Significance
Meiotic crossovers (COs) are required for accurate chromosome segregation and mix parental genomes to generate genetically diversified gametes. The distribution of COs among chromosomes is not random. In most organisms, at least one CO is formed between each chromosome pair. However, the molecular mechanisms controlling CO distribution are not well understood. Here, we found that Arabidopsis AtFANCD2, AtFANCM, AtFIGL1, and AtRMI1 promote normal distribution of Class I CO to maintain CO assurance in addition to their previously described roles in regulating Class II CO number. These factors, alone or in combination, appear to control ∼50% of the variation between wild-type and random distributions. These discoveries provide genetic insights into understanding of the regulation of CO distribution among chromosomes.
Keywords: crossover distribution, crossover assurance, FANCD2, FANCM
Abstract
During meiosis, crossovers (COs) are typically required to ensure faithful chromosomal segregation. Despite the requirement for at least one CO between each pair of chromosomes, closely spaced double COs are usually underrepresented due to a phenomenon called CO interference. Like Mus musculus and Saccharomyces cerevisiae, Arabidopsis thaliana has both interference-sensitive (Class I) and interference-insensitive (Class II) COs. However, the underlying mechanism controlling CO distribution remains largely elusive. Both AtMUS81 and AtFANCD2 promote the formation of Class II CO. Using both AtHEI10 and AtMLH1 immunostaining, two markers of Class I COs, we show that AtFANCD2 but not AtMUS81 is required for normal Class I CO distribution among chromosomes. Depleting AtFANCD2 leads to a CO distribution pattern that is intermediate between that of wild-type and a Poisson distribution. Moreover, in Atfancm, Atfigl1, and Atrmi1 mutants where increased Class II CO frequency has been reported previously, we observe Class I CO distribution patterns that are strikingly similar to Atfancd2. Surprisingly, we found that AtFANCD2 plays opposite roles in regulating CO frequency in Atfancm compared with either in Atfigl1 or Atrmi1. Together, these results reveal that although AtFANCD2, AtFANCM, AtFIGL1, and AtRMI1 regulate Class II CO frequency by distinct mechanisms, they have similar roles in controlling the distribution of Class I COs among chromosomes.
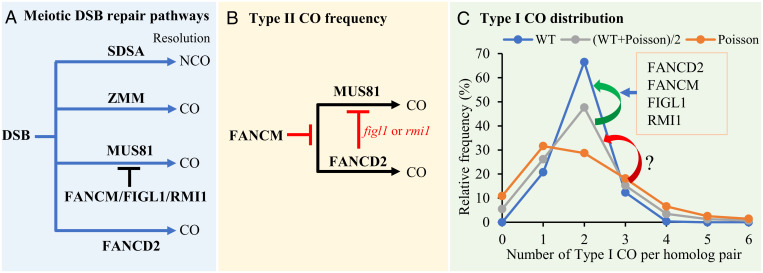
Meiosis is a specialized cell division process that includes two rounds of chromosome segregation following a single round of premeiotic DNA replication and is essential for sexual reproduction in most eukaryotes. During meiosis, homologous recombination (HR) is employed to repair double strand breaks (DSBs) catalyzed by the SPO11 transesterase, yielding crossovers (COs) or noncrossovers (1). In most eukaryotes, meiotic COs are required to ensure the faithful segregation of homologous chromosomes (homologs). COs create new haplotypes, which in turn generate phenotypic diversity among offspring (2). Numerous studies in multiple species have shown that the number and distribution of COs are tightly regulated (3, 4). For instance, the model plant Arabidopsis thaliana has about 10 COs during each meiosis (5) that are nonrandomly distributed along and among chromosomes. Despite the small number of COs, each of the five pairs of Arabidopsis homologs experiences at least one CO—a phenomenon known as CO assurance (6). A second regulatory mechanism, CO interference, inhibits closely spaced double COs (7). However, the molecular mechanisms controlling CO distribution are elusive.
Most eukaryotes have two kinds of COs (1): Class I COs that are sensitive to interference and Class II COs which are interference insensitive. In wild-type (WT) Arabidopsis, 85 to 90% of COs belong to Class I and are mediated by the ZMM group of proteins (Zip1-4, Mer3, and Msh4-5) and MLH1/3 (8–13). Class II COs can be generated by at least two parallel pathways in Arabidopsis (14–16), which depend on either the structure-specific endonuclease AtMUS81 (15, 16) or a homolog of Fanconi Anemia Complementation Group D2 (AtFANCD2) (14). The homolog of the Holliday junction resolvase GEN1 is required for the formation of Class II CO in rice (17) but not in Arabidopsis (18), suggesting that Class II CO pathways have diverged between monocots and dicots. In addition to pro–Class II CO factors, Arabidopsis has at least three anti–Class II CO mechanisms: the AtFANCM helicase and its cofactors AtMHF1 and AtMHF2 (19, 20); members of BLM-TOP3-RMI1 (BTR) complex AtRECQ4A, AtRECQ4B, AtTOP3α, and AtRMI1 (21, 22); and AtFIGL1 AAA-ATPase and its interacting protein AtFLIP1 (23, 24). Interestingly, although total CO frequency increases and Class I CO numbers remain unchanged when these anti–Class II CO pathways are perturbed, univalents are still occasionally observed, suggesting that CO assurance has also been weakened (22, 23).
In this study, we investigated the influence of AtFANCD2, AtFANCM, AtFIGL1, and AtRMI1 on the distribution of Class I COs among chromosomes. We demonstrate that these factors have a role in promoting a non-Poisson distribution of Class I COs among chromosomes.
Results
WT Non-Poisson Distribution of Class I COs Requires AtFANCD2 but Not AtMUS81.
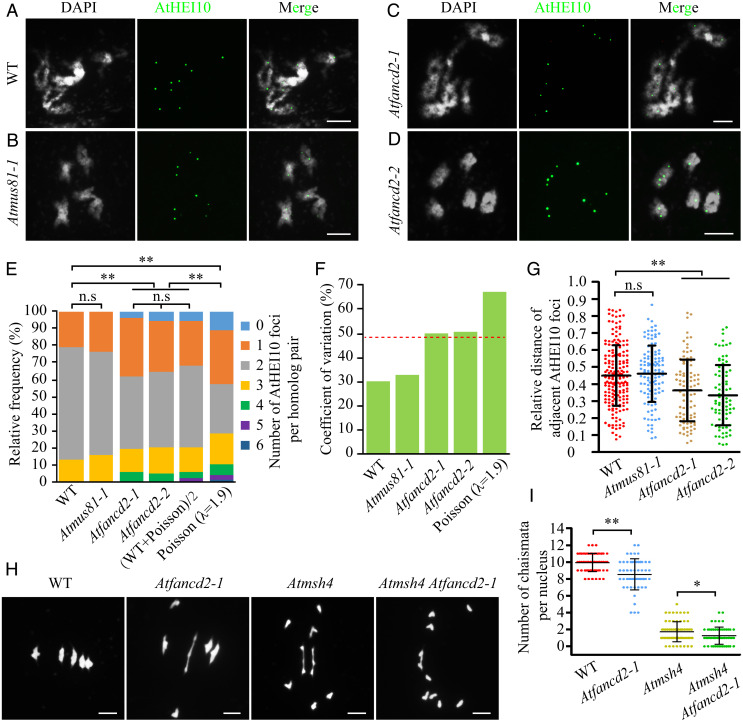
In Arabidopsis, the distribution of total COs among chromosomes deviates from a Poisson distribution (10, 25). To characterize the distribution of Class I COs among chromosomes specifically, we immunostained diakinesis male meiocytes using an anti-AtHEI10 antibody that marks only Class I COs (9, 26) (Fig. 1 A–D). WT meiocytes have an average of 1.92 AtHEI10 foci (n = 275; range 1 to 4) per bivalent, and the distribution is markedly centralized toward the modal value of 2 foci (Fig. 1E). This distribution deviates significantly from the Poisson expectation (P < 0.01) (Fig. 1E). The Poisson distribution is a distribution of independent events occurring at a constant mean frequency (expressed as λ here) in a fixed number of intervals (in this case, bivalents) (12). Based on the Poisson expectation, ∼11% of the bivalents should lack AtHEI10 foci (range 0 to 6), with a coefficient of variation (CV, SD divided by the mean) of 67% compared to the CV of 30% observed in WT (Fig. 1F).
Fig. 1.
Normal distribution of Class I COs requires AtFANCD2 but not AtMUS81. (A–D) Immunostaining of AtHEI10 at diakinesis in WT, Atmus81-1, Atfancd2-1, and Atfancd2-2, respectively. DAPI-stained chromosomes are shown in white, and AtHEI10 foci are shown in green. (E) Distributions of the number of AtHEI10 foci on homologous chromosome pairs (bivalents) in WT, Atmus81-1, Atfancd2-1, and Atfancd2-2, compared to (WT + Poisson)/2 and Poisson expectations. n.s., not significant, **P < 0.01 χ2 test. (F) CVs in the distribution of AtHEI10 foci per homolog pairs in WT, Atmus81-1, Atfancd2-1, Atfancd2-2, and the Poisson expectation. The red dash line indicates the mean of the WT and Poisson CVs. (G) The distance between adjacent AtHEI10 foci in WT, Atmus81-1, Atfancd2-1, and Atfancd2-2 expressed as a percentage of bivalent length. Error bars indicate SD. n.s., not significant, **P < 0.01 two-tailed Mann–Whitney U test. (H) Meiotic chromosome morphologies of WT, Atfancd2-1, Atmsh4, and Atmsh4 Atfancd2-1 at metaphase I. (I) The number of chiasmata per nucleus in WT, Atfancd2-1, Atmsh4, and Atmsh4 Atfancd2-1, based on metaphase I bivalent conformations. Error bars indicate SD. *P < 0.05, **P < 0.01 two-tailed Mann–Whitney U test. (Scale bars, 5 μm.)
Class I COs frequencies do not decrease in Atfancd2-1, but 13% of metaphase I meiocytes have univalents (homologous chromosomes not held together by a CO) (14). Consistent with those observations, the number of AtHEI10 foci per cell in Atfancd2-1 (9.14 ± 1.9) is similar to WT (9.58 ± 1.26, P = 0.23) (SI Appendix, Fig. S1), but their distribution differs from WT and Poisson (Fig. 1E) (P values are listed in SI Appendix, Table S1). In Atfancd2-1, the range of AtHEI10 foci numbers per bivalent increases to 0 to 5. Compared with the WT distribution, the percentage of bivalents with two AtHEI10 foci decreases from 66.5 to 42.8% in Atfancd2-1 with coordinate increases in the proportions of other classes. Notably, 3.9% of Atfancd2-1 chromosomes lack AtHEI10 foci. We observed a similar alteration of CO distribution using another Class I CO marker AtMLH1, in Atfancd2-1 (SI Appendix, Fig. S2). Overall, the CO distribution in Atfancd2 is more diffuse than WT but more centralized than a Poisson distribution. A second allele, Atfancd2-2, has a similar distribution of AtHEI10 foci (Fig. 1E and SI Appendix, Table S1). By contrast, another pro–Class II CO factor mutant Atmus81-1 has an AtHEI10 foci distribution indistinguishable from WT (Fig. 1E and SI Appendix, Table S1). Intriguingly, although the distribution of Class I COs in Atfancd2 mutants deviates from both WT and Poisson expectations it does not deviate significantly from the average of the WT and Poisson distributions [designated as (WT + Poisson)/2] (Fig. 1E and SI Appendix, Table S1). Similarly, the CVs of the Atfancd2 mutants (50.4 and 50.9%) are close to the average of the WT and Poisson CVs (48.7%) (Fig. 1F). These data imply that AtFANCD2 accounts for 50% of the difference between the WT distribution of Class I COs among chromosomes and a Poisson distribution.
We analyzed the effect of AtFANCD2 on the distance between adjacent Class I COs relative to the total length of the chromosomes as previously reported (27). The interfoci distance is significantly smaller in Atfancd2-1 (0.36 ± 0.18, n = 95 pairs of AtHEI10 foci in 21 cells; P < 0.01) and Atfancd2-2 (0.33 ± 0.18, n = 85 pairs of AtHEI10 foci in 21 cells; P < 0.01) compared to WT (0.45 ± 0.18, n = 194 pairs of AtHEI10 foci in 37 cells) but not in Atmus81-1 (0.46 ± 0.17, n = 119 pairs of AtHEI10 foci in 27 cells; P = 0.58) (Fig. 1G). This suggests that Class I COs are positioned more closely in Atfancd2 than in WT, indicating weaker CO interference. A previous study reported a 14% reduction of COs in Atfancd2-1 based on cytological analysis of chiasmata (14). Differentiating closely spaced chiasmata can be challenging, and our results suggest that inter-CO spacing may be reduced in Atfancd2 mutants, so we performed a similar analysis in an Atmsh4 background, which should eliminate ZMM-dependent Class I COs (Fig. 1H). We confirmed the previous difference between WT and Atfancd2-1 (WT, 9.95 ± 1.08, n = 56; Atfancd2-1, 8.55 ± 1.83, n = 53, P < 0.01) and a significant reduction of chiasmata in Atmsh4 Atfancd2-1 (1.27 ± 1.03, n = 56, P < 0.05) compared to Atmsh4 (1.74 ± 1.19, n = 69) (Fig. 1I).
AtFANCM, AtFIGL1, and AtRMI1 Are Similar to AtFANCD2 in Regulating Class I CO Distribution.
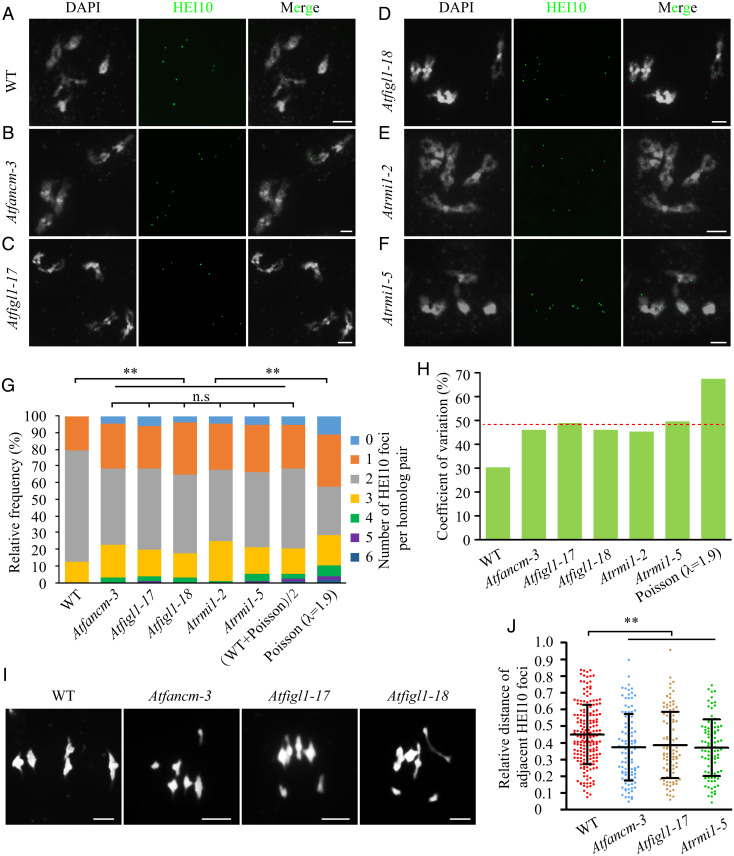
We previously demonstrated that FANCM is required for the normal distribution of Class I COs in both Lactuca sativa and Arabidopsis (28). In addition, the observation of meiotic univalents in Atfigl1 and Atrmi1 mutants (22, 23) suggests that despite elevated CO frequencies, COs are not appropriately distributed in these backgrounds. Here, we analyze the role of AtFANCM, AtFIGL1, and AtRMI1 in controlling Class I CO distribution. The Atfancm-3, Atrmi1-2, and Atrmi1-5 mutant alleles have been described previously (22, 29), and the positions of T-DNA (transfer DNA) insertions in two Atfigl1 alleles (designated as Atfigl1-17 and Atfigl1-18) are shown in SI Appendix, Fig. S3. Consistent with previous studies (19, 22, 24), AtFANCM, AtFIGL1, and AtRMI1 have no obvious effect on the frequency of Class I COs (SI Appendix, Fig. S1). The chromosomal distributions of AtHEI10 foci in Atfigl1-17, Atfigl1-18, Atrmi1-2, Atrmi1-5, and Atfancm-3 are similar to (WT + Poisson)/2, and all of them differ significantly with both WT and Poisson distributions (Fig. 2 A–G and SI Appendix, Table S1). In these mutants, 4 to 6% of homolog pairs lack AtHEI10 foci, and the percentage of bivalents with two foci declines to 43 to 48% relative to 67% in WT (Fig. 2G). The CVs in these mutants are also close to the average of the WT and Poisson CVs (Fig. 2H). These data suggest that AtFANCD2, AtFANCM, AtFIGL1, and AtRMI1 play similar roles in controlling the distribution of Class I CO among chromosomes. Therefore, the univalents previously described in Atfigl1-1 and Atrmi1-5 mutants (22, 23) may be a result of the aberrant Class I CO distribution. Similarly, we found univalent at frequencies of 6.67% (n = 3/45) in Atfancm-3, 13.46% (n = 7/52) in Atfigl1-17, and 13.89% (n = 5/36) in Atfigl1-18 (Fig. 2I) at metaphase I. Consistent with the inter-CO distance phenotype observed in Atfancd2-1, the distances between adjacent AtHEI10 foci in Atfancm-3 (0.37 ± 0.20, n = 109 pairs of AtHEI10 foci in 23 cells), Atfigl1-17 (0.39 ± 0.20, n = 96 pairs of AtHEI10 foci in 25 cells), and Atrmi1-5 (0.37 ± 0.17, n = 94 pairs of AtHEI10 foci in 22 cells) are shorter than that in WT (P < 0.01 for Atfancm-3 versus WT, Atfigl1-17 versus WT, and Atrmi1-5 versus WT) (Fig. 2J).
Fig. 2.
Normal distribution of Class I COs requires AtFANCM, AtFIGL1, and AtRMI1. (A–F) Immunostaining of AtHEI10 at diakinesis in WT, Atfancm-3, Atfigl1-17, Atfigl1-18, Atrmi1-2, and Atrmi1-5. DAPI-stained chromosomes are shown in white, and AtHEI10 foci are shown in green. (G) Distributions of the number of AtHEI10 foci on homologous chromosome pairs (bivalents) in WT, Atfancm-3, Atfigl1-17, Atfigl1-18, Atrmi1-2, and Atrmi1-5, compared to (WT + Poisson)/2, and Poisson expectations. n.s., not significant, **P < 0.01 χ2 test. (H) CVs in the distributions of AtHEI10 foci per homolog pairs in WT, Atfancm-3, Atfigl1-17, Atfigl1-18, Atrmi1-2, Atrmi1-5, and the Poisson expectation. The red dash line indicates the average of the WT and Poisson CVs. (I) Meiotic chromosome morphologies of WT, Atfancm-3, Atfigl1-17, and Atfigl1-18 at metaphase I. (Scale bars, 5 μm.) (J) The distance between adjacent AtHEI10 foci in WT, Atfancm-3, Atfigl1-17, and Atrmi1-5, expressed as a percentage of bivalent length. Error bars indicate SD. **P < 0.01 two-tailed Mann–Whitney U test.
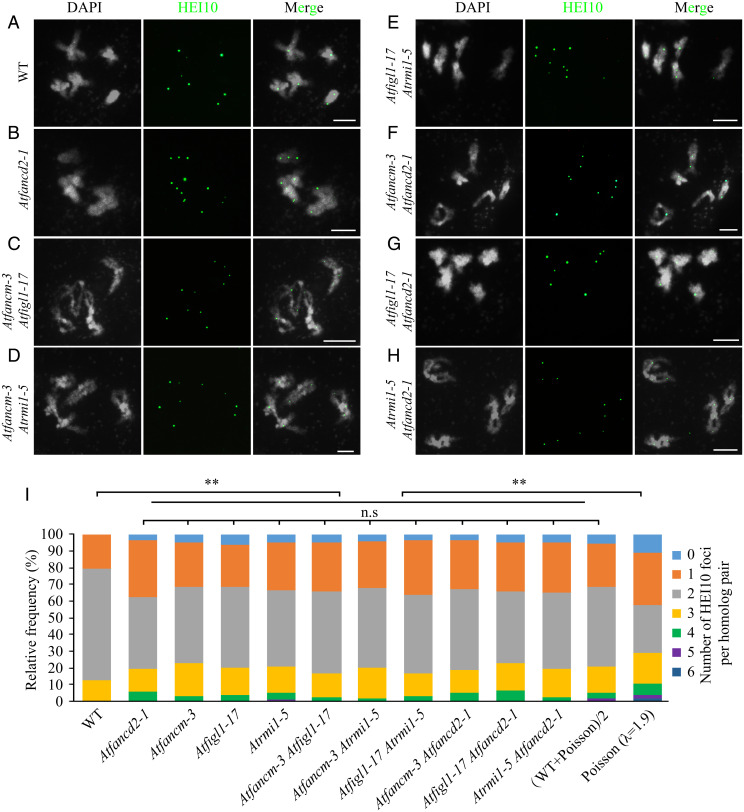
The striking similarity in how Atfancd2, Atfancm, Atfigl1, and Atrmi1 mutants influence the distribution of Class I CO among chromosomes indicates that these proteins might have convergent roles in regulating meiotic CO distribution. To test this hypothesis, we compared the distribution of AtHEI10 foci in double mutants and the corresponding single mutants (Fig. 3 A–H). As shown in Fig. 3I, there is no discernable difference between the distributions observed in any of the single mutants compared to any of the double mutant combinations, or with (WT + Poisson)/2, but each of the single mutants and each of the double mutants have distributions that significantly deviate from both WT and Poisson distributions. These data suggest that these factors act in a similar manner to regulate the distribution Class I CO among chromosomes.
Fig. 3.
AtFANCD2, AtFANCM, AtFIGL1, and AtRMI1 regulate the distribution of Class I CO among chromosomes similarly. (A–H) Immunostaining of AtHEI10 at diakinesis in WT, Atfancd2-1, Atfancm-3 Atfigl1-17, Atfancm-3 Atrmi1-5, Atfigl1-17 Atrmi1-5, Atfancm-3 Atfancd2-1, Atfigl1-17 Atfancd2-1, and Atrmi1-5 Atfancd2-1. DAPI-stained chromosomes are shown in white, and AtHEI10 foci are shown in green. (I) Distributions of the number of AtHEI10 foci on homologous chromosome pairs (bivalents) in WT, Atfancd2-1, Atfancm-3 Atfigl1-17, Atfancm-3 Atrmi1-5, Atfigl1-17 Atrmi1-5, Atfancm-3 Atfancd2-1, Atfigl1-17 Atfancd2-1, and Atrmi1-5 Atfancd2-1, compared to (WT + Poisson)/2 and Poisson expectations. n.s., not significant, **P < 0.01 χ2 test. (Scale bars, 5 μm.)
AtFANCM, AtFIGL1, and AtRMI1 Regulate Class I CO Distribution Independent of Promoting the Formation of AtFANCD2-Dependent CO.
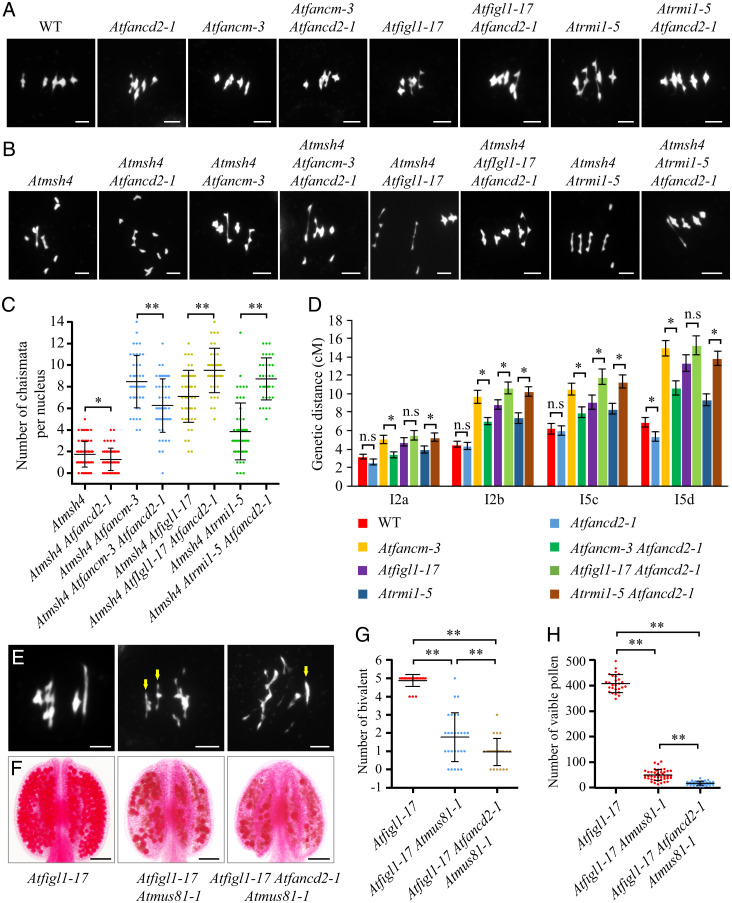
As AtFANCD2, AtFANCM, AtFIGL1, and AtRMI1 all act to regulate the distribution Class I CO among chromosomes similarly, we tested whether AtFANCM, AtFIGL1, and AtRMI1 are required for the formation of AtFANCD2-dependent COs. We first estimated the number of AtFANCD2-dependent chiasmata on metaphase I bivalents (Fig. 4 A and B). As shown in Fig. 4C, the number of chiasmata decreases from 1.74 (±1.19, n = 69) in Atmsh4 to 1.27 (±1.03, n = 56, P < 0.05) in Atmsh4 Atfancd2. We observed a larger reduction from 8.47 (±2.39, n = 43) chiasmata in Atmsh4 Atfancm-3 to 6.26 (±2.45, n = 47, P < 0.01) in Atmsh4 Atfancm-3 Atfancd2-1, indicating that the number of AtFANCD2-dependent chiasmata increases from 0.47 in Atmsh4 to 2.21 in Atmsh4 Atfancm-3. We validated the alteration of AtFANCD2-dependent CO frequency by analyzing four genetic intervals (I2a, I2b, I5c, and I5d) with the fluorescent tagged lines (FTLs) system (30). As shown in Fig. 4D and SI Appendix, Table S2, disruption of AtFANCD2 in WT results in a general trend down but only reaches statistical significance in one interval I5d. By comparison, map distances in all four intervals are significantly decreased in Atfancm-3 Atfancd2-1 double mutants compared to Atfancm-3. Both chiasma counting and FTL data suggest the existence of AtFANCD2-dependent COs in Atfancm-3. Thus, in addition to suppressing AtMUS81-dependent COs, AtFANCM may also limit AtFANCD2-dependent COs. To examine the influence of AtFIGL1 and AtRMI1 on the number of AtFANCD2-dependent CO, we conducted similar analyses in Atfigl1-17 and Atrmi1-5 backgrounds. Intriguingly, we show that AtFANCD2 mutation leads to a 34% increase in chiasma frequency in Atmsh4 Atfigl1-17 and a 127% increase in Atmsh4 Atrmi1-5 backgrounds (Atmsh4 Atfigl1-17, 7.11 ± 2.37, n = 47, Atmsh4 Atfigl1-17 Atfancd2-1, 9.51 ± 2.02, n = 37, P < 0.01; Atmsh4 Atrmi1-5, 3.85 ± 2.61, n = 48, Atmsh4 Atrmi1-5 Atfancd2-1, 8.73 ± 1.91, n = 33, P < 0.01) (Fig. 4C). FTL analyses also showed that mutating AtFANCD2 significantly increases CO frequency in Atfigl1-17 or Atrmi1-5 backgrounds (Fig. 4D and SI Appendix, Table S2). These results demonstrate that some of the extra COs in an Atfancm background are AtFANCD2 dependent, but unexpectedly, AtFANCD2 appears to inhibit some extra CO formation in Atfigl1 and Atrmi1 backgrounds. These disparate phenotypes indicate a complex regulation of hyperrecombinant states. Furthermore, consistent with previous reports (14, 19, 22, 24), tetrad analysis of two pairs of intervals showed reduced interference in Atfancm-3, Atfigl1-17, and Atrmi1-5 compared to WT (SI Appendix, Fig. S4) but not between Atfancd2-1 and WT (SI Appendix, Fig. S4).
Fig. 4.
Genetic interactions between Atfancd2-1 and Atfancm-3, Atfigl1-17, and Atrmi1-5, as measured by CO frequency. (A) DAPI-stained chromosome spreads at metaphase I in WT, Atfancd2-1, Atfancm-3, Atfancm-3 Atfancd2-1, Atfigl1-17, Atfigl1-17 Atfancd2-1, Atrmi1-5, and Atrmi1-5 Atfancd2-1. (B) DAPI-stained chromosome spreads at metaphase I in Atmsh4-1, Atmsh4-1 Atfancd2-1, Atmsh4-1 Atfancm-3, Atmsh4-1 Atfancm-3 Atfancd2-1, Atmsh4-1 Atfigl1-17, Atmsh4-1 Atfigl1-17 Atfancd2-1, Atmsh4-1 Atrmi1-5, and Atmsh4-1 Atrmi1-5 Atfancd2-1. (C) Number of chiasmata per nucleus in Atmsh4, Atmsh4 Atfancd2-1, Atmsh4-1 Atfancm-3, Atmsh4-1 Atfancm-3 Atfancd2-1, Atmsh4-1 Atfigl1-17, Atmsh4-1 Atfigl1-17 Atfancd2-1, Atmsh4-1 Atrmi1-5, and Atmsh4-1 Atrmi1-5 Atfancd2-1, based on metaphase I bivalent conformations. Error bars indicate SD. *P < 0.05, **P < 0.01 two-tailed Mann–Whitney U test. (D) Map distances in centiMorgans (cM) in four genetic intervals from WT, Atfancd2-1, Atfancm-3, Atfancm-3 Atfancd2-1, Atfigl1-17, Atfigl1-17 Atfancd2-1, Atrmi1-5, and Atrmi1-5 Atfancd2-1 plants. Error bars indicate SEM. n.s., not significant, * significant, P value (one-tailed distribution) calculated using Z-score generated by the Stahl Lab Online Tools (https://elizabethhousworth.com/StahlLabOnlineTools/). (E) DAPI-stained chromosome spreads at metaphase I in Atfigl1-17, Atfigl1-17 Atmus81-1, and Atfigl1-17 Atfancd2-1 Atmus81-1. The arrowheads indicate bivalents in Atfigl1-17 Atmus81-1 and Atfigl1-17 Atfancd2-1 Atmus81-1. (F) Alexander red–stained anthers in Atfigl1-17, Atfigl1-17 Atmus81-1, and Atfigl1-17 Atfancd2-1 Atmus81-1. (G and H) Statistical analyses of the number of bivalents per metaphase I nucleus and the number of viable pollen grain per anther in Atfigl1-17, Atfigl1-17 Atmus81-1, and Atfigl1-17 Atfancd2-1 Atmus81-1. **P < 0.01 two-tailed Mann–Whitney U test. (Scale bars in A, B, and E, 5 μm; in F, 100 μm.)
In Atmsh4 Atfigl1-17 Atfancd2-1 and Atmsh4 Atrmi1-5 Atfancd2-1, both Class I COs and AtFANCD2-dependent COs are absent, suggesting that the remaining COs are AtMUS81 dependent. Atrmi1 but not Atfigl1 alleles are synthetic lethal in combination with Atmus81 (22, 24), so we tested whether the increased COs in Atfigl1-17 Atfancd2-1, as compared to Atfigl1-17, are dependent on AtMUS81. We observed chromosome fragments, multivalents, and bivalents in Atfigl1-17 Atmus81-1 and Atfigl1-17 Atmus81-1 Atfancd2-1 at metaphase I. The number of countable bivalents in these mutant backgrounds is negatively correlated with the severity of the meiotic defects. We observed 4.88 ± 0.32 (n = 25) bivalents at metaphase I in Atfigl1-17, 1.78 ± 1.31 (n = 27) in Atfigl1-17 Atmus81-1, and 0.96 ± 0.73 (n = 23) in Atfigl1-17 Atmus81-1 Atfancd2-1 (Fig. 4 E and G; P < 0.01 two-tailed Mann–Whitney U test). Consistent with these observations, we found that the number of viable pollens per anther decreased from 408 ± 34 (n = 27) in Atfigl1-17 to 49.07 ± 20.8 (n = 44) in Atfigl1-17 Atmus81-1 and 17.03 ± 7.67 (n = 33) in Atfigl1-17 Atfancd2-1 Atmus81-1. (Fig. 4 F and H; P < 0.01 two-tailed Mann–Whitney U test). These results support the idea that more recombination intermediates depend on AtMUS81 for resolution in Atfigl1-17 Atfancd2-1 versus Atfigl1-17. Together, our data suggest that AtFANCD2’s role as a pro–Class II CO factor is dependent on a context in which AtFIGL1 and AtRMI1 are active, but the role of all these factors in promoting a WT non-Poisson distribution of Class I COs among the chromosomes is not similarly context dependent, indicating that the two phenotypes (CO frequency and CO distribution) are separable.
Discussion
A Genetic Pathway in Regulating the Distribution of Interference-Sensitive CO.
We present a model describing meiotic CO frequency and the distribution of Type I CO among chromosomes (Fig. 5). The first part of the model summarizes prior studies that delineated the resolution of DSB repair intermediates into non-COs by the synthesis-dependent strand annealing pathway or COs by other pathways. The majority of COs are interference-sensitive (Class I) events processed by the ZMM class of proteins. A small set of COs are not sensitive to interference (Class II) and are mediated by AtMUS81 or AtFANCD2. AtMUS81-dependent COs are inhibited by AtFANCM, AtFIGL1, and AtRMI1 (Fig. 5A). Here, we show that, in addition to suppressing AtMUS81-dependent COs, AtFANCM also inhibits AtFANCD2-dependent COs (Fig. 5B). Moreover, we propose that AtFANCD2 has a conditional role in limiting Class II COs when AtFIGL1 or AtRMI1 are inactivated (Fig. 5B). It has been reported that the distribution of total COs among Arabidopsis chromosomes deviates from a Poisson distribution (10, 25). We show that AtFANCD2, AtFANCM, AtFIGL1, and AtRMI1 have a similar function that promotes the non-Poisson distribution of Class I COs among chromosomes. Interestingly, disrupting any single factor or any combination of two factors results in an altered Class I CO distribution that is an average of the WT state and a Poisson distribution. This is a strikingly consistent phenotype across mutant backgrounds and suggests that AtFANCD2, AtFANCM, AtFIGL1, and AtRMI1 are converging on a common mechanism to control roughly half of the variance between the WT and Poisson distributions (Fig. 5C).
Fig. 5.
A proposed model for the regulation of CO frequency and distribution. (A) Meiotic DSBs can be repaired through synthesis-dependent strand annealing pathway as noncrossovers or ZMM pathway, as Type I CO or MUS81, and FANCD2 pathways as Type II CO, and FANCM, FIGL1, and RMI1 have distinct roles in limiting MUS81-dependent CO. (B) Besides limiting the formation of MUS81-dependent CO, our data show that AtFANCM also limits AtFANCD2-dependent CO and AtFANCD2 limits AtMUS81-dependent CO in Atfigl1 or Atrmi1 background. (C) The distribution of meiotic Class I COs among homologous chromosome pairs in WT plants (blue line) deviates significantly from a Poisson distribution (orange line) in which COs are positioned independently of one another. Based on our genetic analysis, the activities of AtFANCD2, AtFANCM, AtFIGL1, and AtRMI1 appear to control ∼50% of the difference between the WT and Poisson distributions. Mutations in any of these factors results in a distribution that is intermediate between WT and random Poisson (gray line). Factors that regulate the remaining 50% of the difference are yet to be determined. Arrowheads indicate the transition between different distribution patterns.
FANCD2, FANCM, FIGL1, and RMI1 function interdependently to facilitate HR-mediated DNA repair in vertebrates’ somatic cells (31–37). In humans, FANCM is part of a core complex with multiple functions, including the monoubiquitination of FANCD2 that participates in repair of DNA interstrand cross-links (31–33). In addition, FANCM directly interacts with TOP3a and RMI1 in BTR complex to bridge FA and BTR pathways (34), and the BTR complex was shown to promote the activation of FA pathway (35). Human FIGNL1-associated protein KIAA0146/SPIDR binds to BLM, the homologs of AtRECQ4A/B, to participate in somatic HR (36, 37). However, during Arabidopsis meiotic recombination, AtFANCD2 promotes a subset of Class II COs while AtFANCM, AtFIGL1, and AtRMI1 limit Class II CO by different mechanisms (14, 19, 22, 24). As described above (Fig. 3), we show that they play a similar role in controlling the distribution of Class I COs. However, our genetic analyses reveal that AtFANCD2 has opposing roles in mediating CO number in Atfancm compared with either in Atfigl1 or Atrmi1. The consistency in regulating CO distribution and the dissimilarity in controlling CO frequency among AtFANCD2, AtFANCM, AtFIGL1, and AtRMI1 suggest that CO distribution and CO frequency are controlled separately.
It has been reported that E1 enzyme of the neddylation complex AtAXR1 (38) and kinesin AtPSS1 control Class I CO distribution in Arabidopsis (39). In contrast to our findings, Ataxr1 and Atpss1 have an additional serious defect in homologs synapsis, suggesting that the altered distribution of Class I CO in these two mutants might be the result of aberrant chromosome configuration (38, 39). Unlike the Class I CO distributions we observed in Atfancd2-1, Atfancm-3, Atfigl1-17, or Atrmi1-5 (and their double mutant combinations), those seen in Atpss1 (39) and Ataxr1 (38) do not appear to be similar to a Poisson or (WT + Poisson)/2 distribution supporting the idea that they influence CO distribution in a distinct manner.
The Possible Relationship among CO Interference and CO Assurance in Arabidopsis.
Reduced CO interference in Atfancm, Atfigl1, and Atrmi1 has been reported in previous studies based on FTL analyses (19, 22, 24) and this study (SI Appendix, Fig. S4). However, FTL analysis uses total CO numbers (30) rather than focusing specifically on Class I COs. The decreased distance between adjacent AtHEI10 foci, in Atfancm, Atfigl1, and Atrmi1 (Fig. 2J) suggests weaker interference between Class I COs. Our data also show similar decreases in AtHEI10 interfoci distances when both the number of Class II COs increase and decrease, thus the distribution of Class I COs does not seem to be correlated with the frequency of Class II COs. The reduced interference of Class I CO in Atfancd2, Atfancm, Atfigl1, and Atrmi1 is not accompanied by a change in Class I CO frequency (SI Appendix, Fig. S1), which is different from the situation in budding yeast top2 mutants in which reduced interference is coincident with increased Class I CO numbers (40). The data in Arabidopsis suggests that the regulation of CO interference strength is at least partially independent of the regulation of Class I CO frequency. Nonetheless, since interference reduces the likelihood of multiple Class I COs on the same bivalent, reduced interference elevates the potential for multi-CO bivalents. And, if the total number of Class I CO remains unchanged, the probability of bivalents lacking Class I COs also increases, which may influence CO assurance.
Previous studies and our results in Arabidopsis show that each bivalent typically experiences at least one Class I CO, suggesting that the CO assurance is facilitated primarily by Class I COs (39) in WT. In Atfancm-3, Atfigl1-17, and Atrmi1-5, ∼5% of homolog pairs lack Class I COs because of abnormal distribution, and although these mutants all have higher Class II CO frequencies compared to WT, univalent can be observed occasionally. This suggests that Class II COs are less efficient at promoting CO assurance than Class I COs. CO interference likely contributes to CO assurance by dispersing Class I COs along the chromosome and limiting the number of Class I COs per bivalent, which in turn contributes to a non-Poisson distribution of COs among chromosomes.
In total, our data show that Arabidopsis AtFANCD2, AtFANCM, AtFIGL1, and AtRMI1 have a role in regulating the distribution of Class I CO among chromosomes. These factors account for ∼50% of the difference between the observed WT distribution and a theoretical Poisson distribution. These results provide better understanding to the molecular mechanism in controlling CO distribution and number. Given the relatively conserved meiotic process and the universal existence of homologous genes of these factors in vertebrates, we believe that the mechanisms we described here may be relevant in other species such as human meiosis.
Materials and Methods
Plant Materials and Growth Conditions.
A. thaliana mutants used in this study are as follows: Atfancd2-1 (SALK_113293) (14), Atfancd2-2 (GK-209C01) (14), Atmus81-1 (SALK_107515) (16), Atmsh4 (SALK_136296) (10), Atfancm-3 (SALK_151218) (28), Atfigl1-17 (SALK_066994), Atfigl1-18 (SAIL_563_C03), Atrmi1-2 (SALK_094387) (22), and Atrmi1-5 (SALK_005449) (22). Mutations were confirmed by PCR using the following primers: Atfancd2-1: Atfancd2-1LP, Atfancd2-1RP, and LBb1.3; Atfancd2-2: Atfancd2-2LP, Atfancd2-2RP, and GABItest1; Atmus81-1: Atmus81-1LP, Atmus81-1RP, and LBb1.3; Atmsh4: Atmsh4LP, Atmsh4RP, and LBb1.3; Atfancm-3: Atfancm-3LP, Atfancm-3RP, and LBb1.3; Atfigl1-17: Atfigl1-17LP, Atfigl1-17RP, and LBb1.3; Atfigl1-18: Atfigl1-18LP, Atfigl1-18RP, and LB3; Atrmi1-2: Atrmi1-2LP, Atrmi1-2RP, and LBb1.3; and Atrmi1-5: Atrmi1-5LP, Atrmi1-5RP, and LBb1.3. The sequences of these primers are listed in SI Appendix, Table S3. Higher-order mutants were generated by crossing single mutants. Plants were grown on soil in growth rooms at 22 °C under long-day conditions with 16-h light and 8-h dark.
Cytological Analysis of Meiosis.
Fixation of Arabidopsis inflorescences and chromosome spreads of male meiocytes were performed as described (41). Chromosome spreads were used to observed chromosome morphologies after staining with DAPI or immunostaining with Anti-AtHEI10 and Anti-MLH1 antibodies as previously described (28, 42, 43). Images were captured with a Zeiss Axio Scope A1 microscope (Zeiss). The distances between adjacent AtHEI10 foci were measured along individual chromosomes using Image Tool and calculated as a percent of the total length of the bivalent.
Measuring CO Frequency.
Chiasmata were counted in male meiocytes at metaphase I (44). Rod bivalents were interpreted as representing a chiasma(ta) on one chromosome arm and a ring bivalent as having chiasmata on both arms (44). FTL data collection, calculation of genetic distances, and statistical analyses were performed according to Berchowitz and Copenhaver (45) and using the Stahl Lab Online tools (https://elizabethhousworth.com/StahlLabOnlineTools/).
Statistical Methods.
Microsoft Excel 2019 (Microsoft) was used to calculate mean and SD, prepare bar and stacked graphs, and perform χ2 tests. GraphPad Prism 5 was used to prepare scatter diagrams and perform Mann–Whitney U tests.
Supplementary Material
Acknowledgments
We greatly appreciate Professor Raphaël Mercier at Institut National de la Recherche Agronomique (INRA), Centre de Versailles-Grignon (France), for providing MLH1 antibody and the Arabidopsis Biological Resource Center (ABRC) at Ohio State University for providing the Arabidopsis mutant seeds. This work was supported by National Natural Science Foundation of China (31925005 and 31870293) and Fudan University. J.H. and G.P.C. were supported by a grant from the US NSF (IOS-1844264).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107543118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M., The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66, 297–327 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Wang Y., Engineering stable heterosis. J. Genet. Genomics 46, 1–3 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Lam I., Keeney S., Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 7, a016634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray S., Cohen P. E., Control of meiotic crossovers: From double-strand break formation to designation. Annu. Rev. Genet. 50, 175–210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Copenhaver G. P., Meiotic recombination: Mixing it up in plants. Annu. Rev. Plant Biol. 69, 577–609 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Jones G. H., The control of chiasma distribution. Symp. Soc. Exp. Biol. 38, 293–320 (1984). [PubMed] [Google Scholar]
- 7.Berchowitz L. E., Copenhaver G. P., Genetic interference: Don’t stand so close to me. Curr. Genomics 11, 91–102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chelysheva L., et al. , Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 3, e83 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chelysheva L., et al. , The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8, e1002799 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins J. D., Armstrong S. J., Franklin F. C. H., Jones G. H., The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev. 18, 2557–2570 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins J. D., et al. , AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 55, 28–39 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Mercier R., et al. , Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3, whereas the other one is not. Curr. Biol. 15, 692–701 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Jackson N., et al. , Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 25, 1315–1323 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurzbauer M. T., et al. , Arabidopsis thaliana FANCD2 promotes meiotic crossover formation. Plant Cell 30, 415–428 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins J. D., Buckling E. F., Franklin F. C., Jones G. H., Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54, 152–162 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Berchowitz L. E., Francis K. E., Bey A. L., Copenhaver G. P., The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3, e132 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C., et al. , Resolvase OsGEN1 mediates DNA repair by homologous recombination. Plant Physiol. 173, 1316–1329 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olivier M., et al. , The structure-specific endonucleases MUS81 and SEND1 are essential for telomere stability in Arabidopsis. Plant Cell 28, 74–86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crismani W., et al. , FANCM limits meiotic crossovers. Science 336, 1588–1590 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Girard C., et al. , FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 42, 9087–9095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Séguéla-Arnaud M., et al. , Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl. Acad. Sci. U.S.A. 112, 4713–4718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Séguéla-Arnaud M., et al. , RMI1 and TOP3α limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res. 45, 1860–1871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandes J. B., et al. , FIGL1 and its novel partner FLIP form a conserved complex that regulates homologous recombination. PLoS Genet. 14, e1007317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girard C., et al. , AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 11, e1005369 (2015). Correction in: PLoS Genet. 11, e1005448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wijeratne A. J., Chen C., Zhang W., Timofejeva L., Ma H., The Arabidopsis thaliana PARTING DANCERS gene encoding a novel protein is required for normal meiotic homologous recombination. Mol. Biol. Cell 17, 1331–1343 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K., et al. , The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 8, e1002809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambing C., Kuo P. C., Tock A. J., Topp S. D., Henderson I. R., ASY1 acts as a dosage-dependent antagonist of telomere-led recombination and mediates crossover interference in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 117, 13647–13658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., et al. , Fanconi anemia ortholog FANCM regulates meiotic crossover distribution in plants. Plant Physiol. 186, 344–360 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knoll A., et al. , The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell 24, 1448–1464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francis K. E., et al. , Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 3913–3918 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Che R., Zhang J., Nepal M., Han B., Fei P., Multifaceted Fanconi anemia signaling. Trends Genet. 34, 171–183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meetei A. R. M. A., et al. , A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37, 958–963 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobeck A., Stone S., Landais I., de Graaf B., Hoatlin M. E., The Fanconi anemia protein FANCM is controlled by FANCD2 and the ATR/ATM pathways. J. Biol. Chem. 284, 25560–25568 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deans A. J., West S. C., FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol. Cell 36, 943–953 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Panneerselvam J., et al. , BLM promotes the activation of Fanconi Anemia signaling pathway. Oncotarget 7, 32351–32361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wan L., et al. , Scaffolding protein SPIDR/KIAA0146 connects the Bloom syndrome helicase with homologous recombination repair. Proc. Natl. Acad. Sci. U.S.A. 110, 10646–10651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan J., Chen J., FIGNL1-containing protein complex is required for efficient homologous recombination repair. Proc. Natl. Acad. Sci. U.S.A. 110, 10640–10645 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jahns M. T., et al. , Crossover localisation is regulated by the neddylation posttranslational regulatory pathway. PLoS Biol. 12, e1001930 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duroc Y., et al. , The kinesin AtPSS1 promotes synapsis and is required for proper crossover distribution in meiosis. PLoS Genet. 10, e1004674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L., et al. , Topoisomerase II mediates meiotic crossover interference. Nature 511, 551–556 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Cheng Z., Lu P., Timofejeva L., Ma H., “Molecular Cell Biology of Male Meiotic Chromosomes and Isolation of Male Meiocytes in Arabidopsis thaliana” in Flower Development, J. L. Riechmann and F. Wellmer, Eds. (Humana Press, New York, 2014). [DOI] [PubMed] [Google Scholar]
- 42.Armstrong S. J., Sanchez-Moran E., Franklin F. C. H., Cytological analysis of Arabidopsis thaliana meiotic chromosomes. Methods Mol. Biol. 558, 131–145 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Chelysheva L., et al. , An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: Immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 129, 143–153 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Sanchez Moran E., Armstrong S. J., Santos J. L., Franklin F. C. H., Jones G. H., Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 9, 121–128 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Berchowitz L. E., Copenhaver G. P., Fluorescent Arabidopsis tetrads: A visual assay for quickly developing large crossover and crossover interference data sets. Nat. Protoc. 3, 41–50 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.