Abstract
Since the introduction of antiviral compounds such as lamivudine and famciclovir in the treatment schedules of patients with chronic hepatitis B virus (HBV) infection, the accumulation of a variety of mutations in the HBV polymerase gene has been observed. The selection of these mutations is generally considered the cause of viral nonresponsiveness and treatment failure. Therefore, the detection of these mutations is of clinical importance. Previously genotyped HBV strains isolated from treated and untreated patients were amplified with primers specific for the HBV polymerase region from amino acids 465 to 562. Amplified products were cloned into plasmid vectors. The clones were used as reference strains. A set of 38 highly specific oligonucleotide probes covering three different codon positions, L528M, M552V/I, and V/L/M555I, were selected. These probes were applied as 19 different lines on a membrane strip. The strips were then hybridized with PCR fragments from the reference panel, revealing the amino acids at the three codon positions simultaneously for each clone. PCR products generated from two patients infected with HBV genotypes A and C, respectively, and treated with nucleoside analogs were analyzed on these strips. A gradual increase in genetic HBV polymerase complexity was observed in follow-up samples compared to that in pretreatment samples. Additional analysis of HBV polymerase DNA fragments in recombinant plasmid clones demonstrated the existence of (i) clones with double mutations, (ii) clones with single mutations at either codon 528, 552, or 555, and (iii) the simultaneous occurrence of two or more viral populations within one sample. This line probe assay detected the complex quasispecies nature of HBV and provided some insight into the dynamics of resistance mutations.
Successful antiviral therapy in chronic hepatitis B virus (HBV)-infected patients with active wild-type virus replication is characterized by clearance of HBV DNA from the blood circulation. This is followed by clearance of hepatitis B e antigen (HBeAg) and seroconversion to positivity for anti-HBe antibodies. Unfortunately, the disappearance of HBV DNA is not always followed by HBeAg seroconversion, and quantitative HBeAg measurements were suggested to have predictive value for the outcome of antiviral therapy (7). Currently, the two licensed agents for treatment of chronic hepatitis B are alpha interferon and lamivudine [(−) 2′-deoxy-3′-thiacytidine]. Lamivudine has been shown to have antiviral activity by inhibiting viral DNA synthesis, while alpha interferon has antiviral and immunomodulatory properties. Both drugs may be given as monotherapy or combination therapy (10, 14). Another compound with direct antiviral action, famciclovir [9-(4-hydroxy-3 hydroxymethyl-but-1-yl) guanine], is currently undergoing clinical evaluation (12).
Several independent reports illustrated the development of famciclovir- and lamivudine-resistant HBV isolates (for reviews, see references 2 and 27). Sequence analysis revealed the emergence of a specific mutation in the tyrosine-methionine-aspartate-aspartate (YMDD) motif of the HBV polymerase (HBpol) gene, whereby the methionine is replaced by either an isoleucine or a valine. Another important mutation site is located 24 amino acids (aa) upstream from the YMDD motif, whereby a leucine (L) replaces a methionine (M). Overviews of the mutations observed previously following lamivudine or famciclovir treatment and the consequences of viral fitness are available (2, 3, 6, 8, 9, 10–17, 20, 25).
In the present study, viral isolates from a random selection of untreated patients as well as from two patients with documented antiviral treatment failure were used to optimize a line probe assay (LiPA). This assay allows antiviral resistance testing at three different aa positions (positions 528, 552, and 555) in the HBpol of viral populations from HBV-infected individuals.
MATERIALS AND METHODS
Patients.
The two patients included in this study were selected on the basis of the following criteria: (i) at least three samples were available, i.e., prior to therapy, during therapy, and at the time of viral breakthrough during therapy; (ii) the patients had virological (viral load) and biochemical (alanine aminotransferase [ALT] levels) evidence of treatment failure; and (iii) the virus from each patient belonged to a different genotype. The following HBV genotypes were included: genotype A (patient A) and genotype C (patient C). Follow-up samples from antiviral agent-treated patients infected with other genotypes were not available, although for most of the genotypes cross-sectional samples could be analyzed (data not shown). HBV genotypes were determined by a research version of the HBV genotyping LiPA (26).
HBV DNA purification and amplification.
HBV DNA was isolated from serum or plasma by using the commercially available High Pure PCR Template Preparation Kit (Boehringer Mannheim, Brussels, Belgium). Purified DNA of the HBpol region was amplified by a nested PCR approach. In a total reaction volume of 50 μl, 10 μl of DNA was added to 5 μl of 10× buffer, 0.4 μl of 10 mM deoxynucleoside triphosphates, 10 pmol of each primer, and 1 U of Taq polymerase (Stratagene Europe, Amsterdam, The Netherlands), and the mixture was brought to volume with high-pressure liquid chromatography-grade H2O. The HBpol region was amplified by using the following primer combinations: outer sense primer HBPr134 (5′-TGCTGCTATGCCTCATCTTC-3′), outer antisense primer HBPr135 (5′-CA(G/A)AGACAAAAGAAAATTGG-3′), nested sense primer HBPr75 (5′-CAAGGTATGTTGCCCGTTTGTCC-3′), and nested antisense primer HBPr 94 (5′-GG(T/C)A(A/T)AAAGGGACTCA(A/C)GATG-3′). The thermocycling profile consisted of annealing at 45°C, extension at 72°C, and denaturation at 94°C for 30 s each. The outer PCR contained 40 cycles; the nested reaction contained 35 cycles. Nested amplification products were (primers included) 341 bp long, analyzed on a 2% agarose gel, and visualized by staining with ethidium bromide. The LiPA experiments used primers biotinylated at their 5′ ends.
Plasmid cloning and DNA purification.
Two microliters of the amplification product was mixed with 1 μl of the pretreated plasmid (EcoRV site of pGemT; Promega, Leiden, The Netherlands) and ligated with Ready to Go T4 ligase (Pharmacia, Leusden, The Netherlands). After transformation in competent Escherichia coli strains, single recombinant clones were selected and plasmid DNA was purified with the Qiaprep 96 Turbo BioRobot kit (Qiagen, Leusden, The Netherlands). Inserts from recombinant clones were amplified by PCR with either plasmid-derived primers or the nested HBV primers. The inserts were used either for sequencing or for LiPA analysis.
Sequence analysis.
Double-stranded sequences were obtained directly from biotinylated PCR products or, in the case of recombinant clones, by using vector-derived sequencing primers as described previously (21).
LiPA strip preparation and test performance.
Specific probes were designed by considering the parameters percent G+C content, probe length, ionic strength of the hybridization buffer, and temperature of incubation. These specific probes were applied to nitrocellulose membranes, followed by reverse hybridization of the biotinylated PCR fragment in a LiPA format, incubation with streptavidin-alkaline phosphatase, and color development. Details about the probe optimization phase, LiPA strip production, and reverse hybridization have been published previously (21, 22, 26).
RESULTS
Alignment of the seven different genotypic HBpol aa sequences.
Figure 1 shows a partial alignment of aa sequences obtained from samples stored in GenBank. The overlapping reading frame of the hepatitis B surface antigen (HBsAg) is indicated for one genotype A strain. The majority of the HBpol variability was found in the region between aa 480 and 510, a region that partially overlaps the variable HBsAg extracellular loop (aa 99 to 159) (19). HBpol resistance mutations, however, were mostly found in highly conserved regions, domain B (aa 511 to 537) and domain C (aa 548 to 558) (18). These two regions encode two transmembrane-spanning helices (aa 160 to 184 and aa 189 to 209) in the overlapping HBsAg open reading frame (ORF). Figure 1 also shows that the three target codons for the test design (codons 528, 552, and 555) were located in conserved areas of the polymerase. Hence, apart from a few type-specific polymorphisms, only little variability is expected on the nucleotide level for the seven different genotypes (23). Of importance, natural variability for the known drug resistance motifs exists at codons 521 (V and G), 555 (V, M, and L), 561 (T and S), and 567 (S and F) (Fig. 1).
FIG. 1.
HBV amino acid alignment of GenBank sequences. Sequences, indicated with their accession numbers, were grouped according to their genotypes. Genotype G (strain FR1) is a recently discovered genotype (23). On top, the overlapping HBsAg ORF is shown. The HBpol numbering is genotype dependent. Numbering for each genotype is indicated. Positions of previously described drug resistance are indicated with an exclamation point. Amino acids are indicated with the single-letter nomenclature; X, translational stop.
Recombinant plasmid clones encoding the different wild-type and mutant strains.
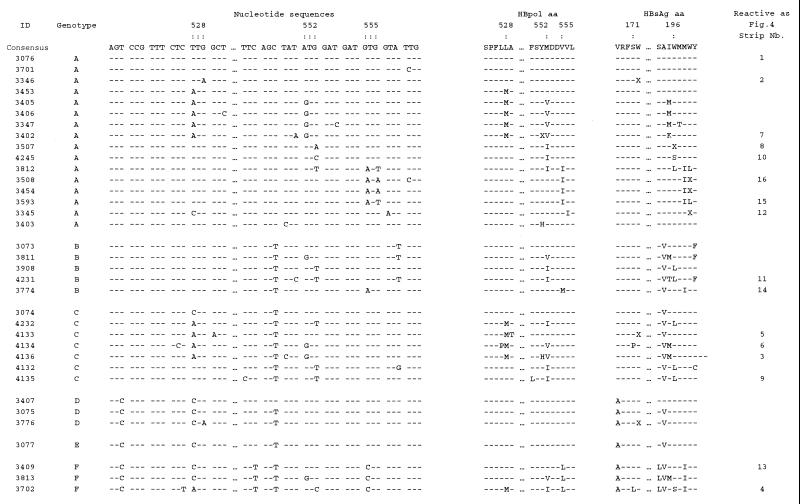
On the basis of the information presented in Fig. 1, a panel of recombinant clones representing these different genotypes in HBsAg was created (26). To further complete the panel, PCR fragments obtained during virological failure from several patients in cross-sectional studies (data not shown) were cloned into plasmids. The clones contained lamivudine and famciclovir drug resistance motifs. Figure 2 shows the relevant details for 35 recombinant clones with different genetic constitutions. Figure 2 provides details for a selection of recombinant clones taken (i) from a variety of untreated individuals whose isolates showed so-called wild-type motifs L528, M552, and V/L/M555 and (ii) from several patients who were treated with lamivudine and whose isolates showed so-called mutant motifs M528, V/I552, and I555. Codon degeneration was observed at all three codon positions: L528 presented as TTG, TTA, CTG, and CTA; I552 presented as ATA, ATT and ATC; and I555 presented as ATT and ATA. Similar to the data in Fig. 1, M555 (ATG) was a very typical motif found in an untreated genotype B context, and L555 (CTG) was found in a genotype F context. Figure 2 also shows the predicted aa variability that is observed in the overlapping HBsAg reading frame.
FIG. 2.
Detailed presentation of the recombinant HBV reference plasmid panel. ID, clone identification number. Codon numbering is according to genotype A numbering. Three periods indicate a gap introduced between codons 529 and 548; sequences in this gap are not relevant for the design of the assay. The figure also shows the amino acid alignments in both the HBpol and HBsAg ORFs. Amino acids are indicated with the single-letter nomenclature; X, translational stop. The selected LiPA probes were based on this panel of clones; the positions of probes are not indicated. The strips with reactivity correspond to the strips shown in Fig. 4. Nb., number.
LiPA for HBV drug resistance.
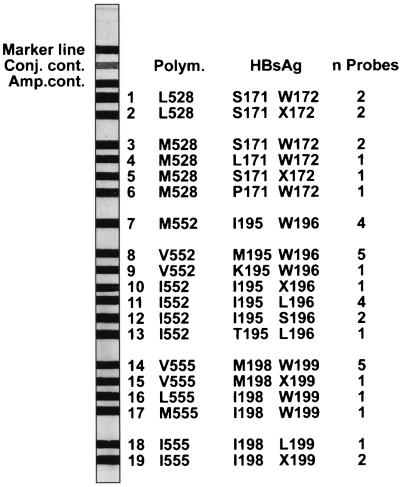
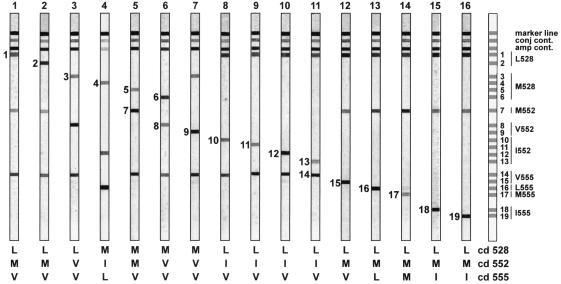
The panel of recombinant clones was used to design and validate specific probes for codon positions 528, 552, and 555. The probes covered both the wild-type and mutant motifs for the different genotypes and polymorphisms. A total of 38 probes were optimized and subsequently pooled according to the information presented in Fig. 2. This resulted in a strip with 19 different probe lines (Fig. 3). These strips were then incubated with biotinylated PCR products from the recombinant clones. Figure 4 illustrates the specific reactivity from the different clones with the probes on the strip. Figure 4 also shows the simultaneous detection at codons 528, 552, and 555 of a wild-type or a mutant codon in one experiment.
FIG. 3.
Design of the HBV drug resistance research strip. Conj. cont., conjugate control; Amp. cont, amplification control. The strip contains a total of 19 probe lines; n probes equal the total amount of probes on each line, for a total of 38 probes. Interpretation of HBpol or HBsAg, amino acid for the polymerase or HBsAg ORF applied on this probe line, respectively. Several probes that were designed for different nucleotide polymorphisms (Polym.) but in which an amino acid change was not introduced were pooled and applied on one line.
FIG. 4.
HBV LiPA strip results illustrating the reactivity of each independent probe line. Results are obtained with a selection of the reference panel shown in Fig. 2. Reactive lines are indicated with their number. A strip showing the relative positions of all lines on the strip is indicated on the left. Interpretation for each codon is given below each strip. conj cont., conjugate control; amp cont., amplification control.
Patient follow-up samples on the HBV LiPA for drug resistance.
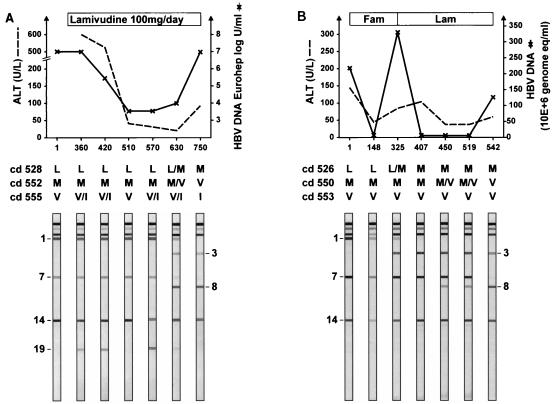
Figure 5 shows the LiPA reactivities of the follow-up samples from two different patients. A high degree of genetic complexity was observed within these samples. The samples from patient A, who had been treated with lamivudine, showed a mixture of V555I (line 14 and 19) that was transiently present on days 360, 420, and 570 and weakly present on day 630. Figure 3 shows reactivity at line 19, which correlates with a translational stop at HBsAg codon 199. This motif disappeared after the emergence of a mutation at codons 552 and 528. Also, on day 630, mixtures of mutations at both codon 528 (Fig. 5, lines 1 and 3) and codon 552 (Fig. 5, lines 7 and 8) were observed. Wild-type motifs L528 and M552 disappeared at day 750, which corresponded to viral breakthrough. The samples from patient C, who was treated with famciclovir followed by lamivudine, showed that the wild type L528 (Fig. 5, line 1) coexisted with a mutant M528 codon (Fig. 5, line 3) at day 325; however, this wild-type motif was not detected from day 407 onward. Progressive detection of a mutation (Fig. 5, line 8) over the wild type (Fig. 5, line 7) was observed at codon 552 from day 450. From day 542 onward a pure double mutation, M528 plus V552, was present. In this HBV strain, no changes at codon 555 were observed.
FIG. 5.
Monitoring of two patients infected with HBV with drug resistance. (A) patient A; (B) patient C. Days of follow-up are indicated on the x axis. The interpretation of the reactivity pattern on each strip is given for the three codons (indicated as cd). Fam, famciclovir; Lam, lamivudine.
Clonal analysis of follow-up samples.
A total of seven plasma samples were available from patient A at different time points over a 750-day period of antiviral treatment. Amplification products were cloned, and 158 recombinant clones were retained for LiPA analysis (Table 1). Except for I555, there was no evidence of selection of mutations at codons 528 and 552 between day 1 and day 570 in a total of 99 clones. At day 630, the majority of the clones had double mutations (M528 plus V552). However, clones with single mutations (L/V/V and M/M/V) were marginally present; these could be interpreted as the remnants of the intermediate forms for double-mutation selection. The emergence of resistance occurred within a period of 60 days, between days 570 and 630.
TABLE 1.
Clonal analysis of follow-up samples
| Patient and day | No. of clones with the following mutationsa:
|
|||||
|---|---|---|---|---|---|---|
| Total | L/M/V | L/M/I | L/V/V | M/M/V | M/V/V | |
| Patient A | ||||||
| 1 | 9 | 9 | 0 | 0 | 0 | 0 |
| 360 | 8 | 4 | 4 | 0 | 0 | 0 |
| 420 | 9 | 3 | 6 | 0 | 0 | 0 |
| 510 | 18 | 18 | 0 | 0 | 0 | 0 |
| 570 | 55 | 23 | 32 | 0 | 0 | 0 |
| 630 | 46 | 2 | 1 | 1 | 2 | 40 |
| 750 | 13 | 0 | 0 | 0 | 0 | 13 |
| Total | 158 | 59 | 43 | 1 | 2 | 53 |
| Patient C | ||||||
| 1 | 5 | 5 | 0 | 0 | 0 | 0 |
| 148 | 6 | 6 | 0 | 0 | 0 | 0 |
| 325 | 11 | 1 | 0 | 0 | 10 | 0 |
| 407 | 9 | 0 | 0 | 0 | 9 | 0 |
| 450 | 12 | 1 | 0 | 0 | 11 | 0 |
| 519 | 23 | 0 | 0 | 0 | 3 | 20 |
| 542 | 6 | 0 | 0 | 0 | 0 | 6 |
| Total | 72 | 13 | 0 | 0 | 33 | 26 |
528/552/555.
Samples were collected from patient C at seven different time points. Seventy-two clones were generated from the samples and analyzed. Due to the treatment schedule with famciclovir, the clone with the single mutation (M528) existed as the majority population at day 325. The mutant with M528 plus V552 mutations was detected as the major population only after the introduction of lamivudine at day 325 (Fig. 5 and Table 1). The selection of the mutant with the M552 mutation occurred in a maximum period of 69 days (between days 450 and 519).
DISCUSSION
This article describes a new LiPA application for genotypic monitoring of antiviral drug resistance during HBV therapy. The current selection of 38 probes covers three codon positions (positions 528, 552, and 555) and was applied on one strip in 19 different probe lines. As shown in Fig. 4, the current selection of probes was very specific in detecting the corresponding amplicon and, in addition, was found to be useful for monitoring the emergence of drug resistance during antiviral treatment (Fig. 5).
As many different numbering systems for indicating resistant codon positions exist, comparative analysis is difficult. This has given rise to a need for standardization. The basis for this confusion lies in the nature of the different genotypes. Our previous studies showed that genotype A has a pandemic distribution but is most abundant in the Western world, while all other genotypes are more or less geographically restricted (26). Compared to genotype A, alignment of complete genomes from the different genotypes show (i) a 6-nucleotide deletion at the carboxy-terminal part of the hepatitis B core antigen in genotypes B, C, D, E, F, and G; (ii) a 33-nucleotide deletion in the amino-terminal part of PreS1 in genotype D; and (iii) a 3-nucleotide deletion at the amino terminus of PreS1 in genotypes E and G (23). All three variations are located within the ORF of the polymerase gene and thus influence the codon numbering. Therefore, as a general rule for codon numbering and on the basis of the genotype A polymerase structure for domains B and C, genotypes B, C, and F have an aa numbering of −2, genotype D has an aa numbering of −13, and genotypes E and G have an aa numbering of −3. However, in routine diagnostics the genotype of the HBV strain under investigation is commonly unknown, and since genotype A has a pandemic distribution, we have used the genotype A numbering throughout this report. The numbering for the other genotypes is included in Fig. 1 for clarity.
LiPA is a convenient tool for clonal analysis and illustrates the dynamics of the virus under drug pressure in vivo. The most interesting clones that emerged during the course of this study were (i) those that showed stop codons in the overlapping HBsAg ORF and (ii) those that had a single mutation (M528 or V552). Careful analysis of Fig. 2 showed that in the 35 clones selected from clinical samples, a total of eight stop codons were present. Seven of these eight clones had this translational stop in the HBsAg ORF, and when compared to the consensus wild-type sequences, all seven clones showed a G-to-A variation. This high G-to-A mutation rate was previously recognized in HBV strains and was explained as a lentivirus hallmark (5). Typically, these events might occur during reverse transcriptase reactions as a consequence of changes in the intracellular pools of dTTP versus dCTP and might arise as a consequence of a local depletion of dCTP (5). Furthermore, it is expected that viral sequences showing these translational stops in the HBsAg region will be incapable of synthesizing a complete functional HBsAg protein since these HBsAg mutations (W172X, W196X, and W199X, respectively) are located in the third and fourth transmembrane regions of the protein (19). Such variants are detectable on this LiPA strip (Fig. 4, strips 2, 5, 10, 15, and 19). As shown for patient A, the variant with the W199X mutation (line 19) was always present as a mixture with the wild type (line 14), probably because the wild type is needed for production of intact HBsAg. Clonal analysis showed that this strain might take up approximately 60% (32 of 55 clones; Table 1) of the circulating virus at day 570. The V555I/X199 mutation might also be clinically relevant (6, 16). Pichoud et al. (16) showed that the mutant with this mutation has a decreased replication capacity, does not produce HBsAg, and is resistant to penciclovir but remains sensitive to lamivudine. The translational stop observed in the HBpol region at codon 551 (ID3402; Fig. 2) might be either a real sequence present in the viral quasispecies or possibly an amplification artifact.
Two clones with an M528 plus M552 combination of mutations and one clone with the L528 plus V552 mutations were selected from patient A (Table 1). Although it is impossible to detect mutants with the latter combination of mutations as the major quasispecies in the clinical samples, they must have importance in the way in which the mutant with the double M528 plus V552 mutation is formed. The results from a transient transfection cell culture assay (1) showed that there was an increasing resistance (expressed as multifold increases in the 50% inhibitory concentration for 2.2.15 cells) in the series from the wild type (sensitive) → L528M (18-fold) → M552V (153-fold or 333-fold [9]) → M552I = L528M plus M552V = L528M/M552I (>10,000-fold). Superimposition of these in vitro data on our findings supports the hypothesis that the possible route to obtaining high-level resistance starts gradually with the selection of a mutant with a single mutation (L528M or M552V), nearly immediately followed by the selection of a mutant with a second mutation. Both mutants with single mutation might exist next to each other but have very short half-lives because they emerged and were removed nearly completely from the total population in the 60-day (days 570 to 630) window period.
Patient C was sequentially treated with famciclovir and lamivudine. A sequential selection of a mutant with the M528 mutation followed by selection of a mutant with the double M528 plus V552 mutation was observed. Indeed, the pressure created by lamivudine therapy on a mutant virus with the single M528 mutation (from day 325) resulted in a rapid selection of the mutant with a double mutation at day 450 (approximately 17 weeks), but viral breakthrough occurred only at day 542, or 31 weeks of therapy. The duration of successful lamivudine therapy was therefore considerably less compared to that for patient A. This result confirmed the findings that the M528 mutation is a risk factor for lamivudine resistance breakthrough (24). Additionally, the appearance of the mutant with the V552 mutation preceded the increase in viral load by at least 112 days. This finding is consistent with previously observed data (4), but here the change is observed in the context of sequential therapy.
During the course of this study, we selected 38 highly specific probes, applied the probes on a LiPA strip, and showed the usefulness of this probe selection for viral monitoring during antiviral therapy. Probes were applied in such a way that meaningful information from both the HBpol and the HBsAg regions could be deduced immediately after strip hybridization. Sensitive detection of viral mixtures accompanied by clonal analysis might help (i) unravel the dynamics of emerging HBV resistance and (ii) improve the monitoring of antiviral treatment.
ACKNOWLEDGMENT
We thank Mimi Healy (Innogenetics, Atlanta, Ga.) for helpful discussions.
REFERENCES
- 1.Allen M I, Deslauriers M, Andrews C W, Tipples G A, Walters K A, Tyrell D L J, Brown N, Condreay L D The Lamivudine Clinical Investigation Group. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology. 1998;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 2.Bartholmeusz A, Schinazi R F, Locarnini S A. Significance of mutations in the hepatitis B virus polymerase selected by nucleoside analogues and implications for controlling chronic disease. Viral Hep Rev. 1998;4:167–187. [Google Scholar]
- 3.Buti M, Jardi R, Cotrina M, Rodriguez-Frias F, Esteban R, Guardia J. Transient emergence of hepatitis B variants in a patient with chronic hepatitis B resistant to lamivudine. J Hepatol. 1998;28:510–513. doi: 10.1016/s0168-8278(98)80327-9. [DOI] [PubMed] [Google Scholar]
- 4.Chayama K, Suzuki Y, Kobayashi M, Kobayashi M, Tsubota A, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, Saitoh S, Murashima N, Ikeda K, Kumada H. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology. 1998;27:1711–1716. doi: 10.1002/hep.510270634. [DOI] [PubMed] [Google Scholar]
- 5.Gunther S, Sommer G, Plikat U, Iwanska A, Wain-Hobson S, Will H, Meyerhans A. Naturally occurring hepatitis B virus genomes bearing the hallmarks of retroviral G→A hypermutation. Virology. 1997;18:104–108. doi: 10.1006/viro.1997.8676. [DOI] [PubMed] [Google Scholar]
- 6.Günther S, van Breunig F, Santantonio S, Jung M-C, Gaeta G B, Fisher L, Sterneck M, Will H. Absence of mutations in the YMDD motif/B region of the hepatitis B virus polymerase in famciclovir therapy failure. J Hepatol. 1999;30:749–754. doi: 10.1016/s0168-8278(99)80124-x. [DOI] [PubMed] [Google Scholar]
- 7.Heijtink R A, Kruining J, Honkoop P, Kuhns M C, Hop W C, Osterhaus A D, Schalm S W. Serum HBeAg quantitation during antiviral therapy for chronic hepatitis B. J Med Virol. 1997;53:282–287. [PubMed] [Google Scholar]
- 8.Honkoop P, Niesters H G M, de Man R A M, Osterhaus A D M E, Schalm S W. Lamivudine resistance in immunocompetent chronic hepatitis B. J Hepatol. 1997;26:1393–1395. doi: 10.1016/s0168-8278(97)80476-x. [DOI] [PubMed] [Google Scholar]
- 9.Ladner S K, Miller T J, King R W. The M539V polymerase variant of human hepatitis B virus demonstrates resistance to 2′-deoxy-3′-thiacytidine and a reduced ability to synthesize viral DNA. Antimicrob Agents Chemother. 1998;42:2128–2131. doi: 10.1128/aac.42.8.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai C L, Chien R N, Leung N W Y, Chang T T, Guan R, Tai D I, Ng K-Y, Wu P C, Dent J C, Barber J, Stephenson S L, Gray D F. One year trial of lamivudine for chronic hepatitis B. N Engl J Med. 1998;339:61–68. doi: 10.1056/NEJM199807093390201. [DOI] [PubMed] [Google Scholar]
- 11.Ling R, Mutimer D, Ahmed M, Boxall E H, Elias E, Dusheiko G M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 12.Main J, Brown J L, Howells C, Galassini R, Crossay M, Karayiannis P, Georgiou P, Atkinson G, Thomas H C. A double blind, placebo controlled study to assess the effect of famciclovir on virus replication in patients with chronic hepatitis B virus infection. J Viral Hep. 1996;3:211–215. doi: 10.1111/j.1365-2893.1996.tb00098.x. [DOI] [PubMed] [Google Scholar]
- 13.Melagari M, Scaglioni P P, Wands J R. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology. 1998;27:628–633. doi: 10.1002/hep.510270243. [DOI] [PubMed] [Google Scholar]
- 14.Mutimer D, Naoumov N, Honkoop P, Marinos G, Ahmed M, de Man R, McPhillips P, Johnson M, Williams R, Elias E, Schalm S. Combination alpha-interferon and lamivudine therapy for alpha-interferon-resistant chronic hepatitis B infection: results of a pilot study. J Hepatol. 1998;28:923–929. doi: 10.1016/s0168-8278(98)80338-3. [DOI] [PubMed] [Google Scholar]
- 15.Niesters H G M, Honkoop P, Haagsma E B, de Man R A, Schalm S W, Osterhaus A D M E. Identification of more than one mutation in the hepatitis B virus polymerase gene arising during prolonged lamivudine treatment. J Infect Dis. 1998;177:1382–1385. doi: 10.1086/517819. [DOI] [PubMed] [Google Scholar]
- 16.Pichoud C, Seignères B, Wang Z, Trèpo C, Zoulim F. Transient selection of a hepatitis B virus polymerase gene mutant associated with a decreased replication capacity and famciclovir resistance. Hepatology. 1998;29:230–237. doi: 10.1002/hep.510290119. [DOI] [PubMed] [Google Scholar]
- 17.Pillay D, Bartholomeusz A, Cane P, Mutimer D, Schinazi R F, Locarnini S. Mutations in the hepatitis B virus DNA polymerase associated with antiviral resistance. Int Antivir News. 1998;6:167–169. [Google Scholar]
- 18.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prange R, Streeck R E. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Severini A, Liu X Y, Wilson J S, Tyrrell D L. Mechanism of inhibition of duck hepatitis B virus polymerase by (−)-β-l-2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1995;39:1430–1435. doi: 10.1128/aac.39.7.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuyver L, Wyseur A, van Arnhem W, Hernandez F, Maertens G. A second-generation line probe assay for hepatitis C virus. J Clin Microbiol. 1996;34:2259–2266. doi: 10.1128/jcm.34.9.2259-2266.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stuyver L, Wyseur A, Rombout A, Louwagie J, Scarcez T, Verhofstede C, Rimland D, Schinazi R F, Rossau R. Line probe assay for the rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuyver, L., S. De Gendt, C. Van Geyt, F. Zoulim, M. Fried, R. F. Schinazi, and R. Rossau. Characterization of a new hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol., in press. [DOI] [PubMed]
- 24.Tillmann H L, Trautwein C, Bock T, Böker K H W, Jäckel E, Glowienka M, Oldhafer K, Bruns I, Gauthier J, Condreay L D, Raab H-R, Manns M P. Mutational pattern of hepatitis B virus on sequential therapy with famciclovir and lamivudine in patients with hepatitis B virus reinfection occurring under HBIg immunoglobulin after liver transplantation. Hepatology. 1999;30:244–256. doi: 10.1002/hep.510300141. [DOI] [PubMed] [Google Scholar]
- 25.Tipples G A, Ma M M, Fisher K P, Bain V G, Kneteman N M, Tyrell D L J. Mutations in HBV RNA-dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]
- 26.Van Geyt C, De Gendt S, Rombaut A, Wyseur A, Maertens G, Rossau R, Stuyver L. A line probe assay for hepatitis B virus genotypes. In: Schinazi R F, Sommadossi J P, Thomas H, editors. Therapies of viral hepatitis. London, United Kingdom: International Medical Press; 1998. pp. 139–145. [Google Scholar]
- 27.Zoulim F, Trépo C. Drug therapy for chronic hepatitis B: antiviral efficacy and influence of hepatitis B virus polymerase mutations on the outcome of therapy. J Hepatol. 1998;29:151–168. doi: 10.1016/s0168-8278(98)80191-8. [DOI] [PubMed] [Google Scholar]