Significance
The vast majority of autoimmune diseases are polygenic, and causal loci uncovered by genetic-mapping studies explain only a minority of the heritable contribution to trait variation. Multiple explanations for this missing heritability include rare meaningful variants, rare copy number variations or deletions, epistasis, epigenetics, disease heterogeneity, and rare or infrequent variants that segregate within individual families (even within monozygotic twins). Here we demonstrate that experimental models of spontaneous autoimmune diseases may be invaluable tools to map rare germline variants impacting disease susceptibility traits. We identified a variant of the dual-specificity phosphatase 10 encoding gene that accelerates disease in an autoimmune type 1 diabetes model, the nonobese diabetic mouse.
Keywords: autoimmunity, type 1 diabetes, NOD mouse, genetic mapping
Abstract
Insulin-dependent or type 1 diabetes (T1D) is a polygenic autoimmune disease. In humans, more than 60 loci carrying common variants that confer disease susceptibility have been identified by genome-wide association studies, with a low individual risk contribution for most variants excepting those of the major histocompatibility complex (MHC) region (40 to 50% of risk); hence the importance of missing heritability due in part to rare variants. Nonobese diabetic (NOD) mice recapitulate major features of the human disease including genetic aspects with a key role for the MHC haplotype and a series of Idd loci. Here we mapped in NOD mice rare variants arising from genetic drift and significantly impacting disease risk. To that aim we established by selective breeding two sublines of NOD mice from our inbred NOD/Nck colony exhibiting a significant difference in T1D incidence. Whole-genome sequencing of high (H)- and low (L)-incidence sublines (NOD/NckH and NOD/NckL) revealed a limited number of subline-specific variants. Treating age of diabetes onset as a quantitative trait in automated meiotic mapping (AMM), enhanced susceptibility in NOD/NckH mice was unambiguously attributed to a recessive missense mutation of Dusp10, which encodes a dual specificity phosphatase. The causative effect of the mutation was verified by targeting Dusp10 with CRISPR-Cas9 in NOD/NckL mice, a manipulation that significantly increased disease incidence. The Dusp10 mutation resulted in islet cell down-regulation of type I interferon signature genes, which may exert protective effects against autoimmune aggression. De novo mutations akin to rare human susceptibility variants can alter the T1D phenotype.
Type one diabetes (T1D) is an autoimmune disease in which autoreactive T lymphocytes destroy insulin-producing β-cells of the islets of Langerhans (1). The disease occurs spontaneously in nonobese diabetic (NOD) mice, an inbred strain (NOD/Shi), first reported in 1980 by Makino et al. in Japan by selective breeding of outbred cataract-prone ICR:Jcl mice at the Shionogi Research Laboratories (2).
NOD mice were initially distributed by the Central Institute for Experimental Animals, Japan (NOD/ShiJcl). Substrains were thus bred over the world. We started our colony at the Hôpital Necker in Paris (NOD/Nck) in 1986 (3). The other currently most-studied NOD substrains are NOD/ShiJcl (Central Institute for Experimental Animals, Japan), NOD/ShiLtJ (The Jackson Laboratory) and the NOD/ShiLtDvs substrain recently derived from it (4), NOD/MrkTac, and NOD/BomTac (Taconic Europe).
Breakdown of self-tolerance in NOD mice and infiltration of the islets of Langerhans by mononuclear cells (i.e., insulitis) begins at 3 wk of age, eventually causing the destruction of β-cells that precedes hyperglycemia, occurring by 3 mo, when ∼70% of the insulin-secreting β-cell mass has been destroyed (1). The etiology of T1D, either in humans or NOD mice, is only partially understood, although compelling data point to the involvement of both genetic and environmental factors (4–8). The disease is polygenic, with a large portion of disease risk in both mice and humans attributed to class II major histocompatibility complex (MHC)/human leukocyte antigen (HLA) alleles together with the contribution of numerous other loci (6, 7). However, identification of the genes of interest in the candidate loci is incomplete, and part of the genetic risk remains unexplained (9).
Results and Discussion
Generation and Whole-Genome Sequencing of High-Incidence NOD/NckH and Low-Incidence NOD/NckL Sublines.
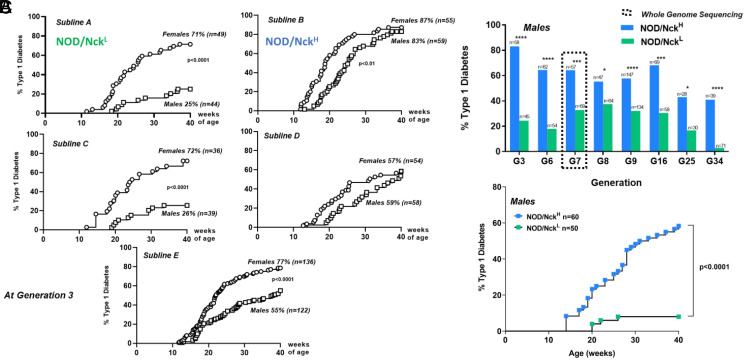
To explore novel experimental designs addressing missing heritability in T1D, we took advantage of a restart of our colony from a single pair of NOD/Nck mice rederived by cesarean delivery. In breeding the colony, we noted as before that disease incidence widely differed between litters (of ∼8 to 12 mice per litter). We hypothesized that if this phenotypic variation in litters presenting high or low T1D incidence could be permanently fixed in sublines of NOD/Nck mice generated through brother–sister breeding, causal gene variants could be mapped. We initially established five sublines, which indeed differed in T1D incidence and after three generations we concentrated on the two showing the most significant difference. In these NOD/NckH (high incidence) and NOD/NckL (low incidence) mice (Fig. 1A) the phenotypic variation was stably maintained for over 30 generations, as assessed by comparing incidence and age of onset of T1D at equivalent generations (Fig. 1B and SI Appendix, Fig. S1A). This phenotypic variation was observed both in females and males. As the outcome was more obvious in males, these were preferentially used. We confirmed that the difference in T1D incidence in NOD/NckH and NOD/NckL mice rederived from embryos frozen at generation 7 was comparable to the original G7 mice (Fig. 1C and SI Appendix, Fig. S1B).
Fig. 1.
Diabetes incidence in sublines of the NOD/Nck strain. (A) Incidence of T1D at generation 3 in five distinct sublines (A to E) established from the NOD/Nck strain by brother–sister mating of mice within individual litters. For further studies we concentrated on sublines A and B, which we named NOD/NckL (for low incidence) and NOD/NckH (for high incidence). (B) Incidence of T1D in NOD/NckH and NOD/NckL male mice followed from generation 3 up to generation 34. ****P < 0.0001, ***P < 0.001, *P < 0.05. (C) Incidence of T1D in NOD/NckH and NOD/NckL male mice derived from embryos frozen at generation 7 and revitalized. Actuarial survival curves were compared using the log-rank (Mantel–Cox) statistical test.
Genetic drift is one major factor explaining phenotypic differences within inbred strains (4, 10–13). Therefore, at generation 7 we sequenced the whole genome of four individuals of each subline. Sequencing data were generated to an average mapped read depth of 31.4× per individual and variants were then called from the alignment against the mm10 reference genome (C57BL/6J; GRCm38). The filtering strategy for the identification of the putative variants used both manual steps encompassing coding and noncoding regions, which identified subline-specific single-nucleotide variants (SNVs) and small insertions–deletions (indels) (SI Appendix, Table S1) in addition to a more systematic automated analysis (14) also extended by the search for structural variants. In total, 118 mutations validated in 10 to 15 additional individuals by Ion Torrent sequencing were used for mapping the early T1D onset (Dataset S1).
Interestingly, we found that the validated coding mutations were private to either NOD/NckH or NOD/NckL, since they were not found in the NOD/ShiLtJ bred since 1988 at The Jackson Laboratory (15). Six of the variants were present in insulin-dependent diabetes (Idd) genetic regions: five noncoding SNVs in Paqr8, Spsb1, Nox4, Aaed1, and a 3' UTR in Rgs16, and a nonsynonymous missense SNV in H2-Q4 (7).
Identification of Causative Mutations in NOD/NckH and NOD/NckL: Using Age of Onset of T1D as a Quantitative Trait and Automated Meiotic Mapping (AMM).
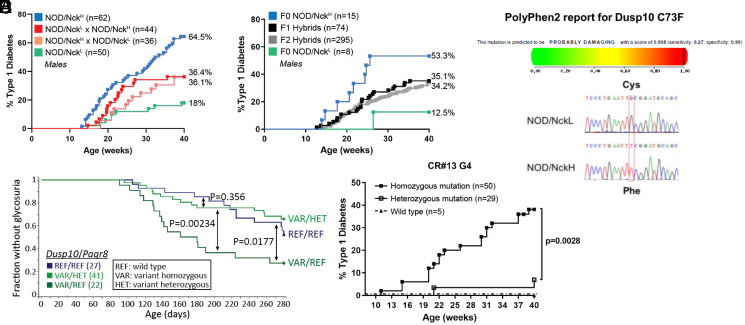
To identify causative mutations, we mapped the age of T1D onset as a quantitative trait in a large cohort of F2 hybrid mice (294 males and 316 females) obtained by crossing NOD/NckH and NOD/NckL parents and intercrossing their progeny (Fig. 2 A and B). Importantly, T1D incidence in F1 hybrids was intermediate to that of parental F0 NOD/NckH and NOD/NckL mice (Fig. 2 A and B) and it was similar whether the dam was from one or the other subline, indicating that there was no subline-specific parental origin effect on the phenotype transmission (Fig. 2A). F2 hybrids showed a similar incidence and age of onset of T1D as F1 hybrids (Fig. 2B).
Fig. 2.
Variants of Dusp10 and Paqr8 responsible for differences in T1D incidence between NOD/NckH and NOD/NckL sublines. (A) Incidence of T1D in F1 hybrids obtained by crossing NOD/NckH and NOD/NckL at generation 6 compared to parental lines. (B) Incidence of T1D is shown in F1 hybrids obtained by crossing NOD/NckH and NOD/NckL at generation 24 and in F2 hybrids obtained by intercross of F1 hybrids. These F2 hybrid mice were used for automated meiotic mapping. (C) Kaplan–Meier curves of glycosuria onset in F2 mice with the indicated Dusp10 and Paqr8 genotypes are presented. As compared to wild-type F2 individuals expressing control alleles for Dusp10 and Paqr8 (REF/REF, blue line), F2 mice expressing the Dusp10 mutation in the homozygous state (VAR/REF, dark green line) showed significant acceleration of T1D development (recessive effect). Moreover, heterozygosity for the Paqr8 mutation (VAR/HET, light green line) abolishes the recessive Dusp10 effect, rescuing mice from early onset T1D (dominant effect of Paqr8 mutation). (D) (Upper) PolyPhen-2 score of Dusp10 mutation = 0.998/1, classified as probably damaging. (Lower) DNA sequence chromatogram of the region of Dusp10 containing the identified SNV. (E) A second mutant allele of Dusp10 was generated on the NOD/NckL background using CRISPR-Cas9 technology. T1D incidence in male wild-type or CR#13 mice carrying the homozygous and the heterozygous Dusp10Y86X allele. The CR#13 founder was backcrossed to a particular NOD/NckL subline expressing the wild-type Paqr8 allele, developed by serendipity from the original NOD/NckL subline at generation 16. Note that incidence of T1D in male NOD/NckH and NOD/NckL mice at generation 34 concurrent to generation 4 of CR#13 was, respectively, 53 and 2%.
We combined this traditional breeding approach for quantitative trait locus (QTL) analysis with a validated method for variant genotyping (Ion Torrent sequencing using barcoded libraries) and analyzed the data using software (Linkage Analyzer) to test the probability of single-locus associations with phenotypes using recessive, semidominant (additive), and dominant transmission models (14). For this particular study, a second mode of analysis was developed in which epistatic effects of mutations at all loci were tested to identify modifications of phenotypic effects by mutations at all other loci.
When single-locus linkage analysis was performed, the early age of T1D onset in the NOD/NckH line was mapped to a recessive missense mutation of NOD/NckH origin in Dusp10, encoding the dual specificity phosphatase 10 (DUSP10) (Fig. 2C). Subsequently, an epistatic effect was detected indicating a dominant inhibitory effect of a noncoding polymorphism of NOD/NckL origin in progestin and adipoQ receptor family member VIII (Paqr8) on the Dusp10 variant allele (Fig. 2C). Importantly, cumulative T1D incidence curves in F2 Dusp10 homozygous variant mice with either zero or one Paqr8 variant allele (Fig. 2C) strictly reflected the disease incidence curves in the parental F0 sublines (Fig. 2B) tested concurrently.
The Dusp10 nonsynonymous SNV is a G-to-T transversion (Fig. 2D) generating a cysteine-to-phenylalanine substitution at position 73 of the DUSP10 protein. The mutation occurs within the first N-terminal domain, which is predicted to be intrinsically disordered (SI Appendix, Fig. S2) (16, 17). PolyPhen-2 classifies this mutation as probably damaging (Fig. 2D). We found highly conserved orthologs of DUSP10 in representatives of all vertebrate classes, including the lamprey Petromyzon marinus, and the amino acid sequences flanking mouse Dusp10 Cys73 were identical in the vertebrates examined (SI Appendix, Fig. S3).
Validating the Causative Effect of the Dusp10 Mutation in the Increased T1D Incidence Observed in NOD/NckH Mice.
A second mutant allele of Dusp10 was generated in NOD/NckL mice using the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 technology (18). We expanded a line (CR#13) in which deletion of a single nucleotide resulted in the nonsense mutation Y86X, a premature STOP (SI Appendix, Fig. S2). To reduce the risk of off-target effects while eliminating the dominant inhibitory effect of the Paqr8 polymorphism on the Dusp10 variant allele, the CR#13 founder was backcrossed to a particular NOD/NckL subline expressing the wild-type Paqr8 allele, rather than the epistatic variant (Fig. 2E). CR#13 mice homozygous for the Dusp10Y86X mutation showed a T1D incidence similar to that of NOD/NckH mice (Fig. 2E).
Mechanistic Immunological Studies in NOD/NckH or NOD/NckL Mice.
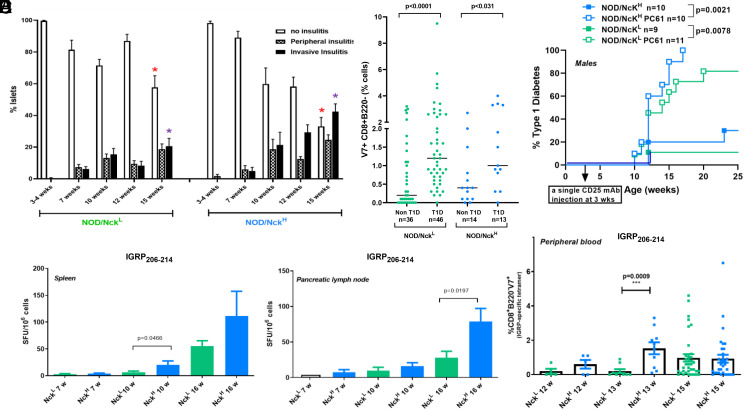
Proportions of major immune cell subsets did not differ between the two sublines (SI Appendix, Fig. S4). Monitoring of immunopathological parameters characteristic of the autoimmune process disclosed a clear time shift in the kinetics of some events, which occurred earlier in NOD/NckH as compared to NOD/NckL mice, such as progression of insulitis (Fig. 3A and SI Appendix, Fig. S5) and increase in number of diabetogenic T lymphocytes in spleen, pancreatic lymph nodes, and peripheral blood (Fig. 3 B–D) (19). In addition, we found no intrinsic differences when comparing diabetogenic lymphocytes (SI Appendix, Fig. S6), regulatory T lymphocytes (Fig. 3E and SI Appendix, Fig. S7), or the timing of islet antigen presentation (SI Appendix, Fig. S8) between NOD/NckL and NOD/NckH mice.
Fig. 3.
Islet infiltration and pathogenic T lymphocytes in NOD/NckH and NOD/NckL. (A) Insulitis was monitored in NOD/NckH and NOD/NckL male mice. The proportion of insulitis-free islets and of islets presenting peripheral or invasive insulitis were counted on hematoxylin and eosin stained tissue sections; 60 to 100 islets were counted per pancreas from 9 to 20 individuals. Results are expressed as mean ± SEM. At 15 wk of age in NOD/NckH mice a significant decrease (red asterisk) in the proportion of normal noninfiltrated islets and a significant increase (purple asterisk) of islets with invasive insulitis was observed compared to NOD/NckL (Mann–Whitney U test, P < 0.0094 and P < 0.0029, respectively). (B) Pathogenic CD8+ T cells specific for the β-cell IGRP206–214 epitope were enumerated in the spleen and pancreatic lymph nodes of NOD/NckH and NOD/NckL male mice using an IFNγ ELIspot. Spot forming units (SFU)/1 × 106 cells per lymphoid organ were counted and data were expressed as mean ± SEM of organs from three to nine individuals. Statistical differences were assessed using the Mann–Whitney U test. (C) Pathogenic CD8+ T cells specific for the β-cell–specific IGRP206–214 epitope were enumerated in peripheral blood of NOD/NckH and NOD/NckL male mice using the NRP-V7 mimotope (8). Data are expressed in individual mice as % NRP-V7+ cells within the CD8+B220− gate and also presented as mean ± SEM of the analyzed individuals. Statistical differences were assessed using the Mann–Whitney U test. (D) Pathogenic CD8+NRP-V7+ T cells were enumerated in peripheral blood of NOD/NckH and NOD/NckL males and females at 12 to 25 wk of age. Results were separated to show for each subline mice that developed diabetes (T1D) versus those that did not (non-T1D). Data were expressed as in C. Statistical differences were assessed using the Mann–Whitney U test. (E) Incidence of T1D in NOD/NckH and NOD/NckL mice following administration at 3 wk of age of 500 µg of the CD25 monoclonal antibody (PC61) that targets thymic-derived regulatory T cells. Actuarial survival curves were compared using the log-rank (Mantel–Cox) statistical test.
Zhang et al. established C57BL/6 mice homozygous for a robust knockout allele of Dusp10 (20). Dusp10-deficient cells showed increased innate immune responses, i.e., production of greatly enhanced levels of proinflammatory cytokines by peritoneal macrophages upon TLR2, TLR3, and TLR4 stimulation, which we did not find in NOD/NckH mice expressing the Dusp10 variant (SI Appendix, Fig. S9). Dusp10-deficient C57BL/6 mice showed increased resistance to experimental autoimmune encephalomyelitis (EAE) (20). This finding in apparent conflict with our present data in NOD/NckH mice expressing the Dusp10 variant and higher T1D incidence may be related to the different genetic background and/or to the fact that EAE is not a spontaneous autoimmune disease like T1D but is induced upon administration of the autoantigen in the presence of adjuvant.
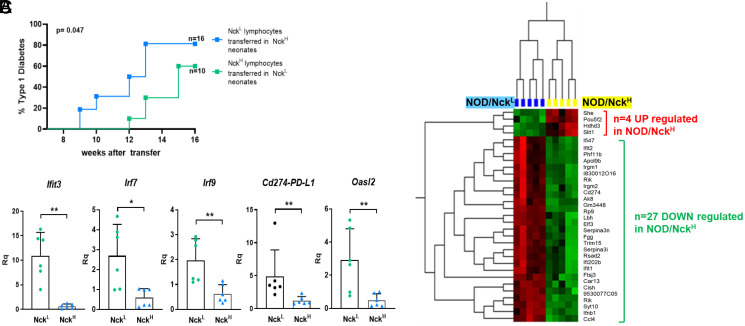
To address whether the difference in T1D incidence driven by the Dusp10 mutation resulted from an impact on the immune system, or on the target of disease (namely the pancreatic islets), we used an adoptive transfer model (21). Splenic T lymphocytes from overtly diabetic NOD/NckH or NOD/NckL mice were transferred into the reciprocal newborn recipients. NOD/NckL recipients were less sensitive to T1D transfer than NOD/NckH recipients arguing for an impact of the Dusp10 mutation on the islets of Langerhans (Fig. 4A).
Fig. 4.
The NOD/NckH Dusp10 variant acts within islets to confer susceptibility to T1D and results in down-regulation of type I interferon signature genes. (A) Incidence of T1D in female NOD/NckH and NOD/NckL recipients after adoptive transfer of diabetogenic lymphocytes of the reciprocal genotype at 24 to 48 h after birth. Groups of experimental and control (sham injected) recipients were equally distributed within the litters. NOD/NcKL recipients were less sensitive to T1D transfer than NOD/NckH recipients. (B) Comparative transcriptome analysis of purified islets of Langerhans from NOD/NckH and NOD/NckL mice at 3 wk of age. Identification of 27 down-regulated genes in NOD/NckH mice that included various type I interferon signature genes. (C) The differential expression of type I interferon signature genes was validated by qPCR targeting Cd274 (PD-L1) and four additional well-known type I interferon signature genes (Ifit3, Irf7, Irf9, and Oasl2). **P < 0.01, *P < 0.05.
It is an open question whether NOD mice are differentially sensitive to an immune response perhaps at the β-cell level once it has begun. We therefore performed transcriptome analysis of pancreatic islets from 3-wk-old NOD/NckH mice, before onset of insulitis, which showed down-regulation of various type I interferon signature genes (ISGs) compared to expression in NOD/NckL islets, including Cd274 that encodes programmed death ligand 1 (PD-L1) (Fig. 4B). These results were validated by qPCR (Fig. 4C). Type I IFN signature is a critical marker of early stages of T1D in NOD mice for which triggering stimuli are ill defined (22–24). It may appear counterintuitive that ISGs are down-regulated in NOD/NckH mice that express high disease incidence. This is however reminiscent of data showing that one well-established type I IFN inducer, the TLR3 ligand polyinosinic-polycytidylic acid (poly I:C), provides full protection from T1D when injected into young NOD mice (25). As the PD1/PD-L1 axis is an important checkpoint in T1D (26), the increased expression of PD-L1 as part of ISGs is a plausible functional explanation for the delayed kinetics of insulitis in NOD/NckL mice, although presently we cannot rule out other mechanisms.
Our results have three major implications. First, we identified a single nonsynonymous nucleotide change in the DUSP10 encoding gene and provide evidence that it directly affects the phenotype of a complex autoimmune disease. Second, our data may illustrate the rapidity of appearance and fixation of de novo germline mutations that can alter a complex phenotype such as T1D, although one cannot exclude that the variants were already present in a heterozygous form in the two initial founders. This suggests that the T1D phenotype has a large genomic footprint. Finally, these results offer avenues to tackle the problem of missing heritability in autoimmune diabetes. For studies using mice, they highlight the potential of applying random germline mutagenesis and a forward genetic strategy (14) to identify novel genes that modify T1D. For studies in humans, they point to the importance of searching for rare variants in well-defined subgroups of patients as successfully done for other autoimmune and inflammatory diseases (27, 28).
Materials and Methods
Mice.
NOD/Nck mice were bred and housed under specific pathogen-free conditions at the Hôpital Necker-Enfants Malades animal facility (agreement: C751515). Animals were fed ad libitum with an irradiated VRF1 diet (Special Diets Services) with fresh autoclaved water.
This study was carried out in strict accordance with the recommendations of European Directives (2010/63/UE) and institutional guidelines (INSERM, Faculté Paris Descartes). The protocols were approved by the Ethical Committee of Paris Descartes University and the French Ministry of Education and Research (PROJET No. 2019092612242506–V3 APAFiS #25948).
Diabetes Monitoring.
Mice were monitored for diabetes weekly by testing for glycosuria using colorimetric Diabur-Test 5000 strips (Roche). Overt diabetes was confirmed by testing for fasting glycemia (>250 mg/dL) (Accu-Check; Roche).
Insulitis Scoring.
Pancreata were collected, fixed in 4% formaldehyde, and paraffin embedded. Serial 5-μm sections cut at 60-μm intervals were hematoxylin and eosin stained and at least 80 islets per pancreas and/or 200 islets per group of mice were scored. Mononuclear cell infiltration of the islets was graded as follows: no insulitis (intact islets); periinsulitis (infiltration remaining confined to the periphery of the islets); and invasive/destructive insulitis.
Detection of IFNγ Producing Autoreactive CD8+ T Cells by a T-Cell Enzyme Linked ImmunoSpot (EliSpot).
Microplates (96-well PVDF plates, Millipore) and an IFNγ capture and detection antibody pair (U-CyTech Biosciences) were used. Splenocytes were cultured in the presence of IL-2 at 2.5 × 105 per well. For lymph nodes, 0.3 × 105 cells were used. Irradiated (35 Gy, X-Ray Source) splenocytes 2 × 105 were added per microwell. Peptides used were InsB15-23 and IGRP206-214 (7 μM). As a positive control the CD3 antibody (clone 2C11; 1 µg/mL) was used. After a 20-h culture in RPMI medium supplemented with 5% fetal calf serum (FCS), IFNγ was detected using biotinylated anti-IFNγ capture antibody, alkaline phosphatase conjugated ExtrAvidin (Sigma-Aldrich), and 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (Sigma-Aldrich). Air-dried plates were read and spots counted using an AID reader (Autoimmun Diagnostika). Data were expressed as the mean of triplicate wells minus the average number of spots in negative control wells (cells cultured without peptides) of spot-forming cells per 106 cells.
Tetramer Staining.
Peripheral blood collected from the retroorbital sinus using heparinized capillary tubes was incubated with H-2Kd tetramers bearing the peptide NRP-V7 or an irrelevant peptide (TUM) for 1 h on ice. Tetramers were produced in the P.S. laboratory (Calgary University). After a washing step, surface staining was performed at 4 °C in staining buffer (PBS 1×–FCS 1%–sodium azide 0.1%) using anti-CD8-APC (clone 53-6.7) and anti-CD45R-PerCP (clone RA3-6B2) (both from BD Biosciences). Red blood cells were lysed at room temperature using FACS Lysing Solution (BD Biosciences) before cell suspensions were washed prior to acquisition. Data were acquired on a FACSCanto II flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star). NRP-V7 tetramer-positive cells were expressed as percentage of B220−CD8+ cells minus the percentage of TUM tetramer-positive cells.
In Vivo Tracking of CD4+ BDC 2.5 T Cells.
Naïve CD4+ T cells were isolated from spleens and lymph nodes of BDC-2.5 transgenic NOD mice using the Mouse CD4 Naïve T Cell Isolation Kit (BioLegend). Naïve T cells were further selected positively by treatment with anti-CD62L biotinylated antibodies (BD Pharmingen) followed by anti-biotin magnetic beads and sorted using MACS technology (Miltenyi Biotec). After carboxyfluorescein diacetate succinimidyl ester-labeling (5 μM), 1 × 106 cells were injected intravenously into 25-d-old NOD/NckH and NOD/NckL recipients. On day 3 after transfer, pancreatic, mesenteric, and axillary lymph nodes were collected and cells were stained for flow cytometry analysis.
Adoptive Transfer of Diabetes into Neonates.
Within 24 to 48 h after birth neonates subjected to hypothermic anesthesia (4 min at −20 °C) were injected via the periorbital superficial vein under microscopical control with 0.05 mL cell suspension at the appropriate concentration (20 × 106 B cell–depleted splenocytes from diabetic donors). Groups of experimental and control recipients (NOD/NckH and NOD/NckL newborns sham injected with saline) were equally distributed within the litters used. Here, to ask if the mutation in the recipient’s islet of Langerhans cells can impact T1D, NOD/NckH and NOD/NckL neonates were injected with 20 × 106 B cell–depleted lymphocytes from overtly diabetic NOD/NckL and NOD/NckH mice, respectively.
Isolation of Pancreatic Islets.
Pancreata were perfused with a solution of collagenase P (0.6 mg/mL; Roche), dissected free from surrounding tissues, and then digested at 37 °C for 13 min. After extensive washes, islets were purified by hand picking.
Flow Cytometry.
Single-cell suspensions were prepared from lymphoid organs by and from different tissues. Prior to staining, cells were incubated with a Fixable Viability Dye (eBioscience) and then with 2.4G2 antibody for FcγRII/III blocking (BD Pharmingen). Surface staining was performed at 4 °C in staining buffer (PBS 1×–FCS 2%–EDTA 5 mM). Intracellular staining of cytokines was performed using Fixation Buffer (BioLegend) and Intracellular Staining Perm Wash Buffer (BioLegend). Transcription factors were stained using the Foxp3 TF Staining Buffer Set (eBioscience). See the list of antibodies below. Data were acquired on a FACSFortessa flow cytometer (BD Biosciences) and analyzed using FlowJo version 10 software (Tree Star).
Antibodies used for staining were biotin or fluorochrome-labeled antibodies to murine CD11c (HL3), B220 (RA3-6B2), CD8 (53-6.7), CD103 (2E7), CD80 (19-10A1), CD86 (GL1), CD40 (3/23), I-Ak (10-3.6; crossing with the MHC class II from NOD mice, I-Ag7), CD11b (M1/70), ICAM-1 (3E2), OX40L (RM134L), CCR7 (4B12), CCR9 (9B1), TCR (H57-597), CD4 (GK1.5), CD19 (1D3), CD25 (PC61), DX5, and streptavidin-pacific blue were obtained from PharMingen BD. Antibodies to PD-L1 (MIH5), GITRL (eBioYGL386), CD73 (eBioTY/11.8), and CD39 (24DMS1) were purchased from eBisocience.
Whole-Genome Sequencing and Variant Identification.
High molecular weight genomic DNA was prepared from the liver of four animals of each NOD/Nck subline. The sequencing core facility of the Institut de Génétique et de Biologie Moléculaire et Cellulaire in Strasbourg performed eight paired-end runs on the HiSEq. 2500 platform (Illumina). Primary analysis was performed using CASAVA v1.8.2 (Illumina). A minimum of 1,308,000 reads was obtained for each of the eight samples (range: 1,308,440,124 to 1,971,179,220 reads). Sequencing data were mapped using BWA (v 0.7.10) to align against the mm10 reference genome (C57BL/6J; GRCm38) allowing a maximum of three mismatches and a maximal length of 50 nucleotides for indels. On average, 78% of reads could be aligned against mm10 with a mean coverage of 31.3 (see below).
Using the Genome Analysis Toolkit (GATK) and SAMtools, we found an average of 7.3 million SNVs and small indels differentiating the NOD/Nck and the C57BL/6J strains using single-sample variant calling.
In a first step alignment of sequencing data to mouse reference genome (GRCm38, mm10) variant calling and automated filtering were performed by collaborators (M. Dumas and B. Jost, Plateforme Biopuces et Séquençage, IGBMC, Strasbourg, France) with particular attention to variants in noncoding regions using GATK v2.5-2 Unified Genotyper and Samtools mpileup v0.1.18. Merge of identified variants and annotation were performed using GATK CombineVariants and SnpEff v3.3c, respectively. Using the GATK and SAMtools we found an average of 7.3 million SNVs and small indels differentiating the NOD/Nck and the C57BL/6J strains using single-sample variant calling.
Private variants present in the four datasets of one subline and absent from the four datasets of the other subline were then selected using in-house Perl scripts. To reduce the number of false positive calls and lessen the burden for validation, variants located in intergenic regions were excluded. Variants located in protein coding regions and/or within Idd intragenic regions were then selected. Identified variants were curated manually using IGV2.3 viewer (Broad Institute), revealing a high number of false positive calls. Following this strategy 11 subline-specific variants were identified for NOD/NckL and 9 for NOD/NckH (SI Appendix, Table S1). These variants were validated by Sanger sequencing of genomic DNA from the same eight males used to perform the sequencing and also from 10 to 15 NOD/NckL and 9 for NOD/NckH individuals of further generations.
In a second step at the University of Texas Southwestern Medical Center, to prepare for AMM, filtering was repeated by means of joint variant calling using the four samples of each subline. Using this jointly called data, variants from each subline were compared. They were then filtered for those variants unique to each subline and heterozygous calls were excluded (allelic ratio >0.875). A quality score of 80 was used as a filtering threshold for jointly called variants. Additionally, 34 SVs differentiating the NOD/Nck and C57BL/6J strains were identified using speedSeq SV (v 0.1.0). In addition to the SVs, 168 SNVs and small indels were chosen for validation via Ion Torrent sequencing to give genomic linkage using 20 Mb as a cutoff. A total of 118 variants were validated in 10 to 15 additional mice and used for AMM. A total of 77.3% of the mouse genome was in linkage with at least one of the 118 sequence variants, using a distance cutoff criterion of 20 Mb.
AMM.
A panel of 118 validated variant loci distinguishing NOD/NckH and Nod/NckL sublines was used to map the high diabetes incidence phenotype in F2 mice. The F2 animals generated by two-generation intercrosses were monitored for overt diabetes for 40 wk to determine the age of diabetes onset. Genomic DNA was extracted from tail snips and used as a template for multiplexed targeted amplification of the 118 loci using custom Ampliseq primers. Amplification products were barcoded to correspond to individual mice prior to sequencing using an Ion PGM (Life Technologies). Following phenotypic screening, AMM using recessive, additive, and dominant models of inheritance was performed for each variant using the Linkage Analyzer program as previously described (14). Kaplan–Meier plots of phenotypic data and Manhattan plots of linkage data were generated using the Linkage Explorer program (14). The P values of association between genotype and phenotype were calculated based on Kaplan–Meier analysis of time of onset of T1D, as related to zygosity for each of the mutations using a likelihood ratio test from a generalized linear model or generalized linear mixed effect model and Bonferroni correction applied.
Generation of Genetically Modified NOD/Nck Mice Using CRISPR-Cas9.
NOD/NckL females (generation 32) were superovulated by administration of pregnant mare serum gonadotropin at day 0 (5 U) and human chorionic gonadotropin at day 2 with a 44-h interval (5 U), before being mated with NOD/NckL males (generation 32). Because NOD females were found to give more eggs at 8 wk of age than at 4 wk of age (data from F.V.), 8- to 10-wk-old mice were used as zygote donors. Zygote collection, fertilized egg microinjection, and transfer in the oviduct of pseudogestant foster mothers were performed by collaborators (Plateforme de Transgénèse, F.L.-V., Institut Pasteur, Paris). The design of the single-guide RNA for the knockin and knockout experiments, and the selection of the homology-directed repair (HDR) donor template for the knockin experiment, were performed in collaboration with the laboratory of B.B., University of Texas Southwestern. All the reagents were from Integrated DNA Technologies (IDT): crRNA (IDT Alt-R CRISPR-Cas9 crRNA), tracrRNA (IDT Alt-R CRISPR-Cas9 tracrRNA), HDR donor template (IDT Ultramer DNA oligo), and Cas9 (IDT Alt-R S.p. HiFi Cas9 nuclease 3NLS). For genotyping of pups, DNA was extracted from tail snips using Genomic DNA purification kit (Qiagen) and proteinase K (Roche). PCR was performed by using the forward Xf and reverse Xr primers; and Accuprime Pfx DNA polymerase (Invitrogen) and the PCR products, after purification using the QIAquick PCR purification kit (Qiagen), were analyzed by sequencing using Xf primer (Plateforme de Séquençage, Eurofins).
Transcriptome, Gene Expression Profiling.
Agilent SurePrint G3 Mouse Gene Expression 8 × 60 K Microarrays (Agilent Technologies) were used for microarray experiments on purified CD4+CD25−CD62L− spleen lymphocytes, colon, ileon, and islets of Langerhans from NOD/NckH and NOD/NckL mice. Five replicas were prepared for the various cell/tissue preparations. Cell and islet pellets and tissues were lysed using SuperAmpTM lysis buffer (Miltenyi Biotech) and stored at −80 °C until all samples were collected. Preparation of RNA, amplification of RNA, sample hydridization (Agilent whole mouse genome oligo microarrays) scanning and data acquisition were performed by Miltenyi Biotech. RNA quality and purity were assessed on the Agilent Bioanalyzer 2100 (Agilent Technologies). Microarray probe fluorescence signals produced by the Agilent Feature Extraction (AFE) image analysis software were converted to expression values using Bioconductor Agi4 × 44PreProcess package with a custom annotation package.
Gene Expression Analysis.
RNA was isolated from cell suspensions using TRIzol reagent (Invitrogen, Thermo Fisher Scientific) and purified according to the manufacturer’s instructions using the RNeasy Mini Kit (Qiagen). Samples were reverse transcribed into complementary DNA using reagents from Promega (A3800; Promega). The qPCR for relevant genes was performed using a 7700 Sequence Detector (TaqMan; Perkin-Elmer Applied Biosystems, Thermo Fisher Scientific) and TaqMan-validated oligonucleotides.
Statistical Analyses.
All statistics were performed using GraphPad Prism version 6 software. Cumulative actuarial diabetes incidence was calculated according to the Kaplan–Meier method. Incidence curves were compared using the log-rank (Mantel–Cox) test. Comparison between means was performed using the nonparametric Mann–Whitney U test for other experiments, unless otherwise specified in the figure legend. Data are presented as means ± SEM. A P value <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We are indebted to Dr. Alicia Pérez-Arroyo, Mrs. Laurène Magne, Mme. Céline Keime, and Mr. Bernard Jost for their support of this work. Funding was provided by NIH Grant R01 AI125581 (B.B.); NIH Grant U19 AI100627 (B.B.); European Research Council Advanced Grant, Hygiene No. 250290 (J.-F.B.); Institut National de la Santé et de la Recherche Scientifique (L.C.); Fondation Day Solvay (L.C.); and the Lyda Hill Foundation (B.B.).
Footnotes
Reviewers: M.J.J., The Hospital for Sick Children, Toronto, Ontario; T.W.H.K., St. Vincent’s Institute of Medical Research; and L.S.W., University of Oxford.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112032118/-/DCSupplemental.
Data Availability
Transcriptome raw data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession no. GSE183286. All other data are available in the main text or the supplementary information.
References
- 1.Bach J. F., Insulin-dependent diabetes mellitus as an autoimmune disease. Endocr. Rev. 15, 516–542 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Makino S., et al., Breeding of a non-obese, diabetic strain of mice. Jikken Dobutsu 29, 1–13 (1980). [DOI] [PubMed] [Google Scholar]
- 3.Hunger R. E., et al., Inhibition of submandibular and lacrimal gland infiltration in nonobese diabetic mice by transgenic expression of soluble TNF-receptor p55. J. Clin. Invest. 98, 954–961 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simecek P., et al., Genetic analysis of substrain divergence in non-obese diabetic (NOD) mice. G3 (Bethesda) 5, 771–775 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bach J. F., The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 347, 911–920 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Concannon P., et al., Type 1 Diabetes Genetics Consortium, Genome-wide scan for linkage to type 1 diabetes in 2,496 multiplex families from the Type 1 Diabetes Genetics Consortium. Diabetes 58, 1018–1022 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridgway W. M., et al., Gene-gene interactions in the NOD mouse model of type 1 diabetes. Adv. Immunol. 100, 151–175 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Yurkovetskiy L., et al., Gender bias in autoimmunity is influenced by microbiota. Immunity 39, 400–412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pociot F., Type 1 diabetes genome-wide association studies: not to be lost in translation. Clin. Transl. Immunology 6, e162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar V., et al., C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science 342, 1508–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poltorak A., et al., Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 282, 2085–2088 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Simon M. M., et al., A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 14, R82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baxter A. G., et al., Genetic basis for diabetes resistance in NOD/Wehi mice. Eur. J. Immunogenet. 20, 409–417 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Wang T., et al., Real-time resolution of point mutations that cause phenovariance in mice. Proc. Natl. Acad. Sci. U.S.A. 112, E440–E449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keane T. M., et al., Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao X., Tong L., Crystal structure of the MAP kinase binding domain and the catalytic domain of human MKP5. Protein Sci. 16, 880–886 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y. Y., Wu J. W., Wang Z. X., A distinct interaction mode revealed by the crystal structure of the kinase p38α with the MAPK binding domain of the phosphatase MKP5. Sci. Signal. 4, ra88 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Cong L., et al., Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trudeau J. D., et al., Prediction of spontaneous autoimmune diabetes in NOD mice by quantification of autoreactive T cells in peripheral blood. J. Clin. Invest. 111, 217–223 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., et al., Regulation of innate and adaptive immune responses by MAP kinase phosphatase 5. Nature 430, 793–797 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Bendelac A., Carnaud C., Boitard C., Bach J. F., Syngeneic transfer of autoimmune diabetes from diabetic NOD mice to healthy neonates. Requirement for both L3T4+ and Lyt-2+ T cells. J. Exp. Med. 166, 823–832 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrero J. A., Calderon B., Towfic F., Artyomov M. N., Unanue E. R., Defining the transcriptional and cellular landscape of type 1 diabetes in the NOD mouse. PLoS One 8, e59701 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colli M. L., et al., An integrated multi-omics approach identifies the landscape of interferon-α-mediated responses of human pancreatic beta cells. Nat. Commun. 11, 1–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos-Rodríguez M., et al., The impact of proinflammatory cytokines on the β-cell regulatory landscape provides insights into the genetics of type 1 diabetes. Nat. Genet. 51, 1588–1595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aumeunier A., et al., Systemic toll-like receptor stimulation suppresses experimental allergic asthma and autoimmune diabetes in NOD mice. PLoS One 5, e11484 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ansari M. J., et al., The programmed death-1 (PD-1) pathway regulates autoimmune diabetes in nonobese diabetic (NOD) mice. J. Exp. Med. 198, 63–69 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium IMSG, Low-frequency and rare-coding variation contributes to multiple sclerosis risk. Cell 175, 1679–1687.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gettler K., et al.; UK IBD Genetics Consortium, National Institute of Diabetes, Digestive and Kidney Diseases Inflammatory Bowel Disease Genetics Consortium, Common and Rare Variant Prediction and Penetrance of IBD in a Large, Multi-ethnic, Health System-based Biobank Cohort. Gastroenterology 160, 1546–1557 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome raw data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession no. GSE183286. All other data are available in the main text or the supplementary information.