Abstract
In a search for methods for subtyping of Bartonella henselae in clinical samples, we amplified and sequenced a 701-bp region in the 3′ end of the ftsZ gene in 15 B. henselae isolates derived from cats and humans in the United States and Europe. The ftsZ sequence variants that were discovered were designated variants Bh ftsZ 1, 2, and 3 and were compared with 16S rRNA genotypes I and II of the same isolates. There was no ftsZ gene variation in the strains of 16S rRNA type I, all of which were Bh ftsZ 1. The type II strains constituted two groups, with nucleotide sequence variation in the ftsZ gene resulting in amino acid substitutions at three positions, one of which was shared by the two groups. One 16S rRNA type II isolate had an ftsZ gene sequence identical to those of the type I strains. Variants Bh ftsZ 1 and 2 were detected in tissue specimens from seven Swedish patients with diagnoses such as chronic multifocal osteomyelitis, cardiomyopathy, and lymphadenopathy. Patients with similar clinical entities displayed either Bh ftsZ variant. The etiological role of B. henselae in these patients was supported by positive Bartonella antibody titers and/or amplification and sequencing of a part of the B. henselae gltA gene. B. henselae ftsZ gene sequence variation may be useful in providing knowledge about the epidemiology of various B. henselae strains in clinical samples, especially when isolation attempts have failed. This report also describes manifestations of atypical Bartonella infections in Sweden.
The genus Bartonella comprises four species established as human pathogens, namely, B. henselae, B. quintana, B. elizabethae, and B. bacilliformis. B. henselae was first isolated in 1992 (22) and has been implicated in the etiology of cat scratch disease (CSD) (23), bacillary angiomatosis (BA) (14), hepatic peliosis (30), endocarditis (10), and fever and bacteremia (29); B. quintana has been implicated in the etiology of trench fever (fever, rash, bone pain, and splenomegaly) (32), BA (14), and endocarditis (21); B. elizabethae has been implicated in the etiology of endocarditis (7); and B. bacilliformis has been implicated in the etiology of Carrion's disease, which consists of an acute hemolytic anemia (Oroya fever), followed by the emergence of vascular proliferative lesions (verruga peruana) resembling those seen in BA (3). Less common clinical manifestations caused by Bartonella species include osteomyelitis (24), myocarditis (11), fever of unknown origin in children (12), and Parinaud's oculoglandular syndrome (3).
The fastidious nature of this organism is a problem in clinical practice since isolation usually is difficult and time-consuming, consequently delaying the patient's diagnosis. Therefore, diagnosis is reliant upon serological methods, currently, the indirect immunofluorescence assay (IFA) or Western blotting, or the detection of the bacterial DNA in tissue specimens by PCR and sequencing. The drawbacks of IFA include problems with cross-reactivity between different species of the same genus and between more distantly related species of other genera (17, 18). A species-specific serological method is not available. Consequently, detection to the species level requires additional methods. Also, variations in the antigenicities between B. henselae isolates, which might result in false-negative results for serum samples (8), and a chronic form of B. quintana infection with the absence of seroconversion have been reported (5).
The use of the cell division protein FtsZ as a means of differentiating Bartonella species was recently described (13). The ftsZ gene encodes a protein that plays an important role in bacterial cell division. In contrast to most FtsZ proteins currently characterized in other bacteria, the FtsZ proteins of different Bartonella species are nearly twice as large, with an additional part at the C-terminal end (13, 20). A comparison of the Bartonella ftsZ genes in the region encoding the C-terminal region of the protein shows that it has a higher degree of sequence divergence than the part of the gene encoding the N-terminal region. Padmalayam et al. (20) showed that sera from patients with verruga peruana (systemic bartonellosis) caused by B. bacilliformis strongly react with epitopes located in the C-terminal part of the protein. Amino acid sequence differences in this part of the gene might, consequently, mirror antigenic variants. Kelly et al. (13) used the 3′-end sequence variations between B. henselae, B. quintana, and B. bacilliformis to design species-specific PCR assays.
Humans have been shown to acquire infection from cats (B. henselae) or via insect vectors (B. quintana and B. bacilliformis) (3). However, the epidemiology of Bartonella infections remains incompletely understood. For example, questions such as whether different subspecies have divergent pathogenic potentials need to be elucidated. To achieve this, techniques that allow subtyping of strains in clinical samples are required. Sander et al. (27) compared different DNA fingerprinting techniques for molecular typing of B. henselae isolates derived from cultures of blood from domestic cats. Three main variants among the cat isolates were identified. These data suggest wide genetic variation among B. henselae strains.
When isolation fails, subtyping of Bartonella strains is sometimes more difficult. Bergmans et al. (4) used the 16S rRNA gene and 16S to 23S rRNA gene spacer region in studying DNA extracted from pus aspirates and lymph nodes from patients with CSD. They concluded that two different variants of B. henselae were predominant in immunocompetent patients with CSD in The Netherlands, namely, type I and type II. It has been suggested that genotype I could be more pathogenic for humans than genotype II and that the two genotypes are unevenly distributed geographically (26). Because of the high degree of conservation in the 16S rRNA gene, there is always a risk of amplifying non-Bartonella DNA. In order to achieve a higher degree of specificity for Bartonella detection, genes with greater degrees of sequence divergence are desired targets for PCR amplification, e.g., gltA (19), htrA (2), and ftsZ (13). However, these genes have not been reported to be able to differentiate different strains within the species B. henselae.
We have investigated the possibility of using the Bartonella ftsZ gene as a target for PCR amplification and subsequent sequencing of DNA extracted from clinical specimens as a means of subtyping of strains within the species B. henselae. We have amplified and sequenced a part of the ftsZ gene in 15 different B. henselae isolates, and these data were correlated to 16S rRNA genotypes I and II of the same isolates and compared to the ftsZ sequences discovered in clinical samples from seven Swedish patients.
(This study was presented in part at the First Congress of the European Society for Emerging Infections, 13 to 16 September 1998, Budapest, Hungary.)
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. henselae strains 3507, 3508, 3509, 3750, 3883, 3884, 4271, 4272, 5249, URLLY8 (Marseille), FR97/K7, FR96/BK3, FR96/BK77, and FR96/BK78, and type strain Houston-1 (ATCC 49882) were kindly provided by R. Regnery, Centers for Disease Control and Prevention, Atlanta, Ga. (Table 1). The bacterial strains were grown for 2 weeks on Columbia blood agar containing 5% whole horse blood at 35°C in 5% CO2.
TABLE 1.
Nucleotide sequences of 16S rRNAs and ftsZ genes amplified from the B. henselae isolates studied
| Isolate | Species | Strain | Source | Geographical origin | Additional strain designation | Reference | 16S rRNA type | Bh ftsZ variant |
|---|---|---|---|---|---|---|---|---|
| B. henselae | Houston-1 | HIVa patient's blood | United States | ATCC 49882 | 22 | I | 1 | |
| 3508 | B. henselae | SA-2 | CSD pat | United States | R1330 | I | 1 | |
| 3509 | B. henselae | CA-1 | HIV patient's blood | United States | R1173 | I | 1 | |
| 3507 | B. henselae | Tiger 2 | Cat blood | United States | R1556 | II | 1 | |
| 3750 | B. henselae | Houston-2 | United States | I | 1 | |||
| 3883 | B. henselae | Patient's blood | United States | I | 1 | |||
| 3884 | B. henselae | Patient's blood | United States | I | 1 | |||
| 4271 | B. henselae | Cat blood | United States | I | 1 | |||
| 4272 | B. henselae | Cat blood | United States | I | 1 | |||
| 5249 | B. henselae | United States | II | 2 | ||||
| URLLY8 | B. henselae | Marseille | Endocarditis patient's blood | France | R9064 | 8 | II | 2 |
| B. henselae | FR97/K7ITS | Cat blood | Germany | R14472 | 27 | I | 1 | |
| B. henselae | FR96/BK77 | Cat blood | Germany | R14473 | 25 | II | 3 | |
| B. henselae | FR96/BK3 | Cat blood | Germany | R14476 | 25 | II | 3 | |
| B. henselae | FR96/BK78 | Cat blood | Germany | R14479 | 25 | II | 3 |
HIV, human immunodeficiency virus.
Clinical samples.
Tissue and blood samples from six Swedish patients and a blood sample from a cat belonging to a patient with Parinaud's oculoglandular syndrome were obtained.
Extraction of DNA from bacterial strains and clinical samples.
Total genomic DNA was extracted from the B. henselae strains and from the blood and tissue samples with the Qiagen QIAamp Tissue Kit (QIAGEN, Inc., Chatsworth, Calif.) according to the manufacturer's instructions. Between 10 and 25 mg of tissue and 200 μl of whole blood were used. The samples were strictly handled separately under sterile conditions to avoid the risk of cross contamination between different samples. DNA was precipitated in all samples with ethanol for additional purification. A total of 100 μl of elution buffer was used to resuspend the precipitated DNA.
Oligonucleotide primers.
See Table 2 for the oligonucleotide primers used in the study.
TABLE 2.
Sequences and positions of oligonucleotides used for 16S rRNA, ftsZ, and gltA gene amplifications
| Oligonucleotide name | Oligonucleotide sequencea | Target organism | Target gene | Nucleotide position (direction)b | Reference or source |
|---|---|---|---|---|---|
| 16SF | AGAGTTTGATCCTGG(CT)TCAG | Eubacteria | 16S rRNA | 10 (→) | 4 |
| 16SR | CTTTACGCCCA(AG)TAA(AT)TCCG | Eubacteria | 16S rRNA | 521 (←) | 4 |
| Bh ftsZ 965.p | GTATTCGCGAAGAAGTGGATGC | Bartonella spp. | ftsZ | 965 (→) | This study |
| Bh ftsZ 1393.n | GCGAACTACGGCTTACTTGC | B. henselae | ftsZ | 1393 (←) | This study |
| Bh ftsZ 1247.p | CGGTTGGAGAGCAGTTTCGTC | B. henselae | ftsZ | 1247 (→) | This study |
| Bh ftsZ 1754.n | CGACGTGGAACATAAACAGA | Bartonella spp. | ftsZ | 1754 (←) | This study |
| BHCS212.p | GTTATCCTATTGACCAA | Bartonella spp. | gltA | 212 (→) | 11 |
| BHCS613.n | TATTCTTCACAAGGAAC | Bartonella spp. | gltA | 613 (←) | 11 |
| BHCS510.p | AACTCTTGCCGCTATGG | Bartonella spp. | gltA | 510 (→) | 11 |
| BHCS897.n | CCAAAACCCATAAGGCG | Bartonella spp. | gltA | 897 (←) | 11 |
Bases in parentheses are mixed at one position.
Arrows indicate direction of primers (→, forward; ←, reverse).
PCR amplification.
PCR amplifications were performed with a PCR Master kit from Boehringer Mannheim Scandinavia AB (Bromma, Sweden). The 50-μl reaction mixture consisted of the template, forward and reverse primers (10 pmol/primer), 25 μl of the PCR master mixture and distilled water of PCR grade up to 50 μl, giving a final concentration of 1× PCR buffer, 2.5 U of Taq DNA polymerase in 0.005% Brij 35, dATP, dCTP, dGTP, and dTTP each at a concentration of 0.2 mM, 10 mM Tris-HCl, 50 mM KCl, and 1.5 mM MgCl2. A Perkin-Elmer GeneAmp 9600 thermocycler was used for all amplifications.
For 16S rRNA gene amplification the parameters consisted of 3 min at 95°C, followed by 10 cycles of 95°C for 20 s, 50°C for 1 min, and 72°C for 1 min and 30 s, and the annealing temperature was lowered by 1°C per cycle, finally reaching 40°C. This was followed by 40 cycles at 95°C for 20 s, 40°C for 1 min, and 72°C for 1 min and 30 s, with a 5-min extension at 72°C. The products were stored at 4°C until they were further processed.
For amplification of the ftsZ genes from the isolates, the parameters consisted of 3 min at 94°C, followed by 40 cycles at 94°C for 1 min, 49°C for 1 min, and 72°C for 1 min, followed by a 5-min extension at 72°C. The products were stored at 4°C until they were further processed.
Seminested PCR protocols for amplification of the Bartonella gltA and ftsZ genes were applied for the clinical samples. In the first reaction, the protocol for the 16S rRNA amplification described above was used. Primers Bh ftsZ 965.p and Bh ftsZ 1754.n were used for ftsZ amplification, and primers BHCS212.p and BHCS897.n were used for gltA amplification. The products were stored at 4°C until they were further processed. In the second reaction, 1 μl of the PCR product from the first reaction was used as the template for two separate seminested PCRs, in which the program for amplification of the ftsZ genes of the isolates described above was used. For ftsZ gene amplification, primer pairs Bh ftsZ 965.p–Bh ftsZ 1393.n and Bh ftsZ 1247.p–Bh ftsZ 1754.n were used, and for gltA amplification primer pairs BHCS212.p-BHCS613.n and BHCS510.p-BHCS897.n were used. The products were stored at 4°C until they were further processed.
Measures were taken to prevent PCR carryover contamination. Different rooms were used for preparation of the template DNA, mixing of the PCR reagents except the template, addition of the template for the first reaction, and addition of the PCR product for the subsequent reaction. Single PCR tubes and filter tips were used, and protective gloves were changed repeatedly. All reactions comprised tests for positive and negative controls, which were treated the same as the clinical samples.
Gel electrophoresis and purification of PCR products.
PCR products were electrophoresed through a 1% agarose gel containing ethidium bromide in Tris-borate buffer. The DNA was detected on a UV transilluminator and photographed.
Single PCR products of the expected size compared to a DNA size marker were purified by using the QIAQUICK PCR purification system from QIAGEN, Inc., by following the manufacturer's instructions.
DNA sequencing.
The nucleotide sequences on both strands were obtained by the methods of Sanger et al. (28) with a model ABI 310 Genetic Analyzer (Perkin-Elmer Corp., Norwalk, Conn.). A DNA sequencing kit with AmpliTaq DNA Polymerase, FS, for the BigDye Terminator Cycle Sequencing Ready Reaction protocol (Perkin-Elmer, Applied Biosystems, Warrington, Great Britain) was used for the cycle sequencing reaction.
Analysis of sequence data and construction of phylogenetic trees.
DNA analyses were performed with the software packages ABI Prism DNA Sequencing Analysis Software, version 3.0 (Perkin-Elmer Applied Biosystems, Foster City, Calif.), Sequencher (Gene Codes Corporation, Ann Arbor, Mich.), and the National Center for Biotechnology Information's sequence homology search program BLAST (1).
Partial primary ftsZ sequences of the three variants, Bh ftsZ 1, 2, and 3, were aligned with each other and with the corresponding ftsZ sequences of B. bacilliformis and B. quintana published previously by using version W of the CLUSTAL multisequence alignment program (31). A total of 500 bootstrap samples were produced by using the Phylo Win package (9). A matrix of evolutionary distances was derived from each bootstrap alignment by using the assumptions of Jukes and Cantor.
IFA.
Serum samples were analyzed by IFA for immunoglobulin G reactivity against crude antigens of B. henselae Houston-1 (ATCC 49882), B. quintana OK 90-268, and B. elizabethae R2798 as described previously (11).
Nucleotide sequence accession numbers.
The nucleotide sequences of the 701-bp fragment of the B. henselae ftsZ gene have been deposited in the GenBank database under the following accession numbers: B. henselae 3508, representing Bh ftsZ variant 1, AF161249; B. henselae URLLY8 (Marseille), representing Bh ftsZ variant 2, AF161250; and B. henselae FR96/BK77, representing Bh ftsZ variant 3, AF161251.
RESULTS
B. henselae isolates. (i) 16S rRNA gene sequence variation.
PCR amplification of part of the 16S rRNA gene with the broad-range PCR primers 16SF and 16SR resulted in single products of the expected size. These products were purified and sequenced. Weak amplification products were detected on a few occasions for the negative controls. Purification and sequencing of these PCR products did not result in any readable sequence data. The existence of two variants according to the 16S rRNA sequence analysis was confirmed. Nine of 15 strains belonged to type I, and the remaining 6 belonged to type II (Table 1). The results were consistently repeated.
(ii) ftsZ gene sequence variation.
The PCR amplification resulted in single products, which were purified and sequenced. Positive controls were always positive. Amplification products were never detected for the negative controls. Three ftsZ variants among the B. henselae isolates were revealed. The results were consistently repeated. The variants were designated Bh ftsZ 1, 2, and 3 and were given the accession numbers AF161249, AF161250, and AF161251, respectively. Nucleotide sequences differed at position 1185 (A [Bh ftsZ 1]→T [Bh ftsZ 2 and 3]), position 1404 (G [Bh ftsZ 1]→A [Bh ftsZ 2]), position 1467 (G [Bh ftsZ 1]→T [Bh ftsZ 2 and 3]), and position 1537 (C [Bh ftsZ 1]→T [Bh ftsZ 3]). The sequence of Bh ftsZ 1 is identical to the previously published corresponding partial sequence of B. henselae Houston-1 (accession no. AF061746) (13).
(iii) FtsZ amino acid sequence variation.
In comparison with the previously published ftsZ sequence of B. henselae Houston-1, point mutations were discovered in the ftsZ gene at four nucleotide positions. Three of these positions resulted in amino acid substitutions, two of which were different between Bh FtsZ 2 and 3 and one of which was shared, as follows: amino acid position 432, methionine (Bh FtsZ 1)→isoleucine (Bh FtsZ 2); position 453, glutamine (Bh FtsZ 1)→histidine (Bh FtsZ 2 and 3); and position 477, proline (Bh FtsZ 1)→serine (Bh FtsZ 3).
(iv) Phylogeny of partial Bartonella ftsZ sequences.
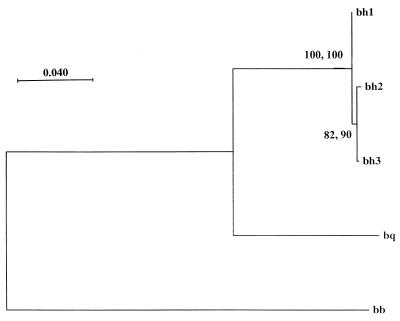
A phylogenetic tree was constructed from the distance matrix by the neighbor-joining method. This tree was compared with a phylogenetic tree inferred by using the maximum parsimony method (Fig. 1). An Intra-B. henselae phylogeny was not quite statistically supported (82 and 90% of 500 bootstrap samples for the neighbor-joining and maximum parsimony methods, respectively), although the neighbor-joining and maximum parsimony methods gave identical topologies. Within the genus, three well-supported branches were identified for B. henselae, B. quintana, and B. bacilliformis on the basis of previously published sequence data corresponding to the partial B. henselae ftsZ sequence within the region of PCR primer design.
FIG. 1.
Distance matrix tree derived from a 701-bp fragment of the ftsZ sequences of B. bacilliformis, B. quintana, and three B. henselae ftsZ variants. The values at the nodes indicate the percentages of 500 bootstraps (neighbor-joining and maximum parsimony methods) and the support for each branch. The lengths of the vertical lines are not significant. bh1, bh2, and bh3, Bh ftsZ 1, 2, and 3 variants, respectively; bq, B. quintana; bb, B. bacilliformis. The scale in the right upper corner represents evolutionary distance.
Clinical samples.
Clinical, epidemiological, and demographic data, specimen source, diagnostic test results, treatment, and outcome data for seven Swedish patients are summarized in Table 3.
TABLE 3.
Age on admission or death, gender, clinical presentation, cat contact, demographic data, specimen source, diagnostic test results, and treatment and outcome data for seven Swedish patientsa
| Age (yr), gender (reference) | Clinical diagnosis or signs | Cat contact | Origin of tissue sample | gltA sequence | 16S rRNA type | Bh ftsZ variant | Date of serum sample (mo/day/yr) | Antibody titer
|
Treatment | Outcome | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B.h. | B.e. | B.q. | ||||||||||
| 37, M (33) | ARVC, autopsy finding | Former cat owner | Myocardium from right ventricle wallb | B.h. | ND | 1 | 1/5/99 | <64 | <64 | <64 | ||
| 32, M (33) | ARVC (endomyocardial biopsy; presence of late potentials) | Former cat owner | Whole blood | Neg | ND | 2 | 1993 | 256 | 256 | <64 | Tetracycline, 200 mg daily, was started | |
| 1/98 | 256 | 256 | <64 | |||||||||
| 6/10/98 | <64 | 128 | <64 | |||||||||
| 02/17/99 | <64 | <64 | <64 | |||||||||
| 8, M | Chronic multifocal osteomyelitis | Cat scratch 4 mo before onset of symptoms | Bone biopsy specimen from the left sternoclavicular joint | Neg | ND | 1 | 3/20/98 | 128 | <64 | <64 | Erythromycin, 250 mg twice daily, 8 wk | Signs of recovery before treatment |
| 4/17/98 | 256 | <64 | <64 | |||||||||
| 5/19/98 | 64 | <64 | <64 | |||||||||
| 6/18/98 | <64 | <64 | <64 | |||||||||
| 12, F | Chronic multifocal osteomyelitis | Close contact with kittens 2 mo before onset of symptoms | Bone biopsy specimen from right sternoclavicular joint | B.h. | ND | 2 | 2/18/99 | <64 | <64 | <64 | Clarithromycin, 250 mg twice daily, 8 wk | Complete recovery within 4 wk |
| 5/10/99 | <64 | <64 | <64 | |||||||||
| 14, M | Lymph node hyperplasia; abdominal pain; granulocytopenia and trombocytopenia developed | Regular cat contact | Enlarged axillary lymph node | B.h. | ND | 1 | 11/13/98 | <64 | <64 | <64 | (i) T/S, 14 days; (ii) Clarithromycin, 250 mg twice daily, 8 wk | Subjective improvement, but persisting granulocytopenia and trombocytopenia |
| 17, F | Lymph node hyperplasia and anemia | Cat owner | Hyperplastic abdominal lymph node | Neg | ND | 2 | 08/17/98 | <64 | <64 | <64 | Excision of abdominal enlarged lymph gland | Complete recovery |
| 10/7/98 | <64 | <64 | <64 | |||||||||
| 46, F | Parinaud's oculoglandular syndrome of the cat owner | Cat owner | Cat blood | B.h. | ND | 1 | Cat owner, 1/4/99; 5/4/99; cat, 2/1/99 | <64 | <64 | Azitromycin, 500 mg on day 1, 250 mg on days 2 to 5 | Symptoms gradually resolving after 2 days | |
| <64 | <64 | |||||||||||
| 256 | 256 | |||||||||||
Abbreviations: ND, not determined; B.h., B. henselae; B.q., B. quintana; B.e., B. elizabethae, T/S, trimethoprim-sulfamethoxazole; M, male; F, female; ARVC, arrhythmogenic right ventricular cardiomyopathy; neg, negative. At the time of admission all patients lived in Central Sweden.
PCR products were obtained by both PCR assays from the following tissue specimens: (i) left heart ventricle (myocardium and epicardium); (ii) right heart ventricle (superficial myocardium and epicardium); (iii) right heart ventricle (myocardium); (iv) vena cava and right atrium; (v) left atrioventricular (mitral) valve; (vi) papillary muscle adjacent to specimen 5; (vii) lymph node from the proximity of the tracheal bifurcation. Subsequent sequencing of the amplified products gave identical results.
DISCUSSION
In the study described in this report we characterized partial sequences of the 16S rRNA and ftsZ genes of 15 B. henselae isolates. The sequences were aligned to detect nucleotide sequence differences, and the results from the 16S rRNA and ftsZ sequence alignments were correlated. We confirmed the existence of two different 16S rRNA gene variants as previously reported by others (4). The ftsZ gene analysis displayed somewhat more complex results. B. henselae Houston-1, 3508, 3509, 3750, 3883, 3884, 4271, 4272, and FR97/K7 had identical sequences and thus constituted one ftsZ variant (Bh ftsZ 1). However, strain 3507, a type II isolate according to 16S rRNA analysis, was also a Bh ftsZ 1 variant. The remaining type II isolates constituted two Bh ftsZ variants. Thus, isolates 5249 and URLLY8 (Marseille) represented a second variant (Bh ftsZ 2), and German strains FR96/BK3, FR96/BK77, and FR96/BK78 represented a third variant (Bh ftsZ 3). 16S rRNA types I and II and variants Bh ftsZ 1 and 2 were identified in B. henselae strains from both the United States and Europe and were recovered from both humans and cats. Bh ftsZ 3 was discovered only among three German isolates derived from cat blood.
Clinical samples from seven Swedish patients with cardiomyopathy (n = 2), osteomyelitis (n = 2), lymphadenopathy (n = 2), and Parinaud's oculoglandular syndrome (n = 1) were analyzed for signs of Bartonella infection. Among these, four specimens contained a Bh ftsZ 1 variant and three specimens contained a Bh ftsZ 2 variant. Either ftsZ variant was found to cause cardiomyopathy, osteomyelitis, and lymphadenopathy. The serological response to Bartonella antigens was low or negative in the patient sera tested. Antigenic variation among B. henselae strains (8) and low-grade challenge of the infecting agent leading to evasion of the host's immune defense, low antibody response, and chronic Bartonella infection (5, 16) are possible explanations for the positive PCR results and negative serology seen in our patient material.
The B. henselae ftsZ gene region encoding the C-terminal part of the protein was chosen in this study since it is more variable in interspecies comparisons than the 5′-end region. Also, antigenic epitopes responsible for the elicited immune response to the B. bacilliformis FtsZ protein are located in the C-terminal part of the protein, as demonstrated by Padmalayam et al. (20). The nucleotide sequence positions that differ between the Bh ftsZ 1, 2, and 3 variants (Fig. 1) resulted in different amino acids at three positions, one of which was common for Bh ftsZ 2 and 3. Only one of four base differences was a silent substitution. The large percentage of base differences that result in amino acid substitutions and the shared substitutions of many isolates raise the question of whether the amino acid sequence variation might be due to a selective pressure, e.g., from the reservoir host's immune response, and also whether the C-terminal part of the B. henselae FtsZ protein has antigenic properties in humans.
The 16S rRNA genotypes I and II are assumed to represent two different lineages of B. henselae because of the high degree of conservation of the 16S rRNA gene. This is also compatible with the Bartonella ftsZ phylogeny. The intra-Bartonella architecture of trees inferred from ftsZ sequence data by using both distance matrix and parsimony methods had statistically well-supported lineages within the genus with the same topologies (Fig. 1). Interestingly, isolate 3507 displayed a type II 16S rRNA sequence, whereas it contained a Bh ftsZ 1 variant. This might suggest a lateral transfer of genes or convergent evolution. In a study by Sander et al., (27), different fingerprinting methods were applied to the examination of the genetic heterogeneity of B. henselae isolates. Four main variants were proposed. Three of the isolates from that study, as well as reference strain B. henselae Houston-1, were included in our study. We confirmed the results of 16S rRNA typing. The Bh ftsZ 3 variants, comprising strains FR96/BK3, FR96/BK77, and FR96/BK78, were also grouped together in their study. However, FR97/K7 and B. henselae Houston-1 were indiscernible in our study, with identical 16S rRNA and ftsZ sequences, whereas they diverged into two variants in the German study.
In this study, we have also used the ftsZ gene as a target for a diagnostic PCR with clinical samples, and we have identified B. henselae as the etiologic agent in two immunocompetent patients with cardiomyopathy and two immunocompetent patients with multifocal osteomyelitis. These are unusual manifestations of Bartonella infections. Holmberg et al. (11) amplified and sequenced the gltA gene of B. quintana from the myocardial tissue of a 60-year-old Swedish male with fatal myocarditis. Others have also reported on myocarditis associated with Bartonella infections (15; E. B. Breitschwerdt, C. Atkins, and T. Brown, First Int. Conf. Bartonella as Emerging Pathogens, Poster 3, 1999). Chronic recurrent multifocal osteomyelitis is a disease of unknown etiology, is characterized by recurrent attacks of subacute inflammation involving multiple skeletal regions, and usually pursues a self-limiting course (6). Osteomyelitis occurs as a complication of CSD in 0.3% of patients and may affect any bone (3). In a previous report B. henselae has been verified as the etiologic agent of the osteolytic lesions (24).
A larger collection of isolates is needed to confirm our results. An increasing number of B. henselae isolates is expected to be available for further studies. We suggest that the ftsZ gene sequence variation in the 3′ end may prove to be a useful tool for study of the intraspecies phylogeny and epidemiology of B. henselae.
ACKNOWLEDGMENTS
We thank Jill Clarridge, Lars Engstrand, Russel Regnery, and Anna Sander for generously providing B. henselae isolates and Siv Andersson for help with phylogenetic analysis. Petra Råsten Almqvist is thanked for help with the autopsy of patient 1.
A grant from the Swedish Foundation for International Cooperation in Research and Higher Education (STINT) is gratefully acknowledged.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson B, Sims K, Regnery R, Robinson L, Schmidt M J, Goral S, Hager C, Edwards K. Detection of Rochalimaea henselae DNA in specimens from cat scratch disease patients by PCR. J Clin Microbiol. 1994;32:942–948. doi: 10.1128/jcm.32.4.942-948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson B E, Neuman M A. Bartonella spp. as emerging human pathogens. Clin Microbiol Rev. 1997;10:203–219. doi: 10.1128/cmr.10.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmans A M, Schellekens J F, van Embden J D, Schouls L M. Predominance of two Bartonella henselae variants among cat-scratch disease patients in The Netherlands. J Clin Microbiol. 1996;34:254–260. doi: 10.1128/jcm.34.2.254-260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouqui P, La Scola B, Roux V, Raoult D. Chronic Bartonella quintana bacteremia in homeless patients. N Engl J Med. 1999;340:184–189. doi: 10.1056/NEJM199901213400303. [DOI] [PubMed] [Google Scholar]
- 6.Chow L T, Griffith J F, Kumta S M, Leung P C. Chronic recurrent multifocal osteomyelitis: a great clinical and radiologic mimic in need of recognition by the pathologist. APMIS. 1999;107:369–379. doi: 10.1111/j.1699-0463.1999.tb01567.x. [DOI] [PubMed] [Google Scholar]
- 7.Daly J S, Worthington M G, Brenner D J, Moss C W, Hollis D G, Weyant R S, Steigerwalt A G, Weaver R E, Daneshvar M I, O'Connor S P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drancourt M, Birtles R, Chaumentin G, Vandenesch F, Etienne J, Raoult D. New serotype of Bartonella henselae in endocarditis and cat-scratch disease. Lancet. 1996;347:441–443. doi: 10.1016/s0140-6736(96)90012-4. . (Erratum, 347:842.) [DOI] [PubMed] [Google Scholar]
- 9.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 10.Hadfield T L, Warren R, Kass M, Brun E, Levy C. Endocarditis caused by Rochalimaea henselae. Hum Pathol. 1993;24:1140–1141. doi: 10.1016/0046-8177(93)90196-n. [DOI] [PubMed] [Google Scholar]
- 11.Holmberg M, McGill S, Ehrenborg C, Wesslén L, Hjelm E, Darelid J, Blad L, Engstrand L, Regnery R, Friman G. Evaluation of human seroreactivity to Bartonella species in Sweden. J Clin Microbiol. 1999;37:1381–1384. doi: 10.1128/jcm.37.5.1381-1384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs R F, Schutze G E. Bartonella henselae as a cause of prolonged fever and fever of unknown origin in children. Clin Infect Dis. 1998;26:80–84. doi: 10.1086/516256. [DOI] [PubMed] [Google Scholar]
- 13.Kelly T M, Padmalayam I, Baumstark B R. Use of the cell division protein FtsZ as a means of differentiating among Bartonella species. Clin Diagn Lab Immunol. 1998;5:766–772. doi: 10.1128/cdli.5.6.766-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koehler J E, Quinn F D, Berger T G, LeBoit P E, Tappero J W. Isolation of Rochalimaea species from cutaneous and osseous lesions of bacillary angiomatosis. N Engl J Med. 1992;327:1625–1631. doi: 10.1056/NEJM199212033272303. [DOI] [PubMed] [Google Scholar]
- 15.Kordick D L, Brown T T, Shin K, Breitschwerdt E B. Clinical and pathologic evaluation of chronic Bartonella henselae or Bartonella clarridgeiae infection in cats. J Clin Microbiol. 1999;37:1536–1547. doi: 10.1128/jcm.37.5.1536-1547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosoy M Y, Regnery R L, Kosaya O I, Childs J E. Experimental infection of cotton rats with three naturally occurring Bartonella species. J Wildl Dis. 1999;35:275–284. doi: 10.7589/0090-3558-35.2.275. [DOI] [PubMed] [Google Scholar]
- 17.La Scola B, Raoult D. Serological cross-reactions between Bartonella quintana, Bartonella henselae, and Coxiella burnetii. J Clin Microbiol. 1996;34:2270–2274. doi: 10.1128/jcm.34.9.2270-2274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurin M, Eb F, Etienne J, Raoult D. Serological cross-reactions between Bartonella and Chlamydia species: implications for diagnosis. J Clin Microbiol. 1997;35:2283–2287. doi: 10.1128/jcm.35.9.2283-2287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norman A F, Regnery R, Jameson P, Greene C, Krause D C. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–1803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padmalayam I, Anderson B, Kron M, Kelly T, Baumstark B. The 75-kilodalton antigen of Bartonella bacilliformis is a structural homolog of the cell division protein FtsZ. J Bacteriol. 1997;179:4545–4552. doi: 10.1128/jb.179.14.4545-4552.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raoult D, Fournier P E, Drancourt M, Marrie T J, Etienne J, Cosserat J, Cacoub P, Poinsignon Y, Leclercq P, Sefton A M. Diagnosis of 22 new cases of Bartonella endocarditis. Ann Intern Med. 1996;125:646–652. doi: 10.7326/0003-4819-125-8-199610150-00004. . (Erratum, 127:249, 1997.) [DOI] [PubMed] [Google Scholar]
- 22.Regnery R L, Anderson B E, Clarridge J E D, Rodriguez-Barradas M C, Jones D C, Carr J H. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol. 1992;30:265–274. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Regnery R L, Olson J G, Perkins B A, Bibb W. Serological response to “Rochalimaea henselae” antigen in suspected cat-scratch disease. Lancet. 1992;339:1443–1445. doi: 10.1016/0140-6736(92)92032-b. [DOI] [PubMed] [Google Scholar]
- 24.Robson J M, Harte G J, Osborne D R, McCormack J G. Cat-scratch disease with paravertebral mass and osteomyelitis. Clin Infect Dis. 1999;28:274–278. doi: 10.1086/515102. [DOI] [PubMed] [Google Scholar]
- 25.Sander A, Buhler C, Pelz K, von Cramm E, Bredt W. Detection and identification of two Bartonella henselae variants in domestic cats in Germany. J Clin Microbiol. 1997;35:584–587. doi: 10.1128/jcm.35.3.584-587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sander A, Posselt M, Bohm N, Ruess M, Altwegg M. Detection of Bartonella henselae DNA by two different PCR assays and determination of the genotypes of strains involved in histologically defined cat scratch disease. J Clin Microbiol. 1999;37:993–997. doi: 10.1128/jcm.37.4.993-997.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sander A, Ruess M, Bereswill S, Schuppler M, Steinbrueckner B. Comparison of different DNA fingerprinting techniques for molecular typing of Bartonella henselae isolates. J Clin Microbiol. 1998;36:2973–2981. doi: 10.1128/jcm.36.10.2973-2981.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slater L N, Welch D F, Hensel D, Coody D W. A newly recognized fastidious gram-negative pathogen as a cause of fever and bacteremia. N Engl J Med. 1990;323:1587–1593. doi: 10.1056/NEJM199012063232303. [DOI] [PubMed] [Google Scholar]
- 30.Slater L N, Welch D F, Min K W. Rochalimaea henselae causes bacillary angiomatosis and peliosis hepatis. Arch Intern Med. 1992;152:602–606. [PubMed] [Google Scholar]
- 31.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vinson J W, Varela G, Molina-Pasquel C. Trench fever. 3. Induction of clinical disease in volunteers inoculated with Rickettsia quintana propagated on blood agar. Am J Trop Med Hyg. 1969;18:713–722. [PubMed] [Google Scholar]