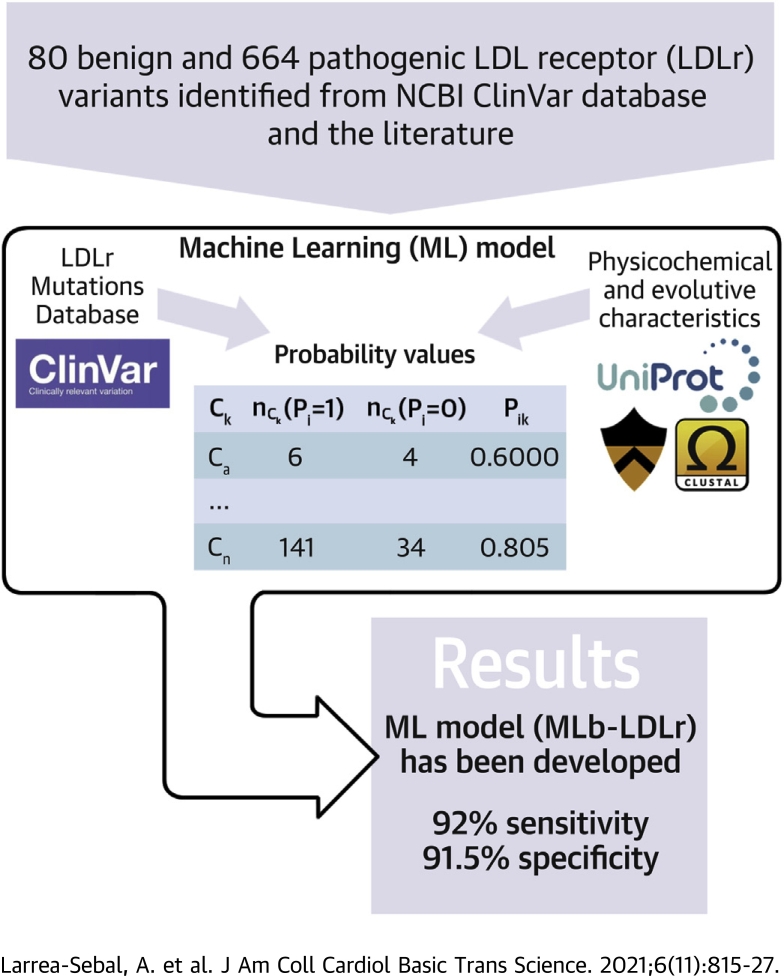
Visual Abstract
Key Words: familial hypercholesterolemia, LDL receptor, machine learning software, pathogenicity, prediction
Abbreviations and Acronyms: ANN, artificial neural network; AUROC, area under the receiver operating curve; EGS, expert-guided selection; ESEA, Excel Solver Evolutionary algorithm; FH, familial hypercholesterolemia; LDA, linear discriminant analysis; LDL, low-density lipoprotein; LDLr, low-density lipoprotein receptor; LNN, linear neural networks; ML, machine learning; MLb-LDLr, machine-learning–based low-density lipoprotein receptor software; MLP, multilayer perceptron; RBF, radial basis function; UTR, untranslated region
Highlights
-
•
A machine-learning model has been developed to improve accuracy on predicting the activity of missense LDLr mutations.
-
•
ClinVar was used as database, and the model function was defined by using specific characteristics of the LDLr.
-
•
A high-score prediction ML model with specificity of 92.5% and sensitivity of 91.6% has been developed to predict pathogenicity of LDLr variants.
-
•
Implementation of high-predicting capacity software constitutes a valuable approach for assessing pathogenicity of LDLr variants to help in the early diagnosis and management of FH disease.
-
•
An open-access predictive software (MLb-LDLr) is provided to the scientific community.
Summary
Untreated familial hypercholesterolemia (FH) leads to atherosclerosis and early cardiovascular disease. Mutations in the low-density lipoprotein receptor (LDLr) gene constitute the major cause of FH, and the high number of mutations already described in the LDLr makes necessary cascade screening or in vitro functional characterization to provide a definitive diagnosis. Implementation of high-predicting capacity software constitutes a valuable approach for assessing pathogenicity of LDLr variants to help in the early diagnosis and management of FH disease. This work provides a reliable machine learning model to accurately predict the pathogenicity of LDLr missense variants with specificity of 92.5% and sensitivity of 91.6%.
Familial hypercholesterolemia (FH) is the most common autosomal dominant disorder with an estimated prevalence between 1:200 and 1:250 (1). FH is characterized by an elevated concentration of low-density lipoprotein (LDL) cholesterol in plasma as a consequence of a defective catabolism of LDL particles (2). The progression of FH is asymptomatic and normally is not detected until advanced stages of the disease when long-term exposure to LDL induces the development of atheroma plaques and increases the risk of cardiovascular diseases (3,4). Despite the high incidence of FH, <1% of the patients are properly diagnosed, and consequently, the implementation of early intervention programs to prevent plasma LDL accumulation and long-term associated problems is limited (5,6).
Mutations in the LDL receptor gene (LDLr) are the most common genetic cause of FH, accounting for more than 90% of the cases (7). To date, more than 3,000 LDLr genetic variants have been described and submitted to the ClinVar database. Among them, missense variants, resulting from single nucleotide substitution, are the most frequent ones (8). Single nucleotide variations can affect (pathogenic) or not (benign) protein structure and function; however, only a reduced number of variants have been functionally validated and proven to be pathogenic (9). Cascade screening and in vitro functional characterization are the most reliable methodologies to validate pathogenicity of LDLr variants (10). Nevertheless, both methods have their own limitations: in vitro functional validation is laborious and time-consuming, whereas cascade screening requires patient clinical data availability and a high number of patients (11). Lately, the use of computational tools has been extended to many fields of medical sciences, including development of predictive software to assess the pathogenicity of protein variants (12,13). Given the high frequency of the mutations found in the LDLr gene, developing specific software to predict LDLr pathogenicity would allow a rapid and systematic characterization of pathogenic variants.
Taking advantage of the large number of missense LDLr variants already characterized and annotated in the ClinVar database, the aim of this work has been to develop an advanced machine learning (ML) algorithm to accurately predict the pathogenicity of LDLr missense variants. To do so, 7 characteristics of the protein have been considered in order to obtain a high-score prediction. The introduction of a ML algorithm provides a predictive model with a specificity of 92.5% and a sensitivity of 91.6%, which shows high accuracy in predicting both pathogenic and benign variants. Here, we provide an open access machine learning–based LDLr predictive software (MLb-LDLr) for the scientific community to predict the pathogenicity of missense LDLr variants (14). Ultimately, the data presented here will help clinicians and researchers interpret the effect of any missense mutation in the LDLr gene.
Methods
Dataset
To date, more than 3,000 LDLr variants have been annotated in the ClinVar database (May 25, 2021). These variants are divided into 6 subclasses according to the type of the mutation: frameshift, missense, nonsense, splice site, noncoding RNA, and untranslated region (UTR). In order to develop an efficient ML diagnostic tool, each subclass must be analyzed individually because a given feature could be important for one subclass, but not relevant for others, taking into account their very different effect. However, most of the subclasses have a reduced number of variants, thus prediction of protein activity is not possible due to the limited available information. In addition to limited data availability, the nature of some mutation subclasses leads to a deleterious effect in most of the variants (ie, frameshift and nonsense mutations) or almost always a benign effect (noncoding RNA and UTR). Therefore, the lack of enough variants with the contrary effect does not allow obtaining an accurate ML model if included. On the other hand, the existence of large amount of both pathogenic and benign LDLr missense variants whose activity has been validated allows the development of a ML predictive software to predict the pathogenicity of these variants.
More than 1,200 missense variants are already annotated in ClinVar, which are distributed according to their pathogenicity as follows: 7 benign, 58 likely benign, 284 of uncertain significance, 239 of conflictive interpretation, 568 likely pathogenic, and 248 pathogenic, although some of these variants have been included into more than 1 subclass. In order to ease the classification, benign and likely benign variants were grouped into a benign subclass (n = 65), and pathogenic and likely pathogenic variants into a pathogenic subclass (n = 639). ClinVar is a great source to evaluate the pathogenicity of LDLr variants, and although most of the used variants have multiple submitters to support their pathogenicity, 1 limitation of the database is that in some cases, there is only 1 submitter, and a few of them have no assertion. Therefore, this has been taken into consideration in the training and validation of the model.
In addition, an exhaustive bibliographic search allowed to ascertaining the effect of variants with conflictive interpretation or uncertain significance and their inclusion in the benign or pathogenic categories. This process allowed increasing the number of variants with a reliable diagnosis. In sum, the data set consisted of 80 benign and 664 pathogenic LDLr variants.
Model definition
A classification model to predict the probability p(Pi = 1)pred of pathogenicity (Pi) of the ith protein variant was defined. Applying these criteria, the model fits the objective function f(Pi)obs. The function f(Pi)obs = 1 when Pi = pathogenic and f(Pi)obs = 0 when Pi = benign. The output of the model is the scoring function f(Pi)calc. This function gets real values and consequently cannot be compared directly to f(Pi)obs. As a consequence, f(Pi)calc was transformed into the searched probability scale values p(Pi = 1)pred. Using these probability values, the predicted classification of each protein variant f(Pi)pred can be obtained. The predicted classification f(Pi)pred = 1 (Pi = pathogenic) when p(Pi = 1)pred >0, otherwise f(Pi)pred = 0 (Pi = benign). Both linear and nonlinear ML models were trained. The general formula for the linear ML model is shown in Equation 1.
| (1) |
The coefficient e0 is the independent term, and ek>0 are the coefficients for each input variable Pik. These coefficients ek>0 quantify the influence (weight) given to each characteristic on the overall pathogenicity. In order to fit the objective function f(Pi)obs, we used as input variable Pik parameters. These Pik parameters are the probabilities with which pathogenic protein variants in the data set present a given value of the characteristic Ck within a given range. We encoded Pik values of each protein variant into the quantitative vector Pik = [Pi1, Pi2, Pi3, … Pi7]. The specific characteristics studied were: C1 = conservation of the substituted residue, C2 = charge change, C3 = original amino acid, C4 = substituting amino acid, C5 = amino acid hydrophobicity change, C6 = amino acid size change, and C7 = affected domain. These characteristics were further divided into 2 different subgroups: physicochemical (C2, C5, C6, and C7) or biological-evolutive (C1, C3, and C4). The values of each characteristic Ck of the 3 continuous variables (C1, C5, and C6) were split into 5 mutually exclusive and equal intervals or classes (c). On the other hand, the values of each characteristic Ck of the discrete variables (C2, C3, C4, and C7) were split into different classes. Characteristic selection and obtention process are shown in the Supplemental Appendix and Supplemental Tables S1 to S4. Ck variables were transformed into Pik probability values according to Equation 2.
| (2) |
In Equation 2, nCk(Pi = 1) is the number of pathogenic protein variants (Pi = 1) with values of Ck within the class c. By analogy, in this formula, nCk(Pi = 0) is the number of benign protein variants (Pi = 0) with values of Ck within the class c. Pik values are shown in Supplemental Table S5.
Model training and validation
As mentioned before, training was performed in both linear and nonlinear models. In the case of the ML linear models, different algorithms were used to define the ek coefficients. The linear ML algorithms used were linear discriminant analysis (LDA) (parametric), linear neural networks (LNN) (nonparametric) from STATISTICA data analysis software system, version 6.0 (StatSoft, Inc.) and Excel Solver Evolutionary algorithm (ESEA) (parametric) (15). In the case of nonlinear models, 2 types of nonlinear artificial neural network (ANN) algorithms were used: multilayer perceptron (MLP) (parametric) and radial basis function (RBF) (nonparametric) (STATISTICA, data analysis software system, version 6.0, StatSoft, Inc.). These models were chosen because they allow testing different types of ML mechanisms, linear and nonlinear, parametric and nonparametric. Linear and parametric models create lineal equations formed by coefficients such as weight or threshold, whereas the nonparametric ones are more complex and use other types of equations.
In order to train/validate the model, the dataset was split into 2 subsets using the variable subset = T (training series) and subset = V (validation series). Variants in training series were used to obtain Pik values and to train the ML models. Variants in validation series were used neither to obtain Pik values nor to train the model. The variants were assigned to training or validation series randomly. Three-quarters of the pathogenic variants (n = 499) were used for training and the remaining (n = 166) to validate the model. In the case of benign variants, two-thirds (n = 54) were used in the training group, and the remaining (n = 26) were used in the validation group due to the limited number of annotated variants.
Variable selection and optimization
In a first stage, linear and ANN ML algorithms from STATISTICA were run with different variable selection strategies: forward stepwise, backward stepwise, etc. Next, ESEA strategy was used. ESEA maximized f(Pi)calc function and increased the number of correctly predicted variants, modifying ek coefficients (see the details in the Supplemental Appendix). The maximum and minimum limits established for weights and threshold were ek>0max = 0.2 and ek>0min = 0.001, and e0max = 1 and e0min = 0.1, respectively. These limit values were set up arbitrarily. In order to obtain a balanced relation between the correctly predicted pathogenic and benign variants, the objective function Equation 3 was described as follows:
| (3) |
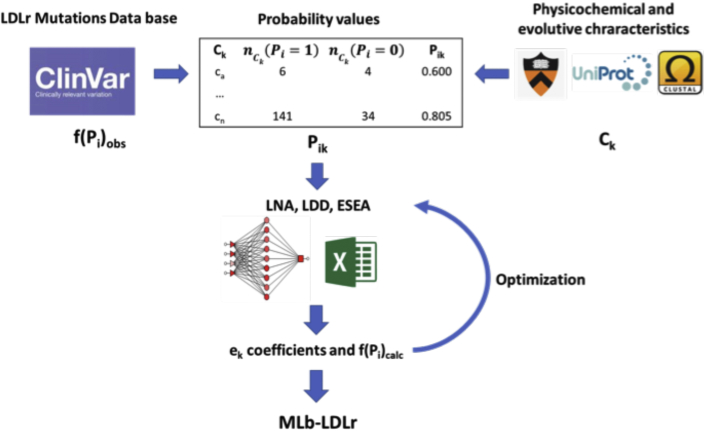
Next, ESEA expert-guided selection (EGS) strategy was used to optimize ek values. EGS was carried out as follows: Once the best fitting weights and threshold were established through ESEA, some Pik values were adjusted to improve the number of correctly predicted variants. Consequently, EGS strategy included a reparameterization of some Pik values, thus increasing the number of correct predictions; see the details in the Supplemental Appendix and Supplemental Table S6. The general workflow of the model is shown in Figure 1.
Figure 1.
General Workflow of the Model
Clinical significance of characterized LDLr variants (f(Pi)obs) and values of physicochemical and evolutive characteristics (Ck) were obtained from several software and databases. Pathogenicity probability values (Pik) were calculated using the relation between pathogenic and benign variants in each characteristic. Then, different ML models (LNN, LDA, ESEA) were applied to obtain the weight of each characteristic on the overall pathogenicity and the threshold that divides pathogenic and benign variants (ek coefficients). These values were later used to calculate a pathogenicity score of each variant (f(Pi)calc). Finally, some f(Pi)calc values were manually modified, and the ML models were applied again in order to optimize ek values. The resulting coefficients were used on MLb-LDLr software. ESEA = Excel Solver Evolutionary algorithm; LDA = linear discriminant analysis; LNN = linear neural network; ML = machine learning; MLb-LDLr = machine-learning–based low-density lipoprotein receptor software.
Statistical analysis
Accuracy of the model was tested by 4 statistic parameters: sensitivity, specificity, positive predictive value, and negative predictive value. The sensitivity and the specificity values refer to the percentage of correctly predicted pathogenic and benign variants, respectively. These parameters were calculated in both training and validation sets.
In addition, random bootstrapping training and validation subsets of the same sample size with replacement were used to test the sampling distribution. One thousand bootstrapped samples were tested, and the statistics previously mentioned were presented with 95% confidence intervals.
Experimental model validation
In vitro functional characterization of ClinVar-annotated conflictive LDLr variants.
LDLr variant selection
Thirteen LDLr variants with conflictive or uncertain interpretations on ClinVar were selected to experimentally validate the accuracy of MLb-LDLr and other predictive software. The selection criteria were based on the disparity of the prediction of the most used pathogenicity-predictive software. Hence, not only the functional characterization of conflictive LDLr variants was performed, but also software accuracy was assessed when facing a hard to predict variant. In addition, the selected variants have been described in patients with elevated levels of LDL cholesterol. Descriptions of the studied variants and in silico predictions are shown in Table 1.
Table 1.
Selected LDLr Variants for In Vitro Characterization and Their Pathogenicity Prediction
| LDLr Variant | MLb-LDLr | PolyPhen-2 | SIFT | SFIP-MutID | MutationTaster | CADD |
|---|---|---|---|---|---|---|
| p.(Phe200Cys) | Pathogenic | Pathogenic | Benign | Pathogenic | Pathogenic | Pathogenic |
| p.(Asn297His) | Pathogenic | Benign | Benign | Pathogenic | Pathogenic | Pathogenic |
| p.(Trp305Ser) | Pathogenic | Pathogenic | Benign | Pathogenic | Pathogenic | Pathogenic |
| p.(Leu371Pro) | Pathogenic | Pathogenic | Benign | Pathogenic | Benign | Pathogenic |
| p.(Gly373Ala) | Pathogenic | Pathogenic | Benign | Benign | Pathogenic | Pathogenic |
| p.(Arg449Gly) | Pathogenic | Benign | Benign | Pathogenic | Pathogenic | Benign |
| p.(Ile473Asn) | Benign | Benign | Pathogenic | Pathogenic | Pathogenic | Benign |
| p.(Val578Ile) | Benign | Pathogenic | Benign | Pathogenic | Benign | Benign |
| p.(Asp638His) | Pathogenic | Pathogenic | Benign | Pathogenic | Pathogenic | Pathogenic |
| p.(Met652Thr) | Benign | Benign | Pathogenic | Pathogenic | Pathogenic | Pathogenic |
| p.(Pro683Leu) | Pathogenic | Pathogenic | Benign | Pathogenic | Benign | Benign |
| p.(Asp700Asn) | Pathogenic | Pathogenic | Benign | Pathogenic | Pathogenic | Benign |
| p.(Ala705Pro) | Benign | Benign | Benign | Pathogenic | Pathogenic | Pathogenic |
CADD = Combined Annotation-Dependent Depletion software; MLb-LDLr = machine-learning–based low-density lipoprotein receptor software; PolyPhen-2 = Polymorphism Phenotyping v2 software; SFIP-MutID = structure-based functional impact prediction for mutation identification; SIFT = Sorting Intolerant From Tolerant software.
CHO-LDLΔ7 cell culture and transfection
The CHO-ldlΔ7 cell line was cultured in Dulbeccós modified eagle medium low glucose, 1 g/L (GE Healthcare) supplemented with 10% fetal bovine serum, 2 mmol/L L-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin. Ten thousand CHO-ldlΔ7 cells were plated into 96-well culture plates and transiently transfected with the plasmids carrying wild-type LDLr or Ex3_4del, c.599T>G p.(Phe200Cys), c.889A>C p.(Asn297His), c.914G>C p.(Trp305Ser), c.1112T>C p.(Lau371Pro), c.1118G>C p.(Gly373Ala), 1345A>G p.(Arg449Gly), c.1418T>A p.(Ile473Asn), 1732G>A p.(Val578Ile), 1912G>C p.(Asp638His), c.1955T>C p.(Met652Thr), c.2049C>T p.(Pro683Leu), c.2098G>A p.(Asp700Asn), and c.2113G>C p.(Ala705Pro) LDLr variants using Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific). Transfected cells were maintained in culture for 48 hours to achieve maximal LDLr expression.
Quantification of LDLr activity
LDLr activity was determined by flow cytometry in CHO-ldlΔ7 cells transfected with plasmids encoding the LDLr variants as previously described (16). Transfected CHO-ldlΔ7 cells were grown in 96-well culture plates. Forty-eight hours after transfection, cells were incubated 4 hours at 37 °C with 20 μg/mL fluorescein isothiocyanate–LDL. Cells were then washed twice in phosphate-buffered saline and incubated with phosphate-buffered saline 5% EDTA for 10 min. To determine the amount of internalized LDL, Trypan blue solution (Sigma-Aldrich) was added directly to the samples to a final concentration of 0.2% (v/v) to extinguish the extracellular signal by dynamic quenching of the noninternalized LDLr-LDL complexes (17). Fluorescence intensities were measured by flow cytometry in a CytoFlex cytometer (Beckman Coulter) according to the manufacturer’s instructions. For each sample, fluorescence of 10,000 events was acquired for data analysis, and the results were expressed as the mean fluorescence.
Ethical approval
All methods were carried out in accordance with relevant guidelines and regulations. This study was approved by the research ethics committee from the University of the Basque Country (Comité de Ética en la Investigación y la Práctica Docente de la Universidad del País Vasco/Euskal Herriko Unibertsitatea, CEID/IIEB).
Results
Computational model for pathogenicity prediction
Once the weights of each characteristic in the overall pathogenicity (ek>0) and the pathogenicity threshold (e0) were calculated (Supplemental Table S7), pathogenicity prediction of missense LDLr variants was assessed following the equation model shown in Equation 4.
| (4) |
Where NTraining = 552, NValidation = 192, NTotal = 744, χ2 = 349, and P < 0.05. The model classifies correctly 91.2% of pathogenic variants (454 of 498) and 90.7% of benign variants (49 of 54) on training, and 92.8% of pathogenic (154 of 166) and 96.2% of benign (25 of 26) on validation. The positive predictive values are 98.9% and 99.9% on training and validation, respectively, and negative predictive values are 52.7% and 67.6% on training and validation, respectively.
The obtained results show the predictive ability of the model, which is able to classify LDLr variants into benign or pathogenic with an accuracy higher than 90% in training and validation. The similarity between specificity and sensitivity parameters of MLb-LDLr is explained by obtaining the variables through the ESA algorithm, where balance on the percentage of correctly predicted pathogenic and benign variants was prioritized instead of better overall score in only 1 category (Equation 3). However, this process was performed only with training variants, so the balance on validation ones is a sign of the homogeneity of the training/validation data set division.
Comparative analysis of the predictive accuracy of ML-based methodologies
The predictive accuracy of 5 ML models was tested: 3 linear models (LDA, LNN, and ESEA) and 2 nonlinear ones from ANN (MLP and RBF) (Table 2). LDA shows a much lower specificity and a slightly higher sensitivity than ESEA. By contrast, MLP, RBF, and LNN show a slightly higher specificity but much lower sensitivity than ESEA.
Table 2.
Comparison of ESEA With Other ML Models
| Model | Set | Sp | Sn | PPV | NPV | Technique |
|---|---|---|---|---|---|---|
| ESEA | Training | 90.7 | 91.1 | 98.9 | 52.7 | ESEA |
| Validation | 96.2 | 92.8 | 99.4 | 67.6 | ||
| LDA | Training | 44.4 | 96.2 | 94.1 | 55.8 | LDA |
| Validation | 50.0 | 95.1 | 92.4 | 61.9 | ||
MLP 7:7-9-1:1 
|
Training | 92.3 | 70.3 | 98.9 | 25.3 | BP100, CG20, CG0b |
| Validation | 100 | 65.1 | 100 | 30.9 | ||
RBF 4:4-9-1:1 
|
Training | 90.7 | 66.1 | 98.5 | 22.5 | KM, KN, PI |
| Validation | 92.3 | 66.3 | 98.2 | 30.0 | ||
LNN 7:7-1:1 
|
Training | 92.6 | 71.2 | 98.8 | 26.1 | PI |
| Validation | 100 | 66.9 | 100 | 32.1 |
Green indicates positive input. Red indicates negative input.
BP = backpropagation; CG = conjugated gradient; ESEA = Excel Solver Evolutionary algorithm; KM = K-means; KN = K-nearest neighbor; LDA = linear discriminant analysis; LNN = linear neural network; ML = machine learning; MLP = multilayer perceptron; NPV = negative predictive value; PI = pseudoinversion; PPV = positive predictive value; RBF = radial basis function; Sn = sensitivity; Sp = specificity.
MLb-LDLr software
The best ML model found with ESEA was implemented into user-friendly software denominated MLb-LDLr. MLb-LDLr was developed using several libraries: Python (3.8.5), Click (7.1.2), Flask (1.1.2), Gunicorn (20.0.4), Itsdangerous (1.1.0), Jinja2 (2.11.2), MarkupSafe (1.1.1), and Werkzeug (1.0.1). The code and the database used for the software are available at GitHub under Creative Commons CC0 license.
MLb-LDLr software uses its own algorithm to give pathogenicity predictions of every single LDLr missense variant. f(Pi)calc score values were relativized to a maximum and the pathogenicity threshold. The final percentage value is obtained relativizing the score to 100 (5).
| (5) |
The maximum possible value (Max(f(Pi = 1)calc)) is obtained when Pi1 = Pi2 =…= Pi7 = 1 (Equation [4]). The max value gets only positive values because during the ESEA optimization, we used the restriction ek >0. Because it is not possible for any LDLr missense variants to reach that value (f(Pi=1)calc = 0.752) using this model, the maximum value was set in f(Pi = 1)calc = 0.74 in order to obtain more accurate results. The software visualizes the final probability values (p(Pi = 1)% or p(Pi = 0)%) in a 50-100 scale. Whether a variant is predicted as a benign (f(Pi = 1)calc < e0), the software visualizes p(Pi = 0)%. This way, the result displayed is always positive and higher than 50% of being pathogenic or benign. The top-5 pathogenic and benign variant predictions on the training and validation groups are shown in Supplemental Table S8. The complete prediction database is shown in Supplemental Table S9, and data used for prediction are available on Figshare.
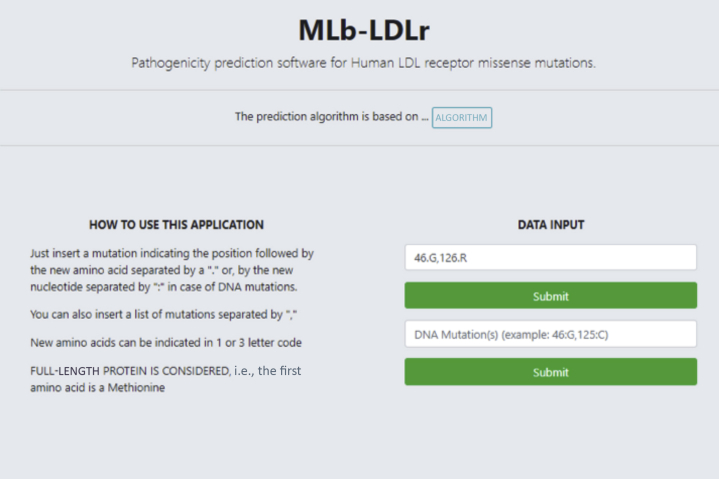
MLb-LDLr interface
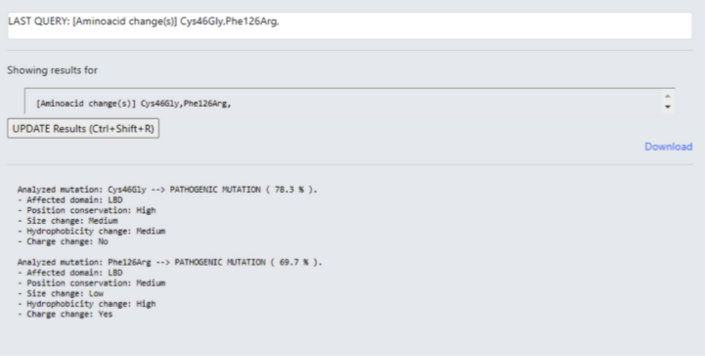
Both DNA or amino acid nomenclature can be used as input, and the software is able to carry out multiple analyses at once (Figure 2). The result summary includes information about the affected domain, the conservation and amino acid size, charge, and hydrophobicity change. The information of the last 4 characteristics is given in “low,” “medium,” or “high” format (Figure 3), according to Supplemental Table S6 probability table values.
Figure 2.
Interface of MLb-LDLr
The variants to be predicted can be introduced on amino acid or nucleotide code and only the position and the substituting residue is necessary. LDL = low-density lipoprotein; MLb-LDLr = machine-learning–based low-density lipoprotein receptor software.
Figure 3.
MLb-LDLr Results Display
Results display page shows the pathogenicity prediction of the variant and its most important characteristics. The prediction is given on probability values and the characteristics are categorized in ranges, high, medium, and low, depending on their effect on the overall pathogenicity. MLb-LDLr = machine-learning–based low-density lipoprotein receptor software.
Accuracy of MLb-LDLr versus other predictive software
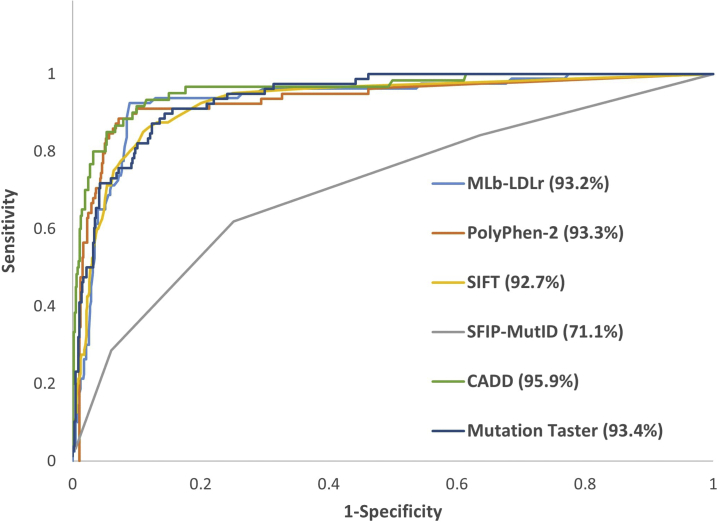
To date, several software programs are available to predict the effect of a given mutation on the protein function. Although most of them are used to predict the functional effect of a mutation in any protein (eg, Polymorphism Phenotyping v2 [PolyPhen-2] [18], Sorting Intolerant From Tolerant [SIFT) [19], Combined Annotation-Dependent Depletion [CADD] [20], MutationTaster [21]), recently, a structure-based software for missense LDLr variants has been developed (SFIP-MutID) (22). Each model uses different databases and techniques for prediction, so their effectiveness can vary. In this work, we have used all missense LDLr variants (744) annotated at ClinVar database to compare the accuracy of different software, with and without bootstrapping resampling. Because EGS optimization process must be done manually, this process has not been carried out on bootstrapped resampling. The main results are shown in Table 3 and Figure 4. The prediction of each variant on the nonbootstrapped sample is shown in Supplemental Table S10.
Table 3.
Comparison of Predictive Software Using ClinVar Database
| Predictive Tool | No Bootstrapping |
Bootstrapping |
||||||
|---|---|---|---|---|---|---|---|---|
| T |
V |
T |
V |
|||||
| Sn | Sp | Sn | Sp | Sn | Sp | Sn | Sp | |
| MLb-LDLra | 90.7 | 91.1 | 92.8 | 96.2 | 87.5 | 91.2 | 85.8 | 81.8 |
| PolyPhen-2 | 92.9 | 79.6 | 94.6 | 96.1 | 93.6 | 83.6 | 93.8 | 83.7 |
| SIFT | 83.9 | 90.7 | 90.3 | 88.5 | 85.4 | 89.9 | 85.5 | 90.5 |
| SFIP-MutIDb | 92.1 | 18.5 | 87.3 | 26.9 | 90.5 | 21.1 | 90.5 | 21.1 |
| CADD | 88.5 | 88.9 | 89.7 | 100 | 88.9 | 92.5 | 89.1 | 92.6 |
| MutationTaster | 95.8 | 66.7 | 95.8 | 84.6 | 96.1 | 72.5 | 96.2 | 73.0 |
Statistics of the original sampling and randomly bootstrapped sampling. One thousand bootstrapped samples were used, and the results are shown with a 95% confidence interval.
The expert-guided strategy optimization process cannot be carried out when bootstrapping, decreasing the accuracy of the model
SFIP-MutID is not able to predict mutation within 1-21 and 715-860 residues.
Figure 4.
Performance of MLb-LDLr in Comparison to Other Software
Several scores are compared by area under the receiver operating curve (AUROC) using the ClinVar database. MLb-LDLr = machine-learning–based low-density lipoprotein receptor software; other abbreviations as in Tables 1 and 2.
Regarding the nonbootstrapped samples, MLb-LDLr is the only software program with all statistic values higher than 90%. MutationTaster shows the top score for sensitivity in both training and validation sets but has the second-worst specificity values. The highest specificity in the training set corresponds to MLb-LDLr, and the highest in the validation set corresponds to CADD.
These results are mostly maintained in bootstrapped samples, MutationTaster having the highest sensitivity values and CADD the highest specificity ones. MLb-LDLr on the other hand shows slightly lower values in most statistics because EGS optimization cannot be performed when bootstrapping samples.
Regarding area under the receiver operating curve (AUROC) values, CADD has the highest score (0.959) followed by MutationTaster (0.934), PolyPhen-2 (0.933), and MLb-LDLr (0.932).
As shown in Supplemental Table S11, 453 of 744 variants are correctly predicted by the 6 analyzed software programs, 181 variants are correctly predicted by 5 software programs, 62 variants are correctly predicted by 4 programs, 27 by 3 software programs, 17 by 2 software programs, and only 4 variants are correctly predicted by 1 or none. These results show that the 60% of the variants annotated in ClinVar are correctly predicted by any of the analyzed software programs, and that the remaining variants except for p.(Ala299Thr) can be correctly predicted by a combination of them. Altogether, this indicates that the available software should be used in combination to improve prediction accuracy.
In vitro functional characterization of conflictive LDLr variants
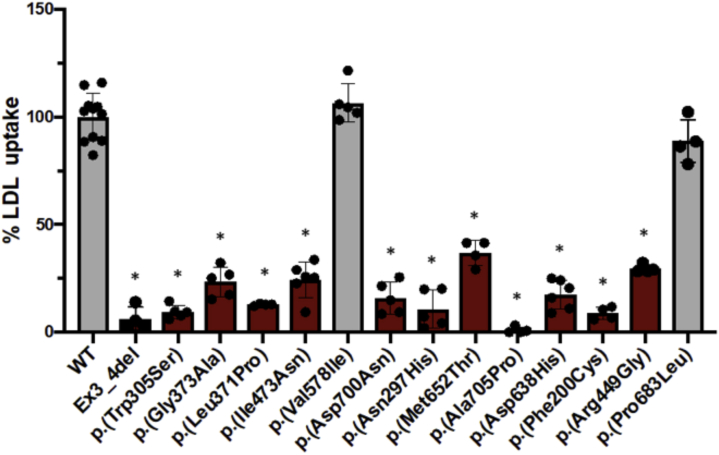
In order to experimentally validate the model, 13 LDLr variants were selected according to their conflictive interpretations in ClinVar and contradictive activity predictions. LDLr activity was assessed as indicated in Methods. As shown in Figure 5, p.(Val578Ile) and p.(Pro683Leu) variants show similar LDLr activity to wild type, indicating that these mutations are not pathogenic. On the contrary, p.(Phe200Cys), p.(Asn297His), p.(Trp305Ser), p.(Lau371Pro), p.(Gly373Ala), p.(Arg449Gly), p.(Ile473Asn), p.(Asp638His), p.(Met652Thr), p.(Asp700Asn), and p.(Ala705Pro) variants showed a reduced activity ranging from 5% to 40% and thus are classified as pathogenic.
Figure 5.
In Vitro Validation of the Model by Assessing LDL Uptake on CHO-ldlΔ7 Cells Transfected With LDLr Variants
LDL uptake was quantified by flow cytometry, as described in Methods. The values represent the mean of at least 4 experiments, error bars represent ±SD. ∗P < 0.01 comparing WT with each variant. WT = wild-type; other abbreviations as in Figure 2.
Activity results were compared with the pathogenicity predictions of each software program. As shown in Table 4, MLb-LDLr shows the second-best accuracy for both pathogenic and benign variants, being the most balanced software. By contrast, PolyPhen-2 and CADD correctly predict barely more than one-half of the pathogenic variants and not a single benign variant. SIFT is the only one correctly predicting the benign variants, but only hits 2 pathogenic ones. MutationTaster correctly predicts 8 pathogenic variants, but no benign ones. Finally, SFIP-MutID has the highest score on pathogenic variants, but does not correctly predict any benign ones. The similarity between the results obtained in Table 4 and the prediction of the ClinVar database shown in Table 3 is noteworthy.
Table 4.
Accuracy of MLb-LDLr, PolyPhen-2, SIFT, SFIP-MutID, CADD, and MutationTaster on 13 Characterized Novel LDLr Variants
| Software | Predicted Pathogenic (n = 11) | Predicted Pathogenic, % | Predicted Benign (n = 2) | Predicted Benign, % | General Accuracy, % |
|---|---|---|---|---|---|
| MLb-LDLr | 8 | 72 | 1 | 50 | 69 |
| PolyPhen-2 | 6 | 54 | 0 | 0 | 46 |
| SIFT | 2 | 18 | 2 | 100 | 30 |
| SFIP-MutID | 10 | 91 | 0 | 0 | 77 |
| CADD | 6 | 54 | 0 | 0 | 46 |
| MutationTaster | 8 | 72 | 0 | 0 | 61 |
Discussion
Alongside the extraordinary growth of newly described LDLr variants brought by the fast development of next-generation sequencing (10), there is developing desire to develop powerful software to accurately assign biological activity roles to LDLr variants. PolyPhen-2, CADD, MutationTaster, and SIFT are some of the most used software packages to predict the effect of a mutation in the LDLr (18, 19, 20, 21). PolyPhen-2 utilizes a combination of sequence-and structure-based attributes for the description of an amino acid substitution; SIFT makes inferences from sequence similarity; CADD is based on evolutive gene factors and uses more than 60 variables; MutationTaster analyzes evolutionary conservation, splice-site changes, loss of protein features, and changes that might affect the amount of mRNA Therefore, as they consider common features of many proteins, when predicting the effect on the activity of LDLr variants the results often disagree. More recently, a specific model for LDLr missense variants based on LDLr structural resolution has been developed (SFIP-MutID), but the model lies in predicting pathogenic mutations rather than predicting benign mutations thus limiting its use (22).
This work sought to develop a ML model to improve accuracy on predicting the activity of missense LDLr mutations. It has previously been shown that combining information obtained from multiple sequence alignment and 3-dimensional protein structure can increase prediction performance (23). Therefore, specific features of the LDLr protein such as conservation of the substituted residue, charge change, the original amino acid, the substituting amino acid, hydrophobicity change, amino acid size change, and the location of the substituted amino acid within the different LDLr domains have been considered to quantify their influence on the overall pathogenicity. After having been represented as quantitative vectors, ESEA has been used to calculate both the weight of each characteristic on the overall pathogenicity and the threshold that determines whether a variant is pathogenic or not. The introduction of ClinVar database in the ML model constitutes an innovative feature about the MLb-LDLr software, which sets it apart from other software. This strategy allowed increasing the predictive power of the MLb-LDLr by integrating ML algorithms, resulting in a specificity of 92.5% and a sensitivity of 91.6%. A major challenge in the MLb-LDLr optimization process was generating a balanced software program able to predict both pathogenic and benign variants with high accuracy. Our results demonstrated the value of combining information, especially the use of the ClinVar database, which provided predictive software with an accuracy higher than 90% in both pathogenic and benign variants.
The bootstrap resampling method is commonly used to test a model with many different training and validation sets. This test is usually carried out automatically, because more than 1,000 groups are required, but the last process on MLb-LDLr software, EGS, must be done manually. Although the optimization process could not be implemented on the results shown in Table 3, these results are quite similar to the ones obtained with no EGS. Therefore, the bootstrapping resampling shows the validity of the training and validation set division, because the results obtained with and without bootstrapping are similar in all analyzed software programs.
All the analyzed software programs, except for CADD, provide pathogenicity predictions according to a specific threshold so that a probability value can be given as a result. By contrast, CADD is based on comparative scores and only provides relative pathogenicity values. This means that CADD relies on AUROC and similar general statistics to obtain an accuracy value; meanwhile, the rest of the software programs have optimized a specific threshold to obtain an optimum accuracy. This could explain why CADD has the highest AUROC values by far, but it is surpassed by MLb-LDLr and PolyPhen-2 on the overall accuracy.
SIFT and PolyPhen-2 are among the predictive software programs with the highest ease of use and speed, which allows for direct batch queries using amino acid and genome coordinates (24). In order to facilitate the use of MLb-LDLr, an interface has been developed, which allows direct input using DNA or amino acid nomenclature as well as querying multiple predictions.
One of the major advantages of MLb-LDLr software relies in the possibility of actualizing the dataset periodically thus including newly annotated variants in ClinVar. This allows continuously increasing database accuracy in order to perform an updated prediction for each new described variant. In the future, some other ML algorithms may be introduced in the MLb-LDLr software to integrate information such as phenotype of the patients carrying pathogenic variants and the most suitable treatment for each of them (25).
It is noteworthy that all the predicted models tested in this work showed an accuracy of over 80% when predicting variants, which indicates that the data accessible in ClinVar are highly accurate, even those with a single submitter to support the pathogenicity. This fact supports the use of the ClinVar database as a reliable source of information regarding pathogenic variants due to the big effort done to correctly assign pathogenicity to variants described so far. These pathogenic LDLr variants have been mostly found in patients with high plasma LDL plasma levels and have been characterized both by cascade screening or in vitro functional assays of the LDLr variants (10,26,27).
As mentioned in the preceding text, the fast development of next-generation sequencing brings to light a great number of new LDLr variants that will favor the emergence of benign ones. Hence, the development of predictive software such as MLb-LDLr with high accuracy in predicting benign variants is crucial. Nowadays, the simultaneous use of several software programs to predict the effect of a given mutation on protein activity is almost mandatory, because the ones with the highest accuracy on pathogenic variants have low accuracy on benign variants.
Study limitations
First, MLb-LDLr software only predicts missense LDLr variants. There are many mutation types depending on their location and effect (frameshift, UTR, splicing site…), and each of them has a different mechanism that should be analyzed individually. Here, only missense variants have been analyzed because they represent a high percentage of total LDLr variants (40%) (8) and more than 50% in the ClinVar database (28,29). Although ClinVar is a great source to evaluate the pathogenicity of LDLr variants, 1 limitation of the database is that in some cases, there is only 1 submitter and a few of them have no assertion. In addition, there are enough cases of both missense pathogenic and benign variants to develop a ML model. Second, some Pik values may not be totally correct due to the lack of enough benign variants in some classes. However, this problem will be fixed, because the number of characterized benign variants will increase. In the meantime, we tried to solve this by applying EGS strategy. The accuracy of the artificial intelligence model using a validation cohort was not confirmed.
Conclusions
Here, we provide a powerful tool to predict the impact of LDLr mutations on protein activity. The strength of MLb-LDLr software relies in its precision with both type of variants, benign and pathogenic, with an estimated hit rate over 90% in both of them. These results highlight the usefulness of MLb-LDLr software as a helping diagnostic tool for clinicians.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: This study developed a machine learning model (MLb-LDLr), which can help to provide a fast diagnosis to FH patients saving time and expenditure for characterizing the pathogenicity of LDLr variants. The MLb-LDLr software developed here can be clinically implemented to refine the diagnosis of FH and to improve disease prognosis. MLb-LDLr software may prove clinically useful and assist clinicians in tailoring precise management and therapy for the patients with FH and provide a novel diagnostic approach to manage FH. This study provides an open access predictive software to the scientific community, to predict the pathogenicity of missense LDLr variants.
TRANSLATIONAL OUTLOOK: The MLb-LDLr software increases the predictive power of previous software used to predict the pathogenicity of LDLr variants and provides an open-access interface to the scientific community and clinicians, which allows direct input using DNA or amino acid nomenclature as well as querying multiple predictions. The strength of MLb-LDLr software relies in its capacity of predicting both benign and pathogenic, with an estimated hit rate over 90%, highlighting the usefulness of MLb-LDLr software as a helping diagnostic tool for clinicians. Collectively, MLb-LDLr is expected to lower the incidence of cardiovascular events by collecting backpropagation data that are unmeasurable by current diagnostic modalities.
Funding Support and Author Disclosures
This study was supported by grants from the Basque Government (Cesar Martin, Grupos Consolidados IT-1264-19). Mr Larrea-Sebal was supported by a FPI grant from Gobierno Vasco (2019–2020). Dr Benito-Vicente was supported by Programa de especialización de Personal Investigador Doctor en la UPV/EHU (2019) 2019-2020. Dr Galicia-Garcia was supported by Fundación Biofísica Bizkaia. Ms Jebari-Benslaiman was supported by grant PIF (2017–2018), Gobierno Vasco. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgement
The authors thank Mikel Larrea for his invaluable help in using some complex processes of Excel.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For an expanded Methods section as well as supplemental tables, please see the online version of this paper.
Contributor Information
Humberto González-Díaz, Email: humberto.gonzalezdiaz@ehu.eus.
César Martín, Email: cesar.martin@ehu.eus.
Appendix
References
- 1.Sjouke B., Kusters D.M., Kindt I., et al. Homozygous autosomal dominant hypercholesterolaemia in the Netherlands: prevalence, genotype-phenotype relationship, and clinical outcome. Eur Heart J. 2015;36:560–565. doi: 10.1093/eurheartj/ehu058. [DOI] [PubMed] [Google Scholar]
- 2.Brown M.S., Goldstein J.L. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232(4746):34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 3.Sharifi M., Rakhit R.D., Humphries S.E., Nair D. Cardiovascular risk stratification in familial hypercholesterolaemia. Heart. 2016;102:1003–1008. doi: 10.1136/heartjnl-2015-308845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ference B.A., Ginsberg H.N., Graham I., et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur Heart J. 2017;38:2459–2472. doi: 10.1093/eurheartj/ehx144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benito-Vicente A., Uribe K.B., Jebari S., Galicia-Garcia U., Ostolaza H., Martin C. Familial hypercholesterolemia: the most frequent cholesterol metabolism disorder caused disease. Int J Mol Sci. 2018;19(11):3426. doi: 10.3390/ijms19113426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordestgaard B.G., Chapman M.J., Humphries S.E., et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease. Eur Heart J. 2013;34:3478–3490. doi: 10.1093/eurheartj/eht273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palacios L., Grandoso L., Cuevas N., et al. Molecular characterization of familial hypercholesterolemia in Spain. Atherosclerosis. 2012;221:137–142. doi: 10.1016/j.atherosclerosis.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 8.Chora J.R., Medeiros A.M., Alves A.C., Bourbon M. Analysis of publicly available LDLR, APOB, and PCSK9 variants associated with familial hypercholesterolemia: application of ACMG guidelines and implications for familial hypercholesterolemia diagnosis. Genet Med. 2018;20:591–598. doi: 10.1038/gim.2017.151. [DOI] [PubMed] [Google Scholar]
- 9.Benito-Vicente A., Uribe K.B., Jebari S., Galicia-Garcia U., Ostolaza H., Martin C. Validation of LDLr activity as a tool to improve genetic diagnosis of familial hypercholesterolemia: a retrospective on functional characterization of LDLr variants. Int J Mol Sci. 2018;19(6):1676. doi: 10.3390/ijms19061676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knowles J.W., Rader D.J., Khoury M.J. Cascade screening for familial hypercholesterolemia and the use of genetic testing. JAMA. 2017;318:381–382. doi: 10.1001/jama.2017.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huijgen R., Kindt I., Defesche J.C., Kastelein J.J.P. Cardiovascular risk in relation to functionality of sequence variants in the gene coding for the low-density lipoprotein receptor: a study among 29 365 individuals tested for 64 specific low-density lipoprotein-receptor sequence variants. Eur Heart J. 2012;33:2325–2330. doi: 10.1093/eurheartj/ehs038. [DOI] [PubMed] [Google Scholar]
- 12.Wainberg M., Merico D., Delong A., Frey B.J. Deep learning in biomedicine. Nat Biotechnol. 2018;36:829–838. doi: 10.1038/nbt.4233. [DOI] [PubMed] [Google Scholar]
- 13.Thusberg J., Olatubosun A., Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 14.MLb-LDLr software. Accessed September 30, 2021. https://www.ehu.eus/es/web/hypercholesterolemia-mechanisms/mlb-ldlr1
- 15.Barati R. Application of Excel solver for parameter estimation of the nonlinear Muskingum models. KSCE J Civ Eng. 2013;17:1139–1148. [Google Scholar]
- 16.Galicia-Garcia U., Benito-Vicente A., Uribe K.B., et al. Mutation type classification and pathogenicity assignment of sixteen missense variants located in the EGF-precursor homology domain of the LDLR. Sci Rep. 2020;10:1727. doi: 10.1038/s41598-020-58734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etxebarria A., Benito-Vicente A., Alves A.C., Ostolaza H., Bourbon M., Martin C. Advantages and versatility of fluorescence-based methodology to characterize the functionality of LDLR and class mutation assignment. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0112677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adzhubei I.A., Schmidt S., Peshkin L., et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:452–457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 22.Guo J., Gao Y., Li X., et al. Systematic prediction of familial hypercholesterolemia caused by low-density lipoprotein receptor missense mutations. Atherosclerosis. 2019;281:1–8. doi: 10.1016/j.atherosclerosis.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Berman H.M., Battistuz T., Bhat T.N., et al. The protein data bank. Acta Crystallogr Sect D Biol Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 24.Dong C., Wei P., Jian X., et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sifrim A., Popovic D., Tranchevent L.C., et al. EXtasy: variant prioritization by genomic data fusion. Nat Methods. 2013;10:1083–1086. doi: 10.1038/nmeth.2656. [DOI] [PubMed] [Google Scholar]
- 26.Etxebarria A., Benito-Vicente A., Palacios L., et al. Functional characterization and classification of frequent low-density lipoprotein receptor variants. Hum Mutat. 2015;36:129–141. doi: 10.1002/humu.22721. [DOI] [PubMed] [Google Scholar]
- 27.Lamiquiz-Moneo I., Civeira F., Mateo-Gallego R., et al. Diagnostic yield of sequencing familial hypercholesterolemia genes in individuals with primary hypercholesterolemia. Rev Esp Cardiol (Engl Ed) 2021;74(8):664–673. doi: 10.1016/j.rec.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Harrison S.M., Riggs E.R., Maglott D.R., et al. Using ClinVar as a resource to support variant interpretation. Curr Protoc Hum Genet. 2016;89:8.16.1–8.16.23. doi: 10.1002/0471142905.hg0816s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landrum M.J., Lee J.M., Benson M., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.