Abstract
With the use of real-time PCR, we developed and evaluated a rapid, sensitive, specific, and reproducible method for the detection of Epstein-Barr virus (EBV) DNA in plasma samples. This method allowed us to screen plasma and serum samples over a range between 100 and 107 copies of DNA per ml using two sample preparation methods based on absorption. A precision study yielded an average coefficient of variation for both methods of less than 12%, with a coefficient of regression for the standard curve of a minimum of 0.98. We detected EBV DNA in 19.2% of plasma samples from immunosuppressed solid-organ transplant patients without symptoms of EBV infections with a mean load of 440 copies per ml. EBV DNA could be detected in all transplant patients diagnosed with posttransplant lymphoproliferative disorder, with a mean load of 544,570 copies per ml. No EBV DNA could be detected in healthy individuals in nonimmunosuppressed control groups and a mean of 6,400 copies per ml could be detected in patients with infectious mononucleosis. Further studies revealed that the inhibitory effect of heparinized plasma could be efficiently removed by use of an extraction method with Celite as the absorbent.
Epstein-Barr virus (EBV) is the etiological agent of infectious mononucleosis (IM) and is also etiologically associated with Burkitt's lymphoma and nasopharyngeal carcinoma. Usually, the virus produces a mild and self-limiting primary infection in childhood. However, as a gamma herpesvirus, it persists for life by a combination of latency in B lymphocytes and chronic replication in oropharyngeal epithelial cells. A serious complication after allogeneic bone marrow or solid-organ transplantation is the development of EBV-related posttransplant lymphoproliferative disorder (PTLD) due to immunosuppressive therapy (5, 8). The condition can be rapidly fatal if it is not diagnosed and treated in an early stage.
Recently, it has been observed that there is a relation between PTLD and the EBV load in plasma or infected peripheral blood lymphocytes, as measured by semiquantitative or competitive PCR assays (3–7, 12, 14, 15). With the advent of real-time TaqMan quantitation and improved sample preparation techniques, the whole process from sample retrieval to quantitative result can be reduced. Furthermore, the dynamic range in which samples can be analyzed quantitatively without dilution has improved considerably (7, 9, 10).
In this paper, we describe the validation of a TaqMan-based assay for the quantitation of EBV DNA in plasma. The assay is based on the linearity of the TaqMan assay, takes into account intra- and interassay variability as well as detection limits, and can be performed in a routine setting, providing quantitative results within less than 6 h. Furthermore, we have evaluated two different extraction methods, not only for EDTA-treated plasma but also for heparin-treated plasma because heparin is known to be inhibitory for PCRs (13, 16).
MATERIALS AND METHODS
Patients and samples.
Serum samples from patients with clinical suspicion of a primary EBV infection (IM; n = 22) and a serological profile positive for an immunoglobulin M response to the variable capsid antigen and negative for anti-EBV nuclear antigen were used for this analysis. These samples were kindly provided by Peter Schroder (Groningen Public Health Laboratory). Plasma samples from bone marrow (n = 5) and solid-organ (n = 5) transplant patients who were diagnosed clinically and histologically with EBV-related PTLD were also enrolled in the present evaluation study. Samples were taken before the start of treatment (including before the start of reduction of immunosuppressive treatment) was initiated. Lymph node biopsy specimens were also obtained from these patients, and the diagnosis was confirmed with the EBER probe. Furthermore, samples from a cohort of randomly selected, EBV-seropositive, solid-organ transplant recipients (kidney, heart, and liver; n = 109) were included for cross-sectional analysis. These patients had no EBV-related disease and were routinely screened for hepatitis markers. Samples from healthy individuals and blood donors (n = 100) without any sign of IM or PTLD were used as a control group. From all patients, EDTA-treated plasma samples were aliquoted and frozen at −80°C within 2 h after collection. Only serum was available from the 22 patients with a primary EBV infection.
Nucleic acid extraction.
For the isolation of EBV DNA from plasma or serum samples, two protocols were used. The first protocol was essentially based on the method described by Boom and coworkers (1). Briefly, 100 μl of plasma was added to 1 ml of buffer 1 (120 g of guanidinium isothiocyanate in 100 ml of 0.1 M Tris [pH 6.4], 22 ml of 0.2 M EDTA [pH 8.0], 2.6 g of Triton X-100). After the addition of 50 μl of Celite solution, the mixture was incubated for 10 min at room temperature and was subsequently centrifuged for 10 s at full speed in a tabletop centrifuge. The pellet was washed twice with buffer 2 (identical to buffer 1 but without Triton X-100 solution and EDTA), twice with 70% ethanol, and once with acetone. The silica pellet was dried at room temperature in a vacuum exsiccator for 10 min, after which the DNA was eluted from the silica by adding 100 μl of RNase- and DNase-free water and was incubated for 10 min at 56°C. After centrifugation at 12,000 × g for 2 min, the supernatant contained the DNA and was ready for use.
The second and commercially available protocol was essentially based on the High Pure Viral Nucleic Acid kit protocol (Roche Diagnostics, Almere, The Netherlands). To compare this method directly with the procedure described above, a 100-μl plasma sample is added to the mixture provided with the kit, and finally, the same volume of 100 μl is eluted. Briefly, a 100-μl plasma sample was added to 100 μl of 6 M guanidine-HCl–10 mM urea–10 mM Tris-HCl–20% (vol/vol) Triton X-100 supplemented with carrier RNA and 800 μg of proteinase K. After incubation for 10 min at 72°C, 50 μl of isopropanol was added and the mixture was transferred to a High Pure filter tube combined with a collection tube. The filter tube was centrifuged at 12,000 × g for 1 min in a standard tabletop centrifuge at room temperature. The filter was washed twice with 450 μl of buffer (20 mM NaCl and 2 mM Tris-HCl [pH 7.5] in ethanol). After placement of a new collection tube under the filter, 100 μl of RNase- and DNase-free water was added to elute the DNA. To reduce the detection level of the assay, the input and elution volumes compared to those used in the original procedure can be changed; that is, the input volume can be increased to 200 μl of plasma and the elution volume can be decreased to 50 μl.
Real-time TaqMan assay.
The PCR primers for the TaqMan assay were selected from the EBV DNA genome and encode the nonglycosylated membrane protein BNRF1 p143 (2, 11). The forward and reverse primers and the probe were designed with Primer Express software (PE Biosystems, Nieuwerkerk aan de IJssel, The Netherlands) and generated a DNA product of 74 bp. The primers used were the EBV p143 forward primer (5′-GGA.ACC.TGG.TCA.TCC.TTG.C and the EBV p143 reverse primers (5′-ACG.TGC.ATG.GAC.CGG.TTA.AT), which were synthesized at Isogen Biosciences (Maarssen, The Netherlands). A fluorogenic probe (5′-CGC.AGG.CAC.TCG.TAC.TGC.TCG.CT) was synthesized by PE Biosystems with a FAM reporter molecule attached to the 5′ end and a TAMRA quencher linked at the 3′ end. The PCR amplification was performed in a 50-μl volume containing 2× TaqMan universal master mixture, 45 pmol of forward primer per μl, 2.5 pmol of reverse primer, 5 pmol of the TaqMan probe, and 10 μl of isolated DNA. All reactions were performed in duplicate. After preparation of the reaction tubes, the whole plate holder was centrifuged at 1,000 × g for 1 min at room temperature in a swingout rotor (Hettich, Rotina 48R; Tuttlingen, Germany) to remove small air bubbles in the vessels. The amplification and detection were performed with an ABI Prism 7700 Sequence Detection System (PE Biosystems). After incubation for 2 min at 50°C with uracil N′-glycosylase to inactivate possible PCR contaminants from former reactions, the reaction tube was incubated for 10 min at 95°C to inactivate the uracil N′-glycosylase and to release the activity of the AmpliTaq Gold DNA polymerase. The PCR cycling program consisted of 42 two-step cycles of 15 s at 95°C and 60 s at 60°C. Real-time measurements were taken, and a threshold cycle (Ct) value for each sample was calculated by determining the point at which the fluorescence exceeded a threshold limit of 0.04. Each run contained several negative controls (no template), a positive control containing a known EBV copy number based on a standard for which the EBV copy number was counted by electron microscopy (EBV EM standard), and a standard dilution curve for plasmid DNA containing the PCR product as an insert (see below). Each specimen was run in duplicate and was considered positive only if both replications were above the threshold limit.
Standardization.
For standardization of the assay, a standard containing 6.68 × 109 EBV particles per ml (EBV B95-8; Advanced Biotechnologies Incorporated, Columbia, Md.), as determined by electron microscopy, was used. Serial half-log dilutions of this standard, ranging from 107 to 10 copies per ml, were made to characterize the linearity, precision, specificity, and sensitivity of the TaqMan assay.
For the preparation of the standard curve for the routine TaqMan runs, the PCR product of 74 bp was directly cloned into a pCRII vector (InVitrogen, Leek, The Netherlands) and was transformed into the appropriate bacterial strain. The colonies that were obtained were prescreened by PCR to confirm the size of the insert. Plasmid DNA was isolated on the Vistra Labstation (Amersham Pharmacia Biotech, The Netherlands) and was isolated in bulk. The standard curve made from the plasmid was calibrated with the EBV EM standards and was routinely made in duplicate with a range equivalent to from 100 to 107 copies per ml. It was shown that the slope for the plasmid standard was not significantly different from the slope obtained with the EBV EM standard (P < 0.0001) (data not shown).
Statistics.
The standard curve was created automatically with ABI 7700 Sequence Detection System software by plotting the Ct values against each standard of known concentration. This Ct value was also used for calculation of the intra- and interassay coefficients of (CVs) variation for the technique. Logarithmic transformation of the readings of the different assays was carried out for comparison of the isolation procedures. x-y scatter diagrams were drawn, and the correlation coefficients (r2) or Spearman correlation (r) was determined and linear regression analysis was done by using the statistical functions of SPSS (version 8.0) software. Student's t test was used for comparison of the EBV DNA copy numbers in each group analyzed.
RESULTS
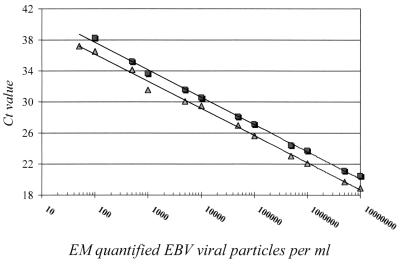
The limit of detection by both extraction methods was determined with half-log dilutions of the EBV EM standard. Both assays were used in an identical format, in which 100 μl of patient material was used as input, while the DNA was eluted in 100 μl of RNase- and DNase-free water. Both assays were able to detect viral DNA over a linear span of between 100 and 107 copies per ml (Fig. 1). Statistical analysis of the standard curves over this range showed that both methods were linear with an r2 value of a minimum of 0.98. Furthermore, the slopes of both standard curves were not significantly different (P < 0.0001; 95% confidence interval, −0.196 to 0.245). However, the average Ct values obtained by the extraction method described by Boom et al. (1) were 1.44 lower over the whole linear range (P < 0.0001) than those obtained with the High Pure Viral Nucleic Acid extraction kit. This shows that the efficiency of the extraction step was better for the method described by Boom et al. (1), as also indicated by the fact that the 50 copies per ml of the dilution were detected in all eight replicates, whereas 50 copies per ml were detected in only one of eight replicates with the adapted High Pure Viral Nucleic Acid extraction kit. Both methods were unable, however, to detect 10 copies of the EBV EM standard per ml used in the formats described above.
FIG. 1.
Standard curve for TaqMan PCR. Serial dilutions of the EBV EM standard ranging from 50 to 107 copies per ml were made. Extraction was performed by the method of Boom et al. (1) (triangles; Spearman correlation coefficient, 0.997) or the method with the High Pure Viral Nucleic Acid kit (squares; Spearman correlation coefficient, 0.998) as a matrix. Equal volumes of input and output material (100 μl) were used. The Ct values, which correspond to the PCR cycle number in which the value is above the threshold limit, are plotted against the calculated number of particles counted by electron microscopy.
A precision study for both extraction methods was performed by evaluating serial half-log dilutions of the EBV EM standard ranging from 50 to 107 copies per ml in originally EBV-negative serum. The Ct values obtained were used for the calculation. The study was carried out over 3 consecutive days, and two sets of independent isolations were performed. A total of eight replicates of the 12 dilutions for both extraction methods were tested on each day. Again, only the method of Boom et al. (1) was able to detect EBV in the sample with 50 copies per ml. The assay exhibited a very good total precision throughout the range of the numbers of EBV DNA copies in the EBV-positive samples, with CVs ranging from 0.7 to 11.7%. The relatively high CV (11.7%) was due to the inability to detect 100 copies per ml in one of the eight replicates by the method with the High Pure Viral Nucleic Acid extraction kit. The average CV for the High Pure Viral Nucleic Acid extraction kit was 2.37% (range, 1.1 to 11.7%), and that for the method of Boom et al. was 1.56% (range, 0.7 to 7.0%). There was no difference in between-day variation and within-run variation or within the independent isolations (data not shown).
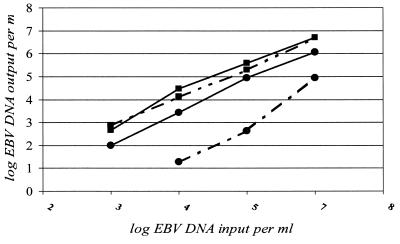
We furthermore evaluated whether both extraction methods were able to remove efficiently the inhibitory effect of heparin on the PCR-based assay. Therefore, four dilutions of the EBV EM standard were made in heparin- or EDTA-treated plasma; the dilutions ranged from 500 to 107 copies per ml. The efficiencies of isolation and amplification by the method of Boom et al. (1) were almost identical whether EDTA-treated or heparin-treated plasma was used (Fig. 2), with the variation being less than twofold. However, with the High Pure Viral Nucleic Acid extraction kit, the efficiency was reduced between 12- and 200-fold when heparin-treated plasma samples were compared with EDTA-treated plasma samples. EBV could not be detected in the heparin-treated plasma sample with 500 copies per ml. Use of EDTA is much easier than the use of heparinase I, which degrades heparin and involves another incubation step (13). This experiment also confirms again that the method of Boom et al. (1) is able to extract the EBV DNA more efficiently than the High Pure Viral Nucleic Acid extraction kit, as indicated by the lower Ct value.
FIG. 2.
Detection of EBV EM standard dilutions made in heparin-treated plasma (dashed line) or EDTA-treated plasma (solid line). From these EBV dilutions, ranging from 500 to 107 copies per ml, DNA was isolated by the extraction method of Boom et al. (1), which uses Celite (■) to absorb the DNA, as well as by the method with the High Pure Viral Nucleic Acid kit (Roche Diagnostic) (●). The extracted DNA was quantified by the TaqMan assay.
The analytical specificity of the assay was determined by analyzing DNAs from other human herpesviruses (herpes simplex virus types 1 and 2, varicella-zoster virus, cytomegalovirus, and human herpesviruses 6, 7, and 8), as well as from other viruses routinely used in the laboratory for DNA or RNA analysis (hepatitis B, C, and G viruses and human papillomavirus). All these samples yielded results below the detection level of 100 copies per ml or a Ct value of 42.
DISCUSSION
To demonstrate that the TaqMan-based assay described here could be used to detect EBV DNA in clinical samples and to determine the baseline value for plasma EBV DNA levels in different groups, we evaluated clinical samples from 100 healthy individuals, 22 patients with IM, a cohort of 109 asymptomatic immunosuppressed solid-organ transplant patients, and 10 patients with a confirmed diagnosis of PTLD. The results are summarized in Table 1. For this evaluation, samples were analyzed in duplicate with the High Pure Viral Nucleic Acid extraction kit, which has a cutoff value of 100 copies per ml. As expected, EBV DNA could not be detected in the plasma of any of the 100 healthy individuals. We were able to detect low levels of EBV DNA in 21 of 109 solid-organ transplant patients (19.2%), with a mean of 440 copies per ml (range, <100 to 12,000 copies per ml). This was, however, not statistically different from the value for the control group (P = 0.19). We could detect a signal for EBV DNA in 16 of 22 samples (72.7%) from IM patients, with a mean value of 6,400 copies per ml (range, <100 to 45,000 copies per ml). This EBV DNA load was significantly higher than that for the control group (P < 0.006).
TABLE 1.
Quantitation of EBV DNA load by TaqMan analysis
| Patient group | No. of patients | % Positive | Mean EBV DNA load (copies/mla [range]) |
|---|---|---|---|
| Healthy donors | 100 | 0 | <100 |
| Solid-organ transplant patients | 109 | 19.2 | 440 (<100–12,000)b |
| IM patients | 22 | 72.7 | 6,400 (<100–45,000)c |
| PTLD patients | 10 | 100 | 544,750 (74,000–3,200,000)d |
All values are averages of two independent experiments.
Not statistically different from healthy control group (P = 0.19).
Statistically different from control group (P < 0.006) and solid organ transplant group (P < 0.0001).
Statistically different from all other groups studied (P < 0.0001).
It has been shown previously that the presence of EBV DNA in plasma is diagnostic for a clinical EBV infection (3, 4, 17). EBV DNA should be absent from the plasma of healthy individuals. Using a sensitive detection method like PCR, however, one is able to detect viral genomes in peripheral blood mononuclear cells (PBMCs) of healthy controls (7, 17; unpublished data). In the study of Kimura et al. (7), a viral load of 315 copies per μg of PBMC DNA was set as a criterion for distinguishing a latent infection from a symptomatic EBV infection or EBV-related disease.
An EBV DNA load could be determined for only 16 of 22 samples from IM patients with a primary infection. It can be concluded from the data of Yamamoto et al. (17) that this is due to the time point of sampling. Also, the group of Kimura et al. (7) did not find a positive EBV DNA signal in PBMCs from all of their IM patients analyzed.
We expected to be able to find active replication of EBV in the group of immunocompromised solid-organ transplant patients due to the immunosuppression. We were able to detect EBV DNA in the plasma of 19.2% of the patients analyzed. None of these patients had clinical signs of an active EBV infection or EBV-related disease. However, one could expect specifically that reactivation of EBV is more likely to occur in this group than in healthy individuals. Our findings indicate that plasma EBV loads of up to 12,000 copies of DNA per ml can easily be detected in our cohort with no indications of an EBV-related disease. However, no data are available on the longitudinal follow-up period required to determine whether several EBV reactivation periods can be detected in this group. We confirm the data of Kimura et al. (7), who also detected EBV DNA in PBMCs from 14% of posttransplant patients with no signs of EBV-related disease.
The group for which the use of a quantitative PCR should be most useful are patients with a diagnosis of PTLD. In the group of 10 transplant patients diagnosed with PTLD but for whom treatment such as reduction of immunosuppressive therapy, or the initiation of antiviral treatment was not initiated, the mean EBV load in plasma was 544,750 copies per ml (range, 74,000 to 3.2 × 106 copies per ml), which is significantly higher than those in the groups mentioned above (P < 0.0001). However, there is a difference between a clinical diagnosis of PTLD and the viral load at which one should be aware that PTLD is developing. Therefore, we suggest that routine monitoring of patients at risk for EBV-related proliferative disease will allow determination of whether there is a progression of this life-threatening disease from a virological point of view.
In summary, by use of the real-time PCR technique described here, an easy-to-use and highly reproducible technique is available for evaluation of the significance of EBV DNA in plasma samples in immunosuppressed patients. Depending on the isolation method used, inhibition by heparin of the amplification reaction can be eliminated. In this study we also confirm the data presented by others (3, 4, 17) that there is a relation between plasma EBV DNA levels and EBV-related diseases. However, active replication could also be detected in patients without clinical EBV-related disease. Future studies must define the cutoff levels at which treatment of patients at risk for PTLD should be initiated. The technique can then be used to monitor the effect of antiviral therapy on EBV, whether this is by infusion of donor T cells, a change of immunosuppressive therapy, or provision of nucleoside analogues to inhibit EBV replication.
ACKNOWLEDGMENTS
We thank Judith Guldemeester for assisting in various parts of this study and Philip Rothbarth (Department of Microbiology, Enschede, The Netherlands), who started initiating the use of molecular techniques for the detection of EBV-related disease in bone marrow transplant patients.
REFERENCES
- 1.Boom R, Sol C J, Heijtink R, Wertheim van Dillen P M, van der Noordaa J. Rapid purification of hepatitis B virus DNA from serum. J Clin Microbiol. 1991;29:1804–1811. doi: 10.1128/jcm.29.9.1804-1811.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cameron K R, Stamminger T, Craxton M, Bodemer W, Honess R W, Fleckenstein B. The 160,000-Mr virion protein encoded at the right end of the herpesvirus saimiri genome is homologous to the 140,000-Mr membrane antigen encoded at the left end of the Epstein-Barr virus genome. J Virol. 1987;61:2063–2070. doi: 10.1128/jvi.61.7.2063-2070.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drouet E, Brousset P, Fares F, Icart J, Verniol C, Meggetto F, Schlaifer D, Desmorat-Coat H, Rigal-Huguet F, Niveleau A, Delsol G. High Epstein-Barr virus serum load and elevated titers of anti-ZEBRA antibodies in patients with EBV-harboring tumor cells of Hodgkin's disease. J Med Virol. 1999;57:383–389. doi: 10.1002/(sici)1096-9071(199904)57:4<383::aid-jmv10>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 4.Gan Y J, Sullivan J L, Sixbey J W. Detection of cell-free Epstein-Barr virus DNA in serum during acute infectious mononucleosis. J Infect Dis. 1994;170:436–439. doi: 10.1093/infdis/170.2.436. [DOI] [PubMed] [Google Scholar]
- 5.Green M, Cacciarelli T V, Mazariegos G V, Sigurdsson L, Qu L, Rowe D T, Reyes J. Serial measurement of Epstein-Barr viral load in peripheral blood in pediatric liver transplant recipients during treatment for posttransplant lymphoproliferative disease. Transplantation. 1998;66:1641–1644. doi: 10.1097/00007890-199812270-00012. [DOI] [PubMed] [Google Scholar]
- 6.Haque T, Thomas J A, Parratt R, Hunt B J, Yacoub M H, Crawford D H. A prospective study in heart and lung transplant recipients correlating persistent Epstein-Barr virus infection with clinical events. Transplantation. 1997;64:1028–1034. doi: 10.1097/00007890-199710150-00015. [DOI] [PubMed] [Google Scholar]
- 7.Kimura H, Morita M, Yabuta Y, Kuzushima K, Kato K, Kojima S, Matsuyama T, Morishima T. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J Clin Microbiol. 1999;37:132–136. doi: 10.1128/jcm.37.1.132-136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limaye A P, Huang M L, Atienza E E, Ferrenberg J M, Corey L. Detection of Epstein-Barr virus DNA in sera from transplant recipients with lymphoproliferative disorders. J Clin Microbiol. 1999;37:1113–1116. doi: 10.1128/jcm.37.4.1113-1116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martell M, Gomez J, Esteban J I, Sauleda S, Quer J, Cabot B, Esteban R, Guardia J. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J Clin Microbiol. 1999;37:327–332. doi: 10.1128/jcm.37.2.327-332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mercier B, Burlot L, Ferec C. Simultaneous screening for HBV DNA and HCV RNA genomes in blood donations using a novel TaqMan PCR assay. J Virol Methods. 1999;77:1–9. doi: 10.1016/s0166-0934(98)00075-5. [DOI] [PubMed] [Google Scholar]
- 11.Meyohas M C, Marechal V, Desire N, Bouillie J, Frottier J, Nicolas J C. Study of mother-to-child Epstein-Barr virus transmission by means of nested PCRs. J Virol. 1996;70:6816–6819. doi: 10.1128/jvi.70.10.6816-6819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orentas R J. Determination of Epstein-Barr virus (EBV) load by RT-PCR and cellular dilution. Mol Cell Probes. 1998;12:427–430. doi: 10.1006/mcpr.1998.0207. [DOI] [PubMed] [Google Scholar]
- 13.Poli F, Cattaneo R, Crespiatico L, Nocco A, Sirchia G. A rapid and simple method for reversing the inhibitory effect of heparin on PCR for HLA class II typing. PCR Methods Appl. 1993;2:356–358. doi: 10.1101/gr.2.4.356. [DOI] [PubMed] [Google Scholar]
- 14.Rowe D T, Qu L, Reyes J, Jabbour N, Yunis E, Putnam P, Todo S, Green M. Use of quantitative competitive PCR to measure Epstein-Barr virus genome load in the peripheral blood of pediatric transplant patients with lymphoproliferative disorders. J Clin Microbiol. 1997;35:1612–1615. doi: 10.1128/jcm.35.6.1612-1615.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telenti A, Marshall W F, Smith T F. Detection of Epstein-Barr virus by polymerase chain reaction. J Clin Microbiol. 1990;28:2187–2190. doi: 10.1128/jcm.28.10.2187-2190.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willems M, Moshage H, Nevens F, Fevery J, Yap S H. Plasma collected from heparinized blood is not suitable for HCV-RNA detection by conventional RT-PCR assay. J Virol Methods. 1993;42:127–130. doi: 10.1016/0166-0934(93)90184-s. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto M, Kimura H, Hironaka T, Hirai K, Hasegawa S, Kuzushima K, Shibata M, Morishima T. Detection and quantification of virus DNA in plasma of patients with Epstein-Barr virus-associated diseases. J Clin Microbiol. 1995;33:1765–1768. doi: 10.1128/jcm.33.7.1765-1768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]