Abstract
The yeast Saccharomyces cerevisiae has four genes, MCK1, MDS1 (RIM11), MRK1, and YOL128c, that encode glycogen synthase kinase 3 (GSK-3) homologs. The gsk-3 null mutant, in which these four genes are disrupted, shows temperature sensitivity, which is suppressed by the expression of mammalian GSK-3β and by an osmotic stabilizer. Suppression of temperature sensitivity by an osmotic stabilizer is also observed in the bul1 bul2 double null mutant, and the temperature sensitivity of the bul1 bul2 double null mutant is suppressed by multiple copies of MCK1. We have screened rog mutants (revertants of gsk-3) which suppress the temperature sensitivity of the mck1 mds1 double null mutant and found that two of them, rog1 and rog2, also suppress the temperature sensitivity of the bul1 bul2 double null mutant. Bul1 and Bul2 have been reported to bind to Rsp5, a hect (for homologous to E6-associated-protein carboxyl terminus)-type ubiquitin ligase, but involvement of Bul1 and Bul2 in protein degradation has not been demonstrated. We find that Rog1, but not Rog2, is stabilized in the gsk-3 null and the bul1 bul2 double null mutants. Rog1 binds directly to Rsp5, and their interaction is dependent on GSK-3. Furthermore, Rog1 is stabilized in the npi1 mutant, in which RSP5 expression levels are reduced. These results suggest that yeast GSK-3 regulates the stability of Rog1 in cooperation with Bul1, Bul2, and Rsp5.
Glycogen synthase kinase 3 (GSK-3) was originally characterized as a serine/threonine kinase that phosphorylates and inactivates glycogen synthase and subsequently has been demonstrated to be identical to protein kinase FA, which activates ATP-Mg-dependent type 1 protein phosphatase (29). GSK-3 is now implicated in the regulation of several physiological responses in mammalian cells by phosphorylating many substrates, including neuronal cell adhesion molecules, neurofilaments, synapsin I, tau, transcription factors, adenomatous polyposis coli gene products, β-catenin, and cyclin D1 (10, 18, 25, 29, 34, 49). The cDNAs of GSK-3α and GSK-3β in mammals have been isolated, and they encode protein kinases with molecular masses of 51 and 47 kDa, respectively (48). GSK-3 is highly conserved through evolution and plays a fundamental role in cellular responses. For example, Xenopus GSK-3 regulates axis formation during early development (15, 53). The Drosophila zeste-white3/shaggy gene product is structurally and functionally homologous to GSK-3β (35) and is required at several developmental stages during fruit fly embryogenesis for correct embryogenic segmentation (27, 39). The Dictyostelium homolog of GSK-3 has been found to be important for cellular differentiation (14). In Schizosaccharomyces pombe, the skp1+ gene product is a homolog of GSK-3 and regulates cytokinesis (28).
In Saccharomyces cerevisiae, there are four genes, MCK1, MDS1 (RIM11), MRK1, and YOL128c, which encode homologs of mammalian GSK-3. MCK1 acts in the transcription of IME1 at the beginning of meiosis and also in the chromosomal segregation processes at mitosis (26, 38). Mds1 (Rim11) is suggested to induce expression of meiotic genes by promoting complex formation between Ime1 and Ume6 transcriptional factors (4, 33). Thus, S. cerevisiae GSK-3s appear to play important roles in both meiosis and mitosis, although their molecular mechanisms are not elucidated in detail. Further, it is possible that they have additional functions, since mammalian GSK-3 has multiple substrates and functions (29).
Recently it has been clarified that mammalian GSK-3 triggers ubiquitination and subsequent degradation of proteins, such as β-catenin and cyclin D1 (1, 10). In general, degradation of proteins by the ubiquitin-proteasome pathway involves a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) (7). An E3 enzyme is the component of the ubiquitin-conjugating system that is generally thought to be the most directly involved in substrate recognition. It has been shown that the F-box protein Cdc4 forms a complex with Skp1 and Cdc53 in budding yeast and that this multiprotein complex (SCF complex) functions as an E3 ubiquitin ligase, which catalyzes the ubiquitination of the cyclin-dependent kinase inhibitor Sic1 in combination with the ubiquitin-conjugating enzyme Ubc3 (Cdc34) (11, 40). The ubiquitination of Sic1 requires its phosphorylation by Cdc28 (11). βTrCP (FWD1), a mammalian F-box protein, associates with β-catenin and stimulates its ubiquitination and degradation (22). The phosphorylation of β-catenin by GSK-3β is necessary for its ubiquitination (1). The phosphorylation of cyclin D1 by GSK-3β is also required for its ubiquitination (10), but which type of E3 is involved is not known.
The hect (for homologous to the E6-AP [E6-associated protein] carboxyl terminus) domain defines another family of E3, including E6-AP and Nedd4 in mammals (7). E6-AP is required along with the human papillomavirus E6 oncoproteins for the ubiquitination and degradation of p53 (17). Nedd4 targets the kidney epithelial sodium channel (36, 42). In contrast to the functional interaction between protein kinases and the SCF-type E3 enzymes, it has not been shown whether the phosphorylation of targets is necessary for the degradation by the hect-type E3 enzyme. S. cerevisiae RSP5 encodes a hect-type E3. Rsp5 has been shown to be involved in the downregulation of uracil permease (Fur4), general amino acid permease (Gap1), maltose permease (Mal61), plasma membrane H+-ATPase, and the large subunit of RNA polymerase II (Rpb1) (2, 9, 16, 24). On the other hand, Bul1 and Bul2 have been shown to form a complex with Rsp5 (50, 51). Since the bul1 bul2 double null mutant shows phenotypes similar to those of the rsp5-101 recessive mutant, Bul1 and Bul2 are thought to facilitate Rsp5 function. However, whether Bul1 and Bul2 affect protein degradation and why the bul1 bul2 mutant and the rsp5-101 mutant show common phenotypes have not been revealed. To clarify the relationship between yeast GSK-3, Bul1 and Bul2, and Rsp5, we have screened mutants which suppress the temperature sensitivity of the mck1 mds1 double null mutant. Here we report a protein, named Rog1 (derived from a revertant of gsk-3), the disruption of which also suppresses the temperature sensitivity of the bul1 bul2 double null and the rsp5-101 mutants. Further, Rog1 is stabilized in the gsk-3, the bul1 bul2, and the rsp5 mutants, and it binds directly to Rsp5. These results suggest that the stability of Rog1 is regulated by GSK-3, Bul1 and Bul2, and Rsp5.
MATERIALS AND METHODS
Materials and chemicals.
Yeast strains YAT2-1C and YHY009K (50) were kindly given by H. Yashiroda (The Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). 24346c and 27038a (41) were generously provided by B. André (Université Libre de Bruxelles-Campus, Brussels, Belgium). Single-copy plasmid vectors pRS314-GAL-myc-BS and pRS316-GAL-HA-BS were from K. Tanaka (Hokkaido University, Sapporo, Japan). pHY06, pHY20 (51), and pHY22 (50) were from Y. Kikuchi (University of Tokyo, Tokyo, Japan). pAT525 was generously given by A. Toh-e (University of Tokyo). Glutathione S-transferase (GST) fusion proteins and maltose-binding protein (MBP) fusion proteins were purified from Escherichia coli according to the manufacturer's instructions. 32Pi, [35S]methionine, and [35S]cysteine were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). Other materials and chemicals were obtained from commercial sources.
Strains and genetic manipulations.
S. cerevisiae strains used in this study are listed in Table 1. KA31 (19), W303 (43), and 24346c (41) were used as wild-type strains. Strains KA31a and KA31α were derived from KA31. Strains YTA002K and YTA003W were isolated by disruption of genes as described below. Strains YTA004K and YTA011K were isolated as revertants of YTA002K as described below. Strains YTA005K and YTA006K were isolated from a backcross of strain YTA004K with strain KA31α. Strain YTA101K was created by crossing strain YHY009 with strain YTA006K, sporulating the resulting diploid, and isolating a Leu+ Trp+ haploid cell from the diploid cell, spores of which were divided 2:2 for auxotrophy of leucine. Strain YTA102K was created by crossing strain YAT2-1C with strain YTA005K, sporulating the resulting diploid, and isolating a Leu+ segregant which possessed temperature sensitivity suppressed by expression of RSP5 with the pHY22 plasmid (50). W303a was derived from W303. Media and methods for mating, sporulation, tetrad analysis, and transformation were described previously (37). YPGlycerol and YPEthanol were prepared by substitution of 2% glycerol and 2% ethanol, respectively, for 2% glucose in YPD medium (37). YPAcetate was prepared as described previously (31).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Background or reference |
|---|---|---|
| KA31 | MATa/MATα his3/his3 leu2/leu2 ura3/ura3 trp1/trp1 | 19 |
| KA31a | MATa his3 leu2 ura3 trp1 | KA31 (this study) |
| KA31α | MATα his3 leu2 ura3 trp1 | KA31 (this study) |
| YTA002K | MATa his3 leu2 ura3 trp1 mck1::TRP1 mds1::HIS3 | KA31 (this study) |
| YHY009K | MATa his3 leu2 ura3 trp1 bul1::TRP1 bul2::ura3::LEU2 | KA31 (50) |
| YTA004K | MATa his3 leu2 ura3 trp1 mck1::TRP1 mds1::HIS3 rog1::LEU2 | KA31 (this study) |
| YTA005K | MATa his3 leu2 ura3 trp1 rog1::LEU2 | KA31 (this study) |
| YTA006K | MATα his3 leu2 ura3 trp1 rog1::LEU2 | KA31 (this study) |
| YTA101K | MATa his3 leu2 ura3 trp1 bul1::TRP1 bul2::ura3::LEU2 rog1::LEU2 | KA31 (this study) |
| YTA011K | MATa his3 leu2 ura3 trp1 mck1::TRP1 mds1::HIS3 rog2::LEU2 | KA31 (this study) |
| YAT2-1C | MATα his3 leu2 ura3 trp1 rsp5-101 | KA31 (51) |
| YTA102K | MATα his3 leu2 ura3 trp1 rsp5-101 rog1::LEU2 | KA31 (this study) |
| W303 | MATa/MATα his3/his3 leu2/leu2 ura3/ura3 trp1/trp1 | 43 |
| W303a | MATa his3 leu2 ura3 trp1 | W303 (this study) |
| YTA003W | MATα his3 leu2 ura3 trp1 mck1::TRP1 mds1::HIS3 mrk1 yol128c::LEU2 | W303 (this study) |
| 24346c | MATa ura3 | 41 |
| 27038a | MATa ura3 npi1 | 41 |
Plasmid constructions.
All fragments amplified by PCR were produced by using genomic DNA of strain W303a cells as a template, and correct sequences were confirmed by sequence analysis. pTA021 (YCp50-MCK1) was constructed by inserting the PCR fragment of nucleotides −809 to +1618 of MCK1, including the region around the MCK1 open reading frame (ORF), into the blunt-ended BamHI site of YCp50 (23). pTA022 (YEp24-MCK1) was created by ligation of the blunt-ended 3.0-kb HindIII-SalI fragment of pTA021 including MCK1 to the blunt-ended BamHI site of YEp24 (3). pTA023 (pYES2-GSK-3β) was constructed by inserting the 1.4-kb BclI-EcoRV fragment containing the human GSK-3β gene from the pBSSK/GSK-3β plasmid (18) into the BamHI and the blunt-ended XhoI sites of pYES2. pTA023 was cut by SphI, blunt ended by Klenow fragment, and then cut by KpnI, and the 1.4-kb fragment containing GSK-3β was inserted between the KpnI and PvuII sites of pKT10 (44), yielding pTA024 (pKT10-GSK3β). pTA025 (pRS314-GAL-myc-Rog1) and pTA026 (pRS316-GAL-HA-Rog1) were constructed by inserting the BglII-SmaI PCR fragment of the ROG1 ORF (from the first ATG to the stop codon), produced with the primers 5′-TTGAGATCTATGTCTCTGACACCAAC-3′ and 5′-AAACCCGGGTCATTGTACCAAATCAC-3′, into the BamHI and SmaI sites of pRS314-GAL-myc-BS and pRS316-GAL-HA-BS, respectively. pTA025 was cut with EcoRI and SmaI and inserted between the EcoRI and SmaI sites of pRS316-GAL-HA-BS, yielding pTA027 (pRS316-GAL-myc-Rog1). pTA028 (pKT10-myc-Rog1) was created by inserting the 2.1-kb EcoRI-SmaI fragment of pTA025 into the EcoRI and PvuII sites of pKT10. pTA009 (pRS314-GAL-myc-Rog2) was constructed by inserting the BamHI-SmaI PCR fragment of the ROG2 ORF, produced with the primers 5′-GTGGGATCCATGGGCCGTGATATATG-3′ and 5′-TTTCCCGGGATCTGTTAAGAAATGTG-3′, into the BamHI and SmaI sites of pRS314-GAL-myc-BS. pTA029 was cut with EcoRI and SmaI, and the 0.6-kb EcoRI-SmaI fragment was inserted between the EcoRI and PvuII sites of pKT10, yielding pTA030 (pKT10-myc-Rog2).
For expression of fusion proteins in E. coli, pHY22 was cut with PstI, blunt ended, and cut with HindIII, and the 3.8-kb fragment including nucleotide +37 to the region of RSP5 corresponding to the C terminus was inserted into the blunt-ended SacI and the HindIII sites of pMAL-c2 (18), yielding pTA041. The PCR fragment of the ROG1 ORF was inserted between the XbaI and BamHI sites of pGEX-KG (18), yielding pTA042.
To create pTA003 (mck1::TRP1), the 0.9-kb EcoRI-BamHI fragment of pJJ246 (20) including TRP1 was blunt ended and inserted between the EcoRV and the blunt-ended BamHI sites of pTA002, in which the XbaI-BamHI PCR fragment containing nucleotides −809 to −1 of MCK1 and the BamHI-SalI PCR fragment containing nucleotides +1 to +1618 of MCK1 were inserted into the XbaI and SalI sites of pTA001, which is created by self-ligation of the BamHI-cut pBluescript KS(+) (Toyobo Co., Osaka, Japan) followed by Klenow fragment treatment. To create pTA005 (mds1::HIS3), the 1.8-kb BamHI fragment of pJJ215 (20) including HIS3 was blunt ended and inserted between the blunt-ended BamHI and the NruI sites of pTA004, in which the PstI-BamHI PCR fragment containing nucleotides −1000 to −1 of MDS1 and the BamHI-XhoI PCR fragment containing nucleotides +1 to +1657 of MDS1 were inserted into the PstI and XhoI sites of pTA001. To create pTA007 (yol128c::LEU2), the blunt-ended 2.0-kb BamHI-SalI fragment of pJJ250 (20) including LEU2 was blunt ended and inserted between the blunt-ended BamHI and the EcoRI sites of pTA006, in which the PstI-BamHI PCR fragment containing nucleotides −1019 to −1 of YOL128c and the BamHI-SalI PCR fragment containing nucleotides +1 to +1424 of YOL128c were inserted into the PstI and SalI sites of pTA001. pTA009 (mrk1::URA3) was created as follows. The 1.6-kb NruI-SmaI fragment of YCp50 including URA3 was inserted into the blunt-ended XhoI site of pAT525 (45), which possessed tandem repeats on both sides of the XhoI site, yielding TAp700. The 2.4-kb blunt-ended HindIII fragment of TAp700 including URA3 between the tandem repeats was inserted into the blunt-ended BglII site of pTA008, in which the PstI-BglII PCR fragment containing nucleotides −1000 to −1 of MRK1 and the BglII-XhoI PCR fragment containing nucleotides +1 to +2278 of MRK1, including the intron, were inserted into the PstI and XhoI sites of pTA001, yielding pTA009.
Disruption of GSK-3 genes.
Cells of strains KA31 and W303 were transformed with pTA003 cut with SalI, and the disruptions of the MCK1 gene of Trp+ transformants were confirmed by Southern blotting analysis (51). Then the MCK1/mck1::TRP1 diploids were transformed with pTA005 cut with SmaI, and the disruptions of the MDS1 gene of His+ transformants were confirmed by Southern blotting analysis. The MCK1/mck1::TRP1 MDS1/mds1::HIS3 diploid cells derived from KA31 were sporulated, and an mck1::TRP1 mds1::HIS3 haploid segregant was named YTA002K. The MCK1/mck1::TRP1 MDS1/mds1::HIS3 diploid cells derived from W303 were transformed with pTA007 cut with PstI and SalI, and the disruptions of the YOL128c gene of Leu+ transformants were confirmed by Southern blotting analysis. Then the MCK1/mck1::TRP1 MDS1/mds1::HIS3 YOL128c/yol128c::LEU2 diploid cells were transformed with pTA009 cut with XhoI, and the disruptions of the MRK1 gene of Ura+ transformants were confirmed by Southern blotting analysis. From spores of the MCK1/mck1::TRP1 MDS1/mds1::HIS3 YOL128c/yol128c::LEU2 MRK1/mrk1::URA3 diploid cells, an mck1::TRP1 mds1::HIS3 yol128c::LEU2 mrk1::URA3 haploid segregant was isolated. Strain YTA003W was obtained by selecting a clone, which formed a colony on a synthetic complete medium (SC) plate containing 0.1% 5-fluoro-orotic acid as the result of the pop-out of the URA3 gene, from mck1::TRP1 mds1::HIS3 yol128c::LEU2 mrk1::URA3 haploid cells incubated for several days on a YPD plate.
Isolation of suppressor mutations of the mck1 mds1 double null mutants.
The strain YTA002K (mck1::TRP1 mds1::HIS3) cells were transformed with Leu+ plasmids from the insertion library (6) cut with NotI. Transformants were selected on SC-Leu plates at 30°C, patched on SC-Leu plates, and incubated at 30 and 37°C for 2 days. Among 3,000 transformants, 16 strains which showed temperature-sensitive growth were crossed with the KA31α cells, and growth at 37°C of the Trp+ His+ Leu+ segregants and that of the Trp+ His+ Leu− segregants were compared. The Trp+ His+ segregants from 5 of the 16 strains showed Leu+-dependent growth at 37°C. The Trp− His− Leu+ segregants from the five strains were further crossed with the YHY009K (bul1::TRP1 bul2::LEU2) cells. Then four Trp+ Leu+ segregants from the spores, in which two of the four segregants were Leu+ and the other two were Leu−, were compared with the YHY009K cells for growth at 37°C. The segregants from two of the five strains grew better than YHY009K, and for these two strains DNA sequences at the insertion sites were determined as described previously (6). LEU2 fragments were inserted just before nucleotide +75 of the YGL144c ORF in strain YTA004K and at nucleotide +244 of the YOR253w ORF in strain YTA011K.
Immunoblot analysis and immunoprecipitation.
Yeast cells were grown in 25 ml of selective media to a density of 0.5 × 107 to 1 × 107 cells/ml, collected by centrifugation, washed twice with lysis buffer (50 mM Tris-HCl [pH 7.5], 0.3 M mannitol, 0.1 M KCl, 1 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg of antipain per ml, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 μg of pepstatin A per ml) (52), and resuspended in 100 μl of ice-cold lysis buffer. The cells were broken by using a vortex mixer containing glass beads for 5 min at 4°C. Then 100 μl of lysis buffer was added and the homogenate was again mixed vigorously. The homogenate was centrifuged at 14,000 × g for 5 min, and the supernatant was recovered and centrifuged again at 14,000 × g for 5 min. The resultant supernatant fraction was used as the cell lysates. The cell lysates were probed with the antihemagglutinin (anti-HA) and anti-myc antibodies (21). For the immunoprecipitation assay, the lysates were incubated with the anti-HA or anti-myc antibody and protein A-Sepharose and then precipitated by centrifugation. The immunoprecipitates were washed five times with the lysis buffer.
Pulse-chase analyses and in vivo labeling.
Cells containing pKT10-myc-Rog1 or pKT10-myc-Rog2 were grown overnight in minimal medium (MV medium) in which all sulfate salts were replaced by chloride salts supplemented with 100 μM (NH4)2SO4 (32). Exponentially growing cells were harvested, resuspended in fresh MV medium, and labeled with 10 μCi of [35S]methionine and [35S]cysteine per unit of optical density at 600 nm (OD600) for 20 min at 30°C. Then cells were washed four times; resuspended in 4 ml of fresh MV medium containing 1 mM (NH4)2SO4, 0.004% methionine, and 0.003% cysteine to an OD600 of 0.7; and further incubated at 37°C. The chase was terminated by the addition of 2 ml of ice-cold 30 mM sodium azide and rapid chilling on ice. The immunoprecipitation of myc-tagged proteins was performed as described previously (46). Briefly, the cells were centrifuged and incubated for 10 min on ice with 40 μl of 1.85 M NaOH–1% 2-mercaptoethanol. Proteins were then precipitated by adding 40 μl of 50% trichloroacetic acid. The precipitate was centrifuged at 12,000 × g for 5 min, resuspended in 30 μl of the sample buffer without 2-mercaptoethanol and then with 20 μl of 1 M Tris base, and heated at 37°C for 10 min. TNET buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100) was added, and the insoluble material was removed by centrifugation at 12,000 × g for 30 min. The supernatant was incubated overnight at 4°C with the anti-myc antibody and protein A-Sepharose. The immunoprecipitates were probed with the anti-myc antibody, followed by radioluminography with the Storm system (Amersham Pharmacia Biotech).
For the metabolic labeling with 32Pi, exponentially growing cells with an OD600 of 0.25 in low-phosphate medium were harvested, resuspended in 0.5 ml of fresh low-phosphate medium, and labeled with 150 μCi of 32Pi at 37°C for 3 h. After incubation, the cells were lysed and immunoprecipitated with the anti-myc antibody.
Other procedures.
Nucleotide sequences were determined using a Thermo Sequenase premixed cycle sequencing kit (Amersham Pharmacia Biotech). Southern hybridization was performed as previously described (51). In the measurement of the protein concentration of cell lysates, the dye-binding assay was performed (5) using the protein assay kit of Bio-Rad Laboratories (Hercules, Calif.).
RESULTS
The gsk-3 null mutant shows temperature-sensitive growth, which is suppressed by expression of mammalian GSK-3β and by an osmotic stabilizer.
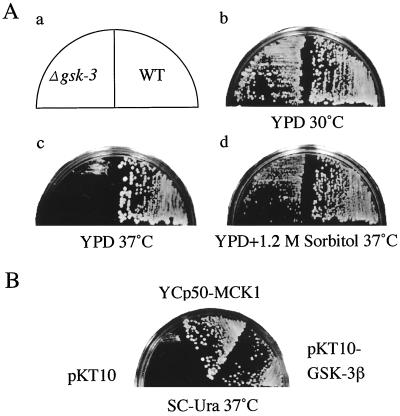
To investigate cellular functions of GSK-3 in yeast, we disrupted all four genes encoding GSK-3 homologs. The mck1 mds1 mrk1 yol128c quadruple null mutant (the gsk-3 null mutant) was not lethal and showed temperature sensitivity at 37°C as reported previously (13) (Fig. 1Ac). Interestingly, human GSK-3β expressed under the control of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter suppressed the temperature-sensitive phenotype of the gsk-3 null mutant as observed with expression of MCK1 (Fig. 1B), suggesting some functional conservation between mammalian GSK-3β and yeast GSK-3 homologs. We also found that the temperature sensitivity of the gsk-3 null mutant was suppressed when the medium contained the osmotic stabilizer sorbitol (Fig. 1Ad).
FIG. 1.
Suppression of the temperature sensitivity of the gsk-3 null mutant by an osmotic stabilizer and expression of mammalian GSK-3β. (A) The strains W303a (wild type [WT]) and YTA003W (Δgsk-3) were streaked on YPD plates (b and c) or a plate containing YPD plus 1.2 M sorbitol (d) as indicated in panel a and incubated at 30°C (b) or 37°C (c and d) for 3 days. (B) YTA003W was transformed with pKT10 vector, pTA024 (pKT10-GSK3β), and pTA021 (YCp50-MCK1), streaked on an SC-Ura plate, and incubated at 37°C for 3 days.
MCK1 is a multicopy suppressor of the bul1 bul2 double null mutant.
Several temperature-sensitive mutants, including the plc1, the bul1 bul2, and the rsp5 mutants, were reported to be suppressed by an osmotic stabilizer (51, 52). We examined genetic interactions between these mutants and yeast GSK-3. The temperature sensitivity of the bul1 bul2 double null mutant was suppressed by multiple copies of MCK1 (Fig. 2), whereas the plc1 (52) and the rsp5-101 mutants were not (data not shown). These results indicated that BUL1 and BUL2 interacted genetically with MCK1. Bul1 and Bul2 were previously shown to form a complex with Rsp5, a hect-type ubiquitin ligase, and genetic analysis of growth phenotype suggested that they facilitate the function of Rsp5 (50, 51). Multiple copies of UBC4, which are suggested to work in protein degradation coordinately with RSP5 (9), also partially suppressed the temperature sensitivity of the gsk-3 null mutant (data not shown). These results implied a functional involvement of yeast GSK-3 in Rsp5-dependent protein degradation.
FIG. 2.
Suppression of the temperature sensitivity of the bul1 bul2 double null mutant by multiple copies of MCK1. Strain YHY009K (Δbul1 Δbul2) was transformed with YEp24 vector, pTA022 (YEp24-MCK1), and pHY06 (YCp-BUL1), streaked on an SC-Ura plate, and incubated at 37°C for 3 days.
Mutations in ROG1 and ROG2 suppress the temperature-sensitive phenotypes of both the bul1 bul2 and the gsk-3 mutants.
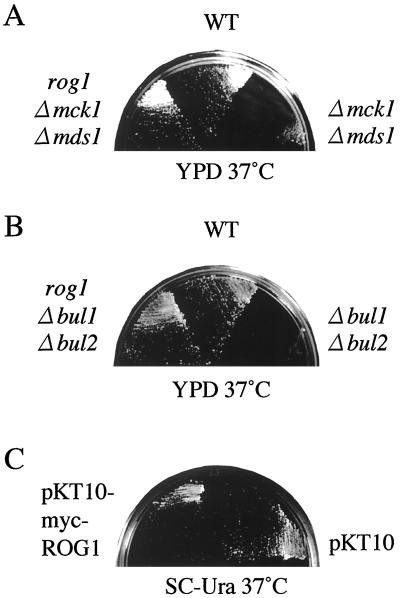
Genetical interaction between BUL1 and BUL2 and MCK1 suggested that they were involved in the protein degradation mediated by Rsp5. If the temperature sensitivities of the bul1 bul2 and the gsk-3 mutants are caused by accumulation of a protein which is degraded dependently on Bul1 and Bul2 and GSK-3, mutations of the degradation-targeted protein may suppress the temperature sensitivities of the bul1 bul2 and the gsk-3 mutants. To find out the target protein of degradation, we screened common revertants of both the bul1 bul2 and the gsk-3 mutants. Using a random insertion library, we screened insertional mutations which suppressed the temperature sensitivity of the mck1 mds1 double null mutants (30). The mck1 mds1 double null mutants were used in this screening since the double mutant was easier to use for tetrad analysis for confirmation of a library-dependent suppression than the quadruple mutant, and it showed a phenotype similar to that of the gsk-3 null mutant as to temperature sensitivity. By screening 3,000 insertion mutants, five revertants were obtained. Two of these, rog1 (Fig. 3) and rog2 (data not shown), also suppressed the temperature sensitivity of the bul1 bul2 double null mutant. By sequence analysis, ROG1 and ROG2 were revealed to be YGL144c and YOR253w, respectively, using a Saccharomyces genome database. Rog1 contains a lipase-like motif, and the rog1 mutant was reported to show alteration in lipid metabolism (8). Rog2 did not show any significant homology with other proteins. Three other rog mutations that did not suppress the temperature-sensitive phenotype of the bul1 bul2 double null mutant were not further investigated.
FIG. 3.
Suppression of the temperature sensitivities of both the bul1 bul2 double null mutant and the mck1 mds1 double null mutant by rog1 (A and B); temperature sensitivity conferred by overexpression of ROG1 (C). (A) KA31a (wild type [WT]), YTA002K (Δmck1 Δmds1), and YTA004K (rog1 Δmck1 Δmds1) were streaked on a YPD plate and incubated at 37°C for 3 days. (B) KA31a (WT), YHY009K (Δbul1 Δbul2), and YTA101K (rog1 Δbul1 Δbul2) were streaked on a YPD plate and incubated at 37°C for 3 days. (C) KA31a was transformed with pKT10 or pTA028 (pKT10-myc-ROG1), streaked on an SC-Ura plate, and incubated at 37°C for 3 days.
The degradation of Rog1 is dependent on Bul1 and Bul2 and GSK-3.
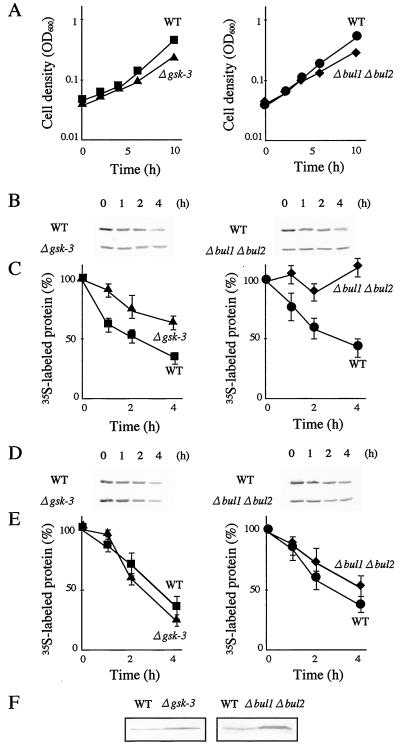
We next examined whether Bul1 and Bul2 and GSK-3 regulated the stability of Rog1 and Rog2. Since the YCp-Rog1-HA plasmid, possessing the double HA epitope-tagged Rog1 under the control of the original promoter, produced only a low level of protein in yeast cells, detection of labeled Rog1-HA was difficult. Therefore, we used pKT10-myc-Rog1 and pKT10-myc-Rog2 plasmids in which Rog1 and Rog2, respectively, tagged with triple myc epitope at the N terminus, were expressed under the control of the GAPDH promoter, and pulse-chase analyses of Rog1 and Rog2 were performed at 37°C. Overexpression of ROG1 from the GAPDH promoter conferred slow growth at 37°C (Fig. 3C) but did not completely block cell growth (Fig. 4A). Overexpression of ROG2 did not inhibit cell growth (data not shown). Pulse-chase analyses were done for 4 h, since the gsk-3 null and the bul1 bul2 double null mutants grew at the same rate as wild-type cells until 4 h after temperature shift to 37°C (Fig. 4A), although these mutants were temperature sensitive. The amounts of labeled myc-Rog1 were reduced to 34.4% in wild-type cells for 4 h, whereas labeled myc-Rog1 amounts were reduced only to 64.9% in the gsk-3 null mutant cells (Fig. 4B and C). In contrast, labeled myc-Rog2 was reduced in the gsk-3 null mutant cells (25.3%, 4 h) at a rate similar to that in wild-type cells (37.3%, 4 h) (Fig. 4D and E). In the bul1 bul2 double null mutant, myc-Rog1 was definitively stable (109%, 4 h) compared with the result in wild-type cells (44.1%, 4 h) (Fig. 4B and C). myc-Rog2 also showed a little more stability in the bul1 bul2 mutant after 4 h (54.7%) than in wild-type cells (36.5%) (Fig. 4D and E). These results showed that Rog1 was degraded dependently on Bul1 and Bul2 and also on GSK-3, whereas degradation of Rog2 was independent on GSK-3. Consistent with these results, overproduced Rog1 was accumulated in the gsk-3 and the bul1 bul2 null cells more than in wild-type cells (Fig. 4F). Rog2 was not accumulated in these mutants (data not shown). Disruption of ROG1 by replacement of the genomic ORF of ROG1 by the LEU2 marker resulted in the same phenotypes as those of the insertional mutation in the ROG1 locus shown in Fig. 3A and B (data not shown), suggesting that the loss of function of Rog1 suppressed the growth defects in both of the bul1 bul2 and the mck1 mds1 double null mutants.
FIG. 4.
Dependence of Rog1 degradation on Bul1 and Bul2 and GSK-3. (A) Growth curves of strains W303a (wild type [WT]), YTA003W (Δgsk-3), KA31a (WT), and YHY009K (Δbul1 Δbul2) carrying pTA028 (pKT10-myc-Rog1). Exponentially growing cells in MV medium at 30°C were shifted to 37°C at 0 h, and OD600 was measured at the indicated times. W303a (WT), YTA003W (Δgsk-3), KA31a (WT), and YHY009K (Δbul1 Δbul2) carrying pTA028 (pKT10-myc-Rog1) (B and C) or pTA030 (pKT10-myc-Rog2) (D and E) were pulse-labeled with [35S]methionine and [35S]cysteine for 20 min and chased by cold methionine and cysteine for 0, 1, 2, and 4 h. Cells were lysed and myc-Rog1 was immunoprecipitated with the anti-myc antibody, followed by radioluminography (B and D). The relative amounts of 35S-labeled proteins at each of the times in panels B and D were expressed as the percentages of 35S-labeled myc-Rog1 at time zero in panels C and E, respectively. The results shown are the representative results (B and D) or the means ± standard errors (C and E) of three independent experiments. (F) Exponentially growing cells with an OD600 of 0.5 of strains W303a (wild type [WT]), YTA003W (Δgsk-3), KA31a (WT), and YHY009K (Δbul1 Δbul2) carrying pTA026 (pRS316-GAL-HA-Rog1) in SG-Ura at 30°C were harvested, and the whole lysates were probed with the anti-HA antibody.
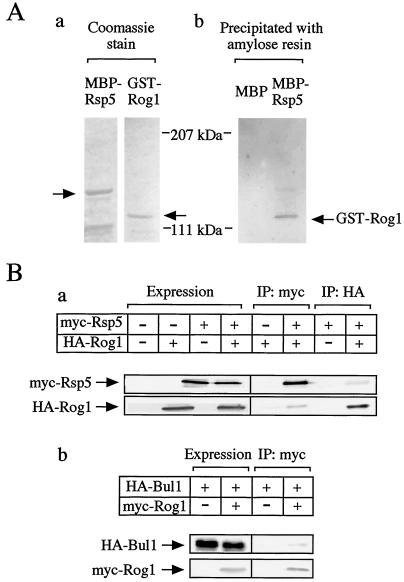
Rog1 binds to Rsp5.
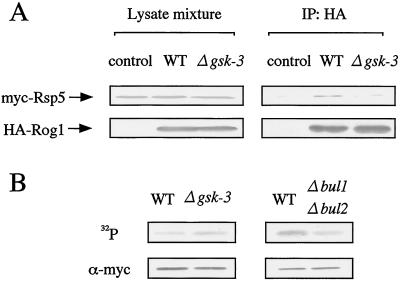
Since Bul1 and Bul2 and GSK-3 appeared to control degradation of Rog1, we examined the physical interaction between Rsp5 and Rog1 to clarify whether Rsp5 is involved in the degradation of Rog1. GST-Rog1 purified from E. coli was incubated with MBP-Rsp5 or MBP immobilized to amylose resin (Fig. 5A). GST-Rog1 was coprecipitated with MBP-Rsp5 but not with MBP (Fig. 5A). These results indicate that Rsp5 binds directly to Rog1. Wild-type yeast cells coexpressing myc-Rsp5 (pHY22) (50) and HA-Rog1 (pGAL1-HA-Rog1) were lysed, and the lysates were immunoprecipitated with the anti-HA or anti-myc antibody. HA-Rog1 was detected in the myc-Rsp5 immune complex and myc-Rsp5 was detected in the HA-Rog1 immune complex (Fig. 5Ba), suggesting that Rsp5 forms a complex with Rog1 in intact cells. HA-Bul1 was also coimmunoprecipitated with myc-Rog1 (Fig. 5Bb), suggesting ternary complex formation between Bul1, Rsp5, and Rog1 in intact cells, since Bul1 was shown to form a complex with Rsp5 (50, 51).
FIG. 5.
Complex formation of Rog1, Rsp5, and Bul1. (A) Binding of Rog1 to Rsp5 in vitro. (a) MBP-Rsp5 and GST-Rog1 purified from E. coli were subjected to gel electrophoresis followed by Coomassie brilliant blue staining. The arrows show the positions of MBP-Rsp5 and GST-Rog1. (b) The purified GST-Rog1 was incubated with MBP-Rsp5 or MBP immobilized to amylose resin at 4°C for 1 h, and the precipitates were probed with the anti-GST antibody. (B) Binding of Rog1 to Rsp5 in intact cells. (a) Cell lysates of W303a (wild type [WT]) cells carrying both pHY22 (myc-Rsp5) and pTA026 (HA-Rog1) or one of the two were directly probed with or immunoprecipitated (IP) with the anti-myc or the anti-HA antibody. The immunoprecipitates were probed with the same antibodies. (b) Cell lysates of W303a (WT) carrying both pHY20 (HA-Bul1) and pTA025 (myc-Rog1) or pHY20 alone were directly probed with the anti-myc and anti-HA antibodies or immunoprecipitated with the anti-myc antibody. The immunoprecipitates were probed with the anti-myc and anti-HA antibodies.
Rog1 is stable in the npi1 mutant.
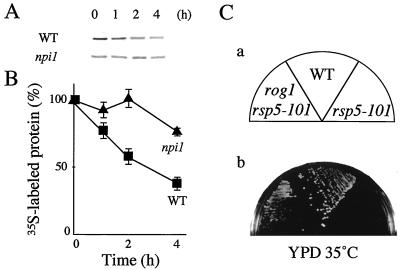
Since Rsp5 interacted with Rog1, we examined whether the degradation of Rog1 was dependent on Rsp5. As Rsp5 is an essential protein, we used the npi1 mutant, in which expression of RSP5 was reduced by mutation in a promoter region of the genomic RSP5 (41). Pulse-chase analyses showed that labeled myc-Rog1 was reduced to 38.4% in wild-type cells after 4 h, whereas labeled myc-Rog1 was reduced only to 76.9% in the npi1 cells (Fig. 6A and B). Furthermore, rog1 suppressed the temperature sensitivity of the rsp5-101 mutant (Fig. 6C). Therefore, Rsp5 is involved in the regulation of the degradation of Rog1.
FIG. 6.
Degradation of Rog1 in the npi1 mutant and suppression of the temperature sensitivity of the rsp5-101 mutant by rog1. (A) Pulse-chase analyses of myc-Rog1 were performed as described in the legend for Fig. 4. Pulse-labeled myc-Rog1 in strain 24346c (wild type [WT]) or 27038a (npi1) carrying pTA028 (pKT10-myc-Rog1) was immunoprecipitated and subjected to radioluminography. (B) The relative amounts of radiolabeled myc-Rog1 at each of the times in panel A were expressed as the percentages of 35S-labeled myc-Rog1 at time zero. The results shown are the representative results (A) or the means ± standard errors (B) of two independent experiments. (C) KA31a (WT), YTA2-1C (rsp5-101), and YTA102K (rog1 rsp5-101) cells were streaked on a YPD plate and incubated at 35°C for 2 days.
GSK-3 controls the interaction between Rog1 and Rsp5.
How does GSK-3 control the degradation of Rog1? When the lysates of wild-type cells expressing myc-Rsp5 (pHY22) were incubated with those expressing HA-Rog1 (pGAL1-HA-Rog1), myc-Rsp5 was coimmunoprecipitated with HA-Rog1 (Fig. 7A). However, myc-Rsp5 was coimmunoprecipitated with HA-Rog1 less effectively when the lysates of the gsk-3 null cells were used (Fig. 7A). Since the amount of Rog1 was larger in the gsk-3 null mutant than in wild-type cells as shown in Fig. 4F, the cell lysates used here were prepared to normalize the levels of HA-Rog1. This result suggests that GSK-3 promotes complex formation between Rsp5 and Rog1.
FIG. 7.
Regulation of the complex formation of Rog1 with Rsp5 by GSK-3. (A) Lysates from cells carrying pHY22 (myc-Rsp5) and cells transformed with pTA026 (HA-Rog1) (wild type [WT] and Δgsk-3) or transformed with pRS316-GAL-HA-BS vector (control) were prepared. As the amount of HA-Rog1 in the gsk-3 null mutant was more than that in wild-type cells, the levels of HA-Rog1 in the lysates of both cells were normalized. The lysates expressing myc-Rsp5 and HA-Rog1 individually from the wild-type or the gsk-3 null cells were mixed and the aliquots were probed with the anti-myc and anti-HA antibodies. The mixture of the lysates was immunoprecipitated (IP) with the anti-HA antibody, and then the immunoprecipitates were probed with the anti-myc and anti-HA antibodies. (B) In vivo phosphorylation of Rog1. W303a (WT), YTA003W (Δgsk-3), KA31a (WT), and YHY009K (Δbul1 Δbul2) carrying pTA028 (pKT10-myc-Rog1) were labeled with 32Pi at 37°C for 3 h, and the normalized amount of myc-Rog1 (lower panel) was subjected to autoradiography (upper panel). The results shown are representative of two independent experiments.
Mammalian GSK-3 is known to promote the degradation of some substrates by direct phosphorylation (1, 10). To test whether GSK-3 phosphorylates Rog1, in vivo phosphorylation analysis was performed. Cells of the wild type and the gsk-3 null and the bul1 bul2 double null yeasts were labeled with 32Pi, and myc-Rog1 was immunoprecipitated and subjected to autoradiography. Interestingly, the phosphorylation level of Rog1 was not decreased but rather increased in the gsk-3 null cells compared to that in wild-type cells (Fig. 7B). In the bul1 bul2 double null mutant, where GSK-3 existed and the degradation system was probably impaired, not the highly phosphorylated but the lowly phosphorylated form of Rog1 was accumulated (Fig. 7B). These results suggest that GSK-3 induces the dephosphorylation of Rog1.
rog1 suppresses the growth defects of the bul1 bul2 double null mutant on nonfermentable carbon sources.
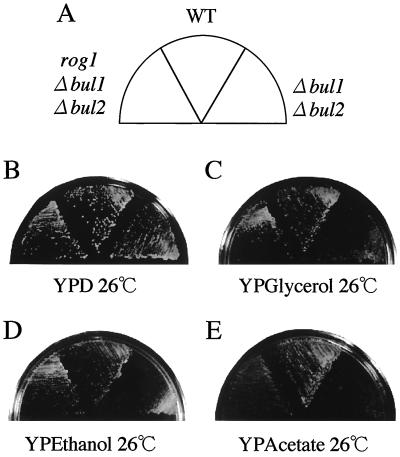
The bul1 bul2 double null mutant was reported to show growth defects not only at high temperature but also on glycerol medium (51). Interestingly, rog1 suppressed the growth defects of the bul1 bul2 double null mutant on nonfermentable carbon sources, such as glycerol, ethanol, and acetate media (Fig. 8). Since respiration is necessary for growth on these media, Rog1 may play a role in the regulation of respiratory functions.
FIG. 8.
Suppression of the growth defects of the bul1 bul2 double null mutant on nonfermentable carbon sources by rog1. KA31a (wild type [WT]), YHY009K (Δbul1 Δbul2), and YTA101K (rog1 Δbul1 Δbul2) were streaked on YPD, YPGlycerol, YPEthanol, and YPAcetate plates as indicated in panel A and incubated at 26°C for 2 days (B and C) or 4 days (D and E).
DISCUSSION
Phenotypes of the gsk-3 null mutant.
The gsk-3 null mutant was not lethal and showed temperature sensitivity, like the mck1 single null mutant (30). Comparison between temperature sensitivities of every single, double, triple, and quadruple null mutant of the four yeast GSK-3 homologs showed that temperature sensitivity was severe when both MCK1 and MDS1 (RIM11) were disrupted (data not shown). The quadruple null mutant was only a little sicker than the mck1 mds1 double null mutant at high temperatures. Therefore, MCK1 and MDS1 (RIM11) may play an important role in growth at high temperature among the four GSK-3 homologs. Mammalian GSK-3β suppresses the temperature sensitivity of the yeast gsk-3 null mutant, suggesting that the functions of GSK-3 are evolutionarily conserved.
Degradation of Rog1.
We have identified two rog mutations (rog1 and rog2) which suppress the temperature sensitivity of both the mck1 mds1 and the bul1 bul2 double null mutants. Bul1 and Bul2 have been shown to form a complex with Rsp5, and the PY motif of Bul1 is necessary for its binding to Rsp5 (50). Since a mutation of the PY motif of Bul1 does not overcome growth defects of the bul1 bul2 double null mutant, Bul1 and Bul2 may function through their binding to Rsp5. Rsp5 is a member of the hect-type E3 enzymes and plays a role in the degradation of several proteins in yeast (2, 7, 9, 16, 24). However, no evidence that Bul1 and Bul2 are involved in the protein degradation has been reported. The SCF-type E3 enzymes recognize their substrates only when the substrates are phosphorylated (7, 11, 22, 40, 47). However, whether protein degradation by Rsp5, a hect-type E3, requires the phosphorylation of the target protein is not known. The degradation of Rog1 is inhibited in the gsk-3 null mutant, while Rog2 is degraded independently of GSK-3. Rog1 is also stabilized in the bul1 bul2 double null mutant, unlike Rog2. These results indicate that GSK-3 and Bul1 and Bul2 regulate the stability of Rog1 specifically. Further, Rog1 is stable in the npi1 mutant (the rsp5 mutant) and rog1 suppresses the temperature sensitivity of the rsp5-101 mutant. These results clearly demonstrate that the stability of Rog1 is regulated by GSK-3, Bul1 and Bul2, and Rsp5 and suggest that these proteins functionally interact. Since overexpression of ROG1 confers slow growth at 37°C, the accumulation of Rog1 may be one of the causes for the temperature sensitivity of the gsk-3 null and the bul1 bul2 double null mutants. However, it is possible that other, unknown, factors besides the elevated level of Rog1 cause the temperature sensitivity of these mutants, as wild-type cells overexpressing ROG1 grow a little faster than the mutants (compare Fig. 3A or B with C).
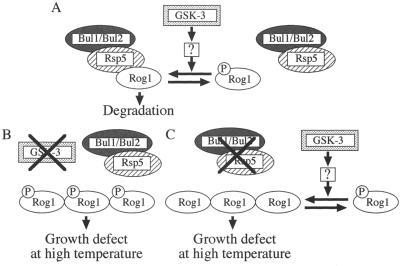
In mammalian cells, GSK-3 phosphorylates β-catenin, and β-TrCP, a component of the SCF-type E3 enzyme, recognizes the phosphorylated β-catenin, resulting in its ubiquitination (22). Our results show that Rog1 binds directly to Rsp5 and is coimmunoprecipitated with Rsp5 from the lysates when GSK-3 is present, suggesting that GSK-3 is involved in protein degradation by the hect-type E3 enzyme. How does GSK-3 control complex formation between Rog1 and Rsp5? One possibility is that GSK-3 phosphorylates Rog1, thereby increasing the binding of Rog1 to Rsp5. However, as Mck1 does not phosphorylate Rog1 in vitro (data not shown) and the phosphorylation of Rog1 is increased in the gsk-3 null mutant, it is not likely that the phosphorylation of Rog1 by GSK-3 directly triggers the degradation of Rog1. We propose a model, shown in Fig. 9, that explains the results of this study. GSK-3 promotes degradation of Rog1 by facilitating complex formation between Rog1 and Rsp5, probably through the dephosphorylation of Rog1. GSK-3 may activate a phosphatase to dephosphorylate Rog1, and the unphosphorylated form of Rog1 may be recognized by Rsp5 (Fig. 9A). In the gsk-3 null mutant, the phosphorylated form of Rog1 is accumulated since it is hard to form a complex with Rsp5 (Fig. 9B). In the bul1 bul2 double null or the rsp5 mutant, the unphosphorylated form is accumulated (Fig. 9C). Isolation of a protein that induces the GSK-3-dependent dephosphorylation of Rog1 will reveal the mechanism.
FIG. 9.
Possible model of functional interaction between GSK-3, Bul1 and Bul2, and Rsp5. (A) Usually, GSK-3 promotes dephosphorylation of Rog1, which is recognized by Bul1 and Bul2 and Rsp5, resulting in the degradation of Rog1. (B and C) Both the phosphorylated form of Rog1, which accumulates in the gsk-3 null mutant (B), and the dephosphorylated form of Rog1, which accumulates in the bul1 bul2 double null or the rsp5 mutant (C), may inhibit cell growth at 37°C.
How do Bul1 and Bul2 control the degradation of Rog1? In our coimmunoprecipitation analysis, Rog1 was coimmunoprecipitated with Rsp5 independently of Bul1 and Bul2 (data not shown). It is not likely that Bul1 and Bul2 promote the binding of Rog1 to Rsp5. Therefore, Bul1 and Bul2 may control an activity of Rsp5. Taken together with the observations that MCK1 suppresses the temperature sensitivity of the bul1 bul2 mutant, it is conceivable that a complex of Bul1 or Bul2 and Rsp5 recognizes and ubiquitinates Rog1, which is modulated indirectly by GSK-3, leading to the degradation of Rog1.
Functions of Rog1.
The bul1 bul2 and the rsp5 mutants show growth defects on glycerol medium (50), where the respiratory functions of mitochondria are necessary for growth. BUL1 and RSP5 are reported to be involved in the transmission of mitochondria from a mother cell to a daughter cell (12). Although whether protein degradation is important for the process of inheritance of mitochondria is not known, this might be one of the reasons why the bul1 bul2 and the rsp5 mutants exhibit growth defects on nonfermentable carbon sources. Since the rog1 mutation suppressed growth defects of the bul1 bul2 double null mutant not only at a high temperature but also on nonfermentable carbon sources, Rog1 may affect mitochondrial functions, including inheritance.
ACKNOWLEDGMENTS
We are grateful to B. André, A. Toh-e, Y. Kikuchi, K. Tanaka, and H. Yashiroda for donating plasmids and yeast strains. We also thank A. Toh-e, Y. Kikuchi, K. Tanaka, and H. Yashiroda for helpful discussion.
This work was supported by grants-in-aid for scientific research and for scientific research on priority areas from the Ministry of Education, Science, and Culture, Japan (1998, 1999), and by grants from the Yamanouchi Foundation for Research on Metabolic Disorders (1998, 1999) and the Uehara Memorial Foundation (1998).
REFERENCES
- 1.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaudenon S L, Huacani M R, Wang G, Mcdonnell D P, Huibregtse J M. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6972–6979. doi: 10.1128/mcb.19.10.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botstein D, Fallo S C, Stewart S E, Brennan M, Scherer S, Stinchcomb D T, Struhl K, Davis R W. Sterile host yeast (SHY); a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979;8:17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- 4.Bowdish K S, Yuan H E, Mitchell A P. Analysis of RIM11, a yeast protein kinase that phosphorylates the meiotic activator IME1. Mol Cell Biol. 1994;14:7909–7919. doi: 10.1128/mcb.14.12.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Burns N, Grimwade B, Ross-Macdonald P B, Choi E-Y, Finberg K, Roeder G S, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 7.Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daum G, Tuller G, Nemec T, Hrastnik C, Balliano G, Cattel L, Milla P, Rocco F, Conzelmann A, Vionnet C, Kelly D E, Kelly S, Schweizer E, Schuller H-J, Hojad U, Greiner E, Finger K. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast. 1999;15:601–614. doi: 10.1002/(SICI)1097-0061(199905)15:7<601::AID-YEA390>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 9.de la Fuente N, Maldonado A M, Portillo F. Glucose activation of the yeast plasma membrane H+-ATPase requires the ubiquitin-proteasome proteolytic pathway. FEBS Lett. 1997;411:308–312. doi: 10.1016/s0014-5793(97)00721-7. [DOI] [PubMed] [Google Scholar]
- 10.Diehl J A, Cheng M, Roussel M F, Sherr C J. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 12.Fisk H A, Yaffe M P. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1199–1208. doi: 10.1083/jcb.145.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajji K, Clotet J, Arino J. Disruption and phenotypic analysis of seven ORFs from the left arm of chromosome XV of Saccharomyces cerevisiae. Yeast. 1999;15:435–441. doi: 10.1002/(SICI)1097-0061(19990330)15:5<435::AID-YEA367>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 14.Harwood A J, Plyte S E, Woodgett J, Strutt H, Kay R R. Glycogen synthase kinase 3 regulates cell fate in Dictyostelium. Cell. 1995;80:139–148. doi: 10.1016/0092-8674(95)90458-1. [DOI] [PubMed] [Google Scholar]
- 15.He X, Saint-Jeannet J-P, Woodgett J R, Varmus H E, Dawid I B. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature. 1995;374:617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- 16.Hein C, Springael J-Y, Volland C, Haguenauer-Tsapis R, André B. NPI1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 17.Huibregtse J M, Scheffner M, Howley P M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irie K, Nomoto S, Miyajima I, Matsumoto K. SGV1 encodes a CDC28/cdc2-related kinase required for a Gα subunit-mediated adaptive response to pheromone in S. cerevisiae. Cell. 1991;65:785–795. doi: 10.1016/0092-8674(91)90386-d. [DOI] [PubMed] [Google Scholar]
- 20.Jones J S, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi A, Williams L T. Regulation of interaction of ras p21 with RalGDS and Raf-1 by cyclic AMP-dependent protein kinase. J Biol Chem. 1996;271:588–594. doi: 10.1074/jbc.271.1.588. [DOI] [PubMed] [Google Scholar]
- 22.Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama K-I, Nakayama K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of β-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo C-L, Campbell J L. Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8-1 mutation. Mol Cell Biol. 1983;3:1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucero P, Lagunas R. Catabolite inactivation of the yeast maltose transporter requires ubiquitin-ligase npi1/rsp5 and ubiquitin-hydrolase npi2/doa4. FEMS Microbiol Lett. 1997;147:273–277. doi: 10.1111/j.1574-6968.1997.tb10253.x. [DOI] [PubMed] [Google Scholar]
- 25.Mandelkow E-M, Drewes G, Biernat J, Gustke N, Van Lint J, Vandenheede J R, Mandelkow E. Glycogen synthase kinase-3 and the Alzheimer-like state of microtubule-associated protein tau. FEBS Lett. 1992;314:315–321. doi: 10.1016/0014-5793(92)81496-9. [DOI] [PubMed] [Google Scholar]
- 26.Neigeborn L, Mitchell A P. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 1991;5:533–548. doi: 10.1101/gad.5.4.533. [DOI] [PubMed] [Google Scholar]
- 27.Perrimon N, Smouse D. Multiple functions of a Drosophila homeotic gene, zeste-white 3, during segmentation and neurogenesis. Dev Biol. 1989;135:287–305. doi: 10.1016/0012-1606(89)90180-2. [DOI] [PubMed] [Google Scholar]
- 28.Plyte S E, Feoktistova A, Burke J D, Woodgett J R, Gould K L. Schizosaccharomyces pombe skp1+ encodes a protein kinase related to mammalian glycogen synthase kinase 3 and complements a cdc14 cytokinesis mutant. Mol Cell Biol. 1996;16:179–191. doi: 10.1128/mcb.16.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plyte S E, Hughes K, Nikolakaki E, Pulverer B J, Woodgett J R. Glycogen synthase kinase-3: functions in oncogenesis and development. Biochim Biophys Acta. 1992;1114:147–162. doi: 10.1016/0304-419x(92)90012-n. [DOI] [PubMed] [Google Scholar]
- 30.Puziss J W, Hardy T A, Johnson R B, Roach P J, Hieter P. MDS1, a dosage suppressor of an mck1 mutant, encodes a putative yeast homolog of glycogen synthase kinase 3. Mol Cell Biol. 1994;14:831–839. doi: 10.1128/mcb.14.1.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rieger K-J, Aljinovic G, Lazowska J, Pohl T M, Slonimski P P. A novel nuclear gene, CBT1, essential for mitochondrial cytochrome b formation: terminal processing of mRNA and intron dependence. Curr Genet. 1997;32:163–174. doi: 10.1007/s002940050262. [DOI] [PubMed] [Google Scholar]
- 32.Rothblatt J, Schekman R. A hitchhiker's guide to analysis of the secretory pathway in yeast. Methods Cell Biol. 1989;32:3–36. doi: 10.1016/s0091-679x(08)61165-6. [DOI] [PubMed] [Google Scholar]
- 33.Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3β to the APC-β-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 35.Ruel L, Bourouis M, Heitzler P, Pantesco V, Simpson P. Drosophila shaggy kinase and rat glycogen synthase kinase-3 have conserved activities and act downstream of Notch. Nature. 1993;362:557–560. doi: 10.1038/362557a0. [DOI] [PubMed] [Google Scholar]
- 36.Schild L, Lu Y, Gautschi I, Schneeberger E, Lifton R P, Rossier B C. Identification of a PY motif in the epithelial Na channel subunits as a target sequence for mutations causing channel activation found in Liddle syndrome. EMBO J. 1996;15:2381–2387. [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman F, Fink G R, Hicks J M. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 38.Shero J H, Hieter P. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1) Genes Dev. 1991;5:549–560. doi: 10.1101/gad.5.4.549. [DOI] [PubMed] [Google Scholar]
- 39.Simpson P, El Messal M, Moscoso del Prado J, Ripoll P. Stripes of positional homologies across the wing blade of Drosophila melanogaster. Development. 1988;103:391–401. [Google Scholar]
- 40.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 41.Springael J-Y, André B. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1253–1263. doi: 10.1091/mbc.9.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 43.Sutton A, Immanuel D, Arndt K T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka K, Nakafuku M, Tamanoi F, Kaziro Y, Matsumoto K, Toh-e A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol. 1990;10:4303–4313. doi: 10.1128/mcb.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toh-e A. Construction of a marker gene cassette which is repeatedly usable for gene disruption in yeast. Curr Genet. 1995;27:293–297. doi: 10.1007/BF00352095. [DOI] [PubMed] [Google Scholar]
- 46.Volland C, Garnier C, Haguenauer-Tsapis R. In vivo phosphorylation of the yeast uracil permease. J Biol Chem. 1992;267:23767–23771. [PubMed] [Google Scholar]
- 47.Winston J T, Strack P, Beer-Romero P, Chu C Y, Elledge S J, Harper J W. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woodgett J R. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 1990;9:2431–2438. doi: 10.1002/j.1460-2075.1990.tb07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang S-D, Song J-S, Hsieh Y-T, Liu H-W, Chan W-H. Identification of the ATP · Mg-dependent protein phosphatase activator (FA) as a synapsin I kinase that inhibits cross-linking of synapsin I with brain microtubules. J Protein Chem. 1992;11:539–546. doi: 10.1007/BF01025031. [DOI] [PubMed] [Google Scholar]
- 50.Yashiroda H, Kaida D, Toh-e A, Kikuchi Y. The PY-motif of Bul1 protein is essential for growth of Saccharomyces cerevisiae under various stress conditions. Gene. 1998;225:39–46. doi: 10.1016/s0378-1119(98)00535-6. [DOI] [PubMed] [Google Scholar]
- 51.Yashiroda H, Oguchi T, Yasuda Y, Toh-e A, Kikuchi Y. Bul1, a new protein that binds to the Rsp5 ubiquitin ligase in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3255–3263. doi: 10.1128/mcb.16.7.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoko-o T, Kato H, Matsui Y, Takenawa T, Toh-e A. Isolation and characterization of temperature-sensitive plc1 mutants of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1995;247:148–156. doi: 10.1007/BF00705644. [DOI] [PubMed] [Google Scholar]
- 53.Yost C, Torres M, Miller J R, Huang E, Kimelman D, Moon R T. The axis-inducing activity, stability, and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]