Abstract
The hypothalamus plays a central role in the integrated regulation of feeding and energy homeostasis. The hypothalamic arcuate nucleus (ARC) contains a population of neurons that express orexigenic and anorexigenic factors and is thought to control feeding behavior via several neuronal circuits. In this study, a comparative proteomic analysis of low-fat control diet- (LFD-) and high-fat diet- (HFD-) induced hypothalamic ARC was performed to identify differentially expressed proteins (DEPs) related to changes in body weight. In the ARC in the hypothalamus, 6621 proteins (FDR < 0.01) were detected, and 178 proteins were categorized as DEPs (89 upregulated and 89 downregulated in the HFD group). Among the Gene Ontology molecular function terms associated with the DEPs, protein binding was the most significant. Fibroblast growth factor receptor substrate 2 (Frs2) and SHC adaptor protein 3 (Shc3) were related to protein binding and involved in the neurotrophin signaling pathway according to Kyoto Encyclopedia of Genes and Genomes analysis. Furthermore, high-precision quantitative proteomic analysis revealed that the protein profile of the ARC in mice with HFD-induced obesity differed from that in LFD mice, thereby offering insight into the molecular basis of feeding regulation and suggesting Frs2 and Shc3 as novel treatment targets for central anorexigenic signal induction.
1. Introduction
Obesity is a common issue worldwide and a cause of various disorders, including hypertension, diabetes, and cardiovascular disorders [1]. Several neural circuits in the brain, including the hypothalamus, solitary nucleus, midbrain, and limbic system, are involved in the maintenance of body weight and appetite [2]. Leptin and insulin regulate feeding behavior and energy homeostasis in the hypothalamus [3, 4], and the arcuate nucleus (ARC) of the hypothalamus is involved in feeding and energy expenditure [5].
The ARC is anatomically located at the lateral wall of the third ventricle and lies directly above the pituitary gland upon the median eminence in the hypothalamus. Several types of neurons in the feeding circuit of the ARC have been discovered, including those expressing agouti-related protein/neuropeptide Y (agouti-related protein [AgRP]/NPY) or proopiomelanocortin (POMC) derivatives [6, 7]. Orexigenic AgRP neurons are negatively regulated by adipocyte-derived leptin, whereas anorexigenic POMC neurons are triggered by leptin to release mainly α-MSH derivatives that affect nuclei, including the paraventricular nucleus (PVN) of the brain [8, 9]. Aside from several peptides and neural pathways, the molecular mechanisms underlying the regulation of the feeding circuit in the ARC have not been fully elucidated. Considering its pivotal role in food intake and satiety, we hypothesized that proteome profiling of the ARC would uncover critical circuits related to obesity. Although proteomic analyses of the hypothalamus in a high-fat diet- (HFD-) induced obesity (DIO) model have been performed [9], few studies have focused specifically on the ARC.
In this study, we characterized the ARC proteome in order to identify protein alterations in mice exposed to an HFD and provide new insights into the molecular basis of appetite regulation and energy metabolism.
2. Materials and Methods
2.1. Animals
Male C57BL/6 mice (7 weeks old; 20–25 g; n = 10) were purchased from Orient Bio (Seongnam, Korea) and maintained at 23 ± 1°C under a 12 h light/dark cycle starting at 8:00 A.M., with ad libitum access to the designated type of food and water in a breeding room. Body weights were measured weekly.
2.2. Ethics Statement
This study was carried out in accordance with the Guide for the Care and Use of Laboratory Animals of Seoul National University Hospital. The protocol was approved by the Institutional Animal Care and Use Committee of the Seoul National University Hospital (protocol 18-0154-S1A1).
2.3. DIO Model
All mice underwent a week of adjustment to the housing environment and were fed a chow diet. The mice were then randomly assigned to two groups (n = 5/group) for the 8-week experimental period. One group was fed a low-fat control diet (LFD; rodent diet with 10% of energy from fat and 3.8 kcal/g), and the other was fed an HFD (60% of energy from fat and 5.2 kcal/g) ad libitum for 8 weeks (D12450B and D12492, respectively; Research Diets, Brunswick, NJ, USA). Mean body weights were measured weekly in both groups to evaluate the reliability of the DIO model.
2.4. Sample Preparation
After 8 weeks, each mouse was anesthetized with a ketamine-xylazine mixture (80–120 mg/kg) and intracardially perfused with sterilized cold saline and 4% paraformaldehyde. Brains were extracted, snap-frozen over liquid nitrogen, and stored at −80°C. A punch biopsy was performed, as described previously, to excise ARC tissue samples from the brain [10, 11], and the samples were deep-frozen immediately after sampling.
2.5. Protein Quantification
Each ARC sample was collected, as described previously [10, 11], and left and right samples were pooled and subjected to BCA/Bradford peptide assay for peptide extraction before quantification. Peptide sequences in the extracts were profiled in depth using a quadrupole-orbitrap mass spectrometer (Q Exactive Plus; Thermo Fisher Scientific, Waltham, MA, USA). Peptides were identified using the MaxQuant database (Max Planck Institute of Biochemistry, Munich, Germany) and assigned with a false discovery rate (FDR) < 1% [12]. The level of peptide expression was calculated using intensity-based absolute quantification (iBAQ) [13]. Peptides identified in at least one sample were included in the final list of valid proteins, which was imported to the Perseus platform (Max Planck Institute of Biochemistry) for statistical analyses and detection of differentially expressed proteins (DEPs).
2.6. Functional Pathway Analysis
The list of DEPs was uploaded via the STRING 11.0 tool to the STRING database (http://string-db.org) to generate a protein-protein interaction network (ELIXIR Core Data Resource) [14], and DEPs were searched against the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.kegg.jp/kegg/) and Gene Ontology (GO) (http://geneontology.org/) databases to evaluate functional enrichment. The overall characteristics of DEPs were examined by GO enrichment analysis [15] using the Biological Networks Gene Ontology (BiNGO) plug-in of Cytoscape [16]. Biochemical pathways involving two or more DEPs were determined [17].
2.7. Statistical Analysis
Bodyweight data are shown as means ± standard error of the mean and analyzed with two-way repeated measures analysis of variance implemented in SPSS (v.24.0; IBM Corp., Armonk, NY, USA).
For proteomic analysis for each protein, iBAQ intensities were transformed to log2 values, and missing values were imputed based on the Gaussian distribution. We then performed width adjustment (a process to equalize the quartiles of intensity values between the samples) for normalization. Student's t-tests were used for comparisons between the normalized iBAQ intensities of the HFD and LFD groups, and fold change values (defined as the ratio of the normalized mean iBAQ intensities of group HFD to group LFD) were then calculated. A DEP was defined as a protein with a p < 0.05 and either a fold change > 1.5 (upregulated protein) or <1.0/1.5 (downregulated protein). A volcano plot was generated for the DEPs based on p values and fold changes.
3. Results
3.1. Mouse Model of HFD-Induced Obesity
After unrestricted administration of an HFD for 8 weeks, a DIO model was successfully established. From 3 weeks (LFD vs. HFD: 28.06 ± 0.18 g vs. 31.16 ± 0.86 g, F = 12.501) to 8 weeks (LFD vs. HFD: 33.38 ± 0.64 g vs. 44.12 ± 0.62 g, F = 102.399) after the adjustment period, the mean weight in the HFD group was significantly higher than that in the LFD group (Figure 1(a)). After 15 weeks, the HFD group showed a significantly higher body weight than the LFD group (p < 0.0001) (Figure 1(b)).
Figure 1.
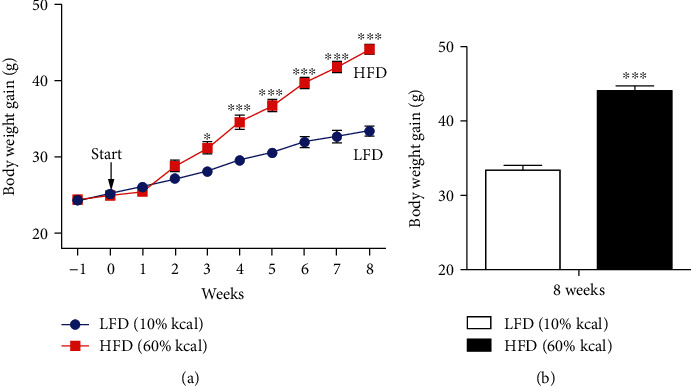
Mean change in body weight. (a) Mean body weights in the LFD (n = 5) and HFD (n = 5) groups were measured weekly from the adjustment period to the end of the experiment. (b) The mean body weight in the HFD group was significantly higher (p < 0.001) than that in the LFD group at the week of sacrifice (8 weeks after the adjustment period). ∗p < 0.05, ∗∗∗p < 0.001, two-way repeated measures analysis of variance. LFD: low-fat control diet; HFD: high-fat diet.
3.2. Protein Quantification
Left and right ARC tissue samples were pooled and used for peptide extraction and quantification. In total, 6,621 proteins were identified in the LFD and HFD groups. A volcano plot was generated to visualize the proteins and display the distribution and significant proteins (determined by −log10p values for the mean normalized iBAQ intensity). In total, 336 proteins were identified at a threshold p value of 0.05. After filtering with the fold change threshold, 178 proteins were identified as DEPs, including 89 upregulated DEPs (fold change ≥ 1.5) and 89 downregulated DEPs (fold change ≤ 0.66) (Figure 2(a), Table S1). Hierarchical clustering revealed a distinct proteomic signature in the hypothalamus under HFD and LFD. The normalized iBAQ intensities of the 178 DEPs and 6,621 valid proteins for each sample (five samples each for the LFD and HFD groups) were visualized using a heatmap (Figures 2(b) and 2(c)).
Figure 2.
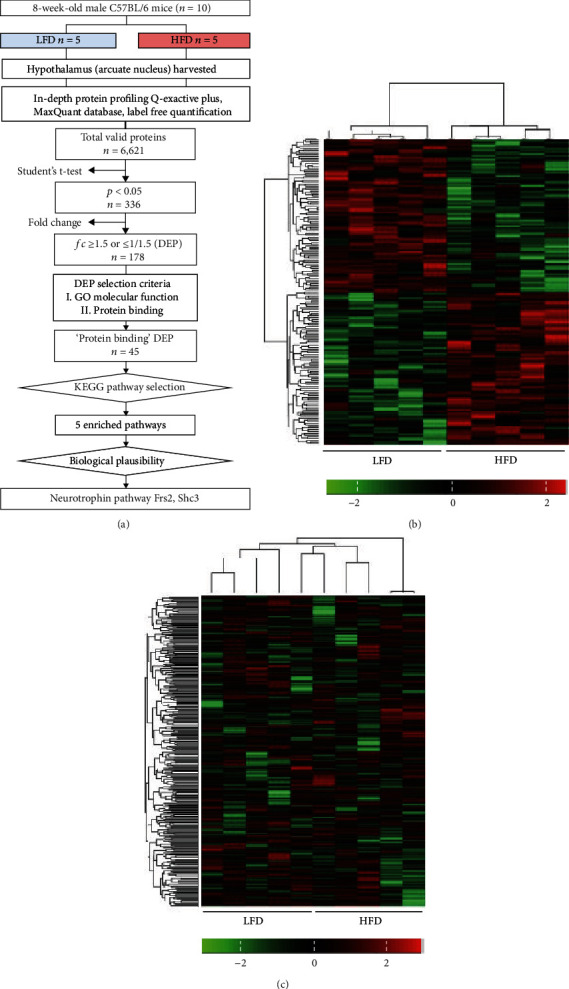
Pipeline and heatmap of DEPs. (a) A total of 178 proteins were differentially expressed between the two models. Heatmap of normalized iBAQ intensity values of (b) each DEP (total 178) and (c) each valid protein (total 6,621). Scale bars are shown on the bottom right corner of each image. Left columns represent the values from the LFD group, and right columns are from the HFD group. Hierarchical clustering analysis is based on Euclidean distances. DEP: differentially expressed protein; LFD: low-fat control diet; HFD: high-fat diet.
3.3. iBAQ Analysis of the Neurotrophin Signaling Pathway
We evaluated a candidate pathway (the neurotrophin signaling pathway) based on previous results demonstrating its important roles in neurogenesis, axogenesis, and synaptic plasticity in the hypothalamus and feeding and energy imbalances due to obesity [18, 19]. Among the DEPs in our analysis, we detected two proteins (fibroblast growth factor (FGF) receptor substrate 2 (Frs2) and SHC adaptor protein 3 (Shc3)) involved in the neurotrophin signaling pathway (Figure 3(a)). The intensity-based quantification confirmed that Frs2 and Shc3 levels were lower in the HFD group than in the control group (Figures 3(b) and 3(c)).
Figure 3.
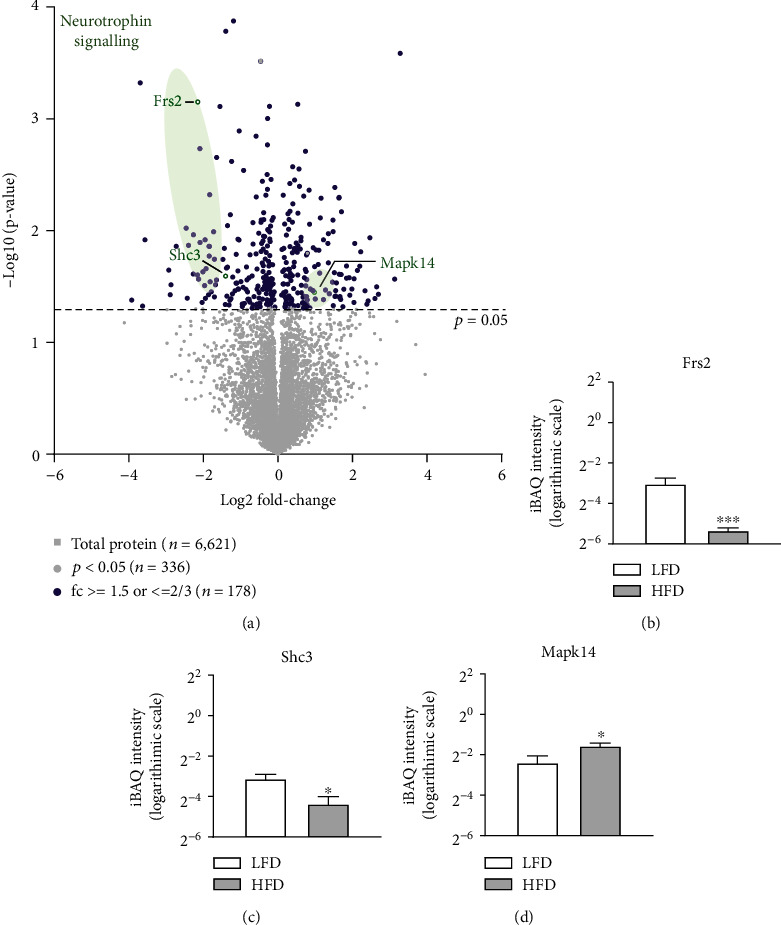
Volcano plot of DEPs. (a) The statistical significance (−log10p value between the mean iBAQ intensities of the LFD and HFD groups) is plotted against the difference in expression intensity (log2 fold change between the mean iBAQ intensities of the LFD and HFD groups) in a volcano plot. Green dots represent Frs2 and Shc3 in the neurotrophin signaling pathway. Data were filtered in accordance with the cut-off value of p < 0.05 and a fold change ≥ 1.5 or ≤1.0/1.5. Bar graphs indicate the mean and SEM of the iBAQ intensity (logarithmic scale) in each group of proteins. (b) Frs2 (p = 7.1 × 10−4, fold change = 0.26), (c) Shc3 (p = 0.03, fold change = 0.38), and (d) Mapk14 (p = 0.0753, fold change = 1.4146). LFD: low-fat control diet; HFD: high-fat diet; iBAQ: intensity-based absolute qualification; SEM: standard error of the mean; Shc3: SHC adaptor protein 3; Frs2: fibroblast growth factor receptor substrate 2; Mapk14: mitogen-activated protein kinase 14. ∗p < 0.05.
3.4. KEGG Pathway Analysis
The DEPs were further evaluated by KEGG pathway analysis, with a KEGG pathway diagram generated after deleting unrelated molecules and redundant data. Among 45 DEPs related to “protein binding,” five KEGG pathways that included ≥3 DEPs related to this term were further explored. The threshold was set to three DEPs, because the number of KEGG pathways was high (n = 16) when the threshold was set to two DEPs, which increased the risk of false-positive results. The five selected KEGG pathways and their corresponding genes were as follows: mitogen-activated protein kinase (MAPK) signaling pathway (Mapk14, Map3k12, and Rps6ka4), thermogenesis (Mapk14, Frs2, Prkab2, and Uqcrh), proteoglycans in cancer (Mapk14, Frs2, and Lum), Salmonella infection (Mapk14, Arpc5, and Wasf2), and neurotrophin signaling pathway (Mapk14, Frs2, and Shc3). The roles of the MAPK signaling pathway and the thermogenesis pathway are already clearly established; therefore, these pathways were excluded from further analyses. In addition, proteoglycans in cancer and Salmonella infection pathways were excluded, because they were highly unlikely to have therapeutic value in obesity. Accordingly, the neurotrophin signaling pathway (including Mapk14, Frs2, and Shc3) was selected as a key pathway for further analyses (Figure 4). The modified pathway diagram suggests that these proteins might suppress axonal outgrowth (Figure 5).
Figure 4.
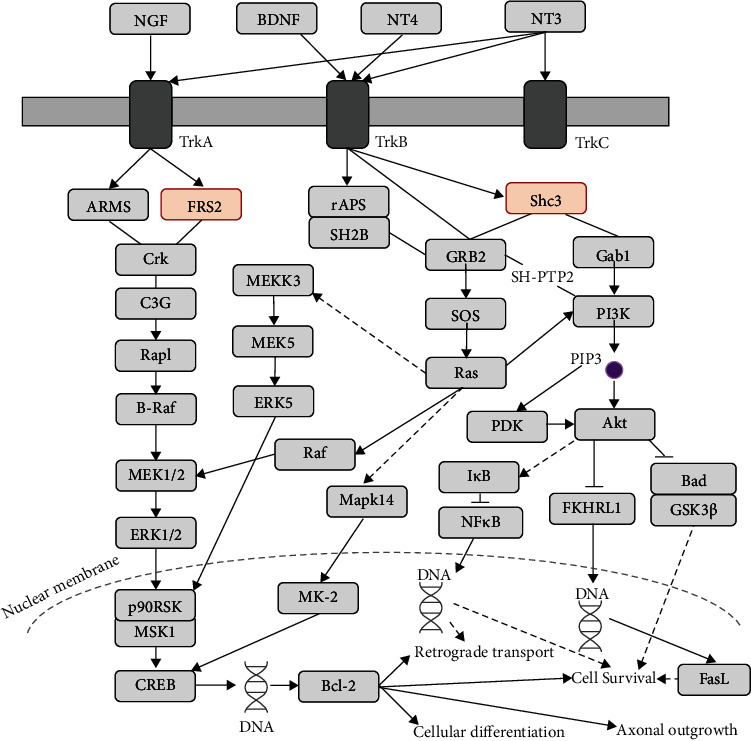
Neurotrophin signaling pathway and corresponding DEPs. “Neurotrophin signaling pathway” within the overall “full” KEGG pathway. DEP: differentially expressed protein; NGF: nerve growth factor; BDNF: brain-derived neurotrophic factor; NT3,4: neurotrophin 3,4; TrkA/B/C: neurotrophic receptor tyrosine kinase 1, 2, and 3; ARMS: ankyrin repeat-rich membrane-spanning protein; FRS2: fibroblast growth factor receptor substrate 2; CREB: CAMP-responsive element-binding protein; GRB2: growth factor receptor-bound protein 2; SOS: Son of Sevenless; Mapk14: mitogen-activated protein kinase 14; MK-2: mitogen-activated protein kinase-activated protein kinase 2; Gab1: GRB2-associated binding protein 1; PI3K: phosphoinositide-3-kinase; PIP3: phosphatidylinositol-3,4,5-trisphosphate; PDK: 3-phosphoinositide-dependent protein kinase-1; Bad: BCL2-associated agonist of cell death; GSK3β: glycogen synthase kinase 3 beta; FasL: Fas ligand.
Figure 5.
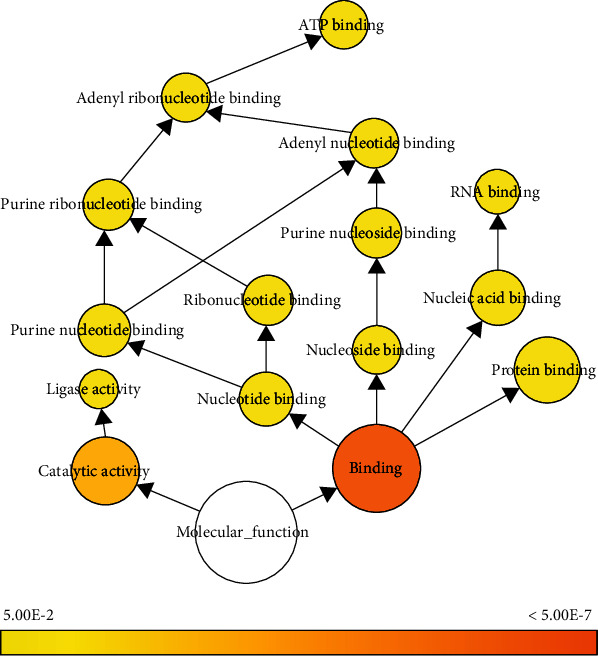
Molecular functions according to GO pathways associated with DEPs and visualized using the Biological Networks GO plug-in. Colored nodes correspond to GO terms that were significant according to a p = 0.05. Node sizes represent the number of DEPs related to the corresponding GO term. GO: Gene Ontology; DEP: differentially expressed gene.
3.5. GO Functional Enrichment Analysis
According to GO enrichment analysis and the BiNGO network (Tables 1–3), DEPs were enriched for various GO terms and particularly in two categories: biological processes and cellular components (Figures S1 and S2). BiNGO enabled graphical visualization of several DEPs and p values for each GO term (Figure 4). The results showed that enriched terms in the molecular function category included “binding” (p = 3.9 × 10−7, 90 DEPs) and catalytic activity (p = 6.9 × 10−4, 47 DEPs) (Figure 4). Compared with catalytic activity, “binding” included more DEPs and was more highly significant. Among subgroups of the “binding” term, “protein binding” (p = 0.02, 45 DEPs) was the most highly significant and included the most DEPs. The DEPs related to “protein binding” were further evaluated for involvement in KEGG pathways, revealing one pathway related to the hypothalamic ARC (i.e., the neurotrophin signaling pathway) (Figure 4).
Table 1.
GO pathway analysis—biological process.
| Term (GO ID) | Reference | Observed | Expected | F.E. | p value | FDR |
|---|---|---|---|---|---|---|
| Negative regulation of protein kinase activity by regulation of protein phosphorylation (0044387) | 8 | 3 | 0.06 | 47.78 | 7.32E − 05 | 3.99E − 02 |
| mRNA catabolic process (0006402) | 119 | 7 | 0.93 | 7.49 | 5.97E − 05 | 3.37E − 02 |
| RNA metabolic process (0016070) | 1234 | 25 | 9.69 | 2.58 | 1.56E − 05 | 1.03E − 02 |
| Nucleic acid metabolic process (0090304) | 1770 | 34 | 13.89 | 2.45 | 1.58E − 06 | 1.39E − 03 |
| Nucleobase-containing compound metabolic process (0006139) | 2194 | 41 | 17.22 | 2.38 | 1.55E − 07 | 3.05E − 04 |
| Organic cyclic compound metabolic process (1901360) | 2618 | 46 | 20.55 | 2.24 | 1.92E − 07 | 3.37E − 04 |
| Organic substance metabolic process (0071704) | 6707 | 89 | 52.64 | 1.69 | 1.16E − 08 | 4.56E − 05 |
| Metabolic process (0008152) | 7224 | 98 | 56.7 | 1.73 | 1.94E − 10 | 1.54E − 06 |
| Cellular nitrogen compound metabolic process (0034641) | 2791 | 46 | 21.91 | 2.1 | 9.02E − 07 | 8.90E − 04 |
| Nitrogen compound metabolic process (0006807) | 5727 | 79 | 44.95 | 1.76 | 2.98E − 08 | 9.41E − 05 |
| Cellular metabolic process (0044237) | 6339 | 93 | 49.75 | 1.87 | 1.01E − 11 | 1.59E − 07 |
| Cellular process (0009987) | 14067 | 143 | 110.41 | 1.3 | 1.40E − 07 | 3.15E − 04 |
| Heterocycle metabolic process (0046483) | 2331 | 42 | 18.3 | 2.3 | 3.27E − 07 | 5.17E − 04 |
| Cellular aromatic compound metabolic process (0006725) | 2405 | 41 | 18.88 | 2.17 | 1.96E − 06 | 1.55E − 03 |
| Primary metabolic process (0044238) | 6275 | 88 | 49.25 | 1.79 | 1.00E − 09 | 5.28E − 06 |
| Macromolecule metabolic process (0043170) | 5065 | 70 | 39.75 | 1.76 | 4.34E − 07 | 5.71E − 04 |
| Nucleobase-containing compound catabolic process (0034655) | 234 | 13 | 1.84 | 7.08 | 7.58E − 08 | 2.00E − 04 |
| Aromatic compound catabolic process (0019439) | 292 | 13 | 2.29 | 5.67 | 8.47E − 07 | 8.92E − 04 |
| Cellular nitrogen compound catabolic process (0044270) | 272 | 13 | 2.13 | 6.09 | 3.94E − 07 | 5.66E − 04 |
| Organic cyclic compound catabolic process (1901361) | 323 | 13 | 2.54 | 5.13 | 2.48E − 06 | 1.87E − 03 |
| Heterocycle catabolic process (0046700) | 278 | 13 | 2.18 | 5.96 | 4.99E − 07 | 6.06E − 04 |
| Cellular macromolecule metabolic process (0044260) | 3967 | 58 | 31.14 | 1.86 | 1.31E − 06 | 1.22E − 03 |
| rRNA processing (0006364) | 199 | 9 | 1.56 | 5.76 | 3.84E − 05 | 2.25E − 02 |
| Cellular component organization or biogenesis (0071840) | 5284 | 67 | 41.47 | 1.62 | 2.29E − 05 | 1.45E − 02 |
| rRNA metabolic process (0016072) | 208 | 10 | 1.63 | 6.13 | 8.53E − 06 | 6.13E − 03 |
| ncRNA metabolic process (0034660) | 419 | 15 | 3.29 | 4.56 | 1.67E − 06 | 1.39E − 03 |
| ncRNA processing (0034470) | 343 | 12 | 2.69 | 4.46 | 2.35E − 05 | 1.43E − 02 |
| Cellular biosynthetic process (0044249) | 1984 | 34 | 15.57 | 2.18 | 1.46E − 05 | 1.00E − 02 |
| Biosynthetic process (0009058) | 2157 | 34 | 16.93 | 2.01 | 9.15E − 05 | 4.67E − 02 |
| G protein-coupled receptor signaling pathway (0007186) | 1849 | 0 | 14.51 | <0.01 | 5.21E − 07 | 5.88E − 04 |
| Sensory perception of chemical stimulus (0007606) | 1225 | 0 | 9.61 | <0.01 | 8.20E − 05 | 4.32E − 02 |
GO: Gene Ontology; FDR: false discovery rate; F.E.: fold enrichment.
Table 2.
GO pathway analysis—cellular component.
| Term (GO ID) | Reference | Observed | Expected | F.E. | p value | FDR |
|---|---|---|---|---|---|---|
| Small-subunit processome (0032040) | 37 | 5 | 0.29 | 17.22 | 1.82E − 05 | 3.01E − 03 |
| Preribosome (0030684) | 81 | 6 | 0.64 | 9.44 | 6.13E − 05 | 7.16E − 03 |
| Protein-containing complex (0032991) | 5312 | 64 | 41.69 | 1.54 | 1.69E − 04 | 1.59E − 02 |
| Nucleolus (0005730) | 808 | 19 | 6.34 | 3 | 2.58E − 05 | 3.66E − 03 |
| Nuclear lumen (0031981) | 3906 | 60 | 30.66 | 1.96 | 1.22E − 07 | 3.02E − 05 |
| Intracellular organelle lumen (0070013) | 4314 | 64 | 33.86 | 1.89 | 1.31E − 07 | 2.88E − 05 |
| Organelle lumen (0043233) | 4315 | 64 | 33.87 | 1.89 | 1.31E − 07 | 2.61E − 05 |
| Membrane-enclosed lumen (0031974) | 4315 | 64 | 33.87 | 1.89 | 1.31E − 07 | 2.37E − 05 |
| Cellular anatomical entity (0110165) | 18664 | 170 | 146.49 | 1.16 | 2.31E − 08 | 6.56E − 06 |
| Organelle (0043226) | 12317 | 140 | 96.68 | 1.45 | 1.14E − 11 | 2.27E − 08 |
| Intracellular organelle (0043229) | 11971 | 135 | 93.96 | 1.44 | 2.06E − 10 | 2.05E − 07 |
| Intracellular (0005622) | 13708 | 146 | 107.59 | 1.36 | 3.61E − 10 | 2.39E − 07 |
| Nucleus (0005634) | 6832 | 79 | 53.62 | 1.47 | 6.93E − 05 | 7.24E − 03 |
| Intracellular membrane-bounded organelle (0043231) | 10296 | 119 | 80.81 | 1.47 | 7.41E − 09 | 2.45E − 06 |
| Membrane-bounded organelle (0043227) | 11045 | 127 | 86.69 | 1.46 | 1.05E − 09 | 5.21E − 07 |
| Intracellular nonmembrane-bounded organelle (0043232) | 4175 | 55 | 32.77 | 1.68 | 5.81E − 05 | 7.20E − 03 |
| Nonmembrane-bounded organelle (0043228) | 4194 | 55 | 32.92 | 1.67 | 6.20E − 05 | 6.84E − 03 |
| Mitochondrion (GO:0005739) | 1798 | 30 | 14.11 | 2.13 | 1.22E − 04 | 1.22E − 02 |
| Cytoplasm (0005737) | 10948 | 126 | 85.93 | 1.47 | 1.10E − 09 | 4.37E − 07 |
| Cytosol (0005829) | 3534 | 50 | 27.74 | 1.8 | 2.57E − 05 | 3.92E − 03 |
| Nucleoplasm (0005654) | 3331 | 47 | 26.14 | 1.8 | 4.51E − 05 | 5.97E − 03 |
GO: Gene Ontology; FDR: false discovery rate; F.E.: fold enrichment.
Table 3.
GO pathway analysis—molecular function.
| Term (GO ID) | Reference | Observed | Expected | F.E. | p value | FDR |
|---|---|---|---|---|---|---|
| RNA binding (0003723) | 1091 | 23 | 8.56 | 2.69 | 1.92E − 05 | 2.96E − 02 |
| Binding (0005488) | 13336 | 133 | 104.67 | 1.27 | 8.63E − 06 | 1.99E − 02 |
| Transmembrane signaling receptor activity (0004888) | 2135 | 2 | 16.76 | 0.12 | 7.22E − 06 | 3.33E − 02 |
GO: Gene Ontology; FDR: false discovery rate; F.E.: fold enrichment.
4. Discussion
This study identified DEPs in the ARC between mice with HFD-induced obesity and mice fed an LFD. Glutamate transporters (e.g., GLT and GLAST) are among the exceptionally abundant and important proteins in the central nervous system (~>2.5% of the brain protein mass exists in the form of glutamate transporters) and critical for the function of glutamatergic signaling (the balance between glutamate excitation and GABA inhibition is particularly important and relevant for body mass control mechanisms in the hypothalamus). Although proteomic changes in the hypothalamus of an obesity model have been studied [9], our comparison of the ARC in LFD and HFD groups was novel. Moreover, in-depth profiling of the global proteome targeting the ARC using the punch biopsy method revealed various proteins that have not previously been reported in studies of obesity or energy metabolism.
Recent studies suggest that tumor necrosis factor α and interleukin 1β, especially in AgRP-producing neurons, might contribute to inflammation-induced anorexia during acute inflammatory conditions [20]. Additionally, single-cell RNA sequencing of the hypothalamic ARC after HFD-induced obesity demonstrated selective changes in AgRP neurons via neuron-astrocyte interactions, which contributed to the exaggerated sympathoexcitation observed in obese rats [21, 22]. These effects might be mediated by the regulatory effects of leptin on astrocytic glutamate transporters within the ARC of the hypothalamus [23].
GO enrichment analysis identified binding and catalytic activity as key molecular functions. Notably, the DEPs related to protein binding were involved in the neurotrophin signaling pathway. These results were consistent with those of previous reports indicating that the neurotrophin signaling pathway contributes to neurogenesis, axogenesis, and synaptic plasticity in the hypothalamus and dysfunctions in feeding and energy balance due to obesity [18, 19]. “Binding” was a highly significant term and included a number of DEPs, and the subgroup “protein binding” was further identified as a significant molecular function. DEPs associated with “protein binding,” particularly Frs2 and Shc3, were involved in the neurotrophin signaling pathway. This pathway is generally involved in retrograde transport, cell survival, cellular differentiation, and axonal outgrowth of neurons and triggered by the binding of extracellular signaling molecules, such as nerve growth factor, brain-derived neurotrophic factor (BDNF), and neurotrophins 4 and 3. Furthermore, a previous study reported that the distribution of tropomyosin receptor kinase B- (TrkB-) expressing neurons is altered in response to an HFD in the mediobasal hypothalamus [24].
These results showed that Frs2 and Shc3 levels and the related neurotrophin pathway were altered in the ARC of a mouse model of HFD-induced obesity. Neurotrophins, which mediate receptors and signaling molecules, play multiple roles in survival and development [25]. FRS2 functions by transmitting FGF and/or neurotrophin signaling during dorsal forebrain development, with a critical role in the formation of the hippocampal dentate gyrus [26]. Additionally, ARCTrkB neurons regulate food intake by projecting to BDNF-expressing neurons in the PVN [27].
A previous study reported that BDNF deficiency signaled a reduced number of axonal projections of ARCTrkB to PVN, subsequently inducing hyperphagia [28]. SHC molecules and neurotrophins bind to Trks and the neurotrophin receptor [25, 29]. In the present study, the DEPs included Frs2 and Shc3, which represent novel candidate pharmaceutical targets for obesity based on their close links to pathways involved in obesity. Furthermore, these two proteins identified by in-depth profiling of the ARC using the punch biopsy method have not previously been reported in studies of obesity or energy metabolism. However, the central effects of Frs2 or Shc3 remain unknown. To fully understand their roles in the regulation of energy expenditure and food intake, further studies are needed, such as an intracerebroventricular injection study along with analyses of hourly and daily food intake and metabolic parameters.
5. Conclusions
We identified 178 DEPs in the ARC of a reliable mouse model of HFD-induced obesity. These DEPs play key roles in protein binding and the neurotrophin signaling pathway and include Frs2 and Shc3 as novel candidates involved in appetite and energy metabolism.
Acknowledgments
We thank Ki Soon Dan and Hye-Yeon Kim from the Proteomics & Biomarker Lab (Seoul National University College of Medicine, Seoul, Korea) for their contributions to the proteomic analysis. We also thank Min Sun Kim for providing technical assistance. This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korean Government (MSIT) (grant no. NRF-2018R1A5A2025964) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant no. 2017R1A6A3A1131814) and the Seoul National University Hospital Research Fund (grant no. 26-2016-0030).
Contributor Information
Cherl NamKoong, Email: nkcfe@snu.ac.kr.
Hyung Jin Choi, Email: hjchoi@snu.ac.kr.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Chang Yeon Kim and Jang Ho Ahn contributed equally to this study.
Supplementary Materials
Figure S1: GO pathway—biological process. Figure S2: GO pathway—cellular component.
Table S1: DEPs and iBAQ intensity values in the HFD group relative to the LFD group.
References
- 1.Scherer P. E., Hill J. A. Obesity, diabetes, and cardiovascular Diseases. Circulation Research . 2016;118(11):1703–1705. doi: 10.1161/CIRCRESAHA.116.308999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith J. K. Exercise, obesity and CNS control of metabolic homeostasis: a review. Frontiers in Physiology . 2018;9:p. 574. doi: 10.3389/fphys.2018.00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jennings J. H., Rizzi G., Stamatakis A. M., Ung R. L., Stuber G. D. The inhibitory circuit architecture of the lateral hypothalamus orchestrates feeding. Science . 2013;341(6153):1517–1521. doi: 10.1126/science.1241812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hommel J. D., Trinko R., Sears R. M., et al. Leptin receptor signaling in midbrain dopamine neurons regulates feeding. Neuron . 2006;51(6):801–810. doi: 10.1016/j.neuron.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 5.Joly-Amado A., Cansell C., Denis R. G. P., et al. The hypothalamic arcuate nucleus and the control of peripheral substrates. Best Practice & Research Clinical Endocrinology & Metabolism . 2014;28(5):725–737. doi: 10.1016/j.beem.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Aponte Y., Atasoy D., Sternson S. M. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nature Neuroscience . 2011;14(3):351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhan C., Zhou J., Feng Q., et al. Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. Journal of Neuroscience . 2013;33(8):3624–3632. doi: 10.1523/JNEUROSCI.2742-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui H., Lopez M., Rahmouni K. The cellular and molecular bases of leptin and ghrelin resistance in obesity. Nature Reviews Endocrinology . 2017;13(6):338–351. doi: 10.1038/nrendo.2016.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manousopoulou A., Koutmani Y., Karaliota S., et al. Hypothalamus proteomics from mouse models with obesity and anorexia reveals therapeutic targets of appetite regulation. Nutrition & Diabetes . 2016;6(4, article e204) doi: 10.1038/nutd.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim M.-S., Pak Y. K., Jang P.-G., et al. Role of hypothalamic Foxo1 in the regulation of food intake and energy homeostasis. Nature Neuroscience . 2006;9(7):901–906. doi: 10.1038/nn1731. [DOI] [PubMed] [Google Scholar]
- 11.Palkovits M. Punch sampling biopsy technique. Methods in Enzymology . 1983;103:368–376. doi: 10.1016/S0076-6879(83)03025-6. [DOI] [PubMed] [Google Scholar]
- 12.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature Protocols . 2016;11(12):2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 13.Schwanhäusser B., Busse D., Li N., et al. Global quantification of mammalian gene expression control. Nature . 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 14.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research . 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Research . 2019;47(D1):D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maere S., Heymans K., Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics . 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Research . 2019;47(D1):D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitre M., Mariga A., Chao M. V. Neurotrophin signalling: novel insights into mechanisms and pathophysiology. Clinical Science . 2017;131(1):13–23. doi: 10.1042/CS20160044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.An J. J., Kinney C. E., Tan J.-W., Liao G.-Y., Kremer E. J., Xu B. TrkB-expressing paraventricular hypothalamic neurons suppress appetite through multiple neurocircuits. Nature Communications . 2020;11(1):p. 1729. doi: 10.1038/s41467-020-15537-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaves F. M., Mansano N. S., Frazão R., Donato J., Jr. Tumor necrosis factor α and interleukin-1β acutely inhibit AgRP neurons in the arcuate nucleus of the hypothalamus. International Journal of Molecular Sciences . 2020;21(23):p. 8928. doi: 10.3390/ijms21238928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng G., Morselli L. L., Wagner V. A., et al. Single-nucleus RNA sequencing of the hypothalamic arcuate nucleus of C57BL/6J mice after prolonged diet-induced obesity. Hypertension . 2020;76(2):589–597. doi: 10.1161/HYPERTENSIONAHA.120.15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X., Zheng H. Leptin-mediated sympathoexcitation in obese rats: role for neuron-astrocyte crosstalk in the arcuate nucleus. Frontiers in Neuroscience . 2019;13:p. 1217. doi: 10.3389/fnins.2019.01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barkholt P., Rigbolt K. T. G., Falkenhahn M., et al. Global transcriptome analysis of rat hypothalamic arcuate nucleus demonstrates reversal of hypothalamic gliosis following surgically and diet induced weight loss. Scientific Reports . 2019;9(1, article 16161) doi: 10.1038/s41598-019-52257-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramalho A. F., Bombassaro B., Dragano N. R., et al. Dietary fats promote functional and structural changes in the median eminence blood/spinal fluid interface—the protective role for BDNF. Journal of Neuroinflammation . 2018;15(1):p. 10. doi: 10.1186/s12974-017-1046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bibel M., Barde Y. A. Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes & Development . 2020;14(23):2919–2937. doi: 10.1101/gad.841400. [DOI] [PubMed] [Google Scholar]
- 26.Nandi S., Alvina K., Lituma P. J., Castillo P. E., Hebert J. M. Neurotrophin and FGF Signaling Adapter Proteins, FRS2 and FRS3, Regulate Dentate Granule Cell Maturation and Excitatory Synaptogenesis. Neuroscience . 2018;369:192–201. doi: 10.1016/j.neuroscience.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu B., Xie X. Neurotrophic factor control of satiety and body weight. Nature Reviews Neuroscience . 2016;17(5):282–292. doi: 10.1038/nrn.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao G.-Y., Bouyer K., Kamitakahara A., et al. Brain-derived neurotrophic factor is required for axonal growth of selective groups of neurons in the arcuate nucleus. Molecular Metabolism . 2015;4(6):471–482. doi: 10.1016/j.molmet.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakazawa T., Nakano I., Sato M., Nakamura T., Tamai M., Mori N. Comparative expression profiles of TrK receptors and Shc-related phosphotyrosine adapters during retinal development: potential roles of N-Shc/ShcC in brain-derived neurotrophic factor signal transduction and modulation. Journal of Neuroscience Research . 2002;68(6):668–680. doi: 10.1002/jnr.10259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: GO pathway—biological process. Figure S2: GO pathway—cellular component.
Table S1: DEPs and iBAQ intensity values in the HFD group relative to the LFD group.
Data Availability Statement
The data used to support the findings of this study are included within the article.


