Significance
Cancer cell–intrinsic IFN-I responses can have opposite effects in response to DNA damage, either facilitating or inhibiting cytotoxicity. We found that PD-L1 regulates these seemingly paradoxical effects, inhibiting the cytotoxic acute response to high levels of IFN-I but sustaining the protective chronic response to constitutive low levels of IFN-I. This revealed facet of PD-L1 activity operates independently of the cancer cell–extrinsic immune system. Our improved understanding of the functions of PD-L1 in cancer informs more effective strategies to target PD-L1 and block both its cell-intrinsic and its immune-dependent functions.
Keywords: programmed death ligand 1 (PD-L1), type I interferon (IFN-I), DNA damage resistance, cGAS-STING pathway
Abstract
Programmed death ligand 1 (PD-L1), an immune-checkpoint protein expressed on cancer cells, also functions independently of the immune system. We found that PD-L1 inhibits the killing of cancer cells in response to DNA damage in an immune-independent manner by suppressing their acute response to type I interferon (IFN; IFN-I). In addition, PD-L1 plays a critical role in sustaining high levels of constitutive expression in cancer cells of a subset of IFN-induced genes, the IFN-related DNA damage resistance signature (IRDS) which, paradoxically, protects cancer cells. The cyclic GMP-AMP synthase-stimulator of the IFN genes (cGAS-STING) pathway is constitutively activated in a subset of cancer cells in the presence of high levels of PD-L1, thus leading to a constitutive, low level of IFN-β expression, which in turn increases IRDS expression. The constitutive low level of IFN-β expression is critical for the survival of cancer cells addicted to self-produced IFN-β. Our study reveals immune-independent functions of PD-L1 that inhibit cytotoxic acute responses to IFN-I and promote protective IRDS expression by supporting protective chronic IFN-I responses, both of which enhance the resistance of cancer cells to DNA damage.
Programmed death ligand 1 (PD-L1) is an immune checkpoint protein expressed on cancer cells that inhibits T cell function by binding to programmed cell death protein 1 (PD-1) on T cells (1). Recent studies have revealed that PD-L1 also functions in an immune-independent manner, protecting cancer cells from radiation, chemotherapy, and exposure to high levels of interferon-β (IFN-β) in a cancer cell–intrinsic manner, independently of the immune system (2–6). PD-L1 increases resistance to radiation or cisplatin by stabilizing NBS1, BRCA1, and other proteins that facilitate the repair of DNA damage (5). PD-L1 also diminishes IFN-β cytotoxicity by inhibiting an IFN-β–induced STAT3/caspase-7–dependent pathway (6). Interestingly, radiation, chemotherapy, and IFN-β all increase the expression of PD-L1, suggesting that PD-L1 provides acquired resistance to these exogenous therapeutic agents (2–6). In some cancer cells resistant to DNA damage, the basal levels of PD-L1 are constitutively high (2–4). PD-L1 expression in cancer cells is increased by several different mechanisms, including gene amplification and translocation, transcriptional activation by oncogenic proteins (MYC, HIF-1α, STAT3), posttranscriptional regulation by microRNAs, and posttranslational modulation (1, 7).
Recent studies have revealed that the responses to DNA damage are regulated by type I IFN (IFN-I) in cancer cells (8–10). In response to cytotoxic levels of DNA damage, IFN-I is induced in cancer cells and the resulting acute IFN-I response contributes to killing these cells. Anthracyclines (e.g., doxorubicin) stimulate IFN-I induction through Toll-like receptor 3 (TLR3), promoting cancer cell death through the expression of chemokine (C-X-C motif) ligand 10 (CXCL10) (9). Cytotoxicity induced by ionizing radiation (IR) is correlated with IFN-β expression, and inhibiting the response to IFN-β by using neutralizing antibodies or knocking out the IFN-I receptor decreases cancer cell death in response to IR (10). Our previous work showed that most of the IFN-stimulated genes (ISGs) that encode cytotoxic proteins are induced in the acute phase of IFN signaling (11). On the other hand, chronic stimulation with low doses of IFN-β contributes to resistance to DNA damage. The chronic responses to IFN-β increase the levels of U-ISGF3 (ISG factor 3 lacking tyrosine phosphorylation of STATs 1 and 2), which induce the expression of about a quarter of the ISGs, the IFN-related DNA damage resistance signature (IRDS) (12). Chronic IFN-I responses and elevated IRDS expression are often observed in cancer cells exposed to repeated or prolonged radiation or chemotherapy, which correlates with acquired resistance to the therapy (8, 10, 13). Some intrinsic factors also induce chronic IFN-I responses in cancer cells, which might be responsible for their intrinsic resistance to DNA damage. For example, dysfunction of Ataxia-telangiectasia mutated (ATM), a central component of the DNA repair machinery, results in constitutive IFN-β expression through the cytosolic DNA-sensing stimulator of IFN genes (STING) pathway (14), and loss of tumor-suppressor p53 function leads to IFN-β expression through a double-stranded (ds)RNA-dependent pathway, reducing the cytotoxicity of the DNA damaging agent doxorubicin (15).
We have investigated cancer cell–intrinsic functions of PD-L1, which suppress cytotoxic acute responses to IFN-I and promote protective chronic responses through IRDS expression, thus enhancing the resistance of cancer cells to DNA damage independently of the immune system.
Results
PD-L1 Increases the Resistance of Cancer Cells to DNA Damage Independently of the Immune System.
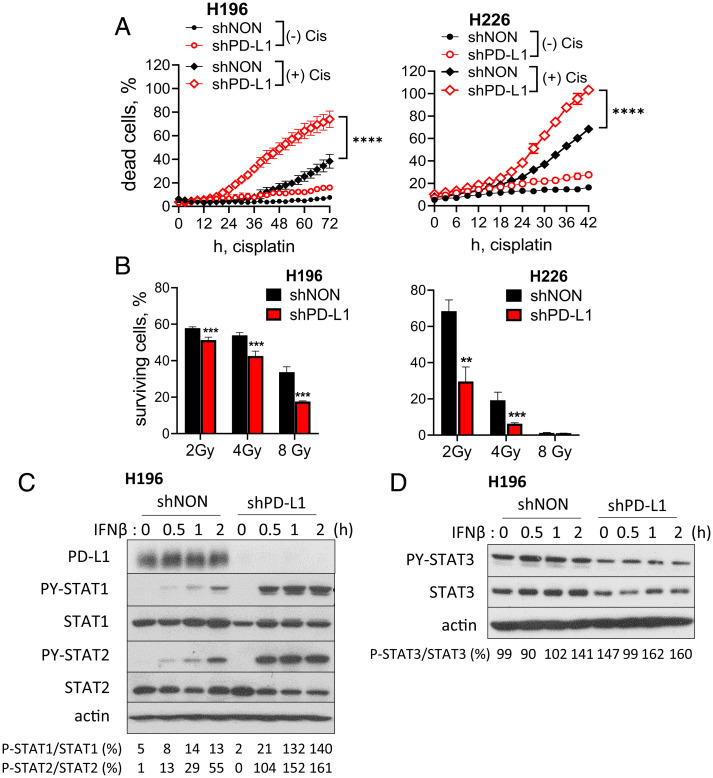
To better understand the immune-independent functions of PD-L1, we used an in vitro model in which only cancer cells are present. We knocked PD-L1 expression down by using short-hairpin (sh)RNA in H196 small cell lung carcinoma (SCLC) and H226 non-SCLC (NSCLC) cells, which express relatively high levels of PD-L1 (the levels of PD-L1 after knockdown [KD] are shown in Fig. 1C and SI Appendix, Fig. S1A). The percentage of dead cells following treatment with cisplatin, assessed by the real-time cell monitoring system, was significantly higher in PD-L1 KD cells (shPD-L1) compared to control cells (shNON) 15 h after cisplatin treatment and at later times (Fig. 1A). In response to IR, the PD-L1 KD cells were substantially more sensitive than control cells at all doses tested (2, 4, and 8 Gy) (Fig. 1B). We conclude that high levels of PD-L1 make cancer cells more resistant to DNA damage in an immune-independent manner.
Fig. 1.
PD-L1 enhances the resistance of cancer cells to DNA damage in an immune-independent manner. (A) PD-L1 was knocked down using shRNA (shPD-L1). H196 or H226 cells were treated with 20 or 40 μM of cisplatin, respectively. The number of dead cells was monitored every 3 h using the IncuCyte ZOOM real-time cell monitoring system, and percentages of dead cells over the total number of cells at the time of cisplatin treatment were calculated at each time point. Means of triplicates ± SD are shown. ****P < 0.0001 in two-way ANOVA. (B) H196 or H226 cells were treated with IR (2, 4, or 8 Gy) and cultured for 7 or 14 d, respectively. Total nucleic acid from surviving cells was measured to determine the fraction of surviving cells. The percentage of surviving irradiated cells over unirradiated cells was calculated at each dose. Means of quadruplicates ± SD, **P < 0.01; ***P < 0.001 in the t test. (C and D) H196-shPD-L1 or -shNON cells were treated with IFN-β (300 IU/mL) for 30 min, 1 h, or 2 h, and the levels of the indicated proteins were assessed by the Western method. The densitometry analyses were performed using ImageJ and the ratio (%) of phospho STAT/total STAT is presented at the bottom. shPD-L1, PD-L1 KD cells; shNON, nontarget-shRNA transduced control cells. Representative data, reproduced at least three times, are shown.
High Levels of PD-L1 Inhibit the Acute Response to IFN-β.
DNA damage induces IFN-β expression, which helps to kill cancer cells in response to radiation or chemotherapy (9, 10). To understand the role of PD-L1 in the acute response to IFN-β, in which cytotoxic genes are induced, we treated PD-L1 KD and control H196 cells with IFN-β for 2 h (Fig. 1C). The IFN-β–induced phosphorylation of STAT1 and STAT2 is substantially higher in PD-L1 KD cells, showing that high levels of PD-L1 suppress the acute IFN-I response. Gato-Cañas et al. (6) have shown that PD-L1 decreases IFN-β cytotoxicity by inhibiting an IFN-β–induced STAT3/caspase-7–dependent pathway in mouse melanoma cells, but we observed that the levels of STAT3 or PY-STAT3 were not increased by PD-L1 KD in H196 cells in the presence or absence of IFN-β (Fig. 1D). We confirmed these results in HME (normal mammary epithelial) and HeLa (cervical cancer) cells after increasing PD-L1 expression using lentiviral constructs. When we treated the cells with IFN-β, the phosphorylation of STAT1 and STAT2 was suppressed by high levels of PD-L1 (SI Appendix, Fig. S1B). These results show that PD-L1 inhibits the acute response to IFN-β, which contributes to cancer cell death in response to radiation and chemotherapy.
PD-L1 Expression Is Correlated with IRDS Gene Expression in Cancer Cells.
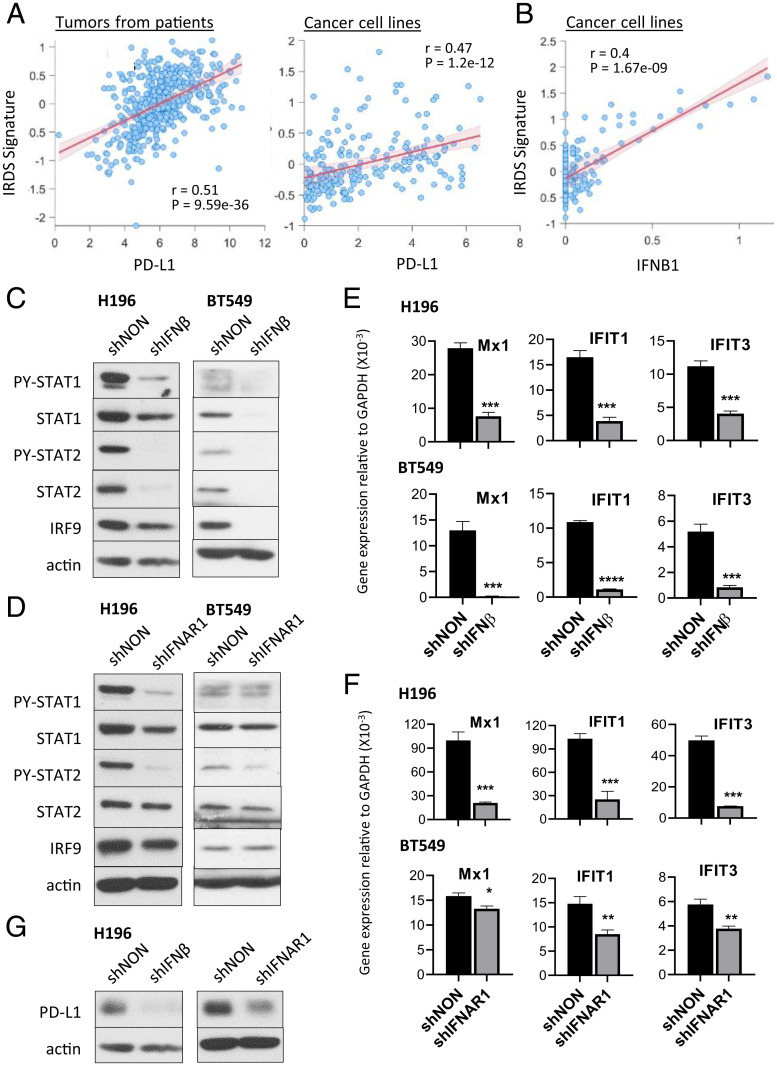
The IRDS, a group of genes highly up-regulated in various types of cancers, correlates with resistance to radiation or chemotherapy (16). The observation that PD-L1 increases resistance to DNA damage prompted us to investigate whether a possible correlation between PD-L1 levels and IRDS expression in cancer cells. Data from patients with lung adenocarcinoma, from The Cancer Genome Atlas (TCGA), reveal that the expression of PD-L1 (CD274) is significantly positively correlated with the expression of the IRDS signature (Fig. 2 A, Left). To ensure that this conclusion is not due to contaminating immune cells, we repeated the analysis in lung cancer cell lines, finding the same result (Fig. 2 A, Right). We validated this observation further by analyzing RNA-sequencing (RNA-seq) data from 60 diverse human cancer cell lines (NCI-60) (17), comparing the expression of PD-L1 and the representative IRDS genes OAS1, OAS2, Mx1, IFIT1, and IFIT3. Thirteen (22%) of these cell lines express high levels of PD-L1. Among these, six (46%) express high levels of IRDS genes (SI Appendix, Fig. S2-1A). These cell lines originated from breast cancer (BT549), colon cancer (HT29), melanoma (LOX IMVI), NSCLC (H226), and kidney cancer (RXF393 and A498), indicating that the coelevation of PD-L1 and IRDS genes is not limited to specific types of cancer.
Fig. 2.
IRDS genes are induced by chronic low levels of IFN-β, originating from cancer cells and correlated with PD-L1 expression. (A) Correlation of PD-L1 (CD274) expression levels with IRDS signature score in tumors from lung adenocarcinoma patients (Left) or lung cancer cell lines (Right). Significance determined by Spearman rank correlation coefficient for all comparisons. (B) Correlation of IFN-β (IFNB1) expression levels with IRDS signature score in lung cancer cell lines. Significance determined by Spearman rank correlation coefficient for all comparisons. (C and D) Chronic IFN-I response signatures (PY-STAT1, PY-STAT2, total STAT1, STAT2, and IRF9) were assessed by the Western method in H196 or BT549 cells after KD of IFN-β (C) or IFNAR1 (D). (E and F) The expression of IRDS genes (Mx1, IFIT1, and IFIT3) was assessed using qRT-PCR after the KD of IFN-β (E) or IFNAR1 (F). Means of triplicates ± SD **P < 0.01; ***P < 0.001; ****P < 0.0001 in t test. (G) The level of PD-L1 protein was assessed by the Western method in H196 cells after KD of IFN-β or IFNAR1. shIFNβ or shIFNAR1, IFN-β or IFNAR1 KD cells; shNON, control cells transduced with nontargeted shRNA. Representative data, reproduced more than three times, are shown.
We further validated the data by qPCR, also including H196 cells, which are not included in the NCI-60 panel but express high levels of IRDS genes and PD-L1 (SI Appendix, Fig. S2-1 B and C). Eight cancer cell lines, including MDA-MB-231, express high levels of PD-L1, but not the IRDS genes (SI Appendix, Fig. S2-1 B and C), showing only about a half of cancer cells expressing high PD-L1 express high levels of IRDS genes. To confirm that the positive correlation between PD-L1 and IRDS can be generalized across different cancer types, we analyzed the relationship across cell lines from 19 different cancers (SI Appendix, Fig. S2-1D) as well as patient tumors from 32 solid tumor types (from the TCGA) (SI Appendix, Fig. S2-1E). The IRDS score was significantly positively correlated with PD-L1 in most of the cancer cell lines analyzed (15 of 19 cancer types) as well as in patient tumors (30 of 32 cancer types). Recent studies show that some cancer cells express PD-1 (18–20), but the cancer cell lines used in this study do not (SI Appendix, Fig. S2-1F). We analyzed in the database correlations between PD-1 expression and IRDS score across cell lines from 20 different cancers, finding no significant correlation in most cancer cell lines (SI Appendix, Fig. S2-1G). PD-1 expression was not correlated with PD-L1 expression in cancer cells either (SI Appendix, Fig. S2-1H).
IRDS Genes Are Induced by Chronic Low Levels of IFN-β that Originate from Cancer Cells.
It is not well understood why some cancer cells express high levels of the proteins encoded by the IRDS genes while others do not. Our previous study revealed that IRDS expression is increased by chronic exposure to low doses of IFN-β (12). We found that cancer cell–intrinsic basal levels of IFN-β expression are positively correlated with the expression of IRDS genes in lung cancer cell lines (Fig. 2B), suggesting that cancer-produced IFN-β may be responsible for chronic IFN-I responses and increased IRDS expression in cancer cells. The positive correlation between IFN-β expression and IRDS was also detected in lung tumors (SI Appendix, Fig. S2-2A) and was conserved across multiple cancer types (cell lines from 18 of 19 cancer types, tumors from 30 of 32 cancer types) (SI Appendix, Fig. S2-2 B and C). We confirmed this observation by qRT-PCR, finding that cancer cell lines expressing high levels of both PD-L1 and IRDS genes (BT549, HT29, A498, and H196) constitutively express endogenous IFN-β, while others (MDA-MB-231, HeLa) do not (SI Appendix, Fig. S2-2D). The IFN-β response is required to induce the expression of IFN-α and IFN-λ (21, 22), whose signaling is mediated by ISGF3. We analyzed the correlation between IFN-α and IRDS expression. Our analysis revealed that IFN-α1 was not detected in most cancer cell lines. It was detected only in certain types of patients’ tumors, suggesting that noncancer cells in the tumor microenvironment may express IFN-α1 (SI Appendix, Fig. S2-2F). IFN-α2 was detected in only limited types of cancer cells, among which only four types show a correlation between IFN-α2 and IRDS expression (SI Appendix, Fig. S2-2E). In patients’ tumors, IFN-α1 and -2 correlated with IRDS expression in limited types of cancer (SI Appendix, Fig. S2-2F), suggesting that IFN-α, which originates from the tumor microenvironment, is not the main driver of IRDS expression. Our previous study shows that IFN-λ, whose expression is dependent on IFN-β responses, increases the levels of U-ISGF3 and IRDS expression in hepatocytes (22). We analyzed correlations between IFN-λs (λ1, λ2, and λ3) and IRDS expression, finding positive correlations in most types of cancer cells and patients’ tumors (SI Appendix, Fig. S2-2 G–I). These results show that self-produced IFN-β and IFN-λ are responsible for high levels of IRDS expression in cancer cells.
Since IFN-λ expression is dependent on the response to IFN-β (22), we further investigated the effects of cancer cell-produced IFN-β on IRDS expression. In cancer cells that express high levels of PD-L1/IRDS (e.g., H196 and BT549 cells), we observed that the basal levels of phosphorylated STAT1 and STAT2 are constitutively high in the absence of any exogenous stimulation (shNON control cells in Fig. 2C). When IFN-β or the IFN-I receptor subunit IFNAR1 was knocked down (the levels of KD are shown in SI Appendix, Fig. S2-2 J and K), the basal levels of phosphorylated STAT1 and STAT2 were reduced (Fig. 2 C and D; it is of note that these figures are from long-exposure films because the basal levels of P-STAT1/2 are significantly lower compared to IFN-β–induced P-STAT1/2 shown in Fig. 1C). The decreased chronic IFN-I responses also down-regulate total levels of the U-ISGF3 components STAT1, STAT2, and IRF9 (Fig. 2 C and D), since the corresponding genes are ISGs (12). Increased levels of STAT1, STAT2, and IRF9, driven by chronic exposure to low doses of IFN-β, are required to induce a high level of IRDS expression (12). The expression of the IRDS genes (Mx1, IFIT1, IFIT3, OAS1, and OAS2) was down-regulated by KD of IFN-β or IFNAR1 (in H196 and BT549 cells in Fig. 2 E and F and SI Appendix, Fig. S2-2 L and M; in HT29 and A498 cells in SI Appendix, Fig. S2-2 N and O), showing that endogenous IFN-β is responsible for their high levels of expression. PD-L1 expression is also down-regulated by decreased IFN-β or IFNAR1 in H196 cells, but not in BT549 cells (Fig. 2G and SI Appendix, Fig. S2-2P), suggesting that endogenous IFN-β is responsible for the high levels of PD-L1 expression in some cancer cells, but not all.
Constitutive Cancer Cell–Intrinsic IFN-β Is Synthesized through the cGAS-STING Pathway.
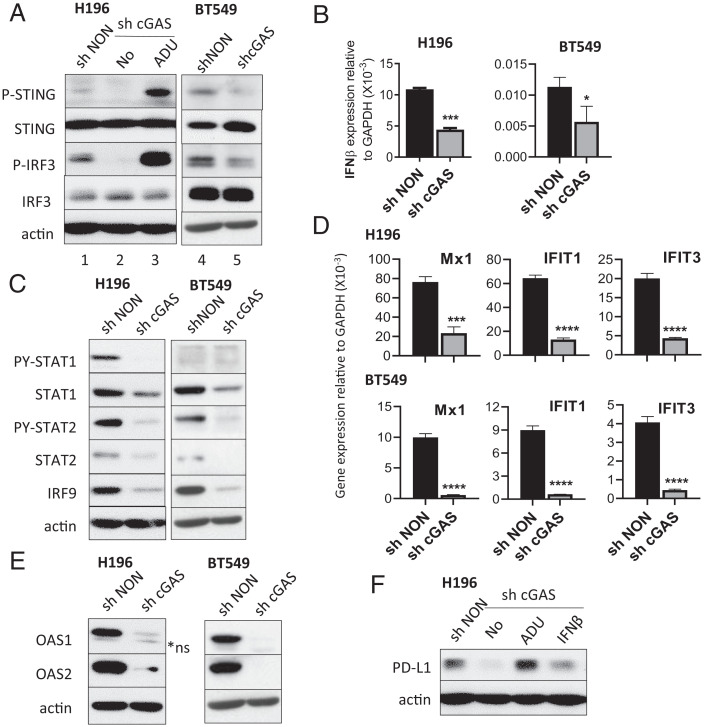
The cyclic GMP-AMP synthase (cGAS)-STING pathway is activated by cytoplasmic DNA, which may come from an infecting DNA virus or damaged nuclear or mitochondrial DNA (8, 23). Several studies have shown that IRDS expression is induced by the cGAS-STING pathway following DNA damage from prolonged or repeated radiation or treatment with chemotherapeutic drugs, leading to enhanced resistance to the therapies (8, 13). In H196 and BT549 cells constitutively expressing IFN-β and IRDS proteins, we found that STING and its downstream transcription factor IRF3 are constitutively phosphorylated (P-STING on Ser-366, P-IRF3 on Ser-386) without exogenous stimulation (Fig. 3 A, lanes 1 and 4). KD of cGAS (SI Appendix, Fig. S3A), which activates STING by synthesizing cyclic GMP-AMP (cGAMP), suppressed the levels of P-STING and P-IRF3 (Fig. 3 A, lanes 2 and 5) and substantially decreased endogenous IFN-β expression (Fig. 3B and SI Appendix, Fig. S3B). Treatment of cGAS KD cells with the cGAMP analog STING agonist ADU-S100 (24) restored high levels of P-STING and P-IRF3 and induced IFN-β expression (Fig. 3 A, lane 3, and SI Appendix, Fig. S3C). KD of cGAS also decreased the chronic IFN-β response signature (constitutive P-STAT1 and P-STAT2 and elevated total STAT1, STAT2, and IRF9) (Fig. 3C). The expression of IRDS genes was also decreased by cGAS KD (in H196 and BT549 cells in Fig. 3 D and E; in HT29 and A498 cells in SI Appendix, Fig. S3 D and E). PD-L1 expression was decreased by KD of cGAS, which was restored by ADU-S100 or IFN-β, in H196 cells (Fig. 3F). We confirmed this observation by using c-176, a small molecule that inhibits STING activation (25), which decreased constitutive STING activation in H196 cells (SI Appendix, Fig. S3F). c-176 substantially down-regulates chronic IFN-β response signature (PY-STAT1, PY-STAT2, and total STAT1, STAT2, and IRF9) (SI Appendix, Fig. S3G), leading to decreased expression of PD-L1 and IRDS proteins OAS1 and OAS2 in H196 cells (SI Appendix, Fig. S3H). However, KD of cGAS did not decrease PD-L1 expression in BT549 cells (SI Appendix, Fig. S3I). These results indicate that the constitutively activated cGAS/STING/IFN-β pathway sustains high levels of IRDS expression, and also PD-L1 expression in some cancer cells.
Fig. 3.
Constitutive cancer cell–intrinsic IFN-β is synthesized through the cGAS-STING pathway. (A) The levels of P-STING (phosphorylated on Ser-366) and P-IRF3 (phosphorylated on Ser-386) were assessed by the Western method in H196 or BT549 cells after KD of cGAS (sh cGAS, lanes 2 and 5). Lysates of H196-sh cGAS cells treated with 10 μM of ADU-S100 (lane 3) for 48 h were also analyzed. (B and D) The mRNA levels of IFN-β (B) and the IRDS gene products Mx1, IFIT1, and IFIT3 (D) were assessed using qRT-PCR. Means of triplicates ± SD, *P < 0.05; ***P < 0.001; ****P < 0.0001 in t test. (C and E) The levels of indicated proteins were assessed by the Western method. *ns, nonspecific bands (lower band). (F) The levels of PD-L1 protein were assessed in H196-sh cGAS cells untreated or treated with 10 μM of ADU-S100 or 10 IU/mL IFN-β for 48 h. shNON, nontarget shRNA transduced control cells.
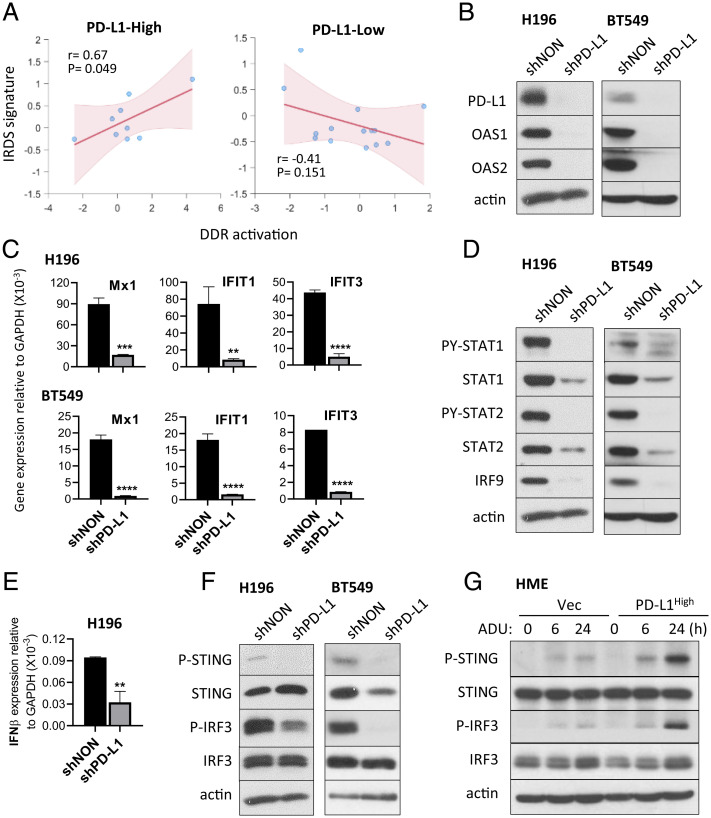
PD-L1 Promotes Constitutive Cancer Cell–Intrinsic STING Activation, Increasing IFN-β and IRDS Expression.
One potential source of constitutive cGAS-STING activation in cancer cells is endogenous DNA damage, which can promote the accumulation of cytosolic DNA. We analyzed the correlation between IRDS expression and basal DNA damage response (DDR) activation (defined by levels of P-CHK1 and P-CHK2) in lung cancer cell lines. Since PD-L1 expression is correlated with IRDS expression (Fig. 2A and SI Appendix, Fig. S2-1), we analyzed this correlation in cancer cells expressing high or low levels of PD-L1. This analysis revealed that levels of IRDS expression are positively correlated with basal levels of DDR activation only when lung cancer cell lines express high levels of PD-L1 (Fig. 4 A, Left), while no significant relationship between DDR activation and IRDS expression was observed in lung cancer cell lines expressing low levels of PD-L1 (Fig. 4 A, Right). These results suggest that high expression of PD-L1 facilitates increasing IRDS expression through the cGAS-STING pathway. In cancer cells expressing high levels of PD-L1/IRDS, KD of PD-L1 substantially decreased IRDS expression (in H196 and BT549 cells in Fig. 4 B and C, in H226 and A498 cells in SI Appendix, Fig. S4). PD-L1 KD also down-regulated chronic IFN-I response signature (PY-STAT1, PY-STAT2, and total STAT1, STAT2, and IRF9) (Fig. 4D). To understand what causes the reduced IFN-β responses by PD-L1 KD, we examined the expression of IFN-β mRNA and the basal levels of P-STING and P-IRF3 proteins (Fig. 4 E and F), finding that KD of PD-L1 substantially decreased the expression of IFN-β mRNA due to the down-regulation of STING and IRF3 phosphorylation. We confirmed the effect of PD-L1 on STING activation in HME cells. Since HME cells express low basal levels of PD-L1 and IRDS expression and show no basal levels of P-STING and P-IRF3, we stimulated HME cells expressing basal (Vec control) or high levels of PD-L1 (PD-L1High) with ADU-S100 to induce the phosphorylation of STING and IRF3 (Fig. 4G). The levels of ADU-S100–induced P-STING and P-IRF3 were substantially higher when PD-L1 expression was elevated compared to control cells. These results show that high levels of PD-L1 play a critical role in sustaining the constitutive-expression of IFN-β and IRDS genes in cancer cells, thus enhancing resistance to DNA damage.
Fig. 4.
PD-L1 promotes constitutive cancer cell–intrinsic STING activation, increasing IFN-β and IRDS expression. (A) Spearman rank correlation coefficient between DDR activation, determined by levels of P-CHK1 and P-CHK2, and IRDS signature expression in PD-L1 high (Left) or PD-L1 low (Right) lung cancer cell lines. Mean PD-L1 (CD274) expression level was used to divide cell lines into PD-L1 high and low. (B–F) Lysates from H196 or BT549 cells after PD-L1 KD (shPD-L1) and nontargeted shRNA-transduced control cells (shNON) were analyzed. The levels of the IRDS proteins (OAS1 and OAS2 in B) and the chronic IFN-I signature (PY-STAT1, PY-STAT2, total STAT1, STAT2, and IRF9 in D) were assessed by the Western method. The mRNA levels of IRDS gene products (Mx1, IFIT1, IFIT3 in C) and IFN-β (E) were assessed using qRT-PCR. Means of triplicates ± SD, **P < 0.01; ***P < 0.001; ****P < 0.0001 in t test. The levels of P-STING (phosphorylated on Ser-366) and P-IRF3 (phosphorylated on Ser-386) were assessed by the Western method (F). (G) HME cells transduced with lentiviral vectors containing PD-L1 cDNA (PD-L1High) and the matching control (Vec) were treated with 10 μM of ADU-S100 for 6 or 24 h. The levels of P-STING or P-IRF3 were assessed by the Western method. Representative data, reproduced at least three times, are shown.
Cancer Cells Expressing Constitutive Endogenous IFN-β Are Addicted to the Self-Produced IFN-β, which Is Critical for Their Survival.
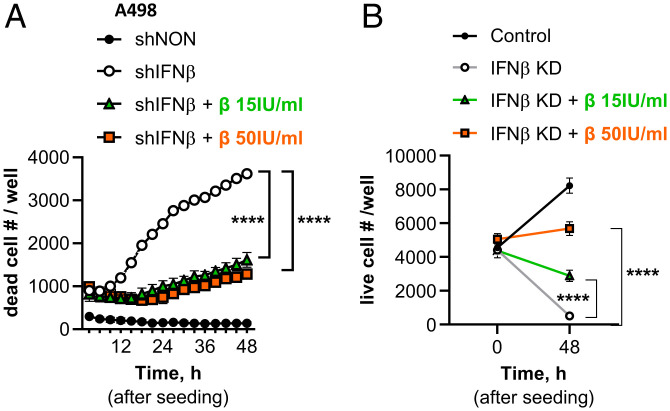
High levels of PD-L1 are critical for inducing constitutive IFN-β expression in some cancer cells that experience endogenous DNA damage (Fig. 4). We observed that cancer cells producing IFN-β did not survive when their IFN-β expression was knocked down (Fig. 5). After 5 d of introducing an shRNA against IFN-β (after selecting transduced cells with puromycin for 3 d), we seeded the same number of control (shNON) and IFN-β KD (shIFNβ) A498 cells and monitored the numbers of dead and live cells. The IFN-β KD cells underwent massive spontaneous apoptosis (Fig. 5A) and most cells died after 2 d (7 d after KD) (Fig. 5). However, when IFN-β KD cells were cultured in media supplemented with IFN-β, their apoptosis was significantly reduced and the number of surviving cells was increased (Fig. 5 and SI Appendix, Fig. S5A). We observed similar results with H196 cells, which are less affected by IFN-β KD compared to A498 cells (SI Appendix, Fig. S5 B and C). We conclude that cancer cells expressing constitutive endogenous IFN-β are addicted to the self-produced IFN-β, requiring it for survival.
Fig. 5.
KD of IFN-β induces apoptosis in cancer cells expressing constitutive endogenous IFN-β. IFN-β mRNA in A498 cells was knocked down by infecting cells with a pseudovirus containing an shRNA against IFN-β (open circles) or nontarget control shRNA (closed circles). IFN-β KD cells were cultured in the absence or presence of recombinant IFN-β (15 or 50 IU/mL) in the media and seeded (5,000 cells per well in 96-well plates) 5 d after pseudovirus infection. The number of apoptotic cells (caspase 3/7 green-positive in A) or live cells (B) was quantified by using the IncuCyte ZOOM real-time cell monitoring system for the next 48 h. Means of triplicates ± SD, ****P < 0.0001 in two-way ANOVA. Representative data, reproduced three times, are shown.
Discussion
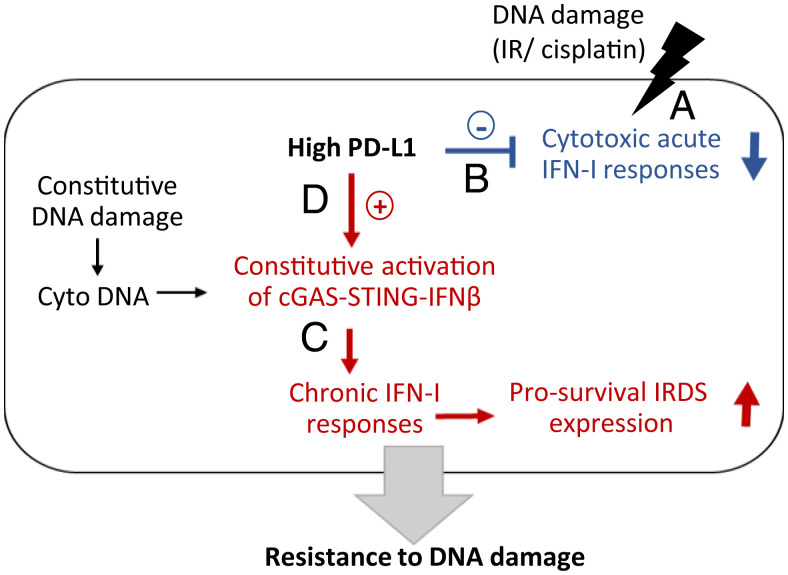
In this study, we have elucidated cancer cell–intrinsic functions of PD-L1 that enhance resistance to DNA damage in an immune-independent manner. In response to DNA damaging agents, such as radiation or cisplatin, IFN-I expression is induced and the acute responses it stimulates contribute to killing cancer cells (Fig. 6). We found that PD-L1 protects cancer cells from DNA damage-induced cell death by two different mechanisms. First, high levels of PD-L1 inhibit acute responses to IFN-I, which help to kill cancer cells in response to DNA damage. Second, PD-L1 sustains constitutive activation of the cGAS-STING pathway and the expression of low levels of IFN-β, which induces prosurvival IRDS expression. In some cancer cells, but not all, the expression of PD-L1 is increased by IFN-β produced by the cancer cells themselves, providing a positive feedback loop. When we analyzed a large number of cancer cell lines (1,376 all types of cancer, 206 lung cancer), 9.2% of each group express high levels of IFN-β/IRDS/PD-L1 (SI Appendix, Fig. S6), suggesting that this working model operates in about 10% of all cancer cells.
Fig. 6.
PD-L1 inhibits cytotoxic acute IFN-I responses and promotes chronic, cancer cell–intrinsic responses to type I interferon, enhancing resistance to DNA damage. (A) Cytotoxic levels of DNA damage induce cancer cell death, which is facilitated by the acute responses to IFN-I that are produced by DNA damage. (B) High levels of PD-L1 inhibit cytotoxic acute responses to IFN-I, reducing cancer cell death in response to DNA damage. (C) Chronic DNA damage in cancer cells constitutively activates the intrinsic cGAS-STING pathway, which produces low levels of IFN-β, increasing IRDS expression, which in turn promotes cancer cell survival. (D) High levels of PD-L1 promote constitutive activation of STING, which is induced by cancer cell–intrinsic damaged DNA.
Our previous study showed that IFN-I increases the expression of cytotoxic genes in the initial acute phase and that their expression returns to basal levels in a prolonged phase of IFN signaling, in which the tyrosine phosphorylation of STATs 1 and 2 has been down-regulated. In the prolonged phase, IFN-induced high expression of U-STATs 1 and 2, which, together with IRF9 (constituting U-ISGF3), drive the transcription of the IRDS subset of ISGs (12). We also found that the levels of U-ISGF3 and IRDS proteins, which are correlated with resistance to DNA damage, are elevated by chronic exposure of cells to low doses of IFN-β (12). These results indicate that the effects of IFN-I are determined by the strength and duration of stimulation; strong and acute IFN-I responses are cytotoxic, whereas weak and chronic IFN-I responses promote cell survival (26). Many studies have shown that the cytotoxicity of DNA damage is related to IFN-I responses. Cytotoxic levels of radiation or chemotherapy induce IFN-I expression in cancer cells, which facilitates cell death (9, 10, 27). On the other hand, cancer cells exposed to prolonged radiation or repeated treatment with drugs that damage DNA acquires resistance to these therapies, accompanied by weak and chronic IFN-I responses and IRDS expression (10, 13, 28). The elevated expression of IRDS genes is a feature of cancer cells resistant to DNA damage (16). From recent studies of others, we learned that PD-L1 expression is also increased by repeated DNA damage (2–4) and enhances resistance to DNA damage (2, 3, 5). We found that PD-L1 is critical for sustaining constitutive IRDS expression in about 10% of all cancer cell lines available in public databases. Our studies and those of others reveal that PD-L1 mediates a self-defense mechanism in cancer cells, protecting them from deleterious IFN-I responses that are induced by cytotoxic DNA damage and potentiating their viability by promoting constitutive IFN-β expression through the cGAS-STING pathway.
We discovered how cancer cells express high levels of IRDS proteins, finding that a subset of these cells produces IFN-β constitutively, through the activity of cell-intrinsic factors, in the absence of exogenous stimulation. Leonova et al. (15) reported that the loss of p53 induces IFN-β by increasing dsRNA expression, thus facilitating the transcription of repetitive elements in the genome. p53 deficiency alone induces low levels of IFN-β, decreasing the cytotoxicity of doxorubicin, a DNA damaging agent, and a combination of p53 deficiency and 5-aza-2′deoxycytidine–induced DNA demethylation leads to extremely high levels of IFN-β that induce massive cell death (15). Coinactivation of the ARF tumor suppressor in addition to p53 helps cells to produce IFN-β and IRDS proteins, promoting long-term proliferation in vitro and tumorigenesis in vivo (29). We found that the cGAS/STING/IFN-β pathway is constitutively activated in a subset of cancer cells (Fig. 3).
Several studies have revealed the presence of cytoplasmic DNA in cancer cells (8, 14). Cytosolic single-stranded DNA was observed in various breast cancer cells and the basal level of cytoplasmic DNA is correlated with IRDS expression and DNA damage resistance (8). Cancer cells constantly experience endogenous DNA damage, producing cytoplasmic DNA (8, 30). Endogenous DNA damage in cancer cells is caused by the collapse of damaged replication forks, and also through damage caused by reactive oxygen species (30). To survive endogenous DNA damage, as well as the exogenous DNA damage caused by therapy, cancer cells develop protective mechanisms. Our recent study showed that the IRDS protein OAS1 inhibits cell death in response to oxidative DNA damage (31), suggesting that constitutive IFN-β and IRDS expression play important roles in enabling cancer cells to survive such damage. Defects in ATM, a DNA repair kinase, increases both cytoplasmic DNA and IFN-I expression (14). Aging is another factor that increases cytoplasmic DNA through the activation of retrotransposons, long-interspersed nuclear elements (LINEs), creating DNA damage in the nucleus and causing LINE cDNA to accumulate in the cytoplasm (32, 33).
Through these diverse mechanisms, the cGAS-STING pathway is constitutively activated in a subset of cancer cells. We found that cancer cells are addicted to self-produced IFN-β, and they initiate spontaneous apoptosis and do not proliferate normally when IFN-β expression is knocked down. Importantly, levels of endogenous DNA damage are correlated with IFN-β and IRDS expression only when cancer cells express high levels of PD-L1. In this study, we measured the levels of IFN-β mRNA to determine the difference in its expression for the following reasons: 1) type I IFN induction is primarily controlled at the transcriptional level (4); and ii) after binding to its receptor, IFN and its receptor proteins are down-regulated through endocytosis and degradation (34). KD of IFNAR1 shows similar effects to knockdown of IFN-β (Fig. 2), suggesting that the IFN-β protein is produced and binds to its receptor to induce downstream events.
Most studies concerning PD-L1 have focused on its immune-dependent functions, since it induces the apoptosis of T cells by binding to PD-1 (35). Recent studies have revealed that PD-L1 protects cancer cells not only from T cells, but also from radiation, chemotherapy, or cytotoxic doses of IFN-β in an immune-independent manner (3, 5, 6). PD-L1 promotes cancer cell proliferation and survival in cells in culture, and tumor growth and metastasis in immune-deficient NSG mice (19, 36). Some cancer cells express PD-1 (18–20), but the cancer cell lines examined in this study do not (SI Appendix, Fig. S2-1F), showing that PD-1 is not involved in the cancer cell–intrinsic PD-L1 functions revealed in this study. Our analyses of public databases show that PD-1 expression is not correlated with IRDS or PD-L1 expression in cancer cells, confirming that PD-1 expression in cancer cells does not contribute to PD-L1 expression or function through IRDS expression. Recent studies have found soluble PD-1 (sPD-1) in the sera of cancer patients at levels higher than those found in healthy individuals (3, 6). Some studies show that PD-1 expression in cancer cells is correlated with resistance to cisplatin or cytotoxic IFN-β (18–20), suggesting the possibility of other PD-1–dependent, but not IRDS-dependent, mechanisms of PD-L1–induced resistance to cytotoxicity. As a cell-surface protein, PD-L1 contains only a short cytoplasmic domain with no canonical signaling motifs (37). Gato-Cañas et al. (6) found that a phylogenetically conserved motif in the cytoplasmic domain of PD-L1 plays a role in inhibiting IFN-β–induced STAT3 activation, which induces caspase 7 expression in B16 mouse melanoma cells. We found that PD-L1 inhibits the phosphorylation of STAT1 and STAT2 in response to IFN-I in H196 small cell lung cancer cells, HME mammary epithelial cells, and HeLa cervical cancer cells (Fig. 1C and SI Appendix, Fig. S1B). PD-L1 did not affect the expression and phosphorylation of STAT3 in response to IFN-β in the cells we have studied (Fig. 1D). The molecular mechanisms by which PD-L1 inhibits the IFN-β–induced phosphorylation of STAT1 and STAT2 and promotes constitutive STING activation in cancer cells remain to be investigated.
Our study revealed that PD-L1 promotes constitutive STING activation, followed by constitutive IFN-β and IRDS expression. How PD-L1 facilitates STING activation is not yet known, but considering that STING is on the endoplasmic reticulum, PD-L1 needs to be inside the cell to activate it. Indeed, several studies have shown that PD-L1 is present in both the cytoplasm and the nuclei of cancer cells, as well as on the cell membrane, and have proposed that intracellular PD-L1 is correlated with poor prognosis (5, 20, 38–42). Gao et al. (42) showed that the translocation of PD-L1 from the cell membrane to the nucleus is regulated by p300-mediated acetylation and HDAC2-dependent deacetylation of PD-L1. Nuclear PD-L1 plays an important role in sister chromatid cohesion, helping cancer cell proliferation, colony formation in vitro, and tumor growth in vivo (41). The presence of PD-L1 in the nuclei of circulating cancer cells correlates with poor prognosis in colorectal and prostate cancer (40). What drives the nuclear translocation of PD-L1 in cancer cells is not well understood. Ghebeh et al. (39) showed that doxorubicin down-regulates the expression of PD-L1 on the cell surface and up-regulates its nuclear expression in breast cancer cells. Some studies show the correlation between the presence of cytoplasmic PD-L1 and poor prognosis in cancer. The positivity of cytoplasmic PD-L1 is correlated with the risk of recurrence at all stages of thyroid cancer (38). Tu et al. (5) revealed that cytoplasmic PD-L1 promotes DNA repair by stabilizing mRNAs encoding DNA damage-response proteins, inhibiting cancer cell death in response to DNA damage.
Our study and those of others have shown that down-regulation of PD-L1 sensitizes cancer cells to radiation and chemotherapy. Another recent study from our group reveals that PD-L1 is degraded by the ubiquitin E3 ligase FBXO22, whose activity is inhibited by cyclin-dependent kinase 5 (CDK5) (43). Inhibition of CDK5 down-regulates PD-L1 and sensitizes cancer cells to radiation and cisplatin. Tu et al. (5) showed that H1A, a PD-L1 antibody that they developed, promotes PD-L1 degradation by abrogating its interaction with CMTM6, thus inhibiting cancer cell–intrinsic PD-L1 functions. Unlike KD of PD-L1, different effects of anti–PD-L1 antibodies have been reported, probably because each antibody binds to a different epitope, leading to different effects on PD-L1 function. Two studies show that immune-independent PD-L1 functions were inhibited by specific anti–PD-L1s (5, 6), but another study found that these functions were augmented by a different anti–PD-L1 (19). It has not been investigated whether clinically approved anti–PD-L1 (atezolizumab, durvalumab, and avelumab), which are developed to block the interaction with PD-1 to reactivate anticancer immunity, can abolish the immune-independent functions of PD-L1, and thus enhance the cytotoxicity of radiation or chemotherapy. The addition of cytotoxic doses of exogenous IFN-β to clinical regimens may promote the efficacy of combination treatment with anti–PD-L1 and radiation or chemotherapy because IFN-β will enhance cell death in response to DNA damage. Monoclonal antibodies targeting both PD-1 and PD-L1 are widely used in the clinic for immune checkpoint blockade, which has demonstrated unprecedented efficacy in treating a large number of advanced malignancies (1). However, responses are too often disappointingly low and dependent on patients’ preexisting immune status. To overcome this limitation, many clinical trials are ongoing in which immune checkpoint blockade is combined with other treatments, including radiotherapy, chemotherapy, and other types of immunotherapy (1, 44, 45). Most studies investigate the effects of combination treatments on T cell activity and immune responses. Based on our finding that PD-L1 inhibits the cytotoxicity of cisplatin and radiation in an immune-independent manner, we anticipate that it will be possible to improve current regimens to kill cancer cells more efficiently regardless of the patients’ immune status.
Materials and Methods
Additional details (reagent catalog numbers, buffer components, detailed description of methods, and so forth) are available in SI Appendix, Supplementary Materials and Methods.
Cells.
Human cancer cells and normal mammary epithelial cells were cultured in the media described in SI Appendix, Supplementary Materials and Methods. All cell lines were regularly tested for mycoplasma infection using the MycoAlert Kit (Lonza) and authenticated by using Short Tandem Repeat analysis by Genetica.
Real-Time Cytotoxicity Analyses.
Cells were plated at 5,000 cells per well in 96-well plates. After 20 to 24 h, the cells were treated with cisplatin in the presence of 250 nM of SytoxGreen dye, which does not penetrate live cells. To detect apoptotic cells, 5 μM of caspase 3/7 Green dye was added. Images were acquired every 3 h, using a 10× objective in the green fluorescence channel in the IncuCyte ZOOM image analysis system, automatically quantifying the number of dead cells. The total number of cells was determined by counting SytoxGreen-positive cells after adding 0.5% Triton X-100, which causes 100% cell death. The percentage of dead cells was calculated at each time point as 100× the number of dead cells in response to drugs/total number of cells at the time of drug treatment.
Cell Survival Analyses.
The fraction of surviving cells in response to ionizing radiation was assessed as described previously (46). H196 cells (2,500 cells per well) or H226 cells (500 cells per well) were plated in 24-well plates and irradiated on the following day. Unirradiated and irradiated cells were cultured (H196 for 7 d, H226 for 14 d), following which dead cells were removed by washing the wells with PBS; the remaining live cells were dissolved in 1 M NaOH and the amounts of nucleic acid in the lysates were assessed by measuring the OD260. The percentage of surviving cells was calculated as 100 × OD260 of irradiated cell lysate/OD260 of unirradiated cells.
Subcloning of PD-L1 cDNA and Gene Transfection.
To increase the expression of PD-L1 using a pseudo lentivirus, we subcloned human PD-L1 cDNA (NCBI Ref Seq NM_014143.2) from a Sino Biological construct into pLenti-CMV-puro using the Gateway recombination cloning technology. The construct was verified by nucleotide sequencing. The pseudo lentivirus containing supernatants from HEK 293T cells transfected with pLenti-CMV-puro-PD-L1 were harvested 24 and 48 h after transfection, combined and stored at −80 °C. To express PD-L1, hTERT-HME1 or HeLa cells were infected for 3 consecutive days with pseudo lentivirus and then selected with puromycin.
KDs Using shRNA.
shRNAs in the lentiviral vector pLKO.1-puro against PD-L1, IFNβ, IFNAR1, and cGAS were obtained from Sigma-Aldrich. Cells were infected for two consecutive days with diluted pseudo lentivirus and then selected with puromycin. The extent of KD in the transfected cell pool was monitored by Western analysis (PD-L1) or real-time PCR (IFN-β, IFNAR1, and cGAS). The sequences of shRNA constructs are in SI Appendix, Supplementary Materials and Methods.
Western Analyses.
Cells were lysed with a buffer (250 mM Tris·HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, and 0.1% SDS) containing phosphatase and protease inhibitors. Cell lysates were analyzed by electrophoresis in 8% SDS polyacrylamide gels. Proteins were transferred to PVDF membranes and incubated with 5% nonfat milk for 1 h of blocking, followed by incubation with primary antibodies for 1 to 2 h at room temperature and then with secondary antibodies for 1 h at room temperature. The signal was detected using Western Lightning Plus-ECL. The catalog number and dilution rate of antibodies are described in SI Appendix, Supplementary Materials and Methods.
Real-Time qRT-PCR.
cDNA was synthesized from total RNA treated with DNase I by using a modified manufacturer’s protocol with random hexamers. Real-time PCR was performed with Bullsyeye EvaGreen qPCR master mix (MidSci) in a Light Cycler 480 II (Roche). The PCR protocol is initial activation at 95 °C for 10 min, 50 cycles at 95 °C for 15 sec, and 60 °C for 1 min. Ct values were converted into relative gene-expression levels compared to that of an internal control gene, GAPDH, using the ΔΔCt method. Each PCR run also included nontemplate controls containing all reagents except cDNA, which generated no amplification. The specificity was confirmed by analysis of the melting curves of the PCR products. The sequences of primers are available in SI Appendix, Supplementary Materials and Methods.
In Silico Analyses.
Gene and protein expression data for cell lines were acquired from the Cancer Cell Line Encyclopedia (PMID:31068700), release 21Q1, available at depmap.org. Gene-expression data from TCGA were downloaded using the TCGA data portal (https://portal.gdc.cancer.gov/) from the Pan-Caner Atlas release (April 2018). The IRDS gene-expression signature was acquired from Weichselbaum et al. (16). IRDS gene expression score was determined as the average z-normalized, log-transformed, expression value of all genes in the IRDS signature. DNA damage activation score was taken as the average value of z-normalized phospho-S345 CHK1 and phospho-T68 CHK2. Relationships between different variables were determined either by regression or Spearman rank correlation coefficient.
Statistical Analyses.
Statistical significance was determined using Student’s two-tailed t test or two-way ANOVA (with Bonferroni's multiple comparisons test), and Spearman rank correlation coefficient, as indicated.
Supplementary Material
Acknowledgments
This study was supported by Grants R21CA252387 (to H.C.), R03CA215941 (to H.C.), and P01CA062220 (to G.R.S.) from the National Cancer Institute. Additional support is from Grant IRG91-022-18 to the Case Comprehensive Cancer Center from the American Cancer Society (sub-award to H.C.) and Grant K99CA240689 from the National Cancer Institute (to D.J.M.).
Footnotes
Reviewers: L.A.D., Memorial Sloan Kettering Cancer Center; P.J.H., Hudson Institute of Medical Research.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112258118/-/DCSupplemental.
Data Availability
All study data are included in the main text and SI Appendix.
References
- 1.Sun C., Mezzadra R., Schumacher T. N., Regulation and function of the PD-L1 checkpoint. Immunity 48, 434–452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulz D., et al. , Increased PD-L1 expression in radioresistant HNSCC cell lines after irradiation affects cell proliferation due to inactivation of GSK-3beta. Oncotarget 10, 573–583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan F., et al. , Elevated cellular PD1/PD-L1 expression confers acquired resistance to cisplatin in small cell lung cancer cells. PLoS One 11, e0162925 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ock C. Y., et al. , Changes in programmed death-ligand 1 expression during cisplatin treatment in patients with head and neck squamous cell carcinoma. Oncotarget 8, 97920–97927 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tu X., et al. , PD-L1 (B7-H1) competes with the RNA exosome to regulate the DNA damage response and can be targeted to sensitize to radiation or chemotherapy. Mol. Cell 74, 1215–1226.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gato-Cañas M., et al. , PDL1 signals through conserved sequence motifs to overcome interferon-mediated cytotoxicity. Cell Rep. 20, 1818–1829 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Dong P., Xiong Y., Yue J., Hanley S. J. B., Watari H., Tumor-intrinsic PD-L1 signaling in cancer initiation, development and treatment: Beyond immune evasion. Front. Oncol. 8, 386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdal E., Haider S., Rehwinkel J., Harris A. L., McHugh P. J., A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 31, 353–369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sistigu A., et al. , Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat. Med. 20, 1301–1309 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Widau R. C., et al. , RIG-I-like receptor LGP2 protects tumor cells from ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 111, E484–E491 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheon H., Stark G. R., Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc. Natl. Acad. Sci. U.S.A. 106, 9373–9378 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheon H., et al. , IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 32, 2751–2763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaston J., et al. , Intracellular STING inactivation sensitizes breast cancer cells to genotoxic agents. Oncotarget 7, 77205–77224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Härtlova A., et al. , DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42, 332–343 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Leonova K. I., et al. , p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc. Natl. Acad. Sci. U.S.A. 110, E89–E98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weichselbaum R. R., et al. , An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc. Natl. Acad. Sci. U.S.A. 105, 18490–18495 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinhold W. C., et al. , CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 72, 3499–3511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu D., Ao X., Yang Y., Chen Z., Xu X., Soluble immune checkpoints in cancer: Production, function and biological significance. J. Immunother. Cancer 6, 132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jalali S., et al. , Reverse signaling via PD-L1 supports malignant cell growth and survival in classical Hodgkin lymphoma. Blood Cancer J. 9, 22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y., Chen W., Xu Z. P., Gu W., PD-L1 distribution and perspective for cancer immunotherapy-blockade, knockdown, or inhibition. Front. Immunol. 10, 2022 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erlandsson L., et al. , Interferon-beta is required for interferon-alpha production in mouse fibroblasts. Curr. Biol. 8, 223–226 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Sung P. S., et al. , Roles of unphosphorylated ISGF3 in HCV infection and interferon responsiveness. Proc. Natl. Acad. Sci. U.S.A. 112, 10443–10448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L., Wu J., Du F., Chen X., Chen Z. J., Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corrales L., et al. , Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haag S. M., et al. , Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Cheon H., Borden E. C., Stark G. R., Interferons and their stimulated genes in the tumor microenvironment. Semin. Oncol. 41, 156–173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ranoa D. R., et al. , Cancer therapies activate RIG-I-like receptor pathway through endogenous non-coding RNAs. Oncotarget 7, 26496–26515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khodarev N. N., et al. , Signal transducer and activator of transcription 1 regulates both cytotoxic and prosurvival functions in tumor cells. Cancer Res. 67, 9214–9220 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Forys J. T., et al. , ARF and p53 coordinate tumor suppression of an oncogenic IFN-β-STAT1-ISG15 signaling axis. Cell Rep. 7, 514–526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyman C., Kanaar R., DNA double-strand break repair: All’s well that ends well. Annu. Rev. Genet. 40, 363–383 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Kondratova A. A., et al. , Suppressing PARylation by 2',5'-oligoadenylate synthetase 1 inhibits DNA damage-induced cell death. EMBO J. 39, e101573 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasior S. L., Wakeman T. P., Xu B., Deininger P. L., The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357, 1383–1393 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon M., et al. , LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. 29, 871–885.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zanin N., Viaris de Lesegno C., Lamaze C., Blouin C. M., Interferon receptor trafficking and signaling: Journey to the cross roads. Front. Immunol. 11, 615603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong H., et al. , Tumor-associated B7-H1 promotes T-cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 8, 793–800 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Clark C. A., et al. , Tumor-intrinsic PD-L1 signals regulate cell growth, pathogenesis, and autophagy in ovarian cancer and melanoma. Cancer Res. 76, 6964–6974 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong H., Zhu G., Tamada K., Chen L., B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 5, 1365–1369 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Chowdhury S., et al. , Programmed death-ligand 1 overexpression is a prognostic marker for aggressive papillary thyroid cancer and its variants. Oncotarget 7, 32318–32328 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghebeh H., et al. , Doxorubicin downregulates cell surface B7-H1 expression and upregulates its nuclear expression in breast cancer cells: Role of B7-H1 as an anti-apoptotic molecule. Breast Cancer Res. 12, R48 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satelli A., et al. , Potential role of nuclear PD-L1 expression in cell-surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. Sci. Rep. 6, 28910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu J., et al. , Regulation of sister chromatid cohesion by nuclear PD-L1. Cell Res. 30, 590–601 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y., et al. , Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat. Cell Biol. 22, 1064–1075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De S., et al. , The ubiquitin E3 ligase FBXO22 degrades PD-L1 and sensitizes cancer cells to DNA damage. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.2112674118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaretsky J. M., et al. , Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med. 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sade-Feldman M., et al. , Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 8, 1136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De S., Cipriano R., Jackson M. W., Stark G. R., Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res. 69, 8035–8042 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.