Abstract
Early detection of human immunodeficiency virus (HIV) in blood and blood products can be achieved by a sensitive nucleic acid amplification-based assay. We report on the performance of a PCR-based qualitative assay that detects both HIV type 1 (HIV-1) and HIV-2 with a sensitivity of 20 to 50 copies/ml. The assay has a specificity of 99.6% and an inhibition rate of 1.7%. One milliliter of sample is processed with a manifold system and Qiagen columns, and one-third of the extracted sample is used for PCR amplification. An internal control sequence, which is processed and amplified with each sample, monitors for amplification inhibition. Samples are reverse transcribed and are then amplified by reverse transcription-coupled PCR, after which HIV-1- and HIV-2-specific probes are hybridized to the amplified products. Following hybridization, samples are detected in the LCx instrument by microparticle enzyme immunoassay techniques. The detection system has an automated inactivation step that controls for PCR contamination. The HIV-1/2 qualitative RNA assay detects HIV-1 group M subtypes A, B, C, D, E, F, and G and group O. Testing of several HIV-1 seroconversion panels has demonstrated that the HIV-1/2 qualitative RNA assay detects HIV infection on the average of 6 days before p24 antigen can be detected and 11 days before antibodies can be detected.
The first serological assay for detection of human immunodeficiency virus (HIV) type 1 (HIV-1) was implemented in 1985, 3 years after the recognition of HIV as the causative agent of AIDS (6, 17, 33). In 1986, a second HIV type, HIV-2, was isolated from patients with AIDS in West Africa (11), which led to the implementation of simultaneous screening for HIV-1 and HIV-2 (5, 41). In the following years, continuous improvements of the serological assays, along with donor education and deferral procedures, have greatly reduced the risks of transfusion-acquired HIV infection (16, 25, 39, 46). However, a residual risk that is extremely small still exists and remains a source of public concern over the safety of blood and blood products.
The residual risk of transfusion-acquired infection is attributed primarily to donations during the antibody-negative, preseroconversion window, the period between infection and detectable seroconversion. For HIV-1, that period is about 3 to 4 weeks for the contemporary serological tests (8, 9). Rare cases of HIV transmissions from seronegative blood and organ donors have been reported in the literature (42, 45; F. Coutlee, G. Delage, F. Lamothe, S. Cassol, and F. Decary, Lancet 340:59, 59, 1992, Letter). That residual risk led to investigations into newer technologies for direct detection of the virus to further close the HIV window. Several studies demonstrated the detection of HIV-1 p24 antigen prior to the detection of antibodies (4, 9, 43), which led to the implementation of p24 antigen screening, in addition to antibody screening, in the United States (19).
Direct detection of the virus can also be achieved by nucleic acid amplification techniques, which enable detection of the viral genome. Previous studies reported the detection of HIV RNA approximately 1 week earlier than detection of p24 antigen and HIV DNA and about 2 weeks earlier than antibody detection (9, 28, 31, 32). Here, we report on the performance of a PCR assay for the detection of both HIV-1 and HIV-2 RNAs. This multiplex assay for HIV-1 and HIV-2 (HIV-1/2) RNA detection is designed to detect both HIV-1 group M subtypes and group O. An internal control is incorporated to verify RNA isolation, amplification, and detection for each specimen.
MATERIALS AND METHODS
Primers, probes, and RNA transcripts.
Two distinct sets of primers and probes were designed for HIV-1 and HIV-2 detection. Both HIV-1- and HIV-2-specific primer and probe sequences were targeted against conserved sequences located in the pol genes of the HIV-1 and HIV-2 genomes, respectively (23). The internal control primer and probe sequences were selected from regions in the pumpkin hydroxypyruvate reductase gene. HIV-1 transcripts (subtypes A to F) were generated from plasmids containing a fragment from the HIV-1 pol gene (provided by Steve Wolinski, Northwestern University, Chicago, Ill.). The plasmids were linearized with the restriction enzymes BamHI (for subtypes A, C, D, and F), HindIII (subtype E), and XbaI (subtype B) (Gibco BRL, Rockville, Md.). The RNAs were transcribed with the Ambion MegaScript T7 transcription kit (Ambion, Inc., Austin, Tex.) to yield a 1,693-base sequence. The HIV-2 transcript, of 1,643 bases, was produced by in vitro transcription of an XbaI-linearized plasmid that contained a fragment from the HIV-2 polymerase gene. The internal control transcript, of 1,242 bases, was obtained by in vitro transcription of a BssSI-linearized plasmid containing a 900-bp fragment of the pumpkin hydroxypyruvate reductase gene (provided by B. R. Andersen, University of Wisconsin-Parkside, Kenosha). All transcript RNA was purified from plasmid DNA by digesting the transcript mixture with RNase-free DNase followed by two phenol-chloroform-isoamyl alcohol extractions and one chloroform-isoamyl alcohol extraction. Unincorporated deoxynucleoside triphosphates were removed by passing the transcript over 5 Prime→3 Prime Select Spin (5 Prime-3 Prime Inc., Boulder, Colo.) columns. RNA transcripts were quantitated by digesting 0.8 μg of RNA sample per μl with 100 μg of snake venom phosphodiesterase (Boehringer Mannheim, Indianapolis, Ind.) per ml plus 30 U of calf intestinal alkaline phosphatase (Gibco BRL) per ml in 1× hyperchromicity buffer (8 mM C4H6O4Mg · 4H2O, 3 mM MgCl2, 40 mM K2C2H3O2, 8 mM Tris-acetate [pH 7.5]) at 32°C for 24 h. After digestion, the sample was heated for 10 min at 75°C. The nucleoside concentration was determined by measuring the optical density at 260 nm.
Sample preparation.
Samples were processed with a modified Qiagen QIAmp sample preparation kit (Qiagen, Chatsworth, Calif.). For each sample processed, 1 ml of plasma was added to 1 ml of Qiagen AL lysis buffer containing the internal control transcript. A total of 104 copies of internal control transcript was added to each reaction mixture. One hundred twenty-five microliters of Qiagen protease solution was then added to each sample. The samples were heated for 10 min at 70°C. One milliliter of 100% ethanol was added to each sample. Each sample was then transferred to a Qiagen QIAamp spin column on a vacuum manifold to which a vacuum of 20 to 25 in. of Hg was applied. The columns were washed two times with 0.5 ml of Qiagen AW1 buffer, followed by two washes with 0.5 ml of Qiagen AW2 buffer. The columns were transferred to collection tubes, and the tubes were centrifuged at 13,000 × g for 4 min to remove the remaining wash buffer. After transferring the columns to new collection tubes, the nucleic acids were eluted by the addition of 170 μl of RNase-free water to each column and centrifugation at 13,000 × g for 4 min. The extracted RNA was stored frozen for up to 1 week before amplification.
Amplification.
The master reagent mixture contained 100 μl of the following at pH 8.5: 100 mM bicine (Calbiochem, San Diego, Calif.), 164 mM K2C2H3O2, 66 mM KOH, 0.02 mg of acetylated bovine serum albumin (Gibco BRL) per ml, 0.2 mM EDTA, 0.4 mg of NaN3 per ml, 16% molecular biology-grade glycerol (Gibco BRL), 1,700 μM deoxynucleoside triphosphates (Amersham Pharmacia, Picataway, N.J.), 300 nM HIV-1- and HIV-2-specific forward primers, 400 nM HIV-1- and HIV-2-specific reverse primers, 30 nM (each) HIV-1- and HIV-2-specific probes, 260 nM internal control primers, 18 nM internal control probe, and 11 U of rTth (Roche Molecular, Branchburg, N.J.). Preassembly of a master reagent mixture that includes all the components, including probes, eliminates the need to open the tubes after amplification and reduces potential contamination risk. Fifty microliters of processed specimen and 50 μl of a 14 mM MnCl2 solution were added to each 100 μl of reaction mixture. Samples were vortexed and placed in a Perkin-Elmer 480 DNA Thermal Cycler (Perkin-Elmer, Foster City, Calif.) and were cycled by using the following parameters: 1 cycle consisting of 30 min at 60°C and 2 min at 94°C, followed by 43 cycles consisting of 30 s at 94°C, 40 s at 60°C, and 30 s at 72°C, followed by 1 cycle consisting of 15 min at 97°C, followed by a 4°C soak for a minimum of 5 min.
HIV capture and detection.
After amplification, the tubes were transferred to the Abbott LCx analyzer (Abbott Laboratories, Abbott Park, Ill.) where a signal was generated by automated microparticle enzyme immunoassay detection (15). The HIV-1- and HIV-2-specific reverse primers were labeled with adamantane on the 5′ end, yielding adamantane-labeled amplification products (amplicon) following the amplification cycles. The amplicons were hybridized to the carbazole-labeled HIV-1- or HIV-2-specific probes, and the hybrids were then captured by anticarbazole antibody-coated microparticles in the LCx instrument. The labeled amplicon was then fluorometrically detected with an alkaline phosphatase-labeled antiadamantane conjugate which reacts with the substrate 4-methylumbelliferyl phosphate (MUP).
Internal control capture and detection.
The internal control probe was labeled with two distinct haptens: carbazole on the 5′ end and adamantane on the 3′ end. When amplification failed to occur, the internal control probe was captured with the anticarbazole-coated microparticles in the LCx instrument and was detected with a beta-galactosidase–antiadamantane conjugate which reacts with the substrate 4-methylumbelliferyl galactopyranoside (MUG) to produce a fluorescent molecule. When amplification of the internal control target did occur, the probe was degraded by the 5′ exonuclease activity of the recombinant Tth (rTth), which separates the capture and detection haptens, leading to a decrease in signal indicating that amplification had occurred. After the detection step, amplified products are chemically destroyed by automated addition of copper(II) phenanthroline and hydrogen peroxide to prevent contamination (1, 22).
Data analysis.
Samples were evaluated for amplification efficiency and HIV positivity. The internal control signal was used to evaluate amplification efficiency, with a decrease in the MUG signal of 70% or greater from the background MUG signal being indicative of efficient amplification (2). The MUP signal generated indicated amplification of HIV sequences present in the sample, with a positive sample defined as having a signal/cutoff ratio (S/CO) greater than 1.
RESULTS
Sensitivity.
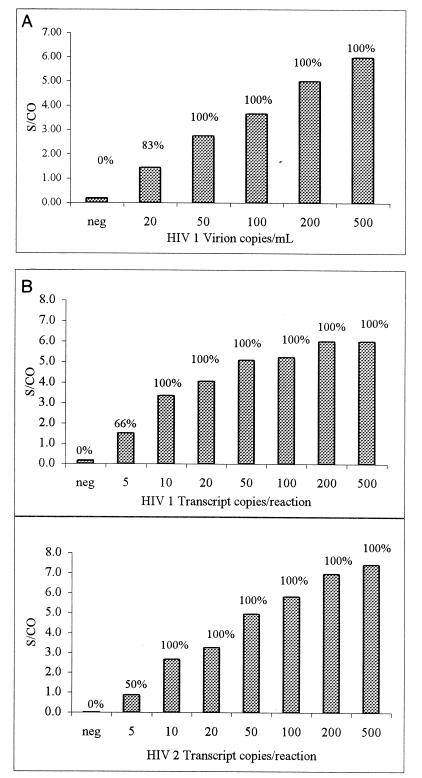
The analytical sensitivity of the assay for HIV-1 detection was assessed by using quantitated HIV-1 virion panels. In vitro-cultured HIV-1 virions were quantified by using the Viral Quality Assurance (Viral Quality Assurance Laboratory, Rush Presbyterian-St Luke, Chicago, Ill.) standards and were further diluted in anti-HIV-1 and anti-HIV-2 antibody-negative and HIV-1 PCR-negative plasma to the concentrations shown in Fig. 1A. One milliliter of each panel member along with an internal control was processed by using the modified Qiagen system described in the Materials and Methods section. Each panel member was tested in replicates of six. One-third of the processed samples (equivalent to about 0.3 ml of plasma) was amplified and was then detected in the LCx analyzer as described above. As few as 20 copies per ml, which is equivalent to 6 copies per reaction mixture, were detectable. The mean S/CO for the six replicates at this level was 1.5, and five of the six replicates were positive, i.e., S/CO of >1.0. For the panel member with 50 copies per ml, which is equivalent to 15 copies per reaction mixture, all the replicates were positive, with a mean S/CO of 2.8 (Fig. 1A).
FIG. 1.
(A) Analytical sensitivity of the LCx HIV-1/2 qualitative RNA assay with virion panels. HIV-1 virion panels quantitated against viral quality assurance standards were diluted in negative human plasma to the indicated concentrations. One milliliter of each sample was processed as described in Materials and Methods, and one-third of each of the processed samples (0.3 ml plasma equivalent) was tested. Six replicates of each concentration were tested. Percent positive samples are indicated above the bars. An S/CO of >1 is considered a positive result. (B) Analytical sensitivity of the LCx HIV-1/2 qualitative RNA assay with transcript panels. HIV-1 and HIV-2 transcripts quantitated by hyperchromicity were tested with the indicated number of copies per reaction mixture as described in the Materials and Methods section. Six replicates of each concentration were tested. Percent positive samples are indicated above the bars. An S/CO of >1 is considered a positive result.
Analytical sensitivities for detection of HIV-1 and HIV-2 were also assessed with HIV-1 and HIV-2 RNA transcripts, since a quantitative assay for quantitation of HIV-2 virions in plasma is not available. HIV-1 and HIV-2 transcripts were generated as described above and were diluted to the concentrations shown in Fig. 1B. Since the transcripts were not processed through sample preparation, an internal control transcript was added to the HIV transcripts prior to addition to the reaction mixture. The transcripts were amplified and were then detected in the LCx analyzer as described above. Six replicates at each concentration were tested. For HIV-1, 5 copies per reaction mixture could be detected in four of six replicates (66%), while 10 copies or more per reaction mixture could be detected 100% of the time (Fig. 1B). These results are consistent with the data shown in Fig. 1A for detection of HIV-1 virions after sample processing, suggesting that no significant loss of RNA occurs during sample processing. Sensitivity for detection of HIV-2 was very similar to that for detection of HIV-1; 5 copies per reaction mixture could be detected in three of six replicates (50%), while 10 copies or more per reaction mixture were detected 100% of the time (Fig. 1B).
Detection of HIV seroconversion samples.
Twenty-five seroconversion panels (Boston Biomedica Inc., Boston, Mass., and Bioclinical Partners Inc., Franklin, Mass.) consisting of 151 total members were tested by the qualitative assay for detection of HIV-1 and HIV-2 RNA (HIV-1/2 qualitative RNA assay) to determine the earliest date of detection. Due to the limited quantities of the samples, 0.2 ml of each panel member was processed through Qiagen columns and was eluted with 120 μl of water, and 50 μl of the eluate was tested per reaction mixture. This is equivalent to 0.083 ml of plasma sample, in contrast to the 0.3 ml of plasma equivalent that was used in the experiment described above. Table 1 lists the commercial seroconversion panels tested and the bleed day at which the individual became positive by the HIV-1/2 qualitative RNA assay, a p24 antigen detection assay (HIV AG-1 monoclonal EIA; Abbott Laboratories), and an HIV-1/2 antibody detection assay (HIV1/HIV2 third-generation antibody test; Abbott Laboratories). In general, RNA detection preceded antigen detection, which in turn preceded antibody detection. For 22 of 25 panels, detection of HIV infection by the RNA test preceded detection by the antigen test, and for 24 of 25 panels, RNA detection preceded antibody detection.
TABLE 1.
Evaluation of the LCx HIV-1/2 qualitative RNA assay with seroconversion panels
| Panel no. | First day of positivity
|
Viral load on first day of positivity measured by an HIV quantitative test
|
||||
|---|---|---|---|---|---|---|
| HIV-1/2 qualitative RNA assay | Antigen detection test | Antibody detection test | HIV-1/2 qualitative RNA assay | Antigen detection test | Antibody detection test | |
| PRB916 | 9 | 15 | 30 | NTa | NT | NT |
| PRB923 | 35 | 37 | 47 | NT | NT | NT |
| PRB926 | 2 | 7 | 27 | NT | NT | NT |
| PRB929 | 14 | 18 | >21 | NT | NT | NT |
| PRB930 | 0 | 3 | 7 | NT | NT | NT |
| PRB931 | 15 | 28 | 28 | 3 × 104 | 2 × 106 | 2 × 106 |
| PRB932 | 27 | 27 | 27 | 4 × 105 | 4 × 105 | 4 × 105 |
| PRB934 | 0 | 0 | 7 | 4 × 105 | 4 × 105 | 4 × 105 |
| PRB935 | 24 | 28 | 43 | 4 × 103 | 7 × 105 | 5 × 105 |
| PRB936 | 5 | 12 | 19 | 4 × 102 | >8 × 105 | >8 × 105 |
| PRB937 | 7 | 14 | 21 | 4 × 102 | 1 × 105 | >8 × 105 |
| PRB939 | 0 | 7 | 89 | 2 × 104 | 8 × 105 | 2 × 104 |
| PRB941 | 4 | >18 | 18 | 3 × 103 | NAb | 7 × 104 |
| PRB942 | 9 | 14 | >14 | 5 × 103 | >8 × 105 | NA |
| PRB943 | 5 | 12 | 14 | 5 × 103 | >8 × 105 | >8 × 105 |
| PRB944 | 0 | 2 | 14 | 7 × 103 | 8 × 104 | 6 × 105 |
| PRB945 | 0 | 3 | 13 | 7 × 102 | >8 × 105 | >8 × 105 |
| PRB946 | 4 | 7 | >11 | 3 × 104 | 7 × 105 | NA |
| PRB947 | 0 | 9 | 9 | 1 × 104 | 5 × 105 | 5 × 105 |
| PRB948 | 20 | 23 | >23 | 3 × 104 | 6 × 105 | NA |
| PRB951 | 8 | 8 | 19 | 2 × 105 | 1 × 105 | 2 × 105 |
| PRB952 | 7 | 10 | 14 | 1 × 103 | 3 × 105 | 1 × 105 |
| 6243 | 18 | 27 | 32 | 1 × 103 | 4 × 105 | 3 × 105 |
| 6244 | 25 | 28 | 33 | 5 × 103 | 3 × 103 | >8 × 105 |
| 6245 | 60 | 66 | 73 | 2 × 103 | 3 × 105 | >8 × 105 |
NT, not tested.
NA, not available.
For most of the seroconversion panels tested, the viral loads of the samples, measured by a quantitative HIV-1 RNA assay (Amplicor Monitor, version 1.0; Roche Diagnostic Systems, Somerville, N.J.), which has a low limit of detection of 400 copies per ml, was provided by the vendors. Table 1 lists the viral load for each sample in the panel on the day that the HIV-1/2 qualitative RNA assay, the antigen detection test, and the antibody detection tests were first positive. All samples with viral loads of 400 copies per ml or above were detected by the HIV-1/2 qualitative RNA assay. In contrast, a viral load of 80,000 copies per ml or above was needed for the detection of the p24 antigen.
To assess the impact of the HIV-1/2 qualitative RNA test on closure of the window period, only a select number of the panels were considered for the calculations. Only panels that had series of samples that were frequently obtained from the same individual (bleeds of less than 1-week intervals) and panels that had samples with positive results by all three tests were included. The inclusion of panels that did not have samples for frequent bleeds may have resulted in an overestimation of the reduction of window period, such as with panel PRB935, or, conversely, to an underestimation, such as with panel PRB932 (Table 1). Panels that did not have bleeds in which virus could be detected by the antigen or antibody detection tests were not included, since without the date of the first positive result, accurate estimation of the window period could not be made. For example, for panel PRB941 the antigen detection test was still negative at day 18, which was the last day that a sample was tested, yet the sample was positive by the antibody detection test at that time. For four of the panels (panels PRB929, PRB942, PRB946, and PRB948) the last samples tested were still negative by the antibody test. The results for the HIV-1/2 qualitative RNA assay, the p24 antigen detection test, and the HIV-1 and HIV-2 antibody detection tests for each sample for the nine selected panels is shown in Table 2. The day since the first bleed at which each test becomes positive is highlighted. Table 3 lists the number of days between the earliest detection by the RNA assay versus the antigen assay, antigen assay versus the antibody assay, and the RNA assay versus the antibody assay for those nine panels. The RNA detection test detected HIV a mean of 6 days (range, 2 to 10 days) earlier than the p24 antigen detection assay and a mean of 11 days (range, 7 to 14 days) earlier than the antibody detection assay.
TABLE 2.
Evaluation of seroconversion panels with frequently obtained samples
| Panel and panel member | No. of days since first bleed | S/CO
|
||
|---|---|---|---|---|
| HIV-1/2 RNA qualitative assay | Monoclonal antigen detection test | HIV-1/2 antibody detection test | ||
| PRB936 | ||||
| 1a | 0 | 0.3 | 0.1 | |
| 2 | 5 | 2.9b | 0.3 | 0.1 |
| 3 | 7 | 5.6 | 0.4 | 0.1 |
| 4 | 12 | 5.8 | 13 | 0.1 |
| 5 | 14 | 5.8 | >31.7 | 0.1 |
| 6 | 19 | 6.2 | >31.7 | 1.5 |
| PRB937 | ||||
| 1 | 0 | 0.2 | 0.3 | 0.1 |
| 2 | 7 | 4.1 | 0.3 | 0.1 |
| 3 | 9 | 5.7 | 0.3 | 0.1 |
| 4 | 14 | 5.6 | 2.1 | 0.1 |
| 5 | 16 | 6.2 | 3.2 | 0.1 |
| 6 | 21 | 5.8 | 13.6 | 1 |
| PRB943 | ||||
| 1 | 0 | 0.1 | 0.4 | 0.1 |
| 2 | 5 | 4.7 | 0.4 | 0.1 |
| 3 | 7 | 4.7 | 0.7 | 0.1 |
| 4 | 12 | 6 | 10.6 | 0.2 |
| 5 | 14 | 6.5 | 25.6 | 2.5 |
| PRB944 | ||||
| 1 | 0 | 4.5 | 0.5 | 0.1 |
| 2 | 2 | 4.3 | 1 | 0.1 |
| 3 | 7 | 5.4 | 6.6 | 0.1 |
| 4 | 9 | 4 | 7 | 0.6 |
| 5 | 14 | 4.9 | 5.8 | 11.7 |
| PRB945 | ||||
| 1 | 0 | 0.2 | 0.4 | 0.1 |
| 2 | 3 | 1.7 | 0.4 | 0.1 |
| 3 | 7 | 4.7 | 0.7 | 0.1 |
| 4 | 13 | 4.9 | 4 | 2.9 |
| PRB952 | ||||
| 1 | 0 | 0.1 | 0.4 | 0.2 |
| 2 | 7 | 2 | 0.5 | 0.2 |
| 3 | 10 | 3.7 | 2.3 | 0.2 |
| 4 | 14 | 3.3 | 1.2 | 1 |
| 6243 | ||||
| 4 | 12 | 0.6 | 0.49 | 0.43 |
| 5 | 18 | 2.2 | 0.49 | 0.43 |
| 6 | 20 | 4.6 | 0.49 | 0.44 |
| 7 | 25 | 5.5 | 0.93 | 0.47 |
| 8 | 27 | 6.1 | 1.04 | 0.53 |
| 9 | 32 | 6.2 | 0.88 | 4.39 |
| 6244 | ||||
| 10 | 22 | 0.5 | 0.44 | 0.37 |
| 11 | 25 | 3.8 | 0.46 | 0.38 |
| 12 | 26 | 4.5 | 0.51 | 0.37 |
| 13 | 28 | 5.2 | 1.33 | 0.41 |
| 14 | 33 | 5.6 | 18.33 | 2.17 |
| 6245 | ||||
| 4 | 55 | 0.2 | 0.47 | 0.36 |
| 5 | 60 | 3.1 | 0.53 | 0.38 |
| 6 | 66 | 5.7 | 1.44 | 0.47 |
| 7 | 69 | 5.6 | 4.91 | 0.41 |
| 8 | 73 | 6.1 | 11.1 | 1.32 |
Panel member 1 was not supplied by the vendor.
Boldface numbers represent the first positive result by each test.
TABLE 3.
Window reduction by the LCx HIV-1/2 qualitative RNA assay
| Panel no. | No. of days between earliest detection times by the following assaysa:
|
||
|---|---|---|---|
| RNA→Ag | Ag→Ab | RNA→Ab | |
| PRB936 | 7 | 7 | 14 |
| PRB937 | 7 | 7 | 14 |
| PRB943 | 7 | 2 | 9 |
| PRB944 | 2 | 12 | 14 |
| PRB945 | 10 | 0 | 10 |
| PRB952 | 3 | 4 | 7 |
| BP6243 | 9 | 5 | 14 |
| BP6244 | 3 | 5 | 8 |
| BP6245 | 6 | 7 | 13 |
| Mean | 6 | 5.4 | 11.4 |
| Range | 2–10 | 0–12 | 7–14 |
RNA, LCx HIV-1/2 qualitative RNA assay; Ag, monoclonal antigen detection test; Ab, HIV-1/2 antibody detection test.
Subtype detection.
Detection of HIV-1 subtypes by the HIV-1/2 qualitative RNA assay was assessed with samples from HIV-positive patients from Uganda, HIV isolates from cultures, and transcript RNAs of HIV-1 isolates of different subtypes. Twenty-one samples were obtained from HIV-positive patients from Uganda, with the samples containing subtype A (n = 8), subtype C (n = 3), and subtype D (n = 10) HIV-1 isolates. Each sample was processed, and 0.25 ml of plasma equivalent of each sample was tested by the HIV-1/2 qualitative RNA assay. They were all positive, with S/COs ranging from 5.6 to 7.7, all well above the cutoff value of 1.0 (Table 4).
TABLE 4.
Testing of samples from Uganda containing HIV-1 subtypes A, C, and D
| Subtype | Sample no. | S/CO |
|---|---|---|
| A | 225.692 | 6.6 |
| 134.447 | 7.7 | |
| 135.544 | 6.4 | |
| 171.004 | 6.8 | |
| 204.987 | 5.6 | |
| 225.706 | 7.2 | |
| 230.568 | 7.3 | |
| 264.643 | 6.8 | |
| C | 225.745 | 6.7 |
| 108.45 | 7.3 | |
| 165.396 | 7.0 | |
| D | 225.796 | 7.2 |
| 205.586 | 6.2 | |
| 127.566 | 6.4 | |
| 130.555 | 7.5 | |
| 134.46 | 7.1 | |
| 171.404 | 6.1 | |
| 182.09 | 7.7 | |
| 191.013 | 7.5 | |
| 234.179 | 7.4 | |
| 401.383 | 7.7 |
To further assess the ability of the HIV-1/2 qualitative RNA assay to detect HIV-1 isolates of different subtypes, 21 HIV isolates from cultures consisting of isolates of subtypes A (n = 4), Thai B (n = 3) D (n = 3), E (n = 4), F (n = 6), and G (n = 1) were quantitated by branched DNA assay (version 2.0; Chiron Corp., Emersville, Calif.) (15). Each sample was diluted in RNase-free water so that it contained 100 and 20 copies per reaction mixture. All isolates except for an F isolate (isolate FBr 112) were positive at 20 copies per reaction mixture. At 100 copies per reaction mixture, all 21 isolates were positive (Table 5). Since commercially available assays for quantitative HIV RNA detection cannot accurately quantitate virus in specimens containing HIV-1 isolates of group O, the two group O-containing specimens were diluted 1,000- and 10,000-fold and were tested. Tests with group O-containing specimens at each dilution were positive.
TABLE 5.
Subtype detection by the LCx HIV-1/2 qualitative RNA assay
| Subtype and isolates | Result with the following no. of copies/reaction mixturea:
|
|
|---|---|---|
| 20 | 100 | |
| A | ||
| 312 | + | + |
| 327 | + | + |
| 419 | + | + |
| 422 | + | + |
| B | ||
| Thai 1273 | + | + |
| Thai 1600 | + | + |
| Thai 50788 | + | + |
| D | ||
| 306 | + | + |
| 308 | + | + |
| 418 | + | + |
| E | ||
| 155 | + | + |
| 577 | + | + |
| 1102 | + | + |
| 50436 | + | + |
| F | ||
| Br41 | + | + |
| Br57 | + | + |
| Br58 | + | + |
| Br59 | + | + |
| Br97 | + | + |
| Br112 | − | + |
| G, 3671 | + | + |
| O | ||
| 11897755A | +b | +c |
| HAM112 | +b | +c |
+, mean S/CO of >1; −, mean S/CO of <1.
Result with a 1:1,000 dilution.
Result with a 1:10,000 dilution.
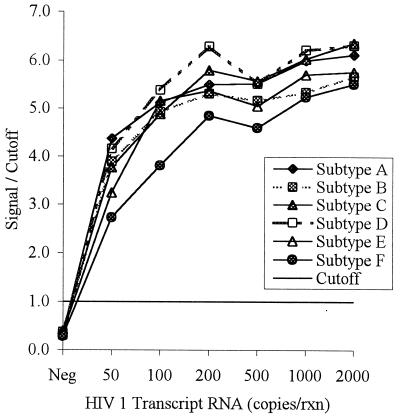
To better assess the analytical sensitivity of the HIV-1/2 qualitative RNA assay for its ability to detect different HIV-1 subtypes, HIV-1 clones of subtypes A through F were generated. The clones were subsequently sequenced to verify their identities and determine the number of mismatches with the HIV-1-specific primers and probe. The number of mismatches for the forward primer ranged from 0 to 2, and the number of mismatches for the reverse primer and the probe ranged from 0 to 1. These clones were transcribed in vitro, and their RNAs were purified and quantitated as described earlier. The RNA was diluted to between 50 and 2,000 copies per reaction mixture and tested (Fig. 2). RNAs from all subtypes were detectable at 50 copies per reaction mixture (equivalent to 170 copies per ml), which was the lowest level tested.
FIG. 2.
Sensitivity of the LCx HIV-1/2 qualitative RNA assay with HIV-1 subtype A to F clones. Cloned fragments containing a region of the pol genes from HIV-1 subtypes A to F were transcribed in vitro. The transcripts were quantitated by hyperchromicity, diluted to the indicated concentrations, and tested by the HIV-1/2 qualitative RNA assay. An S/CO of >1 is considered a positive result. rxn, reaction.
Specificity and inhibition rate.
Seven hundred fifty-one HIV-negative samples were tested by the HIV-1- and HIV-2-specific assay to assess the specificity of the assay. Three samples tested positive initially, resulting in an initial false-positive result frequency of 0.4% and an assay specificity of 99.6%. These samples were retested in duplicate and each was negative upon repeat testing, resulting in a confirmed false-positive result frequency of 0%.
The same seven hundred fifty-one HIV-negative samples were also evaluated for inhibition frequency. The HIV-1/2 qualitative RNA assay contains an internal control that detects amplification inhibition, thus preventing a sample from being called falsely negative due to inhibition. Thirteen samples (1.7%) were initially inhibited. These samples, which were stored frozen for up to 1 week, were retested in duplicate, and none were inhibited, resulting in a repeat inhibition frequency of 0%.
DISCUSSION
The value of amplification-based assays in improving the safety of the blood supply will depend on the extent to which these assays can identify HIV-positive units of blood that escape detection by current serological screening methods due to the window period between infection and the time at which current assays can detect the infection. We demonstrate in this report that the multiplex HIV-1/2 qualitative RNA assay detects HIV RNA before the p24 antigen test and the antibody detection tests do for a majority of samples in the seroconversion panels tested. Analysis of well-characterized panels with frequently obtained samples from the same individual suggests that the PCR assay detects HIV RNA approximately 6 days before the p24 antigen detection test does and 11 days before antibody detection assays do. These results are consistent with previous reports in which the reduction in the preseroconversion window period was estimated with a mathematical model. In that analysis, viral RNA detection preceded p24 antigen detection by 5 days and seroconversion by 11 days (9).
The ability of nucleic acid-based amplification assays to further reduce the window period is due to the high degrees of sensitivity of such assays. Analysis of seroconversion panels in this study indicated that a viral load greater than 80,000 copies per ml was required for p24 antigen detection. In contrast, less than 100 copies of RNA per ml was sufficient for detection of the viral genome by the RNA detection assay. In the multiplex HIV-1/2 qualitative RNA assay, HIV-1 can be detected 100% of the time when it is present at 50 copies per ml and 83% of the time when it is present at 20 copies per ml. While similar sensitivities for HIV-1 RNA detection have been reported for other amplification-based assays, none have incorporated HIV-2 RNA detection in their assays (30, 37).
HIV-2 has been found predominantly in West Africa and to a lesser extent in Portugal and France. Despite its rare occurrence in the United States, serological screening for HIV-2, in addition to serological screening for HIV-1, has been implemented in an effort to increase the safety of blood and blood products (5). Similarly, once the RNA assays are implemented, the requirement to test for both HIV-1 and HIV-2 RNAs could be imminent. Despite its overall similarities to HIV-1, HIV-2 shares only 60% overall nucleotide homology with HIV-1, even in the most conserved genes, gag and pol (20). Therefore, a distinct set of PCR primers was needed to detect the HIV-2 RNA in the HIV-1/2 qualitative RNA assay. Multiplexing in PCR, that is, detection of several markers by the use of multiple, distinct primer sets, has been shown to be a challenging task (7, 36). Multiple primers tend to cause a loss of sensitivity, mainly due to the formation of primers-dimers and extension products of nonspecifically paired primers. These nonspecific reactions compete for and deplete the limiting reagents. In the multiplex HIV-1/2 qualitative RNA assay described here, these difficulties were overcome by the meticulous choice of primers and cycling conditions, without compromising sensitivity for HIV-1 detection. The multiplex assay for HIV-1 and HIV-2 detection has a sensitivity of 20 to 50 copies per ml for HIV-1, which is similar to the sensitivities of ultrasensitive versions of assays that detect only HIV-1 (22, 30, 37). Sensitivity is also critical for detection of HIV-2, especially since lower HIV-2 RNA levels in plasma have been demonstrated (29, 41). The multiplex HIV-1/2 qualitative RNA assay can detect 10 copies of HIV-2 per reaction mixture, which is equivalent to 34 copies per ml.
The sample processing procedure described above uses about 0.3 ml of plasma equivalent. One milliliter of sample is processed and one-third of the processed sample is used for the multiplex HIV-1/2 qualitative RNA assay. The remaining two-thirds can be used for any other hepatitis or retrovirus detection assays, giving the flexibility of running three distinct assays from a single processed sample. Alternatively, all of the processed sample can be used for the HIV-1/2 qualitative RNA assay to further increase the sensitivity for detection of HIV-1 and HIV-2 by eluting the entire sample from the column in a smaller volume.
The ability of an HIV-1-specific PCR assay to detect diverse subtypes is a critical requirement. Until recently, data on subtype distribution indicated that in Europe and the United States, infections primarily occurred with subtype B. The subtype pattern is changing quite drastically in Europe. Reports from Germany, England, and France on the prevalence of non-subtype B infections indicate that it ranges from 10 to 30% (18, 40). In some selected populations, such as French soldiers deployed overseas, the prevalence of non-subtype B infections is as high as 63% (26). Worldwide, subtype B accounts for only 16% of infections, while subtype C accounts for 36%, subtype A accounts for 23%, subtype D accounts for 13%, and subtype E accounts for 7%. Group O, which was originally found in Cameroon and Gabon (13, 27), has now been identified in the United States and Europe as well (34).
All the nucleic acid detection assays developed so far have been based primarily on subtype B strains from North America and Europe. The sensitivities of these tests for detection of divergent strains were evaluated in several studies (3, 14, 22, 35, 44). Evaluation of the HIV-Monitor, version 1.0, and the nucleic acid sequence-based amplification (NASBA) assays revealed that 56 and 44% of subtype A-containing samples tested were negative by these assays, respectively (3). This poor sensitivity was likely due to the number of mismatches in the selected primers. It was reported that a decrease in amplification efficiency occurs with five to six mutations in the primer-binding region (10, 24), and this amount of divergence occurs most frequently with subtypes A and E. Similar results were reported when samples from Uganda, where subtypes A and D predominate, were tested with the Amplicor HIV-1 DNA test; the sensitivity for the samples from Uganda was 74% (22). Replacement of the primer-probe set selected from the gag gene with a primer-probe set from the better conserved pol gene increased the sensitivity of the DNA detection assay to 98% (22). Similarly, Respess et al. (35) reported 100% sensitivity for detection of subtypes A and E with pol-based primers but only 87 and 67% sensitivities, respectively, when gag-based primers were used. Moreover, with gag-based primers, none of the five group O isolates were detected, while with the pol-based primers all five were detected (35). It has recently been reported that detection of subtypes A and E was significantly improved with the Monitor, version 1.5, assay, which uses primers different from the primers used for version 1.0 of the assay (44). Group O-containing specimens were still not detectable with the Monitor, version 1.5, assay. The branched DNA assay, which uses 49 specific probes, has been shown to be more robust to the diversity of different subtypes of the M group (14).
For the multiplex HIV-1/2 qualitative RNA assay described in this report, the HIV-1-specific primers and probe were chosen from the highly conserved pol region of the genome after an analysis of 29 available pol sequences from the database which included the pol sequences of group M subtypes A, B, and D and group O isolates (23). The probe and the reverse primer had no mismatches or a single base mismatch with the sequences of all subtypes including group O. The forward primer had no mismatches or a single mismatch with subtype A, B, and D sequences and four mismatches with group O sequences. Our study with samples from Uganda demonstrates that the HIV-1/2 qualitative RNA assay detects all (100%) isolates of subtypes A, C, and D. Moreover, to ensure that even very low levels of diverse subtypes could be detected, 21 HIV isolates consisting of subtypes A, Thai B, D, E, F, and G from cultures were diluted to 100 copies per reaction mixture; they were detected 100% of the time. Also two group O isolates diluted 1,000- and 10,000-fold were detected by the multiplex HIV-1/2 qualitative RNA assay. To better assess the analytical sensitivity of the HIV-1/2 qualitative RNA assay for the detection of multiple subtypes, one isolate each of subtypes A, B, C, D, E, and F was cloned to generate transcripts. These transcripts were quantitated by an independent and accurate method (12), and all subtypes were positive when they were present at 50 copies per reaction mixture, which is equivalent to 170 copies per ml.
Several levels of specificity are built into the HIV-1/2 qualitative RNA assay: specific primers for amplification of the target, a specific probe to which the amplified target can hybridize, and specific immunoassay reagents for detection of the amplified target-probe complex. In nucleic acid-based tests, false-positive results that affect “assay specificity” are usually caused by contamination with low levels of target sequences combined with the high degrees of sensitivity of the tests and do not necessarily reflect true assay specificity. The system described in this report includes several measures that control for contamination, such as the use of a single reaction vessel, which does not require opening after amplification, and the automatic chemical destruction of the amplified product after detection. These measures were previously shown to be extremely effective (1, 21). Of the 751 HIV-negative samples tested by the HIV-1/2 qualitative RNA assay, three samples tested positive initially, resulting in an initial frequency of false positivity of 0.4%. Upon retesting in duplicate, all three were negative, resulting in a confirmed frequency of false positivity of 0%.
The internal control included in the HIV-1/2 qualitative RNA assay is critical to achieving a high degree of confidence in negative values. The internal control, which is added prior to sample processing, controls for sample recovery, impurities in extracted samples that may inhibit the amplification reaction, and technical errors during the testing process. A negative result for the internal control indicates that the test result may be falsely negative, and such a sample should be retested. A positive result for the internal control validates the test result and ensures that a negative result is truly negative (38). Of the 751 samples tested by the HIV-1/2 qualitative RNA assay, 13 samples were initially inhibited (1.7%); that is, the result for the internal control was negative. Without inclusion of an internal control, these samples would have been called negative and would not have been identified as inhibited.
In conclusion, we have described a qualitative multiplex assay for the detection of HIV-1 and HIV-2 RNAs in plasma. The LCx HIV-1/2 qualitative RNA assay detects HIV RNA in samples prior to p24 antigen detection and before seroconversion and antibody detection. Furthermore, HIV-1 subtypes A, B, C, D, E, and F and group O were detected with high degrees of sensitivity. This assay demonstrated excellent specificity due to effective contamination control measures and the use of highly selective primers and probes. Moreover, the inclusion in each reaction mixture of an internal control that controls for sample inhibition ensures that the negative specimens are truly negative.
REFERENCES
- 1.Abravaya K, Hu H, Khalil O. Strategies to avoid amplicon contamination. In: Lee H H, Morse S, Olsvik O, editors. Nucleic acid amplification technologies. Cambridge, Mass: Biotechniques Books, Eaton Publishing; 1997. pp. 125–133. [Google Scholar]
- 2.Abravaya K, Cao J, Flanders R, Macioszeck J, Muldoon S, Robinson J. Research investigations into the suitability of the LCx platform for detection of bloodborne viruses. Infusion Ther Transfusion Med. 1998;25:170–177. [Google Scholar]
- 3.Alaeus A, Lidman K, Sonnerborg A, Albert J. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS. 1997;11:859–865. doi: 10.1097/00002030-199707000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Allain J, Laurian Y, Paul D A, Senn D Members of the AIDS-Haemophilia French Study Group. Serological markers in early stages of human immunodeficiency virus infection in haemophiliacs. Lancet. 1986;ii:1233–1236. doi: 10.1016/s0140-6736(86)92673-5. [DOI] [PubMed] [Google Scholar]
- 5.Ayres L, Avillez F, Garcia-Benito A, Deinhardt F, Gurtler L, Denis F, Leonard G, Ranger S, Grob P, Joller-Jemelka H, Hess G, Seidl S, Flacke H, Simon F, Brun-Vezinet F, Sondag D, Andre A, Hampl H, Schoen R, Stramer S, Troonen H. Multicenter evaluation of a new recombinant enzyme immunoassay for the combined detection of antibody to HIV-1 and HIV-2. AIDS. 1990;4:131–138. doi: 10.1097/00002030-199002000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Barre-Sinoussi F, Chermann J C, Rey F. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immunodeficiency syndrome. Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 7.Bej A K, Mahbubani M H, Miller R, DiCesare J L, Haff L, Atlas R M. Multiplex PCR amplification and immobilized capture proves for detection of bacterial pathogens and indicators in water. Mol Cell Probes. 1990;4:353–365. doi: 10.1016/0890-8508(90)90026-v. [DOI] [PubMed] [Google Scholar]
- 8.Busch M, Stramer S. The efficiency of HIV p24 antigen screening of US blood donors: projections versus reality. Infusion Ther Transfusion Med. 1998;25:194–197. [Google Scholar]
- 9.Busch M P, Lee L L L, Satten G A, Henrard D R, Farzadegan H, Nelson K E, Read S, Dodd R Y, Petersen L R. Time course of detection of viral and serologic markers preceding human immunodeficiency virus type 1 seroconversion: implications for screening of blood and tissue donors. Transfusion. 1995;35:91–97. doi: 10.1046/j.1537-2995.1995.35295125745.x. [DOI] [PubMed] [Google Scholar]
- 10.Christopherson C, Sninsky J, Kwok S. The effects of internal primer-template mismatches on RT-PCR: HIV-1 model studies. Nucleic Acids Res. 1997;25:654–658. doi: 10.1093/nar/25.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clavel F, Guetard D, Brun-Vezinet F, Chamaret S, Rey M, Santos-Ferreira M O, Laurent A G, Dauguet C, Katlama C, Rouzioux C, Klatzmann D, Champalimaud J L, Montagnier L. Isolation of a new human retrovirus from West African patients with AIDS. Science. 1986;233:343–346. doi: 10.1126/science.2425430. [DOI] [PubMed] [Google Scholar]
- 12.Collins M, Zayati C, Detmer J J, Daly B, Kolberg J A, Cha T, Irvine B D, Tucker J, Urdea M S. Preparation and characterization of RNA standards for use in quantitative branched DNA hybridization assays. Anal Biochem. 1995;226:120–129. doi: 10.1006/abio.1995.1199. [DOI] [PubMed] [Google Scholar]
- 13.DeLeys R B, Vanderborght B, Vanden Haesevelde M, Heyndrickx L, van Geel A, Wauters C, Bernaerts R, Saman E, Nijs P, Willems B, Taelman H, van der Groen G, Plot P, Tersmette T, Huisman J G, Van Heuverswyn H. Isolation and partial characterization of an unusual human immunodeficiency retrovirus from two persons of West-Central African origin. J Virol. 1990;64:1207–1216. doi: 10.1128/jvi.64.3.1207-1216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunne A L, Crowe S M. Comparison of branched DNA and reverse transcriptase polymerase chain reaction for quantifying six different HIV-1 subtypes in plasma. AIDS. 1997;11:126–127. [PubMed] [Google Scholar]
- 15.Fiore M D, Mitchell J E, Doan T, Winter G, Grandone C, Zeng K, Haraden R, Smith J, Harria K, Leszczynski J, Berry D, Sanord S, Barnes G, Scholnick A, Ludington K. The Abbott IMx automated benchtop immunochemistry analyzer system. Clin Chem. 1988;34:1726–1732. [PubMed] [Google Scholar]
- 16.Gallarda J L, Henrard D R, Liu D, Harrington S, Stramer S L, Valinsky J, Wu P. Early detection of antibody to human immunodeficiency virus type 1 by using an antigen conjugate immunoassay correlates with the presence of immunoglobulin M antibody. J Clin Microbiol. 1992;30:2379–2384. doi: 10.1128/jcm.30.9.2379-2384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallo R C, Salahuddin S Z, Popovic M, Shearer G M, Kaplan M, Haynes B F, Palker T J, Redfield R, Oleske J, Safaj B, White G, Foster P, Markham P D. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–502. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 18.Gurtler L G. The impact of HIV subtypes and variants on the stability of HIV screening assays. Infusion Ther Transfusion Med. 1998;25:198–199. [Google Scholar]
- 19.Hewlett I K, Epstein J S. Food and Drug Administration conference on the feasibility of genetic technology to close the HIV window in donor screening. Transfusion. 1997;37:346–351. doi: 10.1046/j.1537-2995.1997.37397240219.x. [DOI] [PubMed] [Google Scholar]
- 20.Hu D J, Dondero T J, Rayfield M A, George J R, Schochetman G, Jaffe H W, Luo C, Kalish M L, Weniger B G, Pau C, Schable C A, Curran J W. The emerging genetic diversity of HIV. JAMA. 1996;275:210–216. [PubMed] [Google Scholar]
- 21.Hu H Y, Burczak J D, Leckie G W, Ray K A, Muldoon S, Lee H H. Analytic performance and contamination control methods of a ligase chain reaction DNA amplification assay for detection of Chlamydia trachomatis in urogenital specimens. Diagn Microbiol Infect Dis. 1996;24:71–76. doi: 10.1016/0732-8893(95)00272-3. [DOI] [PubMed] [Google Scholar]
- 22.Jackson J B, Piwowar E M, Parsons J, Kataaha P, Bihibwa G, Onecan J, Kabengera S, Kennedy S D, Butcher A. Detection of human immunodeficiency virus type 1 (HIV-1) DNA and RNA sequences in HIV-1 antibody-positive blood donors in Uganda by the Roche AMPLICOR assay. J Clin Microbiol. 1997;35:873–876. doi: 10.1128/jcm.35.4.873-876.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroeger, P. E., K. Abravaya, C. A. Esping, J. J. Gorzowski, R. J. Hoenle, and J. J. Moore. October 1999. Nucleic acid primers and probes for detecting HIV-1 and HIV-2. U.S. patent 5,962,665.
- 24.Kwok S, Kellogg D E, McKinney N, Spasic D, Goda L, Levenson C, Sninsky J J. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 1990;18:999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lackritz E M, Satten G A, Aberle-Grasse J, Dodd R Y, Raimondi V P, Janssen R S, Lewis W F, Notari IV E P, Petersen L R. Estimated risk of transmission of the human immunodeficiency virus by screened blood in the United States. N Engl J Med. 1995;333:1721–1725. doi: 10.1056/NEJM199512283332601. [DOI] [PubMed] [Google Scholar]
- 26.Lasky M, Perret J, Peeters M, Bibollet-Ruche F, Liegeois F, Patrel D, Molinier S, Gras C, Delaporte E. Presence of multiple non-B subtypes and divergent subtype B strains of HIV-1 in individuals infected after overseas deployment. AIDS. 1997;11:43–51. doi: 10.1097/00002030-199701000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Loussert-Ajaka I, Chaix M L, Korber B, Letourneur F, Gomas E, Allen E, Ly T D, Brun-Vezinet F, Simon F, Saragosti S. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J Virol. 1995;69:5640–5649. doi: 10.1128/jvi.69.9.5640-5649.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loussert-Ajaka I, Descamps D, Simon F, Brun-Vezinet F, Ekwalanga M, Saragosti S. Genetic diversity and HIV detection by polymerase chain reaction. Lancet. 1995;346:912–913. doi: 10.1016/s0140-6736(95)92762-x. [DOI] [PubMed] [Google Scholar]
- 29.Loussert-Ajaka I, Simon F, Farfara I, Descamps D, Collin G, Brun-Vezinet F. Detection of circulating human immunodeficiency virus type 2 in plasma by reverse transcription polymerase chain reaction. Res Virol. 1995;146:409–414. doi: 10.1016/0923-2516(96)80900-9. [DOI] [PubMed] [Google Scholar]
- 30.McDonough S H, Giachetti C, Yang Y, Kolk D P, Billyard E, Mimms L. High throughput assay for the simultaneous or separate detection of human immunodeficiency virus (HIV) and hepatitis type C virus (HCV) Infusion Ther Transfusion Med. 1998;25:164–169. [Google Scholar]
- 31.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J Clin Microbiol. 1994;32:292–300. doi: 10.1128/jcm.32.2.292-300.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen L R, Satten G A, Dodd R, Busch M, Kleinman S, Grindon A, Lenes B the HIV Seroconversion Study Group. Duration of time from onset of human immunodeficiency virus type 1 infectiousness to development of detectable antibody. Transfusion. 1994;34:283–289. doi: 10.1046/j.1537-2995.1994.34494233574.x. [DOI] [PubMed] [Google Scholar]
- 33.Popovic M, Sarngadharan M G, Read E, Gallo R C. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- 34.Rayfield M A, Sullivan P, Banden C L, Britvan L, Otten R A, Pau C P, Pieniazek D, Subbarao S, Simon P, Schable C A, Wright A C, Ward J, Schochetman G. HIV-1 group O virus identified for the first time in the United States. Emerg Infect Dis. 1996;2:209–212. doi: 10.3201/eid0203.960307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Respess R A, Butcher A, Wang H, Chaowanachan T, Young N, Shaffer N, Mastro T D, Biryahwaho B, Downing R, Tanuri A, Schechter M, Pascu R, Zekeng L, Kaptue L, Gurtler L, Eberle J, Ellenberger D, Fridlund C, Rayfield M, Kwok S. Detection of genetically diverse human immunodeficiency virus type 1 group M and O isolates by PCR. J Clin Microbiol. 1997;35:1284–1286. doi: 10.1128/jcm.35.5.1284-1286.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez E R, Nasim S, Hsia J, Sandin R L, Ferreira A, Hilliard B A, Ross A M, Garrett C T. Cardiac myocytes and dendritic cells harbor human immunodeficiency virus in infected patients with and without cardiac dysfunction: detection by multiplex, nested polymerase chain reaction in individually microdissected cells from right ventricular endomyocardial biopsy tissue. Am J Cardiol. 1991;68:1511–1520. doi: 10.1016/0002-9149(91)90288-v. [DOI] [PubMed] [Google Scholar]
- 37.Rosenstraus M, Gutekunst K, Herman S, Soviero S, Sun R, Yang J Q, Spadoror J. Improved COBAS AMPLICOR™ viral assays as a basis for minipools screening of viruses in blood or plasma. Infusion Ther Transfusion Med. 1998;25:153–159. [Google Scholar]
- 38.Rosenstraus M, Wang Z, Chang S, DeBonville D, Spadoro J P. An internal control for routine diagnostic PCR: design, properties, and effect on clinical performance. J Clin Microbiol. 1998;36:191–197. doi: 10.1128/jcm.36.1.191-197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreiber G B, Busch M P, Kleinman S H, Korelitz J J. The risk of transfusion-transmitted viral infections. N Engl J Med. 1996;334:1685–1690. doi: 10.1056/NEJM199606273342601. [DOI] [PubMed] [Google Scholar]
- 40.Simon F, Loussert-Ajaka I, Damond F, Saragosti S, Barin F, Brun-Vezinet F. HIV type 1 diversity in northern Paris, France. AIDS Res Hum Retrovir. 1996;12:1427–1431. doi: 10.1089/aid.1996.12.1427. [DOI] [PubMed] [Google Scholar]
- 41.Simon F, Matheron S, Tamalet C, Loussert-Ajaka I, Bartczak S, Pepin J M, Dhiver C, Gamba E, Elbim C, Gastaut J A, Saimot A G, Brun-Vezinet F. Cellular and plasma viral load in patients infected with HIV-2. AIDS. 1993;7:1411–1417. doi: 10.1097/00002030-199311000-00002. [DOI] [PubMed] [Google Scholar]
- 42.Simonds R J, Holmberg S D, Hurwitz R L, Coleman T R, Bottenfield S, Conley L J, Kohlenberg S H, Castro K G, Dahan B A, Schable C A, Rayfield M A, Rogers M F. Transmission of human immunodeficiency virus type 1 from a seronegative organ and tissue donor. N Engl J Med. 1992;326:726–732. doi: 10.1056/NEJM199203123261102. [DOI] [PubMed] [Google Scholar]
- 43.Stramer S L, Heller J S, Coombs R W, Parry J V, Ho D D, Allain J. Markers of HIV infection prior to IgG antibody seropositivity. JAMA. 1989;262:64–69. [PubMed] [Google Scholar]
- 44.Triques K, Coste J, Perret J L, Segarra C, Mpoudi E, Reynes J, Delaporte E, Butcher A, Dreyer K, Herman S, Spadoro J, Peeters M. Efficiencies of four versions of the AMPLICOR HIV-1 MONITOR test for quantification of different subtypes of human immunodeficiency virus type 1. J Clin Microbiol. 1999;37:110–116. doi: 10.1128/jcm.37.1.110-116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward J W, Holmberg S D, Allen J R, Cohn D L, Critchley S E, Kleinman S H, Lenes B A, Ravenholt O, Davis J R, Quinn M G, Jaffe H W. Transmission of human immunodeficiency virus (HIV) by blood transfusions screened as negative for HIV antibody. N Engl J Med. 1988;318:473–477. doi: 10.1056/NEJM198802253180803. [DOI] [PubMed] [Google Scholar]
- 46.Zaaijer H L, Exel-Oehlers P V, Kraaijeveld T, Altena E, Lelie P N. Early detection of antibodies to HIV-1 by third-generation assays. Lancet. 1992;340:770–772. doi: 10.1016/0140-6736(92)92303-w. [DOI] [PubMed] [Google Scholar]