Abstract
It has long been accepted that Shiga toxin (Stx) only exists in Shigella dysenteriae serotype 1. However, in recent decades, the presence of Shiga toxin genes (stx) in other Shigella spp. have been reported. We screened 366 Shigella flexneri strains from Alberta, Canada (2003 to 2016) for stx and 26 positive strains were identified. These isolates are highly related with the majority originating from the Dominican Republic and three isolates with Haiti origin. Both phylogenetic and spanning tree analysis of the 26 Alberta and 29 stx positive S. flexneri originating from the U.S., France, Canada (Quebec) and Haiti suggests that there are geographic specific distribution patterns (Haiti and Dominican Republic clades). This study provides the first comprehensive whole genome based phylogenetic analysis of stx positive S. flexneri strains as well as their global transmission, which signify the public health risks of global spreading of these strains.
Keywords: Shigella flexneri, Shiga toxin, phage, phylogenomic, global transmissions
1. Introduction
Shigella is a genus of Gram-negative bacterium that can be transmitted to humans through contaminated food, water, or direct/indirect contact with an infected person. The natural reservoirs of Shigella are typically human and non-human primates, although Shigella infections have been reported in other animals, e.g., chicken and calves [1,2,3,4]. Shigellosis can result from a relatively low infective dose between 10 to 100 organisms and the patient can exhibit symptoms such as diarrhea, fever, stomach pain, nausea, and vomiting [5,6]. In the past century, Shigellosis has decreased greatly through improved sanitation. However, currently Shigella is still one of the most important foodborne pathogens both in developing [6] and developed countries [7,8]. It was estimated that Shigella causes 188 million cases of disease and 164,300 deaths per year globally [5].
The genus Shigella is comprised of four major species including Shigella dysenteriae, Shigella flexneri, Shigella sonnei, and Shigella boydii. A large number of virulence factors have been identified in Shigella spp. [9], including Shiga toxin (Stx) which is commonly found in S. dysenteriae serotype 1 and closely resembles Stx in Shiga toxin-producing Escherichia coli (STEC). In STEC, Stx can be categorized into two main groups, Stx1 and Stx2. Once it enters the host cells, the A subunit can remove an adenine from 28S rRNA, and therefore, inhibit protein synthesis resulting in cell death [10,11]. Based on DNA sequence and biological activity, stx1 has three subtypes stx1a, stx1c, and stx1d while stx2 has seven subtypes, stx2a to stx2g [12].
In recent decades, clinical isolates of Stx carrying Shigella strains from other Shigella spp. were continuously observed. The first published case of stx positive non-S. dysenteriae serotype 1 strain was reported in Germany [13], where a S. sonnei strain was isolated from a patient with recent travel to Ukraine. To date, stx positive S. sonnei, S. flexneri, and S. dysenteriae serotype 4 had been identified in patients from countries such as the U.S. [14,15,16,17,18], Canada [19], Germany [13], Hungary [20], France [21], Finland [22], and Haiti [23]. At present, only one Canadian province (Quebec) has reported cases of non-S. dysenteriae serotype 1 stx positive Shigella.
In 2015, an enteric outbreak was declared by Alberta Health Services, AB, Canada and two epidemiologically linked cases were identified to be infected with Shigella flexneri. Both cases experienced bloody diarrhea and the molecular testing performed for one of the cases as a result of enrollment to a prospective acute gastroenteritis study [24] (APPETITE) indicated the presence of stx genes from the stool sample. Further submission of the stool samples from the two cases by public health resulted in the isolation of Shigella flexneri and both strains confirmed to carry the stx gene and the expression of the toxin was also confirmed by immunoassay. Consequently, this study was initiated, and the objectives were to screen for Shiga toxin positive isolates from Shigella flexneri collected from 2003 to 2016 as related to confirmation and typing of clinical isolates, reference testing service provided by Provincial Laboratory for Public Health (ProvLab) in Alberta, Canada and to further characterize these strains.
2. Results
2.1. stx1-Producing S. flexneri Identified in Clinical Isolates
The stx1 gene was identified in 26/366 (S1 to S26) S. flexneri isolates (Table 1) archived in Alberta from 2003 to 2016. All 26 S. flexneri isolates carried the stx1a gene subtype and expressed the toxin as detected by the SHIGA TOXIN QUIK CHEK™ assay.
Table 1.
stx1 positive S. flexneri strains and associated demographic data.
| Strain Name in This Study | Strains Names in NCBI Database | Country Where Shigellosis Was Diagnosed | Travel History | Year of Diagnosis | Strain Name in This Study | Strains Names in NCBI Database | Country Where Shigellosis Was Diagnosed | Travel History | Year of Diagnosis |
|---|---|---|---|---|---|---|---|---|---|
| S1 | S1 | Canada | NO a | 2003 | SN20 | BS982 | USA | NA b | 2008 |
| S2 | S2 | Canada | Dominican Republic | 2003 | SN21 | BS937 | USA | Haiti | 2010 |
| S3 | S3 | Canada | NO a | 2003 | SN22 | BS942 | USA | NA b | 2010 |
| S4 | S4 | Canada | Dominican Republic | 2003 | SN23 | BS974 | USA | Haiti | 2001 |
| S5 | S5 | Canada | Dominican Republic | 2003 | SN259 | BS1023 | France | Dominican Republic | 2005 |
| S6 | S6 | Canada | NO a | 2003 | SN260 | BS1022 | France | Dominican Republic | 2004 |
| S7 | S7 | Canada | NO a | 2004 | SN261 | BS1025 | France | Haiti | 2008 |
| S8 | S8 | Canada | Dominican Republic | 2004 | SN262 | BS1044 | France | Dominican Republic | 2005 |
| S9 | S9 | Canada | Dominican Republic | 2004 | SN263 | BS1021 | France | Haiti | 2003 |
| S10 | S10 | Canada | Dominican Republic | 2004 | SN264 | BS1057 | Haiti | Haiti | 2013 |
| S11 | S11 | Canada | Dominican Republic | 2004 | SN267 | BS1039 | Haiti | Haiti | 2013 |
| S12 | S12 | Canada | Dominican Republic | 2005 | SN268 | BS1059 | Haiti | Haiti | 2014 |
| S13 | S13 | Canada | Dominican Republic | 2005 | SN269 | BS1060 | Haiti | Haiti | 2014 |
| S14 | S14 | Canada | NO a | 2005 | SN314 | SH200 | Canada | NA b | 2014 |
| S15 | S15 | Canada | Dominican Republic | 2007 | SN315 | SH199 | Canada | Haiti | 2014 |
| S16 | S16 | Canada | Dominican Republic | 2007 | SN339 | BS989 | USA | NA b | 2013 |
| S17 | S17 | Canada | Dominican Republic | 2007 | SN341 | BS972 | USA | NA b | 2012 |
| S18 | S18 | Canada | NO a | 2008 | SN343 | BS968 | USA | Haiti | 2010 |
| S19 | S19 | Canada | Dominican Republic | 2008 | SN349 | BS973 | USA | NA b | 2012 |
| S20 | S20 | Canada | Dominican Republic | 2008 | SN351 | BS971 | USA | Haiti | 2011 |
| S21 | S21 | Canada | NO a | 2009 | SN352 | BS951 | USA | NA b | 2005 |
| S22 | S22 | Canada | Turks and Caicos Islands | 2010 | SN4 | BS1042 | France | Dominican Republic | 2005 |
| S23 | S23 | Canada | NO a | 2010 | SN5 | BS1045 | France | Dominican Republic | 2007 |
| S24 | S24 | Canada | NO a | 2015 | SN6 | BS1024 | France | French Guiana | 2005 |
| S25 | S25 | Canada | NO a | 2015 | SN7 | BS1041 | France | Dominican Republic | 1999 |
| S26 | S26 | Canada | NO a | 2015 | SN8 | BS1043 | France | Haiti | 2005 |
| SN9 | BS1046 | France | Dominican Republic | 2008 | |||||
| SN18 | BS988 | USA | Haiti | 2012 | |||||
| SN19 | BS938 | USA | NA b | 2012 |
a—NO means the patient had no recent travel history; b—NA means the patient’s travel history is not available.
2.2. Sporadic Cases of stx1 Positive S. flexneri in Alberta, Canada
Among the 366 cases infected with S. flexneri with isolates archived in Alberta, 76 had travelled to the Caribbean area such as Cuba, Haiti, Dominican Republic, Mexico, El Salvador; 15 of the 26 stx1 positive S. flexneri cases were travel-related and they all had recent travelled to the Dominican Republic except one who had visited the Turks and Caicos Islands. The remaining 11 cases had no recent travel and also had no knowledge of contact with persons who have had recent travel history.
2.3. Location of stx1 Gene in S. flexneri Strains
In a report of Shigella infections related to travel to Hispaniola, it was indicated that the stx1 gene of these S. flexneri was located on a 62 kb φPOC-J13 phage [17]. This phage carries the stx1 gene and was inserted on the S. flexneri chromosome [17]. Whole genome sequence (WGS) analysis showed that the same phage was carried by all of our 26 stx1 positive S. flexneri isolates with a sequence similarity of 99.9% except that strain S1 had a 528 bp deletion and strain S3 had a 180 bp insertion within the phage. In additon, our PCR results of the insertion site showed the φPOC-J13 phage was inserted on the same chromosome site as demonstrated by Gray et al. [17]. All 26 isolates showed 100% sequence identity in the stx1 gene. The general genome characteristics are shown in Table S3.
2.4. Relatedness of Travel and Non-Travel stx1 S. flexneri Strains
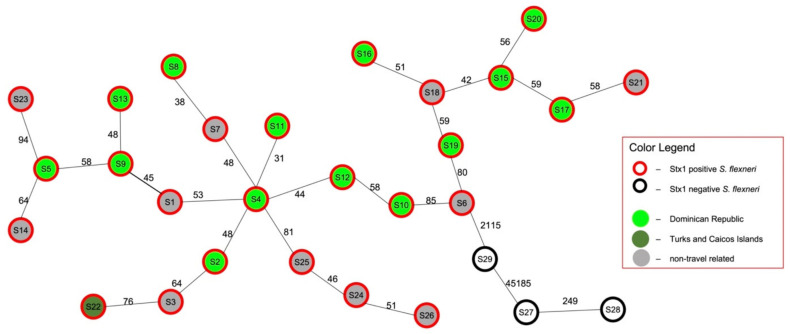
Core genome-based SNP analysis was performed to evaluate the extent of relatedness between the 26 stx1 positive S. flexneri strains in Alberta. These isolates differed from each other from 31 (S4 and S11) to 170 (S20 and S26) SNPs at the core genome level (3.6 MB) (Table S4) with a median SNP difference of 104. Data from a minimum spanning tree further illustrated that closely related strains with less SNP differences clustered together (Figure 1) regardless of the patient travel history. The three stx1 negative S. flexneri (S27, S28 and S29) clustered separately from the 26 stx1 positive S. flexneri group, and they differ by a distance of >2115 SNPs. A pairwise whole genome analysis was also performed on these 26 strains and genome similarities ranged from 99.3% to 99.7% (Table S5).
Figure 1.
Core genome SNP based minimum spanning tree of 29 S. flexneri strains isolated in Alberta, Canada. S. flexneri strains were isolated from patients that had recent travel to Dominican Republic (light green in-filled circles), Turks and Caicos Islands (dark green in-filled circles), and non-travel history (grey in-filled circles). The red color of the outer circle represents stx1 positive S. flexneri while the ones with black outer circle are stx1 negative S. flexneri. Numbers on lines indicate core genome SNP differences between adjacent strains.
2.5. Relatedness of stx1 Positive S. flexneri Strains from Different Countries
The core genome SNPs analysis on sequences of 55 stx1 positive (26 from Alberta and 29 from the NCBI genome database) and 3 stx1 negative S. flexneri strains (S27, S28, and S29) were performed to examine the relatedness of Alberta isolates with global isolates from NCBI (Figure 2). The 29 NCBI sequences were from cases diagnosed in the US, France, Canada (Quebec), and Haiti along with their travel histories (Table 1). Interestingly, cases with travel history all indicated to have travelled to or reside in the Caribbean countries, including the Dominican Republic, Haiti, French Guiana, and Turks and Caicos Islands. The pairwise SNP difference analysis (Table S6) demonstrated that the 55 stx1 positive S. flexneri strains were closely related and the SNPs differences varied from 0 (S25 and S26) to 189 (SN21 and SN339) SNPs with a core genome size of 3.6 MB.
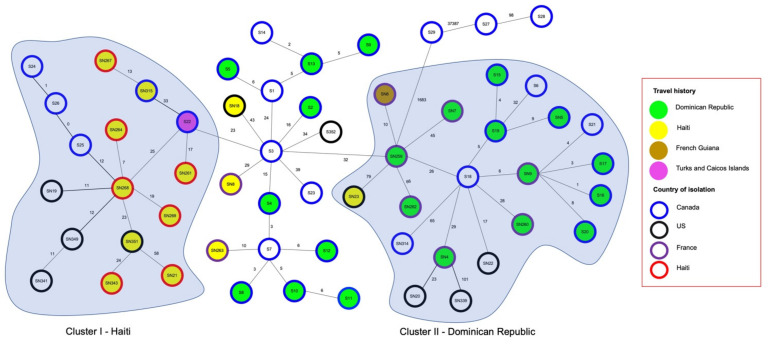
Figure 2.
Core genome SNP based minimum spanning tree of stx1 positive S. flexneri strains isolated from four countries and their related travel history. These strains were isolated from patients in Canada (blue outer circles), U.S. (black outer circles), France (purple outer circles), and Haiti (red outer circles). The travel history is depicted by the filled colors of the circles with green (Dominican Republic), yellow (Haiti), light brown (French Guiana), and purple (Turks and Caicos Islands). Country specific Clusters I and II are shaded in light blue. Numbers on lines indicate core genome SNP differences between adjacent strains.
In the minimum spanning tree (Figure 2), two country specific clusters (I and II) of stx1 positive S. flexneri were identified based on the case’s country of residency or recent travel destinations. In Cluster I, all strains with known travel history were isolated from cases in or who had visited Haiti except S22, which was isolated from a case who travelled to Turks and Caicos Islands. In comparison, all strains within Cluster II were isolated from cases who travelled to the Dominican Republic, except one that visited Haiti (SN23) and one that visited French Guiana (SN6).
2.6. Bayesian Phylogenomic Analysis of stx1 Positive S. flexneri from Different Countries
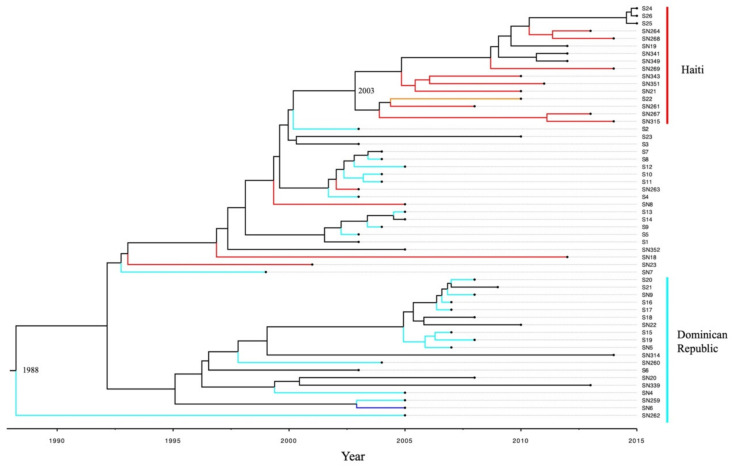
In order to investigate the country related evolution of all 55 stx1 positive S. flexneri strains as revealed in the spanning tree analysis, a Bayesian phylogenomic analysis was performed. As illustrated in Figure 3, stx1 positive S. flexneri isolates formed two clusters based on their source countries including Cluster I—Haiti and Cluster II—Dominican Republic. Within Cluster II—Dominican Republic, 16 strains had source country data, among which only two strains [one Haiti (SN23) and one French Guiana (SN6)] were not from the Dominican Republic. In Cluster—Haiti, only one Turks and Caicos Islands strain (SN22) was observed while the other nine strains were all from Haiti. Root reference (Figure 3) suggested that the most common ancestor of the stx1 positive S. flexneri most likely first emerged in 1988 [95% credible interval (CI): 1984–1992] in the Dominican Republic. A Haiti Cluster arose in 2003 with 95% CI of (2001, 2004). Strains S24, S25, and S26 were from Canadian cases with no travel or contact with people with recent travel. However, their isolates grouped within the Haiti Cluster (Figure 2 and Figure 3) suggesting the origin might be from Haiti.
Figure 3.
Bayesian phylogenetic tree based on 55 stx1 positive S. flexneri strains. Two country specific Haiti and Dominican Republic were formed. The branches were colored according to the strain’s country of isolation or the respective case’s recent travel country. Branches colored with red represents Haiti strains while cyan represents Dominican Republic strains. The branches colored with orange and blue represent Turks Caicos Island and French Guiana strains, respectively. The most common ancestor of stx1 positive S. flexneri strains emerged in 1988. The Haiti Clade arose in 2003.
2.7. Genome Evolution of stx1 Positive S. flexneri Strains
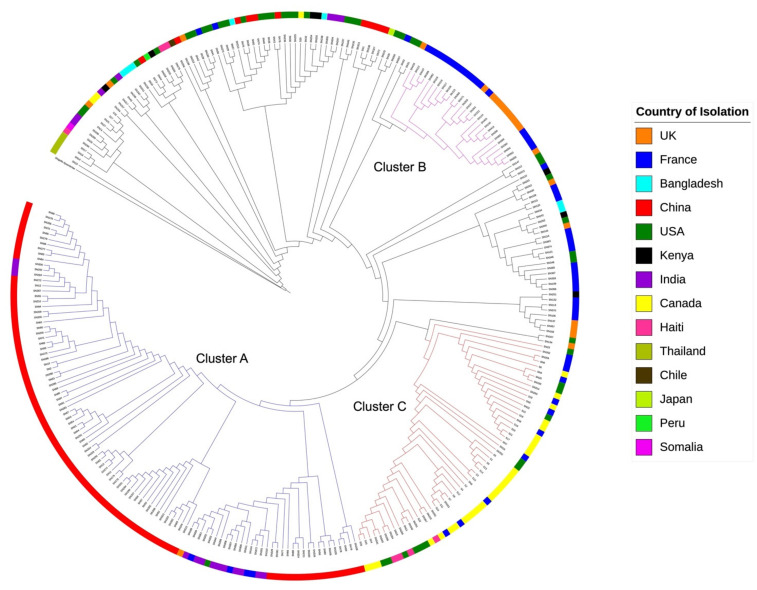
A maximum likelihood phylogenetic tree was constructed (Figure 4) to understand the evolutionary relationship of S. flexneri, including stx negative and stx positive strains. This tree was based on 57,889 SNPs (core genome size of 1.7 MB) identified among the 296 S. flexneri genomes originated from 14 different countries. Overall, strains from the same or geographically close countries clustered closely together (Figure 4). For example, in Cluster A, 94.3% of strains came from two Asian countries, China (87.0%, n = 86) and India (13%, n = 13). It was also observed that Cluster B strains (n = 27) were all from the UK and France. All 55 stx1 positive S. flexneri strains clustered in a single clade (Cluster C) in the phylogenetic tree suggesting that the stx1 positive S. flexneri strains are highly related and share a common ancestor.
Figure 4.
Phylogenetic tree based on 297 S. flexneri genomes. S. dysenteriae was used as the outgroup and branch length was not represented in this tree. The colors of circle represent 14 different countries where the S. flexneri were isolated. Three geographic specific clusters were identified, and their branches were colored in blue, red, and purple respectively. Strains in Cluster A were mostly isolated in China and India. Most of strains in Cluster B were from UK and France. In Cluster C consisted of all stx1 positive S. flexneri strains.
3. Discussion
Evolution of pathogens can give rise to highly virulent strains as illustrated by the 2011 STEC O104:H4 outbreak in Germany. That outbreak strain originated from an enteroaggregative E. coli (EAEC) but later acquiring a Shiga-toxin-encoding phage and antibiotic resistance genes making it highly pathogenic [25]. Although uncommon, identification of stx positive S. flexneri strains have been reported by several studies [17,19]. Therefore, understanding their global circulation and evolution is of great public health importance. This study provides the first comprehensive whole genome-based analysis of stx positive S. flexneri strains obtained globally.
In this investigation, 26/366 S. flexneri strains isolated between 2003 and 2016 in Alberta were found carrying Shiga toxins. Among the 366 S. flexneri cases, 23 cases (data not shown) had recent travel to the Dominican Republic and 60.9% (14/23) of the strains isolated from these cases carried the stx1 gene. A similar study by Gray et al. [23] also found 57.1% (4/7) of S. flexneri isolates cultured from cases in Haiti tested positive for stx1. No severe clinical symptoms such as hemolytic urrmic syndrome were observed for stx1 positive S. flexneri infection as reported [23]. The stx1 carried by the clinical isolates in this study belong to stx1a which is not as virulent as other subtypes, such as stx2a [26]. Currently, it is still unknown whether acquisition of this stx1 phage has increased the pathogenicity of these Shigella strains; however, it is evident that stx1 positive S. flexneri has exceptionally high prevalence rates (60.9% observed in Alberta cases returning from Dominican Republic and 57.1% observed in residents in Haiti as shown above) suggesting the stx1 positive S. flexneri strains have established in Alberta and spread locally.
When compared to sequence of stx1 positive S. flexneri strains reported in other countries (Figure 2), we have observed no apparent clustering of strains from the same country of diagnoses. Instead, strains isolated from cases residing in or had recent travel to the same country (i.e., Dominican Republic) clustered closely to each other. For example, two strains (S16 and SN9) were almost identical (1 SNP difference) across their core genome, albeit the cases were diagnosed in Canada and France (Figure 2), and they all have travelled to the Dominican Republic. Results for pairwise whole genome similarity analysis (Table S5) further support that all stx1 positive S. flexneri strains are extremely similar, with genome similarities ranging from 99.3% to 99.7%. This provides strong evidence that most of the Alberta clinical cases caused by stx1 positive S. flexneri were related to international travel to the Caribbean countries highlighting the public health risk of global transmission.
Based on the travel information collected, 12 of the 26 infections in the Canadian cases were not travel-related and had no acknowledgement of personal contact to people returning from the Caribbean countries, suggesting the possibility of secondary transmission in Canada. This has substantial public health implications because of the potential of having carriers of these strains or unidentified source, e.g., imported food, in Alberta. In a similar U.S. study, some Stx-producing S. sonnei strains circulating in south California may have been introduced from Mexico but then established themselves locally [10]. Therefore, under appropriate conditions, this endemic Caribbean S. flexneri may become endemic in a new geographic region, causing sporadic infections or even outbreaks. Even if the stx1 positive S. flexneri were not establishing endemicity in an area outside of the Caribbean, its phage φPOC-J13 carrying the stx1 gene can be passed on to the other bacteria thorugh horizonal gene transfer [26] and cause public health problems. The stx1 carrying φPOC-J13 phages can infect and lysogenize E. coli and other Shigella strains, such as S. dysentariae, S. boydii, and S. sonnei [17,23,27]. Once brought into a new geographic area by international travelers, this phage may find its way to establish themselves in local pathogenic strains. Under this circumstance, a more pathogenic strain may evolve and cause severe outbreaks similar to the E. coli O104 outbreak started in Germany.
Similar to the findings of Fogolari et al. [27], our spanning tree and Bayesian phylogenetic analyses demonstrated that stx1 positive S. flexneri have developed country specific subclades (the Haiti Clade and the Dominican Republic Clade). Haiti and Dominican Republic are located on the same island of Hispaniola whose size is 76,192 km2, therefore, the existence of two country-specific clades is unexpected. Although sharing the same island, the two countries have considerable differences with regard to economy, geography, and demography, etc. [28,29]. It has been found that people of different ethnic groups have unique gut microbial profiles which might be caused by their differences in diet, genetics, cultural habits, and socioeconomic status [30]. There is a great difference in the ethnicity of the population between Haiti and Dominican Republic [28,29], and therefore the S. flexneri strains from the two country specific clades may have adapted themselves to survive in these two different populations.
Although it has been demonstrated that the φPOC-J13 can integrate in E. coli or other S. flexneri strain in laboratory conditions [17], all 55 φPOC-J13 carrying S. flexneri isolated from cases fall into the same clade on the phylogenetic tree (296 S. flexneri strains) but was not observed in other clades of the tree. This result indicates that the φPOC-J13 can only be steadily transferred to certain types of S. flexneri populations as bacteria have developed several phage resistance mechanisms to prevent phage transfection [31]. This unique clustering allows us to suspect that the common ancestor of the φPOC-J13 carrying S. flexneri strains might have lost their ability to resist φPOC-J13 transfection, which leads to stable integration of φPOC-J13. In the phylogenetic tree (Figure 4), we have also observed clades with abundant Asian S. flexneri strains or UK-France strains. This result implicates that the transmission of some S. flexneri strains are limited to geographically close regions, which is also the case for various pathogens [32,33,34]. However, this geographical boundary will be diminishing with the global spread of pathogens facilitated by increasing global travel and trades.
4. Conclusions
In summary, we have identified 26 stx1 positive S. flexneri strains among 366 clinical Shigella isolates from 2003 to 2016 in Alberta, Canada. The limitation of this study is the failure to obtain clinical information of all the S. flexneri cases and the outcome of the disease. The majority of the 26 stx1 positive S. flexneri strains originated from Dominican Republic while three of them may have Haiti origin based on genomic anaylsis. These strains were observed to have geographic specific distribution patterns as Haiti and Dominican Republic specific clades. However, with the capability of the transferring of the stx gene on the phage to other strains, and with increase in international travel, it can facilitate the global spread and cause an alarm in public health, therefore systematic surveillance of stx1 positive S. flexneri strains is of high importance.
5. Materials and Methods
5.1. Bacterial Strains
This study included all archived S. flexneri (n = 366) clinical strains referred to ProvLab for confirmation and typing from 2003 to 2016 in Alberta, Canada. These isolates were retrieved from 10% skim milk stored at −80 °C, cultured on sheep blood agar plates [BAP] (Dalynn Biologicals, Calgary, AB, Canada) at 37 °C for 16 h.
5.2. Identification of Shiga Toxin-Producing S. flexneri
Real time PCR amplification of stx1 and stx2 genes as a multiplex assay was used for stx screening as previously described [35]. Sequences of primers and probes for PCR are shown in Table S1. Subtyping of stx1 positive isolates was performed by conventional PCR as described by Scheutz et al. [12]. The presence of the toxin was confirmed by SHIGA TOXIN QUIK CHEK™ (TechLab, Blacksburg, VA, USA), a commercial enzyme immunoassay. The procedure was carried out as per manufacturer’s instruction.
To verify the insertion site of the phage φ POC-J13 [14] into the genome of stx positive S. flexneri, two sets of PCRs were designed using Primer 3 [36]. Each set has a primer targeting the phage φPOC-J13 region and another primer targeting upstream or downstream of insertion sites based on the S. flexneri genome sequence (Figure S1 and Table S1). The final volume of each PCR reaction was 20 μL, containing 10 μL of 2X SYBR® Green PCR Master Mix (Life Technologies, Carlsbad, CA, USA), and 900 nM of each primer. Real-time PCR was performed in an ABI 7500 Fast system (Applied Biosystems, Foster City, CA, USA) with the following conditions: 95 °C for 10 min, 40 cycles of 95 °C for 15 s, 60 °C for 1 min.
5.3. Whole Genome Sequencing and Pairwise Whole Genome Similarity Analysis
WGS was performed on all stx positive S. flexneri strains identified in this study along with three stx negative S. flexneri strains. Genomic DNA was extracted from overnight cultures using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA). Sequencing libraries were prepared using the Nextera XT kit (Illumina, San Diego, CA, USA), and WGS was carried out on the Illumina MiSeq platform. Trimmomatic Version 0.38 [37] was used to trim the low-quality reads of each genome. De novo assembly was performed using SPAdes Version 3.9.1 [38] and contigs smaller than 500 bp were removed. Pairwise whole genome similarity analysis was performed using REALPHY 1.12 [39] where strain S1 was randomly selected as reference.
5.4. Core Genome SNP Analysis and Minimum Spanning Tree
The core genome of all stx positive S. flexneri and three stx negative strains were analysed using REALPHY 1.12 [39]. One of the stx positive S. flexneri strains was randomly selected as reference, and genome sequences of all other strains were then mapped to the reference genome to identify their core genome. The core genome was analyzed by MEGA X Version 10.1.0 [40] to calculate their pairwise SNP differences. In addition, a minimum spanning tree was generated using Phyloviz [41] based on core genome SNPs differences.
Another core genome SNP analysis was performed with an extended number of S. flexneri strains by inclusion of sequence data of stx positive S. flexneri strains (n = 29) whose genome sequences were obtained from the NCBI genome database. Their epidemiological data were also collected through NCBI or their corresponding publications. The same analysis settings were used as described in the previous paragraph.
5.5. Bayesian Phylogenetic Analysis of stx Positive S. flexneri Strains from Different Countries
Core genomes of all stx positive S. flexneri strains identified in this study and 29 strains from NCBI database were analyzed using REALPHY 1.12 [39]. Recombination sequences in the core genome were predicted and removed using Gubbins [42]. The core genome SNP alignment was subjected to Bayesian evolutionary analysis using BEAST Version 2.6 [43]. For the BEAST analysis, HKY substitution model, strict molecular clock, constant population size model was selected. The chain length was set to 100 million and sampling was set to every 10,000 iterations. The tip dates were defined as the year of isolation. The BEAST tree was annotated using TreeAnnotater from the BEAST package, visualized using FigTree Version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/), and annotated using iTOL [44,45].
5.6. Phylogenomic Analysis of S. flexneri
To understand the evolutionary trajectory of these stx positive S. flexneri isolates, a maximum likelihood (ML) phylogenetic tree was constructed. This analysis included 267 S. flexneri genomes downloaded from the NCBI database (Table S2) and sequence of 29 S. flexneri strains identified in this study. One S. dysenteriae genome (Accession No.: NC_007606.1) from NCBI was used as an outgroup. The core genome was called, and recombination sequences were removed as described in the previous paragraph. The phylogenetic tree was constructed using RAxML Version 8.2.4 with GTRGAMMA option and visualized using the Interactive Tree of Life (iTOL) [44,45].
Acknowledgments
We thank Public Health Agency of Canada—National Microbiology Laboratory, Winnipeg, MB, Canada for providing next genome sequencing; Alberta Precision Laboratories-ProvLab for supporting this study by contributing the study strains.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/toxins13110755/s1, Table S1: PCR primers used in this study. Table S2: NCBI accession numbers of S. flexneri strains used in this study. Table S3: general genomic characteristics of S. flexneri strains. Table S4: SNP differences based on the core genome of 26 stx positive S. flexneri strains from Canadian cases. Table S5: pairwise genome similarity stx1 positive S. flexneri strains. Table S6: SNP differences based on the core genome of 55 stx positive S. flexneri strains. Figure S1: location of primers for confirming the specific insertion of phage φPOC-J13 in the chromosome of S. flexneri.
Author Contributions
Conceptualization, L.C. and S.Z.; methodology, L.C., S.Z., B.D.P., J.S., Y.T.K.Y. and C.F.; software, S.Z. and V.L.; validation, S.Z.; formal analysis, S.Z.; investigation, L.C. and S.Z.; resources, L.C., P.F. and S.D.; data curation, L.C. and S.Z.; writing—original draft preparation, L.C. and S.Z.; writing—review and editing, L.C., S.Z., B.D.P., J.S., P.F., S.D.,V.L., Y.T.K.Y., C.F., S.B.F., B.E.L. and X.-L.P.; visualization, S.Z.; supervision, L.C.; project administration, L.C.; funding acquisition, L.C. and S.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Alberta Health Services Residual Funds. B.D.P. was supported by a Collaborative Research Innovation Opportunity Grant from Alberta Innovates; grant number 20140161. S.B.F. is supported by the Alberta Children’s Hospital Foundation Professorship in Child Health and Wellness. J.S. was the recipient of Undergraduate Research Initiative (URI) Stipend, University of Alberta; Y.T.K.Y. was the recipient of Alberta Innovates Health Solutions summer studentship. S.Z. was partially supported by the National Natural Sciences Foundation of China while completing the genomic analysis; grant number 82073514.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
Some stx positive S. flexneri isolates were identified from Alberta, Canada (2003 to 2016) from patients with travel history to the Dominican Republic. Geographic specific distribution patterns (Haiti and Dominican Republic clades) were observed through genomic analysis of stx positive S. flexneri strains obtained globally.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhu Z., Wang W., Cao M., Zhu Q., Ma T., Zhang Y., Liu G., Zhou X., Li B., Shi Y., et al. Virulence factors and molecular characteristics of Shigella flexneri isolated from calves with diarrhea. BMC Microbiol. 2021;21:214. doi: 10.1186/s12866-021-02277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi R., Yang X., Chen L., Chang H.T., Liu H.Y., Zhao J., Wang X.W., Wang C.Q. Pathogenicity of Shigella in chickens. PLoS ONE. 2014;9:e100264. doi: 10.1371/journal.pone.0100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao M., Wang W., Zhang L., Liu G., Zhou X., Li B., Shi Y., Zhu Z., Zhang J. Epidemic and molecular characterization of fluoroquinolone-resistant Shigella dysenteriae1 isolates from calves with diarrhea. BMC Microbiol. 2021;21:6. doi: 10.1186/s12866-020-02050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu G.Z., Chen H.Y., Si H.B., Deng L.X., Wei Z.Y., Yuan L., Kuang X.H. Phenotypic and molecular characterization of TEM-116 extended-spectrum beta-lactamase produced by a Shigella flexneri clinical isolate from chickens. FEMS Microbiol. Lett. 2008;279:162–166. doi: 10.1111/j.1574-6968.2007.01017.x. [DOI] [PubMed] [Google Scholar]
- 5.Kotloff K.L., Riddle M.S., Platts-Mills J.A., Pavlinac P., Zaidi A.K.M. Shigellosis. Lancet. 2018;391:801–812. doi: 10.1016/S0140-6736(17)33296-8. [DOI] [PubMed] [Google Scholar]
- 6.Taneja N., Mewara A. Shigellosis: Epidemiology in India. Indian J. Med. Res. 2016;143:565–576. doi: 10.4103/0971-5916.187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowen A., Hurd J., Hoover C., Khachadourian Y., Traphagen E., Harvey E., Libby T., Ehlers S., Ongpin M., Norton J.C., et al. Importation and Domestic Transmission of Shigella sonnei Resistant to Ciprofloxacin—United States, May 2014–February 2015. MMWR Morb. Mortal. Wkly. Rep. 2015;64:318–320. [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J.Y., Henao O.L., Griffin P.M., Vugia D.J., Cronquist A.B., Hurd S., Tobin-D’Angelo M., Ryan P., Smith K., Lathrop S., et al. Infection with Pathogens Transmitted Commonly Through Food and the Effect of Increasing Use of Culture-Independent Diagnostic Tests on Surveillance—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2012–2015. MMWR Morb. Mortal. Wkly. Rep. 2016;65:368–371. doi: 10.15585/mmwr.mm6514a2. [DOI] [PubMed] [Google Scholar]
- 9.Mattock E., Blocker A.J. How Do the Virulence Factors of Shigella Work Together to Cause Disease? Front. Cell Infect. Microbiol. 2017;7:64. doi: 10.3389/fcimb.2017.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryan A., Youngster I., McAdam A.J. Shiga Toxin Producing Escherichia coli. Clin. Lab. Med. 2015;35:247–272. doi: 10.1016/j.cll.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Melton-Celsa A.R. Shiga Toxin (Stx) Classification, Structure, and Function. Microbiol. Spectr. 2014;2:2–4. doi: 10.1128/microbiolspec.EHEC-0024-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheutz F., Teel L.D., Beutin L., Pierard D., Buvens G., Karch H., Mellmann A., Caprioli A., Tozzoli R., Morabito S., et al. Multicenter Evaluation of a Sequence-based Protocol for Subtyping Shiga toxins and Standardizing Stx Nomenclature. J. Clin. Microbiol. 2012;50:2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beutin L., Strauch E., Fischer I. Isolation of Shigella sonnei Lysogenic for a Bacteriophage Encoding Gene for Production of Shiga toxin. Lancet. 1999;353:1498. doi: 10.1016/S0140-6736(99)00961-7. [DOI] [PubMed] [Google Scholar]
- 14.Lamba K., Nelson J.A., Kimura A.C., Poe A., Collins J., Kao A.S., Cruz L., Inami G., Vaishampayan J., Garza A., et al. Shiga Toxin 1-Producing Shigella sonnei Infections, California, United States, 2014–2015. Emerg. Infect. Dis. 2016;22:679–686. doi: 10.3201/eid2204.151825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams C., Vose A., Edmond M.B., Lyckholm L. Shigella sonnei and Hemolytic Uremic Syndrome: A Case Report and Literature Review. IDCases. 2017;8:6–8. doi: 10.1016/j.idcr.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter C.C., Fierer J., Chiu W.W., Looney D.J., Strain M., Mehta S.R. A Novel Shiga Toxin 1a-Converting Bacteriophage of Shigella sonnei With Close Relationship to Shiga Toxin 2-Converting Pages of Escherichia coli. Open Forum Infect. Dis. 2016;3:ofw079. doi: 10.1093/ofid/ofw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray M.D., Lampel K.A., Strockbine N.A., Fernandez R.E., Melton-Celsa A.R., Maurelli A.T. Clinical isolates of Shiga toxin 1a-producing Shigella flexneri with An Epidemiological Link to Recent Travel to Hispaniola. Emerg. Infect. Dis. 2014;20:1669–1677. doi: 10.3201/eid2010.140292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta S.K., Strockbine N., Omondi M., Hise K., Fair M.A., Mintz E. Emergence of Shiga toxin 1 genes within Shigella dysenteriae Type 4 Isolates from Travelers Returning from the Island of Hispanola. Am. J. Trop. Med. Hyg. 2007;76:1163–1165. doi: 10.4269/ajtmh.2007.76.1163. [DOI] [PubMed] [Google Scholar]
- 19.Bekal S., Pilon P.A., Cloutier N., Doualla-Bell F., Longtin J. Identification of Shigella flexneri isolates carrying the Shiga toxin 1-producing gene in Quebec, Canada, linked to travel to Haiti. Can. J. Microbiol. 2015;61:995–996. doi: 10.1139/cjm-2015-0538. [DOI] [PubMed] [Google Scholar]
- 20.Nogrady N., Kiraly M., Borbas K., Toth A., Paszti J., Toth I. Antimicrobial Resistance and Genetic Characteristics of Integron-Carrier Shigellae Isolated in Hungary (1998–2008) J. Med. Microbiol. 2013;62:1545–1551. doi: 10.1099/jmm.0.058917-0. [DOI] [PubMed] [Google Scholar]
- 21.Gray M.D., Lacher D.W., Leonard S.R., Abbott J., Zhao S., Lampel K.A., Prothery E., Gouali M., Weill F.X., Maurelli A.T. Prevalence of Shiga toxin-producing Shigella Species Isolated from French Travellers Returning from the Caribbean: An Emerging Pathogen with International Implications. Clin. Microbiol. Infect. 2015;21:765.e9–765.e14. doi: 10.1016/j.cmi.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyholm O., Lienemann T., Halkilahti J., Mero S., Rimhanen-Finne R., Lehtinen V., Salmenlinna S., Siitonen A. Characterization of Shigella sonnei Isolate Carrying Shiga Toxin 2-Producing Gene. Emerg. Infect. Dis. 2015;21:891–892. doi: 10.3201/eid2105.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray M.D., Leonard S.R., Lacher D.W., Lampel K.A., Alam M.T., Morris J.G., Jr., Ali A., LaBreck P.T., Maurelli A.T. Stx-Producing Shigella Species From Patients in Haiti: An Emerging Pathogen With the Potential for Global Spread. Open Forum Infect. Dis. 2015;2:ofv134. doi: 10.1093/ofid/ofv134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freedman S.B., Lee B.E., Louie M., Pang X.L., Ali S., Chuck A., Chui L., Currie G.R., Dickinson J., Drews S.J., et al. Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE): Epidemiology, emerging organisms, and economics. BMC Pediatr. 2015;15:89. doi: 10.1186/s12887-015-0407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasko D.A., Webster D.R., Sahl J.W., Bashir A., Boisen N., Scheutz F., Paxinos E.E., Sebra R., Chin C.S., Iliopoulos D., et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 2011;365:709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruger A., Lucchesi P.M. Shiga Toxins and stx Phages: Highly Diverse Entities. Microbiology. 2015;161:451–462. doi: 10.1099/mic.0.000003. [DOI] [PubMed] [Google Scholar]
- 27.Fogolari M., Mavian C., Angeletti S., Salemi M., Lampel K.A., Maurelli A.T. Distribution and Characterization of Shiga Toxin Converting Temperate Phages Carried by Shigella flexneri in Hispaniola. Infect. Genet. Evol. 2018;65:321–328. doi: 10.1016/j.meegid.2018.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The World Factbook–Dominican Republic. [(accessed on 1 December 2020)]; Available online: https://www.cia.gov/library/publications/the-world-factbook/geos/dr.html.
- 29.The World Factbook–Haiti. [(accessed on 1 December 2020)]; Available online: https://www.cia.gov/library/publications/the-world-factbook/geos/ha.html.
- 30.Liu W., Zhang J., Wu C., Cai S., Huang W., Chen J., Xi X., Liang Z., Hou Q., Zhou B., et al. Unique Features of Ethnic Mongolian Gut Microbiome Revealed by Metagenomic Analysis. Sci. Rep. 2016;6:34826. doi: 10.1038/srep34826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labrie S.J., Samson J.E., Moineau S. Bacteriophage Resistance Mechanisms. Nat. Rev. Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 32.Davies T.J., Pedersen A.B. Phylogeny and Geography Predict Pathogen Community Similarity in Wild Primates and Humans. Proc. Biol. Sci. 2008;275:1695–1701. doi: 10.1098/rspb.2008.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franz E., Rotariu O., Lopes B.S., MacRae M., Bono J.L., Laing C., Gannon V., Soderlund R., van Hoek A., Friesema I., et al. Phylogeographic Analysis Reveals Multiple International Transmission Events Have Driven the Global Emergence of Escherichia coli O157:H7. Clin. Infect. Dis. 2018;69:428–437. doi: 10.1093/cid/ciy919. [DOI] [PubMed] [Google Scholar]
- 34.Lemey P., Rambaut A., Drummond A.J., Suchard M.A. Bayesian Phylogeography Finds Its Roots. PLoS Comput. Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bugarel M., Beutin L., Martin A., Gill A., Fach P. Micro-array for the Identification of Shiga toxin-producing Escherichia coli (STEC) Seropathotypes Associated with Hemorrhagic Colitis and Hemolytic Uremic Syndrome in Humans. Int. J. Food Microbiol. 2010;142:318–329. doi: 10.1016/j.ijfoodmicro.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 36.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3—New Capabilities and Interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertels F., Silander O.K., Pachkov M., Rainey P.B., van Nimwegen E. Automated Reconstruction of Whole-Genome Phylogenies from Short-Sequence Reads. Mol. Biol. Evol. 2014;31:1077–1088. doi: 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nascimento M., Sousa A., Ramirez M., Francisco A.P., Carrico J.A., Vaz C. PHYLOViZ 2.0: Providing Scalable Data Integration and Visualization for Multiple Phylogenetic Inference Methods. Bioinformatics. 2017;33:128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- 42.Croucher N.J., Page A.J., Connor T.R., Delaney A.J., Keane J.A., Bentley S.D., Parkhill J., Harris S.R. Rapid Phylogenetic Analysis of Large Samples of Recombinant Bacterial Whole Genome Sequences Using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchene S., Fourment M., Gavryushkina A., Heled J., Jones G., Kuhnert D., De Maio N., et al. BEAST 2.5: An Advanced Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letunic I., Bork P. Interactive Tree Of Life (iTOL): An Online Tool for Phylogenetic Tree Display and Annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 45.Letunic I., Bork P. Interactive Tree Of Life v2: Online Annotation and Display of Phylogenetic Trees Made Easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.