dominant negative effect of HEBbm is most likely reflecting a real function of HEB in the T-cell lineage. In the absence of the wild-type HEB gene, escalating phenotypes were found due to sequential introduction of one and two copies of HEBbm. Zero copy (for HEBko/ko mice), one copy (for HEBbm/ko mice), and two copies (for HEBbm/bm mice) of HEBbm render a partial block in ISP and DP development, a complete block before SP, and a complete block before ISP and DP, respectively. This result suggests that HEB is required for T-cell development in at least three distinct developmental stages: DN, ISP, and the transition from DP to SP. It is not clear whether the dimerization partners of HEB in these three developmental stages are the same. We provided biochemical evidence that this HEBbm protein is capable of dimerizing with E47 proteins and consequently reduces the DNA-binding ability of E47. It is formally possible that the dominant negative effect of HEBbm proteins could result from their ability to dimerize with an unknown bHLH protein partner instead of E2A. However, the existence of such a bHLH protein is not supported by the EMSA analysis with CD4-3 E-box sites.
E2A-HEB heterodimer activity is associated with lineage differentiation but not commitment.
Results from this study do not exclude the possibility that E2A homodimers may play a separate role prior to the DN3 stage, as has been implicated in the study of E2A knockout mice (3). Bain et al. have reported a proportional increase of DN1 thymocytes in mice carrying mutant alleles of E2A (4), which indicates that E2A may be needed for T-cell lineage commitment. Presumably, the HEBbm allele can effectively function as a dominant negative gene only in developmental stages where it is highly expressed. In fact, B-cell development is only mildly affected by the HEBbm allele, which is consistent with observations that E2A is expressed at a much higher level than HEB in the B-cell lineage (data not shown). The strong block in thymopoiesis at the DN3 stage suggests that E2A-HEB heterodimers are specifically required for lineage differentiation rather than commitment. It is plausible that a switch from E2A homodimers to E2A-HEB heterodimers may have occurred after T-cell lineage commitment.
Are E2A homodimers and E2A-HEB heterodimers functionally equivalent?
It will be important to determine the functional specificity of individual E-protein dimers. E2A protein homodimers have been suggested to provide tissue-specific function in regulating lymphoid- and B-cell-specific genes (3, 7, 9, 19, 31, 34). T-cell-restricted heterodimerization between E2A and HEB may be necessary for eliminating E2A homodimers, which would otherwise lead to activation of B-cell-specific genes. This argument is supported by detailed characterization of E2Aheb mice whose E2A gene is replaced by HEB (47). Due to gene replacement, these mice express no E2A proteins but an excess amount of HEB proteins. We have shown that B-cell deficiency induced by E2A knockout is only partially rescued by the addition of extra copies of HEB. Interestingly, the thymic phenotype of E2Aheb resembles that of E2Ako rather than that of the wild-type animals (data not shown). Therefore, HEB homodimers and E2A-HEB heterodimers are not equally efficient in supporting T-cell development, even if they were produced to the same level.
Possible roles for E2A-HEB heterodimers both before and after TCRβ gene rearrangement.
The defect in V(D)J recombination at the TCRβ gene locus indicates a possible role of E2A-HEB heterodimers in regulating TCRβ gene expression and/or rearrangement. This result substantiates recent work by Tripathi et al. showing that the TCRβ gene rearrangement is critically dependent on two E-box sites present in the TCRβ gene enhancer (37). However, our finding did not rule out the possibility that the defect in TCRβ gene rearrangement is due to altered expression of other genes, such as RAG1 and RAG2, required for normal TCRβ gene activation and rearrangement. Similarly, the results from the AND transgenic rescue experiment may also be due to lack of proper expression of CD3 or other TCR components needed for proper function of the AND transgene. To address these possibilities, we have carried out reverse transcriptase-PCR analysis on sorted DN3 cells to evaluate the expression of selected genes relevant to thymocyte development. This preliminary study shows that the expression of RAG1, p56lck, SLP-76, IL-7R, TdT, and pre-Tα are the same in HEBbm and wild-type mice; RAG2, CD3δ, and CD3γ are decreased in HEBbm mice; and CD3ɛ expression is increased in HEBbm mice (data not shown). Although this analysis did not provide a simple answer to the question, these results clearly indicate that E-proteins may regulate multiple genes in T-cell development.
The use of E2A-HEB heterodimers instead of E2A homodimers in the T-cell lineage may provide one possible means for T-cell-specific gene expression. This partially explains why TCR genes but not Ig genes are activated in the T-cell lineage while Ig genes but not TCR genes are activated in the B-cell lineage. Further tests are needed to determine if E2A-HEB heterodimers are sufficient to activate the TCRβ gene locus.
ACKNOWLEDGMENTS
We thank Jenifer Hanrahan and Reshma Rangwala for assistance in making constructs, Mike Cook for assistance in flow cytometry analysis, and Lihua Pan, Jenifer Hanrahan, Steve Greenbaum, Curtis Bradney, and Michael Krangel for critical reading of the manuscript.
This work has been supported by the Leukemia Society of America Scholarship, the Whitehead Scholarship, and NIH grants (R01CA72433 and R01GM59638) to Y.Z.
REFERENCES
- 1.Bain G, Gruenwald S, Murre C. E2A and E2-2 are subunits of B-cell-specific E2-box DNA-binding proteins. Mol Cell Biol. 1993;13:3522–3529. doi: 10.1128/mcb.13.6.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bain G, Maandag E C, Izon D J, Amsen D, Kruisbeek A M, Weintraub B C, Krop I, Schlissel M S, Feeney A J, van Roon M, van der Valk M, te Riele H P J, Berns A, Murre C. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 3.Bain G, Maandag E C R, te Riele H P, Feeney A J, Sheehy A, Schlissel M, Shinton S A, Hardy R R, Murre C. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- 4.Bain G, Engel I, Maandag E C R, te Reile H P J, Voland J R, Sharp L L, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in α/β T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barndt R J, Dai M F, Zhuang Y. A novel role for HEB downstream or parallel to the pre-TCR signal during alpha/beta thymopoiesis. J Immunology. 1999;163:3331–3343. [PubMed] [Google Scholar]
- 6.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 7.Benezra R. An intermolecular disulfide bond stabilizes E2A homodimers and is required for DNA binding at physiological temperatures. Cell. 1994;79:1057–1067. doi: 10.1016/0092-8674(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 8.Blom B, Heemskerk M H, Verschuren M C, van Dongen J J, Stegmann A P, Bakker A Q, Couwenberg F, Res P C, Spits H. Disruption of alpha beta but not of gamma delta T cell development by overexpression of the helix-loop-helix protein Id3 in committed T cell progenitors. EMBO J. 1999;18:2793–2802. doi: 10.1093/emboj/18.10.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi J K, Shen C P, Radomska H S, Eckhardt L A, Kadesch T. E47 activates the Ig-heavy chain and TdT loci in non-B cells. EMBO J. 1997;15:5014–5021. [PMC free article] [PubMed] [Google Scholar]
- 10.Dudley E C, Petrie H T, Shah L M, Owen M J, Hayday A C. T cell receptor beta chain gene rearrangement and selection during thymocyte development in adult mice. Immunity. 1994;1:83–93. doi: 10.1016/1074-7613(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 11.Fehling H J, von Boehmer H. Early αβ T-cell development in the thymus of normal and genetically altered mice. Curr Opin Immunol. 1997;9:263. doi: 10.1016/s0952-7915(97)80146-x. [DOI] [PubMed] [Google Scholar]
- 12.Georgopolos K, Moore D D, Wang J-H, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 13.Godfrey D I, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244. [PubMed] [Google Scholar]
- 14.Heemskerk M H, Blom B, Nolan G, Stegmann A P, Bakker A Q, Weijer K, Res P C, Spits H. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J Exp Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J-S, Olson E N, Kingston R E. HEB, a helix-loop-helix protein related to E2A and ITF2 that can modulate the DNA-binding ability of myogenic regulatory factors. Mol Cell Biol. 1992;12:1031–1042. doi: 10.1128/mcb.12.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iavarone A, Garg P, Lasorella A, Hsu J, Israel M A. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 17.Kadesch T. Helix-loop-helix proteins in the regulation of immunoglobulin gene transcription. Immunol Today. 1992;13:31–36. doi: 10.1016/0167-5699(92)90201-h. [DOI] [PubMed] [Google Scholar]
- 18.Kaye J, Hsu M-L, Sauron M-E, Jameson S C, Gascoigne N R J, Hedrick S M. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 1989;341:746. doi: 10.1038/341746a0. [DOI] [PubMed] [Google Scholar]
- 19.Kee B L, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D, Peng D C, Sun X H. Massive apoptosis of thymocytes in T-cell-deficient Id1 transgenic mice. Mol Cell Biol. 1999;19:8240–8253. doi: 10.1128/mcb.19.12.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein E S, Simmons D M, Swanson L W, Rosenfeld M G. Tissue-specific RNA splicing generates an ankyrin-like domain that affects the dimerization and DNA-binding properties of a bHLH protein. Genes Dev. 1993;7:55–71. doi: 10.1101/gad.7.1.55. [DOI] [PubMed] [Google Scholar]
- 22.Molina T J, Kishihara K, Siderovski D P, van Ewijk W, Narendran A, Timms E, Wakeham A, Paige C J, Hartmann K U, Veillette A, Davidson D, Mak T W. Profound block in thymocyte development in mice lacking p56lck. Nature. 1992;357:161. doi: 10.1038/357161a0. [DOI] [PubMed] [Google Scholar]
- 23.Mombaerts P, Anderson S J, Perlmutter R M, Mak T W, Tonegawa S. An activated lck transgene promotes thymocyte development in RAG-1 mutant mice. Immunity. 1994;1:261. doi: 10.1016/1074-7613(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 24.Morrow M A, Mayer E W, Perez C A, Adlam M, Siu G. Overexpression of the helix-loop-helix protein Id2 blocks T cell development at multiple stages. Mol Immunol. 1999;36:491–503. doi: 10.1016/s0161-5890(99)00071-1. [DOI] [PubMed] [Google Scholar]
- 25.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 26.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Interactions between heterodimeric helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 27.Okamura R M, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 28.Pivniouk V, Tsitsikov E, Swinton P, Rathbun G, Alt F W, Geha R S. Impaired viability and profound block in thymocyte development in mice lacking the adaptor protein SLP-76. Cell. 1998;94:229–238. doi: 10.1016/s0092-8674(00)81422-1. [DOI] [PubMed] [Google Scholar]
- 29.Roberts J L, Lauzurica P, Krangel M S. Developmental regulation of VDJ recombination by the core fragment of the T cell receptor alpha enhancer. J Exp Med. 1997;185:131–140. doi: 10.1084/jem.185.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawada S, Littman D R. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993;13:5620. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlissel M, Voronova A, Baltimore D. Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 1991;5:1367–1376. doi: 10.1101/gad.5.8.1367. [DOI] [PubMed] [Google Scholar]
- 32.Shen C-P, Kadesch T. B-cell-specific DNA binding by an E47 homodimer. Mol Cell Biol. 1995;15:4518–4524. doi: 10.1128/mcb.15.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinkai Y, Koyasu S, Nakayama K-I, Murphy K, Loh D Y, Reinherz E L, Alt F W. Restoration of T cell development to RAG-2-deficient mice by functional TCR transgenes. Science. 1993;259:822. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- 34.Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 35.Sun X-H. Constitutive expression of the Id1 genes impairs mouse B cell development. Cell. 1994;79:893–900. doi: 10.1016/0092-8674(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 36.Ting C N, Olson M C, Barton K P, Leiden J M. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 37.Tripathi R K, Mathieu N, Spicuglia S, Payet D, Verthuy C, Bouvier G, Depetris D, Mattei M G, Hempel W M, Ferrier P. Definition of a T-cell receptor β gene core enhancer of V(D)J recombination by transgenic mapping. Mol Cell Biol. 2000;20:42–53. doi: 10.1128/mcb.20.1.42-53.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Meerwijk J P, Bluthmann H, Steinmetz M. T-cell specific rearrangement of T-cell receptor beta transgenes in mice. EMBO J. 1990;9:1057. doi: 10.1002/j.1460-2075.1990.tb08210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Oers N S, Lowin-Kropf B, Finlay D, Connolly K, Weiss A. Alpha beta T cell development is abolished in mice lacking both Lck and Fyn protein tyrosine kinases. Immunity. 1996;5:429–436. doi: 10.1016/s1074-7613(00)80499-9. [DOI] [PubMed] [Google Scholar]
- 40.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald H R, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 41.Voronova A, Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci USA. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J H, Nichogiannopoulou A, Wu L, Sun L, Sharpe A H, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 43.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Turner D, Rupp R, Hollenberg S, Zhuang Y, Lassar A. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W, Sommers C L, Burshtyn D N, Stebbins C C, DeJarnette J B, Trible R P, Grinberg A, Tsay H C, Jacobs H M, Kessler C M, Long E O, Love P E, Samelson L E. Essential role of LAT in T cell development. Immunity. 1999;10:323–332. doi: 10.1016/s1074-7613(00)80032-1. [DOI] [PubMed] [Google Scholar]
- 45.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 46.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and HEB. Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhuang Y, Bardnt R J, Pan L, Kelley R, Dai M. Functional replacement of the mouse E2A gene with a human HEB cDNA. Mol Cell Biol. 1998;18:3340–3349. doi: 10.1128/mcb.18.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]