Abstract
Poly(ADP-ribose) polymerase (PARP) is a DNA binding zinc finger protein that catalyzes the transfer of ADP-ribose residues from NAD+ to itself and different chromatin constituents, forming branched ADP-ribose polymers. The enzymatic activity of PARP is induced upon DNA damage and the PARP protein is cleaved during apoptosis, which suggested a role of PARP in DNA repair and DNA damage-induced cell death. We have generated transgenic mice that lack PARP activity in thymocytes owing to the targeted expression of a dominant negative form of PARP. In the presence of single-strand DNA breaks, the absence of PARP activity correlated with a strongly increased rate of apoptosis compared to cells with intact PARP activity. We found that blockage of PARP activity leads to a drastic increase of p53 expression and activity after DNA damage and correlates with an accelerated onset of Bax expression. DNA repair is almost completely blocked in PARP-deficient thymocytes regardless of p53 status. We found the same increased susceptibility to apoptosis in PARP null mice, a similar inhibition of DNA repair kinetics, and the same upregulation of p53 in response to DNA damage. Thus, based on two different experimental in vivo models, we identify a direct, p53-independent, functional connection between poly(ADP-ribosyl)ation and the DNA excision repair machinery. Furthermore, we propose a p53-dependent link between PARP activity and DNA damage-induced cell death.
Poly(ADP-ribose) polymerase (PARP) is rapidly activated during the cellular response to DNA damage and is part of a large network of safeguard mechanisms protecting cells from genotoxic damage (7, 8, 14, 15, 21). Upon DNA damage, PARP binds to DNA at the sites where double-strand breaks emerge and catalyzes the formation of branched ADP-ribose polymers from NAD+. PARP has a molecular mass of 116 kDa and contains two zinc fingers located at the N terminus next to a bipartite nuclear localization signal sequence which form a DNA binding moiety that functions as a sensor for single- and double-strand DNA breaks. The central part of the enzyme contains the automodification domain, and the C terminus harbors the NAD+ binding site. Physiological substrates for polymer attachment are DNA binding proteins such as histones and PARP itself (7).
The localization of PARP and its activity to modify chromatin constituents after genotoxic stress have prompted speculations about its function in DNA repair, DNA recombination, and the maintenance of genomic integrity (15, 37). In addition, PARP function has been associated with apoptosis because it is cleaved during apoptosis into two subfragments of 89 and 24 kDa (5, 44). To shed more light on the role of PARP and its role in apoptosis and DNA repair, several experimental approaches have been followed. One that involves the inhibition of PARP has been addressed by the use of either antisense strategies or 3-aminobenzamide, which is a competitor for NAD+ (25, 41, 42, 44), or by the expression of the DNA binding domain (DBD) of PARP, which acts as a dominant negative inhibitor (16, 17, 18, 29, 35, 39). With the latter strategy, the activity of PARP to synthesize poly(ADP-ribose) is inhibited by interfering with the ability of endogenous PARP to bind to DNA strand breaks. Experiments with 3-aminobenzamide or the PARP DBD have produced similar results and demonstrated that the absence of PARP activity renders cells more susceptible to apoptosis upon DNA damage either by irradiation or by treatment with alkylating agents such as N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), which predominantly induces DNA single-strand breaks. However, these experiments did not provide direct evidence for a role of PARP or its activity in DNA repair and did not clarify the molecular basis of a function of PARP in apoptosis. The second approach to elucidate the mechanisms that mediate the observed effects of PARP used gene targeting. To date, three groups have inactivated the PARP gene in mice but have obtained different results (8, 26, 51, 52). One group reported enhanced sensitivity of these mice toward alkylating agents and a decreased DNA excision repair rate of PARP null cells (8, 49). In this experiment only the coding region for the second zinc finger was disrupted, leaving the potential to express a truncated PARP protein with residual function. However, further analysis showed that the fourth exon of the PARP gene, which comprises the DBD and nuclear localization signal, was partially disrupted and not expressed in the PARP null mice generated by de Murcia et al. (8). The second group did not observe any effect on DNA damage-induced apoptosis or DNA repair; however, no analysis of DNA break resealing was done in the latter study (51, 52), leaving the question of PARP function unresolved. In addition, the role that p53 and its effectors play in DNA damage-induced apoptosis or DNA repair with regard to PARP function has not been established.
To clarify the role of PARP and p53 in DNA repair and DNA damage-induced apoptosis, we have chosen an approach which allows inhibition of PARP activity in vivo in a particular cell type. We used the lck promoter to express the PARP DBD in T cells of transgenic mice and were able to successfully block poly(ADP-ribosyl)ation in these cells. We report here that the absence of PARP activity clearly enhances apoptosis upon DNA damage. We demonstrate that susceptibility to apoptosis is mediated through the activation of p53 and an accelerated upregulation of Bax. Further, we demonstrate that DNA excision repair is almost completely inhibited in T cells with inactive PARP and that this lack of repair capacity is independent of p53.
MATERIALS AND METHODS
Generation of transgenic mice.
The construct used to generate lck-PARP DBD transgenic mice is shown schematically in Fig. 1a. The construct was obtained by inserting a fragment encoding the human PARP (hPARP) DBD into the lck vector. The vector contained the proximal lck promoter and the genomic sequence of the human growth hormone (hGH) gene in a pBluescript backbone and has been used extensively for the generation of transgenic mice with expression targeted to the T-cell compartment. The construct was freed from backbone sequences, purified, and injected into fertilized mouse oocytes essentially as described elsewhere (12). The fertilized mouse oocytes were derived from matings between (C57BL/6 × C3H)F1 mice. Successful integration of the injected DNA was monitored by Southern analysis of tail tip DNA as described elsewhere (12). All transgenic mouse lines were maintained by breeding the obtained founders for three or more generations with inbred C57BL/6 animals. Preparation of genomic DNA from tail tips and DNA blotting were performed as described elsewhere (12, 36). The PARP probe was the 1.2-kb PARP DBD cDNA fragment (14).
FIG. 1.
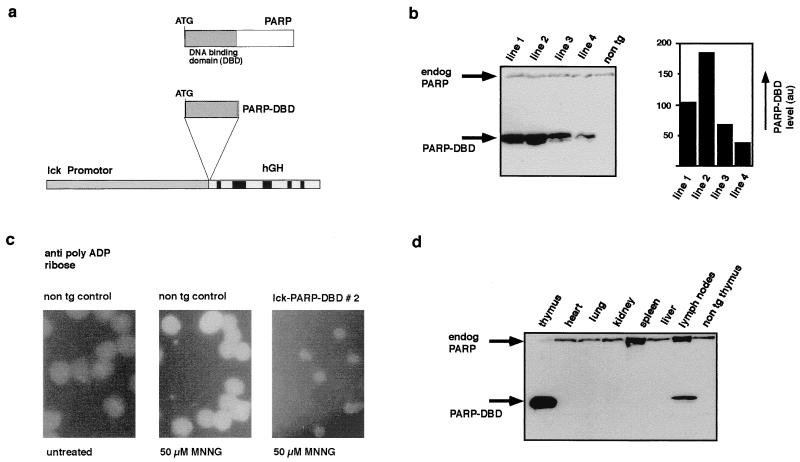
Introduction of a functionally active dominant negative mutant of PARP into the germ line of transgenic mice. (a) Schematic representation of the construct used to express the DBD of PARP in T cells of transgenic mice. (b) Expression of the lck-PARP DBD transgene in extracts from thymi of mice from four different transgenic lines (lck-PARP DBD lines 1, 2, 3, and 4). The level of transgene expression was measured by direct scanning of the ECL-treated membrane after blotting with a video camera and subsequent analysis by the AIDA software (Raytest, Straubenhardt, Germany) on a Fuji phosphorimager. PARP DBD expression was found to be higher in lines 1 and 2 than in lines 3 and 4. The PARP DBD protein was detected with the monoclonal antibody CII10, which recognizes the hPARP transgene and the endogenous mPARP. Relative expression levels are given in arbitrary units (au) obtained through the scanning procedure. (c) Dominant negative effect of the expression of the PARP DBD in T cells. Thymocytes from wild-type and lck-PARP DBD transgenic mice were treated with MNNG, and the formation of poly(ADP-ribose) was followed by immunofluorescence. Compared to nontransgenic (non tg) controls, thymocytes from lck-PARP DBD transgenic mice lack detectable polymer formation after MNNG treatment. (d) Expression level of the PARP DBD in different organs of a transgenic mouse of lck-PARP DBD line 2. As a control, an extract from a nontransgenic thymus was loaded. The PARP DBD protein was detected with the monoclonal antibody CII10, which recognizes the hPARP transgene and the endogenous mPARP. The relative amount of PARP DBD protein expression in splenocytes is detectable but very low (data not shown).
Cell extracts and immunoblotting.
Extracts from thymi were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto membranes, and the hPARP DBD protein or the intact and cleaved mouse PARP (mPARP) was detected with mouse monoclonal antibody CII10, against the DBD, and a secondary antibody carrying horseradish peroxidase. hPARP DBD protein alone was probed with mouse monoclonal antibody FI23 and the appropriate secondary antibody. Enzymatic activity was detected using an ECL kit (Amersham) according to the specifications of the manufacturer. Equal and homogeneous transfer of proteins from the gel to membranes was routinely controlled by Ponceau staining of the membrane. For extract preparation, thymocytes were explanted from animals upon autopsy in 5 ml of phosphate-buffered saline (PBS), collected by centrifugation, and lysed in 100 μl of extraction buffer (50 mM HEPES [pH 7.8], 20 mM NaF, 1 mM sodium orthovanadate, 1 mM sodium molybdate, 450 mM NaCl, 0.2 mM EDTA, 25% glycerol, 1 μg of aprotinin/μl, 1 μg of leupeptin/μl, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 1% NP-40). The lysate was cleared by centrifugation; the supernatant was separated by SDS-PAGE and transferred onto a nitrocellulose membrane. The membrane was blocked for 1 h in PBS with 3% skim milk and then incubated overnight in blocking solution containing the appropriate antibodies at 0.3 μg/ml. Blots were developed with appropriate secondary antibodies and ECL detection reagents (Amersham) as proposed by the manufacturer.
Antibody staining procedures and quantification of apoptotic cells.
Single-cell suspensions were prepared as described previously (30) at the time of autopsy from thymus in PBS supplemented with 0.5% fetal calf serum (staining solution). Cells were washed in this solution and incubated on ice for 30 min with antibodies directly conjugated with fluorochromes. For propidium iodide labeling and quantification of cell death, cells were washed once with cold PBS and fixed with ethanol overnight at −20°C. The cells were again washed and then incubated in propidium iodide (2 μg/ml; Sigma) in PBS-RNase (10 μg/ml) for 30 min at room temperature and analyzed by fluorescence-activated cell sorting (FACS).
Detection of poly(ADP-ribose).
To analyze poly(ADP-ribose) formation, cells from thymi of wild-type and lck-PARP DBD transgenic mice were sedimented for 15 min onto polylysine (Sigma)-pretreated coverslips, washed with PBS, and treated with 50 μM MNNG (Sigma)-PBS supplemented with 1 mM CaCl2 at 37°C for 20 min. After being washed with PBS, the cells were fixed with 10% ice-cold trichloroacetic acid and subjected to PARP-specific immunofluorescence as described elsewhere (18).
DNA fragmentation assay.
Isolated thymocytes were treated with 40 μM MNNG or X irradiation (2 Gy); after incubation for 4 h, the cells were harvested and lysed with proteinase K for 2 h at 56°C. Proteins were extracted with phenol-chloroform, and the DNA was precipitated with 3 volumes of 100% ethanol. After a wash with 70% ethanol, the pellet was resolved in Tris-EDTA buffer, and DNA content was measured at 260 nm. DNA was subjected to agarose gel electrophoresis and analyzed by ethidium bromide staining under a standard UV transilluminator.
Comet assay.
A 100-μl aliquot of a cell suspension was mixed with 500 μl of 0.75% agarose and spread on microscope slides precovered with 0.1% agarose. After gelling at 0°C, the cells were lysed in a 2.5% SDS solution (pH 9.5) for 15 min. Following a 5-min wash in distilled water, electrophoresis was carried out in Tris-borate-EDTA buffer (pH 8.3) for 5 min on a modified flat-bed electrophoresis apparatus with an electric field of 2.5 V/cm (current, 15 to 18 mA). Under these conditions, the DNA of each cell forms a comet-shaped structure with a head and a tail portion. The tail size correlates with the induced DNA damage and chromatin unfolding. DNA repair can be estimated by the shrinkage of the comet tail over time. To examine the individual comets, the slides were stained with a fluorescent dye (propidium iodide). For the measurement of the comets, a fluorescence microscope was coupled with an intensified target camera, and an interactive digital image analysis system was established (2, 3, 31).
Transient transfection assays.
EL-4 T cells (2 × 107) were electroporated at 950 μF and 200 V with 2 μg of human MDM2 (hMDM2) or mutant MDM2-luciferase reporter plasmid (53) and 13 μg of lck-PARP DBD expression construct or empty lck vector as a control. After 14 h, the transfected cells were treated for 20 min with 40 μM MNNG in PBS, then transferred into fresh medium, and incubated for another 4 h before luciferase activity was determined. Other portions of transfected cells were either irradiated with 2 Gy and then incubated in medium for 4 h or treated for 2 h with 50 μM etoposide. After treatment, extracts were prepared and luciferase activity was determined with a luminometer.
RESULTS
We have generated transgenic mice that overexpress the DBD of PARP, which comprises the N terminus of the PARP protein from amino acids 1 to 376 (Fig. 1), with the intention to block the enzymatic activity of the endogenous PARP in a dominant negative manner. It has been demonstrated that the expression of this PARP DBD can efficiently block PARP activity in cell lines (16, 17, 18, 29, 35, 39). Thus, the human cDNA coding for the PARP DBD was placed between the proximal lck promoter and genomic sequences derived from the hGH gene (Fig. 1a) to target expression of the transgene to the T-lymphoid lineage. Mice of two transgenic lines (lck-PARP DBD lines 1 and 2) expressed the PARP DBD transgene at highest levels in the thymus (Fig. 1b) and other T-cell-containing tissues such as spleen and lymph nodes, but not in other organs (Fig. 1d), and at levels significantly higher than the endogenous PARP level (Fig. 1b and d); these mice were chosen for further investigation. To test if the overexpression of this dominant negative PARP mutant can inhibit the enzymatic activity of the endogenous PARP, thymocytes from line 2 transgenic mice were treated with the monofunctional alkylating agent MNNG, which causes DNA single-strand breaks. Cells were then checked for the formation of ADP-ribose polymer by immunofluorescence analysis. This experiment demonstrated clearly the absence of PARP activity in the transgenic thymocytes compared to wild-type thymocytes, indicating the functionality of the transgene and the validity of the chosen approach (Fig. 1c). Similar results were obtained for cells from transgenic line 1 but not for the lines 3 and 4 (data not shown), which express lower levels of the PARP DBD, suggesting that a threshold level is necessary for the DBD to exert its dominant negative effect on the endogenous PARP.
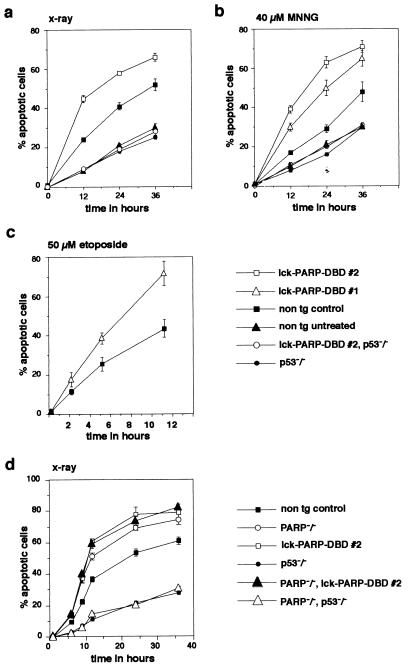
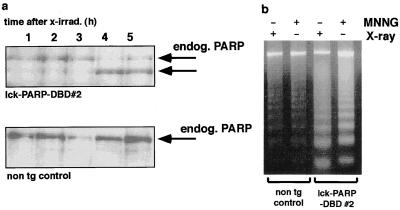
Next we tested the cell death rates of thymocytes from lck-PARP DBD transgenic mice upon the induction of DNA damage by treatment with MNNG or after a single 2-Gy dose of X irradiation, both of which predominantly cause DNA single-strand breaks. This was measured by counting the percentage of cells that contain sub-G1 amounts of propidium iodide-stained DNA after ethanol fixation by FACS (6, 34, 47). Comparing thymocytes from lck-PARP DBD transgenic animals with cells from nontransgenic controls, we found that PARP DBD expression correlated with a significantly higher cell death rate upon both irradiation and MNNG treatment (Fig. 2a and b). Similar results were obtained with etoposide (Fig. 2c). The presence of nucleosomal DNA degradation and cleavage of endogenous PARP (Fig. 3) confirmed that the stimuli used indeed caused apoptotic cell death. As extracts from the same number of cells were loaded per lane, the higher sensitivity of PARP DBD-expressing thymocytes to DNA damage is again reflected by larger amounts of degraded DNA as well as by the earlier cleavage of endogenous PARP (Fig. 3). In contrast, this enhanced sensitivity for DNA damage-induced apoptosis by the PARP DBD was completely lost in thymocytes from mice that carried the lck-PARP DBD transgene and lacked the p53 gene (Fig. 2a and b). The cell death rate of cells from these combinatorial mutant mice after treatment was indistinguishable from rates obtained from untreated nontransgenic controls or from treated p53-deficient thymocytes (Fig. 2a and b).
FIG. 2.
Thymocytes expressing PARP DBD or lacking the PARP gene are more sensitive to apoptosis upon DNA damage only in the presence of p53. (a to c) Thymocytes were explanted from lck-PARP DBD transgenic mice and from nontransgenic (non tg) controls and were either irradiated (2 Gy) (a) or treated with MNNG (40 μM) (b) or etoposide (50 μM) (c). Before treatment and at different time points thereafter, samples were analyzed for the percentage of apoptotic cells. Shown are mean values with standard deviations representative of three independent sets of experiments. Each data point represents an average value with standard deviation obtained from measurements of three different mice. (d) Thymocytes were explanted from PARP null mice (PARP−/−), from mutant mice that are PARP null and express the PARP DBD (PARP−/−, lck-PARP-DBD) from mice that are deficient for both PARP and p53 (PARP−/−, p53−/−), and from nontransgenic controls and were irradiated with a single pulse of 2 Gy. Before treatment and at different times thereafter, samples were analyzed for the percentage of apoptotic cells. Shown are mean values with standard deviations representative of three independent sets of experiments. Each data point represents an average value with standard deviation obtained from measurements of three different mice.
FIG. 3.
Accelerated apoptosis in thymocytes from lck-PARP DBD transgenic mice is reflected by earlier cleavage of endogenous PARP and increased DNA laddering. (a) To monitor endogenous PARP cleavage, thymocytes of lck-PARP DBD transgenic mice and nontransgenic (non tg) littermates were cultured and either left untreated or irradiated with a single dose of 2 Gy. Samples were taken at the indicated time points after the stimulus and were analyzed by Western blotting. Equal loading of each lane was controlled by Ponceau staining (not shown). (b) To confirm that both X irradiation and treatment with MNNG induce apoptosis, genomic DNA prepared from thymocytes of lck-PARP DBD transgenic and nontransgenic control mice after treatment with MNNG was analyzed on agarose gels. The presence of nucleosomal DNA degradation indicates apoptosis.
Next, we compared the sensitivity of thymocytes from lck-PARP DBD transgenic mice upon a single dose (2 Gy) of X irradiation with that of cells from PARP-deficient mice generated by gene targeting (51, 52) as well as combinatorial mutant mice that lack both PARP and p53 or lack PARP but express the PARP DBD transgene (Fig. 2d). Cells from PARP-deficient mice showed the same sensitivity for cell death as cells from lck-PARP DBD mice and cells from mice lacking PARP and expressing the DBD transgene (Fig. 2d). In addition, the absence of p53 rescued thymocytes from apoptosis even in the absence of PARP (Fig. 2d). This demonstrates that thymocytes expressing PARP DBD cannot be distinguished from PARP null cells with regard to a heightened sensitivity for p53-dependent apoptosis in response to DNA damage. Furthermore, we have investigated whether the inactivation of PARP can interfere with DNA repair processes in our transgenic model. The formation of single-strand breaks and subsequent strand break resealing was stimulated by treatment of cells with MNNG or by X irradiation (2 Gy). The repair rate (i.e., resealing of broken DNA ends and chromatin condensation) of these cells was measured by the comet assay (2, 3, 31), which represents one of the most sensitive methods available to quantify DNA repair activity in living cells. Allowing different time periods for repair after the initial stimulus, thymocytes were embedded in agarose, spread on microscope slides, lysed in SDS buffer (pH 9.5), subjected to single-cell gel electrophoresis in buffer (pH 8.3), and stained with propidium iodide. Under these conditions, the DNA of those cells that harbor the induced DNA strand breaks migrates toward the anode and appears as a comet. The total fluorescence of an individual comet is measured, and the fluorescence signals of the head and tail portions are calculated. The tail/head ratio of fluorescence is used as a measure of the individual DNA damage by incision/unwinding and for the resealing-condensation activity.
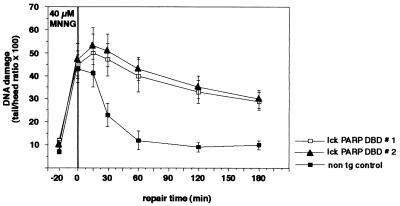
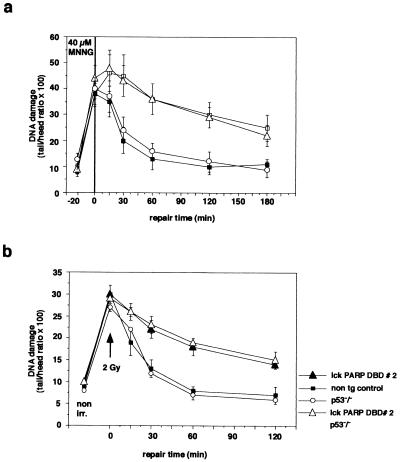
Clearly, thymocytes from both line 1 and line 2 of lck-PARP DBD transgenic mice displayed a significant inhibition of DNA repair activity upon MNNG treatment over a time period of 180 min compared to cells from nontransgenic controls (Fig. 4). Strikingly, this block of DNA resealing was not affected by the absence of p53 in cells from lck-PARP DBD transgenic/p53 null mice (Fig. 5a). Cells from p53 null mice with intact PARP activity showed wild-type repair kinetics (Fig. 5a). In a second set of experiments, the comet assay was performed on thymocytes that were irradiated with 2 Gy. Immediately after this radiation pulse, repair kinetics were taken over a period of 120 min. Similarly to the MNNG-treated cells, thymocytes from lck-PARP DBD transgenic mice showed a strong reduction in the ability to reseal DNA strand breaks and to condense chromatin structure upon X irradiation (Fig. 5b). Again, this reduction did not appear to depend on p53, since irradiated thymocytes from lck-PARP DBD/p53 null mice showed the same degree of repair inhibition as cells from lck-PARP DBD with an intact p53 gene (Fig. 5b). In contrast, thymocytes from p53-deficient mice that did not carry the lck PARP DBD transgene repaired DNA lesions at a wild-type rate (Fig. 5b).
FIG. 4.
Inhibition of poly(ADP-ribosyl)ation inhibits DNA strand break resealing upon MNNG treatment or X irradiation. Comet assays were performed to assess the effects of PARP DBD expression on the resealing of DNA strand breaks. Thymocytes of lck-PARP DBD transgenic mice and age-matched nontransgenic (non tg) control littermates were prepared, treated with 40 μM MNNG for 20 min, stained with propidium iodide, and analyzed for comet formation. Given are average values with standard deviations from 40 cells per sample and time point against elapsed time after the stimulus. In each case, a representative of three independent experiments is depicted. For measurement of the comets, a fluorescence microscope was coupled with an intensified target camera, and an interactive digital image analysis system was established.
FIG. 5.
Influence of p53 and PARP on DNA resealing after induction of damage by MNNG treatment and X irradiation. (a) Comet assay to compare DNA repair kinetics after MNNG treatment between cells from lck-PARP DBD mice, p53-deficient animals, mice that are both p53 deficient and carry an lck-PARP DBD transgene, and age-matched nontransgenic (non tg) littermate controls. (b) Comet assay to compare DNA repair kinetics after X irradiation with a single dose of 2 Gy between cells from lck-PARP DBD mice, p53-deficient animals, mice that are both p53 deficient and carry an lck-PARP DBD transgene, and age-matched nontransgenic littermate controls. Given are average values with standard deviations from 40 cells per sample and time point against elapsed time after the stimulus. In each case, a representative of three independent experiments is depicted. For measurement of the comets, a fluorescence microscope was coupled with an intensified target camera, and an interactive digital image analysis system was established.
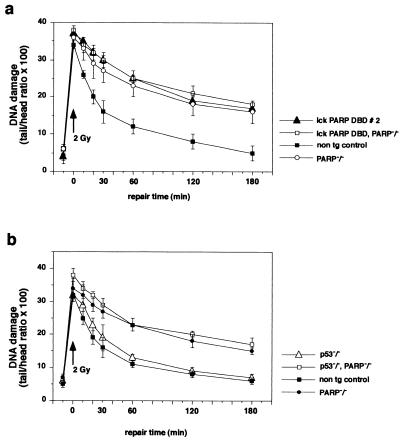
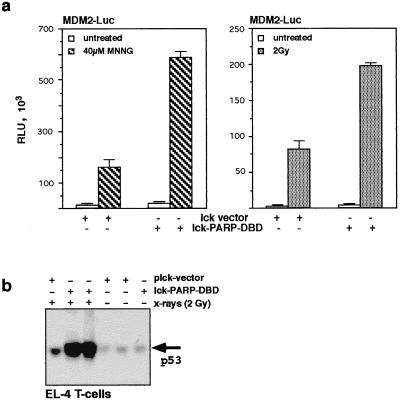
We next wished to investigate whether cells that lack PARP function due to the overexpression of the dominant negative PARP mutant behave differently from cells of mice that are PARP deficient due to gene targeting. We found that thymocytes lacking PARP function due to either gene disruption or DBD expression showed significantly inhibited repair kinetics (Fig. 6). In addition, the lack of p53 did not alter DNA repair under the conditions used regardless of whether PARP was functional (Fig. 6). Expression of a PARP DBD in PARP null thymocytes still showed unaltered inhibition of DNA repair (Fig. 6a). This suggests that the expression of a PARP dominant negative mutant is comparable to the lack of PARP due to gene disruption with regard to DNA single-strand break repair and the requirements of p53 in this process. The finding that lack of PARP activity leads to a sensitization of cells for apoptosis upon DNA damage in a p53-dependent manner prompted us to test the effect of the dominant negative PARP mutant on the activity and the expression of p53 in T cells. To measure p53 activity, we transiently transfected a reporter gene construct containing a luciferase gene driven by the 350-bp fragment of the hMDM2 intronic promoter that includes the p53 response element (53) into EL-4 T cells. After transfection, the cells were either irradiated with 2 Gy or treated with 40 μM MNNG for 20 min, harvested after 4 h, and checked for luciferase activity. Transfection of the lck-PARP DBD construct stimulated transcription from the reporter a further two- to fourfold after MNNG treatment or γ irradiation compared to cells that were transfected with the empty lck vector and were treated likewise (Fig. 7a and b). Western blot analysis showed that the block of poly(ADP-ribosyl)ation through the PARP DBD mutant protein provoked a much higher induction of p53 expression levels in lck-PARP DBD-transfected EL-4 cells than is reached in irradiated cells transfected with the vector control (Fig. 6b). This demonstrated that inhibition of poly(ADP-ribosyl)ation could potentiate p53 activity significantly by allowing or stimulating a strongly increased accumulation of p53 protein levels (23, 42, 43).
FIG. 6.
Influence of p53 and PARP deficiency on DNA resealing after induction of damage by X irradiation. (a) Comet assay to compare DNA repair kinetics after X irradiation with a single dose of 2 Gy between cells from nontransgenic (non tg) controls, PARP null mice (PARP−/−), lck-PARP DBD mice, and mutant mice that both lack the PARP gene and express the PARP DBD transgene (lck-PARP DBD, PARP−/−). (b) Comparison of DNA repair kinetics from PARP null mice (PARP−/−), p53-deficient animals (p53−/−), mice that are both p53 deficient and PARP null (p53−/−, PARP−/−), and age-matched nontransgenic littermate controls. Given are average values with standard deviations from 40 cells per sample and time point against elapsed time after the stimulus. In each case, a representative of three independent experiments is depicted. For measurement of the comets, a fluorescence microscope was coupled with an intensified target camera, and an interactive digital image analysis system was established.
FIG. 7.
Expression and activity of p53 upon DNA damage depend on poly(ADP-ribosyl)ation. (a) Luciferase activity (in relative light units [RLU]) after transient transfection of EL-4 T cells with the lck-driven PARP DBD (lck-PARP-DBD) or the empty lck vector. Transcriptional transactivation of the MDM2 reporter was measured in untreated cells or upon treatment with MNNG (left) or after X irradiation (2 Gy; right). Given are average values with standard deviations from three independent transfections. (b) Immunoblot analysis of p53 expression levels. EL-4 T cells were transiently transfected with either the empty lck vector or the lck-PARP DBD expression construct and left untreated or irradiated (2 Gy). After 4 h of exposure to the stimulus, cell extracts were prepared and analyzed by Western blotting. Equal loading of each lane was controlled by Ponceau staining (not shown).
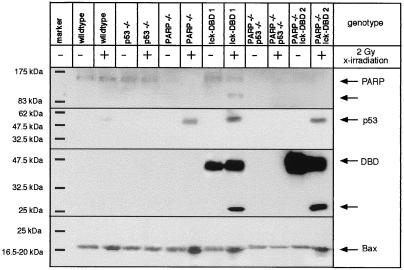
A number of potential direct or indirect downstream effectors of p53 have been identified, among them the proapoptotic protein Bax (27, 28). Thymocytes from lck-PARP DBD transgenic, p53−/−, and PARP−/− mice, combinatorial lck-PARP DBD/PARP−/− and PARP−/−/p53−/− mutants, and nontransgenic controls were X irradiated with a single dose of 2 Gy and checked for p53 and Bax protein expression and N-terminal PARP/DBD cleavage by Western blotting. p53 as well as Bax expression is slightly induced in wild-type thymocytes 3 h after 2-Gy irradiation (Fig. 8). However, thymocytes with a p53 wild-type gene but disrupted PARP function, due to either gene targeting or DBD expression, showed higher levels of p53 and Bax protein 3 h after 2-Gy irradiation compared to wild-type controls (Fig. 8). This effect was not observed in thymocytes that were p53 deficient. This suggests that one effect of the loss of PARP function is a premature activation and accumulation of p53, which in turn may provoke an accelerated upregulation of Bax protein levels (23, 27, 43). In addition, the p53-dependent increase of Bax protein level is accompanied by an accelerated cleavage of endogenous PARP or of both endogenous PARP and PARP DBD in those thymocytes which expressed the transgene (Fig. 8).
FIG. 8.
Expression levels of PARP, the PARP DBD transgene, p53, and Bax upon X irradiation in transgenic, null, and combinatorial mutant mice. The immunoblot shows expression levels of the proteins in thymocytes from the indicated mice 3 h after X irradiation (2 Gy). Equal loading of each lane was controlled by Ponceau staining (not shown).
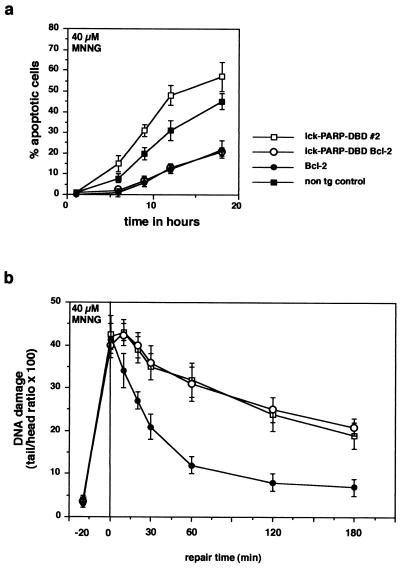
Next, we wanted to test whether the higher rate of apoptosis and the lack of DNA repair in PARP DBD-expressing cells are influenced by the antiapoptotic factor Bcl-2. To this end, the lck-PARP DBD transgenic mice were crossed with Eμ bcl-2 transgenic animals, which express high levels of Bcl-2 in all lymphoid cells, which protect them from various apoptotic stimuli with the exception of Fas triggering (13, 40). Thymocytes from animals carrying both lck-PARP DBD and Eμ bcl-2 alleles were fully protected from DNA damage-induced death by 40 μM MNNG (Fig. 9a) or 2-Gy irradiation (not shown). The effect of the PARP DBD was eliminated, and the cells from doubly transgenic animals were protected to the same degree as cells expressing only Bcl-2. However, the presence of Bcl-2 did not alter repair kinetics of thymocytes treated with MNNG or with 2-Gy X irradiation (not shown) and did not affect the inhibitory activity of PARP DBD on DNA repair (Fig. 9b).
FIG. 9.
High levels of Bcl-2 block sensitization of thymocytes for apoptosis by PARP DBD but do not interfere with DNA repair. (a) Thymocytes were explanted from lck-PARP DBD, Eμ bcl-2, or double-transgenic mice and from nontransgenic (non tg) controls and were treated with MNNG (40 μM). Before treatment and at different times thereafter, samples were analyzed for the percentage of apoptotic cells. Shown are mean values with standard deviations representative of three independent sets of experiments. (b) Comet assay to compare DNA repair kinetics after MNNG treatment between cells from lck-PARP DBD mice, Eμ bcl-2 animals, and mice carry both an lck-PARP DBD transgene and an Eμ bcl-2 transgene (cells from age-matched nontransgenic littermate controls behaved like cells from Eμ bcl-2 mice [not shown]). Given are average values with standard deviations from 40 cells per sample and time point against elapsed time after the stimulus. In each case, a representative of three independent experiments is depicted.
DISCUSSION
Our finding that thymocytes from PARP DBD-expressing transgenic mice have a drastically reduced repair rate of DNA single-strand breaks (Fig. 3) is in complete agreement with data generated by Trucco et al. with fibroblasts from their PARP-deficient mice (49). In contrast, the Wang group did not find alterations in the expression of reporter genes from damaged plasmid DNA transfected in cells derived from their PARP null mice (51). Although the authors conclude from this that PARP is not involved in DNA repair, their experimental design may have been not sensitive enough to detect a role of PARP in DNA single-strand break repair. The fact that comet assays with cells from the PARP null mice of Trucco et al. (49) and with thymocytes from our PARP DBD transgenic mice both show drastically reduced DNA repair indicates that expression of a PARP DBD in transgenic mice creates a phenotype that is identical to the one found in animals with targeted deletions of the PARP gene. Moreover, our own experiments in which we compare thymocytes from PARP null mice from Wang et al. (51) with thymocytes from our lck-PARP DBD mice also showed a similar strong reduction of DNA repair rates. In addition, our data are similar to a number of experimental findings with PARP inhibitors or with PARP antisense strategies (9). Thus, we conclude that our strategy to inactivate only one function of PARP, namely, its enzymatic activity, and leave its DNA binding ability intact by expressing the PARP DBD was successful in generating a bona fide alternative knockout model. This model not only allows study of a distinct PARP function but may also serve to resolve discrepancies that have arisen between different PARP knockout models generated by gene targeting (8, 51, 52).
The higher rate of DNA damage-induced apoptosis that we observe in cells that lack PARP activity is in agreement with findings from PARP null mice generated by the de Murcia group (8) but is at variance with the first results on PARP-deficient mice obtained by Wang et al. (51). However, Wang and coworkers reported in a subsequent study a significantly increased sensitivity of PARP null animals toward alkylating agents without looking at a particular cell type (52). It has been demonstrated (references 4, 8, 22, and 52 and this study) that different cell types of PARP knockout mice behave differently with regard to apoptosis and necrosis. It is possible that fibroblasts or myeloid cells may need an early burst of poly(ADP-ribosyl)ation to undergo apoptosis (44). However, in our study using thymocytes, the situation is very likely to be different, and the absence of poly(ADP-ribosyl)ation clearly sensitizes cells for apoptosis regardless of whether the lack of PARP activity is due to a targeted knockout of the PARP gene or expression of the PARP DBD.
Our data demonstrate a strict requirement of p53 for accelerated cell death after DNA damage in PARP DBD-expressing thymocytes and show that Bcl-2 overexpression inhibits this sensitization. Mice that express a PARP DBD but lack p53 allowed us to study the role of p53 and PARP in the DNA excision repair process of single-strand breaks and to clarify the question of whether a functional connection exists between p53 activation and poly(ADP-ribosyl)ation. Comet assays demonstrated that the inactivation of PARP and the loss of the ability to react to DNA damage with poly(ADP-ribosyl)ation dramatically reduce the DNA repair capacity of cells regardless of whether p53 or Bcl-2 is present. This was true for thymocytes from PARP null mice and from lck-PARP DBD transgenic mice. Both stimuli used here, X irradiation and MNNG treatment, are known to induce the DNA excision repair pathways in cells (18, 29, 50). Our results suggest that these DNA excision repair processes require poly(ADP-ribosyl)ation but are completely independent of the presence or activation of Bcl-2 or p53. This is a novel and unexpected finding which makes a role of p53 in base excision repair processes very unlikely, while it is well established that p53 is required for other repair pathways (45, 50). As expected, Bcl-2 acts downstream of DNA damage and excision repair as an antiapoptotic molecule that does not protect from damage but inhibits apoptosis, most likely by counteracting Bax-induced apoptosis.
The upregulation of p53 expression and activity and the accelerated induction of Bax expression strictly correlated with the expression of the PARP DBD but also with the absence of PARP in cells from PARP null mice and thus with the lack of poly(ADP-ribosyl)ation. This offers an explanation for our findings that thymocytes from lck-PARP DBD mice and from PARP null mice undergo apoptosis much more rapidly than cells with intact PARP. The cause for this upregulation of p53 and Bax and for the observed sensitization for apoptosis by PARP inhibition could be the lack of DNA repair and an increased persistence of DNA strand breaks in cells with no PARP activity. However, we cannot rule out that the PARP DBD used here has other, unknown functions besides blocking poly(ADP-ribosyl)ation that may interfere with apoptosis and DNA repair. Although it has not been demonstrated unequivocally, PARP is thought to dissociate from DNA after automodification and the modification of other chromatin constituents (1, 32, 33, 38). It is possible that the PARP DBD molecule remains bound to DNA (46) and blocks DNA break resealing, which would exclude a direct role of PARP or poly(ADP-ribose) in the repair process. However, in light of our findings with cells from PARP knockout mice, we consider this unlikely and favor a model with a direct role of PARP and the poly(ADP-ribose) polymer in DNA damage-induced apoptosis and DNA break rejoining for several reasons.
The published PARP knockout study that analyzed DNA resealing in fibroblasts clearly demonstrated an inhibition of DNA repair in the absence of PARP (49). The experiments presented here, using thymocytes from PARP-deficient mice or PARP DBD-expressing mice, as well as experiments where PARP was depleted by antisense RNA expression (9) confirm this. All of these findings suggest a dependence of DNA resealing on the ADP-ribose polymer since in different experimental strategies lack of repair correlates with the absence of polymer. In addition, Lee et al. (19, 20) showed that PARP could act as an inhibitor of simian virus 40 DNA replication in vitro when only the DNA polymerase α-primase (monopolymerase) system was used. This inhibition was due to binding of PARP to the ends of nascent DNA chains rather than to its ability to synthesize poly(ADP-ribose). However, when a dipolymerase system including the proliferating cellular nuclear antigen (PCNA) and polymerase δ was used, PARP was displaced from the strand breaks and both leading- and lagging-strand synthesis could occur (10). Interestingly, it was shown recently that both PCNA and polymerase δ participate in DNA base excision repair (48). Thus, it is conceivable that a PARP DBD molecule is replaced from a DNA break by the base excision repair machinery. Furthermore, we show that p53 levels as well as Bax levels rise well above control levels in PARP DBD-expressing thymocytes as well as in PARP-negative thymocytes upon DNA damage. It is generally accepted that DNA strand breaks serve as a sensor for the induction of p53 expression. Hence, it is unlikely that the PARP DBD remains irreversibly bound to DNA strand breaks since this may interfere with the strong induction of p53 expression levels observed here.
Taken together, the data presented here provide direct experimental evidence for a connection between PARP activity and DNA base excision repair that is independent of p53 and for a second link between PARP and DNA damage-induced apoptosis that requires p53 and is inhibited by Bcl-2. Our findings favor a model that would integrate poly(ADP-ribosyl)ation and the ADP-ribose polymer itself into the signaling of DNA damage-induced cell death and the execution of DNA base excision repair and place it as a regulatory element upstream of p53 and Bax. All experimental data that support this model were gathered in primary murine thymocytes. Although it is possible that this model holds generally, we cannot rule out that alternative mechanisms prevail in other cell systems.
ACKNOWLEDGMENTS
We thank R. Perlmutter for the lck-hGH expression vector and W. Deppert for the intact and mutant hMDM2-luciferase reporter gene constructs. We are indebted to X.-Z. Wang, Lyon, France, for the gift of PARP null mice.
This work was supported by grant Mo 435/9-1 from the Deutsche Forschungsgemeinschaft and by the Fond der chemischen Industrie.
REFERENCES
- 1.Althaus F R, Hofferer L, Kleczkowska H E, Malanga M, Naegeli H, Panzeter P L, Realini C A. Histone shuttling by poly ADP-ribosylation. Mol Cell Biochem. 1994;138:53–59. doi: 10.1007/BF00928443. [DOI] [PubMed] [Google Scholar]
- 2.Böcker W, Bauch T, Müller W U, Streffer C. Technical report: image analysis of comet assay measurements. Int J Radiat Biol. 1997;72:449–460. doi: 10.1080/095530097143220. [DOI] [PubMed] [Google Scholar]
- 3.Böcker W, Rolf W, Bauch T, Müller W U, Streffer C. Automated comet assay analysis. Cytometry. 1999;35:134–144. [PubMed] [Google Scholar]
- 4.Burkart V, Wang Z-Q, Radons J, Heller B, Herceg Z, Stingl L, Wagner E F, Kolb H. Mice lacking the poly(ADP-ribose) polymerase gene are resistant to pancreatic beta-cell destruction and diabetes development induced by streptozocin. Nat Med. 1999;5:314–319. doi: 10.1038/6535. [DOI] [PubMed] [Google Scholar]
- 5.D'Amours D, Germain M, Orth K, Dixit V M, Poirier G G. Proteolysis of poly(ADP-ribose) polymerase by caspase 3: kinetics of cleavage of mono(ADP-ribosyl)ated and DNA-bound substrates. Radiat Res. 1998;150:3–10. [PubMed] [Google Scholar]
- 6.Darzynkiewicz Z, Bruno S, DelBino G, Gorzyca W, Hotz M A, Lassota P, Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- 7.de Murcia G, Menissier de Murcia J. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem Sci. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 8.de Murcia J M, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver F J, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding R, Pommier Y, Kang V H, Smulson M. Depletion of poly(ADP-ribose) polymerase by antisense RNA expression results in a delay in DNA strand break rejoining. J Biol Chem. 1992;267:12804–12812. [PubMed] [Google Scholar]
- 10.Eki T, Hurwitz J. Influence of poly(ADP-ribose) polymerase on the enzymatic synthesis of SV40 DNA. J Biol Chem. 1991;266:3087–3100. [PubMed] [Google Scholar]
- 11.Enoch T, Norbury C. Cellular responses to DNA damage: cell cycle checkpoints, apoptosis and the roles of p53 and ATM. Trends Biochem Sci. 1995;20:426–430. doi: 10.1016/s0968-0004(00)89093-3. [DOI] [PubMed] [Google Scholar]
- 12.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo, a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 13.Huang D C, Hahne M, Schroeter M, Frei K, Fontana A, Villunger A, Newton K, Tschopp J, Strasser A. Activation of Fas by FasL induces apoptosis by a mechanism that cannot be blocked by Bcl-2 or Bcl-x(L) Proc Natl Acad Sci USA. 1999;96:14871–14876. doi: 10.1073/pnas.96.26.14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson S P. The recognition of DNA damage. Curr Opin Genet Dev. 1996;6:12–25. doi: 10.1016/s0959-437x(96)90005-2. [DOI] [PubMed] [Google Scholar]
- 15.Jeggo P A. DNA repair: PARP—another guardian angel? Curr Biol. 1998;8:R49–R51. doi: 10.1016/s0960-9822(98)70032-6. [DOI] [PubMed] [Google Scholar]
- 16.Küpper J H, de Murcia G, Bürkle A. Inhibition of poly(ADP-ribosyl)ation by overexpressing the poly(ADP-ribose) polymerase DNA-binding domain in mammalian cells. J Biol Chem. 1990;265:18721–18724. [PubMed] [Google Scholar]
- 17.Küpper J H, Müller M, Bürkle A. Trans-dominant inhibition of poly(ADP-ribosyl)ation potentiates carcinogen induced gene amplification in SV40-transformed Chinese hamster cells. Cancer Res. 1996;56:2715–2717. [PubMed] [Google Scholar]
- 18.Küpper J H, Müller M, Jacobson M K, Tatsumi-Miyajima J, Coyle D L, Jacobson E L, Bürkle A. trans-dominant inhibition of poly(ADP-ribosyl)ation sensitizes cells against γ-irradiation and N-methyl-N′-nitro-N-nitrosoguanidine but does not limit DNA replication of a polyomavirus replicon. Mol Cell Biol. 1995;15:3154–3163. doi: 10.1128/mcb.15.6.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S H, Eki T, Hurwitz J. Synthesis of DNA containing the simian virus 40 origin of replication by the combined action of DNA polymerases alpha and delta. Proc Natl Acad Sci USA. 1989;86:7361–7365. doi: 10.1073/pnas.86.19.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S H, Kwong A D, Ishimi Y, Hurwitz J. Studies on the DNA elongation inhibitor and its proliferating cell nuclear antigen-dependent control in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1989;86:4877–4881. doi: 10.1073/pnas.86.13.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindahl T, Satoh M S, Poirier G G, Klungland A. Post translational modification of poly (ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 22.Love S, Barber R, Wilcock G K. Increased poly(ADP-ribosyl)ation of nuclear proteins in Alzheimer's disease. Brain. 1999;122:247–253. doi: 10.1093/brain/122.2.247. [DOI] [PubMed] [Google Scholar]
- 23.Lu X, Lane D P. Differential induction of transcriptionally active p53 following UV or ionising radiation: defects in chromosome instability syndromes? Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 24.Malanga M, Pleschke J M, Kleczkowska H E, Althaus F R. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J Biol Chem. 1998;273:11839–11843. doi: 10.1074/jbc.273.19.11839. [DOI] [PubMed] [Google Scholar]
- 25.Masutani M, Nozaki T, Wakabayashi K, Sugimura T. Role of poly(ADP-ribose) polymerase in cell-cycle checkpoint mechanisms following gamma-irradiation. Biochimie. 1995;77:462–465. doi: 10.1016/0300-9084(96)88161-2. [DOI] [PubMed] [Google Scholar]
- 26.Masutani M, Suzuki K, Kamada N, Watanabe M, Ueda O, Nozaki T, Jishage K, Watanabe T, Sugimoto T, Nakagama H, Ochiya T, Sugimura T. Poly(ADP-ribose) polymerase gene disruption conferred mice resistant to streptozotocin-induced diabetes. Proc Natl Acad Sci USA. 1999;96:2301–2304. doi: 10.1073/pnas.96.5.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 28.Miyashita T, Krajewski S, Krajewska M, Wang H G, Lin H K, Liebermann D A, Hoffman I B, Reed J C. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene. 1994;6:1799–1805. [PubMed] [Google Scholar]
- 29.Molinete M, Vermeulen W, Bürkle A, Menissier-de Murcia J, Küpper J H, Hoeijmakers J H, de Murcia G. Overproduction of the poly(ADP-ribose) polymerase DNA-binding domain blocks alkylation-induced DNA repair synthesis in mammalian cells. EMBO J. 1993;12:2109–2117. doi: 10.1002/j.1460-2075.1993.tb05859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Möröy T, Grzeschiczek A, Petzold S, Hartmann K U. Expression of a pim-1 transgene accelerates lymphoproliferation and inhibits apoptosis in lpr/lpr mice. Proc Natl Acad Sci USA. 1993;90:10734–10738. doi: 10.1073/pnas.90.22.10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller W U, Bauch T, Streffer C, Niedereichholz F, Böcker W. Comet assay studies of radiation-induced DNA damage and repair in various tumour cell lines. Int J Radiat Biol. 1994;65:315–319. doi: 10.1080/09553009414550371. [DOI] [PubMed] [Google Scholar]
- 32.Panzeter P L, Zweifel B, Malanga M, Waser S H, Richard M, Althaus F R. Targeting of histone tails by poly(ADP-ribose) J Biol Chem. 1993;268:17662–17664. [PubMed] [Google Scholar]
- 33.Realini C A, Althaus F R. Histone shuttling by poly(ADP-ribosylation) J Biol Chem. 1992;267:18858–18865. [PubMed] [Google Scholar]
- 34.Rowan S, Ludwig R L, Haupt Y, Bates S, Lu X, Oren M, Vousden K. Specific loss of apoptotic but not cell-cycle arrest function in a human tumor derived p53 mutant. EMBO J. 1996;15:827–838. [PMC free article] [PubMed] [Google Scholar]
- 35.Rudat V, Küpper J H, Weber K J. Trans-dominant inhibition of poly(ADP-ribosyl)ation leads to decreased recovery from ionizing radiation-induced cell killing. Int J Radiat Biol. 1998;73:325–330. doi: 10.1080/095530098142428. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Satoh M S, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 38.Satoh M S, Poirier G G, Lindahl T. Dual function for poly(ADP-ribose) synthesis in response to DNA breakage. Biochemistry. 1994;33:7099–7106. doi: 10.1021/bi00189a012. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber V, Hunting D, Trucco C, Gowans B, Grunwald D, de Murcia G, Menissier-de Murcia J. A dominant-negative mutant of human poly(ADP-ribose) polymerase affects cell recovery, apoptosis, and sister chromatid exchange following DNA damage. Proc Natl Acad Sci USA. 1995;92:4753–4757. doi: 10.1073/pnas.92.11.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sentman C L, Shutter J R, Hockenbery D, Kanagawa O, Korsmeyer S J. Bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell. 1991;68:879–888. doi: 10.1016/0092-8674(91)90361-2. [DOI] [PubMed] [Google Scholar]
- 41.Shiokawa D, Maruta H, Tanuma S. Inhibitors of poly(ADP-ribose) polymerase suppress nuclear fragmentation and apoptotic-body formation during apoptosis in HL-60 cells. FEBS Lett. 1997;413:99–103. doi: 10.1016/s0014-5793(97)00887-9. [DOI] [PubMed] [Google Scholar]
- 42.Simbulan-Rosenthal C M, Rosenthal D S, Ding R, Bhatia K, Smulson M E. Prolongation of the p53 response to DNA strand breaks in cells depleted of PARP by antisense RNA expression. Biochem Biophys Res Commun. 1998;253:864–868. doi: 10.1006/bbrc.1998.9792. [DOI] [PubMed] [Google Scholar]
- 43.Simbulan-Rosenthal C M, Rosenthal D S, Luo R, Smulson M E. Poly(ADP-ribosyl)ation of p53 during apoptosis in human osteosarcoma cells. Cancer Res. 1999;59:2190–2194. [PubMed] [Google Scholar]
- 44.Simbulan-Rosenthal C M, Rosenthal D S, Iyer S, Boulares A H, Smulson M E. Transient poly(ADP-ribosyl)ation of nuclear proteins and role of poly(ADP-ribose) polymerase in the early stages of apoptosis. J Biol Chem. 1998;273:13703–13712. doi: 10.1074/jbc.273.22.13703. [DOI] [PubMed] [Google Scholar]
- 45.Smith M L, Chen I T, Zhan Q, O'Connor P M, Fornace A J. Involvement of the p53 tumor suppressor in repair of u.v.-type DNA damage. Oncogene. 1995;10:1053–1059. [PubMed] [Google Scholar]
- 46.Smulson M E, Pang D, Jung M, Dimtchev A, Chasovskikh S, Spoonde A, Simbulan-Rosenthal C, Rosenthal D, Yakovlev A, Dritschilo A. Irreversible binding of poly(ADP)ribose polymerase cleavage product to DNA ends revealed by atomic force microscopy: possible role in apoptosis. Cancer Res. 1998;58:3495–3498. [PubMed] [Google Scholar]
- 47.Sofer-Levi Y, Resnitzki D. Apoptosis induced by ectopic expression of cyclin D1 but not cyclin E. Oncogene. 1996;13:2431–2437. [PubMed] [Google Scholar]
- 48.Stucki M, Pascucci B, Parlanti E, Fortini P, Wilson S H, Hübscher U, Dogliotti E. Mammalian base excision repair by DNA polymerases delta and epsilon. Oncogene. 1998;17:835–843. doi: 10.1038/sj.onc.1202001. [DOI] [PubMed] [Google Scholar]
- 49.Trucco C, Oliver F J, de Murcia G, Menissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Freidberg E C, Evans M K, Taffe B G. p53 modulation of TFIIH-associated nucleotide excision repair activity. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 51.Wang Z Q, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner E F. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z Q, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner E F. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 1995;23:2584–2592. doi: 10.1093/nar/23.14.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]