Significance
A protein is a shape-shifter, but it is currently unclear which of the many structures a protein can adopt are relevant for its function. Here, we examine conformations that contain a “cryptic” pocket (i.e., a pocket absent in ligand-free structures). Cryptic pockets have potential utility in drug discovery efforts because they provide a means to target “undruggable” proteins (i.e., proteins lacking known pockets) or enhance rather than inhibit protein function. In this study, we use a combination of thiol-labeling and kinetic assays, NMR, and molecular dynamic simulations to identify the function of the Ω-loop cryptic pocket in β-lactamase enzymes. We find that an open pocket population is beneficial for hydrolysis of the substrate benzylpenicillin.
Keywords: protein dynamics, cryptic pockets, protein evolution
Abstract
Understanding the functional role of protein-excited states has important implications in protein design and drug discovery. However, because these states are difficult to find and study, it is still unclear if excited states simply result from thermal fluctuations and generally detract from function or if these states can actually enhance protein function. To investigate this question, we consider excited states in β-lactamases and particularly a subset of states containing a cryptic pocket which forms under the Ω-loop. Given the known importance of the Ω-loop and the presence of this pocket in at least two homologs, we hypothesized that these excited states enhance enzyme activity. Using thiol-labeling assays to probe Ω-loop pocket dynamics and kinetic assays to probe activity, we find that while this pocket is not completely conserved across β-lactamase homologs, those with the Ω-loop pocket have a higher activity against the substrate benzylpenicillin. We also find that this is true for TEM β-lactamase variants with greater open Ω-loop pocket populations. We further investigate the open population using a combination of NMR chemical exchange saturation transfer experiments and molecular dynamics simulations. To test our understanding of the Ω-loop pocket’s functional role, we designed mutations to enhance/suppress pocket opening and observed that benzylpenicillin activity is proportional to the probability of pocket opening in our designed variants. The work described here suggests that excited states containing cryptic pockets can be advantageous for function and may be favored by natural selection, increasing the potential utility of such cryptic pockets as drug targets.
While it is well established that proteins are dynamic molecules (1), it is often unclear what these dynamics mean for function. An experimentally derived structural snapshot of a protein, such as a crystal structure, is frequently assumed to represent the (highest probability, lowest energy) ground state. This snapshot is also frequently assumed to be the functional state of the protein. In fact, rigidifying the active site or increasing the probability of the ground-state conformation is often used as a design strategy for improving catalytic activity (2, 3). In opposition to this common assumption, there are several compelling examples of functionally relevant excited states (4–9). However, it is still unclear if excited states in general play a role in function.
Here, we consider an important class of excited states that contain a “cryptic” pocket, or a pocket which is absent in the ligand-free, experimentally determined structure(s). These states are of particular interest because of the potential utility of cryptic pockets as drug targets (10). These pockets provide a means to drug otherwise “undruggable” proteins and a means to enhance a desired protein activity rather than just inhibit an undesired one (11, 12). One concern, however, with the use of cryptic pockets as drug targets is that it is uncertain if there is a selective pressure to maintain the existence of a given pocket or if drug binding to that pocket could be trivially evolved away. This is at least partially because it is unknown if excited states containing cryptic pockets are simply a byproduct of the dynamic nature of proteins or if they play a bigger role in protein function.
Despite the many examples of systems which are known to contain cryptic pockets (13–15), their functional relevance remains unclear because these pockets are notoriously difficult to find and study. Identification of a cryptic pocket often requires simultaneous discovery of a ligand that binds to it (16). Fortunately, recent advances in computational and experimental tools allow us to better identify and study these pockets (1, 17). To increase sampling during molecular dynamics simulations, adaptive sampling methods like fluctuation amplification of specific traits (FAST) (18) and replica exchange methods like sampling water interfaces through scaled Hamiltonians (SWISH) (19) have been developed. To analyze these datasets, methods such as Markov state models (MSMs) (20) and exposons (21) have been developed. These computational tools can then be used to inform experimental methods like room temperature crystallography (22), NMR relaxation techniques (23–25), and thiol-labeling assays (26). Previous work using these methods has shown that many different kinds of proteins have cryptic pockets and that these pockets can be targeted with drugs to allosterically affect functional sites (11, 12, 27, 28). However, it is still unclear if cryptic pockets have implications for function in the absence of ligand binding.
To explore the functional relevance of excited states containing cryptic pockets, we consider a set of class A β-lactamases. β-lactamases are enzymes that confer bacteria with antibiotic resistance by hydrolyzing β-lactam antibiotics such as benzylpenicillin and cefotaxime. TEM β-lactamase in particular is an established model system for studying cryptic pockets. TEM has two known and well-characterized cryptic pockets. The first, which was found serendipitously during a drug-screening campaign, is between helices 11 and 12 (16). The second, which was more recently identified in our laboratory (21), forms when the Ω-loop undocks from the protein, so we call this pocket the Ω-loop pocket (Fig. 1). The Ω-loop pocket was discovered in molecular dynamics simulations, confirmed using thiol-labeling experiments, and subsequently shown to exert control over catalysis at the adjacent active site (21). We know the Ω-loop structure is important as it is necessary for the deacylation of β-lactam antibiotics (29), and it has been previously shown that changes in Ω-loop conformations are connected to cefotaxime activity (30, 31).
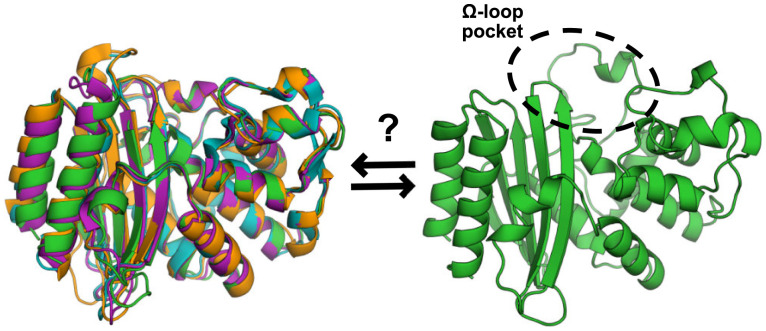
Fig. 1.
The Ω-loop pocket seen in TEM may open in other β-lactamase homologs. The structures of four β-lactamase homologs (Left) overlay well. TEM (Protein Data Base [PDB]: 1xpb) is shown in green, CTX-M-9 (PDB: 1ylj) is shown in cyan, MTB (PDB: 2gdn) is shown in orange, and GNCA (PDB: 4b88) is shown in magenta. The open Ω-loop pocket structure in TEM (Right) was identified in molecular dynamics simulations.
As we have also found that the Ω-loop pocket is present in CTX-M-9 β-lactamase (21), we hypothesize that this pocket may play a role in the enzyme’s function. To test this hypothesis, we first examine if the Ω-loop pocket is conserved across β-lactamase homologs and if the presence of the pocket is correlated with increased activity against classic β-lactam substrates. Here, we mean conservation of the phenomenon of cryptic pocket opening rather than conservation of the specific amino acid identities in that region of the protein. We then use activity data for TEM variants and combine NMR with molecular dynamics to gain insight into how the open Ω-loop pocket affects the hydrolysis reaction for different substrates. Finally, we design mutations to modulate the population of the open Ω-loop pocket to explicitly test whether pocket dynamics are predictive of enzymatic activity.
Results and Discussion
β-lactamase Homologs with Ω-Loop Pockets Hydrolyze Benzylpenicillin More Efficiently.
As a first step to determining if the Ω-loop cryptic pocket seen in TEM β-lactamase is functionally relevant, we examined if this pocket is conserved across β-lactamase homologs. Specifically, we examined MTB, the β-lactamase from Mycobacterium tuberculosis encoded by the blaC gene, and GNCA, the predicted sequence for the last common ancestor of various gram-negative bacteria as determined by ancestral sequence reconstruction (32). Both of these proteins have the same topology as TEM (Fig. 1, Left) but only share about 50% sequence identity (SI Appendix, Fig. S2). MTB is a modern-day homolog that is evolutionarily distant from and exists in a different environment than TEM, while GNCA is a predicted ancestral sequence of TEM (SI Appendix, Fig. S3). Investigating these two homologs allows us to test the conservation of the Ω-loop pocket in two evolutionary directions. We know that the Ω-loop pocket is conserved in at least one homolog, CTX-M-9 (21), and wider conservation would suggest there is a selective pressure to maintain the pocket because it is playing a functional role. However, even if this pocket is not perfectly conserved, we hypothesize that the presence of the pocket may be correlated with enhanced enzyme activity. For example, open Ω-loop conformations could facilitate product release by reducing protein-product contacts. Furthermore, while closed Ω-loop conformations of TEM β-lactamase are predictive of increased cefotaxime activity (30), there are often activity trade-offs in enzymes (32) in which increased activity for one substrate results in decreased activity for another. So, while a closed pocket may be beneficial for cefotaxime activity, an open pocket may be beneficial for a different substrate such as benzylpenicillin.
To determine if the Ω-loop pocket is present in each homolog, we performed thiol-labeling experiments that monitor the solvent exposure of a cysteine residue because of pocket opening. In these experiments, 5,5′-dithiobis-(2-nitrobenzoic acid), or DTNB, is added to the protein sample. DTNB covalently modifies exposed cysteine residues in a reaction that can be monitored as a change in absorbance at 412 nm over time. If a cysteine is buried inside of a pocket but then is exposed to solvent when the pocket opens, we observe an exponential increase in absorbance during the timescale of our experiment. We also ensure that the observed labeling rate is faster than the expected labeling due to protein unfolding, which is calculated from measuring the stability and/or unfolding rate. We have used this method previously to validate known (26) and identify new (21) cryptic pockets in TEM β-lactamase. As MTB and GNCA both have multiple native cysteine residues with one cysteine located in the region of the Ω-loop pocket (Fig. 2 A and B), we added DTNB to each wild-type homolog, monitored the change in absorbance, and used Beer’s law to calculate the number of cysteine residues labeling over time. When cysteine labeling was observed, we individually mutated out each native cysteine to determine which one is labeling in the wild-type protein. For each homolog, we also measured the activity, beginning with the classic substrate benzylpenicillin.
Fig. 2.
Labeling of MTB and GNCA β-lactamases suggests that neither protein open an Ω-loop pocket under the conditions tested. (A and B) Structures of wild-type (WT) MTB (PDB: 2gdn) and WT GNCA (PDB: 4b88) are shown with native cysteine residues highlighted in gray. C69 is located in the region of the Ω-loop pocket for both proteins. (C) The normalized DTNB-labeling trace for WT MTB (orange circles) plateaus at one cysteine labeling. MTB C287S (light orange pentagons) shows significantly reduced labeling. (D) The normalized labeling trace for WT GNCA shows no cysteine labeling.
For the conditions we tested, we find that neither MTB nor GNCA open a pocket in the region of the Ω-loop. The labeling of wild-type MTB is well fit by a single exponential (SI Appendix, Fig. S4) and plateaus at the expected value for one cysteine labeling (Fig. 2C). However, when we mutate out the cysteine residue in the Ω-loop pocket region, C69, the labeling overlays well with the wild-type protein (SI Appendix, Fig. S6). MTB C287S, however, displays significantly reduced labeling, suggesting that this is the cysteine that labels in the wild-type protein. Thus, MTB does not open an Ω-loop pocket under the conditions tested here. Following the same procedure, we do not observe any significant labeling for wild-type GNCA (Fig. 2D). Again, the cysteine residue in the Ω-loop pocket region, C69, does not label, suggesting that GNCA also does not open an Ω-loop pocket under these conditions.
We also find, however, that the β-lactamase homologs with Ω-loop pockets display an increased ability to hydrolyze benzylpenicillin (Table 1). Penicillin-binding proteins, which are unable to hydrolyze benzylpenicillin, lack the Ω-loop entirely (29). The existence of this loop, and E166 in particular, in the MTB and GNCA homologs allows for the deacylation and completed hydrolysis of benzylpenicillin with moderate catalytic efficiency. However, TEM and CTX-M-9 both not only have the Ω-loop but are able to open a pocket in this region of the protein and have a corresponding increase in their ability to hydrolyze benzylpenicillin. Both the overall catalytic efficiency and the catalytic rate are higher for TEM and CTX-M-9 than for MTB and GNCA. As a note, we do not expect the change in magnitude of the pocket open population to perfectly correspond to the change in catalytic efficiency. We observe the equilibrium fluctuations of the apoenzyme during our thiol-labeling experiments while substrate binding modifies the β-lactamase energy landscape during our activity assays. Also, because a more open pocket is beneficial, the increase in activity is not simply due to an increase in proximity of the catalytic residues. So, while the Ω-loop pocket is not conserved across all β-lactamase homologs, the pocket appears to be functionally relevant because benzylpenicillin hydrolysis is increased when the pocket is present.
Table 1.
β-lactamase homologs with open Ω-loop pockets display increased catalytic efficiencies against benzylpenicillin
| Protein | Ω-Loop pocket open population (%) | kcat/KM (sec−1 ⋅ µM−1) | kcat (sec−1) | KM (µM−1) |
| Penicillin-binding proteins | N/A – no Ω-loop | N/A – cannot deacylate penicillin | N/A | N/A |
| GNCA β-lactamase | N/A – no Ω-loop pocket | 0.40 ± 0.02 | 6.1 ± 0.1 | 15.1 ± 0.7 |
| MTB β-lactamase | N/A – no Ω-loop pocket | 0.64 ± 0.01 | 47.0 ± 0.9 | 72.9 ± 0.8 |
| CTX-M-9 β-lactamase | 0.023 ± 0.008 (21) | 9 ± 2 | 250 ± 20 | 27 ± 4 |
| TEM β-lactamase* | 1.1 ± 0.2 (21) | 15.2 ± 0.6 | 411 ± 5 | 27 ± 1 |
N/A, not applicable.
Including an S243C mutation needed for thiol-labeling measurements.
TEM β-lactamase Variants with Higher Probabilities of Ω-Loop Pocket Opening Display Corresponding Increases in Benzylpenicillin Activity.
To further test whether the Ω-loop pocket plays a role in the hydrolysis of benzylpenicillin, we focus the rest of our study on TEM. For TEM variants specifically, we have previously shown that there is a strong correlation between cefotaxime activity and the population of conformations with closed Ω-loop pockets (30). It has been proposed that a larger substrate like cefotaxime (Fig. 3A) might benefit from a more open Ω-loop, as this would expand the volume of the active site (31). However, our above results (Table 1) contrast with this model, as we find that the ability to open an Ω-loop pocket corresponds with increased activity against the smaller substrate. Furthermore, if substrate binding was the only step of the reaction affected by the Ω-loop dynamics, we would expect that the individual rate constants for the acylation of each substrate would remain constant. However, they are different by several orders of magnitude (33–35), suggesting that catalysis, in addition to substrate binding, is important. So, we explore the hypothesis that an open Ω-loop pocket is detrimental for cefotaxime activity and beneficial for benzylpenicillin activity.
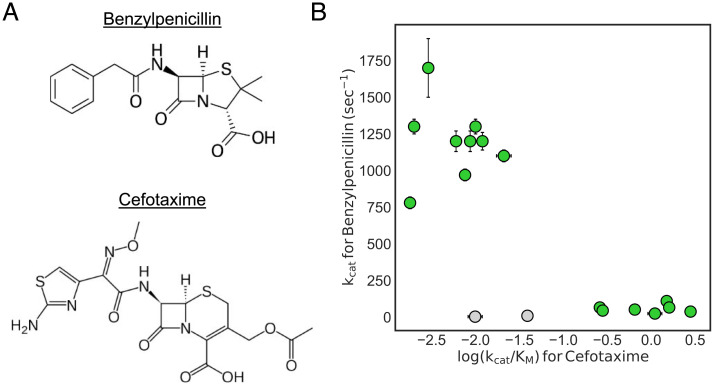
Fig. 3.
There is an inverse correlation between benzylpenicillin (A, Top) and cefotaxime (A, Bottom) activity for TEM variants. (B) Each point represents a different TEM variant. Error bars represent SE values from the fits. The variants shown in gray are R164E/G238S and R164D/G238S, which have decreased activity against both substrates. Data originally reported in ref. 30.
To begin investigating this hypothesis, we reanalyzed data from our previous study (30) in which both benzylpenicillin and cefotaxime activity were measured for a number of TEM variants with mutations at clinically relevant positions. In the study, the benzylpenicillin activities were reported but not explored for their connection to the Ω-loop pocket dynamics. However, because of the correlation found between the Ω-loop closed conformations and cefotaxime activity, variants with decreased cefotaxime activity also have a higher population of conformations with an open Ω-loop pocket. Here, we examine how the catalytic rate for benzylpenicillin correlates to the catalytic efficiency against cefotaxime to determine if there is a trade-off between the two substrates, as this would suggest TEM variants with a higher population of open Ω-loop pocket conformations also have increased benzylpenicillin activity. We examined specifically the catalytic rate for benzylpenicillin (which is described by the acylation and deacylation rates) because we were interested to see if the Ω-loop conformations were correlated with a measure of activity independent of substrate binding. The catalytic efficiency is used for cefotaxime because this is the value we know to be correlated to the population of open Ω-loop conformations.
In fact, we do find a trade-off between benzylpenicillin and cefotaxime activity in the TEM variants examined here. We find that when TEM gains mutations that significantly increase cefotaxime activity, this corresponds to a decrease in catalytic rate for benzylpenicillin (Fig. 3B). As increased cefotaxime activity for these variants is correlated with a lower population of Ω-loop open conformations, the opposite is also true. Variants with a higher population of Ω-loop open conformations are correlated with increased benzylpenicillin activity. A positive correlation between pocket open population and activity provides further evidence for the functional role of excited states containing the cryptic pocket. The variants form two distinct clusters, with the exception of TEM R164E/G238S and R164D/G238S (shown in gray), which display low activity against both substrates. Positions 164 and 238 are known to exhibit negative epistasis with one another (31, 36), so these variants likely have perturbed Ω-loop conformations that are deleterious for all substrates. The correlation between the catalytic efficiencies for both substrates is weaker, and this appears to be due to a compensating decrease in KM for benzylpenicillin as kcat decreases (SI Appendix, Fig. S8). We interpret this to mean that closed Ω-loop conformations, while detrimental for the benzylpenicillin catalytic rate, are generally beneficial for substrate binding.
NMR and Simulations Provide Structural Insight into the TEM Ω-Loop Pocket Open Population without the Need for a Mutation.
To structurally characterize the open Ω-loop pocket population, we used a combination of NMR and molecular dynamics simulations. We performed an NMR relaxation technique called chemical exchange saturation transfer (CEST) (37, 38), which allows us to observe motions on the same timescale as our labeling experiments but without the need for a cysteine mutation. CEST can identify the presence of excited states and the rates of transitioning between states, thereby complementing our thiol-labeling experiments. These NMR experiments also identify which residues contribute to the exchange between the ground state and excited state(s), which we used to inform the collective variable for metadynamics simulations. We then used conformations found during our metadynamics simulations to act as starting conformations for unbiased simulations on the Folding@home distributed computing platform (28, 39). This “adaptive seeding” strategy allows us to access protein motions on the longer timescales of our labeling and NMR experiments while still preserving the thermodynamic and kinetic properties of the system. To analyze our simulation data, we used Correlation of All Rotameric and Dynamical States (CARDS) (40) followed by principal component analysis (PCA) to identify the motions associated with the Ω-loop pocket and pull out exemplar structures for the pocket open and closed populations (SI Appendix, Fig. S10).
Our CEST experiments provide further evidence for the TEM excited states predicted from our simulations and corroborate DTNB-labeling data without the need for a mutation. Exchange was observed for TEM, suggesting the presence of an excited-state population (Fig. 4A). Many of the residues around the Ω-loop pocket report on the exchange between the states, including L169, N170, G236, E240, G242, and S243 (Fig. 4B, circles). Dynamics seen in the 283-loop are consistent with models from room temperature crystallography (31). The population of the excited state was determined to be 1.05 ± 0.03% (kex = 98 ± 6 s−1), which is in excellent agreement with the open pocket population measured using thiol labeling (1.1 ± 0.2%). Furthermore, when we perform the same experiment on TEM with a mutation that abolishes the Ω-loop pocket (R241P, see Fig. 5 below), the dynamics monitored by CEST disappear (Fig. 4B, pentagons) as well as faster timescale dynamics monitored by relaxation dispersion (SI Appendix, Fig. S9). These data suggest that CEST reports on the Ω-loop pocket open population. We then used the residues identified in these experiments as being dynamic on the millisecond-to-second timescale to define the collective variable for metadynamics simulations, which in turn identified seed conformations for unbiased simulations.
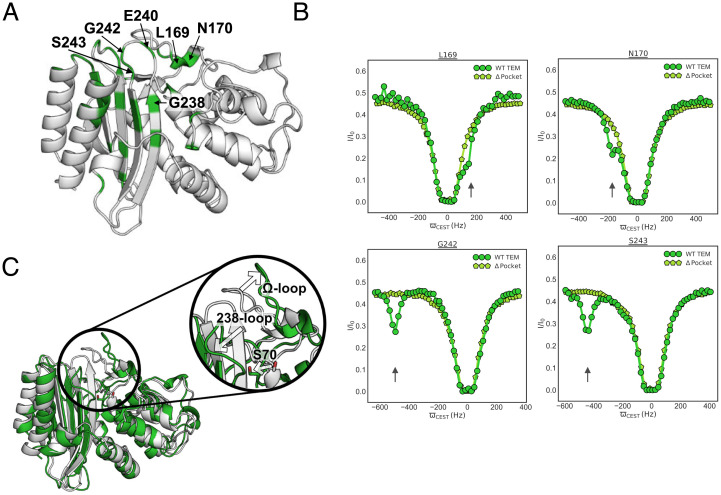
Fig. 4.
Structural insight into the TEM open Ω-loop pocket population identifies conformational changes in the 238-loop and catalytic S70. (A) Highlighted in green on the wild-type (WT) TEM structure (PDB: 1xpb) are the residues showing conformational exchange as established by minor dips in the 15N CEST profiles. Residues around the Ω-loop pocket are annotated. (B) 15N CEST profiles for WT TEM (green circles) and TEM R241P, a variant with no Ω-loop pocket (light green pentagons), are shown for residues in the Ω-loop pocket. Minor dips seen in the WT protein are highlighted with arrows. These experiments informed molecular dynamics simulations that identified the structures shown in C. (C) The closed pocket conformation (white) is similar to the crystal structure. The open pocket conformation (green) has an open Ω-loop, an open 238-loop, and a buried catalytic S70 (shown in sticks).
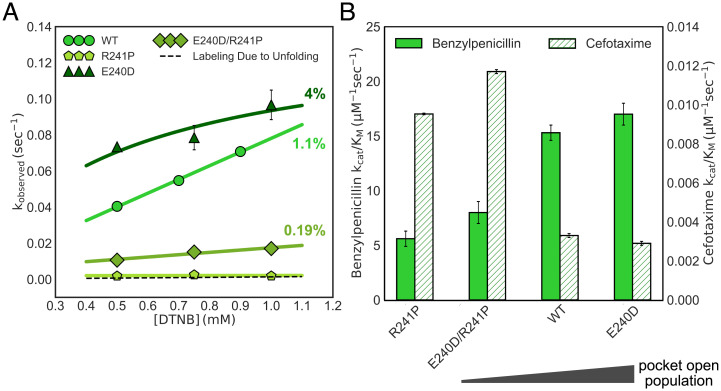
Fig. 5.
Mutations in TEM designed to alter the open Ω-loop pocket population lead to predictable changes in benzylpenicillin and cefotaxime activity. (A) The observed labeling rate as a function of DTNB concentration is shown for wild-type (WT) TEM (circles), TEM E240D (triangles), TEM R241P (pentagons), and TEM E240D/R241P (diamonds). Higher labeling rates are due to a higher open Ω-loop pocket population. The dashed line represents the expected labeling for WT TEM because of the unfolded population. Error bars represent the SD of three measurements. (B) Benzylpenicillin (solid) and cefotaxime (striped) activity is shown for each variant. Error bars are the result of bootstrapping analysis.
An analysis of our simulations shows an Ω-loop pocket open state with an open Ω-loop and open 238-loop (Fig. 4C) in good agreement with the NMR experiments reported here as well as our previous thiol labeling of this pocket (21). We find that the Ω-loop closed conformation looks very similar to the crystal structure as expected. We also find that Ω-loop pocket opening is correlated with a burial of the catalytic serine (S70), making the residue no longer available to bind substrate (SI Appendix, Fig. S11). We observed above that closed Ω-loop pocket conformations were generally beneficial for substrate binding, which is consistent with the closed conformation containing an S70 that is available to bind substrate. These structural results also provide insight into why a higher population of open pocket conformations may be beneficial for a larger substrate and vice versa. Wild-type TEM β-lactamase degrades cefotaxime poorly, and the reaction is rate limited by the acylation rate (33, 34). We reasoned that a more closed Ω-loop pocket would increase protein–substrate contacts, and an available S70 would be beneficial for substrate binding and subsequent acylation. TEM hydrolyzes benzylpenicillin, on the other hand, with an efficiency approaching the diffusion limit (35, 41). In this case, an open Ω-loop pocket may promote product release by reducing protein-product contacts while having a minimal impact on substrate binding. Specifically, we expect a small decrease in acylation rate due to the buried S70 can be tolerated, given the already very fast acylation rate and overall small population of the pocket open conformations.
Variants Designed to Modulate the Dynamics of the Ω-Loop Pocket Predictably Affect Function.
Finally, we explicitly test our model by designing variants to either close or open the Ω-loop pocket, assessing the impact on pocket opening via thiol-labeling experiments, and observing the effects on both benzylpenicillin and cefotaxime hydrolysis functions. We aimed to make mutations in TEM that are not known to be involved in catalysis directly but that would affect the Ω-loop pocket dynamics. Thus, changes in the pocket dynamics resulting in predictable changes in activity would support our understanding of how the TEM Ω-loop pocket is connected to function.
Toward that end, we chose Ω-loop pocket mutations based off of a previous study (42) that measured the effect of every single point mutation in TEM on bacterial fitness by transforming a whole-gene saturated mutagenesis library into Escherichia coli and selecting with either ampicillin or cefotaxime. An E240D mutation led to an increase in fitness in the presence of ampicillin, which we reasoned was due to an increased ability to hydrolyze ampicillin. While a mutation to lysine at this position is known to be clinically important, the charge conserving mutation to aspartic acid at this position has not been seen clinically. We rationalized that mutating this position to an aspartic acid would destabilize closed Ω-loop conformations because of its shorter hydrocarbon chain leading to lower hydrophobicity and reduced ability to screen its charged acid group by positioning it into the solvent. Following this logic, we hypothesized that the E240D mutation opens the Ω-loop pocket in TEM, which in turn increases benzylpenicillin activity. On the other hand, a R241P mutation produced a large positive fitness effect in the presence of cefotaxime. This position is not known to be clinically important, and we reasoned that removal of a charged amino acid might stabilize closed conformations, and the introduction of a proline, which has fewer available dihedral angles, would destabilize open conformations. Thus, we hypothesized that the R241P mutation closes the Ω-loop pocket in TEM, which increases cefotaxime activity and decreases benzylpenicillin activity.
As predicted, we find that the E240D mutation opens the Ω-loop pocket, decreases cefotaxime activity, and increases benzylpenicillin activity (Fig. 5). The thiol-labeling rates for TEM E240D are faster than wild-type, indicating a more open Ω-loop pocket (population = 4 ± 1%, EXX regime). As expected, the catalytic efficiency of E240D for benzylpenicillin increases. The relatively small increase is also expected, as TEM is very close to the diffusion limit for this substrate. We also see a corresponding decrease in cefotaxime activity for this mutant.
We find that the R241P mutation also behaves as expected. The thiol-labeling rates for TEM R241P are much slower than wild-type. In fact, they are on the order of magnitude as those expected for labeling due to protein unfolding, thus suggesting that R241P abolishes the Ω-loop pocket in TEM. As a result, the activity of R241P against benzylpenicillin decreases, and we find a corresponding increase in the catalytic efficiency against cefotaxime, which is consistent with our model and suggests that the mutation does not just simply break the enzyme.
When we introduce the E240D mutation into the background of R241P, we find that the opening of the Ω-loop pocket is rescued (Fig. 5A, population = 0.19 ± 0.01%, EX2 regime). The corresponding benzylpenicillin activity for TEM E240D/R241P is, as expected, higher than that of TEM R241P. It is also lower than that of wild-type TEM, consistent with the stronger effect of the R241P mutation than that of the E240D mutation in the wild-type background. These data support our hypothesis that a higher open Ω-loop pocket population in TEM is correlated with higher benzylpenicillin activity. TEM E240D/R241P also has increased cefotaxime activity compared to wild-type TEM, which is consistent with our model that closed Ω-loop conformations are beneficial for cefotaxime activity. However, the cefotaxime activity of TEM E240D/R241P, which has a low Ω-loop pocket open population, is greater than the cefotaxime activity for TEM R241P, which does not have an Ω-loop pocket. This suggests that, even though closed Ω-loop pocket conformations are beneficial for cefotaxime activity, a low open pocket population may be more advantageous than no pocket at all or that specific closed pocket conformations additionally tune activity. Taken together, the results presented here suggest that the Ω-loop pocket in β-lactamase plays a role in function, as excited states containing an open pocket improve activity.
Conclusions
We hypothesized that β-lactamase excited states play a role in function by increasing enzyme activity. Specifically, we investigated whether excited states containing the Ω-loop cryptic pocket enhance benzylpenicillin activity. We found that the Ω-loop pocket seen in TEM and CTX-M-9 β-lactamase is not conserved in the MTB and GNCA homologs but that homologs with the pocket have higher catalytic efficiencies for the hydrolysis of benzylpenicillin. Focusing our study on TEM β-lactamase, we found that variants with larger open Ω-loop pocket populations also have higher benzylpenicillin catalytic rates. We performed NMR CEST experiments and NMR-guided molecular dynamics simulations to gain structural insight into the TEM open Ω-loop pocket population and rationalize why a higher population of closed Ω-loop conformations might be beneficial for a larger substrate. However, we also acknowledge that the closed Ω-loop conformations may still have an expanded active site, likely because of the 238-loop opening, and can thus better accommodate a larger substrate as crystallographic data suggests for several extended spectrum β-lactamases. Lastly, we designed mutations to modulate the dynamics of the Ω-loop pocket in TEM and observed that the probability of pocket opening is correlated with benzylpenicillin activity. Of course, we cannot say whether formation of the cryptic pocket is causative of the increased benzylpenicillin activity. Future studies will be needed to determine if pocket opening itself is enough to enhance activity by facilitating product release or if broadening of the Ω-loop ensemble in general allows for specific, functional loop conformations to be adopted. That being said, we successfully use our proposed model as a design principle, demonstrating our understanding of how excited states containing the Ω-loop cryptic pocket are connected to β-lactamase function. Our results also provide further evidence for the hypothesis that functionally relevant conformations are sampled during equilibrium fluctuations (i.e., in the apoenzyme). This work demonstrates that cryptic pocket dynamics can be modulated with mutations, setting up future studies to elucidate the sequence determinants of these pockets, and suggests that cryptic pockets may be under positive selective pressure, increasing their potential utility as drug targets.
Materials and Methods
TEM and CTX-M-9 were purified as previously described (21, 30). MTB was purified using a 6x His tag, and GNCA was purified from the periplasm. Thiol-labeling and Michaelis–Menten activity assays were performed as previous described (21). NMR CEST experiments were performed as previously described (43) using 1-s delay alternating with nutation for tailored excitation (DANTE) trains of square pulses and an interpulse delay of 2, 1, and 0.667 ms. CEST profiles were sampled in 51 steps, with increments of 10, 20, and 30 Hz, extending over frequency ranges of 500 (2-ms interpulse delay), 1,000 (1 ms), and 1,500 Hz (0.667 ms), respectively. Simulations were run as previously described (21) using GROMAS and the Amber03 force field. A total of 200 ns of metadynamics simulations were run with gaussians added every 2 ps with a height of 1.0 kJ/mol and a width of 0.05. A total of 220 representative seed conformations were used to collect a total of 100.7 ms of unbiased simulations on the Folding@home distributed computing platform (44). Simulations were analyzed using CARDS (40) and PCA. All methods are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank the community scientists of Folding@home for donating their computing resources. This work was funded by NIH Grant R01GM12400701 (G.R.B.), NSF CAREER Award MCB-1552471 (G.R.B.), Canadian Institutes of Health Research FDN-503573 (L.E.K.), and the Natural Sciences and Engineering Research Council of Canada (L.E.K.). G.R.B. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and a Packard Fellowship for Science and Engineering from the David and Lucile Packard Foundation. L.E.K. holds a Canada Research Chair in Biochemistry.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106473118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. Simulation data are available upon request as there is no standard repository for such data, especially given the size of our data set (at least 256 GB). All processed data and models, specifically MSM data, CARDS data, and structures used for histograms, are found on the Open Source Framework and can be downloaded for free at: https://osf.io/gyxtu/. Previously published data were used for this work (30).
References
- 1.Knoverek C. R., Amarasinghe G. K., Bowman G. R., Advanced methods for accessing protein shape-shifting present new therapeutic opportunities. Trends Biochem. Sci. 44, 351–364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Procko E., et al. , Computational design of a protein-based enzyme inhibitor. J. Mol. Biol. 425, 3563–3575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weitzner B. D., Kipnis Y., Daniel A. G., Hilvert D., Baker D., A computational method for design of connected catalytic networks in proteins. Protein Sci. 28, 2036–2041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen S., et al. , The dynamic conformational landscape of the protein methyltransferase SETD8. eLife 8, e45403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saavedra H. G., Wrabl J. O., Anderson J. A., Li J., Hilser V. J., Dynamic allostery can drive cold adaptation in enzymes. Nature 558, 324–328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraser J. S., et al. , Hidden alternative structures of proline isomerase essential for catalysis. Nature 462, 669–673 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boehr D. D., McElheny D., Dyson H. J., Wright P. E., The dynamic energy landscape of dihydrofolate reductase catalysis. Science 313, 1638–1642 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Xie T., Saleh T., Rossi P., Kalodimos C. G., Conformational states dynamically populated by a kinase determine its function. Science 370, eabc2754 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim S. A., Bolin E. R., Marqusee S., Tracing a protein’s folding pathway over evolutionary time using ancestral sequence reconstruction and hydrogen exchange. eLife 7, e38369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vajda S., Beglov D., Wakefield A. E., Egbert M., Whitty A., Cryptic binding sites on proteins: Definition, detection, and druggability. Curr. Opin. Chem. Biol. 44, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart K. M., et al. , Designing small molecules to target cryptic pockets yields both positive and negative allosteric modulators. PLoS One 12, e0178678 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruz M. A., et al. , Discovery of a cryptic allosteric site in Ebola’s ‘undruggable’ VP35 protein using simulations and experiments. bioRxiv [Preprint] (2020). 10.1101/2020.02.09.940510. Accessed 5 November 2021. [DOI]
- 13.Cimermancic P., et al. , CryptoSite: Expanding the druggable proteome by characterization and prediction of cryptic binding sites. J. Mol. Biol. 428, 709–719 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollingsworth S. A., et al. , Cryptic pocket formation underlies allosteric modulator selectivity at muscarinic GPCRs. Nat. Commun. 10, 3289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schames J. R., et al. , Discovery of a novel binding trench in HIV integrase. J. Med. Chem. 47, 1879–1881 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Horn J. R., Shoichet B. K., Allosteric inhibition through core disruption. J. Mol. Biol. 336, 1283–1291 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Kuzmanic A., Bowman G. R., Juarez-Jimenez J., Michel J., Gervasio F. L., Investigating cryptic binding sites by molecular dynamics simulations. Acc. Chem. Res. 53, 654–661 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmerman M. I., Bowman G. R., FAST conformational searches by balancing exploration/exploitation trade-offs. J. Chem. Theory Comput. 11, 5747–5757 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Comitani F., Gervasio F. L., Exploring cryptic pockets formation in targets of pharmaceutical interest with SWISH. J. Chem. Theory Comput. 14, 3321–3331 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Bowman G. R., Geissler P. L., Equilibrium fluctuations of a single folded protein reveal a multitude of potential cryptic allosteric sites. Proc. Natl. Acad. Sci. U.S.A. 109, 11681–11686 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porter J. R., et al. , Cooperative changes in solvent exposure identify cryptic pockets, switches, and allosteric coupling. Biophys. J. 116, 818–830 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer M., Shoichet B. K., Fraser J. S., One crystal, two temperatures: Cryocooling penalties alter ligand binding to transient protein sites. ChemBioChem 16, 1560–1564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henzler-Wildman K. A., et al. , A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature 450, 913–916 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Stoffel S., Zhang Q. W., Li D. H., Smith B. D., Peng J. W., NMR relaxation dispersion reveals macrocycle breathing dynamics in a cyclodextrin-based rotaxane. J. Am. Chem. Soc. 142, 7413–7424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu D., Mao Y., Gu X., Zhou Y., Long D., Unveiling the “invisible” druggable conformations of GDP-bound inactive Ras. Proc. Natl. Acad. Sci. U.S.A. 118, e2024725118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowman G. R., Bolin E. R., Hart K. M., Maguire B. C., Marqusee S., Discovery of multiple hidden allosteric sites by combining Markov state models and experiments. Proc. Natl. Acad. Sci. U.S.A. 112, 2734–2739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vithani N., et al. , SARS-CoV-2 Nsp16 activation mechanism and a cryptic pocket with pan-coronavirus antiviral potential. Biophys. J. 140, 2280–2889 (2021). [DOI] [PMC free article] [PubMed]
- 28.Zimmerman M. I., et al. , Citizen scientists create an exascale computer to combat COVID-19. SARS-CoV-2 simulations go exascale to predict dramatic spike opening and cryptic pockets across the proteome. Nat. Chem. 13, 651–659 (2021). [DOI] [PMC free article] [PubMed]
- 29.Egorov A., Rubtsova M., Grigorenko V., Uporov I., Veselovsky A., The role of the Ω-loop in regulation of the catalytic activity of TEM-type β-lactamases. Biomolecules 9, 854 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart K. M., Ho C. M., Dutta S., Gross M. L., Bowman G. R., Modelling proteins’ hidden conformations to predict antibiotic resistance. Nat. Commun. 7, 12965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dellus-Gur E., et al. , Negative epistasis and evolvability in TEM-1 β-Lactamase--The thin line between an enzyme’s conformational freedom and disorder. J. Mol. Biol. 427, 2396–2409 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Risso V. A., Gavira J. A., Mejia-Carmona D. F., Gaucher E. A., Sanchez-Ruiz J. M., Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian β-lactamases. J. Am. Chem. Soc. 135, 2899–2902 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Bicknell R., Waley S. G., Single-turnover and steady-state kinetics of hydrolysis of cephalosporins by beta-lactamase I from Bacillus cereus. Biochem. J. 231, 83–88 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saves I., et al. , Mass spectral kinetic study of acylation and deacylation during the hydrolysis of penicillins and cefotaxime by beta-lactamase TEM-1 and the G238S mutant. Biochemistry 34, 11660–11667 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Christensen H., Martin M. T., Waley S. G., Beta-lactamases as fully efficient enzymes. Determination of all the rate constants in the acyl-enzyme mechanism. Biochem. J. 266, 853–861 (1990). [PMC free article] [PubMed] [Google Scholar]
- 36.Giakkoupi P., Tzelepi E., Tassios P. T., Legakis N. J., Tzouvelekis L. S., Detrimental effect of the combination of R164S with G238S in TEM-1 beta-lactamase on the extended-spectrum activity conferred by each single mutation. J. Antimicrob. Chemother. 45, 101–104 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Vallurupalli P., Bouvignies G., Kay L. E., Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc. 134, 8148–8161 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Bouvignies G., Kay L. E., A 2D 13C-CEST experiment for studying slowly exchanging protein systems using methyl probes: An application to protein folding. J. Biomol. NMR 53, 303–310 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Sun X., Singh S., Blumer K. J., Bowman G. R., Simulation of spontaneous G protein activation reveals a new intermediate driving GDP unbinding. eLife 7, e38465 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh S., Bowman G. R., Quantifying allosteric communication via both concerted structural changes and conformational disorder with CARDS. J. Chem. Theory Comput. 13, 1509–1517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardy L. W., Kirsch J. F., Diffusion-limited component of reactions catalyzed by Bacillus cereus beta-lactamase I. Biochemistry 23, 1275–1282 (1984). [DOI] [PubMed] [Google Scholar]
- 42.Stiffler M. A., Hekstra D. R., Ranganathan R., Evolvability as a function of purifying selection in TEM-1 β-lactamase. Cell 160, 882–892 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Yuwen T., Kay L. E., Bouvignies G., Dramatic decrease in CEST measurement times using multi-site excitation. ChemPhysChem 19, 1707–1710 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Shirts M., Pande V. S., COMPUTING: Screen savers of the world unite! Science 290, 1903–1904 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. Simulation data are available upon request as there is no standard repository for such data, especially given the size of our data set (at least 256 GB). All processed data and models, specifically MSM data, CARDS data, and structures used for histograms, are found on the Open Source Framework and can be downloaded for free at: https://osf.io/gyxtu/. Previously published data were used for this work (30).